Abstract
Plant natural products (PNPs) are the main sources of drugs, food additives, and new biofuels and have become a hotspot in synthetic biology. In the past two decades, the engineered biosynthesis of many PNPs has been achieved through the construction of microbial cell factories. Alongside the rapid development of plant physiology, genetics, and plant genetic modification techniques, hosts have now expanded from single-celled microbes to complex plant systems. Plant synthetic biology is an emerging field that combines engineering principles with plant biology. In this review, we introduce recent advances in the biosynthetic pathway elucidation of PNPs and summarize the progress of engineered PNP biosynthesis in plant cells. Furthermore, a future vision of plant synthetic biology is proposed. Although we are still a long way from overcoming all the bottlenecks in plant synthetic biology, the ascent of this field is expected to provide a huge opportunity for future agriculture and industry.
Key words: plant natural products, plant synthetic biology, plant transgenic technology
An emerging synthesis route for plant natural products (PNPs) that utilizes plants has attracted increasing attention alongside the rapid development of plant physiology, genetics, and plant genetic modification techniques. This review introduces recent advances in PNP pathway elucidation and summarizes the progress of engineered PNP biosynthesis in planta.
Introduction
Plants produce numerous natural products with diverse structures and bioactive properties, especially secondary metabolites involved in biological and environmental responses. These plant natural products (PNPs) are widely used in the food, pharmaceutical, and cosmeceutical industries and have a huge commercial value in human society (Liu et al., 2017; Maeda, 2019). However, traditional extraction methods and excessive exploitation could lead to the exhaustion of plant resources and to environmental pollution because of the low abundance of PNPs. The structural complexity of some PNPs often makes total chemical synthesis inefficient (Morrison and Hergenrother, 2014). Therefore, with the boom in synthetic biology and omics-based strategies, great success has been achieved in the production of PNPs in engineered hosts.
Because of their well-defined metabolic background and ease of further genetic manipulation, microbial cells such as Escherichia coli, Saccharomyces cerevisiae, and Corynebacterium glutamicum have been widely used as chassis cells to produce PNPs (Ajikumar et al., 2010; Paddon et al., 2013; Galanie et al., 2015; Li et al., 2018e; Liu et al., 2018; Luo et al., 2019). Standard microbial synthesis strategies include selecting and engineering hosts, planning and reconstructing metabolic pathways, regulating metabolic networks, and protein engineering (Cravens et al., 2019; Pyne et al., 2019; Birchfield and Mcintosh, 2020; Yang et al., 2020). Efficient production of PNPs requires significant effort to artificially modify and engineer microbes, owing to their specific metabolic pathways with very strict regulatory mechanisms. However, reconstruction in heterologous microorganisms is almost impossible for extremely complex or unclear pathways, such as that of paclitaxel, which contains multiple P450 reactions (McElroy and Jennewein, 2018).
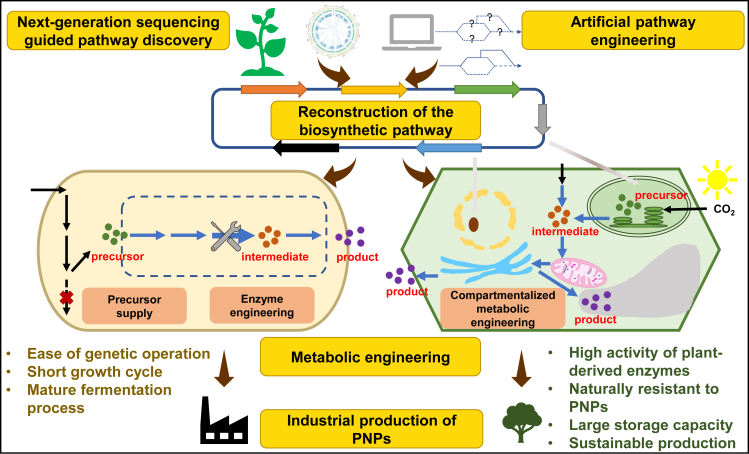
In recent years, increasing progress in the biosynthesis of PNPs in plants has been reported (Table 1). The complex multicellular eukaryotic system has become a topic of significant research interest in synthetic biology (Liu and Stewart, 2015). Engineering of host plants for suspension cell culture/hairy root cultures or heterologous plants with the accumulation of intermediate products seems to be a complementary way to achieve the high-yield production of complex PNPs (Nagegowda and Gupta, 2020). Compared with single-celled microorganisms, plants are better systems for synthesizing PNPs, and their better performance can be attributed to three plant homology aspects (Figure 1): (1) high expression and activity of plant-derived enzymes, (2) richness in various endomembrane systems and organelles that provide the most suitable reaction environment, and (3) high tolerance to the toxicity of PNPs and their intermediates.
Table 1.
List of representative examples of engineered biosynthesis of plant natural products in plant cells
| Classification | Secondary metabolites | Plant host | Strategy and method | References |
|---|---|---|---|---|
| Isoprenoids | Isoprenoids | N. tabacum | Overexpression | (Kumar et al., 2012) |
| Monoterpenoids | Geraniol | N. tabacum | Overexpression | (Vasilev et al., 2014) |
| Geraniol | N. tabacum | Overexpression | (Ritala et al., 2014) | |
| Limonene | M. piperita | Suppression | (Mahmoud et al., 2004) | |
| Mentha oil | M. piperita | Suppression | (Mahmoud and Croteau, 2001) | |
| Sesquiterpenoids | Artemisinin | N. tabacum | Overexpression | (Farhi et al., 2011) |
| Artemisinin | N. tabacum | Compartmentalized metabolic engineering | (Malhotra et al., 2016) | |
| Artemisinin | A. annua | Over expression and Global regulation | (Shen et al., 2018) | |
| Artemisinin | A. annua | Suppression | (Lv et al., 2016b) | |
| Artemisinin | A. annua | Global regulation | (Han et al., 2014b; Lv et al., 2016a; Shen et al., 2016; Tan et al., 2015; Yan et al., 2017) | |
| Artemisinin | P. patens | Overexpression | (Ikram et al., 2017; Ikram et al., 2019) | |
| Artemisinin acid | N. tabacum | COSTREL | (Fuentes et al., 2016) | |
| Patchoulol/santalene | P. patens | Compartmentalized metabolic engineering | (Zhan et al., 2014) | |
| Sesquiterpenoids | N. tabacum | Compartmentalized metabolic engineering | (Wu et al., 2006) | |
| Diterpenoids | Diterpenes | P. patens | Overexpression | (Banerjee et al., 2019) |
| Sclareol | P. patens | Overexpression | (Pan et al., 2015) | |
| Tanshinone | S. miltiorrhiza | Elicitors | (Yang et al., 2018) | |
| Taxadiene | A. thaliana | Compartmentalized metabolic engineering | (Besumbes et al., 2004) | |
| Taxadiene/taxadiene-5-ol | N. benthamiana | Compartmentalized metabolic engineering | (Li et al., 2019) | |
| Taxadiene | P. patens | Overexpression | (Anterola et al., 2009) | |
| Taxol | Taxus spp. cell suspension culture | Plant Cell Fermentation (PCF) Technology | https://phytonbiotech.com/apis/paclitaxel/ | |
| Taxol | Taxus spp. cell suspension culture | Samyang’s plant cell culture technology | https://www.samyangbiopharm.com/eng/ | |
| Triterpenoids | Astragaloside | A. membranaceus hairy root | Elicitors | (Jiao et al., 2016) |
| Diosgenin | N. benthamiana | Pooled screens and Overexpression | (Christ et al., 2019) | |
| Dammarenediol-II | N. tabacum | Overexpression | (Han et al., 2014a) | |
| Ginsenoside | P. ginseng | www.nitto.com | ||
| Protopanaxatriol/protopanaxadiol | N. tabacum | Overexpression | (Chun et al., 2015; Gwak et al., 2019) | |
| Squalene | N. tabacum | Compartmentalized metabolic engineering | (Pasoreck et al., 2016) | |
| Triterpene | N. benthamiana | Combinatorial transformation | (Reed et al., 2017) | |
| Triterpenoids | C. asiatica hairy root | Elicitors | (Baek et al., 2020) | |
| Tetraterpenoids | β-Carotene | Rice | Overexpression | (Ye et al., 2000) |
| β-Carotene | Tomato | Overexpression | (Apel and Bock, 2009) | |
| β-Carotene/carotenoids | Maize | Combinatorial transformation | (Zhu et al., 2008) | |
| Carotenoid | Potato | Overexpression | (Diretto et al., 2007) | |
| Canthaxanthin/astaxanthin | Rice | Overexpression | (Zhu et al., 2018) | |
| GABA | Tomato | CRISPR/Cas9-mediated metabolic engineering | (Nonaka et al., 2017) | |
| Lycopene | Tomato | Suppression | (Li et al., 2018d) | |
| Provitamin A | Tomato | Overexpression | (Wurbs et al., 2007) | |
| Phenylpropanoids | Anthocyanin | A. thaliana | Global regulation | (Xu et al., 2017) |
| Anthocyanin | L. ruthenicum/L. barbarum | Global regulation | (Zong et al., 2019) | |
| Anthocyanin | N. tabacum/A. thaliana | Global regulation | (Appelhagen et al., 2018) | |
| Anthocyanin | Rice | Global regulation | (Zhu et al., 2017) | |
| Anthocyanin | Tomato | CRISPR/Cas9-mediated metabolic engineering | (Cermak et al., 2015) | |
| Anthocyanin | Tomato | Global regulation | (Meng et al., 2014) | |
| Anthocyanin/phenylpropanoid flavonoid derivatives | Tomato | Global regulation | (Tohge et al., 2015) | |
| Anthocyanin | Tomato | Global regulation | (Li et al., 2018a) | |
| Chrysin/wogonin/baicalein | Scutellaria bornmuelleri | Elicitors | (Zg et al., 2020) | |
| Coumarin/phenolic compounds | P. sidoides hairy root | Elicitors | (Yousefian et al., 2021) | |
| Cyanidin 3-O-rutinoside | N. tabacum | Global regulation | (He et al., 2017) | |
| Delphinidin | Chrysanthemum morifolium | Overexpression | (Noda et al., 2017) | |
| Delphinidin | Rosa hybrida | Overexpression | (Katsumoto et al., 2007) | |
| Delphinidin/petunidin | Tomato | Global regulation | (Outchkourov et al., 2018) | |
| Flavonoids | Tomato | Global regulation | (Zhang et al., 2015) | |
| Flavonols | Tomato | Global regulation | (Brown et al., 2015; Zhang et al., 2015) | |
| Isoflavone | Soya bean | CRISPR/Cas9-mediated metabolic engineering | (Zhang et al., 2020a) | |
| Pterostilbene/piceatannol | V. vinifera | Elicitors | (Martinez-Marquez et al., 2018; Martinez-Marquez et al., 2016) | |
| Resveratrol | V. vinifera | Elicitors | (Chu et al., 2017) | |
| Resveratrol/dihydroquercetin | V. vinifera | Global regulation | (Manela et al., 2015) | |
| Silymarin | Silybum marianum | Elicitors | (Gabr et al., 2016) | |
| Alkaloids | Berberine | Coptis japonica/Thalictrum minus cell suspension culture | (Matsubara et al., 1989) | |
| Catharanthine/tabersonine | N. benthamiana | Overexpression | (Caputi et al., 2018) | |
| Etoposide aglycone | N. benthamiana | Overexpression | (Lau and Sattely, 2015) | |
| Hyoscyamine | A. belladonna | CRISPR/Cas9-mediated metabolic engineering | (Zeng et al., 2021) | |
| N-Formyldemecolcine | N. benthamiana | Overexpression | (Nett et al., 2020) | |
| Strictosidine | N. benthamiana | Overexpression | (Miettinen et al., 2014) | |
| Tropane alkaloids | A. belladonna | Global regulation | (Bai et al., 2019) | |
| Tropane alkaloids | A. belladonna | Overexpression | (Zhao et al., 2020) | |
| Tropane alkaloids | Hyoscyamus hairy root | Elicitors | (Hedayati et al., 2020) | |
| Vincristine/vinblastine | C. roseus | Elicitors | (Mekky et al., 2018) | |
| Other PNPs | Dhurrin | N. benthamiana | Compartmentalized metabolic engineering | (de Jesus et al., 2017; Gnanasekaran et al., 2016) |
| Vanillin | C. frutescens | Overexpression | (Chee et al., 2017) | |
| Vitamins | Maize | Combinatorial transformation | (Naqvi et al., 2009) | |
| Vitamin E | Tomato | Overexpression | (Lu et al., 2013) | |
| Vitamin E | Lettuce | Overexpression | (Yabuta et al., 2013) |
Abbreviations: A. belladonna (Atropa belladonna); A. membranaceus (Astragalus membranaceus); C. asiatica (Centella asiatica); C. frutescens (Capsicum frutescens); C. roseus (Catharanthus roseus); L. barbarum (Lycium barbarum); L. ruthenicum (Lycium ruthenicum); M. piperita (Mentha×piperita L.); P. ginseng (Panax ginseng L.); P. sidoides (Pelargonium sidoides); S. miltiorrhiza (Salvia miltiorrhiza); V. vinifera (Vitis vinifera).
