Abstract
Sphingolipids, which comprise membrane systems together with other lipids, are ubiquitous in cellular organisms. They show a high degree of diversity across plant species and vary in their structures, properties, and functions. Benefiting from the development of lipidomic techniques, over 300 plant sphingolipids have been identified. Generally divided into free long-chain bases (LCBs), ceramides, glycosylceramides (GlcCers) and glycosyl inositol phosphoceramides (GIPCs), plant sphingolipids exhibit organized aggregation within lipid membranes to form raft domains with sterols. Accumulating evidence has revealed that sphingolipids obey certain trafficking and distribution rules and confer unique properties to membranes. Functional studies using sphingolipid biosynthetic mutants demonstrate that sphingolipids participate in plant developmental regulation, stimulus sensing, and stress responses. Here, we present an updated metabolism/degradation map and summarize the structures of plant sphingolipids, review recent progress in understanding the functions of sphingolipids in plant development and stress responses, and review sphingolipid distribution and trafficking in plant cells. We also highlight some important challenges and issues that we may face during the process of studying sphingolipids.
Keywords: sphingolipid metabolism, membrane lateral heterogeneity, vesicular and non-vesicular trafficking
This review summarizes recent progress in plant sphingolipid metabolism, diversity, distribution, trafficking, and function. The intersections among these topics are also discussed, then current challenges and issues in the study of plant sphingolipids are highlighted.
Introduction
Since their discovery in the late 19th century, sphingolipids have been recognized as one of the major membrane lipid components in eukaryotic cells and a few bacteria (Thudichum 1884; Karlsson, 1970; Sperling and Heinz, 2003). Containing a fatty acid linked to a long-chain amino alcohol (also known as the sphingoid long-chain base), the ceramide backbone composes the sphingolipid core (Hannich et al., 2011). In animals, enzymes and regulators involved in sphingolipid metabolism have been extensively investigated, and the physicochemical properties of certain sphingolipid structures and trafficking behaviors are also well elucidated (Mauri et al., 2018). Functional studies have revealed that sphingolipids participate in all major cell signaling pathways and that disruptions of their metabolism cause numerous human diseases that have attracted much attention (Hannun and Obeid, 2018). Although still in the primary stage, research on plant sphingolipids has accelerated in the last two decades: there are about 1,000 searchable papers (including no more than 80 reviews) showing a close relationship with plant sphingolipid metabolism, characteristics, or functions.
In eukaryotes, enzymes involved in sphingolipid metabolism are mainly localized in the endoplasmic reticulum (ER) and Golgi apparatus, and the distribution of their products varies in a species-specific and tissue-dependent manner. In this review, we mainly summarize progress in plant sphingolipid metabolism, diversity, distribution, trafficking, and function and discuss their intersections.
Sphingolipid metabolism
Sphingolipid biosynthesis
De novo synthesis of sphingolipids begins with the condensation of serine and palmitoyl-CoA, a reaction catalyzed by serine palmitoyl transferases (SPTs) in the ER (Mandon et al., 1992; Chen et al., 2006). It is notable that precise regulation of SPT activity by poly-peptides named small subunit of SPT (ssSPT) or orosomucoid-like proteins (ORMs, inhibitors of SPT) is necessary for sphingolipid homeostasis (Kimberlin et al., 2013, 2016; Markham et al., 2013). The intermediate 3-ketosphinganine can be reduced by 3-ketodihydrosphingosine reductase (KSR) to form sphinganine (d18:0) (Beeler et al., 1998; Chao et al., 2011), which is also one of the major long-chain bases (LCBs) in animals. In plants such as Arabidopsis, sphingoid base hydroxylases (SBHs) add a third hydroxyl group to the C-4 position of the two-hydroxylated LCB sphinganine to generate phytosphingosine (t18:0) (Figures 1 and 2) (Chen et al., 2008; Sperling et al., 2001). Both d18:0 and t18:0 are then linked to a fatty acid chain (usually 16 to 26 carbons in length) via an amide bond by the ceramide synthase lag one homologues (LOHs) to form ceramides (Ternes et al., 2011; Markham et al., 2011). Among the three identified LOHs, LOH1 and LOH3 show a preference for catalyzing the assembly of very-long-chain fatty acids (VLCFAs) and LCBs, whereas LOH2 is mainly responsible for the synthesis of ceramides with long-chain fatty acids (LCFAs) (Ternes et al., 2011; Markham et al., 2011). The biosynthesis of ceramides in plants, especially t18:0/t18:1 and C24:0/C24:1-ceramides, can also be affected by a putative enzyme named phloem unloading modulator (PLM) (Yan et al., 2019) (Figure 2). The saturated LCBs are then desaturated via sphingoid LCB desaturases (SLDs) at C-8 or via sphingolipid δ-4-desaturase at C-4 to generate d18:1-based, t18:1-based, or d18:2-based ceramides (Michaelson et al., 2009; Chen et al., 2012). Notably, the C-8 to C-9 double bond in LCBs should be either cis or trans in theory, although it is mainly in the trans configuration in wild-type (WT) Arabidopsis tissues (Chen et al., 2012). The ceramides always undergo α-hydroxylation in the fatty acyl chain by fatty acid C-2 hydroxylase (FAH) before modification of their head group (Dunn et al., 2004; Markham and Jaworski 2007; König et al., 2012; Nagano et al., 2012) (Figure 2).
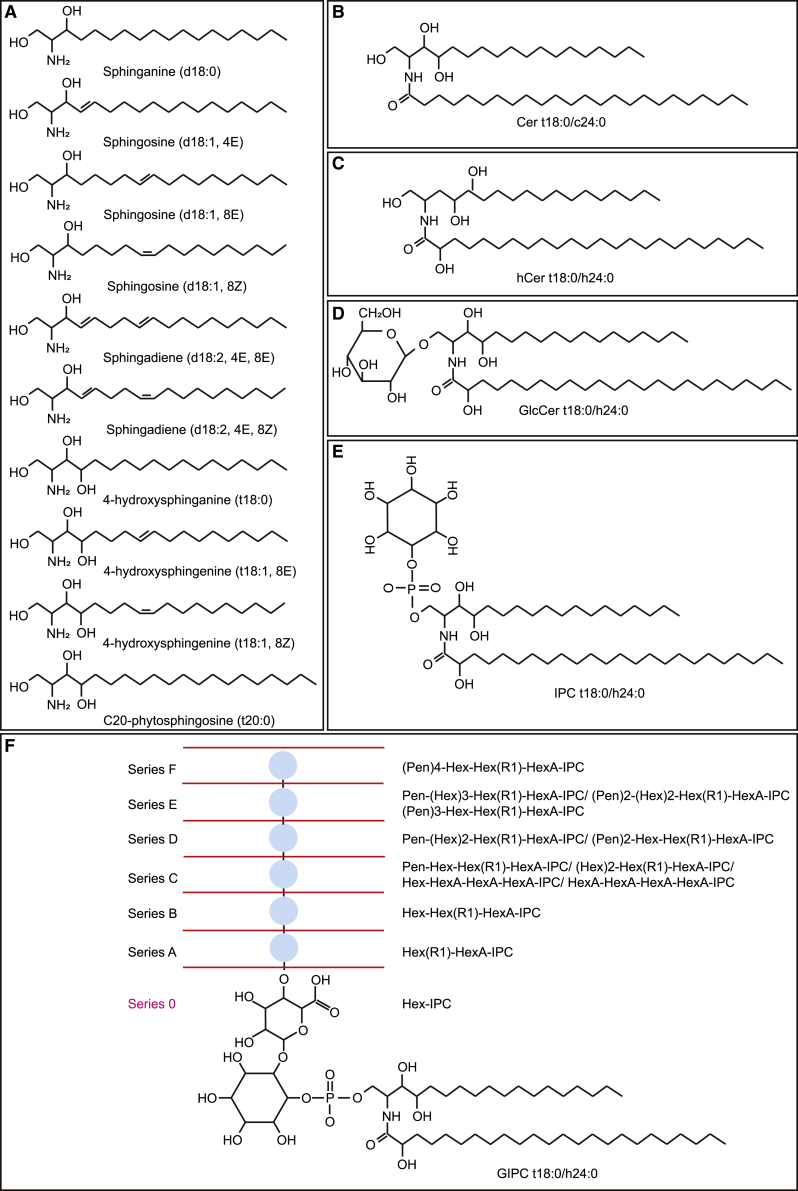
Figure 1.
Plant sphingolipids and diagrams of their structures
(A–F) (modified from Dunn et al., 2004 and Cacas et al., 2013). (A) Sphingoid LCBs. (B) Ceramide backbones. (C) Hydroxyceramides. (D) Glucosylceramides. (E) Inositolphosphoceramides. (F) GIPCs in plants. They can be divided into seven series based on their head groups. Purple cycle with X on it indicates GIPCs with only Hex head groups connected with inositol. R1 refers to OH, NH2, or N(Ac) (Cacas et al., 2013). Cer, ceramides; Hex, hexose; Pen, pentose.
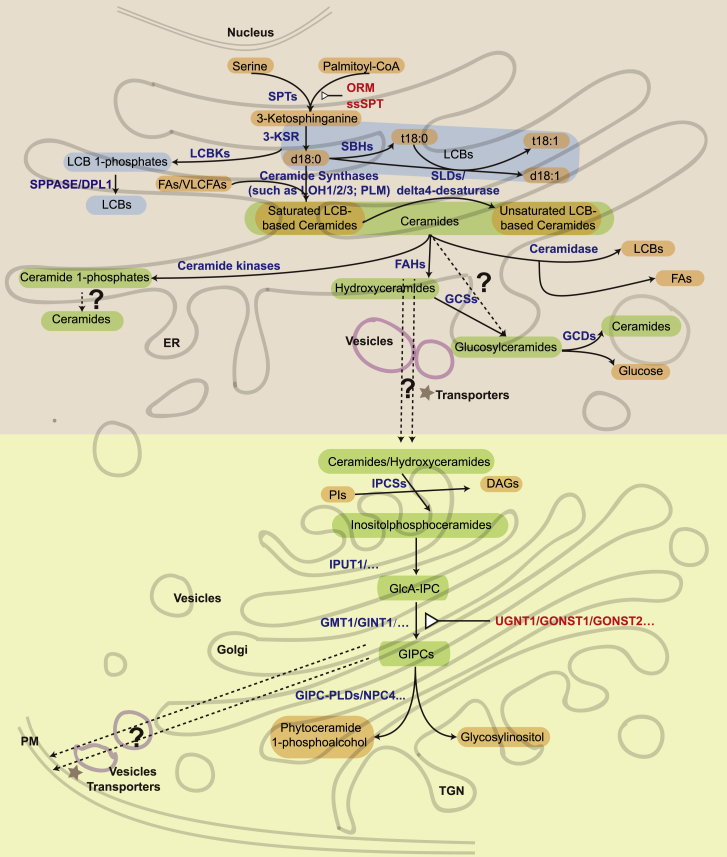
Figure 2.
