Abstract
In trees, stem secondary growth depends on vascular cambium proliferation activity and subsequent cell differentiation, in which an auxin concentration gradient across the cambium area plays a crucial role in regulating the process. However, the underlying molecular mechanism for the establishment of auxin concentration is not fully understood. In this study, we identified two function-unknown MADS-box genes, VCM1 and VCM2, which are expressed specifically in the vascular cambium and modulate the subcellular homeostasis of auxin. Simultaneous knockdown of both VCM1 and VCM2 enhanced vascular cambium proliferation activity and subsequent xylem differentiation. Overexpression of VCM1 suppressed vascular cambium activity and wood formation by regulating PIN5 expression, which tuned the soluble auxin concentration in the vascular cambium area. This study reveals the role of VCM1 and VCM2 in regulating the proliferation activity of the vascular cambium and secondary growth by modulating the subcellular auxin homeostasis in Populus.
Key words: vascular cambium, secondary growth, auxin, xylem, tree
This study demonstrates the function of two MADS-box genes in secondary growth and reveals a molecular mechanism that modulates subcellular auxin homeostasis in the vascular cambium area, which in turn regulates cambium proliferation activity in Populus.
Introduction
Unlike herbaceous plants, woody plants feature perennial secondary growth, which gives rise to radial increments of the stem. Secondary growth is dependent on the activity of the vascular cambium, which divides and produces daughter cells that subsequently differentiate into secondary vascular tissues with phloem cells on the outside and xylem cells on the inside (wood tissue). During this process, a set of sequential events, including cell division, cell fate determination, and cell differentiation, are modulated through a complex of signaling networks (Spicer and Groover, 2010; Campbell and Turner, 2017; Fischer et al., 2019). Studies have reported the involvement of auxin in regulating vascular cambium activity and the secondary growth of stem. Auxin supplied to stem tissues stimulates cambial growth and mediates multiple aspects of xylem development (Bjorklund et al., 2007; Johnsson et al., 2019). Modulations of auxin transport and responsive genes regulate wood formation in the annual growth of trees (Moyle et al., 2002; Schrader et al., 2004; Baba et al., 2011). Auxin-responsive genes respond dynamically to changes in cellular auxin levels in wood-forming tissues (Nilsson et al., 2008). During the secondary growth of stem in trees, indole-3-acetic acid (IAA) and its derivatives form a concentration gradient cross the vascular cambial zone, with an IAA peak in the cambium area (Uggla et al., 1996, 1998). The peak height and the radial width of IAA distribution play a crucial role in determining cambium activity and xylem differentiation during wood formation (Tuominen et al., 1997). However, it is unknown how IAA concentration is maintained and regulated during the secondary growth of stem.
The establishment of cellular auxin concentration can be attributed to polar auxin transport, intracellular auxin homeostasis, and metabolic processes (Park et al., 2017). In Arabidopsis thaliana, it is known that auxin can be translocated by several types of transporters, including pin-formed proteins (PIN), proteins of the auxin transporter protein 1 (AUX1), auxin transporter-like protein (LAX), P-glycoproteins (PGP) of the ATP-binding cassette (ABC) transporter family, and pin-likes (PILS) (Grones and Friml, 2015). These transporters perform their functions in different manners. Most PIN proteins are located at particular plasma membrane regions for directional auxin transport (Petrasek et al., 2006; Wisniewska et al., 2006). However, PIN5 is localized at the endoplasmic reticulum (ER), and it is suggested to control cellular auxin homeostasis by transporting auxin from the cytosol to the ER lumen, which decreases the cellular level of free IAA (Mravec et al., 2009). It is believed that PIN5-dependent transport limits the availability of cytosolic auxin for signaling (Ding et al., 2012). The Populus genome contains three PIN5 homologs, of which two members are highly expressed in the stem, showing localization at the ER (Liu et al., 2014). It is unclear whether the Populus PIN5 homologs function in tuning the cellular auxin level in the stem.
MADS-box transcription factors have active roles in all major developmental processes throughout the entire life cycle of land plants (Gramzow and Theissen, 2010). For example, several MADS-box transcription factor genes are known for their involvement in regulating meristem activity, floral organ identity specification, and organ development (Dornelas et al., 2011). In plants, there is a large family of genes encoding MADS-box transcription factors. The Arabidopsis genome contains 107 MADS-box genes (Parenicova et al., 2003). Seventy-five MADS-box genes are present in rice (Arora et al., 2007). In the Populus genome, there are 105 putative MADS-box genes and 12 pseudogenes with a MADS-box structure (Leseberg et al., 2006). However, the function of most MADS-box genes is unknown.
In our previous work, the regulation of PtrHB4 expression caused serious defects during vascular cambium development in Populus (Zhu et al., 2018). In the defective vascular cambium, the expression of a group of MADS-box genes was extensively modified. In this study, we show evidence demonstrating that two MADS-box genes, VCM1 and VCM2, play a role in regulating the secondary growth of stem in Populus. We carried out a genetic analysis to show that VCM1 and VCM2 affect vascular cambium proliferation activity. Further genomic and cellular analyses revealed that VCM1 and VCM2 regulate PIN5b expression, which in turn modulates the homeostasis of phytohormone auxin. Our study suggests that VCM1 and VCM2 play a role in regulating the secondary growth of stem through PIN5-mediated subcellular auxin transport, which tunes cytosolic auxin levels across the vascular cambium area.