Figure 1.
Biosynthesis of plant natural products in microbial and plant cells.
Even in this emerging research field, many specialized metabolic pathways have been introduced into heterologous plants, and advances in enabling technologies such as omics analysis techniques, plant genetic engineering technology, and analytical techniques have accelerated the development of this field (Naqvi et al., 2010; Bock, 2014; Mortimer, 2019). Even so, whether in microorganisms or plants, the identification of biosynthetic pathways remains the most significant bottleneck in PNP biosynthesis. Accordingly, it is very important to prioritize the reactions and identify the genes involved in the biosynthesis of natural products, especially the genes involved in modification.
In this review, we outline strategies and techniques for the elucidation of PNP biosynthetic pathways and the engineered biosynthesis of PNPs in plant cells. Further progress in plant synthetic biology will not only greatly relieve the pressure on ecological systems but will also facilitate the synthesis of more complex and toxic PNPs. The use of plant cells has become another powerful system for PNP biosynthesis to supplement the deficiencies of microbial chassis.
Elucidation of PNP biosynthetic pathways
Discovery of key catalytic elements in the biosynthetic pathway is the basis of heterologous production of valuable PNPs. To date, only portions of PNP biosynthetic pathways have been completely elucidated. The lack of the key catalytic element is becoming the greatest obstacle to the elucidation of complex PNP biosynthesis, as the complex metabolism of PNPs and limited analytical techniques hinder the identification of metabolic pathways. With advances in next-generation sequencing technology, computational biology, and synthetic biology, numerous gene-mining techniques have been developed.
Next-generation sequencing-guided pathway discovery
Rapid development of high-throughput sequencing makes it possible to unveil the genetic blueprint of many important plant species, as well as the gene variation among individuals at the population level; it has become the basis for in-depth gene mining and investigation of PNP biosynthesis mechanisms (Shang et al., 2014; Guo et al., 2018; Liu et al., 2018; Kang et al., 2020; Tu et al., 2020; Rai et al., 2021). For the analysis of specific metabolic pathways, the transcriptome data of species are often more helpful and easier to obtain. Screening candidate genes that may participate in the synthesis of specific natural products from tens of thousands of genes is always challenging. Hence, it is possible to identify candidate genes by comparative analysis of samples with different metabolite compositions in target pathways; these samples may be from different plant species, accessions, or mutants, or from plants induced by a target-pathway stimulus. Differential transcriptional analysis can be performed on different samples to obtain candidate genes and enable further functional analysis (Winzer et al., 2012; Galanie et al., 2015; Lau and Sattely, 2015; Polturak et al., 2016; Farrow et al., 2018; Kim et al., 2018; Christ et al., 2019; Yang et al., 2019; Nett et al., 2020; Stander et al., 2020; Xie et al., 2020).
Secondary metabolic pathways in microorganisms often exist in the form of gene clusters on chromosomes, which are very helpful for the analysis of metabolite biosynthetic pathways, excavation of biological components, or subsequent engineering design. In recent years, with the in-depth study of plant genomes, biosynthetic genes of some specific plant secondary metabolites have also been found to be arranged in clusters on plant chromosomes. Plant gene clusters are composed of at least three non-homologous genes involved in a distinct chemical pathway (Nutzmann and Osbourn, 2014). Lately, many computational tools have been developed and have facilitated the identification of increasing numbers of predicted plant gene clusters, thereby accelerating the discovery of PNP biosynthetic pathways and the design of synthetic clusters in heterologous hosts (Medema and Osbourn, 2016; Chavali and Rhee, 2018; Arya et al., 2020). These tools include plantiSMASH, PhytoClust, PlantClusterFinder, and MIBiG (Medema et al., 2015; Kautsar et al., 2017; Schlapfer et al., 2017; Topfer et al., 2017). They also provide abundant alternative and modified biological elements for synthetic biology research. The biosynthetic pathway for noscapine, a potential anti-cancer compound, was shown to consist of a 10-gene cluster in opium poppy (Winzer et al., 2012). Guo et al. (2018) reported the draft genome of opium poppy, which enabled them to identify a large 15-gene cluster and obtain information on its evolution. Homology analysis showed that cytochrome P450 and oxidoreductase genes fused to form a STORR gene fusion, which plays a key role in the biosynthesis of morphine in poppy. Cheng et al. (2021) presented a chromosome-level genome assembly for Himalayan yew and revealed that tandem duplication was the driving force for gene family evolution in the genome. This reference genome will serve as a platform for decoding the complete biosynthetic pathway of paclitaxel and for understanding the chemodiversity of taxoids in gymnosperms.
Gene knockdown/knockout-guided pathway verification
At present, the main approach for functional verification of target pathways in their original plants is to inhibit the function of all or part of the enzymes by gene deletion or expression silencing and then to detect changes in secondary metabolites in order to identify the functions of candidate pathway genes. Gene expression silencing technology is a widely used reverse genetics method that can verify the function of plant genes (Winzer et al., 2012; Kamthan et al., 2015; Qua et al., 2015; Zhao et al., 2015, 2016; Ma et al., 2016; Fu et al., 2021). Qua et al. (2015) used virus-induced gene silencing technology to rapidly identify a cytochrome P450 and an alcohol dehydrogenase that completed the pathway from tabersonine to the anti-cancer drug precursor vindoline. Fu et al. (2021) completed chicoric acid biosynthesis in purple coneflower through RNA interference technology for the identification of acyltransferase.
In recent years, with the rise of the CRISPR/Cas9 system, this gene-editing technology has been widely used for functional verification of key enzymes and regulatory elements because of its potency, versatility, and ability to knock out multiple genes simultaneously (Mercx et al., 2017; Montecillo et al., 2020). Compared with previous genome-editing methods, CRISPR/Cas9 enables convenient design with high accuracy and precision, and homozygous mutations at the desired site can be stably inherited by the subsequent generation. Editing plant genomes for transcriptional regulation and enzyme manipulation using CRISPR/Cas9 has been successful in many plants (Lowder et al., 2015; Alagoz et al., 2016; Liu et al., 2017; Feng et al., 2018; Zhu et al., 2018). A gene cluster and two key genes involved in medium-chain acyl sugar accumulation in Solanaceae were discovered by making loss-of-function mutations using the CRISPR/Cas9 system (Fan et al., 2020). This gene-editing technology has been established in rice, corn, and other crops and used to identify the role of various genes (Duan et al., 2016; Ma et al., 2017; Shen et al., 2017; Wang et al., 2017, 2019; Yasui et al., 2017; Huang et al., 2018; Li et al., 2018c; Zhao et al., 2019; Zuo et al., 2019).
Synthetic biology platform-guided pathway reconstruction
Using synthetic biology platforms is a practical approach for pathway elucidation (Ignea et al., 2016; Wang et al., 2020). With the decreasing costs of high-throughput DNA synthesis, an increasing number of alternative enzymes have been codon-optimized, synthesized, and used to complete metabolic pathways. Microbes and model plants are widely used to reconstruct pathways of many important PNPs. E. coli and S. cerevisiae are two kinds of microorganisms with strong biosynthetic ability, and their genetic operation and metabolic modifications have been studied in detail (Borodina and Nielsen, 2014; Blount, 2015). Yeast is a more suitable host as a synthetic biology platform for pathway discovery of complex PNP metabolic pathways because of its eukaryotic properties (Li et al., 2018e; Luo et al., 2019; Srinivasan and Smolke, 2020). Wang et al. (2020) established a platform to elucidate the UGT-mediated glycosylation process of the main triterpene in Panax notoginseng. The creation of such a synthetic biology platform bypassed the difficulties in obtaining intermediates and expressing enzymes in vitro (Wang et al., 2020). Longer PNP biosynthetic pathways are often divided into several biosynthetic modules. The substrates and products of these modules are generally available, which facilitates the identification of unknown enzyme reactions in the module and the detection of target products. One successful case was the construction of a breviscapine synthetic factory in yeast through the use of three modules involving eight genes (Liu et al., 2018). Similarly, candidate pathways of different PNPs such as dihydrosanguinarine, opioids, strictosidine, noscapine, and scopolamine have been successfully elucidated and constructed in microbes using a modular strategy (Fossati et al., 2014; Brown et al., 2015; Galanie et al., 2015; Trenchard and Smolke, 2015; Li and Smolke, 2016; Srinivasan and Smolke, 2020). It is worth noting that in the case of scopolamine biosynthesis, the final strain comprises 34 chromosomal modifications (26 genes, 8 gene disruptions) in six diverse subcellular locations (Srinivasan and Smolke, 2020).
Expression of enzymes in microbial systems often has issues of post-translational modification and requires multigene engineering. Genetic elements in plants, such as cytochrome P450 (Liu et al., 2020), are inefficient when transferred to heterogeneous microorganisms. Therefore, it has become a powerful measure to identify the key enzymes and determine the reaction sequence using plant hosts. Transient expression refers to the transfer of a target gene into cells for a relatively short time. Exogenous DNA introduced into the cells does not integrate with the host DNA but temporarily exists in the cells for transcription and translation, enabling the rapid functional characterization of unknown genes by analysis of the final metabolites (Lau and Sattely, 2015; Caputi et al., 2018; Christ et al., 2019; Nett et al., 2020). Christ et al. (2019) used a pooled-screen approach to identify CYPs that were necessary to catalyze the 5,6-spiroketalization of cholesterol in the biosynthesis of diosgenin in Paris polyphylla and Trigonella foenum-graecum. Combinatorial expression of 29 full-length candidate P. polyphylla CYPs and 33 full-length candidate T. foenum-graecum CYPs in Nicotiana benthamiana led to the construction of a screening pool. The key enzymes were screened by removing one candidate gene at a time from the pool. In the end, several active pairs of CYPs were identified. Recently, an automated high-throughput screening platform was established on the basis of transient expression for screening recombinant protein production, showing that the innovation of various technologies will greatly accelerate the elucidation of PNP biosynthetic pathways (Gengenbach et al., 2020).
Overall, the elucidation of PNP biosynthetic pathways is a laborious process that often requires a combination of various strategies, including but not limited to those mentioned above, such as sequencing technology, bioinformatics analysis, high-throughput screening, and other synthetic biology technologies. For example, Galanie et al. (2015) combined gene mining, enzyme engineering, and metabolic pathway and strain optimization to achieve opiate biosynthesis. A combination of transcriptomics and metabolomics enabled the elucidation of the nearly complete colchicine alkaloid synthesis pathway and the synthesis of a colchicine alkaloid precursor in N. benthamiana (Nett et al., 2020). Gao et al. (2020) reported a biosynthetic intermediate probe-based target identification strategy. Combined with traditional activity-based protein purification and transcriptome analysis, the first flavin adenine dinucleotide-dependent enzyme-catalysed intermolecular [4 + 2] cycloaddition in natural product biosynthesis was discovered, providing a new way to study PNP biosynthetic pathways.
Engineered biosynthesis of PNPs in plant cells
Secondary metabolic pathways of plants are very complex and can be linear, cyclical, or arranged in a three-dimensional grid with many branches (Verpoorte et al., 2000; Farre et al., 2014). Because of the limitations of microbial chassis, such as poor expression of P450 enzymes from plants and poor tolerance to active products, many microbial cell factories cannot produce the final active products or have low yields. Therefore it is very advantageous to perform metabolic engineering transformation in plants, because the whole or partial biosynthetic pathway of many target PNPs already exists. Production of target metabolites can be implemented by overexpression of missing or rate-limiting enzyme genes in the biosynthesis pathway or by blocking competing reactions (Ikram and Simonsen, 2017; Badshah et al., 2018; Nagegowda and Gupta, 2020). It is worth noting that plants have three major genetic systems: nuclear, plastid, and mitochondrial. Some plastids (chloroplasts) are exclusive to plants and produce some specific metabolites, such as terpenoids. Owing to the advantage of plant homology, plant cells are very suitable for the synthesis of some PNPs. Here we introduce recent progress in the engineered biosynthesis of PNPs in plant cells, mainly based on the engineering of different genetic systems (Table 1).
Nuclear genome engineering
Multiple gene co-transformation: Reconstruction of metabolic pathways
Integrating multiple transgenes into the nuclear genome of plants and ensuring stable expression of these genes in future generations have always been the focus of plant synthetic biology. Golden rice is a successful case of plant synthetic biology (Ye et al., 2000). Through Agrobacterium-mediated multigene co-transformation, the entire biosynthetic pathway of β-carotene was introduced into rice endosperm. In subsequent studies, canthaxanthin rice and astaxanthin rice appeared one after another, making rice a dietary supplement to meet daily nutritional needs (Zhu et al., 2018). In addition, rice cell suspension culture is becoming a model for the production of specialized plant metabolites (Arya et al., 2021).
As a model organism, tobacco has been widely used in synthetic biology research. Farhi et al. (2011) achieved heterologous synthesis of artemisinin in Nicotiana tabacum for the first time through co-transformation of several artemisinin synthases and rate-limiting enzymes, which increased the supply of precursors greatly and generated yields of artemisinin as high as 6.8 μg/g dry weight. Forestier et al. (2021) developed an N. benthamiana platform for the production of high-value diterpenoids. The synthesis of plant secondary metabolites is closely related to the regulation of endogenous transcription factors (TFs). There are broad prospects for improving the levels of plant cell secondary metabolites by activating the expression of multiple synthetase genes in plant secondary metabolite synthesis pathways. Appelhagen et al. (2018) achieved stable production of anthocyanin by constitutively expressing the TFs MYB Rosea1 (AmRos1) and bHLH Delila (AmDel) from Antirrhinum majus in N. tabacum suspension cells.
Besides tobacco, other plants also have the potential to serve as excellent hosts. Because of its short life cycle, small genome size, and efficient homologous recombination, Physcomitrella patens has become a promising model organism (Reski et al., 2018; Campos et al., 2020; Decker and Reski, 2020). P. patens was previously used to synthesize taxadiene derived from Taxus, and the yield of taxadiene reached 5 μg/g (Anterola et al., 2009). Since then, owing to its natural tolerance to terpenoids and simple metabolic profile of endogenous terpenoids, P. patens has been widely used for the expression of terpenoid biosynthesis genes and the reconstruction of pathways (Bach et al., 2014; Banerjee et al., 2019). For example, artemisinin was synthesized by directly engineering DNA fragments of five enzymes of the artemisinin synthesis pathway into the moss P. patens without any other modification (Ikram et al., 2017, 2019). The first active ingredient from biotechnologically produced moss is MossCellTec No. 1, which is a highly innovative active compound based on a novel anti-aging concept (https://mibellebiochemistry.com/mosscelltectm-no-1). In addition, the low-scent background characteristics of P. patens make it very suitable for the production of perfume material, such as α/β-santalol, patchoulol, and sclareol (Zhan et al., 2014; Pan et al., 2015). Mosspiration Biotech launched Orbella Moss, a fragrance-producing moss and the first genetically modified fragrant plant (https://www.mosspirationbiotech.com/fragrant-moss/).
The CRISPR/Cas9 system is gradually becoming a powerful tool for plant metabolic engineering (Arya et al., 2020; Montecillo et al., 2020; Sabzehzari et al., 2020; Dey, 2021). For example, anthocyanin content has been shown to accumulate through the introduction of double-strand breaks mediated by CRISPR/Cas9 for a powerful promoter inserted in tomato (Cermak et al., 2015). Also in tomato, the autoinhibitory domain of glutamate decarboxylase, a key enzyme in γ-aminobutyric acid (GABA) biosynthesis, was deleted using CRISPR/Cas9, resulting in 7- to 15-fold increases in GABA yield (Nonaka et al., 2017). A multiplex gene-editing system was used to metabolically engineer the isoflavone content of soya bean. Three genes were targeted simultaneously, and the triple gene mutation efficiency reached 44.44%, confirming the editing efficiency of CRISPR/Cas9 technology (Zhang et al., 2020a). Inhibiting competitive pathway genes can produce more metabolic flow to the target products to improve yield. Lv et al. (2016b) introduced antisense cDNA into Artemisia annua to downregulate branch pathways, resulting in artemisinin accumulation. Li et al. (2018d) used CRISPR/Cas9 multiplexing to simultaneously regulate multiple genes involved in the carotenoid biosynthesis pathway, thus increasing the content of lycopene 5.1-fold in tomato fruit. In the same way, GABA-enriched tomato was developed by manipulating the GABA shunt (Li et al., 2018b). Zeng et al. (2021) generated Atropa belladonna plant lines with high yields of hyoscyamine and without anisodamine and scopolamine via CRISPR/Cas9-based disruption of hyoscyamine-6β-hydroxylase.
Combinatorial transformation: Screening high-yield transgenic plants
With the deepening of research, more and more complex pathways need to be reconstructed in plants. This requires the generation of many transformants with combinations of different candidate genes and expression levels to ensure proper pathway reconstruction and high yields of target products. However, this is difficult to achieve because of the long cycles and demanding operations involved in traditional plant transformation. To overcome these problems, Zhu et al. (2008) proposed a combinatorial nuclear transformation method to dissect and modify the carotenoid biosynthetic pathway in maize to produce multiplex-transgenic plants in a short time. In this process, five carotenogenic genes were constructed in separate plasmids and integrated into the genome at the same locus using biolistic transformation to introduce all plasmids simultaneously. Transgenic plants with distinct metabolic phenotypes that expressed different enzyme combinations were generated to identify the rate-limiting enzymes in the metabolic pathway, providing a unique strategy for the analysis of PNP metabolic pathways (Zhu et al., 2008). The team then used this method to generate transgenic maize plants that could simultaneously increase the production of three vitamins (Naqvi et al., 2009). Results showed that the contents of carotene, ascorbic acid, and folic acid were significantly increased in the endosperm by expression of four transgenes, indicating that multigene transformations could efficiently regulate multiple metabolic pathways at the same time.
Combinatorial transformation is a multifunctional method that can be used to engineer metabolic pathways and control other biochemical, physiological, or developmental processes. Many transformations are performed to create a combinatorial library of transformants, each with a different combination of transgenes with varying copy number and genomic arrangement, which result in diverse expression levels. To date, there is no sign of a limit to the number of transgenes that can be introduced into the plant in one transformation, making this method a powerful form of co-transformation.
Transient expression: Rapid production of target compounds
Stable transformation with good reproducibility has the potential advantage of large-scale production, but transgenic plants are usually the second generation, so their creation is often very laborious (Tyurin et al., 2020). Transient expression can be used to express recombinant protein in batches within a few days and then used to co-transfer candidate genes into plants to rapidly produce target compounds (Lau and Sattely, 2015; Christ et al., 2019; Nett et al., 2020). Many transient expression systems are based on plant viruses. High levels of recombinant protein expression depend on rapid replication and transmission and high expressionyield of these viruses in the plant host (Gleba et al., 2014). The pEAQ vectors have been used to produce a wide variety of proteins in plants. Besides viral proteins and virus-like particles, other types of proteins including active enzymes have also been expressed. Recently, the pEAQ system has been frequently used for PNP production in tobacco. Taxane biosynthesis was carried out in N. benthamiana using Agrobacterium-mediated transient transformation (Li et al., 2019). Transient expression of 16 genes from mayapple in N. benthamiana resulted in the accumulation of the hemotherapeutic drug etoposide at the milligram scale (Schultz et al., 2019). Miettinen et al. (2014) successfully reconstructed the upstream pathway of terpenoid indole alkaloid biosynthesis in N. benthamiana, providing a basis for the development of a variety of therapeutic compounds. Co-infiltration of Agrobacterium strains with different expression constructs is a powerful method of rapidly expressing numerous enzyme combinations (Reed and Osbourn, 2018). Transient combinatorial transformation of multiple P450s generated over 40 oxygenated triterpene analogs in N. benthamiana, many of which had not been reported previously (Reed et al., 2017). Kashkooli et al. (2019) used a combinatorial metabolic engineering approach to generate costunolide and parthenolide derivatives. Calgaro-Kozina et al. (2020) rationally broadened the set of primary metabolites by expressing diverse enzymes, leading to the production of crucifalexins, a novel class of compounds with potent antifungal activity.
These works illustrate the ability to produce high-quality PNPs by transient plant expression technology. They highlight the potential of rapid combinatorial transformation for the synthesis of new PNP derivatives and provide novel ideas for the discovery of new drugs. Moreover, owing to rapid and high-level expression, some transient expression systems have already been used for industrial-scale production of target proteins (Kopertekh and Schiemann, 2019; Huebbers and Buyel, 2021). Recently, a highly efficient Agrobacterium-mediated strategy for transient transformation was created, making this technology a promising application for a variety of plant species (Zhang et al., 2020b).
Plastid engineering
Plastid genetic engineering has attracted much attention due to the plastid’s high expression of genes, accuracy of homologous recombination, and pattern of gene stacking in operons. In addition, its use can avoid environmental safety problems caused by pollen transmission to some extent because of the maternal mode of plastid inheritance in most angiosperm species (Bock, 2014; Yu et al., 2020). However, conventional chloroplast transformation is mainly limited to the Solanaceae (Yu et al., 2020). A recent study reported an efficient system for plastid genetic transformation in Arabidopsis root callus (Ruf et al., 2019). A novel method of plastid transformation using nanoparticles was reported, accelerating the development of plastid genetic transformation technology for other species (Kwak et al., 2019). For plants that cannot undergo chloroplast transformation, horizontal gene transfer mediated by grafting is a promising way to obtain foreign chloroplast genes (Stegemann et al., 2012; Thyssen et al., 2012; Lu et al., 2017). This strategy is helpful for expanding the range of species suitable for plastid engineering.
Plastid engineering for high yield of PNPs
Because there are many copies of the chloroplast genome in each cell, higher levels of protein accumulation can often be achieved from transgenes inserted into the chloroplast genome compared with the nuclear genome, and the operon pattern of chloroplast genes makes expression regulation of the transgene much easier (Jensen and Scharff, 2019). The methylerythritol phosphate (MEP) pathway is unique to plants and takes place in the chloroplast; chloroplast transformation is therefore usually used to increase the yield of isoprenoid compounds (Saxena et al., 2014; Pasoreck et al., 2016). By inserting the whole mevalonate (MVA) pathway into the N. tabacum chloroplast genome, metabolic flux in the isoprenoid biosynthesis pathway was remolded, resulting in the higher accumulation of mevalonate, carotenoids, squalene, sterols, and triacylglycerols (Kumar et al., 2012). Moreover, redirecting the reaction into the plastid has always led to production improvement. One of the successful examples described in tobacco was the targeting of sesquiterpenoid synthase to the plastids. This afforded a 40 000-fold increase in yield (Wu et al., 2006). Through chloroplast-compartmentalized engineering and overexpression of rate-limiting enzymes of isoprenoid pathways, the yield of taxadiene and taxadiene-5α-ol were increased to 56.6 μg/g and 1.3 μg/g fresh weight, respectively, in tobacco (Li et al., 2019).
Plant cytochrome P450s are the largest family of enzymes involved in PNP biosynthesis, and they mainly play the role of monooxygenases in plants (Liu et al., 2020). A new green production plant has been built by coupling P450 enzyme with an in vitro light system or by transforming the P450-dependent pathway into chloroplasts using photosynthetic reducing power to drive the catalytic activity of P450s (Jensen et al., 2011; Nielsen et al., 2013; Lassen et al., 2014; Mellor et al., 2016). Gnanasekaran et al. (2016) expressed two P450 enzymes and one glycosyltransferase in tobacco chloroplasts to construct the biosynthetic pathway of dhurrin. On this basis, three enzymes involved in the pathway were fused with Tat protein to carry out co-localization in plastids, which increased titers of dhurrin 5-fold (de Jesus et al., 2017). Mellor et al. (2019) further investigated the efficacy of four electron carrier proteins fused to CYP79A1 in N. benthamiana. A flavodoxin-like carrier showed the greatest improvement in sequestration of photosynthetic reducing power (nearly 25-fold on a per-protein basis). These studies provide a new strategy for the metabolic engineering of PNPs in plants.
Combined application of chloroplast and nuclear transformation
Under natural conditions, PNPs are synthesized in different compartments of plant cells, and the enzymes of catalytic reactions are located in different organelles, reducing the toxic effects of intermediates on cells (Espinosa-Leal et al., 2018). The strategy of combining nuclear transformation with chloroplast transformation to reorient different reactions in the metabolic pathway often has excellent effects. Artemisinin was successfully biosynthesized in tobacco by engineering the MVA pathway and artemisinin biosynthesis pathway targeted to three different cellular compartments using chloroplast and nuclear transformation. With this approach, the yield of artemisinin reached 0.8 mg/g dry weight (Malhotra et al., 2016). A new synthetic biology strategy called combinatorial supertransformation of transplastomic recipient lines (COSTREL) was developed to achieve the de novo synthesis of artemisinic acid (Fuentes et al., 2016). The core pathway of artemisinic acid biosynthesis was transferred into the chloroplast genome, after which the transplastomic plants were combinatorially supertransformed with cassettes for all additional enzymes known to affect flux through the artemisinin pathway. By screening large populations of COSTREL lines, a plant that could produce more than 120 mg of artemisinic acid per kg of biomass was isolated.