Diagram of sphingolipid metabolism in plants
Enzymes involved in sphingolipid metabolism pathways are in blue characters; regulators are in red characters and are also indicated by the triangular symbol. Dotted line with a question mark means that the details are unknown.
ER, endoplasmic reticulum; TGN, trans-Golgi network; ORMs, orosomucoid-like proteins; 3-KSR, 3-ketodihydrosphingosine reductase; SBHs, sphingoid base hydroxylases; LOH, lag one homologue; PLM, phloem unloading modulator; FAHs, fatty acid C-2 hydroxylases; GCS, glucosylceramide synthase; PI, phosphatidylinositol; DAGs, diacylglycerols; DPL1, phyto-S1P lyase one; SPPASE, phyto-S1P phosphatase; IPCSs, inositolphosphorylceramide synthases; IPUT1, inositol phosphorylceramide glucuronosyltransferase1; GIPCs, glycosyl inositol phosphoceramides; GMT1, GIPC mannosyl-transferase1; GINT1, glucosamine inositol phosphorylceramide transferase one; GONST1/GONST2, Golgi-localized nucleotide sugar transporter 1/2; UGNT1, UDP-N-acetyl-D-glucosamine transporter one; LCBKs, long-chain base kinases; GCDs, glucosylceramidases; GIPC-PLD, GIPC-specific phospholipase D; NPC4, nonspecific phospholipase C4.
Glycosylation of ceramides at the C-1 hydroxyl group produces GlcCers or GIPCs (Luttgeharm et al., 2016). Glucosylceramide synthase (GCS) transfers a glucose residue from UDP-glucose to a hydroxyceramide (hCer) to produce GlcCer via beta-glycosidic linkage (Leipelt et al., 2001; Melser et al., 2010). To synthesize GIPCs, ceramides or hCers must be transported from the ER to the Golgi. In mammalian cells, ceramide transport is mediated by either vesicular trafficking or protein transporters. The identified ceramide transfer protein (CERT) contains a hydrophobic pocket composed of the steroidogenic acute regulatory protein-related lipid transfer (START) domain, which can directly bind ceramides (Hanada et al., 2007). In plants, however, it remains unclear how the ceramides are delivered from the ER to the Golgi. Once transported to the Golgi, the ceramides and hCers are first modified by inositolphosphorylceramide synthases (IPCSs) to add a head group derived from phosphatidylinositol (PI) to form inositolphosphoceramides (IPCs) (Mina et al., 2010). Then, a glucuronic acid (GlcA) moiety is added via α(1,4)-linkage by inositol phosphorylceramide glucuronosyltransferase1 (IPUT1) (Rennie et al., 2014). Further glycosylation of the head group is mediated by GIPC mannosyl transferase1 (GMT1) or glucosamine inositol phosphorylceramide transferase 1 (GINT1) and requires the Golgi-localized nucleotide sugar transporters GONST1 and GONST2 or UDP-N-acetyl-D-glucosamine transporter 1 (UGNT1) (Mortimer et al., 2013; Fang et al., 2016; Ebert et al., 2018; Ishikawa et al., 2018; Jing et al., 2021). GMT1 has been shown to produce mannose- or hexose-mannose-linked GIPCs, whereas GINT1 mainly transfers glucosamine (GlcN) or N-acetyl-glucosamine (GlcNAc) to the GlcA-IPCs (Ishikawa et al., 2018). The Arabidopsis gonst1 mutant shows reduced content of mannose-GIPCs and accumulates more GlcA-IPCs (Mortimer et al., 2013), whereas loss of the UDP-GlcNAc transporter results in a significant GlcN(Ac)-GIPC deficiency (Ebert et al., 2018).
Sphingolipid base phosphorylation
Phosphorylation is an important form of sphingolipid modification and is closely related to sphingolipid function. Phosphorylation can occur at the C-1 OH position of LCBs by long-chain base kinases (LCBKs) to form LCB 1-phosphates (LCB-Ps) (Nishiura et al., 2000; Imai and Nishiura 2005) or at the C-1 OH position of ceramides by ceramide kinases to form ceramide 1-phosphates (C1Ps) (Liang et al., 2003). Dephosphorylation and phosphorylation together create a dynamic balance of LCBs/LCB-Ps and ceramides/C1Ps in cells and affect plant resistance to environmental stresses. The plant LCB-1-P phosphatases, including phyto-S1P phosphatase (SPPASE) and phyto-S1P lyase (DPL1), have been identified, but the enzymes responsible for ceramide-1-phosphate dephosphorylation have not (Liang et al., 2003; Guo and Wang, 2012; Nakagawa et al., 2012; Luttgeharm et al., 2016; Qin et al., 2017).
Sphingolipid degradation
Degradation of sphingolipids has been well investigated in mammalian cells. The pH optima of ceramidases (CDases) can be acidic, neutral, or alkaline (Gault et al., 2010). In Arabidopsis, an alkaline ceramidase (AtACER) has been shown to hydrolyze t18:0-ceramides, and several neutral ceramidases (AtNCERs) can degrade hydroxyceramides into sphingosine and fatty acids (Li et al., 2015; Wu et al., 2015). In addition, an OsCDase in rice with an optimum pH of 5.7–6.0 has been demonstrated to preferentially hydrolyze d18:1Δ4-ceramide over t18:0-ceramide, consistent with the properties of neutral CDases (Mao and Obeid, 2008; Pata et al., 2008). Interestingly, enzyme activity analysis in yeast systems indicates that OsCDase may exhibit reverse ceramidase activity: once induced, the enzyme can increase the content of C26- and C28-phytoceramides (Pata et al., 2008).
In human, glucocerebrosidases that release Cer and glucose from a glucocerebroside are reportedly related to severe diseases such as Gaucher disease (a lysosomal storage disorder caused by mutations of glucocerebrosidase) (van Weely et al., 1993). Their Arabidopsis homolog was recently reported to be glucosylceramidase (GCD) AtGCD3, which preferentially hydrolyzes glucosylceramides that contain long acyl chains. The knockout mutant of GCD3 shows no obvious phenotype compared with the WT (Dai et al., 2020). The author also predicted the existence of another three AtGCDs based on sequence homology analysis (Dai et al., 2020).
In cabbage leaves, a GIPC-specific phospholipase D (GIPC-PLD) was found to cleave at the D position of the ester linkage between inositol and phosphate to produce phytoceramide 1-phosphate (PC1P) (Hasi et al., 2019). The enzyme exhibits a substrate preference for GIPCs that contain two sugars (Hasi et al., 2019). In addition, a nonspecific phospholipase C4 (NPC4) was recently reported to hydrolyze GIPCs during phosphate deficiency (Yang et al., 2021). In general, our understanding of the sphingolipid degradation process in plants is still rudimentary (Figure 2).
Sphingolipid diversity
Sphingolipids are structurally diverse owing to differences in sphingoid bases, fatty acyl chains, and polar head groups (Merrill et al., 2009, see Table 1). Most plant LCBs contain 18 or 20 carbons and differ from each other in the number or position of hydroxyl groups and double bonds or in the conformation of molecules (Figure 1). A high-performance liquid chromatography (HPLC) analysis identified the major plant C18 LCBs, including t18:1(Z), t18:1(E), t18:0, d18:1(Z), d18:1(E), and d18:0 (Chen et al., 2006). Plants also contain traces of another four C18 LCBs (d18:2(4E,8E), d18:2 (4E,8Z), t18:2 [with unknown structure], and a unique d18:1 [with a C10=C11 double bond]), one C20 LCB t20:0, and special d21:1 or d22:1 LCBs (the last two LCBs contain a C7=C8 double bond), which have been validated by either mass spectrometry (MS) or nuclear magnetic resonance (NMR) (Kimberlin et al., 2013; Ngo et al., 2020; Panzenboeck et al., 2020; Wang et al., 2020a). However, the two common LCBs d20:0 and d20:1, which are detected in animals and Saccharomyces cerevisiae, are rarely reported in plant samples (De Castro Levatti et al., 2017; Zhu et al., 2020).
Table 1.
Widely-referenced methods of sphingolipid analysis
| Publications | Characteristics | Organism | Detection methods |
|---|---|---|---|
| Chen et al., 2006 | distinguished six LCBs: t18:1(Z), t18:1(E), t18:0, d18:1(Z), d18:1(E), d18:0 |
A. thaliana | HPLC-MS |
| Markham and Jaworski, 2007 | distinguished eight LCBs with 10 fatty acyl chain (168 species): four free LCBs (d18:0, d18:1, t18:0, t18:1) and four phosphorylation forms 40 ceramides (four LCBs linked to c16:0, c18:0, c20:0. c20:1, c22:0, c22:1, c24:0, c24:1, c26:0, c26:1) 40 hCers; 40 GlcCers with hydroxy fatty acyl chains 40 GIPCs with hydroxy fatty acyl chains |
A. thaliana | HPLC/ESI-MS/MS |
| Buré et al., 2011 | distinguished eight GIPC LCBs: t18:1(8Z); t18:1(8E); t18:0; d18:1(4E); d18:1(8E); d18:2(4E,8Z); d18:2(4E,8E); d18:0 |
A. thaliana (Col-0 and Landsberg erecta) N. tabacum |
GC/MS |
| distinguished 25 GIPCs: Glc-GlcA-IPC t18:0 and t18:1 with hydroxy and non-hydroxy fatty acyl chains | A. thaliana (Col-0 leaves) | MALDI-MS and ESI-MS/MS | |
| distinguished 54 GIPCs: Glc-GlcA-IPC; Hex-Glc-GlcA-IPC; (Ara)2-Hex-Glc-GlcA-IPC; (Ara)3-Hex-Glc-GlcA-IPC; (Ara)4-Hex-Glc-GlcA-IPC: t18:0 and t18:1 with hydroxy and non-hydroxy fatty acyl chains |
A. thaliana (Landsberg erecta cells) | ||
| distinguished 121 GIPC: GlcN-GlcA-IPC; GlcNAc-GlcA-IPC; Hex-GlcN-GlcA-IPC; Hex-GlcNAc-GlcA-IPC; Ara-Hex-GlcN-GlcA-IPC; Ara-Hex-GlcNAc-GlcA-IPC; Ara-(Hex)2-GlcN-GlcA-IPC Ara-(Hex)2-GlcNAc-GlcA-IPC; Ara-(Hex)3-GlcN-GlcA-IPC; Ara-(Hex)3-GlcNAc-GlcA-IPC; (Ara)2-(Hex)3-GlcN-GlcA-IPC; (Ara)2-(Hex)3-GlcNAc-GlcA-IPC. t18:0 and t18:1 with hydroxy and non-hydroxy fatty acyl chains |
N. tabacum | ||
| Kimberlin et al. 2013 | additionally distinguished 40 t20:0-based sphingolipids: t20:0-based Cers, hCers, GlcCers and GIPC | A. thaliana | LC-ESI-MS/MS |
| Cacas et al., 2013 | distinguished GIPCs with different head groups in different plants: Hex-IPC; Hex(R1)-HexA-IPC; Hex-Hex(R1)-HexA-IPC Hex-(HexA)3-IPC; (HexA)4-IPC; PenHexHex(R1)-HexA-IPC (Hex)2Hex(R1)-HexA-IPC; Pen(Hex)2Hex(R1)-HexA-IPC (Pen)2HexHex(R1)-HexA-IPC; Pen(Hex)3Hex(R1)-HexA-IPC (Pen)2(Hex)2Hex(R1)-HexA-IPC; (Pen)3HexHex(R1)-HexA-IPC (Pen)4HexHex(R1)-HexA-IPC |
23 plant species | MALDI-Q-TOF-MS and ESI-MS/MS |
| Tarazona et al., 2015 | detected sphingolipids together with other lipids via an enhanced multiplexed LC-MS platform for broad and in-depth analysis of plant lipids: 31 ceramide species; 24 hexosylceramide species 36 GIPC species |
A. thaliana | UPLC-MS |
| Fang et al., 2016 | detected different monosaccharide compositions of GIPC headgroups, including Ara, GlcN, Gal, Glc, Man, and GlcA | A. thaliana | LC-MS/MS and HPAEC-PAD |
| Cacas et al., 2016 | detected sphingolipids with odd number fatty acyl chains | A. thaliana and N. tabacum | GC-MS |
| detected different monosaccharide compositions of GIPC headgroups | MALDI-TOF-MS | ||
| Ishikawa et al., 2018 | detected 166 sphingolipids with d18:0, d18:1, d18:2, t18:0, t18:1 LCBs | A. thaliana | LC-MS/MS |
| detected 100 sphingolipids (containing odd number fatty acyl chains) | Oryza sativa | ||
| Wang et al., 2020a | Identified a new glycosylsphingolipid with odd number LCBs and fatty acyl chain | Psychotria serpens | NMR and MS (LC-MS, and GC-MS) |
| Ngo et al., 2020 | Identified the new phytosphingolipid markhasphingolipid A | Markhamia stipulata var. canaense V.S. Dang | IR, UV, HR-ESI-MS, HR-ESI-MS/MS and NMR |
| Panzenboeck et al., 2020 | Found four GIPCs containing t18:2 in the Rubus idaeus sample | Lactuca sativa var. capitata nidus tenerimma, Spinacia oleracea, Rubus idaeus and Fragaria | RP-HRMS/MS using the open-source program LDA for automated GIPC assignment |
ESI/MS, electrospray ionization MS; ESI-MS/MS, electrospray ionization tandem MS; MS; HPLC/ESI-MS/MS, high-performance liquid chromatography electrospray ionization tandem MS; GC/MS, gas chromatography-MS; MALDI-(Q)-TOF-MS, matrix-assisted laser desorption ionization (quadrupole) time-of-flight MS; UPLC-MS, ultraperformance liquid chromatography-tandem MS; IR, infrared spectroscopy; UV, ultraviolet; HPAEC-PAD, high-performance anion-exchange chromatography with pulsed amperometric detection; HR-ESI-MS/MS, high-resolution electrospray ionization tandem MS; LDA, an open-source software lipid data analyzer.
Fatty acids with different chain lengths, saturations, or modifications also complicate sphingolipid structures. A widely used protocol based on reversed-phase HPLC separates sphingolipids with 10 nonhydroxy fatty acyl chains (C16:0, C18:0, C20:0, C20:1, C22:0, C22:1, C24:0, C24:1, C26:0, C26:1) and 10 C2-hydroxy fatty acyl chains (Markham and Jaworski, 2007). In addition, a low abundance of odd-chain fatty acids (like C23:0 and c25:0) can also be detected compared with even-chain fatty acids such as C24:0 or C24:1 (Cacas et al., 2016). Different combinations of LCBs and fatty acid chains contribute to the variety of ceramide backbones (~100 species). Glycosylation of head groups in sphingolipids further enriches the structure database of sphingolipids. By contrast, the constituent monosaccharide-ceramides in animals can be glucosylceramides, galactosylceramides, and so on, but they are exclusive glucosylceramides (GlcCers) in plants (Schulze and Sandhoff, 2014; Norberg et al., 1991). The animal disaccharide-ceramides or polysaccharide-ceramides, such as lactosylceramides or globosides, have not been detected in plants (Schulze and Sandhoff, 2014). Compared with GlcCers, GIPCs are present at quite high abundance in plants and in multiple forms (Buré et al., 2011; Cacas et al., 2013; Adem et al., 2021) that are absent in animals. The classic structure of plant GIPCs consists of a ceramide backbone, inositol phosphate, glucuronic acid, and saccharide residues (Figure 1F modified from Cacas et al., 2013). Cacas et al. (2013) identified an unusual GIPC lacking hexuronic acid (HexA) in the algae Chondracanthus acicularis via ESI-MS/MS (Cacas et al., 2013). In this work, they identified an extra 21 polar head structures in GIPCs from 23 different plant species, enlarging the existing database of 54 Arabidopsis GIPCs and 121 tobacco GIPCs provided by Buré et al. (2011). They identified the sugar residues as hexoses (or hexose-R1, R1 representing an OH, NH2, or N(Ac) group) and pentoses (or pentose-R1), but they did not distinguish the isomers of different sugar residues as before (Buré et al., 2011; Cacas et al., 2013). Mortimer et al. (2013) showed that the isomers of GIPC head groups could be arabinose, galactose, mannose, glucose, and so on. In total, based on their polar head variability, the plant GIPCs are generally classified into seven series bearing one to seven saccharide units (series 0–F) (see Figure 1F, Table 1). This classification can cover most of the detectable GIPCs; however, an exception also exists, in that tobacco leaf GIPCs probably contain more than 10 sugar residues (Kaul and Lester, 1975).
Distribution
Sphingolipids only make up a small proportion (0.5%–10%) of the total lipid content in plant cells, depending on cell type and plant species, and this proportion is also strongly influenced by extraction and detection methods. A comparison of sphingolipidomes indicates a striking difference between Arabidopsis leaf and pollen sphingolipids: the sphingolipid LCBs in leaves are mainly t18:1, t18:0, d18:1, and d18:0, whereas they include a large percentage of d18:2 in pollen (Chen et al., 2006; Markham et al., 2006; Luttgeharm et al., 2016). In addition, there are more free LCBs and ceramides in pollen than in leaves (Markham and Jaworski, 2007). Meanwhile, the most abundant sphingolipids in leaves are GIPCs, whereas those in pollen are GlcCers (Luttgeharm et al., 2016). Leaf GIPCs are primarily Hex(OH)-HexA-IPC (series A), with a small amount of Hex-Hex(OH)-HexA-IPC (series B), whereas pollen GIPCs consist mainly of Hex(NAc)-HexA-IPCs (series A), (Pen)3-Hex-Hex(NAc)-HexA-IPCs (series E), (Pen)2-Hex-Hex(NAc)-HexA-IPCs (series D), and trace amounts of Hex-Hex(OH)-HexA-IPCs (series B) (Luttgeharm et al., 2016). Using the published database, we also compared sphingolipids among different plant species. A detailed report on polar heads of GIPCs demonstrates that plants have their own characteristic sphingolipids (Cacas et al., 2013). In addition, differences in the number of sugar residues linked to the IPCs may contribute to plant susceptibility to pathogens that produce necrosis and ethylene-inducing peptide 1-like (NLP) proteins (Lenarčič et al., 2017). The composition and distribution of sphingolipids (free LCBs, Cers/hCers, GlcCers, and GIPCs) also differ among different plants. For example, rice leaf sphingolipids have a higher cis/trans LCB ratio and non-hydroxy fatty acid ratio than Arabidopsis leaf sphingolipids (Ishikawa et al., 2016; Markham et al., 2006).
As lipids, some sphingolipid groups are the major components of membrane lipids. For example, GlcCers and GIPCs constitute about 40% of plant plasma membrane (PM) lipids (Sperling et al., 2005; Vu et al., 2014; Grison et al., 2015; Tarazona et al., 2015) and are enriched in the membrane outer leaflet (Tjellström et al., 2010). Sphingolipids are also relatively abundant in the plant tonoplast, ER, and Golgi, accounting for up to 20% of the total lipids in these membranes (Verhoek et al., 1983; Warnecke and Heinz, 2003; Fouillen et al., 2018). Previous investigations have shown that sphingolipids are highly enriched in plasmodesmata (PDs) and extracellular vesicles (EVs), accounting for over 40% of the membrane lipids in these structures (Grison et al., 2015; Liu et al., 2020b). However, in the chloroplast and mitochondrion, sphingolipids are undetectable or present only at trace levels (Verhoek et al., 1983).
The sphingolipids are not evenly distributed in cellular membrane systems. In the Arabidopsis rosette leaf, the proportions of Cers, hCers, GlcCers, and GIPCs relative to total sphingolipids are 4%, 3%, 37%, and 56% (Markham and Jaworski, 2007). Recently, a published article compared the Arabidopsis leaf plasma and vacuolar membrane sphingolipidomes (Carmona-Salazar et al., 2021). It revealed different sphingolipid compositions in the microsomal membranes (MICs) (which are a heterogeneous population of membrane vesicles from different organelles and the PM), vacuolar membranes (VMs), PMs, and PM-derived detergent-resistant membranes (DRMs) (Carmona-Salazar et al., 2021). In general, the MICs had the lowest sphingolipid content (relative to protein content), and DRMs contained the most sphingolipids. In all membrane fractions, Cers accounted for the smallest proportion, whereas GIPCs were the major constituents (except in VMs) (Carmona-Salazar et al., 2021). Consistent with previous reports, the percentage of GIPCs in PM sphingolipids was ~70% (% mol), whereas that in DRMs was 44% (lower than in the PM), in contrast to data from tobacco (Grison et al., 2015; Cacas et al., 2016; Liu et al., 2020a; Carmona-Salazar et al., 2021). The authors explained this result based on the preference of DRMs for individual sphingolipid species. Combined with other findings, we would like to provide another explanation: Triton X-100 treatment alters the restriction of GIPC-PLDs or other lipases in PMs and results in GIPC degradation to Cers or hCers. We also notice that the VMs contain a large amount of GlcCers, indicating that endo-membranes prefer the simple glycosyl sphingolipid GlcCers to the complex GIPCs. However, the situation is completely reversed in the PM, PD, and EVs: they are enriched in the GIPCs, which are thought to mainly aggregate outside the protoplast (Warnecke and Heinz, 2003; Grison et al., 2015; Liu et al., 2020b).
Overall assessment of sphingolipid profiles also suggests that membrane structures are characterized by sphingolipids with distinct side chains. We have reported that sphingolipids containing saturated LCBs and fatty acyl chains are more abundant in PDs than in PMs and have shown that saturated LCB chains can bind proteins to regulate PD function (Liu et al., 2020a). Coincidentally, Carmona-Salazar et al. (2021) revealed that the saturation of sphingolipids was higher in Arabidopsis leaf DRMs than in the PM fraction, indicating that the DRMs in the PM may function in recruiting specific proteins (Carmona-Salazar et al., 2021).
DRMs and sphingolipid-sterol rafts
Membranes are always nonuniform or asymmetrical lipid bilayers. The ‘rafts’ concept has been proposed to describe clusters of sphingolipids and cholesterol that form platforms for protein attachment within the fluid membranes owing to phase separation; they contribute significantly to membrane lateral heterogeneity (Ahmed et al., 1997; Simons and Ikonen, 1997). Native raft units are small (10–200 nm) and tend to form large raft regions through protein-protein, lipid-lipid, or protein-lipid interactions (Kusumi and Suzuki, 2005; Pike 2006). Among the membrane lipids, sphingolipids exhibit unique properties such as contributing to the molecular order (rigidity) of phospholipid membranes, giving rise to lateral phase separation, interacting with sterols, and forming membrane microdomains (Goñi and Alonso, 2006; Alonso and Goñi, 2018). A method of detergent treatment at low temperature has been commonly used to study the components or functions of sphingolipid-sterol rafts. When treated with detergents at 4°C, cell membranes can be cracked into non-solubilized fractions called DRMs. In plants, these DRMs (mainly extracted upon Triton X-100 treatment) have been shown to contain abundant sphingolipids (especially GIPCs) and sterols, as well as an accumulation of certain proteins (Simon-Plas et al., 2011). However, the properties of detergents have also led to sharp criticisms of the DRM concept, such as the assertion that Triton X-100 actually promotes lipid condensation in the PM (Munro, 2003; Tanner et al., 2011). Even so, DRMs are still considered to be a starting point for the analysis of plant sphingolipid-sterol-enriched membrane microdomains.
Lipid microdomains have been found in the animal ER and Golgi, but they are present only in the Golgi not in ER membranes upon Triton X-100 treatment (Laloi et al., 2007). The lipid clusters can coalesce some proteins, which benefit membrane bending and vesicle fission during trafficking processes, to form stable phases and transfer to the apical PM or elsewhere (Simons and Ikonen, 1997; Gkantiragas et al., 2001; Paladino et al., 2007; Legrand et al., 2019). In plants, Golgi-derived DRMs and PM-derived DRMs have similar GlcCer compositions (Laloi et al., 2007). Depending on the plant species, PM-derived DRMs have approximately 2-fold to 7-fold higher sphingolipid levels (mainly GIPCs) than non-DRM regions in the PM (Simon-Plas et al., 2011). Cacas et al. (2016) showed that Nicotiana tabacum DRMs were more enriched in GIPCs with 2-hydroxylated VLCFAs and polyglycosylated GIPCs compared with the total PM fraction. Consistent with the findings of Carmona-Salazar et al. (2021), they also reported enrichment of t18:n-based GIPCs but did not provide quantitative data for Cers and hCers in DRMs (Cacas et al., 2016; Carmona-Salazar et al., 2021). The relationships between the composition and function of these DRMs in plants require further study.
Sphingolipid trafficking
Ceramides, the backbones of sphingolipids, are synthesized in the ER, and the major plant sphingolipids such as GIPCs are produced in the Golgi. Sphingolipid trafficking and biosynthesis are naturally related. Once synthesized, sphingolipids may undergo translocation, sorting, and transport movements and finally localize to membranes via a sophisticated network. In this section, we mainly introduce sphingolipid movements, including short-distance intramembrane transport/translocation, sorting, and long-distance intermembrane transport (Figure 3), and we discuss factors that affect sphingolipid trafficking (Riboni et al., 2010; Hurlock et al., 2014).
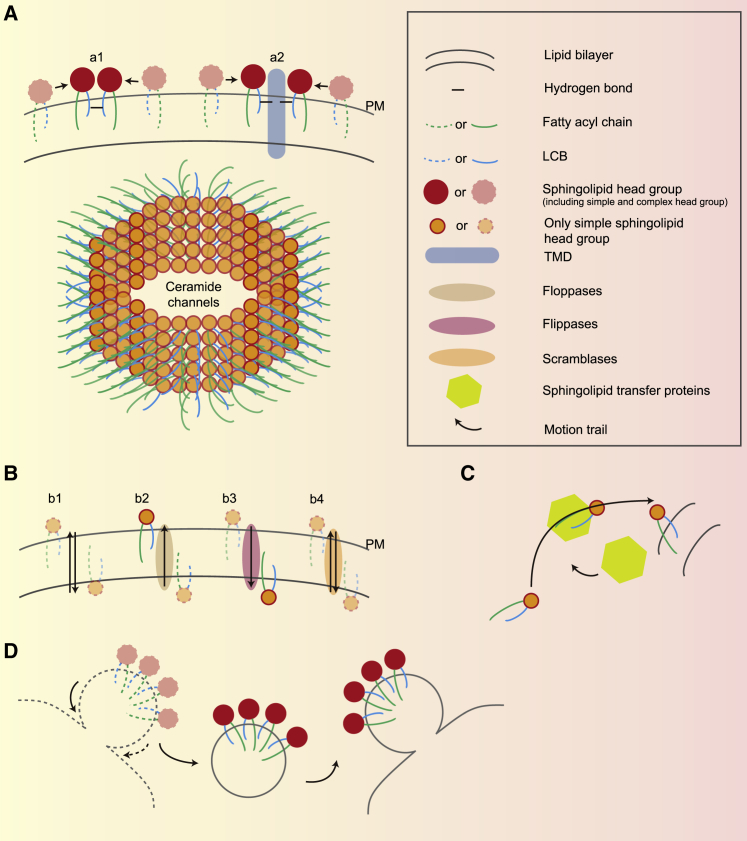
Figure 3.
Sphingolipid movement patterns
(A) The upper lane is a model of sphingolipid lateral diffusion. a1 represents sphingolipids that form phase separation via lipid-lipid interaction; a2 represents sphingolipids that form phase separation via lipid-protein interaction. The lower lane is a ceramide channel, which is a form of large-scale sphingolipid phase separation.
(B) A model of the sphingolipid flip-flop pattern. b1 represents sphingolipid movement that occurs freely; b2 represents sphingolipid movement mediated via floppases; b3 represents sphingolipid movement mediated via flippases; b4 represents sphingolipid movement mediated via scramblase.
(C) A model of sphingolipid transport via protein transporter.
(D) A model of sphingolipid transport via vesicles.
(A) and (B) are intramembrane sphingolipid transport, and (C) and (D) are intermembrane sphingolipid transport.
Intramembrane transport/translocation
In vitro assays of different sphingolipids in different media (e.g., fluid lipid bilayers composed of dipalmitoylphosphatidylcholine or palmitoyloleoylphosphatidylcholine) demonstrate that their lateral diffusion rates vary from 10−10 to 10−7 cm2/s (Riboni et al., 2010). Lateral diffusion contributes to the flexible movement of sphingolipids in bilayers and also provides opportunities for sphingolipid aggregation. In turn, sphingolipid lateral phase separation, which results in membrane lateral heterogeneity through the formation of sphingolipid-enriched domains, restricts lateral diffusion of sphingolipids and is largely regulated by sterols or proteins (Riboni et al., 2010). For example, the single-particle tracking system shows that the sphingolipid GM1 undergoes transient confinement in the PM through interaction within sphingolipids or sterols (Sheets et al., 1997). In addition, the lateral diffusion of sphingolipids can directly mediate sphingolipid flux among cellular organelles through membrane contact sites (Csordás et al., 2006; Grimm, 2012). For instance, the continuous ER attaches to the outer membrane of mitochondria (Csordás et al., 2006) and mediates the direct flow of ceramides to mitochondria (Grimm, 2012). The local excess of ceramides can form ceramide channels and induce mitochondrial outer-membrane permeabilization to proteins (Siskind, 2005; Figure 3A).
Sphingolipid “flip-flop” movement can occur spontaneously or be mediated by facilitators, and the frequency depends on the sphingolipid species. In general, transbilayer movement of sphingolipids with complex head groups takes place more slowly than that of sphingolipids with simple head groups. In model membranes, the animal sphingolipid gangliosides and sphingomyelins (SMs) display little transbilayer movement, whereas GlcCers and Cers can move freely between layers (Buton et al., 2002; Riboni et al., 2010, Figure 3B). Limitation of transbilayer movement ensures membrane asymmetry. It has been proposed that complex animal sphingolipids synthesized on the Golgi luminal side, which are later transported to the PM outer layer, show little translocation to the cytosolic face (Zachowski, 1993; Riboni et al., 2010). Similarly, in plants, the GIPCs are hypothesized to localize exclusively to the apoplastic face, whereas the GlcCers are present at both the apoplastic and cytosolic faces (Cacas et al., 2016). Nonetheless, it remains unknown whether GIPCs never move to membrane cytosolic faces. Facilitators that help with sphingolipid transbilayer movement can be divided into three classes: the flippases (which mediate movement from the outer to inner layer), floppases (inner to outer layer), and scramblases (bidirectional movement). In plants, the flippase that facilitates movement of GlcCers to the cytosolic membrane surface has been reported (Davis et al., 2020), but the floppases and scramblases have not been identified.
Intermembrane transport
The distinct distributions of sphingolipids in membranes raise the question of how sphingolipids are transported between different membranes. Intermembrane movement of sphingolipids generally occurs in two forms: non-vesicular (or protein-mediated) and vesicular transport. In animal cells, the mechanism of the most typical case of the non-vesicular sphingolipid transporter CERT has been well illustrated (Hanada et al., 2003). The protein uses its FFAT motif (two phenylalanines [FF] in an acidic track [AT]) to seek the ER, the START domain binds with Cers, and the PH (pleckstrin homology) domain targets phosphatidylinositol 4-monophosphate-enriched areas of the Golgi, thereby transporting the Cer between the ER and Golgi (Hanada et al., 2003). In addition, animal FAPP2 can mediate the movement of GlcCers from the cis-Golgi to the trans-Golgi (D'Angelo et al., 2007). In plants, the ceramide kinase accelerated cell death 11 (ACD11) and a glycolipid transfer protein (GLTP1) are regarded as possible candidates for sphingolipid transporters (Brodersen et al., 2002; West et al., 2008), although solid evidence is lacking.
Vesicular transport is the dominant mode of sphingolipid intermembrane transport. Cers have been shown to associate with ER-derived vesicles (Kajiwara et al., 2008; Giussani et al., 2009), and the complex sphingolipids synthesized in the Golgi lumina are also packaged into vesicles by uncertain mechanisms (Holthuis and Levine, 2005). In plants, studies have shown that vesicular transport behavior can easily be affected by the sphingolipid biosynthesis process. For example, the TGN secretes VLCFA-sphingolipid-enriched secretory vesicles to assist auxin carrier PIN2 to the apical membrane (Wattelet-Boyer et al., 2016). Inhibition of VLCFA synthesis by Metazachlor can decrease hydroxy-VLCFA (hVLCFA)-based sphingolipids and affect TGN-associated secretory vesicles (SVs) and PIN2 polar localization, indicating that sphingolipid biosynthesis and trafficking are closely related (Wattelet-Boyer et al., 2016). Coincidentally, the Arabidopsis loh1-1 loh3-1 mutant with impaired ceramide synthesis displays disruptions in the endosome transport system and auxin transport (Markham et al., 2011). In addition, animal COP I vesicles that bud from the Golgi have been found to contain low amounts of sterols and stearoyl sphingomyelins, except N-stearoyl sphingomyelin (SM18) (Brügger et al., 2000). The p24 protein, which can recruit COP I vesicles, has been demonstrated to have the specific ability to bind with SM18 (Contreras et al., 2012). Considering that COP I-coated vesicles mediate transport across the TGN to the Golgi and from the Golgi to the ER, the above findings also imply that COP I-dependent vesicle trafficking may be associated with certain sphingolipid (SM18) trafficking. Impairing endosomal trafficking by knocking out Golgi transmembrane 9 superfamily member 2 (TM9SF2) reduces the level of glycosphingolipids such as globotriaosylceramide (Gb3) (Tian et al., 2018). Our previous work also showed that the Arabidopsis tetraspanin 8 mutant contained significantly lower GIPC levels than the WT (Liu et al., 2020b). Although our understanding of them remains rudimentary, factors that affect plant sphingolipid transport deserve further investigation.
Sphingolipid sorting
Enzymes involved in sphingolipid biosynthesis intrinsically form sphingolipid sorting modulators. For example, deletion of Arabidopsis SLDs reduces not only the unsaturation of the LCBs but also the GlcCer/GIPC ratio (Chen et al., 2012). Enzyme activity analysis shows that the AtSLDs are not enzymatically distinct and do not specifically target one class of sphingolipids (Chen et al., 2012). Considering that GCS is ER localized whereas IPCS is Golgi localized, the authors proposed a model in which ceramides containing t18:1Δ8 trans or t18:0 are preferentially shuttled from the ER to the Golgi for GIPC synthesis (Chen et al., 2012). In the sld1 sld2 mutant, increased flux of t18:0-ceramides to the Golgi results in elevated GIPC content (Chen et al., 2012). In addition, proteins that display selectivity to certain sphingolipids may regulate sphingolipid translocation. In our previous work, we found that the plant PD membrane contained more saturated LCB-based sphingolipids and that the PD-localized protein PDLP5 had the specific ability to bind with t18:0-based sphingolipids (Liu et al., 2020a). However, more evidence of facilitators that help to sort sphingolipids and maintain membrane lateral heterogeneity is still needed.
Functions
Membrane components
Sphingolipids are crucial for maintaining the morphology of membrane systems and modulating membrane function. Inhibition of ceramide synthesis by FB1 (fumonisin B1) causes mislocation of auxin carrier proteins by inducing small vesicular structures like dismantled vacuoles or fused vesicles in plant cells (Markham et al., 2011). Downregulation of the SPT subunit LCB2 leads to the destruction of both ER and Golgi membranes in Arabidopsis pollen (Dietrich et al., 2008). Blocking GlcCer biosynthesis by PDMP (D,L-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol) induces Golgi heteromorphosis with swollen surrounding vesicles and, as a result, causes the breakage of the Golgi-dependent protein secretory pathway (Melser et al., 2010). Abundant in the plant cell PM and PD, sphingolipids associate with sterols to build sphingolipid-enriched microdomains and provide locations for protein anchoring, thereby restricting protein diffusion (Simons and van Meer 1988; Grison et al., 2015). Changes in sphingolipid composition of membranes affect the aggregation of PD proteins and finally PD function (Huang et al., 2019; Liu et al., 2020a). In animals, sphingosines or ceramides form specialized channels: sphingosines form channels in the erythrocyte PM with short open lifetimes and small diameters (<2 nm), whereas ceramide channels in mitochondrial outer membranes have longer open times and larger apertures to allow the passage of proapoptotic proteins (Siskind, 2005). Efforts should be made to investigate whether such channel structures exist in plants.
Sphingolipids also act as powerful supporters to help the membrane cope with abiotic stresses. In response to chilling treatment, Arabidopsis plants are stimulated to increase GIPCs and decrease GlcCers (Nagano et al., 2014). Moreover, increasing the saturation of sphingoid long-chain bases impaired plant tolerance to low temperature, as sphingolipid unsaturation may be involved in membrane fluidity and the maintenance of H+-ATPase function in the PM (Chen et al., 2012).
Receptors or sensors
Located in the outer layer of the PM, sphingolipids with sugar head groups may act as surface receptors to sense ectocytic signals. Similar to some animal glycosphingolipids (Smith et al., 2004), plant GIPCs have been found to recognize NLP microbial toxins (Lenarčič et al., 2017). Toxin binding assays show that the NLP toxins have selective binding affinity for series A GIPCs (Figure 1F), which are the major forms of GIPCs in eudicots, providing an explanation for why eudicots are more sensitive than monocots to NLP cytolysins (Lenarčič et al., 2017). In addition to being toxin sensors, plant sphingolipid GIPCs also exhibit the ability to sense salt through direct binding of Na+, subsequently triggering Ca2+ influx (Jiang et al., 2019).
Signaling molecules
Sphingolipids, particularly ceramides, LCBs, or their phosphorylated forms, can act as signaling molecules to regulate plant development and stress responses. In plants, sphingosine-1-phosphate (S1P, also named dihydrosphingosine-1-phosphate or dh-S1P) has been shown to affect stomatal aperture via the abscisic acid (ABA)-mediated signaling pathway (Coursol et al., 2003). However, exogenous application of S1P fails to alter stomatal aperture and guard cell ion channel activities in heterotrimeric G protein alpha-subunit gene knockout plants, which indicates that S1P signaling may be closely related to G protein-coupled signaling pathways (Coursol et al., 2003). In Commelina communis leaves, drought-induced S1P elevation or the application of S1P can trigger a Ca2+ spike and then stomatal closure (Ng et al., 2001). Phytosphingosine phosphate (phyto-S1P) is known to play a similar role in the control of stomatal closure in the Pisum sativum epidermis by elevating the NO content and pH of guard cells (Puli et al., 2016). A short period of cold treatment increases the level of phyto-S1P and ceramide-1-phosphate, suggesting a role for sphingolipids in the rapid signaling response to low temperatures (Cantrel et al., 2011).
Ceramides and free LCBs have been found to induce the hypersensitive response (HR) or programmed cell death (PCD) in plants, whereas their phosphorylated forms show the opposite effect (Liang et al., 2003; Shi et al., 2007; Alden et al., 2011; Lachaud et al., 2011; Saucedo-García et al., 2011). Disruption of sphingolipid biosynthesis always results in a trade-off between plant autotoxicity and defense. Liang et al. (2003) identified the first Arabidopsis ceramide kinase known to affect the balance between ceramides and C1Ps; it could modulate PCD and plant resistance to the bacterial pathogen Pseudomonas syringae pv. tomato (Pst). Shi et al. (2007) reported that free sphingoid bases could trigger PCD, and S1P could rescue the cell death induced by sphingosine but not phyto-sph. Saucedo-García et al. (2011) further reported that LCB-induced PCD signals could be delivered via mitogen-activated protein kinase 6 (MPK6) and function as defense strategies against bacterial pathogens. Qin et al. (2017) then reported that phyto-sph activated jasmonic acid (JA) and phyto-S1P could initiate the salicylic acid (SA) signaling pathway, and together they altered plant resistance to FB1 stress. Inhibition of ceramide synthase by noncontrolled hydroxylated diterpene derivatives causes a severe toxin reaction in Nicotiana attenuata, as well as in herbivores (Li et al., 2021), whereas the controlled hydroxylated diterpene strategy allows plants to gain resistance to their insect herbivores without ceramide-triggered autotoxicity (Li et al., 2021). Infection by the virulent plant pathogen pst DC3000 triggers a temporary increase in free t18:0 in Arabidopsis, whereas the avirulent pst DC3000 avrRpm1 causes sustained upregulation of t18:0 (Peer et al., 2010). Exogenous application of t18:0 or knocking out SLD elevates the expression of disease resistance genes and enhances plant resistance against pst DC3000 (Liu et al., 2020a). Moreover, Arabidopsis mutants with abnormal sphingolipid content exhibit a constitutive elevation of SA content and greater pathogen resistance, indicating a role for sphingolipids as molecular modulators of plant defense (Berkey et al., 2012; Magnin-Robert et al., 2015; Zienkiewicz et al., 2020).
Perspectives
The last two decades have seen impressive research achievements in elucidating the biosynthesis and function of plant sphingolipids; however, there are still obstacles and difficulties to overcome in order to gain a comprehensive understanding of sphingolipids.
Extension of chemical information
Current sphingolipidomic studies rely heavily on liquid chromatography-tandem mass spectrometry (LC-MS/MS) with reference to known compounds; however, there are special sphingolipids that require other analytic techniques, such as markhasphingolipid A, which is identified specifically using NMR. We also noticed that the widely used LC-MS/MS method based on Arabidopsis has limitations in other plant species (Markham and Jaworski, 2007). Ishikawa et al. (2016) established a new MRM library to cover more GIPC structures detectable in rice. However, a recent cotton sphingolipidomics study, which contained only 18 GIPC species, with up to 5% (% mol) of total sphingolipid content (Wang et al., 2020b), indicated that the available MRM databases are still insufficient. Thus, an integrative approach combining high-throughput analyses, precise identification, and broad coverage will be highly valuable for facilitating future research.
Except for some common sphingolipids such as free d18:0 or some of the ceramides, few plant lipids are commercially available. Our recent report provided a method for the chemosynthesis of the major plant sphingolipid LCB t18:1, which contains both t18:18t and t18:18c conformations (Liu et al., 2020a), and purification procedures of either trans- or cis-t18:1 can be improved further. A troublesome problem is that plant GIPCs, always harvested after long extraction times, are a complex mixture with different ceramide backbones and head groups (Buré et al., 2011; Hasi et al., 2020). Experiments using isolated GIPCs leave uncertainties because they rely mainly on the quality of the mixtures, and protocols to chemically synthesize the simple GIPCs have yet to be developed. Thus, sphingolipid research calls for innovation and further development of both chemical and biological methodologies.
Updating of the metabolic pathway
The chemical diversity of sphingolipids contributes to the complexity of composition and function of cellular membranes. Most genes involved in sphingolipid biosynthesis, modification, and degradation in Arabidopsis thaliana have been identified. Although sphingolipids are well studied, black boxes still exist in the sphingolipid metabolism maps. For instance, nearly 200 GIPC species have been described in two reports (Buré et al., 2011; Cacas et al., 2013); however, to date only three enzymes and three sugar transporters involved in the decoration of GIPC head groups have been reported (Figure 2). Research has provided clues that membrane proteins with lipid binding capacity can regulate sphingolipid content. In plants, misexpression of Niemann-Pick disease type C1 (NPC1)-like protein, containing 11 predicted membrane-spanning regions and a putative lipid binding site, causes transgenic Arabidopsis to accumulate sphingolipids with hydroxylated fatty acyls (Feldman et al., 2015). Knocking out one of the Arabidopsis tetraspanin proteins, TET8, which has a potential sphingolipid binding motif and is involved in extracellular vesicle secretion, results in significantly decreased contents of GIPCs (Liu et al., 2020b). Thus, the sphingolipid metabolism pathway must be constantly updated.
Distribution issues
As mentioned earlier, the eukaryotic PMs are asymmetrical because of the different lipid compositions of the inner and outer leaflets, and they tend to exhibit lipid phase separation in one leaflet (Bretscher, 1972; Lin and London, 2015). These observations raise questions about the properties and molecular functions of plant sphingolipids: (1) what is the whole-cellular distribution of sphingolipids in plants? (2) How can lipids in the inner monolayer be distinguished from those in the outer monolayer? (3) How do these lipids stay in separate phases within layers? (4) What is the importance or significance of the heterogeneous lipid distribution?
Current lipidomic analysis of membrane fractions provides primary data for a sphingolipid atlas, and visualization of spatial lipid distribution will contribute more solid evidence. Immunolabeling of certain sphingolipids via antibodies can display the in situ distribution of sphingolipids. In plants, polyglycosylated GIPCs in the tobacco PM have been shown to cluster around the outer layer of the PM fraction using the immune colloidal gold technique (Cacas et al., 2016). Fluorophore-labeled sphingolipids are widely used for lipid imaging in mammalian cells (Makiyama et al., 2015). Fluorescence correlation spectroscopy (FCS) with a stimulated emission depletion (STED) microscope (STED-FCS) can reveal lipid dynamics within the PM (Eggeling et al., 2009). Klitzing et al. (2013) developed secondary ion MS (SIMS) imaging methods for lipids: data pre-collected by bright field microscopy and low-voltage scanning electron microscopy (SEM) were combined with the signal abundance of isotope-labeled mass sphingolipids to produce a distribution map of certain sphingolipids (Klitzing et al., 2013). In addition, single particle tracking systems have been used to evaluate heterogeneous membranes (Komura et al., 2016). However, except for immunolabeling methods, the abovementioned techniques are seldom applied to plants.
Research shows that GIPCs prefer an aggregated status within the ectocytic layer of plant cells (Cacas et al., 2016; Liu et al., 2020b). The lipid-flip model suggests that sphingolipids with more complex head groups have less ability to undergo transbilayer movement (Riboni et al., 2010), indicating that the GIPCs may undergo special and stable transport before targeting to the PM or EVs (Cacas et al., 2016; Liu et al., 2020b). There are not enough studies on this topic, and more evidence is therefore needed to support and explain the maintenance of sphingolipid asymmetrical distribution between bilayers.
Sphingolipid phase separation has been well illustrated based on NMR spectroscopy in mammalian cells. Sphingomyelin molecules have been shown to interact strongly with neighboring lipids via hydrogen bonds and form lateral phase separation within membranes (Boggs, 1987). Grosjean et al. (2015) showed that phytosterols could create an ordered lipid phase through formation of sphingolipid-sterol interacting domains. However, efforts should still be made to explain the reason for and condition of different sphingolipid separated phases in plants.
Proteins interacting with sphingolipids have been shown to function together within certain membrane structures. We have reported that t18:0-based sphingolipids bind the PD-located protein PDLP5 to regulate PD permeability (Liu et al., 2020a). Similar evidence suggests that lipid rafts (or liquid-ordered phase) mediated via Remorin proteins are essential for SA-induced PD closure. However, this work did not include lipidomic analysis of the PD fraction or report the contributions of sphingolipids to PDs (Huang et al., 2019). Thus, more focus should be placed on how their particular structures contribute to membrane fluidity and biophysical order and, finally, to membrane functions.
Trafficking issues
Studies on sphingolipid trafficking in plants are few. It is not known whether the principal factors of sphingolipid sorting are determined by the characteristics of lipids or proteins, nor is it known how sphingolipids complete selective trafficking or what molecular mechanisms underlie the two different transport pathways (protein-mediated and vesicular transport). Fluorophore-labeled sphingolipids have been used to track sphingolipid intermembrane transport in microorganisms and mammalian cells (Hackstadt et al., 1996; van Helvoort et al., 1997; Kenoth et al., 2019). Transgenic plants (usually Arabidopsis) with different tagged-marker proteins involved in lipid trafficking pathways can be used to study sphingolipid transport. In addition, lipidomic analysis of plant submembrane fractions or lipid-protein complexes also provides clues for sphingolipid movements (Grison et al., 2015; Liu et al., 2020a; Carmona-Salazar et al., 2021). However, microscopy techniques (such as single particle tracking) are needed to improve the visualization of sphingolipid trafficking in plants.
Functional studies
Studies on sphingolipid function always revolve around two aspects: the membrane components and bioactive molecules. Cellular membranes are formed from a chemically diverse set of lipids present in various amounts and proportions. High lipid diversity is universal in eukaryotes and is seen from the scale of a membrane leaflet to that of a whole organism, highlighting its importance and suggesting that membrane lipids fulfill many functions. Indeed, alterations in membrane lipid homeostasis are linked to various diseases.
The importance of sphingolipids in membranes is well known, but the factors that affect plant membrane sphingolipid dynamics are still poorly understood. Functional studies of plant membrane sphingolipids start first from the biosynthetic enzymes. The influence of certain sphingolipids on cellular membranes must be investigated carefully; for example, deleting the SLDs in Arabidopsis alters PD sphingolipid composition as well as PD function (Liu et al., 2020a). An unanswered question is how membrane-associated proteins regulate sphingolipids, and the interconnections between different classes of lipids and their coordinated functions can be an exciting area of research. In addition, understanding how membrane sphingolipid dynamics are regulated by sphingolipid metabolism remains important, and lipid trafficking mechanisms among cellular compartments are also a challenging issue to address.
Free sphingolipids, such as ceramides, sphingosine, or their C1-phosphates, are always signaling molecules and serve as second messengers that function in the regulation of stress responses. In animals, these bioactive sphingolipids have been shown to control the balance of cell death/survival, cellular differentiation, senescence, growth, inflammation, immunity, and so forth (Levi et al., 2010; Subei and Cohen, 2015; Czubowicz et al., 2019). Several classes of G protein-coupled receptors (GPCRs) are known to interact with sphingosine-1-phosphate to conduct cellular signals (Levi et al., 2010; Subei and Cohen, 2015). Although plant sphingolipid signaling is associated with heterotrimeric G proteins (Coursol et al., 2003), there is still no direct evidence of plant sphingolipid receptors, such as SIP receptors.
Land plants are sessile and autotrophic organisms; although they do not have a (typical) nervous system, they can respond to their surroundings as readily as animals. Plant sphingolipid homeostasis (including membrane lipids and signal lipids) is easily disturbed by multiple environmental factors (Ng et al., 2001; Cantrel et al., 2011; Chen et al., 2012). We are only beginning to understand why even small changes in lipid structure or composition can have profound effects on crucial biological functions of sphingolipids. Although there are many unknowns, interdisciplinary approaches have begun to reveal novel functions of sphingolipids and their interactions. A detailed picture of sphingolipids will help us to better understand plant growth, development, and adaptation to ever-changing environments.
Funding
This review was supported by the Chinese Academy of Sciences (XDB27020207) and the National Natural Science Foundation of China (31788103, 31690092).
Author contributions
N.-J.L. and X.-Y.C. wrote the manuscript. N.-J.L. prepared the tables and figures. L.-P.H. prepared all the reference lists. J.-J.B. and L.-J.W. contributed to reviewing and editing the manuscript.
Acknowledgments
We appreciate all work in this field and apologize to colleagues whose work is not cited due to space limitations. No conflict of interest declared.
Published: June 29, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
References
- Adem A.A., Belete A., Soboleva A., Frolov A., Tessema E.N., Gebre-Mariam T., Neubert R.H.H. Structural characterization of plant glucosylceramides and the corresponding ceramides by UHPLC-LTQ-Orbitrap mass spectrometry. J. Pharm. Biomed. Anal. 2021;192:113677. doi: 10.1016/j.jpba.2020.113677. [DOI] [PubMed] [Google Scholar]
- Ahmed S.N., Brown D.A., London E. On the origin of sphingolipid/cholesterol-rich detergent-insoluble cell membranes: physiological concentrations of cholesterol and sphingolipid induce formation of a detergent-insoluble, liquid-ordered lipid phase in model membranes. Biochemistry. 1997;36:10944–10953. doi: 10.1021/bi971167g. [DOI] [PubMed] [Google Scholar]
- Alden K.P., Dhondt-Cordelier S., McDonald K.L., Reape T.J., Ng C.K., McCabe P.F., Leaver C.J. Sphingolipid long chain base phosphates can regulate apoptotic-like programmed cell death in plants. Biochem. Biophys. Res. Commun. 2011;410:574–580. doi: 10.1016/j.bbrc.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Alonso A., Goñi F.M. The physical properties of ceramides in membranes. Annu. Rev. Biophys. 2018;47:633–654. doi: 10.1146/annurev-biophys-070317-033309. [DOI] [PubMed] [Google Scholar]
- Beeler T., Bacikova D., Gable K., Hopkins L., Johnson C., Slife H., Dunn T. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Delta mutant. J. Biol. Chem. 1998;273:30688–30694. doi: 10.1074/jbc.273.46.30688. [DOI] [PubMed] [Google Scholar]
- Berkey R., Bendigeri D., Xiao S. Sphingolipids and plant defense/disease: the "death" connection and beyond. Front Plant Sci. 2012;3:68. doi: 10.3389/fpls.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggs J.M. Lipid intermolecular hydrogen bonding: influence on structural organization and membrane function. Biochim. Biophys. Acta. 1987;906:353–404. doi: 10.1016/0304-4157(87)90017-7. [DOI] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Pike H.M., Olszak B., Skov S., Odum N., Jørgensen L.B., Brown R.E., Mundy J. Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 2002;16:490–502. doi: 10.1101/gad.218202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher M.S. Asymmetrical lipid bilayer structure for biological membranes. Nat. New Biol. 1972;236:11–12. doi: 10.1038/newbio236011a0. [DOI] [PubMed] [Google Scholar]
- Brügger B., Sandhoff R., Wegehingel S., Gorgas K., Malsam J., Helms J.B., Lehmann W.D., Nickel W., Wieland F.T. Evidence for segregation of sphingomyelin and cholesterol during formation of COPI-coated vesicles. J. Cell Biol. 2000;151:507–518. doi: 10.1083/jcb.151.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buré C., Cacas J.L., Wang F., Gaudin K., Domergue F., Mongrand S., Schmitter J.M. Fast screening of highly glycosylated plant sphingolipids by tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2011;25:3131–3145. doi: 10.1002/rcm.5206. [DOI] [PubMed] [Google Scholar]
- Buton X., Hervé P., Kubelt J., Tannert A., Burger K.N., Fellmann P., Müller P., Herrmann A., Seigneuret M., Devaux P.F. Transbilayer movement of monohexosylsphingolipids in endoplasmic reticulum and Golgi membranes. Biochemistry. 2002;41:13106–13115. doi: 10.1021/bi020385t. [DOI] [PubMed] [Google Scholar]
- Cacas J.L., Buré C., Furt F., Maalouf J.P., Badoc A., Cluzet S., Schmitter J.M., Antajan E., Mongrand S. Biochemical survey of the polar head of plant glycosylinositolphosphoceramides unravels broad diversity. Phytochemistry. 2013;96:191–200. doi: 10.1016/j.phytochem.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Cacas J.L., Buré C., Grosjean K., Gerbeau-Pissot P., Lherminier J., Rombouts Y., Maes E., Bossard C., Gronnier J., Furt F. Revisiting plant plasma membrane lipids in tobacco: a focus on sphingolipids. Plant Physiol. 2016;170:367–384. doi: 10.1104/pp.15.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrel C., Vazquez T., Puyaubert J., Rezé N., Lesch M., Kaiser W.M., Dutilleul C., Guillas I., Zachowski A., Baudouin E. Nitric oxide participates in cold-responsive phosphosphingolipid formation and gene expression in Arabidopsis thaliana. New Phytol. 2011;189:415–427. doi: 10.1111/j.1469-8137.2010.03500.x. [DOI] [PubMed] [Google Scholar]
- Carmona-Salazar L., Cahoon R.E., Gasca-Pineda J., González-Solis A., Vera-Estrella R., Treviño V., Cahoon E.B., Gavilanes-Ruiz M. Plasma and vacuolar membrane sphingolipidomes: composition and insights on the role of main molecular species. Plant Physiol. 2021;186:624–639. doi: 10.1093/plphys/kiab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao D.Y., Gable K., Chen M., Baxter I., Dietrich C.R., Cahoon E.B., Guerinot M.L., Lahner B., Lü S., Markham J.E. Sphingolipids in the root play an important role in regulating the leaf ionome in Arabidopsis thaliana. Plant Cell. 2011;23:1061–1081. doi: 10.1105/tpc.110.079095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Han G., Dietrich C.R., Dunn T.M., Cahoon E.B. The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell. 2006;18:3576–3593. doi: 10.1105/tpc.105.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Markham J.E., Cahoon E.B. Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 2012;69:769–781. doi: 10.1111/j.1365-313X.2011.04829.x. [DOI] [PubMed] [Google Scholar]
- Chen M., Markham J.E., Dietrich C.R., Jaworski J.G., Cahoon E.B. Sphingolipid long-chain base hydroxylation is important for growth and regulation of sphingolipid content and composition in Arabidopsis. Plant Cell. 2008;20:1862–1878. doi: 10.1105/tpc.107.057851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras F.X., Ernst A.M., Haberkant P., Björkholm P., Lindahl E., Gönen B., Tischer C., Elofsson A., von Heijne G., Thiele C. Molecular recognition of a single sphingolipid species by a protein's transmembrane domain. Nature. 2012;481:525–529. doi: 10.1038/nature10742. [DOI] [PubMed] [Google Scholar]
- Coursol S., Fan L.M., Le S.H., Spiegel S., Gilroy S., Assmann S.M. Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature. 2003;423:651–654. doi: 10.1038/nature01643. [DOI] [PubMed] [Google Scholar]
- Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K.F., Balla T., Mannella C.A., Hajnóczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 2006;174:915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czubowicz K., Jęśko H., Wencel P., Lukiw W.J., Strosznajder R.P. The role of ceramide and sphingosine-1-phosphate in Alzheimer's disease and other neurodegenerative disorders. Mol. Neurobiol. 2019;56:5436–5455. doi: 10.1007/s12035-018-1448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai G.Y., Yin J., Li K.E., Chen D.K., Liu Z., Bi F.C., Rong C., Yao N. The Arabidopsis AtGCD3 protein is a glucosylceramidase that preferentially hydrolyzes long-acyl-chain glucosylceramides. J. Biol. Chem. 2020;295:717–728. doi: 10.1074/jbc.RA119.011274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo G., Polishchuk E., Di Tullio G., Santoro M., Di Campli A., Godi A., West G., Bielawski J., Chuang C.C., van der Spoel A.C. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- Davis J.A., Pares R.B., Palmgren M., López-Marqués R.L., Harper J.F. A potential pathway for flippase-facilitated glucosylceramide catabolism in plants. Plant Signal Behav. 2020;15:1783486. doi: 10.1080/15592324.2020.1783486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Castro Levatti E.V., Toledo M.S., Watanabe Costa R., Bahia D., Mortara R.A., Takahashi H.K., Straus A.H. Leishmania (Viannia) braziliensis inositol phosphorylceramide: distinctive sphingoid base composition. Front Microbiol. 2017;8:1453. doi: 10.3389/fmicb.2017.01453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C.R., Han G., Chen M., Berg R.H., Dunn T.M., Cahoon E.B. Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J. 2008;54:284–298. doi: 10.1111/j.1365-313X.2008.03420.x. [DOI] [PubMed] [Google Scholar]
- Dunn T.M., Lynch D.V., Michaelson L.V., Napier J.A. A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann. Bot. 2004;93:483–497. doi: 10.1093/aob/mch071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert B., Rautengarten C., McFarlane H.E., Rupasinghe T., Zeng W., Ford K., Scheller H.V., Bacic A., Roessner U., Persson S. A Golgi UDP-GlcNAc transporter delivers substrates for N-linked glycans and sphingolipids. Nat. Plants. 2018;4:792–801. doi: 10.1038/s41477-018-0235-5. [DOI] [PubMed] [Google Scholar]
- Eggeling C., Ringemann C., Medda R., Schwarzmann G., Sandhoff K., Polyakova S., Belov V.N., Hein B., von Middendorff C., Schönle A. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- Fang L., Ishikawa T., Rennie E.A., Murawska G.M., Lao J., Yan J., Tsai A.Y., Baidoo E.E., Xu J., Keasling J.D. Loss of inositol phosphorylceramide sphingolipid mannosylation induces plant immune responses and reduces cellulose content in Arabidopsis. Plant Cell. 2016;28:2991–3004. doi: 10.1105/tpc.16.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M.J., Poirier B.C., Lange B.M. Misexpression of the Niemann-Pick disease type C1 (NPC1)-like protein in Arabidopsis causes sphingolipid accumulation and reproductive defects. Planta. 2015;242:921–933. doi: 10.1007/s00425-015-2322-4. [DOI] [PubMed] [Google Scholar]
- Fouillen L., Maneta-Peyret L., Moreau P. ER membrane lipid composition and metabolism: lipidomic analysis. Methods Mol. Biol. 2018;1691:125–137. doi: 10.1007/978-1-4939-7389-7_10. [DOI] [PubMed] [Google Scholar]
- Gault C.R., Obeid L.M., Hannun Y.A. An overview of sphingolipid metabolism: from synthesis to breakdown. Adv. Exp. Med. Biol. 2010;688:1–23. doi: 10.1007/978-1-4419-6741-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giussani P., Brioschi L., Bassi R., Riboni L., Viani P. Phosphatidylinositol 3-kinase/AKT pathway regulates the endoplasmic reticulum to golgi traffic of ceramide in glioma cells: a link between lipid signaling pathways involved in the control of cell survival. J. Biol. Chem. 2009;284:5088–5096. doi: 10.1074/jbc.M808934200. [DOI] [PubMed] [Google Scholar]
- Gkantiragas I., Brügger B., Stüven E., Kaloyanova D., Li X.Y., Löhr K., Lottspeich F., Wieland F.T., Helms J.B. Sphingomyelin-enriched microdomains at the Golgi complex. Mol. Biol. Cell. 2001;12:1819–1833. doi: 10.1091/mbc.12.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goñi F.M., Alonso A. Biophysics of sphingolipids I. Membrane properties of sphingosine, ceramides and other simple sphingolipids. Biochim. Biophys. Acta. 2006;1758:1902–1921. doi: 10.1016/j.bbamem.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Grison M.S., Brocard L., Fouillen L., Nicolas W., Wewer V., Dörmann P., Nacir H., Benitez-Alfonso Y., Claverol S., Germain V. Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell. 2015;27:1228–1250. doi: 10.1105/tpc.114.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S. The ER-mitochondria interface: the social network of cell death. Biochim. Biophys. Acta. 2012;1823:327–334. doi: 10.1016/j.bbamcr.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Grosjean K., Mongrand S., Beney L., Simon-Plas F., Gerbeau-Pissot P. Differential effect of plant lipids on membrane organization: specificities of phytosphingolipids and phytosterols. J. Biol. Chem. 2015;290:5810–5825. doi: 10.1074/jbc.M114.598805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Wang X. Crosstalk between phospholipase D and sphingosine kinase in plant stress signaling. Front Plant Sci. 2012;3:51. doi: 10.3389/fpls.2012.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T., Rockey D.D., Heinzen R.A., Scidmore M.A. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Kumagai K., Tomishige N., Kawano M. CERT and intracellular trafficking of ceramide. Biochim. Biophys. Acta. 2007;1771:644–653. doi: 10.1016/j.bbalip.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- Hannich J.T., Umebayashi K., Riezman H. Distribution and functions of sterols and sphingolipids. Cold Spring Harb Perspect. Biol. 2011;3:a004762. doi: 10.1101/cshperspect.a004762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun Y.A., Obeid L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018;19:175–191. doi: 10.1038/nrm.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasi R.Y., Miyagi M., Morito K., Ishikawa T., Kawai-Yamada M., Imai H., Fukuta T., Kogure K., Kanemaru K., Hayashi J. Glycosylinositol phosphoceramide-specific phospholipase D activity catalyzes transphosphatidylation. J. Biochem. 2019;166:441–448. doi: 10.1093/jb/mvz056. [DOI] [PubMed] [Google Scholar]
- Hasi R.Y., Majima D., Morito K., Ali H., Kogure K., Nanjundan M., Hayashi J., Kawakami R., Kanemaru K., Tanaka T. Isolation of glycosylinositol phosphoceramide and phytoceramide 1-phosphate in plants and their chemical stabilities. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2020;1152:122213. doi: 10.1016/j.jchromb.2020.122213. [DOI] [PubMed] [Google Scholar]
- Holthuis J.C., Levine T.P. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 2005;6:209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- Huang D., Sun Y., Ma Z., Ke M., Cui Y., Chen Z., Chen C., Ji C., Tran T.M., Yang L. Salicylic acid-mediated plasmodesmal closure via Remorin-dependent lipid organization. Proc. Natl. Acad. Sci. U S A. 2019;116:21274–21284. doi: 10.1073/pnas.1911892116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlock A.K., Roston R.L., Wang K., Benning C. Lipid trafficking in plant cells. Traffic. 2014;15:915–932. doi: 10.1111/tra.12187. [DOI] [PubMed] [Google Scholar]
- Imai H., Nishiura H. Phosphorylation of sphingoid long-chain bases in Arabidopsis: functional characterization and expression of the first sphingoid long-chain base Kinase gene in plants. Plant Cell Physiol. 2005;46:375–380. doi: 10.1093/pcp/pci023. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Ito Y., Kawai-Yamada M. Molecular characterization and targeted quantitative profiling of the sphingolipidome in rice. Plant J. 2016;88(4):681–693. doi: 10.1111/tpj.13281. [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Fang L., Rennie E.A., Sechet J., Yan J., Jing B., Moore W., Cahoon E.B., Scheller H.V., Kawai-Yamada M. GLUCOSAMINE INOSITOLPHOSPHORYLCERAMIDE TRANSFERASE1 (GINT1) is a GlcNAc-containing glycosylinositol phosphorylceramide glycosyltransferase. Plant Physiol. 2018;177:938–952. doi: 10.1104/pp.18.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Zhou X., Tao M., Yuan F., Liu L., Wu F., Wu X., Xiang Y., Niu Y., Liu F. Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature. 2019;572:341–346. doi: 10.1038/s41586-019-1449-z. [DOI] [PubMed] [Google Scholar]
- Jing B., Ishikawa T., Soltis N., Inada N., Liang Y., Murawska G., Fang L., Andeberhan F., Pidatala R., Yu X. The Arabidopsis thaliana nucleotide sugar transporter GONST2 is a functional homolog of GONST1. Plant Direct. 2021;5:e00309. doi: 10.1002/pld3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara K., Watanabe R., Pichler H., Ihara K., Murakami S., Riezman H., Funato K. Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol. Biol. Cell. 2008;19:2069–2082. doi: 10.1091/mbc.E07-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson K.A. On the chemistry and occurrence of sphingolipid long-chain bases. Chem. Phys. Lipids. 1970;5:6–43. doi: 10.1016/0009-3084(70)90008-3. [DOI] [PubMed] [Google Scholar]
- Kaul K., Lester R.L. Characterization of inositol-containing phosphosphingolipids from tobacco leaves: isolation and identification of two novel, major lipids: N-acetylglucosamidoglucuronidoinositol phosphorylceramide and glucosamidoglucuronidoinositol phosphorylceramide. Plant Physiol. 1975;55:120–129. doi: 10.1104/pp.55.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenoth R., Brown R.E., Kamlekar R.K. In vitro measurement of sphingolipid intermembrane transport illustrated by GLTP superfamily members. Methods Mol. Biol. 2019;1949:237–256. doi: 10.1007/978-1-4939-9136-5_17. [DOI] [PubMed] [Google Scholar]
- Kimberlin A.N., Majumder S., Han G., Chen M., Cahoon R.E., Stone J.M., Dunn T.M., Cahoon E.B. Arabidopsis 56-amino acid serine palmitoyltransferase-interacting proteins stimulate sphingolipid synthesis, are essential, and affect mycotoxin sensitivity. Plant Cell. 2013;25:4627–4639. doi: 10.1105/tpc.113.116145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberlin A.N., Han G., Luttgeharm K.D., Chen M., Cahoon R.E., Stone J.M., Markham J.E., Dunn T.M., Cahoon E.B. ORM expression alters sphingolipid homeostasis and differentially affects ceramide synthase activity. Plant Physiol. 2016;172:889–900. doi: 10.1104/pp.16.00965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klitzing H.A., Weber P.K., Kraft M.L. Secondary ion mass spectrometry imaging of biological membranes at high spatial resolution. Methods Mol. Biol. 2013;950:483–501. doi: 10.1007/978-1-62703-137-0_26. [DOI] [PubMed] [Google Scholar]
- Komura N., Suzuki K.G., Ando H., Konishi M., Koikeda M., Imamura A., Chadda R., Fujiwara T.K., Tsuboi H., Sheng R. Raft-based interactions of gangliosides with a GPI-anchored receptor. Nat. Chem. Biol. 2016;12:402–410. doi: 10.1038/nchembio.2059. [DOI] [PubMed] [Google Scholar]
- König S., Feussner K., Schwarz M., Kaever A., Iven T., Landesfeind M., Ternes P., Karlovsky P., Lipka V., Feussner I. Arabidopsis mutants of sphingolipid fatty acid α-hydroxylases accumulate ceramides and salicylates. New Phytol. 2012;196:1086–1097. doi: 10.1111/j.1469-8137.2012.04351.x. [DOI] [PubMed] [Google Scholar]
- Kusumi A., Suzuki K. Toward understanding the dynamics of membrane-raft-based molecular interactions. Biochim. Biophys. Acta. 2005;1746:234–251. doi: 10.1016/j.bbamcr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Lachaud C., Da Silva.D., Amelot N., Béziat C., Brière C., Cotelle V., Graziana A., Grat S., Mazars C., Thuleau P. Dihydrosphingosine-induced programmed cell death in tobacco BY-2 cells is independent of H₂O₂ production. Mol. Plant. 2011;4:310–318. doi: 10.1093/mp/ssq077. [DOI] [PubMed] [Google Scholar]
- Laloi M., Perret A.M., Chatre L., Melser S., Cantrel C., Vaultier M.N., Zachowski A., Bathany K., Schmitter J.M., Vallet M. Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiol. 2007;143:461–472. doi: 10.1104/pp.106.091496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand A., Martinez D., Grélard A., Berbon M., Morvan E., Tawani A., Loquet A., Mongrand S., Habenstein B. Nanodomain clustering of the plant protein Remorin by solid-state NMR. Front Mol. Biosci. 2019;6:107. doi: 10.3389/fmolb.2019.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipelt M., Warnecke D., Zähringer U., Ott C., Müller F., Hube B., Heinz E. Glucosylceramide synthases, a gene family responsible for the biosynthesis of glucosphingolipids in animals, plants, and fungi. J. Biol. Chem. 2001;276:33621–33629. doi: 10.1074/jbc.M104952200. [DOI] [PubMed] [Google Scholar]
- Lenarčič T., Albert I., Böhm H., Hodnik V., Pirc K., Zavec A.B., Podobnik M., Pahovnik D., Žagar E., Pruitt R. Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science. 2017;358:1431–1434. doi: 10.1126/science.aan6874. [DOI] [PubMed] [Google Scholar]
- Levi M., Meijler M.M., Gómez-Muñoz A., Zor T. Distinct receptor-mediated activities in macrophages for natural ceramide-1-phosphate (C1P) and for phospho-ceramide analogue-1 (PCERA-1) Mol. Cell Endocrinol. 2010;314:248–255. doi: 10.1016/j.mce.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Li J., Bi F.C., Yin J., Wu J.X., Rong C., Wu J.L., Yao N. An Arabidopsis neutral ceramidase mutant ncer1 accumulates hydroxyceramides and is sensitive to oxidative stress. Front Plant Sci. 2015;6:460. doi: 10.3389/fpls.2015.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Halitschke R., Li D., Paetz C., Su H., Heiling S., Xu S., Baldwin I.T. Controlled hydroxylations of diterpenoids allow for plant chemical defense without autotoxicity. Science. 2021;371:255–260. doi: 10.1126/science.abe4713. [DOI] [PubMed] [Google Scholar]
- Liang H., Yao N., Song J.T., Luo S., Lu H., Greenberg J.T. Ceramides modulate programmed cell death in plants. Genes Dev. 2003;17:2636–2641. doi: 10.1101/gad.1140503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., London E. Ordered raft domains induced by outer leaflet sphingomyelin in cholesterol-rich asymmetric vesicles. Biophys. J. 2015;108:2212–2222. doi: 10.1016/j.bpj.2015.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N.J., Zhang T., Liu Z.H., Chen X., Guo H.S., Ju B.H., Zhang Y.Y., Li G.Z., