Abstract
Phelipanche ramosa is an obligate root-parasitic weed that threatens major crops in central Europe. In order to germinate, it must perceive various structurally divergent host-exuded signals, including isothiocyanates (ITCs) and strigolactones (SLs). However, the receptors involved are still uncharacterized. Here, we identify five putative SL receptors in P. ramosa and show that PrKAI2d3 is involved in the stimulation of seed germination. We demonstrate the high plasticity of PrKAI2d3, which allows it to interact with different chemicals, including ITCs. The SL perception mechanism of PrKAI2d3 is similar to that of endogenous SLs in non-parasitic plants. We provide evidence that PrKAI2d3 enzymatic activity confers hypersensitivity to SLs. Additionally, we demonstrate that methylbutenolide-OH binds PrKAI2d3 and stimulates P. ramosa germination with bioactivity comparable to that of ITCs. This study demonstrates that P. ramosa has extended its signal perception system during evolution, a fact that should be considered for the development of specific and efficient biocontrol methods.
Keywords: α/β-hydrolase, receptor, seed germination stimulant, strigolactones, isothiocyanates, Phelipanche ramosa
Phelipanche ramosa is an obligate root-parasitic weed that threatens major crops in central Europe. This study reports the identification of a strigolactone receptor in P. ramosa, PrKAI2d3, that is involved in the stimulation of seed germination. The high plasticity of PrKAI2d3 allows it to interact with different chemicals, including isothiocyanates and methylbutenolide-OH.
Introduction
Witchweeds (Striga spp.) and broomrapes (Orobanche and Phelipanche spp.) are obligate root-parasitic plants of the Orobanchaceae family that together comprise the most threatening weeds for major domesticated crops worldwide (Parker, 2012). At maturity, a single plant releases up to 100 000 microscopic seeds, resulting in severe soil pollution (Delavault et al., 2017). Seed germination of these obligate parasites requires the strict recognition of host-exuded germination stimulants. Broomrapes and witchweeds are all highly sensitive to strigolactones (SLs) secreted by plants into the rhizosphere at picomolar doses (Brun et al., 2017). The structural core of SLs is a tricyclic lactone, referred to as the ABC part in canonical SLs or as a structural variant in non-canonical SLs. It is invariably connected to an α,β-unsaturated furanone moiety (the D ring) by an enol ether bridge (Supplemental Figure 1A) (Brun et al., 2017). Some germination stimulants are exclusive to specific host-parasite interactions, as illustrated by the unique ability of Phelipanche ramosa to germinate upon sensing isothiocyanates (ITCs). These glucosinolate breakdown products are exuded by rapeseed (Brassica napus) (Auger et al., 2012; Delavault et al., 2017), to which P. ramosa has adapted in a decade (Parker, 2012; Delavault et al., 2017). Broomrapes are increasingly problematic in both intensity and acreage in Europe, North Africa, and Asia, and they are expected to dramatically expand to new territories in the near future (Grenz and Sauerborn, 2007). To date, several physical, cultural, chemical, and biological approaches have been explored to control root-parasitic weeds, but no method has been found completely satisfactory (Fernandez-Aparicio et al., 2016).
In addition to their involvement in germination, SLs strongly stimulate arbuscular mycorrhizal fungi (Glomeromycotina) by promoting mitochondrial metabolism and hyphal branching (Besserer et al., 2008), thereby mediating the establishment of the oldest mutualistic interaction of land plants (Brundrett and Tedersoo, 2018). In addition to their role in rhizosphere signaling, SLs also act as hormones in planta and have pervasive roles throughout plant development (Lopez-Obando et al., 2015). As such, they are perceived by the α/β-hydrolase DWARF14 (D14) in vascular plants (Hamiaux et al., 2012; Waters et al., 2017). Biochemical analyses with recombinant D14 proteins by means of the synthetic SL analog GR24 revealed that SL signal transduction requires GR24 cleavage (Nakamura et al., 2013; Waters et al., 2017). One of the cleavage products, the D ring, may remain covalently attached to the receptor (de Saint Germain et al., 2016; Yao et al., 2016), thereby probably allowing the recruitment of partners for downstream processes (Shabek et al., 2018). The requirement of SL cleavage for signal transduction is still under debate (Shabek et al., 2018; Seto et al., 2019).
In Arabidopsis thaliana, D14 belongs to a small protein family, including KARRIKIN INSENSITIVE2/HYPOSENSITIVE TO LIGHT (AtKAI2/AtHTL), that shares the α/β-hydrolase catalytic triad. However, AtKAI2 regulates AtD14-independent processes such as seed germination and preferentially perceives (−)-GR24, which mimics non-natural SLs, karrikins (Sun et al., 2016), and supposedly an unknown endogenous ligand (Conn and Nelson, 2016; Swarbreck et al., 2019; Villaécija-Aguilar et al., 2019). Interestingly, the KAI2 gene family has expanded during the genome evolution of obligate parasitic plants (Conn et al., 2015). For example, Phelipanche aegyptiaca and Striga hermonthica possess five and eleven KAI2/HTL genes, respectively. Two KAI2 paralogs play a role in the SL response of P. aegyptiaca (Conn et al., 2015), and six S. hermonthica KAI2/HTL proteins are hypersensitive to SLs (Conn et al., 2015; Toh et al., 2015), with ShHTL7 exhibiting the same perception mechanism as D14 (Yao et al., 2017). The remarkable expansion of the KAI2 gene family in Orobanchaceae, along with the capacity of P. ramosa to perceive ITCs, led us to speculate that KAI2 proteins may also perceive other germination stimulants. Here, we characterize PrKAI2d3 as a P. ramosa SL receptor. We demonstrate that it is able to perceive natural SLs by an enzymatically dependent mechanism that contributes to its SL hypersensitivity. We also show that PrKAI2d3 perceives ITCs as well as a wide range of SL analogs that were not found to be bioactive for SL hormonal functions. Together, these results suggest that PrKAI2 proteins evolved as hypersensitive and plastic receptors, enabling the parasitic plant to detect various host exudate metabolites and contributing to its dramatic success.
Results
Identification and gene expression profiles of PrKAI2 homologs
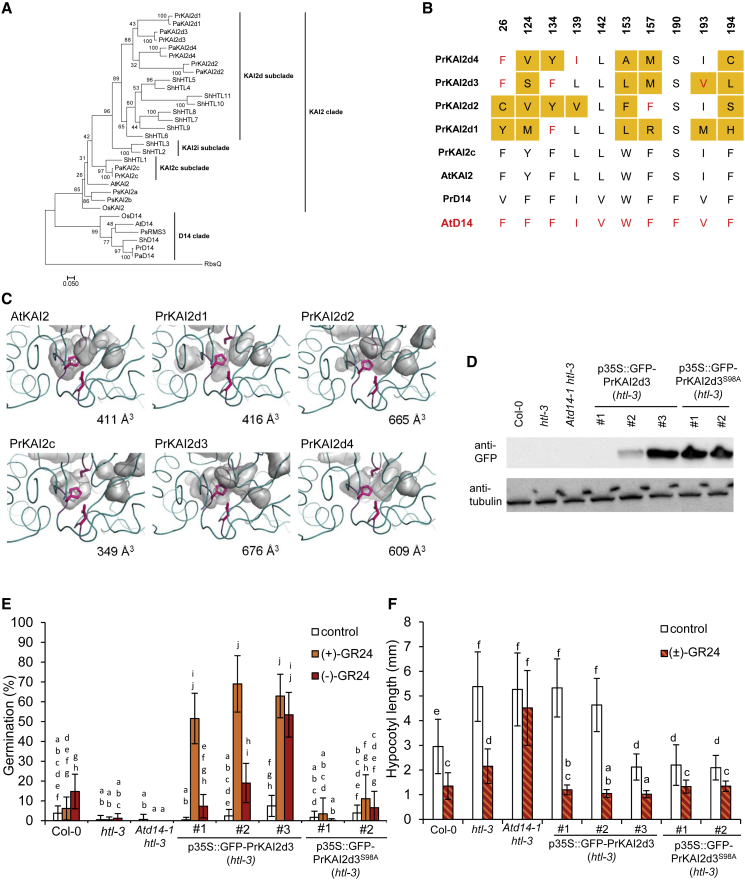
Iterative nucleotide and amino acid BLAST analyses were performed on the recently published transcriptome of the P. ramosa rapeseed strain (Goyet et al., 2017) using known KAI2 and D14 sequences from non-parasitic and parasitic plants as queries (Conn et al., 2015; Toh et al., 2015). Each newly identified sequence was used as a query for new BLAST searches in order to find all complete sequences, and duplicates were eliminated before alignment. One PrD14 and five PrKAI2 putative orthologs were identified, amplified, and re-sequenced from cDNA isolated from germinated seeds of the same P. ramosa strain. Maximum-likelihood (ML) phylogenetic analysis indicated that the predicted PrD14 and PrKAI2 proteins (Figure 1A) and genes (Supplemental Figure 2) belonged to clearly distinct D14 and KAI2 clades. They possessed a conserved catalytic triad and an overall conserved environment of known active-site amino acid residues (Supplemental Figure 3). Despite the poor separation of the non-parasitic and parasitic KAI2 groups, the phylogenetic analysis strongly suggested that none of the retrieved PrKAI2 proteins belonged to the intermediate KAI2i subclade (Figure 1A), as previously described for other members of the Orobanche and Phelipanche genera (Conn et al., 2015). The newly identified PrKAI2 and PrD14 proteins consistently clustered together with P. aegyptiaca sequences. Together with the low levels of genomic divergence reported in Phelipanche (Wicke et al., 2016; Yang et al., 2016), these findings hint at the detection of P. aegyptiaca protein orthologs. Hence, the P. ramosa sequences were renamed according to the current nomenclature (Conn et al., 2015), with one PrKAI2 protein belonging to the conserved KAI2 (PrKAI2c) clade and four PrKAI2 proteins assigned to the divergent KAI2 clade (PrKAI2d1–d4).
Figure 1.
Identification of PrKAI2 putative SL receptors in P. ramosa.
(A) Phylogenetic analysis of KAI2 and D14 amino acid sequences. The phylogenetic tree was constructed with the ML method and 1000 bootstrap replicates using RAxML. The scale bar represents 0.05 substitutions per site. Clades were designated as described previously (Conn et al., 2015).
(B) Amino acid sequence alignment of the active PrKAI2 protein sites. Amino acids that differ from AtKAI2 and those similar to AtD14 are in orange and red, respectively; AtHTL and PrKAI2d3 are in blue and yellow, respectively. A fully expanded alignment can be found in Supplemental Figure 3.
(C) Visual representation of the ligand pockets of the P. ramosa KAI2 proteins. The KAI2 protein sequences were modeled using chain A of the karrikin-bound Arabidopsis KAI2 structure as a template (PDB: 4JYP). The protein structures were generated with PyMOL, and the cavities within the homology models are shown as a semitransparent surface. Catalytic residues (Ser, His, Asp) are shown in stick representation.