Development of non-green plastid transformation
Although we have acquired adequate knowledge about the gene expression mechanisms of chloroplasts in the past few decades, little is known about non-green plastids. Increased β-carotene production in tomato showed the possibility of using chloroplast transformation to construct biochemical pathways for important nutrients in non-green plastids, potentially impacting many metabolic processes in plants (Wurbs et al., 2007; Apel and Bock, 2009; Ahmad et al., 2016). Chromoplasts in mature tomatoes are differentiated from chloroplasts, which are very active in gene expression. This makes plastid transgenic expression in tomato more successful than in other plants (Sidorov et al., 1999; Ruf et al., 2001; Wurbs et al., 2007). Non-green plastids are ideal hosts for recombinant protein expression; they are more suitable for the production and storage of PNPs than chloroplasts without affecting plant adaptability (Mellor et al., 2018). Recent advances have further promoted the efficient expression of transgenes in non-green plastids (Valkov et al., 2011; Zhang et al., 2012; Caroca et al., 2013; Fuentes et al., 2018; Schaub et al., 2018; Yu et al., 2019; Cortese et al., 2021). This improvement in non-green plastid transformation technology accelerates the development of PNP production in edible fruits and also solves the problem of toxic by-product accumulation caused by metabolic engineering transformation in tobacco (Fuentes et al., 2018).
Concluding remarks and perspectives
Secondary metabolites from plants have always been an important source of drugs. Owing to limited resources, we need to establish sustainable routes to supplement the sources of these secondary metabolites. It is important to transform cellular genetic material and metabolic processes into an ideal host and to synthesize PNPs in cells. Although the number of known natural products has exceeded 500 000, the biosynthetic pathways of most natural products have not been identified (Zhang and Schrader, 2017). Therefore, the analysis and biosynthesis of these prodrug compounds have profound significance and considerable prospects.
To date, some synthetic cell factories for PNPs have been successfully established and have reached the level of industrial production. Microbial production of artemisinin, resveratrol, lycopene, and astaxanthin has been successfully commercialized (Mehta et al., 2003; Schmidt et al., 2011; Paddon et al., 2013). Although industrial production of terpenoids in microorganisms has made great advances, microbial production of some specific PNPs still suffers from strong resistance due to pathway complexity, low activity of plant-derived enzymes in the microbial host, and product toxicity (Liu et al., 2017; Maeda, 2019). Use of plants for the synthesis of valuable PNPs has natural advantages. First, plants can better express plant-derived enzymes. Second, some compounds that are toxic to microorganisms, such as alkaloids and phenols, can be stored in plants. Third, the complex organelles and membrane structures of plants allow fine artificial design because plant metabolic pathways are highly localized. Finally, plants synthesize all kinds of high-value natural products through photosynthesis without the need for supplementation with other raw materials, making them low in cost and sustainable. The development of energy crops reduces the competition of land and food crops to the greatest extent, allowing further development of plant cell factories. Using model plants such as tobacco, tomato, rice, and P. patens as platforms for PNP production is becoming very attractive because of their publicly available genome sequences and simplicity of use in biotechnological techniques (Fu et al., 2018).
Even so, it remains challenging to use plants as a chassis for PNP biosynthesis (Sweetlove et al., 2017; Maeda, 2019). First, the genetic and metabolic backgrounds of plants are very complex, with many details still not very clear compared with microorganisms. Second, some enabling technologies for plants are still lacking or inefficient. In addition, the long growth cycle of plants and the difficulty in scaling up to industrial production are always obstacles to plant synthetic biology. Unlike whole plants, the culture cycle of suspension cells is only several days. PNPs could be produced by a process similar to microorganism fermentation using plant suspension cells. In addition, after dedifferentiation of plant cells, a large number of metabolic processes, including secondary metabolism, are silent. This reduces metabolic complexity and may facilitate the systematic regulation of target pathways by genetic engineering, diverse plant endogenous metabolic elicitors, and culture techniques. Some PNPs have already been produced commercially through suspension cell culture and have achieved high yields even without transgenic modification (www.phytonbiotech.com/) (Fujita, 1988; Hibino and Ushiyama, 1999; Wilson and Roberts, 2012; Lange, 2018; Arya et al., 2020). Metabolic engineering of a suspension cell system is expected to be an efficient approach for PNP production in the future. Plant cell culture technology has been widely used for industrial production of active cosmetic ingredients and food additives, such as the novel authorized food ingredients TEUPOL (10P or 50P), ECHINAN 4P, and ACTEOS 10P and the skin care ingredients PhytoCellTec Symphytum and PhytoCellTec nunatak (Georgiev et al., 2018; Krasteva et al., 2021). Some other emerging technologies, such as plant living-root exudation technology–plant milking (https://www.plantadvanced.com/expertises-2/pat-plant-milking), also promote the development of plant cells for PNP biosynthesis. We believe that with the maturity of next-generation sequencing technology, bioinformatics analysis, genetic engineering technology, and other synthetic biology technologies, more and more biosynthetic pathways will be deciphered in medicinal plants. Although there is still a long way to go to overcome the bottlenecks in the development of plant synthetic biology, plant cells will be an excellent alternative candidate for PNP production (Arya et al., 2020; Eljounaidi and Lichman, 2020) and provide a huge opportunity for future agriculture and industry.
Funding
This work was supported by grants from the National Natural Science Foundation of China (grant no. 31901026), the China Postdoctoral Science Foundation (grant no. 2019M661032) and Tianjin Science and technology plan project (grant no. 19PTZWHZ00060).
Author contributions
X.Z. and X.L.: design and writing of the article. T.L. and Y.W.: data curation. Z.L. and H.J.: manuscript revision. All authors contributed to the discussion and approved the final paper.
Acknowledgments
No conflict of interest declared.
Published: August 9, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Contributor Information
Zhichao Li, Email: jiang_hf@tib.cas.cn.
Huifeng Jiang, Email: lizhch@tib.cas.cn.
References
- Ahmad N., Michoux F., Loessl A.G., Nixon P.J. Challenges and perspectives in commercializing plastid transformation technology. J. Exp. Bot. 2016;67:5945–5960. doi: 10.1093/jxb/erw360. [DOI] [PubMed] [Google Scholar]
- Ajikumar P.K., Xiao W.-H., Tyo K.E.J., Wang Y., Simeon F., Leonard E., Mucha O., Phon T.H., Pfeifer B., Stephanopoulos G. Isoprenoid pathway optimization for taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagoz Y., Gurkok T., Zhang B.H., Unver T. Vol. 6. Scientific Reports; 2016. (Manipulating the Biosynthesis of Bioactive Compound Alkaloids for Next-Generation Metabolic Engineering in Opium Poppy Using CRISPR-Cas 9 Genome Editing Technology). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anterola A., Shanle E., Perroud P.-F., Quatrano R. Production of taxa-4(5),11(12)-diene by transgenic Physcomitrella patens. Transgenic Res. 2009;18:655–660. doi: 10.1007/s11248-009-9252-5. [DOI] [PubMed] [Google Scholar]
- Apel W., Bock R. Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. Plant Physiol. 2009;151:59–66. doi: 10.1104/pp.109.140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhagen I., Wulff-Vester A.K., Wendell M., Hvoslef-Eide A.-K., Russell J., Oertel A., Martens S., Mock H.-P., Martin C., Matros A. Colour bio-factories: towards scale-up production of anthocyanins in plant cell cultures. Metab. Eng. 2018;48:218–232. doi: 10.1016/j.ymben.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya S.S., Kumari D.D., Rookes J.E., Cahill D.M., Lenka S.K. Rice cell suspension culture as a model for producing high-value recombinant proteins and plant specialized metabolites. Plant Cell Tissue Organ Cult. 2021;145:463–486. [Google Scholar]
- Arya S.S., Rookes J.E., Cahill D.M., Lenka S.K. Next-generation metabolic engineering approaches towards development of plant cell suspension cultures as specialized metabolite producing biofactories. Biotechnol. Adv. 2020;45:107635. doi: 10.1016/j.biotechadv.2020.107635. [DOI] [PubMed] [Google Scholar]
- Bach S.S., King B.C., Zhan X., Simonsen H.T., Hamberger B. Heterologous stable expression of terpenoid biosynthetic genes using the moss Physcomitrella patens. Methods Mol. Biol. (Clifton N.J.) 2014;1153:257–271. doi: 10.1007/978-1-4939-0606-2_19. [DOI] [PubMed] [Google Scholar]
- Badshah S.L., Ullah A., Ahmad N., Almarhoon Z.M., Mabkhot Y. Increasing the strength and production of artemisinin and its derivatives. Molecules. 2018;23:100. doi: 10.3390/molecules23010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek S., Ho T.T., Lee H., Jung G., Kim Y.E., Jeong C.S., Park S.Y. Enhanced biosynthesis of triterpenoids in Centella asiatica hairy root culture by precursor feeding and elicitation. Plant Biotechnol. Rep. 2020;14:45–53. [Google Scholar]
- Bai F., Li S., Yang C., Zhao T., Zhang T., Lan X., Chen M., Liao Z. Overexpression of the AbSAUR1 gene enhanced biomass production and alkaloid yield in Atropa belladonna. Ind. Crop Prod. 2019;140:111705. [Google Scholar]
- Banerjee A., Arnesen J.A., Moser D., Motsa B.B., Johnson S.R., Hamberger B. Engineering modular diterpene biosynthetic pathways in Physcomitrella patens. Planta. 2019;249:221–233. doi: 10.1007/s00425-018-3053-0. [DOI] [PubMed] [Google Scholar]
- Besumbes O., Sauret-Gueto S., Phillips M.A., Imperial S., Rodriguez-Concepcion M., Boronat A. Metabolic engineering of isoprenoid biosynthesis in Arabidopsis for the production of taxadiene, the first committed precursor of taxol. Biotechnol. Bioeng. 2004;88:168–175. doi: 10.1002/bit.20237. [DOI] [PubMed] [Google Scholar]
- Birchfield A.S., Mcintosh C.A. Metabolic engineering and synthetic biology of plant natural products—a minireview. Curr. Plant Biol. 2020:100163. [Google Scholar]
- Blount Z.D. The unexhausted potential of E. coli. eLife. 2015;4:111705. doi: 10.7554/eLife.05826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. Genetic engineering of the chloroplast: novel tools and new applications. Curr. Opin. Biotechnol. 2014;26:7–13. doi: 10.1016/j.copbio.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Borodina I., Nielsen J. Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol. J. 2014;9:609–620. doi: 10.1002/biot.201300445. [DOI] [PubMed] [Google Scholar]
- Brown S., Clastre M., Courdavault V., O'Connor S.E. De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. U S A. 2015;112:3205–3210. doi: 10.1073/pnas.1423555112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calgaro-Kozina A., Vuu K.M., Keasling J.D., Loque D., Sattely E.S., Shih P.M. Engineering plant synthetic pathways for the biosynthesis of novel antifungals. ACS Cent. Sci. 2020;6:1394–1400. doi: 10.1021/acscentsci.0c00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M.L., Prado G.S., dos Santos V.O., Nascimento L.C., Dohms S.M., da Cunha N.B., Ramada M.H.S., Grossi-de-Sa M.F., Dias S.C. Mosses: versatile plants for biotechnological applications. Biotechnol. Adv. 2020;41:10. doi: 10.1016/j.biotechadv.2020.107533. [DOI] [PubMed] [Google Scholar]