Results
Identification of regulatory genes of vascular cambium activity in Populus
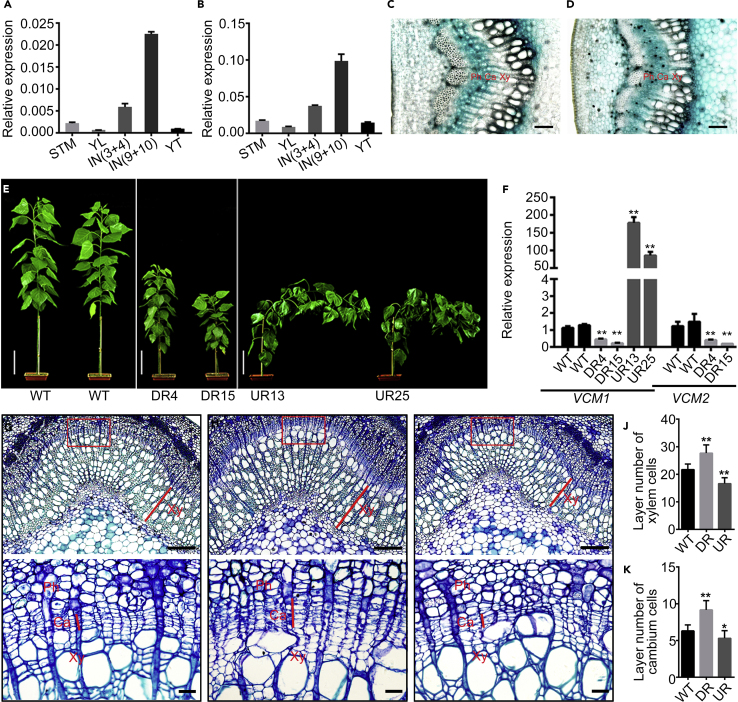
In our previous study, the suppression of PtrHB4 caused defects in the Populus vascular cambium (Zhu et al., 2018). The defective vascular cambium tissue in the PtrHB4-suppressed Populus was subjected to RNA sequencing, and transcriptome analysis revealed that the expression of a group of transcription factor genes was substantially altered (Supplemental Table 1), suggesting that transcriptional regulatory networks were modified in the defective vascular cambium. Further analysis showed that the regulated transcription factors included two homologous function-unknown MADS-box genes, Potri.012G100200 and Potri.015G098400. Interestingly, when they were searched in the Nucleotide Database (https://blast.ncbi.nlm.nih.gov), their homologs were identified primarily in woody plant species (Supplemental Figure 1A). Subsequently, the homologs of Potri.012G100200 and Potri.015G098400 were cloned from Populus deltoides × P. euramericana cv. “Nanlin895” (Supplemental Figure 1B). Quantitative transcript analysis showed that their expression was specific in stem tissues undergoing secondary growth (IN9 and IN10) (Figure 1A and 1B). Thus, the two genes, Potri.012G100200 and Potri.015G098400, were named Vascular Cambium-related MADS 1 and 2 (VCM1 and VCM2), respectively. VCM1 and VCM2 share a 73.3% identity in protein sequence (Supplemental Figure 1C). Next, we cloned VCM1 and VCM2 promoter fragments and transferred the promoter-GUS constructs into Populus to further examine their expression location. GUS activity was detected in the vascular cambium area, including the cambium and developing phloem and xylem cells (Figure 1C and 1D). These results indicate that the expression of VCM1 and VCM2 is closely correlated with the process of secondary growth in Populus.
Figure 1.
VCM1 and VCM2 expression and phenotypes of their transgenic lines.
(A and B) qRT-PCR analysis of VCM1(A) and VCM2(B) expression in different tissues. Values are means ± SD (n = 3 biological replicates). SMT, shoot meristem tissue; YL, young leaf; IN, internode; YT, young roots.
(C and D) GUS activity in transgenic Populus driven by the VCM1 promoter (C) and the VCM2 promoter (D). Ca, cambium; Xy, xylem; Ph, phloem. Scale bar corresponds to 100 μm.
(E) Phenotypes of the wild-type (WT), VCM1 and VCM2 knock-down lines, (DR4 and DR15) and VCM1 overexpression lines (UR13 and UR25). Scale bar corresponds to 10 cm.
(F) qRT-PCR analysis of VCM1 and VCM2 expression in transgenics. Values are means ± SD (n = 3 biological replicates).
(G–I) Cross-section of the 13th internode stained with toluidine blue (above) and enlarged rectangle area (below). (G) WT; (H)VCM1 and VCM2 knock-down lines; (I), VCM1 overexpression lines. Xy, xylem; Ph, phloem; Ca, cambium. Scale bars correspond to 200 μm (50 μm in enlargements).
(J) Number of cambium cell layers (n > 50).
(K) Number of xylem cell layers (n > 50). Plants used for analysis were grown in a phytotron for 3 months. Values are means ± SD. Statistical significance of differences was calculated based on two-tailed, two-sample Student's t-test (∗∗P < 0.01, ∗P < 0.05).
Down- or upregulation of VCM1 and VCM2 expression leads to abnormal secondary growth
To examine the genetic function of VCM1 and VCM2, we carried out knockdowns of their expression in Populus through RNA interference (RNAi) according to our established transformation method (Li et al., 2003). Because VCM1 and VCM2 have high homology, the designed RNAi construct (Supplemental Figure 2A) could not effectively differentiate them and might have suppressed both VCM1 and VCM2. More than 50 independent transgenic lines of the VCM1 and VCM2 downregulation (DR) were generated via RNAi. Examination of the DR transgenics identified different levels of VCM1 and VCM2 DR (Supplemental Figure 2B). Among them, the transgenic plants displayed remarkable phenotypic changes when both VCM1 and VCM2 were substantially downregulated. After screening the transgenics, two transgenic lines, DR4 and DR15, which displayed dramatic suppression of both VCM1 and VCM2 expression (Supplemental Figure 2B), were multiplied through micro-cutting propagation for further characterization. Meanwhile, we overexpressed VCM1 in order to investigate the implications of its upregulation in Populus. More than 30 independent overexpression lines were generated. The transgenic plants showed various levels of VCM1 expression. After the transgenic lines were screened for VCM1 upregulation (UR), two lines, UR13 and UR25, displayed substantial upregulation of VCM1 expression (Supplemental Figure 2C) and phenotypic changes. Thus, UR13 and UR25 were multiplied through micro-cutting propagation and used for characterization along with the DR transgenics.
The DR plants displayed significantly shorter internodes, while the UR plants showed longer internodes (Supplemental Figure 2D and 2G). The leaves of DR plants were smaller, softer, and darker green with smooth margins and shorter petioles compared with those of UR plants (Supplemental Figure 2E and 2H). DR plants showed a dwarfed stature, while UR plants displayed a lodging stem phenotype when grown in a phytotron for 3 months (Figure 1E). The expression of VCM1 and VCM2 was stably downregulated or upregulated in these transgenics after multiple propagations (Figure 1F). In cross-sections, compared with the wild-type (WT) (Figure 1G), the downregulation of VCM1 and VCM2 expression resulted in more xylem tissue formation (Figure 1H), while the upregulation of VCM1 expression inhibited xylem increment (Figure 1I). This is indicated by the cell layers generated in the cambium dividing zone and secondary xylem tissue. In successive cell files, VCM1 and VCM2 DR plants showed more layers of cells in the cambium zone and secondary xylem tissue, while VCM1 UR plants displayed fewer layers of cells produced in secondary growth (Figure 1J and 1K). After growth in a greenhouse for more than 5 months, the stem diameter of DR plants was greater compared with the WT, whereas that of UR plants was smaller (Supplemental Figure 2F and 2I). These results suggest that VCM1 and VCM2 negatively regulate vascular cambium proliferation activity and xylem tissue increment in Populus.