(D–F) Cross-species complementation assays of the htl-3 mutant with P. ramosa KAI2d3 and the catalytic site mutant S98A. (D) PrKAI2d3 protein levels of 4-day-old seedlings transformed with p35S::GFP-PrKAI2d3 or p35S::GFP-PrKAI2d3S98A detected with anti-GFP (top) and anti-tubulin (bottom) antibodies as a loading control. The experiment was repeated twice with comparable results, and one representative replicate is shown. (E) Seed germination after 5 days of growth at 25°C in the dark with DMSO (control), 10 μM (+)-GR24, or 10 μM (−)-GR24 treatment. Transgenes were expressed in the null htl-3 mutant background (Col-0 accession) under the control of the 35S promoter. One representative experiment of 18 wells per condition with an average of 19 seeds/well is shown. (F) Hypocotyl lengths of 4-day-old seedlings grown under continuous red light at 21°C (n = 25) with 10 μM (±)-GR24 treatments. Graphs show the means ± SE of three biological replicates. Statistical groups indicated by letters were determined by the Kruskal–Wallis test with Dunn's post-hoc test, p < 0.001 (E) and p < 0.05 (F).
PrKAI2d3 is a putative SL receptor
By comparing amino acid residues in the PrKAI2 ligand-binding pocket to those from AtD14 and AtKAI2, we found that PrKAI2d3 exhibited the highest similarity to AtD14 (Figure 1B). Predictive models generated with the SWISS-MODEL webserver revealed that the binding pocket of PrKAI2c is smaller than that of AtKAI2 (Figure 1C) and that the PrKAI2d protein binding pockets are wider than those of members of the conserved KAI2 clade, probably because of the absence of large hydrophobic residues (Figure 1C). Among all the divergent proteins, PrKAI2d3 had the largest predicted binding pocket (Figure 1C, Supplemental Figure 4). The levels of all four PrKAI2d transcripts in P. ramosa seeds changed significantly after 6 h of treatment with exogenous (±)-GR24 and 2-PEITC (phenethyl isothiocyanate) (Supplemental Figure 5). By contrast, the transcript levels of PrKAI2c, which is putatively orthologous to AtKAI2, remained unchanged. Based on these data, all the PrKAI2ds may be involved in SL perception and should be considered further for complete biochemical and physiological characterization. In our attempt to characterize all PrKAI2d recombinant proteins, we managed to produce only recombinant PrKAI2d3; the other proteins remained non-soluble under our experimental conditions. As a result, we will focus here on the characterization of PrKAI2d3 at the biochemical and physiological levels.
Because no mutants or simple and rapid transformation methods are available for holoparasitic Orobanchaceae, it is difficult (Fernández-Aparicio et al., 2011) to validate the biological function of PrKAI2d3 directly in P. ramosa. Instead, we assayed the complementation of the Arabidopsis htl-3 mutant phenotype with PrKAI2d3, a previously successful approach (Conn et al., 2015; Toh et al., 2015). To assess the role of the catalytic triad in the perception process, htl-3 was also transformed with a mutated gene that encoded PrKAI2d3S98A. We recorded germination and hypocotyl length phenotypes in three p35S::GFP-PrKAI2d3 lines that displayed various protein levels and two p35S::GFP-PrKAI2d3S98A lines that showed high protein expression (Figure 1D).
In a thermo-inhibition assay (Toh et al., 2012), almost no germination was detected in all lines under control treatments. Germination of Arabidopsis (accession Columbia-0 [Col-0]) seeds was significantly stimulated by 10 μM (−)-GR24 but not by (+)-GR24, whereas none of the enantiomers had an effect on the germination of the htl-3 or Atd14-1/htl-3 mutants (Figure 1E). Germination of all three p35S::GFP-PrKAI2d3 lines was strongly induced by (+)-GR24. Exogenous (−)-GR24 also significantly stimulated germination of these three lines (Figure 1E), and the response amplitude was positively correlated with protein abundance (Figure 1D). Analysis of a fourth p35S::GFP-PrKAI2d3 line showed that even low PrKAI2d3 protein expression increased Arabidopsis sensitivity toward (−)-GR24 100-fold, enabling it to perceive picomolar doses of (+)-GR24 and, hence, to have P. ramosa levels of sensitivity (Figure 2A and 2B, Supplemental Figure 6, Supplemental Table 1). Neither of the two p35S::GFP-PrKAI2d3S98A lines were sensitive to (+)- or (−)-GR24.
Figure 2.
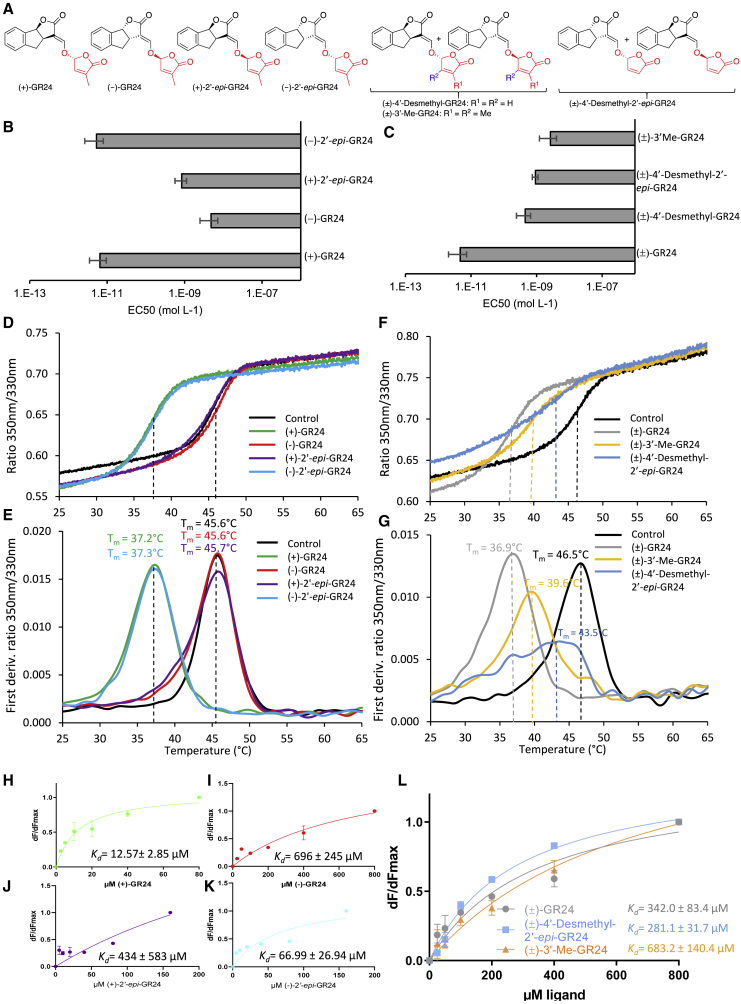
PrKAI2d3 shows stereoselectivity toward GR24 analogs mimicking the SL natural stereoconfigurations and perceives SL analogs without a methyl group on the D ring.
(A) Structures of GR24 analogs.
(B and C) Germination stimulation activity on P. ramosa seeds (EC50) of four stereoisomers and of GR24 analogs harboring variations on the D ring (± SE).
(D–G) Thermostability of PrKAI2d3 at 10 μM in the absence of ligands (black line) or in the presence of various ligands at 100 μM analyzed by nanoDSF. (D) and (F) The changes in fluorescence (F350nm/F330nm ratio) with temperature, and (E) and (G) the first derivatives of the F350nm/F330nm curve against the temperature gradient from which the apparent melting temperature (Tm) was determined for each sample. The experiment was performed twice.
(H–L) SL analogs binding PrKAI2d3 based on intrinsic tryptophan fluorescence. Plots of fluorescence intensity versus (+)-GR24 (H), (−)-GR24 (I), (+)-2′epi-GR24 (J), (−)-2′epi-GR24 (K), and (±)-GR24, (±)-4′-desmethyl-2′-epi-GR24, and (±)-3′-Me-GR24 (L). The change in intrinsic fluorescence was monitored (Supplemental Figure 10) and used to determine the apparent KD values. The plots represent the mean of two replicates, and the experiments were repeated at least three times. The analysis was performed in GraphPad Prism 7.05 software.
Under red light conditions, the hypocotyls of htl-3 and Atd14-1/htl-3 mutants are more elongated than those of Col-0 (Waters et al., 2012) (Figure 1F). Application of (±)-GR24 significantly shortened the hypocotyls of Col-0 and the htl-3 genotypes, but had no effect on those of the Atd14-1/htl-3 plants. This observation corroborates a previous report that Arabidopsis seedling photomorphogenesis is redundantly controlled by D14 and KAI2/HTL (Waters et al., 2012). Interestingly, only hypocotyls of the p35S::GFP-PrKAI2d3 and p35S::GFP-PrKAI2d3S98A lines with the highest protein levels were shorter than those of the htl-3 and Atd14-1/htl-3 mutants under control conditions. However, the hypocotyls were significantly shorter in all transformed lines after treatment with 1 μM (±)-GR24 (Figure 1F). In summary, these data indicate that PrKAI2d3 protein expression rescues the htl-3 mutant phenotypes and that the catalytic Ser98 is essential for germination but not for seedling photomorphogenesis.
A structure-activity relationship study reveals that P. ramosa responds efficiently to SL analogs and mimics with various structures
Because P. ramosa perceives many germination stimulants with highly divergent structures, various receptors may underlie this plasticity (Boyer et al., 2012, 2014). To link SL structural features with P. ramosa seed germination activity, we performed a structure-activity relationship (SAR) study using a rapid bioassay (Pouvreau et al., 2013).
First, we determined the sensitivity of P. ramosa toward several GR24 analogs with varying stereogenic centers (Figure 2A). The lowest EC50 (half maximal effective concentration) values were obtained with (+)-GR24 (6.5 pM) and (−)-2′-epi-GR24 (5.3 pM), whose stereochemistry corresponds to that of natural canonical SLs of the strigol-type and orobanchol-type series, respectively (Supplemental Figure 1A). By contrast, P. ramosa was approximately 100-fold less sensitive to (−)-GR24 and (+)-2′-epi-GR24, whose stereochemistry is not encountered in natural SLs (Figure 2B, Supplemental Figure 1A).
Second, we investigated whether substitutions on the D ring altered the responses of P. ramosa (Boyer et al., 2012). Surprisingly, both GR24 analogs without a methyl group on the D ring, (±)-4′-desmethyl-2′-epi-GR24 and (±)-4′-desmethyl-GR24, significantly stimulated P. ramosa seed germination (EC50 = 0.92 and 0.45 nM), although their EC50 values were 100-fold lower than those of (±)-GR24 (Figure 2C). Notably, none of these substituted GR24 analogs inhibited shoot branching, even at 1 μM, above which SL analogs begin to be toxic and are no longer considered bioactive (Boyer et al., 2012). By contrast, P. ramosa was less sensitive to (±)-3′-methyl-GR24 (EC50 = 2.6 nM), which harbors two methyl groups on the D ring (Figure 2C, Supplemental Figure 7, Supplemental Table 2) and is highly bioactive in repressing pea (Pisum sativum) shoot branching via RMS3/PsD14 (Boyer et al., 2012; de Saint Germain et al., 2016).
Finally, we analyzed the effect of ABC-ring fragment substitution on P. ramosa germination using profluorescent probes in which the ABC ring was replaced by a coumarine moiety (DiFMU) and one, two, or no methyl groups were added to the D ring (GC240, GC242, and GC486, respectively) (de Saint Germain et al., 2016). In another profluorescent probe, the ABC ring was substituted with the fluorescein moiety (Yoshimulactone green (YLG)) (Tsuchiya et al., 2015) (Supplemental Figure 1B). With the exception of DiFMU, all profluorescent probes stimulated P. ramosa germination, although their EC50 values were still 1000- to 10 000-fold lower than those of (±)-GR24 (Supplemental Figure 8A–8C, Supplemental Table 2).
Together, these results demonstrate that the stereochemistry of GR24 analogs is crucial for their bioactivity and determines the sensitivity of P. ramosa toward these germination-inducing substances. The sensitivity of P. ramosa toward desmethyl and 3′-methyl D ring derivatives highlights differences between it and other vascular plants, particularly with regard to signaling via D14 for shoot branching. In addition, the relative bioactivity of the profluorescent SL probes in the stimulation of broomrape germination confirms that ABC rings are not required for bioactivity (Boyer et al., 2014).
SL analogs and mimics interact with PrKAI2d3 according to their germination stimulation activity
The previous results led us to ask whether PrKAI2d3 has the ability to perceive such a large range of structurally divergent compounds and whether its interaction with these molecules is affected by mutations in the catalytic triad. We therefore expressed and purified the PrKAI2d3 and PrKAI2d3S98A proteins in vitro and assessed their ability to interact with SLs and other chemical mediators.
Interactions of PrKAI2d3 with SL analogs and profluorescent SL probes were analyzed with nano differential scanning fluorimetry (nanoDSF) by recording changes in tryptophan fluorescence (350 nm/330 nm ratio). In contrast to classic differential scanning fluorimetry (DSF) (Hamiaux et al., 2012), nanoDSF does not require a dye and highlights interactions that do not induce conformational changes. Analysis of the initial fluorescence ratios revealed that the four GR24 stereoisomers interacted with PrKAI2d3 according to their bioactivity (Figure 2D). However, only (+)-GR24 and (−)-2′-epi-GR24, the most bioactive analogs with natural configurations, induced an 8.5°C decrease in the PrKAI2d3 melting temperature (Figure 2E), consistent with ligand-mediated protein destabilization.
When GR24 analogs with various methyl groups on their D rings were used, all the analogs interacted with and destabilized PrKAI2d3, although their efficiencies were lower than that of (±)-GR24 and especially that of (±)-3′-Me-GR24 (Figure 2F and 2G). Similar shifts in melting temperatures of the PrKAI2d3 protein were observed with DSF, but without destabilization of the mutated PrKAI2d3S98A protein (Supplemental Figure 9). The bioactive profluorescent probe GC242, used as a racemic mixture or as separate pure enantiomers, also induced a shift in the PrKAI2d3 protein melting temperature (Supplemental Figure 8D–8I). These results suggest that enzymatic activity through Ser98 is required to destabilize the protein.
Next, we estimated PrKAI2d3 affinity for GR24 analogs using a tryptophan intrinsic fluorescence assay. Its affinity was higher for (+)-GR24 and (−)-2′-epi-GR24 (KD = 12.57 ± 2.85 μM and 66.99 ± 26.94 μM, respectively) than for (−)-GR24 and (+)-2′-epi-GR24 (KD = 696 ± 245 μM and 434 ± 593 μM, respectively), consistent with their bioactivity in P. ramosa germination (Figure 2H–2K, Supplemental Figure 10). The lower affinity for (±)-3′-Me-GR24 than for (±)-4′-desmethyl-2′-epi-GR24 and (±)-GR24 was consistent with their reduced bioactivity range (Figure 2L). However, the protein affinities were significantly lower (micromolar range) than the observed sensitivity of P. ramosa (picomolar range). Because similar patterns have been reported for S. hermonthica (Tsuchiya et al., 2015), we tested this apparent contradiction by investigating the enzymatic activity of the PrKAI2d3 protein.
PrKAI2d3 enzymatic activity is associated with SL hypersensitivity
First, we tried to visualize PrKAI2d3 hydrolase activity with a generic substrate, para-nitrophenyl acetate (p-NPA). Surprisingly, no hydrolytic activity was detected (Supplemental Figure 12), in contrast to that observed for other SL receptors treated with the same probe (de Saint Germain et al., 2016). When PrKAI2d3 was incubated with (+)-GR24 and (−)-2′-epi-GR24, cleavage activity was unambiguously observed by ultraperformance liquid chromatography (UHPLC)/UV diode array detector (DAD) analysis (Figure 3A). However, unlike AtD14 and AtKAI2, PrKAI2d3 did not cleave (−)-GR24 or (+)-2′-epi-GR24. Interestingly, a residual cleavage activity of PrKAI2d3S98A still occurred with all GR24 isomers but displayed no stereoselectivity, a result that may explain the partial complementation of htl-3 with PrKAI2d3S98A-overproducing constructs (Figure 1G). These data suggest that SL hydrolysis and subsequent signal transduction can occur without an intact catalytic triad.
Figure 3.
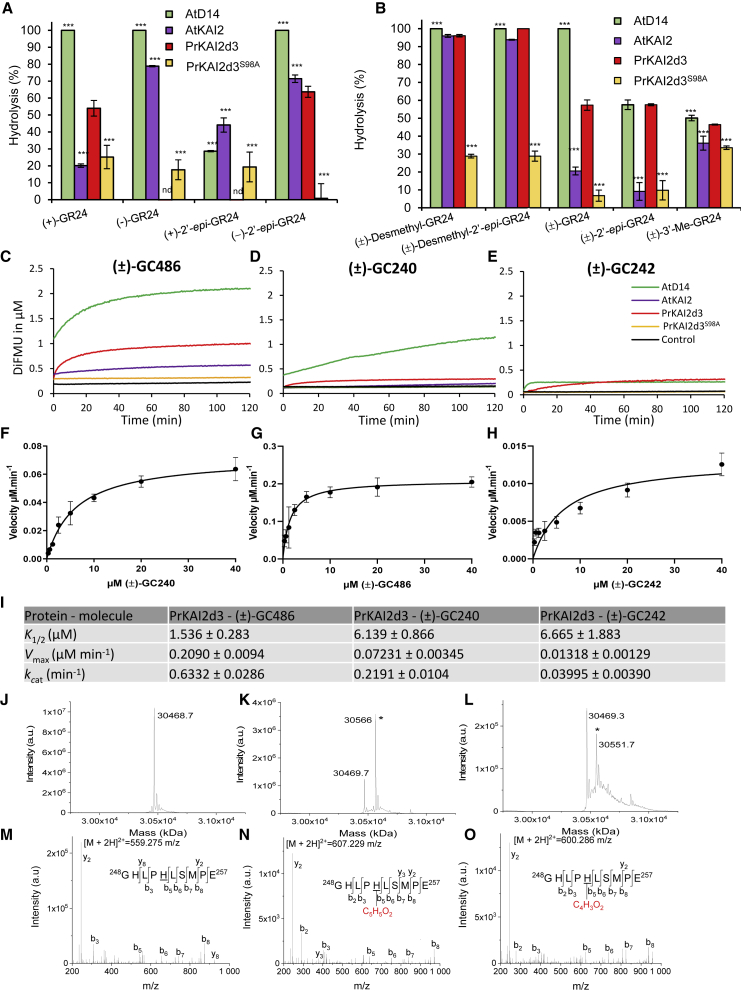
The PrKAI2d3 enzymatic activity improves the SL biological activity.
(A and B) Hydrolysis activity of GR24 isomers and analogs by various proteins. (+)-GR24, (−)-GR24, (+)-2′epi-GR24, and (−)-2′epi-GR24 (A) and (±)-GR24, (±)-4′-desmethyl-GR24, (±)-4′-desmethyl-2′-epi-GR24, and (±)-3′-Me-GR24 (B) at 10 μM were incubated with PrKAI2d3, PrKAI2d3S98A, AtD14, and AtKAI2 at 5 μM for 150 min at 25°C. UPLC-UV (260 nm) analysis was used to detect the remaining amounts of GR24 isomers and analogs. Bars represent the mean value of the hydrolysis rate calculated from the remaining GR24 isomers and analogs, taking into account the hydrolysis in the buffer alone (without protein sample) and quantified using (±)-1-indanol as an internal standard. Error bars represent the SD of three replicates (means ± SD, n = 3). nd, no hydrolysis detected. The asterisks indicate statistical significance from the PrKAI2d3 protein sample as measured by the Kruskal–Wallis test. ∗∗∗p ≤ 0.001 and p > 0.05.
(C–E) Enzymatic kinetics for PrKAI2d3, PrKAI2d3S98A, AtD14, and AtKAI2 proteins incubated with (±)-GC486 (C), (±)-GC240 (D), and (±)-GC242 (E). Progress curves during hydrolysis of the probes monitored (λem 460 nm) at 25°C with the use of 400 nM protein and 20 μM probes. The traces represent one of three replicates, and the experiments were repeated at least twice.
(F–H) Hyperbolic plot of the PrKAI2d3 pre-steady-state kinetics reaction velocity with (±)-GC486 (F), (±)-GC240 (G), and (±)-GC242 (H). The initial velocity was determined with profluorescent probe concentrations from 0.3 μM to 40 μM and with proteins at 400 nM. Error bars represent the SE of three replicates, and the experiments were repeated at least three times.
(I) Kinetics constants of probes toward PrKAI2d3. K1/2 and kcat are pre-steady-state kinetics constants for PrKAI2d3 with different profluorescent probes and represent the mean ± SE of three replicates.
(J–O) MS characterization of covalent PrKAI2d3-ligand complexes. On the left, deconvoluted electrospray mass spectra of PrKAI2d3 before and after addition of different ligands: (±)-GR24 (500 μM) and (±)-GC486 (500 μM). Peaks with an asterisk correspond to PrKAI2d3 covalently bound to a ligand (PrKAI2d3-ligand). The mass increments were measured for different PrKAI2d3-ligand complexes: 96.3 Da (±)-GR24 and 82.4 Da (±)-GC486. Ligand-modified amino acids were identified by nanoLC-MS/MS analyses after Glu-C proteolysis. On the right, fragmentation spectra of unmodified and different ligand-modified peptides. Labeled peaks correspond to the b and y fragments of the double-charged precursor ion displayed at the top. The histidine residue modified by different ligands is underlined.
For the interaction assays, we tested the hydrolysis of analogs with substitutions on the D ring. Desmethyl GR24 isomers were more efficiently hydrolyzed than GR24 by AtKAI2, PrKAI2d3, and PrKAI2d3S98A proteins (Figure 3B). In addition, these three purified proteins and AtD14 also displayed low, but significant, hydrolysis activity toward 3′-Me-GR24. These results indicate that PrKAI2d3 possesses an important hydrolysis capacity toward natural configuration-mimicking GR24, albeit to a lesser extent than AtD14.
Enzymatic kinetic characterization of the purified proteins was performed using bioactive profluorescent probes as substrates. Monitoring the DiFMU fluorescence revealed that PrKAI2d3 hydrolyzed (±)-GC240, (±)-GC242, and (±)-GC486 (Figure 3C–3E). For all probes, we observed a biphasic time course of fluorescence, consisting of a burst phase (also called the initial phase) followed by a plateau phase (or a slow phase for AtD14 incubated with (±)-GC240). In all cases, the plateau did not reach the maximum of the expected product (20 μM) but rather a product concentration closer to the protein concentration (0.4 μM) (Figure 3C–3E). This result implied that PrKAI2d3 may act as a single-turnover enzyme toward all GC probes tested, including GC486, which lacks the 3′-methyl group on the D ring and exhibits Michaelian cleavage kinetics with AtD14 and RMS3/PsD14 proteins (de Saint Germain et al., 2016). The S98A substitution in the catalytic triad drastically reduced (±)-GC240 and (±)-GC242 cleavage, although residual activity toward (±)-GC486 remained statistically significant. These observations confirmed that mutation of the catalytic triad does not fully abolish cleavage activity toward a compound without methyl on the D ring.
For single-turnover enzymes, we defined kcat as the rate constant of the pre-steady-state phase (initial phase) and K1/2 as the probe concentration with half maximal velocity (Vmax) (Figure 3G–3I). The similar K1/2 values of PrKAI2d3 with (±)-GC240 and (±)-GC242 (5.74 μM and 4.60 μM, respectively) confirmed the minor influence of the C3′ methyl chain on substrate affinity. However, differences in Vmax (0.072 M.min−1 and 0.013 M.min−1, respectively) indicated that the C3′ methyl chain reduces the catalytic activity, consistent with the bioactivities of these probes. Higher K1/2 values of PrKAI2d3 for (±)-GC240 than for (±)-GC486 (5.74 μM and 1.53 μM, respectively) and different Vmax values (0.072 M.min−1 and 0.209 M.min−1, respectively) highlighted the importance of the C4′ methyl chain for binding and catalytic affinity, supporting the results of the germination bioassays (Supplemental Figure 8A–8C).
To test the hypothesis that PrKAI2d3 forms a stable intermediate with the D ring, as previously demonstrated for other SL receptors (AtD14, PsD14/RMS3, ShHTL7, and D14) (de Saint Germain et al., 2016; Yao et al., 2016, 2017, 2018b), we incubated two bioactive ligands (±)-GR24 and probe (±)-GC486 with PrKAI2d3 at a pH of 6.8 and recorded mass spectrometry (MS) spectra under denaturing conditions. In all cases, a mass shift occurred corresponding to the D ring covalently bound to the protein (Figure 3J–3L) and specifically attached to His249 of the catalytic triad (Figure 3M–3O).
ITCs interact with PrKAI2d3 and generate a covalent adduct to the catalytic serine
Although PrKAI2d3 perceives structurally diverging SL analogs and mimics that stimulate P. ramosa seed germination, the specificity of the interaction between rapeseed and P. ramosa correlates with the parasite's ability to perceive ITCs, which differ greatly in structure from SLs (Auger et al., 2012). We therefore evaluated the ability of PrKAI2d3 to perceive ITCs. Seeds of P. ramosa were approximately 10 000-fold less sensitive to 2-PEITC and BITC than to (±)-GR24 (Figure 4A–4C). Investigation of the putative interactions between PrKAI2d3 and ITCs by nanoDSF revealed a small shift (1.2°C–2.0°C) in the PrKAI2d3 melting temperature in response to high ITC concentrations, a result that was further confirmed by classic DSF (Figure 4D and 4E, Supplemental Figure 13). These data indicated that ITCs interact with PrKAI2d3 with low affinity, corresponding with a stimulating potential on P. ramosa seeds lower than that of bioactive SLs (Auger et al., 2012) and their analogs. Finally, the apparent melting temperatures of PrKAI2d3 with BITC and 2-PEITC (44.3°C and 45.1°C, respectively) varied from those obtained with (±)-GR24 (36.9°C), suggesting that ITCs may induce a conformational change that differs from GR24-induced destabilization.
Figure 4.
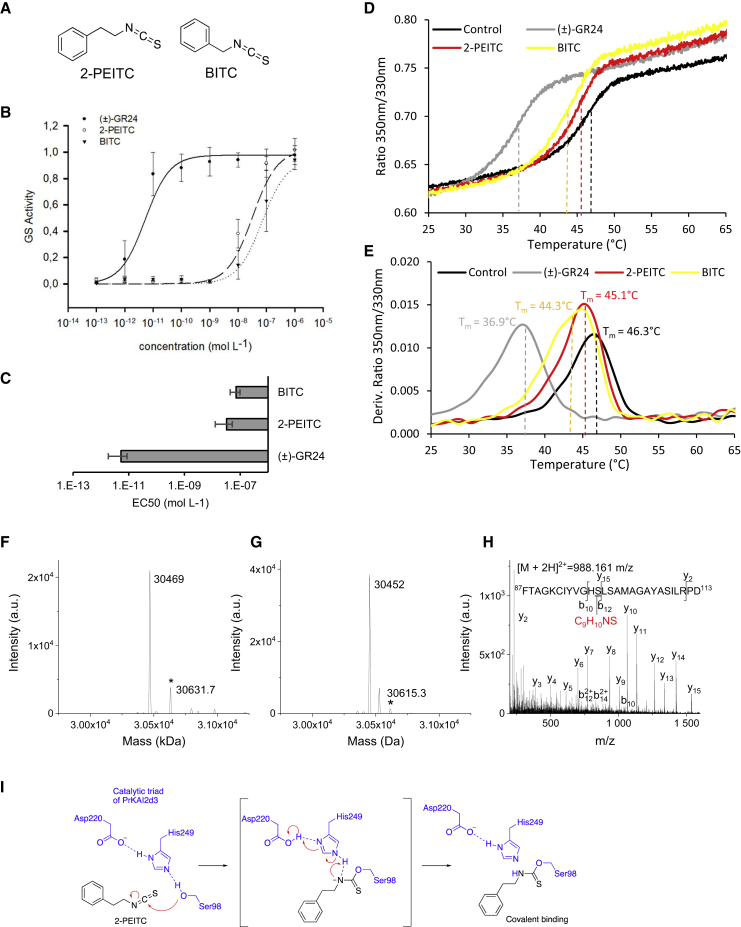
Isothiocyanate germination stimulants are perceived by PrKAI2d3.
(A) Structure of isothiocyanate.
(B and C) Modeled curves of dose-response germination stimulant activities (B) and EC50(C). Data are presented ± SE.
(D and E) Thermostability of PrKAI2d3 at 10 μM in the absence of ligand or in the presence of various ligands at 100 μM analyzed by nanoDSF. Changes in fluorescence (F350nm/F330nm ratio) with temperature (D) and first derivatives of the F350nm/F330nm curve against the temperature gradient from which the apparent melting temperature (Tm) was determined for each sample (E). The experiment was performed twice.
(F–H) MS characterization of covalent PrKAI2d3-ligand complexes. Deconvoluted electrospray mass spectra of PrKAI2d3 (F) and PrKAI2d3S98A(G) after addition of the 2-PEITC ligand (1 mM). Peaks with an asterisk correspond to PrKAI2d3 covalently bound to a ligand (PrKAI2d3-ligand). The mass increments were measured for the PrKAI2d3-2-PEITC complex, 162.7 Da. Ligand-modified amino acids were identified by nanoLC-MS/MS analyses after Glu-C proteolysis. (H) Fragmentation spectra of unmodified and different ligand-modified peptides. Labeled peaks correspond to the b and y fragments of the double-charged precursor ion displayed at the top. The PEITC ligand-modified serine residue is underlined.
(I) Perception mechanism of PEITC.
Because ITCs react easily with nucleophile functions (Satchell et al., 1990), we hypothesized that the interaction of PrKAI2d3 with ITCs may trigger the formation of a covalent adduct. Indeed, a mass shift was detected that correlated with the covalent binding of 2-PEITC to the protein (Figure 4F). After digestion of the PrKAI2d3-2-PEITC complex, the 2-PEITC attachment was localized to a peptide that corresponded to amino acids 87–113 of PrKAI2d3. Tandem MS (MS/MS) data revealed that the 2-PEITC attachment could be on His97 or on the catalytic Ser98 (Figure 4H). Incubation of 2-PEITC with PrKAI2d3S98A allowed us to conclude that Ser98 is the major site for 2-PEITC attachment (Figure 4G). These results hint at a perception mechanism for ITC in which the Ser98 hydroxyl group reacts with isothiocyanate to generate a PrKAI2d3-attached carbamothioate (Figure 4I). However, no 2-PEITC attachment was detected on His249-containing peptides of the catalytic triad. Finally, it is noteworthy that the germination of Arabidopsis Col-0, htl-3, and the three p35S::GFP-PrKAI2d3 complemented lines was insensitive to 2-PEITC (data not shown).
Overall, PrKAI2d3 acts as a receptor for both SL-like molecules and ITCs, both of which are germination stimulants for P. ramosa. These results demonstrate that further research into potential chemical interactors is achievable and may enable the design of control methods.
The search for small-molecule interactors of PrKAI2d3 identified D-OH as a simple and efficient germination stimulant for P. ramosa with a bioactivity similar to that of ITCs
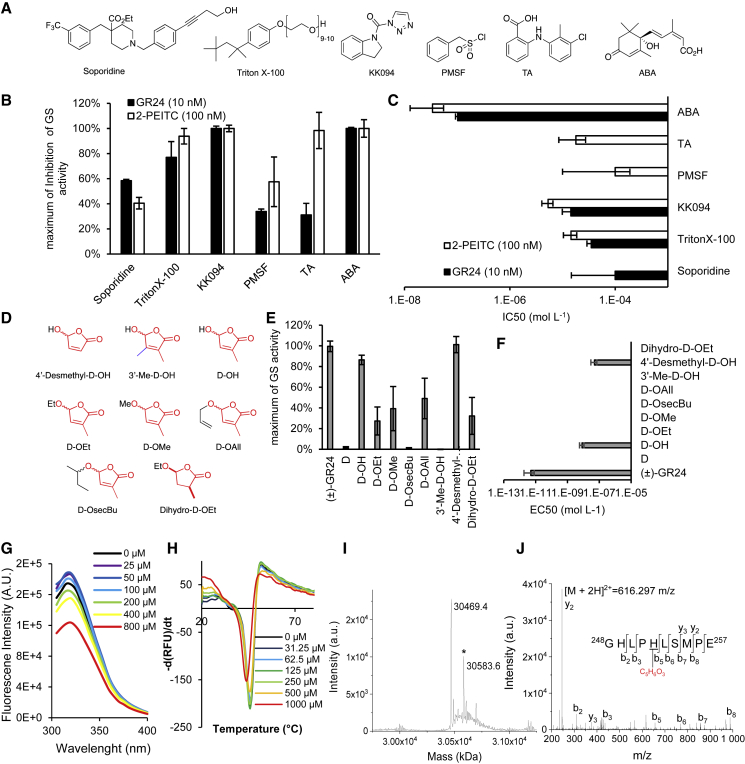
Recently, synthetic inhibitors of D14 and ShHTL7 (the best-characterized SL receptor in S. hermonthica) (Waters, 2019) have been proposed. These include soporidine, KK094, Triton X, and tolfenamic acid (TA) (Figure 5A), all of which are structurally unrelated to SLs. We evaluated the putative inhibitory effects of these compounds, as well as those of the common serine protease inhibitor phenylmethylsulfonyl fluoride (PMSF), on the germination of P. ramosa seeds treated with 10 nM (±)-GR24 or 100 nM 2-PEITC. P. ramosa seeds were clearly hyposensitive to each of these compounds compared with the results obtained in S. hermonthica, and maximum inhibition was obtained at very high concentrations in some instances (Figure 5B and 5C; Supplemental Table 3). Indeed, all half maximum inhibitory concentrations (IC50) fell above the IC50 of abscisic acid (ABA; 100 nM [GR24] and 34 nM [2-PEITC]), a known inhibitor of P. ramosa germination (Zehhar et al., 2002; Lechat et al., 2015). These data indicate that the use of germination inhibitors intended for Striga is relatively irrelevant and unsuitable for P. ramosa biological control.