- Caputi L., Franke J., Farrow S.C., Chung K., Payne R.M.E., Trinh-Don N., Dang T.-T.T., Carqueijeiro I.S.T., Koudounas K., de Bernonville T.D. Missing enzymes in the biosynthesis of the anticancer drug vinblastine in Madagascar periwinkle. Science. 2018;360:1235–1238. doi: 10.1126/science.aat4100. [DOI] [PubMed] [Google Scholar]
- Caroca R., Howell K.A., Hasse C., Ruf S., Bock R. Design of chimeric expression elements that confer high-level gene activity in chromoplasts. Plant J. 2013;73:368–379. doi: 10.1111/tpj.12031. [DOI] [PubMed] [Google Scholar]
- Cermak T., Baltes N.J., Cegan R., Zhang Y., Voytas D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:14. doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali A.K., Rhee S.Y. Bioinformatics tools for the identification of gene clusters that biosynthesize specialized metabolites. Brief. Bioinform. 2018;19:1022–1034. doi: 10.1093/bib/bbx020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee M.J.Y., Lycett G.W., Khoo T.-J., Chin C.F. Bioengineering of the plant culture of Capsicum frutescens with vanillin synthase gene for the production of vanillin. Mol. Biotechnol. 2017;59:1–8. doi: 10.1007/s12033-016-9986-2. [DOI] [PubMed] [Google Scholar]
- Cheng J., Wang X., Liu X., Zhu X., Li Z., Chu H., Wang Q., Lou Q., Cai B., Yang Y. Chromosome-level genome of Himalayan yew provides insights into the origin and evolution of the paclitaxel biosynthetic pathway. Mol. Plant. 2021;14:1199–1209. doi: 10.1016/j.molp.2021.04.015. [DOI] [PubMed] [Google Scholar]
- Christ B., Xu C., Xu M., Li F.-S., Wada N., Mitchell A.J., Han X.-L., Wen M.-L., Fujita M., Weng J.-K. Repeated evolution of cytochrome P450-mediated spiroketal steroid biosynthesis in plants. Nat. Commun. 2019;10:3206. doi: 10.1038/s41467-019-11286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu M., Pedreno M.A., Alburquerque N., Faize L., Burgos L., Almagro L. A new strategy to enhance the biosynthesis of trans-resveratrol by overexpressing stilbene synthase gene in elicited Vitis vinifera cell cultures. Plant Physiol. Biochem. 2017;113:141–148. doi: 10.1016/j.plaphy.2017.02.006. [DOI] [PubMed] [Google Scholar]
- Chun J.-H., Adhikari P.B., Park S.-B., Han J.-Y., Choi Y.-E. Production of the dammarene sapogenin (protopanaxadiol) in transgenic tobacco plants and cultured cells by heterologous expression of PgDDS and CYP716A47. Plant Cell Rep. 2015;34:1551–1560. doi: 10.1007/s00299-015-1806-9. [DOI] [PubMed] [Google Scholar]
- Cortese E., Carraretto L., Baldan B., Navazio L. Arabidopsis photosynthetic and heterotrophic cell suspension cultures. Methods Mol. Biol. (Clifton, N.J.) 2021;2200:167–185. doi: 10.1007/978-1-0716-0880-7_8. [DOI] [PubMed] [Google Scholar]
- Cravens A., Payne J., Smolke C.D. Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 2019;10:2142. doi: 10.1038/s41467-019-09848-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jesus M., Nielsen A.Z., Mellor S.B., Matthes A., Burow M., Robinson C., Jensen P.E. Tat proteins as novel thylakoid membrane anchors organize a biosynthetic pathway in chloroplasts and increase product yield 5-fold. Metab. Eng. 2017;44:108–116. doi: 10.1016/j.ymben.2017.09.014. [DOI] [PubMed] [Google Scholar]
- Decker E.L., Reski R. Mosses in biotechnology. Curr. Opin. Biotechnol. 2020;61:21–27. doi: 10.1016/j.copbio.2019.09.021. [DOI] [PubMed] [Google Scholar]
- Dey A. CRISPR/Cas genome editing to optimize pharmacologically active plant natural products. Pharmacol. Res. 2021;164:105359. doi: 10.1016/j.phrs.2020.105359. [DOI] [PubMed] [Google Scholar]
- Diretto G., Al-Babili S., Tavazza R., Papacchioli V., Beyer P., Giuliano G. Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PLoS One. 2007;2:8. doi: 10.1371/journal.pone.0000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y.-B., Li J., Qin R.-Y., Xu R.-F., Li H., Yang Y.-C., Ma H., Li L., Wei P.-C., Yang J.-B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016;90:49–62. doi: 10.1007/s11103-015-0393-z. [DOI] [PubMed] [Google Scholar]
- Eljounaidi K., Lichman B.R. Nature's chemists: the discovery and engineering of phytochemical biosynthesis. Front. Chem. 2020;8:596479. doi: 10.3389/fchem.2020.596479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Leal C.A., Puente-Garza C.A., Garcia-Lara S. In vitro plant tissue culture: means for production of biological active compounds. Planta. 2018;248:1–18. doi: 10.1007/s00425-018-2910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P., Wang P., Lou Y.-R., Leong B.J., Moore B.M., Schenck C.A., Combs R., Cao P., Brandizzi F., Shiu S.-H. Evolution of a plant gene cluster in Solanaceae and emergence of metabolic diversity. eLife. 2020;9:e56717. doi: 10.7554/eLife.56717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhi M., Marhevka E., Ben-Ari J., Algamas-Dimantov A., Liang Z., Zeevi V., Edelbaum O., Spitzer-Rimon B., Abeliovich H., Schwartz B. Generation of the potent anti-malarial drug artemisinin in tobacco. Nat. Biotechnol. 2011;29:1072–1074. doi: 10.1038/nbt.2054. [DOI] [PubMed] [Google Scholar]
- Farre G., Blancquaert D., Capell T., Van Der Straeten D., Christou P., Zhu C. Engineering complex metabolic pathways in plants. Annu. Rev. Plant Biol. 2014;65:187–223. doi: 10.1146/annurev-arplant-050213-035825. [DOI] [PubMed] [Google Scholar]
- Farrow S.C., Kamileen M.O., Meades J., Ameyaw B., Xiao Y., O'Connor S.E. Cytochrome P450 and O-methyltransferase catalyze the final steps in the biosynthesis of the anti-addictive alkaloid ibogaine from Tabernanthe iboga. J. Biol. Chem. 2018;293:13821–13833. doi: 10.1074/jbc.RA118.004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z.Y., Zhang Z.J., Hua K. A Highly Efficient Cell Division-Specific CRISPR/Cas9 System Generates Homozygous Mutants for Multiple Genes in Arabidopsis. Int. J. Mol. Sci. 2018;19:10. doi: 10.3390/ijms19123925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestier E.C.F., Czechowski T., Cording A.C., Gilday A.D., King A.J., Brown G.D., Graham I.A. Developing a Nicotiana benthamiana transgenic platform for high-value diterpene production and candidate gene evaluation. Plant Biotechnol. J. 2021 doi: 10.1111/pbi.13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossati E., Ekins A., Narcross L., Zhu Y., Falgueyret J.-P., Beaudoin G.A.W., Facchini P.J., Martin V.J.J. Reconstitution of a 10-gene pathway for synthesis of the plant alkaloid dihydrosanguinarine in Saccharomyces cerevisiae. Nat. Commun. 2014;5:3283. doi: 10.1038/ncomms4283. [DOI] [PubMed] [Google Scholar]
- Fu R., Martin C., Zhang Y. Next-generation plant metabolic engineering, inspired by an ancient Chinese irrigation system. Mol. Plant. 2018;11:47–57. doi: 10.1016/j.molp.2017.09.002. [DOI] [PubMed] [Google Scholar]
- Fu R., Zhang P., Jin G., Wang L., Qi S., Cao Y., Martin C., Zhang Y. Versatility in acyltransferase activity completes chicoric acid biosynthesis in purple coneflower. Nat. Commun. 2021;12:1563. doi: 10.1038/s41467-021-21853-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes P., Armarego-Marriott T., Bock R. Plastid transformation and its application in metabolic engineering. Curr. Opin. Biotechnol. 2018;49:10–15. doi: 10.1016/j.copbio.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Fuentes P., Zhou F., Erban A., Karcher D., Kopka J., Bock R. A new synthetic biology approach allows transfer of an entire metabolic pathway from a medicinal plant to a biomass crop. Elife. 2016;5:e13664. doi: 10.7554/eLife.13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y. Industrial-production of shikonin and berberine. Ciba Found. Symposia. 1988;137:228–235. [Google Scholar]
- Gabr A.M.M., Ghareeb H., El Shabrawi H.M., Smetanska I., Bekheet S.A. Enhancement of silymarin and phenolic compound accumulation in tissue culture of Milk thistle using elicitor feeding and hairy root cultures. J. Genet. Eng. Biotechnol. 2016;14:327–333. doi: 10.1016/j.jgeb.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanie S., Thodey K., Trenchard I.J., Interrante M.F., Smolke C.D. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Su C., Du X.X., Wang R.S., Chen S.M., Zhou Y., Liu C.W., Liu X.J., Tian R.Z., Zhang L.Y. FAD-dependent enzyme-catalysed intermolecular 4+2 cycloaddition in natural product biosynthesis. Nat. Chem. 2020;12:620–628. doi: 10.1038/s41557-020-0467-7. [DOI] [PubMed] [Google Scholar]
- Gengenbach B.B., Opdensteinen P., Buyel J.F. Robot cookies—plant cell packs as an automated high-throughput screening platform based on transient expression. Front. Bioeng. Biotechnol. 2020;8:393. doi: 10.3389/fbioe.2020.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev V., Slavov A., Vasileva I., Pavlov A. Plant cell culture as emerging technology for production of active cosmetic ingredients. Eng. Life Sci. 2018;18:779–798. doi: 10.1002/elsc.201800066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleba Y.Y., Tuse D., Giritch A. In: Plant Viral Vectors. Palmer K., Gleba Y., editors. Springer; Berlin Heidelberg: 2014. Plant viral vectors for delivery by agrobacterium; pp. 155–192. [DOI] [PubMed] [Google Scholar]
- Gnanasekaran T., Karcher D., Nielsen A.Z., Martens H.J., Ruf S., Kroop X., Olsen C.E., Motawie M.S., Pribil M., Moller B.L. Transfer of the cytochrome P450-dependent dhurrin pathway from Sorghum bicolor into Nicotiana tabacum chloroplasts for light-driven synthesis. J. Exp. Bot. 2016;67:2495–2506. doi: 10.1093/jxb/erw067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Winzer T., Yang X., Li Y., Ning Z., He Z., Teodor R., Lu Y., Bowser T.A., Graham I.A. The opium poppy genome and morphinan production. Science. 2018;362:343–347. doi: 10.1126/science.aat4096. [DOI] [PubMed] [Google Scholar]
- Gwak Y.S., Han J.Y., Choi Y.E. Production of ginsenoside aglycone (protopanaxatriol) and male sterility of transgenic tobacco co-overexpressing three Panax ginseng genes: PgDDS, CYP716A47, and CYP716A53v2. J. Ginseng Res. 2019;43:261–271. doi: 10.1016/j.jgr.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J.-Y., Wang H.-Y., Choi Y.-E. Production of dammarenediol-II triterpene in a cell suspension culture of transgenic tobacco. Plant Cell Rep. 2014;33:225–233. doi: 10.1007/s00299-013-1523-1. [DOI] [PubMed] [Google Scholar]
- Han J., Wang H., Lundgren A., Brodelius P.E. Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry. 2014;102:89–96. doi: 10.1016/j.phytochem.2014.02.011. [DOI] [PubMed] [Google Scholar]
- He X., Li Y., Lawson D., Xie D.-Y. Metabolic engineering of anthocyanins in dark tobacco varieties. Physiol. Plant. 2017;159:2–12. doi: 10.1111/ppl.12475. [DOI] [PubMed] [Google Scholar]
- Hedayati A., Hosseini B., Palazon J., Maleki R. Improved tropane alkaloid production and changes in gene expression in hairy root cultures of two Hyoscyamus species elicited by silicon dioxide nanoparticles. Plant Physiol. Biochem. 2020;155:416–428. doi: 10.1016/j.plaphy.2020.07.029. [DOI] [PubMed] [Google Scholar]
- Hibino K., Ushiyama K. In: Plant Cell and Tissue Culture for the Production of Food Ingredients. Fu T.-J., Singh G., Curtis W.R., editors. Springer US; Boston, MA: 1999. Commercial production of ginseng by plant tissue culture technology; pp. 215–224. [Google Scholar]
- Huang C., Sun H., Xu D., Chen Q., Liang Y., Wang X., Xu G., Tian J., Wang C., Li D. ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. U S A. 2018;115:E334–E341. doi: 10.1073/pnas.1718058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebbers J.W., Buyel J.F. On the verge of the market - plant factories for the automated and standardized production of biopharmaceuticals. Biotechnol. Adv. 2021;46:18. doi: 10.1016/j.biotechadv.2020.107681. [DOI] [PubMed] [Google Scholar]