VCM1 regulates PIN5b expression
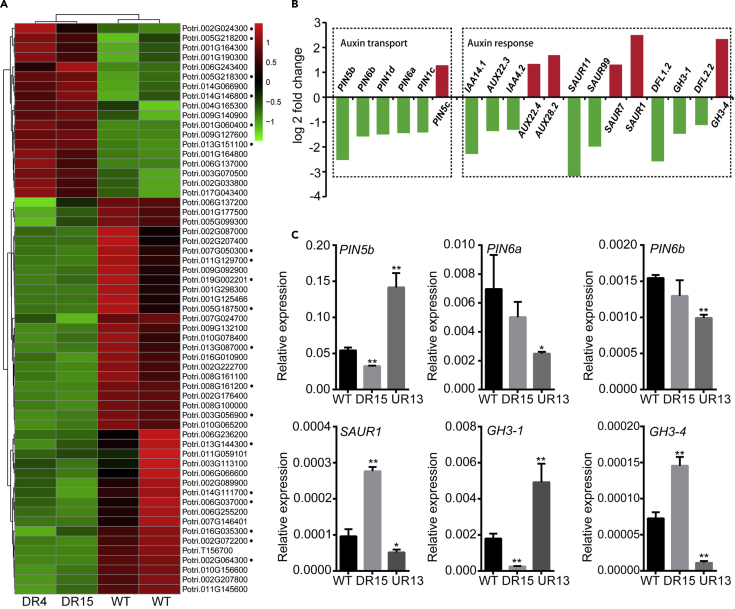
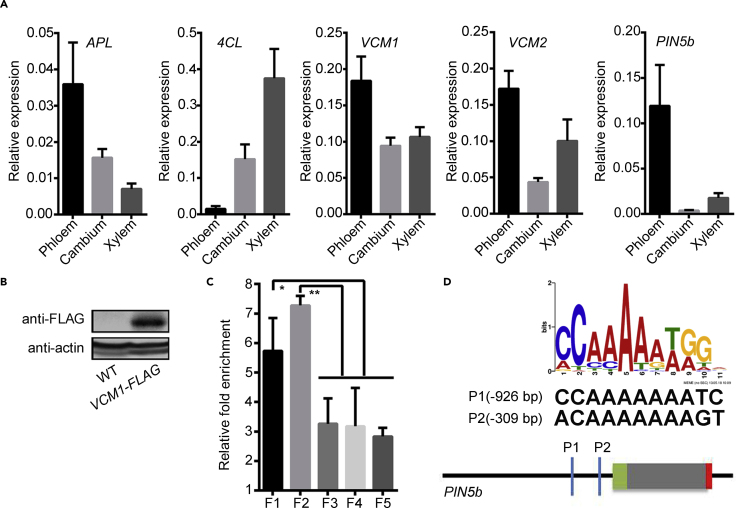
Next, we more closely investigated the processes by which VCM genes affect the secondary growth of Populus. VCM1 and VCM2 were detected to be localized in the nucleus (Supplemental Figure 3) as expected, consistent with their nature as transcription factors. We performed RNA sequencing of the tissue undergoing secondary growth in VCM1 and VCM2 DR plants and the transcriptomes of secondary growth tissues were substantially modified (Supplemental Figure 4A). Compared with WT plants, the top 10 enriched KEGG pathways of differentially expressed genes (DEGs) mainly included plant hormone signaling, mitogen-activated protein kinase signaling, and amino acid metabolism (Supplemental Figure 4B). Particularly, the most prominent changes occurred in genes related to auxin transport and response (Figure 2A and 2B). Given the correlation between auxin and secondary growth, the effect of VCM1 and VCM2 on stem growth could regulate auxin-related genes. Among these genes, PIN5b was substantially downregulated in VCM1 and VCM2 DR plants (Figure 2B). Further analysis confirmed that PIN5b expression was downregulated in DR15 and upregulated in UR13 plants (Figure 2C). However, the other PIN homologs either expressed at much lower levels or showed a discrete expression pattern (Supplemental Figure 4C). In addition, cells from various tissues in the stem were collected using a microdissection laser capture system for transcript analysis. PIN5b, VCM1, and VCM2 showed similar expression profiles and tissue specificity in the vascular cambium area (Figure 3A).
Figure 2.
Expression of auxin-related genes upon the regulation of VCM1 and VCM2.
(A) Heatmap of auxin-related gene expression in the transcriptomes of WT and VCM1 and VCM2 knock-down plants.
(B) Log2 value of differentially expressed auxin transport and responsive genes.
(C) qRT-PCR analysis of the expression of auxin-related genes. Values are means ± SD (n = 3 biological replicates). Statistical significance of differences was calculated based on two-tailed, two-sample Student's t-test (∗∗P < 0.01,∗P < 0.05).
Figure 3.
Expression of VCM1, VCM2, and PIN5b in cells undergoing secondary growth and the binding of VCM1 to PIN5b promoter.
(A) Cells were collected from the phloem, cambium, and xylem using a microdissection laser capture system. The expression of VCM1, VCM2, and PIN5b in cells was measured by qRT-PCR analysis. APL was used as a reference for phloem-preferred expression and 4CL1 as a reference for xylem-preferred expression.
(B) VCM1 was detected in the transgenics expressing VCM1-FLAG and used for ChIP analysis.
(C) Five fragments from the PIN5b promoter and intron (F1: −1209 to −899; F2: −451 to − 188; F3: −1815 to −1519; F4: +1724 to +1856; F5: +1930 to +2123) were examined for enrichment in VCM1-binding precipitation.
(D) Fragments F1 and F2 in the PIN5b promoter region contain possible binding elements P1 and P2, respectively. Values are means ± SD (n = 3 biological replicates). Statistical significance of differences was calculated based on two-tailed, two-sample Student's t-test (∗∗P < 0.01, ∗P < 0.05).
To investigate whether VCM1 directly regulates PIN5 expression, we examined the binding of VCM1 to the PIN5b promoter. VCM1 was fused with a FLAG tag and transferred into Populus. The expressed VCM1 protein was detected in the stem using a FLAG antibody (Figure 3B). Chromatin immunoprecipitation (ChIP) was then carried out to identify VCM1-bound DNA fragments. A ChIP-qPCR assay verified the binding of VCM1 to the fragments in the PIN5b promoter, which contained the predicted binding elements of MADS-boxes (Figure 3C and 3D). Together, these results indicate that VCM1 regulates PIN5 expression presumably by binding to its promoter.
VCM1 regulates cytosolic auxin levels across the vascular cambium area
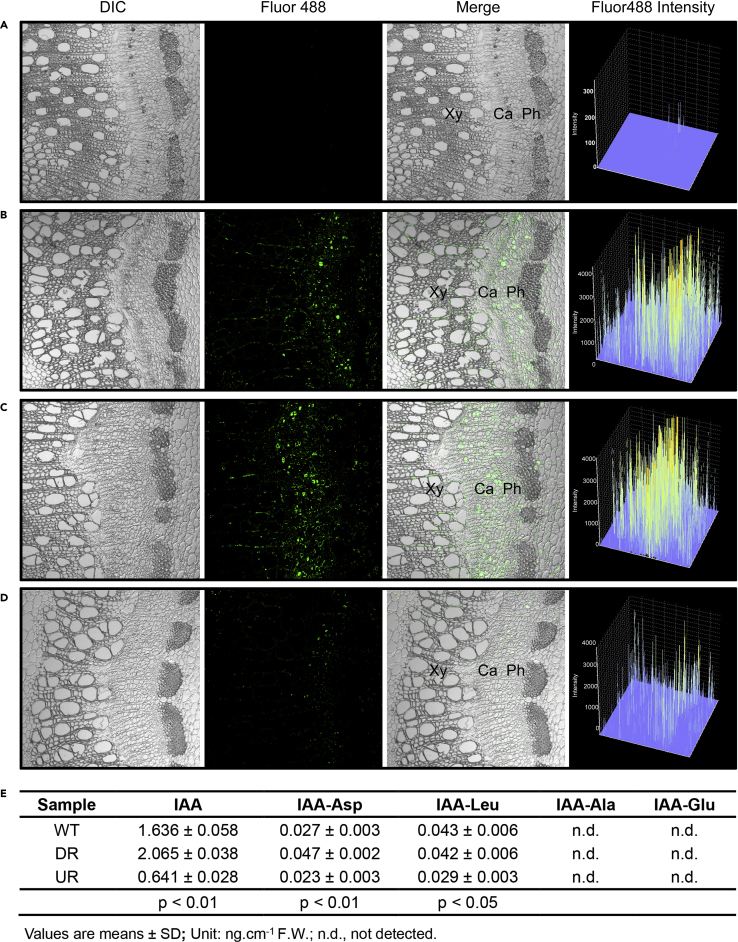
We then investigated whether VCM1 affects cytosolic IAA levels as VCM1 expression regulates PIN5b expression. IAA can be detected by free IAA-specific antibodies in the cytosol (Shi et al., 1993; Avsian-Kretchmer et al., 2002; Marquez-Lopez et al., 2018). Using a specific antibody, we examined cytosolic IAA levels in the vascular cambium area of DR and UR transgenics. IAA levels were higher in DR plants but lower in UR plants compared with the WT (Figure 4A–4D). The difference in IAA levels detected by IAA-specific antibodies was further verified by IAA determination through chemical analysis. Compared with WT plants, the IAA content in the stem was significantly increased in DR plants but decreased in UP plants (Figure 4E). These analyses demonstrate that the modification of VCM1 and VCM2 expression alters cytosolic IAA levels across the vascular cambium area, which in turn regulates cambium proliferation activity and xylem differentiation in the process of secondary growth in Populus.
Figure 4.
Detection of cytosolic IAA in the regulation of VCM1and VCM2.
(A–D) Cytosolic IAA was detected through immune-histochemical fluorescence analysis using IAA-specific antibodies and the fluorescence signal in the examined area is quantified as intensity. IgG served as the negative control. (A) WT; (B)VCM1 and VCM2 knock-down plants; (C)VCM1 overexpression plants; (D) Ca, cambium; Xy, xylem; Ph, phloem.
(E) Chemical determination of IAA and its amino acid conjugates. Values are means ± SD (n = 3 biological replicates). Tissues from the 10th to 14th internodes were collected for analysis. Statistical significance of differences was calculated based on two-tailed, two-sample Student's t-test.
Overexpression of PIN5b represses cambium proliferation activity and modifies cytosolic auxin levels
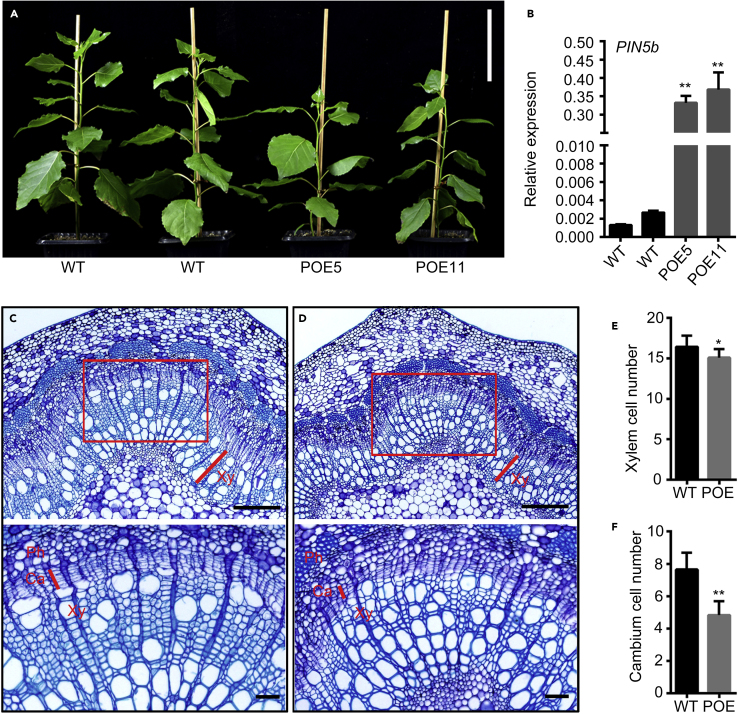
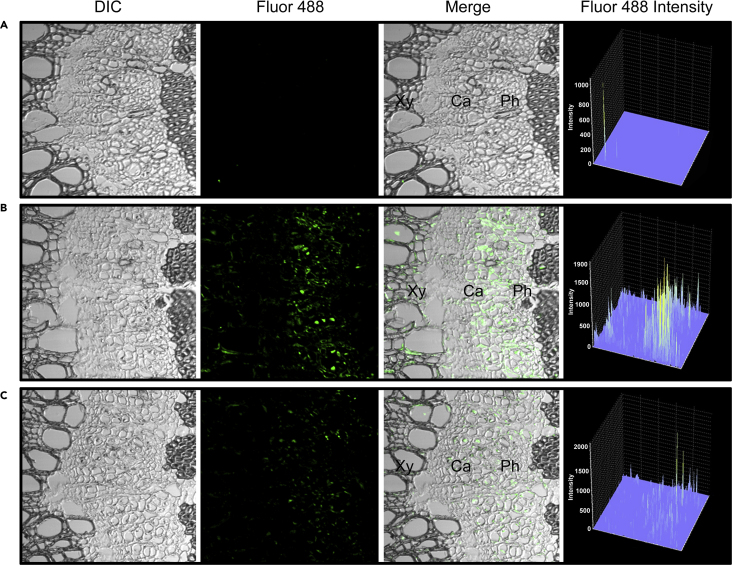
Next, we overexpressed PIN5b in Populus. PIN5b-overexpression plants, POE5 and POE11, were multiply propagated and analyzed in detail. POE5 and POE11 transgenic plants displayed a smaller stature (Figure 5A) and significantly increased PIN5b expression compared with the control (Figure 5B). The stems of the transgenics developed narrower cambium regions with fewer layers of cell proliferation compared with WT plants (Figure 5C and 5D). We then analyzed the cytosolic IAA level in vivo through IAA immunolocalization (Figure 6). In WT plants, a gradient level of cytosolic IAA was detected in the cambium, developing phloem, and developing xylem (Figure 6B), by contrast, the IAA level was reduced in PIN5b-overexpression lines (Figure 6C). These results suggest that PIN5b reduces the cytosolic IAA level in the vascular cambium area in Populus.
Figure 5.
Phenotypes of PIN5b overexpression lines.
(A) WT and PIN5b overexpression lines (POE5 and POE11) were grown in a phytotron for 2 months. Scale bar corresponds to 9 cm.
(B)PIN5b expression. Values are means ± SD (n = 3 biological replicates).
(C and D) Cross-section of the 11th internode stained with toluidine blue (above) and enlarged rectangle area (below). (C) WT; (D)PIN5b overexpression lines. Xy, xylem; Ph, phloem; Ca, cambium. Scale bars correspond to 200 μm (50 μm in enlargements).
(E) Number of xylem cell layers (n > 50).
(F) Number of cambium cell layers (n > 50). Values are means ± SD. Statistical significance of differences was calculated based on two-tailed, two-sample Student's t-test (∗∗P < 0.01, ∗P < 0.05).
Figure 6.
Detection of cytosolic IAA in the PIN5b overexpression line.
(A–C) Cytosolic IAA was detected using IAA-specific antibodies and the fluorescence signal in the examined area was quantified as intensity. (A) Negative control IgG; (B) WT; (C)PIN5b overexpression line. Ca, cambium; Xy, xylem; Ph, phloem.
Discussion
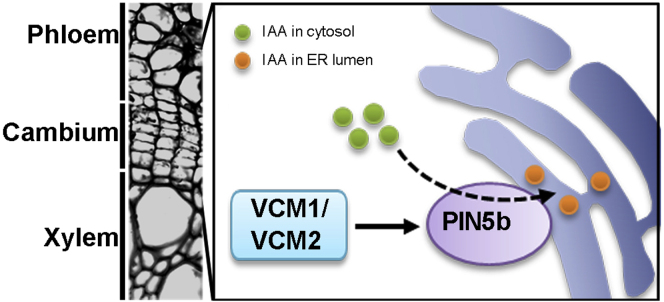
In trees, the secondary growth of stem depends on cambium proliferation activity. During secondary growth, a concentration gradient of active auxin across the vascular cambium area provides hormone signals that regulate cambium activity and secondary xylem differentiation (Sundberg et al., 2000). In this study, we identified two MADS-box genes, VCM1 and VCM2, in Populus, which are specifically expressed in association with secondary growth. Genetic evidence showed that VCM1 and VCM2 function redundantly in the regulation of vascular cambium activity and the formation of secondary xylem tissue. VCM1 and VCM2 likely regulate PIN5b expression across the vascular cambium area and tune the cytosolic IAA level through intracellular auxin transport. This study reveals the mechanism underlying the regulation of auxin concentration across the vascular cambium area, which is essential for the secondary growth of stem in trees (Figure 7).
Figure 7.
A proposed model for the involvement of VCM1and VCM2 in regulating secondary growth.
VCM1 and VCM2 regulate PIN5b expression in the cambium area. PIN5b transports IAA from the cytosol to the ER lumen, thereby tuning the cytosolic IAA concentration. Thus, VCM1 and VCM2 play a critical role in regulating vascular cambium proliferation and secondary growth through modulating auxin homeostasis.
VCM1 and VCM2 regulate the secondary growth of stem in Populus
In plants, MADS-box genes encode a large family of transcription factors that play various roles in diverse developmental processes and in response to stress (Smaczniak et al., 2012; Castelan-Munoz et al., 2019). Although studies have shown the importance of MADS-box genes in regulatory networks, many members of this family are function-unknown. In Populus, 105 putative functional MADS-box genes have been identified in the tree genome (Leseberg et al., 2006). Few of them have known functions in the process of tree development.
Perennial trees feature the secondary growth of stem that depends on vascular cambium proliferation activity. When we analyzed the development of the vascular cambium, we found that the expression of two function-unknown MADS-box genes, VCM1 and VCM2, were modified when the vascular cambium became defective. VCM1 and VCM2 are highly homologous genes in Populus. Interestingly, when the VCM1 or VCM2 sequence was used to BLAST GenBank, matching homologs with high similarity were found primarily in woody plant species. This may indicate a link between the function of VCM1 and VCM2 and woody plant development. Meanwhile, VCM1 and VCM2 were preferentially expressed in tissues undergoing secondary growth. Simultaneous suppression of VCM1 and VCM2 promoted vascular cambium proliferation activity and enhanced the secondary growth of stem. These observations suggest that VCM1 and VCM2 negatively regulate secondary growth in a redundant manner in Populus.
In Populus, secondary growth is perennially maintained, and the underlying regulatory system is presumably different from that of annual plants. The involvement of VCM1 and VCM2 in Populus secondary growth helps to understand how secondary growth is regulated in trees and why it continues over a long period.
PIN5b modulates auxin homeostasis in the vascular cambium area during secondary growth in Populus
Studies have indicated that auxin is involved in vascular tissue formation (Mattsson et al., 1999; Sieburth, 1999; De Rybel et al., 2014). The local maximum concentration of auxin in cells with xylem identity in Arabidopsis root cambium modulates cambium activity and the differentiation of vascular tissues (Smetana et al., 2019). In trees, auxin plays a critical role in the secondary growth of stem (Uggla et al., 1996). Across the vascular cambium area, the auxin concentration shows a gradient distribution (Uggla et al., 1996, 1998). Directional cell-to-cell auxin transport and intracellular auxin compartmentation are two major means of auxin translocation (Park et al., 2017). It is unclear how auxin concentration is regulated, especially in the cambium area, and how local auxin concentration affects the secondary growth of stem. The cellular auxin concentration is regulated by auxin biosynthesis, metabolism, and movement (Park et al., 2017). In this study, we found that intracellular auxin transport plays a role in tuning cellular auxin concentration across the vascular cambium area.
Detection of auxin via immunolocalization at the cellular level has been a useful tool for visualizing free IAA signals in a number of species (Avsian-Kretchmer et al., 2002; Forestan and Varotto, 2013; Marquez-Lopez et al., 2018; Yan et al., 2020). Using specific anti-IAA monoclonal antibodies, we performed IAA immunolocalization in Populus, which allowed us to detect the cellular free IAA level in the cambium, phloem, and developing xylem cells. The results are in good agreement with those obtained by IAA chemical analysis in stem tissues. The overexpression of PIN5b reduced the cytosolic IAA level, presumably through the transporter activity of PIN5b to relocate IAA from the cytosol to the ER lumen. These results reveal that the cellular IAA concentration across the vascular cambium area is tuned by intracellular transport mediated by PIN5b. Because the output of auxin signaling is mainly determined by cytosolic IAA levels (Dharmasiri et al., 2005; Kepinski and Leyser, 2005), intracellular IAA transport may regulate cytosolic auxin concentration, which serves as a signal that modulates secondary growth in trees.
PIN5 transporters are suggested to be localized in the ER, and the subcellular homeostasis of auxin is modulated by the transport of auxin from the cytosol to the ER lumen (Mravec et al., 2009; Ding et al., 2012). Populus PIN5 homologs are preferentially expressed across the area of the vascular cambium and localized in the ER (Liu et al., 2014). A PIN5 homolog has been reported to regulate cellular auxin levels in Populus (Johnsson et al., 2019). In our study, the expression of PIN5b was activated by VCM1 and VCM2 in relation to secondary growth. PIN5b overexpression reduced cambium activity and inhibited secondary growth. This suggests that the tuning of cellular auxin level by PIN5b regulates secondary growth in Populus.