Figure 5.
Identification of antagonists and agonists for P. ramosa seed germination.
(A) Chemical structures of inhibitors of D14 protein and ABA.
(B and C) Comparison of the inhibitory effect on P. ramosa germination stimulation (GS) induced by GR24 (10 nM) or 2-PEITC (100 nM). Maximum inhibition (B) and IC50(C).
(D) Structure of D-OH rings and derivatives.
(E and F) GS activity of D-OH and its derivatives on P. ramosa seeds. Maximum GS activity (E) and EC50(F).
(G) D-OH binding of PrKAI2d3, based on intrinsic tryptophan fluorescence. Plots of fluorescence intensity versus probe concentrations. The change in intrinsic fluorescence was monitored and used to determine the apparent KD values. The plots represent the mean of two replicates, and the experiments were repeated at least three times. The analysis was performed with GraphPad Prism 7.05 software.
(H) Biochemical analysis of the interaction between PrKAI2d3 at 10 μM and D-OH at various concentrations by DSF. Each line represents the average protein melt curve for two technical replicates, and the experiment was performed twice.
(I and J) MS characterization of covalent PrKAI2d3-D-OH complexes. (I) Deconvoluted electrospray mass spectrum of PrKAI2d3 after addition of the D-OH ligand (5 mM). Peaks with an asterisk correspond to PrKAI2d3 covalently bound to D-OH. The mass increments were measured for the PrKAI2d3-D-OH complex, 114.2 Da. Ligand-modified amino acids were identified by nanoLC-MS/MS analyses after Glu-C proteolysis. (J) Fragmentation spectra of unmodified and different ligand-modified peptides. Labeled peaks correspond to the b and y fragments of the double-charged precursor ion displayed at the top. The D-OH ligand-modified histidine residue is underlined.
We therefore looked for specific PrKAI2d3 agonists with the potential for large-scale biocontrol. A serious issue in the search for simple and inexpensive SL analogs and mimics is their low stability owing to the connection of the D ring to a leaving group. As alternatives, molecules have been proposed that carry the D ring only, as it is essential for the high bioactivity of SLs (Takahashi and Asami, 2018). We specifically evaluated the effect of a set of butenolides that contained the D ring (D-OR) only on P. ramosa germination (Figure 5D–5F, Supplemental Figure 14). D-OsecBu and 3′-Me-D-OH were completely inactive, and the P. ramosa sensitivity toward D-OAll, D-OMe, D-OEt, and dihydro-DOEt was low, with EC50 values in the micromolar range. By contrast, 4-Hydroxy-2-methylbut-2-en-4-olide (D-OH) and 4′-desmethyl-D-OH were approximately 10-fold more active than 2-PEITC (Supplemental Table 2). Although D-OH is bioactive in rice (Oryza sativa) at a high concentration (50 μM) (Nakamura et al., 2013), direct injection of D-OH and 3′-Me-D-OH (100 μM) in the stem had no effect on branching control in pea (Supplemental Figure 15).
To determine whether D-OH sensitivity was specific to P. ramosa PrKAI2d3, we reexamined the germination-stimulating activity of D-OH on S. hermonthica. In contrast to previous results (Johnson et al., 1976; Pepperman et al., 1982), D-OH induced S. hermonthica germination (EC50 = 1.2 μM). Moreover, in Arabidopsis lines expressing GFP fused to ShHTL7, the known S. hermonthica SL receptor (Tsuchiya et al., 2015; Xu et al., 2018) was induced by (±)-DOH (EC50 = 3.1 μM) (Supplemental Figure 16, Supplemental Table 1). In summary, our study confirms the perception of D-OH by SL receptors in root-parasitic plants.
DSF and intrinsic fluorescence analyses revealed that D-OH and PrKAI2d3 interacted at high concentrations (Figure 5G and 5H). Incubation of D-OH with PrKAI2d3 at pH 6.8 and MS spectra recorded under denaturing conditions revealed a mass shift corresponding to the covalent binding of the D ring to the protein (Figure 5I). The D-ring attachment could be localized to His249 of the catalytic triad, similar to that observed for the SL analogs (Figure 5J). The observed shift of 114.2 Da (Figure 5I), contrasting with the shift of 96.3 Da observed with (±)-GR24 (Figure 3K), led us to propose a mechanism of complex formation (Supplemental Figure 14F). In this hypothetical mechanism, His247 directly attacks the α,β unsaturated system in D-OH. Unlike the interaction with GR24, this mechanism would not implicate the action of Ser98 because the complex is observed with the mutant protein PrKAI2d3S98A (Supplemental Figure 14E). The ability of PrKAI2d3 to interact with D-OH—as well as various SL analogs, SL-like compounds, and ITCs—highlights the plasticity that permits it to exert significant biological activity through interaction with different structures and emphasizes that all SL hydrolysis products can act as germination stimulants of P. ramosa.
Discussion
Obligate parasitic weeds require host-derived signals to germinate and wither their hosts long before they emerge from the soil, arguably making the early stages of the parasitic life cycle a much better target for control strategies than the later stages (Vurro et al., 2016). An important prerequisite for the design of such methods is an in-depth understanding of how parasites perceive germination stimulants.
In this study, we identified five KAI2 genes in the parasitic plant species P. ramosa. However, we acknowledge that this is arguably an underestimate, as these sequences were obtained from the P. ramosa reference transcriptome (Goyet et al., 2017). It is now necessary to search exhaustively for all KAI2 genes in the forthcoming genomic sequences, which will also help to refine the KAI2 phylogeny in this species and within the Orobanchaceae family (Bythell-Douglas et al., 2017; Conn et al., 2015). We demonstrated that PrKAI2d3 provides P. ramosa with hypersensitivity to rhizosphere SLs, mainly due to its enzymatic activity. Enzymatic data with GR24 analogs [(+)-GR24 and (−)-2′-epi-GR24] and GC probes suggest that PrKAI2d3 acts as a single-turnover enzyme toward profluorescent probes and SL analogs. PrKAI2d3 cleaves these probes and analogs to form a covalent complex between the D ring and the catalytic histidine, as previously described for AtD14 and RMS3, the receptors of SL as a hormonal signal (de Saint Germain et al., 2016). However, this model has recently been challenged (Seto et al., 2019) by the finding that an AtD14 mutant for the catalytic Asp residue can transduce the SL signal, although it is unable to cleave endogenous SL. Hence, it has been concluded that SL cleavage is not required for hormonal SL signaling. Indeed, it is possible that certain SL analogs can be perceived independently of enzymatic activity. We showed that GR24 analogs with non-natural stereochemistry [(−)-GR24 and (+)-2′-epi-GR24] are not cleaved by PrKAI2d3. These uncleaved molecules stimulate P. ramosa seed germination 100-fold less efficiently than GR24 stereoisomers with a natural stereochemistry (Figure 3). Moreover, the affinity (KD) of PrKAI2d3 toward the most bioactive SL analogs, (+)-GR24 and (−)-2′-epi-GR24, is in the micromolar range, several orders lower than the P. ramosa seed sensitivity recorded in the germination bioassays. The KD values for (+)-GR24 and (−)-2′-epi-GR24, which reflect simple binding to the receptor, do not explain this hypersensitivity. Similar differences between in vitro and in vivo analyses were observed for the S. hermonthica SL receptor ShHTL7 (Tsuchiya et al., 2015). Like Striga, P. ramosa may use a similar mechanism of SL perception based on recognition by the active binding pocket, as observed in D14 and its orthologs. However, the hypersensitivity of root-parasitic plants for SLs is unique and could suggest different mechanisms for the perception of hormonal and rhizospheric SLs. Nevertheless, we propose that the SL hypersensitivity of this SL receptor is due to its enzymatic properties, as previously proposed for the femtomolar-range germination stimulant sphynolactone-7 (Uraguchi et al., 2018), which is also aSL mimic.
Interestingly, enzymatic activity is not completely abolished in the PrKAI2d3S98A protein. Significant residual activity is also present in the DAD2S96A protein (Lee et al., 2020). The characterization of a serine protease enzyme in the 1980s demonstrated that the substitution of a catalytic residue by an alanine does not fully abolish enzyme activity (Carter and Wells, 1988). This observation could explain the partial complementation of the htl-3 mutant with constructs that overproduce PrKAI2d3S98A. A similar proposition could also explain why Arabidopsis that overexpress AtD14 with a mutated Asp catalytic residue is able to transduce a branching inhibitor signal, especially the still-untested natural SLs present in Arabidopsis (i.e., non-canonical SLs). However, the His covalent adduct was not detected after incubation of (±)-GR24 with the PrKAI2d3S98A protein (Supplemental Figure 14F), which could also suggest that both enzyme activity and His modification are not critical for SL signal transduction. Notably, p-NPA did not efficiently demonstrate the enzymatic activity of PrKAI2d3 compared with profluorescent SL mimics, pinpointing the weakness of using generic or inappropriate substrates as controls.
Moreover, PrKAI2d3 displays high plasticity that allows it to bind modified SLs such as desmethyl-GR24 isomers or many SL mimics, but in contrast to RMS3 and AtD14, it perceives these compounds similarly to intact SLs. Likewise, desmethyl-YLG was bioactive in Arabidopsis via AtKAI2 (Yao et al., 2018a). Ligand-mediated protein destabilization is not required to perceive SL analogs, but it considerably improves the sensitivity to natural SLs. PrKAI2d3 acts as a single-turnover enzyme for compounds with a D ring that lack a methyl group at the C4′ position, but a methyl group at the C-3′ position led to low interaction with PrKAI2d3. This result is consistent with the low P. ramosa germination stimulation activity of 3′-methyl analogs and mimics, and it corroborates studies conducted in Orobanche cumana, Orobanche minor, P. aegyptiaca, and S. hermonthica (Boyer et al., 2014; Jamil et al., 2019). In other words, P. ramosa may be sensitive to D-ring-modified SL and may perceive diverse SL-related compounds as putative SL degradation products (Yamauchi et al., 2018).
In addition, we established that ITCs, other known germination stimulants, interact with the PrKAI2d3 protein by forming a covalent complex. In P. ramosa, PrKAI2d3 can be considered a germination stimulant receptor for different chemicals, including SLs, SL derivatives and mimics, and also ITCs with completely different structures. Thus, P. ramosa has seemingly optimized its germination capacity by enhancing its ability to perceive various chemical mediators emitted by host plants. The enzymatic activity-dependent ITC perception mechanism suggests that the active catalytic triad may have been conserved to perceive not only SLs but also other germination stimulants that are not yet characterized for P. ramosa. On the other hand, 2-PEITC does not trigger Arabidopsis seed germination, even when complemented with PrKAI2d3. This suggests that Arabidopsis does not possess the necessary machinery for the perception or transduction of ITC signals.