- Ignea C., Athanasakoglou A., Ioannou E., Georgantea P., Trikka F.A., Loupassaki S., Roussis V., Makris A.M., Kampranis S.C. Carnosic acid biosynthesis elucidated by a synthetic biology platform. Proc. Natl. Acad. Sci. U S A. 2016;113:3681–3686. doi: 10.1073/pnas.1523787113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram N.K.B.K., Kashkooli A.B., Peramuna A.V., van der Krol A.R., Bouwmeester H., Simonsen H.T. Stable production of the antimalarial drug artemisinin in the moss Physcomitrella patens. Front. Bioeng. Biotechnol. 2017;5:47. doi: 10.3389/fbioe.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram N.K.B.K., Simonsen H.T. A review of biotechnological artemisinin production in plants. Front. Plant Sci. 2017;8:1966. doi: 10.3389/fpls.2017.01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram N.K.K., Kashkooli A.B., Peramuna A., van der Krol A.R., Bouwmeester H., Simonsen H.T. Insights into heterologous biosynthesis of Arteannuin B and artemisinin in Physcomitrella patens. Molecules. 2019;24:3822. doi: 10.3390/molecules24213822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K., Jensen P.E., Moller B.L. Light-Driven Cytochrome P450 Hydroxylations. ACS Chem. Biol. 2011;6:533–539. doi: 10.1021/cb100393j. [DOI] [PubMed] [Google Scholar]
- Jensen P.E., Scharff L.B. Engineering of plastids to optimize the production of high-value metabolites and proteins. Curr. Opin. Biotechnol. 2019;59:8–15. doi: 10.1016/j.copbio.2019.01.009. [DOI] [PubMed] [Google Scholar]
- Jiao J., Gai Q.Y., Wang W., Lou M., Zu Y.G., Fu Y.J., Ma W. Enhanced astragaloside production and transcriptional responses of biosynthetic genes in Astragalus membranaceus hairy root cultures by elicitation with methyl jasmonate. Biochem. Eng. J. 2016;105:339–346. [Google Scholar]
- Kamthan A., Chaudhuri A., Kamthan M., Datta A. Small RNAs in plants: recent development and application for crop improvement. Front. Plant Sci. 2015;6:17. doi: 10.3389/fpls.2015.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.-H., Pandey R.P., Lee C.-M., Sim J.-S., Jeong J.-T., Choi B.-S., Jung M., Ginzburg D., Zhao K., Won S.Y. Genome-enabled discovery of anthraquinone biosynthesis in Senna tora. Nat. Commun. 2020;11:5875. doi: 10.1038/s41467-020-19681-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkooli A.B., van der Krol A.R., Rabe P., Dickschat J.S., Bouwmeester H. Substrate promiscuity of enzymes from the sesquiterpene biosynthetic pathways from Artemisia annua and Tanacetum parthenium allows for novel combinatorial sesquiterpene production. Metab. Eng. 2019;54:12–23. doi: 10.1016/j.ymben.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Katsumoto Y., Fukuchi-Mizutani M., Fukui Y., Brugliera F., Holton T.A., Karan M., Nakamura N., Yonekura-Sakakibara K., Togami J., Pigeaire A. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 2007;48:1589–1600. doi: 10.1093/pcp/pcm131. [DOI] [PubMed] [Google Scholar]
- Kautsar S.A., Duran H.G.S., Blin K., Osbourn A., Medema M.H. plantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 2017;45:W55–W63. doi: 10.1093/nar/gkx305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim O.T., Um Y., Jin M.L., Kim J.U., Hegebarth D., Busta L., Racovita R.C., Jetter R. A novel multifunctional C-23 oxidase, CYP714E19, is involved in asiaticoside biosynthesis. Plant Cell Physiol. 2018;59:1200–1213. doi: 10.1093/pcp/pcy055. [DOI] [PubMed] [Google Scholar]
- Kopertekh L., Schiemann J. Transient production of recombinant pharmaceutical proteins in plants: evolution and perspectives. Curr. Med. Chem. 2019;26:365–380. doi: 10.2174/0929867324666170718114724. [DOI] [PubMed] [Google Scholar]
- Krasteva G., Georgiev V., Pavlov A. Recent applications of plant cell culture technology in cosmetics and foods. Eng. Life Sci. 2021;21:68–76. doi: 10.1002/elsc.202000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Hahn F.M., Baidoo E., Kahlon T.S., Wood D.F., McMahan C.M., Cornish K., Keasling J.D., Daniell H., Whalen M.C. Remodeling the isoprenoid pathway in tobacco by expressing the cytoplasmic mevalonate pathway in chloroplasts. Metab. Eng. 2012;14:19–28. doi: 10.1016/j.ymben.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S.-Y., Lew T.T.S., Sweeney C.J., Koman V.B., Wong M.H., Bohmert-Tatarev K., Snell K.D., Seo J.S., Chua N.-H., Strano M.S. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 2019;14:447–455. doi: 10.1038/s41565-019-0375-4. [DOI] [PubMed] [Google Scholar]
- Lange B.M. In: Biotechnology of Natural Products. Schwab W., Lange B.M., Wüst M., editors. Springer International Publishing; Cham: 2018. Commercial-scale tissue culture for the production of plant natural products: successes, failures and outlook; pp. 189–218. [Google Scholar]
- Lassen L.M., Nielsen A.Z., Ziersen B., Gnanasekaran T., Moller B.L., Jensen P.E. Redirecting Photosynthetic Electron Flow into Light-Driven Synthesis of Alternative Products Including High-Value Bioactive Natural Compounds. ACS Synth. Biol. 2014;3:1–12. doi: 10.1021/sb400136f. [DOI] [PubMed] [Google Scholar]
- Lau W., Sattely E.S. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science. 2015;349:1224–1228. doi: 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Mutanda I., Wang K., Yang L., Wang J., Wang Y. Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana. Nat. Commun. 2019;10:4850. doi: 10.1038/s41467-019-12879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Wu H., Ding Q., Li H., Li Z., Ding J., Li Y. The heterologous expression of Arabidopsis PAP2 induces anthocyanin accumulation and inhibits plant growth in tomato. Funct. Integr. Genomics. 2018;18:341–353. doi: 10.1007/s10142-018-0590-3. [DOI] [PubMed] [Google Scholar]
- Li R., Li R., Li X.D., Fu D.Q., Zhu B.Z., Tian H.Q., Luo Y.B., Zhu H.L. Multiplexed CRISPR/Cas9-mediated metabolic engineering of gamma-aminobutyric acid levels in Solanum lycopersicum. Plant Biotechnol. J. 2018;16:415–427. doi: 10.1111/pbi.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Zhang L., Wang L., Chen L., Zhao R., Sheng J., Shen L. Reduction of tomato-plant chilling tolerance by CRISPR-cas9-mediated SICBF1 mutagenesis. J. Agric. Food Chem. 2018;66:9042–9051. doi: 10.1021/acs.jafc.8b02177. [DOI] [PubMed] [Google Scholar]
- Li X., Wang Y., Chen S., Tian H., Fu D., Zhu B., Luo Y., Zhu H. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front. Plant Sci. 2018;9:559. doi: 10.3389/fpls.2018.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li S., Thodey K., Trenchard I., Cravens A., Smolke C.D. Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. U S A. 2018;115:E3922–E3931. doi: 10.1073/pnas.1721469115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Smolke C.D. Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat. Commun. 2016;7:12137. doi: 10.1038/ncomms12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Stewart C.N., Jr. Plant synthetic biology. Trends Plant Sci. 2015;20:309–317. doi: 10.1016/j.tplants.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Liu X., Cheng J., Zhang G., Ding W., Duan L., Yang J., Kui L., Cheng X., Ruan J., Fan W. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 2018;9:448. doi: 10.1038/s41467-018-02883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ding W., Jiang H. Engineering microbial cell factories for the production of plant natural products: from design principles to industrial-scale production. Microb. Cell. Fact. 2017;16:125. doi: 10.1186/s12934-017-0732-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhu X., Wang H., Liu T., Cheng J., Jiang H. Discovery and modification of cytochrome P450 for plant natural products biosynthesis. Synth. Syst. Biotechnol. 2020;5:187–199. doi: 10.1016/j.synbio.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder L.G., Zhang D.W., Baltes N.J. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 2015;169:971. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Rijzaani H., Karcher D., Ruf S., Bock R. Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc. Natl. Acad. Sci. U S A. 2013;110:E623–E632. doi: 10.1073/pnas.1216898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Stegemann S., Agrawal S., Karcher D., Ruf S., Bock R. Horizontal transfer of a synthetic metabolic pathway between plant species. Curr. Biol. 2017;27:3034–3041.e3. doi: 10.1016/j.cub.2017.08.044. [DOI] [PubMed] [Google Scholar]
- Luo X., Reiter M.A., d'Espaux L., Wong J., Denby C.M., Lechner A., Zhang Y., Grzybowski A.T., Harth S., Lin W. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature. 2019;567:123–126. doi: 10.1038/s41586-019-0978-9. [DOI] [PubMed] [Google Scholar]
- Lv Z., Wang S., Zhang F., Chen L., Hao X., Pan Q., Fu X., Li L., Sun X., Tang K. Overexpression of a novel NAC domain-containing transcription factor gene (AaNAC1) enhances the content of artemisinin and increases tolerance to drought and Botrytis cinerea in Artemisia annua. Plant Cell Physiol. 2016;57:1961–1971. doi: 10.1093/pcp/pcw118. [DOI] [PubMed] [Google Scholar]
- Lv Z., Zhang F., Pan Q., Fu X., Jiang W., Shen Q., Yan T., Shi P., Lu X., Sun X. Branch pathway blocking in Artemisia annua is a useful method for obtaining high yield artemisinin. Plant Cell Physiol. 2016;57:588–602. doi: 10.1093/pcp/pcw014. [DOI] [PubMed] [Google Scholar]
- Ma L., Zhang D., Miao Q., Yang J., Xuan Y., Hu Y. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 2017;58:863–873. doi: 10.1093/pcp/pcx040. [DOI] [PubMed] [Google Scholar]
- Ma Y., Ma X.H., Meng F.Y., Zhan Z.L., Guo J., Huang L.Q. RNA interference targeting CYP76AH1 in hairy roots of Salvia miltiorrhiza reveals its key role in the biosynthetic pathway of tanshinones. Biochem. Biophysical Res. Commun. 2016;477:155–160. doi: 10.1016/j.bbrc.2016.06.036. [DOI] [PubMed] [Google Scholar]
- Maeda H.A. Harnessing evolutionary diversification of primary metabolism for plant synthetic biology. J. Biol. Chem. 2019;294:16549–16566. doi: 10.1074/jbc.REV119.006132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud S.S., Croteau R.B. Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc. Natl. Acad. Sci. U S A. 2001;98:8915–8920. doi: 10.1073/pnas.141237298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud S.S., Williams M., Croteau R. Cosuppression of limonene-3-hydroxylase in peppermint promotes accumulation of limonene in the essential oil. Phytochemistry. 2004;65:547–554. doi: 10.1016/j.phytochem.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Malhotra K., Subramaniyan M., Rawat K., Kalamuddin M., Qureshi M.I., Malhotra P., Mohmmed A., Cornish K., Daniell H., Kumar S. Compartmentalized metabolic engineering for artemisinin biosynthesis and effective malaria treatment by oral delivery of plant cells. Mol. Plant. 2016;9:1464–1477. doi: 10.1016/j.molp.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manela N., Oliva M., Ovadia R., Sikron-Persi N., Ayenew B., Fait A., Galili G., Perl A., Weiss D., Oren-Shamir M. Phenylalanine and tyrosine levels are rate-limiting factors in production of health promoting metabolites in Vitis vinifera cv. Gamay Red cell suspension. Front. Plant Sci. 2015;6:538. doi: 10.3389/fpls.2015.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Marquez A., Morante-Carriel J.A., Palazon J., Bru-Martinez R. Rosa hybrida orcinol O-methyl transferase-mediated production of pterostilbene in metabolically engineered grapevine cell cultures. New Biotechnol. 2018;42:62–70. doi: 10.1016/j.nbt.2018.02.011. [DOI] [PubMed] [Google Scholar]
- Martinez-Marquez A., Morante-Carriel J.A., Ramirez-Estrada K., Cusido R.M., Palazon J., Bru-Martinez R. Production of highly bioactive resveratrol analogues pterostilbene and piceatannol in metabolically engineered grapevine cell cultures. Plant Biotechnol. J. 2016;14:1813–1825. doi: 10.1111/pbi.12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K., Kitani S., Yoshioka T., Morimoto T., Fujita Y. High-density culture of coptis-japonica cells increases berberine production. J. Chem. Technol. Biotechnol. 1989;46:61–69. [Google Scholar]
- McElroy, C., Jennewein, S. (2018). Biotechnology of Natural Products Schwab, W. Lange, B.M. Wüst, M. pp. 145-185.Springer International Publishing Cham.
- Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K., de Bruijn I., Chooi Y.H., Claesen J., Coates R.C. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M.H., Osbourn A. Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways. Nat. Prod. Rep. 2016;33:951–962. doi: 10.1039/c6np00035e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta B.J., Obraztsova I.N., Cerda-Olmedo E. Mutants and intersexual heterokaryons of Blakeslea trispora for production of beta-carotene and lycopene. Appl. Environ. Microbiol. 2003;69:4043–4048. doi: 10.1128/AEM.69.7.4043-4048.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekky H., Al-Sabahi J., Abdel-Kreem M.F.M. Potentiating biosynthesis of the anticancer alkaloids vincristine and vinblastine in callus cultures of Catharanthus roseus. South Afr. J. Bot. 2018;114:29–31. [Google Scholar]
- Mellor S.B., Behrendorff J.B.Y.H., Nielsen A.Z., Jensen P.E., Pribil M. Non-photosynthetic plastids as hosts for metabolic engineering. Essays Biochem. 2018;62:41–50. doi: 10.1042/EBC20170047. [DOI] [PubMed] [Google Scholar]
- Mellor S.B., Nielsen A.Z., Burow M. Fusion of Ferredoxin and Cytochrome P450 Enables Direct Light-Driven Biosynthesis. ACS Chem. Biol. 2016;11:1862–1869. doi: 10.1021/acschembio.6b00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor S.B., Vinde M.H., Nielsen A.Z., Hanke G.T., Abdiaziz K., Roessler M.M., Burow M., Motawia M.S., Moller B.L., Jensen P.E. Defining optimal electron transfer partners for light-driven cytochrome P450 reactions. Metab. Eng. 2019;55:33–43. doi: 10.1016/j.ymben.2019.05.003. [DOI] [PubMed] [Google Scholar]
- Meng X., Yin B., Feng H.L., Zhang S., Liang X.Q., Meng Q.W. Overexpression of R2R3-MYB gene leads to accumulation of anthocyanin and enhanced resistance to chilling and oxidative stress. Biol. Plant. 2014;58:121–130. [Google Scholar]
- Mercx S., Smargiasso N., Chaumont F., De Pauw E., Boutry M., Navarre C. Inactivation of the beta(1,2)-xylosyltransferase and the alpha(1,3)-fucosyltransferase genes in Nicotiana tabacum BY-2 cells by a multiplex CRISPR/Cas9 strategy results in glycoproteins without plant-specific glycans. Front. Plant Sci. 2017;8:403. doi: 10.3389/fpls.2017.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen K., Dong L.M., Navrot N., Schneider T., Burlat V., Pollier J., Woittiez L., van der Krol S., Lugan R., Ilc T. The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 2014;5:11. doi: 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecillo J.A.V., Chu L.L., Bae H. CRISPR-Cas9 system for plant genome editing: current approaches and emerging developments. Agron. Basel. 2020;10:20–31. [Google Scholar]
- Morrison K.C., Hergenrother P.J. Natural products as starting points for the synthesis of complex and diverse compounds. Nat. Prod. Rep. 2014;31:6–14. doi: 10.1039/c3np70063a. [DOI] [PubMed] [Google Scholar]
- Mortimer J.C. Plant synthetic biology could drive a revolution in biofuels and medicine. Exp. Biol. Med. 2019;244:323–331. doi: 10.1177/1535370218793890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagegowda D.A., Gupta P. Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 2020;294:110457. doi: 10.1016/j.plantsci.2020.110457. [DOI] [PubMed] [Google Scholar]
- Naqvi S., Farre G., Sanahuja G., Capell T., Zhu C., Christou P. When more is better: multigene engineering in plants. Trends Plant Sci. 2010;15:48–56. doi: 10.1016/j.tplants.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Naqvi S., Zhu C., Farre G., Ramessar K., Bassie L., Breitenbach J., Perez Conesa D., Ros G., Sandmann G., Capell T. Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc. Natl. Acad. Sci. U S A. 2009;106:7762–7767. doi: 10.1073/pnas.0901412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett R.S., Lau W., Sattely E.S. Discovery and engineering of colchicine alkaloid biosynthesis. Nature. 2020;584:148–153. doi: 10.1038/s41586-020-2546-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.Z., Ziersen B., Jensen K. Redirecting Photosynthetic Reducing Power toward Bioactive Natural Product Synthesis. ACS Synth. Biol. 2013;2:308–315. doi: 10.1021/sb300128r. [DOI] [PubMed] [Google Scholar]
- Noda N., Yoshioka S., Kishimoto S., Nakayama M., Douzono M., Tanaka Y., Aida R. Generation of blue chrysanthemums by anthocyanin B-ring hydroxylation and glucosylation and its coloration mechanism. Sci. Adv. 2017;3:e1602785. doi: 10.1126/sciadv.1602785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S., Arai C., Takayama M., Matsukura C., Ezura H. Efficient increase of gamma-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 2017;7:7057. doi: 10.1038/s41598-017-06400-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutzmann H.W., Osbourn A. Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 2014;26:91–99. doi: 10.1016/j.copbio.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Outchkourov N.S., Karlova R., Holscher M., Schrama X., Blilou I., Jongedijk E., Simon C.D., van Dijk A.D.J., Bosch D., Hall R.D. Transcription factor-mediated control of anthocyanin biosynthesis in vegetative tissues. Plant Physiol. 2018;176:1862–1878. doi: 10.1104/pp.17.01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C.J., Westfall P.J., Pitera D.J., Benjamin K., Fisher K., McPhee D., Leavell M.D., Tai A., Main A., Eng D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Pan X.-W., Han L., Zhang Y.-H., Chen D.-F., Simonsen H.T. Sclareol production in the moss Physcomitrella patens and observations on growth and terpenoid biosynthesis. Plant Biotechnol. Rep. 2015;9:149–159. [Google Scholar]
- Pasoreck E.K., Su J., Silverman I.M., Gosai S.J., Gregory B.D., Yuan J.S., Daniell H. Terpene metabolic engineering via nuclear or chloroplast genomes profoundly and globally impacts off-target pathways through metabolite signalling. Plant Biotechnol. J. 2016;14:1862–1875. doi: 10.1111/pbi.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polturak G., Breitel D., Grossman N., Sarrion-Perdigones A., Weithorn E., Pliner M., Orzaez D., Granell A., Rogachev I., Aharoni A. Elucidation of the first committed step in betalain biosynthesis enables the heterologous engineering of betalain pigments in plants. New Phytol. 2016;210:269–283. doi: 10.1111/nph.13796. [DOI] [PubMed] [Google Scholar]
- Pyne M.E., Narcross L., Martin V.J.J. Engineering plant secondary metabolism in microbial systems. Plant Physiol. 2019;179:844–861. doi: 10.1104/pp.18.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qua Y., Easson M.L.A.E., Froese J., Simionescu R., Hudlicky T., De Luca V. Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc. Natl. Acad. Sci. U S A. 2015;112:6224–6229. doi: 10.1073/pnas.1501821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A., Hirakawa H., Nakabayashi R., Kikuchi S., Hayashi K., Rai M., Tsugawa H., Nakaya T., Mori T., Nagasaki H. Chromosome-level genome assembly of Ophiorrhiza pumila reveals the evolution of camptothecin biosynthesis. Nat. Commun. 2021;12:405. doi: 10.1038/s41467-020-20508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J., Osbourn A. Engineering terpenoid production through transient expression in Nicotiana benthamiana. Plant Cell Rep. 2018;37:1431–1441. doi: 10.1007/s00299-018-2296-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J., Stephenson M.J., Miettinen K., Brouwer B., Leveau A., Brett P., Goss R.J.M., Goossens A., O'Connell M.A., Osbourn A. A translational synthetic biology platform for rapid access to gram-scale quantities of novel drug-like molecules. Metab. Eng. 2017;42:185–193. doi: 10.1016/j.ymben.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reski R., Bae H., Simonsen H.T. Physcomitrella patens, a versatile synthetic biology chassis. Plant Cell Rep. 2018;37:1409–1417. doi: 10.1007/s00299-018-2293-6. [DOI] [PubMed] [Google Scholar]
- Ritala A., Dong L.M., Imseng N., Seppanen-Laakso T., Vasilev N., van der Krol S., Rischer H., Maaheimo H., Virkki A., Brandli J. Evaluation of tobacco (Nicotiana tabacum L. cv. Petit Havana SR1) hairy roots for the production of geraniol, the first committed step in terpenoid indole alkaloid pathway. J. Biotechnol. 2014;176:20–28. doi: 10.1016/j.jbiotec.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Ruf S., Forner J., Hasse C., Kroop X., Seeger S., Schollbach L., Schadach A., Bock R. High-efficiency generation of fertile transplastomic Arabidopsis plants. Nat. Plants. 2019;5:282–289. doi: 10.1038/s41477-019-0359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S., Hermann M., Berger I.J., Carrer H., Bock R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat. Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- Sabzehzari M., Zeinali M., Naghavi M.R. CRISPR-based metabolic editing: next-generation metabolic engineering in plants. Gene. 2020;759:144993. doi: 10.1016/j.gene.2020.144993. [DOI] [PubMed] [Google Scholar]
- Saxena B., Subramaniyan M., Malhotra K., Bhavesh N.S., Potlakayala S.D., Kumar S. Metabolic engineering of chloroplasts for artemisinic acid biosynthesis and impact on plant growth. J. Biosci. 2014;39:33–41. doi: 10.1007/s12038-013-9402-z. [DOI] [PubMed] [Google Scholar]
- Schaub P., Rodriguez-Franco M., Cazzonelli C.I., Alvarez D., Wuest F., Welsch R. Establishment of an Arabidopsis callus system to study the interrelations of biosynthesis, degradation and accumulation of carotenoids. PLoS One. 2018;13:28. doi: 10.1371/journal.pone.0192158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlapfer P., Zhang P., Wang C., Kim T., Banf M., Chae L., Dreher K., Chavali A.K., Nilo-Poyanco R., Bernard T. Genome-wide prediction of metabolic enzymes, pathways, and gene clusters in plants. Plant Physiol. 2017;173:2041–2059. doi: 10.1104/pp.16.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt I., Schewe H., Gassel S., Jin C., Buckingham J., Huembelin M., Sandmann G., Schrader J. Biotechnological production of astaxanthin with Phaffia rhodozyma/Xanthophyllomyces dendrorhous. Appl. Microbiol. Biotechnol. 2011;89:555–571. doi: 10.1007/s00253-010-2976-6. [DOI] [PubMed] [Google Scholar]
- Schultz B.J., Kim S.Y., Lau W., Sattely E.S. Total biosynthesis for milligram-scale production of etoposide intermediates in a plant chassis. J. Am. Chem. Soc. 2019;141:19231–19235. doi: 10.1021/jacs.9b10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Ma Y., Zhou Y., Zhang H., Duan L., Chen H., Zeng J., Zhou Q., Wang S., Gu W. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science. 2014;346:1084–1088. doi: 10.1126/science.1259215. [DOI] [PubMed] [Google Scholar]
- Shen C., Que Z., Xia Y., Tang N., Li D., He R., Cao M. Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J. Plant Biol. 2017;60:539–547. [Google Scholar]
- Shen Q., Lu X., Yan T., Fu X., Lv Z., Zhang F., Pan Q., Wang G., Sun X., Tang K. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 2016;210:1269–1281. doi: 10.1111/nph.13874. [DOI] [PubMed] [Google Scholar]
- Shen Q., Zhang L.D., Liao Z.H., Wang S.Y., Yan T.X., Shi P., Liu M., Fu X.Q., Pan Q.F., Wang Y.L. The genome of Artemisia annua provides insight into the evolution of Asteraceae family and artemisinin biosynthesis. Mol. Plant. 2018;11:776–788. doi: 10.1016/j.molp.2018.03.015. [DOI] [PubMed] [Google Scholar]
- Sidorov V.A., Kasten D., Pang S.Z., Hajdukiewicz P.T.J., Staub J.M., Nehra N.S. Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- Srinivasan P., Smolke C.D. Biosynthesis of medicinal tropane alkaloids in yeast. Nature. 2020;585:614–619. doi: 10.1038/s41586-020-2650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stander E.A., Sepulveda L.J., Duge de Bernonville T., Carqueijeiro I., Koudounas K., Lemos Cruz P., Besseau S., Lanoue A., Papon N., Giglioli-Guivarc'h N. Identifying genes involved in alkaloid biosynthesis in Vinca minor through transcriptomics and gene co-expression analysis. Biomolecules. 2020;10:1595. doi: 10.3390/biom10121595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann S., Keuthe M., Greiner S., Bock R. Horizontal transfer of chloroplast genomes between plant species. Proc. Natl. Acad. Sci. U S A. 2012;109:2434–2438. doi: 10.1073/pnas.1114076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J., Nielsen J., Fernie A.R. Engineering central metabolism—a grand challenge for plant biologists. Plant J. 2017;90:749–763. doi: 10.1111/tpj.13464. [DOI] [PubMed] [Google Scholar]