VCM1 and VCM2 regulate secondary growth through PIN5b function in intracellular IAA transport
In trees, the secondary growth of stem gives rise to wood formation, and sophisticated transcriptional networks are involved in controlling vascular cambium division and descendant xylem cell differentiation (Demura and Fukuda, 2007; Zhang et al., 2014). A great deal of interest has been invested to identify and construct the hierarchical transcriptional networks involved in the regulation of vascular cambium activity, cell proliferation, xylem differentiation, and cell wall thickening using both Arabidopsis and Populus systems (Hertzberg et al., 2001; Ko et al., 2004; Andersson-Gunneras et al., 2006; Zhang et al., 2019). Although progress has been made in revealing how secondary growth is regulated through transcriptional networks, more studies are needed to fully understand the mechanism of secondary growth. Here, two function-unknown MADS-box transcription factor genes, VCM1 and VCM2, were found to be involved in the regulation of vascular cambium activity and subsequent wood formation in Populus. The downregulation of VCM1 and VCM2 expression resulted in an increase of vascular cambium proliferation activity and wood formation, whereas in the cells of the vascular cambium area, the regulation of VCM1 and VCM2 expression affected cytosolic IAA level and auxin signaling. VCM1/VCM2 and PIN5b displayed the same specificity of expression in the vascular cambium area. Evidence also suggested that VCM1 binds to the PIN5 promoter and regulates its expression. Thus, VCM1/VCM2 and PIN5b may act in a regulatory pathway to tune the level of cytosolic IAA in vascular cambium cells, which in turn serves as a hormone signal to modulate the secondary growth of stem (Figure 7).
Although an IAA concentration gradient is known to exist across the vascular cambium area, more insights into the regulation and establishment of IAA signaling are essential, given its effects on vascular cambium activity, cell proliferation, and xylem differentiation. Our study reveals a transcription regulatory pathway behind the observed phenomenon and an explanation for a better mechanistic understanding of the secondary growth of stem.
Material and methods
Plant material and growth conditions
The Populus tree (Populus deltoides × P. euramericana cv. Nanlin895) used in this study was grown in a phytotron with a light/dark cycle of 14/10 h under 60% relative humidity at 25°C.
Gene sequence analysis
The amino acid sequences of VCM1 and VCM2 were obtained from the Populus genome database (www.phytozome.net/poplar). VCM1 and VCM2 homologs were searched in the NCBI database using BLAST and aligned using CLUSTALW. The phylogenetic relationship was analyzed in MEGA 6.0 (Tamura et al., 2013) using the neighbor-joining method. Bootstrap values were calculated from 1000 trials. Full-length VCM1 and VCM2 coding sequences were cloned from the Populus clone Nanlin895.
Gene expression analysis
Total RNA was isolated from SMT, leaves, different stem internodes, and roots using a total RNA kit following the manufacturer’s instructions (Omega, R6827-02). The first-strand cDNA was synthesized from total RNA using cDNA Synthesis SuperMix (TransGen, AT311-03) and used for the qRT-PCR analysis of gene expression using gene-specific primers (Supplemental Table 2). qRT-PCR was performed using PerfectStart Green qPCR SuperMix (TransGen, AQ601) and a QuantStudio 3 Real-Time PCR Detection System (Thermo Fisher Scientific) according to the manufacturer’s instructions. For the normalization of target gene expression in each sample, Actin2 (Potri.001G309500) was used as an internal control.
Constructs and transformation
Nanlin895 was used for genetic transformation according to the protocol used in our lab (Li et al., 2003). For promoter analysis, ∼2.5 kb fragments from VCM1 and VCM2 promoters were cloned from Populus, respectively. The promoter fragment was cloned into a pCAMBIA2300-GUS vector upstream of uidA (GUS) and transferred to Populus for GUS activity analysis. For the downregulation of VCM, a fragment (245 bp) of VCM2 was constructed to form a hairpin structure that would function as an RNAi suppressor under the control of the CaMV 35S promoter. Because VCM1 and VCM2 share high homology, the RNAi construct may target both of them when it is transferred to Populus for the generation of knock-down lines. Meanwhile, for upregulation, VCM1 was transferred to Populus under the control of the CaMV 35S promoter. On the other hand, the full coding region of PIN5b was subcloned into vector pCAMBIA2300 under the control of the CaMV 35S promoter and transferred to Populus. Agrobacterium strain GV3101 was used for mediating Populus transformation. To determine the subcellular localization of VCM proteins, YFP-VCM1 and YFP-VCM2 fusion proteins were constructed under the control of the CaMV 35S promoter and introduced into tobacco leaves through Agrobacterium-mediated transformation. Primers used for vector construction are listed in Supplemental Table 2.
Histochemical staining
For GUS staining, convenient cross-sections of the stem were incubated in 50% acetone (v/v) for 10 min on ice and then stained in GUS solution (100 mM sodium phosphate [pH 7.0], 10 mM EDTA, 0.5 mM ferrocyanide, 0.1% Triton X-100, 20% methanol, and 2 mM X-Gluc) at 37°C. After staining, sections were cleared by 70% ethanol, washed using ddH2O, cleared by chloral hydrate (7.5% [w/v] gum arabic, 6.05 M chloral hydrate, and 5% glycerol [v/v]), and photographed. For the analysis of stem structure, stem internodes were fixed, paraffin-embedded, sectioned, and observed as described previously (Zhu et al., 2018)
RNA sequencing
Total RNA was isolated from stem internodes using a total RNA kit according to the manufacturer’s instructions (Omega, R6827-02) after a combination of three biological repeats of independent lines of the transgenic and WT plants. After the RNA quality was confirmed, 5 μg of the total RNA was used to construct a cDNA library using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, RS-122-2101) and Agencourt AMPure XP (Beckman Coulter, A63881). The cDNA library was quality controlled using the Agilent 2100 Bioanalyzer and sequenced using the 150 bp paired-end sequencing strategy, which was performed on the Illumina HiSeq X10 platform. Raw data (raw reads) were processed using the NGS QC Toolkit (Patel and Jain, 2012). Clean reads were mapped to the genome of Populus trichocarpa (http://phytozome.jgi.doe.gov/) using the HISAT2 program with default parameters (Kim et al., 2015). Fragments per kilobase per million were calculated using Cufflinks (Roberts et al., 2011). DEGs were identified using the DESeq R package with the estimateSizeFactors and nbinomTest functions. P < 0.05 and fold change >2 or fold change <0.5 were set as the threshold for significantly differential expression. KEGG pathway enrichment analysis of DEGs was performed using R based on the hypergeometric distribution (Kanehisa et al., 2008).
Laser microdissection
Internodes (8th–10th) from WT plants were sampled for stem sectioning. Laser microdissection, RNA extraction, RNA amplification, and qRT-PCR measurement were conducted as described previously (Song et al., 2010). Primers used for PCR analysis are listed in Supplemental Table 2.
ChIP
For the ChIP analysis, stem internodes were sampled from WT and transgenic plants. The transgenics expressing VCM1-FLAG were used to isolate proteins for western blot analysis using an anti-FLAG antibody (Abmart, M2008L). ChIP was performed as described previously (Bowler et al., 2004) using a Magna ChIP A/G Kit (Millipore). The bound DNA fragments were analyzed using qRT-PCR. The primers are listed in Supplemental Table 2.