SLs are very unstable in the soil, especially under basic conditions that produce ABC=CHOH and D-OH derivatives. To date, all SL analogs and mimics possess a D ring connected to an ABC mimic by a hydrolysable function. SL agonist treatments developed to date induce the suicide germination of parasitic plant seeds (Zwanenburg and Pospíšil, 2013). Nevertheless, these agonists are too unstable to be effective under field conditions, even when promising chemicals are designed and applied for S. hermonthica (Uraguchi et al., 2018). In addition, cultures of P. ramosa-infested rapeseed occur mainly in slightly basic soils (Johnson et al., 1981), favoring the formation of ITCs (Auger et al., 2012) and complicating the use of SL mimics as a suicidal germination strategy, especially because of their instability. In sharp contrast, P. ramosa is highly sensitive to D-OH or DesMe-D-OH. As for D-H, no germination activity was detected, and the hemiacetal function seems very important for the interaction with PrKAI2d3. Moreover, the residual bioactivity of D-OR (R ≠ H) may be explained by the hydrolysis of the R group during the bioassay, leading to D-OH. D-OH and DesMe-D-OH are especially interesting for translational research, because both compounds are small, easily synthesized compared to synthetic SLs and SL mimics, and not subject to rapid degradation. As such, D-OH and DesMe-D-OH seem to be promising chemicals for suicide germination in the field; higher active concentrations can compensate for their lower germination activity compared with SL mimics. Moreover, D-OH has the great advantage of being a natural product. The use of SL mimics, in which the portion equivalent to the ABC moiety is typically xenobiotic, increases the risk of potential toxicity or environmental pollution. In contrast to previous studies (Johnson et al., 1976; Pepperman et al., 1982), we found that D-OH is bioactive as a witchweed seed germination stimulant and that it could possibly be used for S. hermonthica biocontrol.
Here, we identified germination inhibitors and demonstrated the involvement of α/β-hydrolases in germination stimulant perception, providing an alternative approach for fighting P. ramosa (Yoneyama, 2016). Novel, more active inhibitors must be designed to control P. ramosa infection without negative effects on host plants and arbuscular mycorrhizal fungi. For this purpose, the identification of the SL receptor(s) in these fungi will undoubtedly be an important milestone. The use of specific inhibitors should be combined with other approaches for integrated management strategies (Pickett et al., 1997), such as chemical suicidal germination with compounds such as D-OH to decrease the parasitic plant seed bank in the soil.
Methods
Preparation of GR24 isomers, probes, and other ligands
See Supplemental methods.
Expression and purification
AtD14 and AtKAI2 were purified and expressed with cleavable GST tags as described previously (de Saint Germain et al., 2016). For PrKAI2d3 expression, the coding sequences from P. ramosa were amplified by PCR using a seed-derived cDNA template and specific primers (Supplemental Table 4) containing a protease cleavage site for tag removal and were subsequently cloned into the pGEXT-4T-3 expression vector. The PrKAI2d3 and PrKAI2d3S98A proteins were purified and expressed as described above.
Site-directed mutagenesis
Site-directed mutagenesis experiments were performed on pGEX-4T-3-PrKAI2d3 (Supplemental Table 4) using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene). Mutagenesis was verified by systematic DNA sequencing.
Plant materials and growth conditions
For pea (P. sativum) and A. thaliana, see Supplemental methods.
Two batches of parasitic plant seeds were used. A population of P. ramosa (L.) Pomel seeds associated with genetic group 1 (P. ramosa 1) was collected in Saint-Martin-de-Fraigneau (France) from broomrape-parasitized winter rapeseed (Brassica napus L.) in 2014 and 2015 (Stojanova et al., 2019). Seeds of S. hermonthica (Delile) Benth. (Sudan, 2007) were provided by Lukas Spichal (Olomouc, Czech Republic). Seeds were surface sterilized and conditioned as described previously (Pouvreau et al., 2013) (darkness; 21°C for P. ramosa and 30°C for S. hermonthica).
Pea shoot branching assay
See Supplemental methods.
Cloning and generation of transgenic lines
For all GFP fusion constructs, cloning was performed by Gateway recombination (Thermo Fisher Scientific). The open reading frame (ORF) of PrKAI2d3 was amplified from P. ramosa cDNA with iProof High-Fidelity DNA Polymerase (Bio-Rad) and the Gateway-specific primers PrKAI2d3_B2_FW and PrKAI2d3_B3_Rev_STOP. The PCR product flanked by attB sites was cloned into pDONR P2R-P3 using BP Clonase II enzyme mix (Invitrogen). The resulting entry vector was used to clone the genes into the destination vector pK7m34GW under the control of the 35S promoter and N-terminally fused with GFP using LR Clonase II Plus enzyme mix (Invitrogen). To construct the catalytic site mutant, pDONR P2R-P3-PrKAI2d3 was mutated with the QuikChange II Site-Directed Mutagenesis Kit (Agilent). The generated clones were checked by sequencing. All primers used for cloning are listed in Supplemental Table 4.
Western blotting
Total protein was extracted from 5-day-old seedlings that had been exposed to white light for 3 h, transferred to darkness for 21 h, and exposed to continuous red light for 4 days. Protein concentrations were determined by the Bradford assay (Bio-Rad). Thirty micrograms of protein extract were separated by SDS-PAGE, transferred onto polyvinylidene fluoride (PVDF) membranes, and treated with horseradish peroxidase (HRP)-conjugated antibodies against GFP (anti-GFP-HRP, 1:10 000, Miltenyi Biotec) or anti-tubulin (mouse monoclonal, 1:10 000, Sigma-Aldrich) and HRP-conjugated anti-mouse antibodies (rabbit polyclonal, 1:10 000, Abcam). The blots were visualized using the Western Lightning Plus Enhanced Chemiluminescence kit (PerkinElmer) and the X-Doc System (Bio-Rad). Precision Plus Protein Dual Color Standards (Bio-Rad) were used as protein size markers.
Arabidopsis hypocotyl elongation assays
Arabidopsis seeds were surface sterilized by consecutive treatments of 70% (v/v) ethanol with 0.05% (w/v) SDS for 5 min and 95% (v/v) ethanol for 5 min, then sown on half-strength Murashige and Skoog medium (Duchefa Biochemie) containing 1% (w/v) agar and supplemented with 1 μM (±)-GR24 (0.01% [v/v] DMSO) or 0.01% (v/v) DMSO only (control). Seeds were stratified at 4°C for 2 days in the dark, exposed to white light for 3 h, transferred to darkness for 21 h, and exposed to continuous red light for 4 days at 21°C. Plates were photographed, and hypocotyl lengths were quantified using ImageJ (http://imagej.nih.gov/ij/).
Arabidopsis germination assays
Arabidopsis seeds were after-ripened for at least 6 weeks before use. Surface-sterilized seeds were incubated in incubation solution (1 mM HEPES buffer; pH 7.5) at a ratio of 10 mg of seeds per milliliter (Lechat et al., 2015). Fifty microliters of seeds (∼20–25 seeds) were distributed on 96-well plates, and 10 μL of germination stimulant [10 μM (+)-GR24, 10 μM (−)-GR24, and 0.1% (v/v) DMSO (control) or 10-fold concentrated D-OH] were added. The final volume was adjusted to 100 μL with incubation solution. Plates were incubated for either 5 days at 25°C in the dark (Brun et al., 2019) or 7 days at 32°C–34°C under constant illumination at a quantum irradiance of 10 μmol m−2 s−1 (Toh et al., 2012; Toh et al., 2015). A seed was considered to have germinated when the radicle protruded from the seed coat.
Germination stimulation activity assay on the seeds of root-parasitic plants
The germination stimulation activity of chemicals on the seeds of parasitic plants was determined as described previously (Pouvreau et al., 2013). Chemicals were suspended in DMSO at 10 mmol L−1, diluted with water to 1 mmol L−1 (water/DMSO; v/v; 9:1), and further diluted from 1 × 10−5 mol L−1 to 1 × 10−12 mol L−1 with water/DMSO (v/v; 9:1). Each compound was applied to conditioned parasitic seeds at a concentration range from 10−13 to 10−6 mol L−1 (water/DMSO; 99:1). One percent (v/v) DMSO was used as a negative control (seed germination <1%), and 1 μmol L−1 (±)-GR24 was used as a positive control (seed germination 72%–87% and 50%–65% for P. ramosa 1 and S. hermonthica, respectively). To avoid variation related to sterilization events, germination percentages are reported as a ratio relative to the positive control (1 μmol L−1 [±]-GR24) included in each germination assay. Each dilution and germination assay was repeated at least three times. For each compound tested, dose-response curves (germination stimulation activity = f(c); germination stimulant activity relative to 1 μmol L−1 (±)-GR24; c, concentration (mol L−1); EC50; and maximum germination stimulant activity) were determined with a four-parameter logistic curve computed with SigmaPlot 10.0.
Enzymatic degradation of GR24 isomers by purified proteins
See Supplemental methods.
Enzymatic assays with profluorescent probes and p-nitrophenyl acetate
These assays were performed as described previously (de Saint Germain et al., 2016) using a TriStar LB 941 Multimode Microplate Reader (Berthold Technologies).
Protein melting temperatures
For DSF, see Supplemental methods.
nanoDSF
Proteins were diluted in PBS (100 mM phosphate, pH 6.8, 150 mM NaCl) to a concentration of ∼10 μM. Ligands were tested at a concentration of 200 μM. The intrinsic fluorescence signal was measured as a function of increasing temperature using a Prometheus NT.48 fluorimeter (NanoTemper Technologies) with 55% excitation light intensity and a 1°C/min temperature ramp. Analyses were performed on capillaries filled with 10 μL of the respective samples. Intrinsic fluorescence signals expressed by the 350 nm:330 nm emission ratio, which increases as proteins unfold, were plotted as a function of temperature (Figures 2D, 2F, and 4D). The plots show one of three independent data collections performed for each protein.
Intrinsic tryptophan fluorescence assays and determination of the dissociation constant KD
These experiments were performed as described previously (de Saint Germain et al., 2016) using a Spark Multimode Microplate Reader (Tecan).
Direct electrospray ionization MS under denaturing conditions and localization of the fixation site of ligands on PrKAI2d3
See Supplemental methods.