- Tan H., Xiao L., Gao S., Li Q., Chen J., Xiao Y., Ji Q., Chen R., Chen W., Zhang L. Trichome and artemisinin regulator 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol. Plant. 2015;8:1396–1411. doi: 10.1016/j.molp.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Thyssen G., Svab Z., Maliga P. Cell-to-cell movement of plastids in plants. Proc. Natl. Acad. Sci. U S A. 2012;109:2439–2443. doi: 10.1073/pnas.1114297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., Zhang Y., Peterek S., Matros A., Rallapalli G., Tandron Y.A., Butelli E., Kallam K., Hertkorn N., Mock H.-P. Ectopic expression of snapdragon transcription factors facilitates the identification of genes encoding enzymes of anthocyanin decoration in tomato. Plant J. 2015;83:686–704. doi: 10.1111/tpj.12920. [DOI] [PubMed] [Google Scholar]
- Topfer N., Fuchs L.-M., Aharoni A. The PhytoClust tool for metabolic gene clusters discovery in plant genomes. Nucleic Acids Res. 2017;45:7049–7063. doi: 10.1093/nar/gkx404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenchard I.J., Smolke C.D. Engineering strategies for the fermentative production of plant alkaloids in yeast. Metab. Eng. 2015;30:96–104. doi: 10.1016/j.ymben.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L., Su P., Zhang Z., Gao L., Wang J., Hu T., Zhou J., Zhang Y., Zhao Y., Liu Y. Genome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis. Nat. Commun. 2020;11:971. doi: 10.1038/s41467-020-14776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurin A.A., Suhorukova A.V., Kabardaeva K.V., Goldenkova-Pavlova I.V. Transient gene expression is an effective experimental tool for the research into the fine mechanisms of plant gene function: advantages, limitations, and solutions. Plants Basel. 2020;9:19. doi: 10.3390/plants9091187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkov V.T., Gargano D., Manna C., Formisano G., Dix P.J., Gray J.C., Scotti N., Cardi T. High efficiency plastid transformation in potato and regulation of transgene expression in leaves and tubers by alternative 5' and 3' regulatory sequences. Transgenic Res. 2011;20:137–151. doi: 10.1007/s11248-010-9402-9. [DOI] [PubMed] [Google Scholar]
- Vasilev N., Schmitz C., Groemping U., Fischer R., Schillberg S. Assessment of cultivation factors that affect biomass and geraniol production in transgenic tobacco cell suspension cultures. PLoS One. 2014;9:e104620. doi: 10.1371/journal.pone.0104620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verpoorte R., van der Heijden R., Memelink J. Engineering the plant cell factory for secondary metabolite production. Transgenic Res. 2000;9:323–343. doi: 10.1023/a:1008966404981. [DOI] [PubMed] [Google Scholar]
- Wang D., Wang J., Shi Y., Li R., Fan F., Huang Y., Li W., Chen N., Huang L., Dai Z. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab. Eng. 2020;61:131–140. doi: 10.1016/j.ymben.2020.05.007. [DOI] [PubMed] [Google Scholar]
- Wang L., Chen L., Li R., Zhao R., Yang M., Sheng J., Shen L. Reduced drought tolerance by CRISPR/Cas9-Mediated SlMAPK3 mutagenesis in tomato plants. J. Agr. Food Chem. 2017;65:8674–8682. doi: 10.1021/acs.jafc.7b02745. [DOI] [PubMed] [Google Scholar]
- Wang R., da Rocha Tavano E.C., Lammers M., Martinelli A.P., Angenent G.C., de Maagd R.A. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 2019;9:1696. doi: 10.1038/s41598-018-38170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson S.A., Roberts S.C. Recent advances towards development and commercialization of plant cell culture processes for the synthesis of biomolecules. Plant Biotechnol. J. 2012;10:249–268. doi: 10.1111/j.1467-7652.2011.00664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T., Gazda V., He Z., Kaminski F., Kern M., Larson T.R., Li Y., Meade F., Teodor R., Vaistij F.E. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- Wu S.Q., Schalk M., Clark A., Miles R.B., Coates R., Chappell J. Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat. Biotechnol. 2006;24:1441–1447. doi: 10.1038/nbt1251. [DOI] [PubMed] [Google Scholar]
- Wurbs D., Ruf S., Bock R. Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J. 2007;49:276–288. doi: 10.1111/j.1365-313X.2006.02960.x. [DOI] [PubMed] [Google Scholar]
- Xie K., Zhang X., Sui S., Ye F., Dai J. Exploring and applying the substrate promiscuity of a C-glycosyltransferase in the chemo-enzymatic synthesis of bioactive C-glycosides. Nat. Commun. 2020;11:5162. doi: 10.1038/s41467-020-18990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.-S., Feng K., Que F., Wang F., Xiong A.-S. A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci. Rep. 2017;7:45324. doi: 10.1038/srep45324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y., Tanaka H., Yoshimura S., Suzuki A., Tamoi M., Maruta T., Shigeoka S. Improvement of vitamin E quality and quantity in tobacco and lettuce by chloroplast genetic engineering. Transgenic Res. 2013;22:391–402. doi: 10.1007/s11248-012-9656-5. [DOI] [PubMed] [Google Scholar]
- Yan T., Chen M., Shen Q., Li L., Fu X., Pan Q., Tang Y., Shi P., Lv Z., Jiang W. Homeodomain Protein 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 2017;213:1145–1155. doi: 10.1111/nph.14205. [DOI] [PubMed] [Google Scholar]
- Yang D., Fang Y., Xia P., Zhang X., Liang Z. Diverse responses of tanshinone biosynthesis to biotic and abiotic elicitors in hairy root cultures of Salvia miltiorrhiza and Salvia castanea Diels f. tomentosa. Gene. 2018;643:61–67. doi: 10.1016/j.gene.2017.11.067. [DOI] [PubMed] [Google Scholar]
- Yang D., Park S.Y., Park Y.S., Eun H., Lee S.Y. Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 2020;38:745–765. doi: 10.1016/j.tibtech.2019.11.007. [DOI] [PubMed] [Google Scholar]
- Yang Y., Li W., Pang J., Jiang L., Qu X., Pu X., Zhang G., Luo Y. Bifunctional cytochrome P450 enzymes involved in camptothecin biosynthesis. ACS Chem. Biol. 2019;14:1091–1096. doi: 10.1021/acschembio.8b01124. [DOI] [PubMed] [Google Scholar]
- Yasui Y., Tanaka W., Sakamoto T., Kurata T., Hirano H.-Y. Genetic enhancer analysis reveals that FLORAL ORGAN NUMBER2 and OsMADS3 Co-operatively regulate maintenance and determinacy of the flower meristem in rice. Plant Cell Physiol. 2017;58:893–903. doi: 10.1093/pcp/pcx038. [DOI] [PubMed] [Google Scholar]
- Ye X.D., Al-Babili S., Kloti A., Zhang J., Lucca P., Beyer P., Potrykus I. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- Yousefian Z., Golkar P., Mirjalili M.H. Production enhancement of medicinally active coumarin and phenolic compounds in hairy root cultures of Pelargonium sidoides: the effect of elicitation and sucrose. J. Plant Growth Regul. 2021;40:628–641. [Google Scholar]
- Yu Q., Barkan A., Maliga P. Engineered RNA-binding protein for transgene activation in non-green plastids. Nat. Plants. 2019;5:486–490. doi: 10.1038/s41477-019-0413-0. [DOI] [PubMed] [Google Scholar]
- Yu Y., Yu P.-C., Chang W.-J., Yu K., Lin C.-S. Plastid transformation: How does it work? Can it Be applied to crops? What can it offer? Int. J. Mol. Sci. 2020;21:4854. doi: 10.3390/ijms21144854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.J., Zhang Q.Z., Jiang C.X., Zheng Y.Y., Zuo Y.W., Qin J.B., Liao Z.H., Deng H.P. Development of Atropa belladonna L. Plants with high-yield hyoscyamine and without its derivatives using the CRISPR/Cas9 system. Int. J. Mol. Sci. 2021;22:10. doi: 10.3390/ijms22041731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zg A., Kb A., Hd B., Asc D. Enhanced flavonoid production in hairy root cultures of Scutellaria bornmuelleri by elicitor induced over-expression of MYB7 and FNSП2 genes - ScienceDirect. Plant Physiol. Biochem. 2020;148:35–44. doi: 10.1016/j.plaphy.2020.01.002. [DOI] [PubMed] [Google Scholar]
- Zhan X., Zhang Y.-H., Chen D.-F., Simonsen H.T. Metabolic engineering of the moss Physcomitrella patens to produce the sesquiterpenoids patchoulol and alpha/beta-santalene. Front. Plant Sci. 2014;5:636. doi: 10.3389/fpls.2014.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Schrader A. TRANSPARENT TESTA GLABRA 1-dependent regulation of flavonoid biosynthesis. Plants (Basel) 2017;6:65. doi: 10.3390/plants6040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Butelli E., Alseekh S., Tohge T., Rallapalli G., Luo J., Kawar P.G., Hill L., Santino A., Fernie A.R. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015;6:8635. doi: 10.1038/ncomms9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ruf S., Hasse C., Childs L., Scharff L.B., Bock R. Identification of cis-elements conferring high levels of gene expression in non-green plastids. Plant J. 2012;72:115–128. doi: 10.1111/j.1365-313X.2012.05065.x. [DOI] [PubMed] [Google Scholar]
- Zhang P., Du H., Wang J., Pu Y., Yang C., Yan R., Yang H., Cheng H., Yu D. Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 2020;18:1384–1395. doi: 10.1111/pbi.13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen M., Siemiatkowska B., Toleco M.R., Jing Y., Strotmann V., Zhang J., Stahl Y., Fernie A.R. A highly efficient agrobacterium-mediated method for transient gene expression and functional studies in multiple plant species. Plant Commun. 2020;1:100028. doi: 10.1016/j.xplc.2020.100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Xu T., Liang Y., Zhao S., Ren L., Wang Q., Dou B. Functional analysis of beta-amyrin synthase gene in ginsenoside biosynthesis by RNA interference. Plant Cell Rep. 2015;34:1307–1315. doi: 10.1007/s00299-015-1788-7. [DOI] [PubMed] [Google Scholar]
- Zhao P.F., You Q.Y., Lei M.G. A CRISPR/Cas9 deletion into the phosphate transporter SlPHO1;1 reveals its role in phosphate nutrition of tomato seedlings. Physiol. Plant. 2019;167:556–563. doi: 10.1111/ppl.12897. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Zhang Y., Wang G., Hill L., Weng J.K., Chen X.Y., Xue H.W., Martin C. A specialized flavone biosynthetic pathway has evolved in the medicinal plant, Scutellaria baicalensis. Sci. Adv. 2016;2:15. doi: 10.1126/sciadv.1501780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Li S., Wang J., Zhou Q., Yang C., Bai F., Lan X., Chen M., Liao Z. Engineering tropane alkaloid production based on metabolic characterization of ornithine decarboxylase in Atropa belladonna. ACS Synth. Biol. 2020;9:437–448. doi: 10.1021/acssynbio.9b00461. [DOI] [PubMed] [Google Scholar]
- Zhu C., Naqvi S., Breitenbach J., Sandmann G., Christou P., Capell T. Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc. Natl. Acad. Sci. U S A. 2008;105:18232–18237. doi: 10.1073/pnas.0809737105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Yu S., Zeng D., Liu H., Wang H., Yang Z., Xie X., Shen R., Tan J., Li H. Development of "Purple Endosperm Rice'' by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol. Plant. 2017;10:918–929. doi: 10.1016/j.molp.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Zeng D., Yu S., Cui C., Li J., Li H., Chen J., Zhang R., Zhao X., Chen L. From golden rice to aSTARice: bioengineering astaxanthin biosynthesis in rice endosperm. Mol. Plant. 2018;11:1440–1448. doi: 10.1016/j.molp.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Zong Y., Zhu X., Liu Z., Xi X., Li G., Cao D., Wei L., Li J., Liu B. Functional MYB transcription factor encoding gene AN2 is associated with anthocyanin biosynthesis in Lycium ruthenicum Murray. BMC Plant Biol. 2019;19:169. doi: 10.1186/s12870-019-1752-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y., Feng F., Qi W., Song R. Dek42 encodes an RNA-binding protein that affects alternative pre-mRNA splicing and maize kernel development. J. Integr. Plant Biol. 2019;61:728–748. doi: 10.1111/jipb.12798. [DOI] [PubMed] [Google Scholar]