IAA immunolocalization and analysis
Based on the previously described method of IAA immunolocalization (Marquez-Lopez et al., 2018), a modification was made for IAA detection in the Populus stem tissue. In brief, stem segments (3 mm in length) from transgenic and WT Populus were prefixed in 3% EDAC (1-Ethyl-3-(3-Dimethylaminopropyl)Carbodiimide Hydrochloride) (ABCONE) in 1× PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4]). To remove the gas from stem tissues, vacuum was applied two to three times to ensure that the stem was completely submerged in the solution. The stem tissues were then stored at 4°C for 30 min. After the samples were fixed in a formaldehyde-acetic acid solution (formaldehyde:glacial acetic acid:70% ethanol [1:1:18]) for 24–48 h, they were dehydrated via a graded ethanol series and embedded in paraffin. The stem samples were sectioned to a thickness of 10 μm using a rotary microtome (Leica RM2235). Sections were incubated with an IAA-specific antibody (Sigma, A0855). Detection was carried out using Alexa Fluor 488 antibody (Abcam, ab150113) following a standard method. For the chemical determination of IAA, fresh internode tissues (from the 10th to the 14th stem internodes) were ground to a fine powder in liquid nitrogen, and 0.5 g of sample was used for the determination of IAA and its amino acid conjugates according to a previously reported method (Matsuda et al., 2005).
Funding
This work was supported by the Ministry of Science and Technology of the People's Republic of China (2016YFD0600104), the National Natural Science Foundation of China(31630014), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27020104).
Author contributions
S.Z., J.S., and L.L. conceived the experiments. S.Z., J.H., Z.L., Y.Z., and J.S. performed the experiments. S.Z. and L.L. analyzed the data and wrote the manuscript.
Acknowledgments
We thank Shuining Yin for her support in fluorescence microscopy and Xiaoli Liu for assistance with ChIP experiments. No conflict of interest declared.
Published: November 23, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental information is available at Plant Communications Online.
Accession numbers
NCBI GenBank accession numbers: VCM1, XM024582264.1; VCM2, XM002321675.3; and PIN5b, XM006375996.2.
Supplemental information
References
- Andersson-Gunneras S., Mellerowicz E.J., Love J., Segerman B., Ohmiya Y., Coutinho P.M., Nilsson P., Henrissat B., Moritz T., Sundberg B. Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J. 2006;45:144–165. doi: 10.1111/j.1365-313X.2005.02584.x. [DOI] [PubMed] [Google Scholar]
- Arora R., Agarwal P., Ray S., Singh A.K., Singh V.P., Tyagi A.K., Kapoor S. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics. 2007;8:242. doi: 10.1186/1471-2164-8-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avsian-Kretchmer O., Cheng J.C., Chen L.J., Moctezuma E., Sung Z.R. Indole acetic acid distribution coincides with vascular differentiation pattern during Arabidopsis leaf ontogeny. Plant Physiol. 2002;130:199–209. doi: 10.1104/pp.003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba K., Karlberg A., Schmidt J., Schrader J., Hvidsten T.R., Bako L., Bhalerao R.P. Activity-dormancy transition in the cambial meristem involves stage-specific modulation of auxin response in hybrid aspen. Proc. Natl. Acad. Sci. U S A. 2011;108:3418–3423. doi: 10.1073/pnas.1011506108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund S., Antti H., Uddestrand I., Moritz T., Sundberg B. Cross-talk between gibberellin and auxin in development of Populus wood: gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 2007;52:499–511. doi: 10.1111/j.1365-313X.2007.03250.x. [DOI] [PubMed] [Google Scholar]
- Campbell L., Turner S. Regulation of vascular cell division. J. Exp. Bot. 2017;68:27–43. doi: 10.1093/jxb/erw448. [DOI] [PubMed] [Google Scholar]
- Castelan-Munoz N., Herrera J., Cajero-Sanchez W., Arrizubieta M., Trejo C., Garcia-Ponce B., Sanchez M.D., Alvarez-Buylla E.R., Garay-Arroyo A. MADS-box genes are key components of genetic regulatory networks involved in abiotic stress and plastic developmental responses in plants. Front. Plant Sci. 2019;10:853. doi: 10.3389/fpls.2019.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., Adibi M., Breda A.S., Wendrich J.R., Smit M.E., Novak O., Yamaguchi N., Yoshida S., Van Isterdael G., Palovaara J. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345:1255215. doi: 10.1126/science.1255215. [DOI] [PubMed] [Google Scholar]
- Demura T., Fukuda H. Transcriptional regulation in wood formation. Trends Plant Sci. 2007;12:64–70. doi: 10.1016/j.tplants.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Ding Z.J., Wang B.J., Moreno I., Duplakova N., Simon S., Carraro N., Reemmer J., Pencik A., Chen X., Tejos R. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012;3:941. doi: 10.1038/ncomms1941. [DOI] [PubMed] [Google Scholar]
- Dornelas M.C., Patreze C.M., Angenent G.C., Immink R.G.H. MADS: the missing link between identity and growth? Trends Plant Sci. 2011;16:89–97. doi: 10.1016/j.tplants.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Fischer U., Kucukoglu M., Helariutta Y., Bhalerao R.P. The dynamics of cambial stem cell activity. Annu. Rev. Plant Biol. 2019;70 70:293–319. doi: 10.1146/annurev-arplant-050718-100402. [DOI] [PubMed] [Google Scholar]
- Forestan C., Varotto S. Auxin immunolocalization in plant tissues. Methods Mol. Biol. 2013;959:223–233. doi: 10.1007/978-1-62703-221-6_15. [DOI] [PubMed] [Google Scholar]
- Gramzow L., Theissen G. A hitchhiker's guide to the MADS world of plants. Genome Biol. 2010;11:214. doi: 10.1186/gb-2010-11-6-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grones P., Friml J. Auxin transporters and binding proteins at a glance. J. Cell Sci. 2015;128:1–7. doi: 10.1242/jcs.159418. [DOI] [PubMed] [Google Scholar]
- Hertzberg M., Aspeborg H., Schrader J., Andersson A., Erlandsson R., Blomqvist K., Bhalerao R., Uhlen M., Teeri T.T., Lundeberg J. A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. U S A. 2001;98:14732–14737. doi: 10.1073/pnas.261293398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson C., Jin X., Xue W.Y., Dubreuil C., Lezhneva L., Fischer U. The plant hormone auxin directs timing of xylem development by inhibition of secondary cell wall deposition through repression of secondary wall NAC-domain transcription factors. Physiol. Plant. 2019;165:673–689. doi: 10.1111/ppl.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kim D., Landmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–U121. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.H., Han K.H., Park S., Yang J.M. Plant body weight-induced secondary growth in Arabidopsis and its transcription phenotype revealed by whole-transcriptome profiling. Plant Physiol. 2004;135:1069–1083. doi: 10.1104/pp.104.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leseberg C.H., Li A.L., Kang H., Duvall M., Mao L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene. 2006;378:84–94. doi: 10.1016/j.gene.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Li L., Zhou Y.H., Cheng X.F., Sun J.Y., Marita J.M., Ralph J., Chiang V.L. Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. U S A. 2003;100:4939–4944. doi: 10.1073/pnas.0831166100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.B., Zhang J., Wang L., Li J.B., Zheng H.Q., Chen J., Lu M.Z. A survey of Populus PIN-FORMED family genes reveals their diversified expression patterns. J. Exp. Bot. 2014;65:2437–2448. doi: 10.1093/jxb/eru129. [DOI] [PubMed] [Google Scholar]
- Marquez-Lopez R.E., Ku-Gonzalez A., Mendez-Hernandez H.A., Galaz-Avalos R.M., Loyola-Vargas V.M. Auxin immunolocalization in Coffea canephora tissues. Methods Mol. Biol. 2018;1815:179–188. doi: 10.1007/978-1-4939-8594-4_11. [DOI] [PubMed] [Google Scholar]
- Matsuda F., Miyazawa H., Wakasa K., Miyagawa H. Quantification of indole-3-acetic acid and amino acid coniugates in rice by liquid chromatography-electrospray ionization-tandem mass spectrometry. Biosci. Biotechnol. Biochem. 2005;69:778–783. doi: 10.1271/bbb.69.778. [DOI] [PubMed] [Google Scholar]
- Mattsson J., Sung Z.R., Berleth T. Responses of plant vascular systems to auxin transport inhibition. Development. 1999;126:2979–2991. doi: 10.1242/dev.126.13.2979. [DOI] [PubMed] [Google Scholar]
- Moyle R., Schrader J., Stenberg A., Olsson O., Saxena S., Sandberg G., Bhalerao R.P. Environmental and auxin regulation of wood formation involves members of the Aux/IAA gene family in hybrid aspen. Plant J. 2002;31:675–685. doi: 10.1046/j.1365-313x.2002.01386.x. [DOI] [PubMed] [Google Scholar]
- Mravec J., Skupa P., Bailly A., Hoyerova K., Krecek P., Bielach A., Petrasek J., Zhang J., Gaykova V., Stierhof Y.D. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–U1127. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- Nilsson J., Karlberg A., Antti H., Lopez-Vernaza M., Mellerowicz E., Perrot-Rechenmann C., Sandberg G., Bhalerao R.P. Dissecting the molecular basis of the regulation of wood formation by auxin in hybrid aspen. Plant Cell. 2008;20:843–855. doi: 10.1105/tpc.107.055798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicova L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee Y., Martinoia E., Geisler M. Plant hormone transporters: what we know and what we would like to know. BMC Biol. 2017;15:93. doi: 10.1186/s12915-017-0443-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel R.K., Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J., Mravec J., Bouchard R., Blakeslee J.J., Abas M., Seifertova D., Wisniewska J., Tadele Z., Kubes M., Covanova M. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312:914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- Roberts A., Trapnell C., Donaghey J., Rinn J.L., Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader J., Moyle R., Bhalerao R., Hertzberg M., Lundeberg J., Nilsson P., Bhalerao R.P. Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. Plant J. 2004;40:173–187. doi: 10.1111/j.1365-313X.2004.02199.x. [DOI] [PubMed] [Google Scholar]
- Shi L., Miller I., Moore R. Immunocytochemical localization of indole-3-acetic-acid in primary roots of Zea mays. Plant Cell Environ. 1993;16:967–973. [Google Scholar]
- Sieburth L.E. Auxin is required for leaf vein pattern in Arabidopsis. Plant Physiol. 1999;121:1179–1190. doi: 10.1104/pp.121.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C., Immink R.G.H., Muino J.M., Blanvillain R., Busscher M., Busscher-Lange J., Dinh Q.D., Liu S.J., Westphal A.H., Boeren S. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proc. Natl. Acad. Sci. U S A. 2012;109:1560–1565. doi: 10.1073/pnas.1112871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smetana O., Makila R., Lyu M., Amiryousefi A., Rodriguez F.S., Wu M.F., Sole-Gil A., Gavarron M.L., Siligato R., Miyashima S. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature. 2019;565:485. doi: 10.1038/s41586-018-0837-0. [DOI] [PubMed] [Google Scholar]
- Song Dongliang, Shen. Li Characterization of cellulose synthase complexes in Populus xylem differentiation. New Phytologist. 2010;187:777–790. doi: 10.1111/j.1469-8137.2010.03315.x. [DOI] [PubMed] [Google Scholar]
- Spicer R., Groover A. Evolution of development of vascular cambia and secondary growth. New Phytol. 2010;186:577–592. doi: 10.1111/j.1469-8137.2010.03236.x. [DOI] [PubMed] [Google Scholar]
- Sundberg B., Uggla C., Tuominen H. Cambial growth and auxin gradients. Cell Mol. Biol. Wood Form. 2000:169–188. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen H., Puech L., Fink S., Sundberg B. A radial concentration gradient of indole-3-acetic acid is related to secondary xylem development in hybrid aspen. Plant Physiol. 1997;115:577–585. doi: 10.1104/pp.115.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uggla C., Mellerowicz E.J., Sundberg B. Indole-3-acetic acid controls cambial growth in Scots pine by positional signaling. Plant Physiol. 1998;117:113–121. doi: 10.1104/pp.117.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uggla C., Moritz T., Sandberg G., Sundberg B. Auxin as a positional signal in pattern formation in plants. Proc. Natl. Acad. Sci. U S A. 1996;93:9282–9286. doi: 10.1073/pnas.93.17.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J., Xu J., Seifertova D., Brewer P.B., Ruzicka K., Blilou I., Rouquie D., Benkova E., Scheres B., Friml J. Polar PIN localization directs auxin flow in plants. Science. 2006;312:883. doi: 10.1126/science.1121356. [DOI] [PubMed] [Google Scholar]
- Yan S.S., Ning K., Wang Z.Y., Liu X.F., Zhong Y.T., Ding L., Zi H.L., Cheng Z.H., Li X.X., Shan H.Y. CsIVP functions in vasculature development and downy mildew resistance in cucumber. PLoS Biol. 2020;18:e3000671. doi: 10.1371/journal.pbio.3000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nieminen K., Serra J.A., Helariutta Y. The formation of wood and its control. Curr. Opin. Plant Biol. 2014;17:56–63. doi: 10.1016/j.pbi.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Zhang J., Eswaran G., Alonso-Serra J., Kucukoglu M., Xiang J., Yang W., Elo A., Nieminen K., Damen T., Joung J.G. Transcriptional regulatory framework for vascular cambium development in Arabidopsis roots. Nat. Plants. 2019;5:1033–1042. doi: 10.1038/s41477-019-0522-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.Y., Song D.L., Xu P., Sun J.Y., Li L.G. A HD-ZIP III gene, PtrHB4, is required for interfascicular cambium development in Populus. Plant Biotechnol. J. 2018;16:808–817. doi: 10.1111/pbi.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.