Phylogenetic analyses
Phylogenetic analyses were performed on 32 D14 and KAI2 sequences, including five P. ramosa sequences obtained in the present study, as well as previously described sequences from P. aegyptiaca, S. hermonthica, P. sativum, A. thaliana, and O. sativa (Arite et al., 2009; Waters et al., 2012; Conn et al., 2015; Toh et al., 2015; de Saint Germain et al., 2016; Carbonnel et al., 2019). The D14 and KAI2 proteins and nucleotide sequences (Supplemental Data 1 and 2, respectively) were aligned using MAFFT (multiple alignment using fast Fourier transform) (Katoh et al., 2019) with the G-INS-i iterative refinement alignment method (Katoh et al., 2005). Sequence alignments (Supplemental Data 3 and 4) were manually trimmed to remove gaps at either end, producing final protein and nucleotide data sets of 262–264 amino acids and 786–792 nucleotides, respectively (Supplemental Data 5 and 6). The α/β hydrolase RbsQ from Bacillus subtilis was used as an outgroup because of its high similarity to KAI2 and D14 and its possession of a conserved catalytic triad (Waters et al., 2012). ML analyses were performed in RAxML (Stamatakis, 2014) with 1000 bootstrap replicates. The best ML tree for the amino acid sequences was inferred with the PROT-GAMMA model and the WAG substitution matrix, and the best ML tree for the nucleotide sequences was inferred with the general time reversible (GTR) model with the gamma rate of heterogeneity among sites. The percentage of trees in which the associated taxa clustered together was indicated next to the branches. The resulting consensus amino acid and nucleotide trees were drawn to scale using MEGA7 (Kumar et al., 2016) with branch lengths representing the number of substitutions per site.
Modeling
P. ramosa KAI2 protein sequences were modeled using the SWISS-MODEL server (http://swissmodel.expasy.org/) (Waterhouse et al., 2018). Models were generated using chain A of the Apo form of the Arabidopsis KAI2 structure (PDB: 4JYP) as a template (Guo et al., 2013). Protein structure figures were generated with PyMOL, and cavities within homology models were visualized using surface mode on the setting “cavities and pockets culled.” Pocket sizes were calculated using the CASTp 3.0 server (Tian et al., 2018) with a probe radius of 1.4 Å. The reported pocket sizes were the Connolly solvent excluded surface volumes of the catalytic pocket.
Expression data
Seeds of A. thaliana complemented lines were imbibed for 24 h in a growth solution (1 mM HEPES buffer, pH 7.5, 0.1 mM Preservative Plant Mixture in sterile water) and treated for 6 h with 10 μM (±)-GR24 or a mock solution. Seeds were dried using a paper tissue, snap-frozen in liquid nitrogen, and ground to a fine powder with a mortar and pestle. RNA was extracted and purified with the RNeasy Plant Mini Kit (Qiagen). Genomic DNA was removed by DNase treatment, and the samples were purified by ammonium acetate (5 M final concentration) precipitation. The iScript cDNA Synthesis Kit (Bio-Rad) was used to reverse transcribe RNA. SYBR Green detection was used for qRT-PCR on a Light Cycler 480 system (Roche). Reactions were performed in triplicate on a 384-well plate in a total volume of 5 μL with a cDNA fraction of 10%. Seeds of P. ramosa were imbibed in the growth solution (see above) for 7 days, then treated for 6 h with 0.1 μM (±)-GR24, 0.1 μM 2-PEITC, or a mock solution. Seeds were dried using a paper tissue, snap-frozen in liquid nitrogen, and ground to a fine powder with a mortar and pestle. RNA was extracted and purified with the PureLink RNA Mini Kit (Invitrogen). Genomic DNA was removed by DNase treatment, and the samples were purified using the RNeasy PowerClean Pro CleanUp Kit (Qiagen). Three hundred nanograms of total RNA were reverse transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). SYBR Green detection was used for qRT-PCR on a CFX Connect Real-Time PCR Detection System (Bio-Rad). Reactions were performed in triplicate on a 96-well plate in a total volume of 20 μL with a cDNA fraction of 25%. Cycle threshold values were obtained and analyzed by the 2−ΔΔCT method (Livak and Schmittgen, 2001). For A. thaliana, the values from three biological replicates and three technical replicates were normalized against those of the seed-specific housekeeping gene At4g12590 (Dekkers et al., 2011). For P. ramosa, the values from three biological replicates and two technical replicates were normalized against those of the EF1-α gene (Lechat et al., 2012). The normalized values were analyzed according to a published model (Rasmussen et al., 2012) using the mixed model procedure (SAS Enterprise).
Statistical analyses
Because deviations from normality have been observed for axillary bud length, hypocotyl length, and germination after SL treatments, the Kruskal–Wallis test was used to test for significant differences between treatment with one compound and treatment with another using R Commander software (version 1.7–3) (Fox, 2016).
Funding
This work was supported by the Institut Jean-Pierre Bourgin's Plant Observatory technological platforms and has benefited from the facilities and expertise of the I2BC proteomic platform (Proteomic-Gif, SICaPS) supported by Infrastructures en Biologie Santé et Agronomie, Ile de France Region, Plan Cancer, CNRS, and Paris-Sud University. The CHARM3AT Labex program (ANR-11-LABX-39) is also acknowledged for its support. A.d.S.G. is the recipient of an AgreenSkills award from the European Union in the framework of the Marie-Curie FP7 COFUND People Program and a fellowship from Saclay Plant Sciences (ANR-17-EUR-0007). A.J. is indebted to the Research Foundation Flanders for a Structural Basic Research fellowship (project 1S15817N) and for a travel grant in the framework of a Tournesol fellowship (project VS04418N).
Author contributions
A.d.S.G., J.-B.P., and F.-D.B. designed the research. G.C. designed and synthesized the profluorescent probes. V.Se. and F.-D.B. synthesized the chemicals. A.d.S.G., A.J., and E.B. produced and purified the proteins. A.d.S.G. and A.J. characterized the proteins and performed the kinetic experiments. A.J., G.B., J.-B.P., and F.-D.B. performed the plant experiments. D.C. performed the mass spectrometry experiments. A.d.S.G., V.St., and F.-D.B. performed the high-pressure liquid chromatography analyses and separations. A.d.S.G., A.J., G.B., J.-B.P., L.B., D.C., K.G., P.S., S.W., S.G., P.D., and F.-D.B. analyzed the data. A.d.S.G., A.J., G.B., J.-B.P., and F.-D.B. wrote the paper. All authors critically revised the manuscript.
Acknowledgments
We thank J.-P. Pillot for the pea plant bioassays, Bruno Baron (Institut Pasteur, France) for access to and help with the nanoDSF experiments, Thomas Larribeau for technical assistance, and Catherine Rameau, Sandrine Bonhomme, and Martine De Cock for their comments on the manuscript. The authors declare no conflict of interest.
Published: February 5, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Arite T., Umehara M., Ishikawa S., Hanada A., Maekawa M., Yamaguchi S., Kyozuka J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009;50:1416–1424. doi: 10.1093/pcp/pcp091. [DOI] [PubMed] [Google Scholar]
- Auger B., Pouvreau J.-B., Pouponneau K., Yoneyama K., Montiel G., Le Bizec B., Yoneyama K., Delavault P., Delourme R., Simier P. Germination stimulants of Phelipanche ramosa in the rhizosphere of Brassica napus are derived from the glucosinolate pathway. Mol. Plant Microbe Interact. 2012;25:993–1004. doi: 10.1094/MPMI-01-12-0006-R. [DOI] [PubMed] [Google Scholar]
- Besserer A., Bécard G., Jauneau A., Roux C., Sejalon-Delmas N. GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol. 2008;148:402–413. doi: 10.1104/pp.108.121400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F.-D., de Saint Germain A., Pillot J.-P., Pouvreau J.-B., Chen V.X., Ramos S., Stévenin A., Simier P., Delavault P., Beau J.-M. Structure-activity relationship studies of strigolactone-related molecules for branching inhibition in garden pea: molecule design for shoot branching. Plant Physiol. 2012;159:1524–1544. doi: 10.1104/pp.112.195826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F.-D., de Saint Germain A., Pouvreau J.-B., Clavé G., Pillot J.-P., Roux A., Rasmussen A., Depuydt S., Lauressergues D., Frei dit Frey N. New strigolactone analogs as plant hormones with low activities in the rhizosphere. Mol. Plant. 2014;7:675–690. doi: 10.1093/mp/sst163. [DOI] [PubMed] [Google Scholar]
- Brun G., Braem L., Thoiron S., Gevaert K., Goormachtig S., Delavault P. Seed germination in parasitic plants: what insights can we expect from strigolactone research? J. Exp. Bot. 2017;69:2265–2280. doi: 10.1093/jxb/erx472. [DOI] [PubMed] [Google Scholar]
- Brun G., Thoiron S., Braem L., Pouvreau J.-B., Montiel G., Lechat M.-M., Simier P., Gevaert K., Goormachtig S., Delavault P. CYP707As are effectors of karrikin and strigolactone signalling pathways in Arabidopsis thaliana and parasitic plants. Plant Cell Environ. 2019;42:2612–2626. doi: 10.1111/pce.13594. [DOI] [PubMed] [Google Scholar]
- Brundrett M.C., Tedersoo L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018;220:1108–1115. doi: 10.1111/nph.14976. [DOI] [PubMed] [Google Scholar]
- Bythell-Douglas R., Rothfels C.J., Stevenson D.W.D., Graham S.W., Wong G.K., Nelson D.C., Bennett T. Evolution of strigolactone receptors by gradual neo-functionalization of KAI2 paralogues. BMC Biol. 2017;15:52. doi: 10.1186/s12915-017-0397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnel S., Torabi S., Griesmann M., Bleek E., Tang Y., Buchka S., Basso V., Shindo M., Wang T.L., Udvardi M. Duplicated KAI2 receptors with divergent ligand-binding specificities control distinct developmental traits in Lotus japonicus. bioRxiv. 2019 doi: 10.1101/754937. [DOI] [Google Scholar]
- Carter P., Wells J.A. Dissecting the catalytic triad of a serine protease. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- Conn C.E., Bythell-Douglas R., Neumann D., Yoshida S., Whittington B., Westwood J.H., Shirasu K., Bond C.S., Dyer K.A., Nelson D.C. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science. 2015;349:540–543. doi: 10.1126/science.aab1140. [DOI] [PubMed] [Google Scholar]
- Conn C.E., Nelson D.C. Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Front. Plant Sci. 2016;6:1219. doi: 10.3389/fpls.2015.01219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A., Clavé G., Badet-Denisot M.-A., Pillot J.-P., Cornu D., Le Caer J.-P., Burger M., Pelissier F., Retailleau P., Turnbull C. An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol. 2016;12:787–794. doi: 10.1038/nchembio.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers B.J.W., Willems L., Bassel G.W., van Bolderen-Veldkamp R.P., Ligterink W., Hilhorst H.W.M., Bentsink L. Identification of reference genes for RT–qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 2011;53:28–37. doi: 10.1093/pcp/pcr113. [DOI] [PubMed] [Google Scholar]
- Delavault P., Montiel G., Brun G., Pouvreau J.B., Thoiron S., Simier P. Communication between host plants and parasitic plants. Adv. Bot. Res. 2017;82:55–82. [Google Scholar]
- Fernandez-Aparicio M., Reboud X., Gibot-Leclerc S. Broomrape weeds. Underground mechanisms of parasitism and associated strategies for their control: a Review. Front. Plant Sci. 2016;7:135. doi: 10.3389/fpls.2016.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Aparicio M., Rubiales D., Bandaranayake P.C.G., Yoder J.I., Westwood J.H. Transformation and regeneration of the holoparasitic plant Phelipanche aegyptiaca. Plant Methods. 2011;7:36. doi: 10.1186/1746-4811-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. Chapman & Hall/CRC Press; New York: 2016. Using the R Commander: A Point-And-Click Interface for R. [Google Scholar]
- Goyet V., Billard E., Pouvreau J.-B., Lechat M.-M., Pelletier S., Bahut M., Monteau F., Spíchal L., Delavault P., Montiel G. Haustorium initiation in the obligate parasitic plant Phelipanche ramosa involves a host-exudated cytokinin signal. J. Exp. Bot. 2017;68:5539–5552. doi: 10.1093/jxb/erx359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenz J.H., Sauerborn J. Mechanisms limiting the geographical range of the parasitic weed Orobanche crenata. Agric. Ecosyst. Environ. 2007;122:275–281. [Google Scholar]
- Guo Y., Zheng Z., La Clair J.J., Chory J., Noel J.P. Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2013;110:8284–8289. doi: 10.1073/pnas.1306265110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C., Drummond R.S.M., Janssen B.J., Ledger S.E., Cooney J.M., Newcomb R.D., Snowden K.C. DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012;22:2032–2036. doi: 10.1016/j.cub.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Jamil M., Kountche B.A., Haider I., Wang J.Y., Aldossary F., Zarban R.A., Jia K.P., Yonli D., Shahul Hameed U.F., Takahashi I. Methylation at the C-3' in D-ring of strigolactone analogs reduces biological activity in root parasitic plants and rice. Front. Plant Sci. 2019;10:353. doi: 10.3389/fpls.2019.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.W., Gowda G., Hassanali A., Knox J., Monaco S., Razavi Z., Rosebery G. The preparation of synthetic analogs of strigol. J. Chem. Soc. Perkin Trans. 1981;1:1734–1743. [Google Scholar]
- Johnson A.W., Rosebery G., Parker C. A novel approach to Striga and Orobanche control using synthetic germination stimulants. Weed Res. 1976;16:223–227. [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinf. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechat M.-M., Pouvreau J.-B., Péron T., Gauthier M., Montiel G., Véronési C., Todoroki Y., Le Bizec B., Monteau F., Macherel D. PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J. Exp. Bot. 2012;63:5311–5322. doi: 10.1093/jxb/ers189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechat M.-M., Brun G., Montiel G., Veronesi C., Simier P., Thoiron S., Pouvreau J.-B., Delavault P. Seed response to strigolactone is controlled by abscisic acid-independent DNA methylation in the obligate root parasitic plant, Phelipanche ramosa L. Pomel. J. Exp. Bot. 2015;66:3129–3140. doi: 10.1093/jxb/erv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.W., Sharma P., Janssen B.J., Drummond R.S.M., Luo Z., Hamiaux C., Collier T., Allison J.R., Newcomb R.D., Snowden K.C. Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners. J. Biol. Chem. 2020;295:4181–4193. doi: 10.1074/jbc.RA119.011509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez-Obando M., Ligerot Y., Bonhomme S., Boyer F.-D., Rameau C. Strigolactone biosynthesis and signaling in plant development. Development. 2015;142:3615–3619. doi: 10.1242/dev.120006. [DOI] [PubMed] [Google Scholar]
- Nakamura H., Xue Y.L., Miyakawa T., Hou F., Qin H.M., Fukui K., Shi X., Ito E., Ito S., Park S.H. Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 2013;4:2613. doi: 10.1038/ncomms3613. [DOI] [PubMed] [Google Scholar]
- Parker C. Parasitic weeds: a world challenge. Weed Sci. 2012;60:269–276. [Google Scholar]
- Pepperman A.B., Connick W.J., Vail S.L., Worsham A.D., Pavlista A.D., Moreland D.E. Evaluation of precursors and analogs of strigol as witchweed (Striga asiatica) seed germination stimulants. Weed Sci. 1982;30:561–566. [Google Scholar]
- Pickett J.A., Wadhams L.J., Woodcock C.M. Developing sustainable pest control from chemical ecology. Agric. Ecosyst. Environ. 1997;64:149–156. [Google Scholar]
- Pouvreau J.-B., Gaudin Z., Auger B., Lechat M.M., Gauthier M., Delavault P., Simier P. A high-throughput seed germination assay for root parasitic plants. Plant Methods. 2013;9:32. doi: 10.1186/1746-4811-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A., Mason M.G., De Cuyper C., Brewer P.B., Herold S., Agusti J., Geelen D., Greb T., Goormachtig S., Beeckman T. Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 2012;158:1976–1987. doi: 10.1104/pp.111.187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchell D.P.N., Satchell R.S., Wassef W.N. The kinetics and mechanism of addition of water and alcohols to p-nitrophenyl isothiocyanate. The effects of added dimethyl sulfoxide. Z. Naturforsch., B: Chem. Sci. 1990;45:1032–1036. [Google Scholar]
- Seto Y., Yasui R., Kameoka H., Tamiru M., Cao M., Terauchi R., Sakurada A., Hirano R., Kisugi T., Hanada A. Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat. Commun. 2019;10:191. doi: 10.1038/s41467-018-08124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabek N., Ticchiarelli F., Mao H., Hinds T.R., Leyser O., Zheng N. Structural plasticity of D3–D14 ubiquitin ligase in strigolactone signalling. Nature. 2018;563:652–656. doi: 10.1038/s41586-018-0743-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanova B., Delourme R., Duffé P., Delavault P., Simier P. Genetic differentiation and host preference reveal non-exclusive host races in the generalist parasitic weed Phelipanche ramosa. Weed Res. 2019;59:107–118. [Google Scholar]
- Sun Y.K., Flematti G.R., Smith S.M., Waters M.T. Reporter gene-facilitated detection of compounds in Arabidopsis leaf extracts that activate the karrikin signaling pathway. Front. Plant Sci. 2016;7:1799. doi: 10.3389/fpls.2016.01799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarbreck S.M., Guerringue Y., Matthus E., Jamieson F.J.C., Davies J.M. Impairment in karrikin but not strigolactone sensing enhances root skewing in Arabidopsis thaliana. Plant J. 2019;98:607–621. doi: 10.1111/tpj.14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi I., Asami T. Target-based selectivity of strigolactone agonists and antagonists in plants and their potential use in agriculture. J. Exp. Bot. 2018;69:2241–2254. doi: 10.1093/jxb/ery126. [DOI] [PubMed] [Google Scholar]
- Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S., Holbrook-Smith D., Stogios P.J., Onopriyenko O., Lumba S., Tsuchiya Y., Savchenko A., McCourt P. Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science. 2015;350:203–207. doi: 10.1126/science.aac9476. [DOI] [PubMed] [Google Scholar]
- Toh S., Kamiya Y., Kawakami N., Nambara E., McCourt P., Tsuchiya Y. Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination. Plant Cell Physiol. 2012;53:107–117. doi: 10.1093/pcp/pcr176. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Yoshimura M., Sato Y., Kuwata K., Toh S., Holbrook-Smith D., Zhang H., McCourt P., Itami K., Kinoshita T. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science. 2015;349:864–868. doi: 10.1126/science.aab3831. [DOI] [PubMed] [Google Scholar]
- Uraguchi D., Kuwata K., Hijikata Y., Yamaguchi R., Imaizumi H., Am S., Rakers C., Mori N., Akiyama K., Irle S. A femtomolar-range suicide germination stimulant for the parasitic plant Striga hermonthica. Science. 2018;362:1301. doi: 10.1126/science.aau5445. [DOI] [PubMed] [Google Scholar]
- Villaécija-Aguilar J.A., Hamon-Josse M., Carbonnel S., Kretschmar A., Schmidt C., Dawid C., Bennett T., Gutjahr C. SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLOS Genet. 2019;15:e1008327. doi: 10.1371/journal.pgen.1008327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vurro M., Prandi C., Baroccio F. Strigolactones: how far is their commercial use for agricultural purposes? Pest Manage. Sci. 2016;72:2026–2034. doi: 10.1002/ps.4254. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T. Spoilt for choice: new options for inhibitors of strigolactone signaling. Mol. Plant. 2019;12:21–23. doi: 10.1016/j.molp.2018.11.012. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Gutjahr C., Bennett T., Nelson D.C. Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 2017;68:291–322. doi: 10.1146/annurev-arplant-042916-040925. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Nelson D.C., Scaffidi A., Flematti G.R., Sun Y.K., Dixon K.W., Smith S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development. 2012;139:1285–1295. doi: 10.1242/dev.074567. [DOI] [PubMed] [Google Scholar]
- Wicke S., Müller K.F., dePamphilis C.W., Quandt D., Bellot S., Schneeweiss G.M. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc. Natl. Acad. Sci. U.S.A. 2016;113:9045. doi: 10.1073/pnas.1607576113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Miyakawa T., Nosaki S., Nakamura A., Lyu Y., Nakamura H., Ohto U., Ishida H., Shimizu T., Asami T. Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat. Commun. 2018;9:3947. doi: 10.1038/s41467-018-06452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi M., Ueno K., Furumoto T., Wakabayashi T., Mizutani M., Takikawa H., Sugimoto Y. Stereospecific reduction of the butenolide in strigolactones in plants. Bioorg. Med. Chem. 2018;26:4225–4233. doi: 10.1016/j.bmc.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Yang Z., Zhang Y., Wafula E.K., Honaas L.A., Ralph P.E., Jones S., Clarke C.R., Liu S., Su C., Zhang H. Horizontal gene transfer is more frequent with increased heterotrophy and contributes to parasite adaptation. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E7010. doi: 10.1073/pnas.1608765113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Mashiguchi K., Scaffidi A., Akatsu T., Melville K.T., Morita R., Morimoto Y., Smith S.M., Seto Y., Flematti G.R. An allelic series at the KARRIKIN INSENSITIVE 2 locus of Arabidopsis thaliana decouples ligand hydrolysis and receptor degradation from downstream signalling. Plant J. 2018;96:75–89. doi: 10.1111/tpj.14017. [DOI] [PubMed] [Google Scholar]
- Yao R., Ming Z., Yan L., Li S., Wang F., Ma S., Yu C., Yang M., Chen L., Li Y. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature. 2016;536:469–473. doi: 10.1038/nature19073. [DOI] [PubMed] [Google Scholar]
- Yao R., Wang F., Ming Z.H., Du X.X., Chen L., Wang Y.P., Zhang W.H., Deng H.T., Xie D.X. ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Res. 2017;27:838–841. doi: 10.1038/cr.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R., Wang L., Li Y., Chen L., Li S., Du X., Wang B., Yan J., Li J., Xie D. Rice DWARF14 acts as an unconventional hormone receptor for strigolactone. J. Exp. Bot. 2018;69:2355–2365. doi: 10.1093/jxb/ery014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama K. Small-molecule inhibitors: weed-control measures. Nat. Chem. Biol. 2016;12:658–659. doi: 10.1038/nchembio.2155. [DOI] [PubMed] [Google Scholar]
- Zehhar N., Ingouff M., Bouya D., Fer A. Possible involvement of gibberellins and ethylene in Orobanche ramosa germination. Weed Res. 2002;42:464–469. [Google Scholar]
- Zwanenburg B., Pospíšil T. Structure and activity of strigolactones: new plant hormones with a rich future. Mol. Plant. 2013;6:38–62. doi: 10.1093/mp/sss141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.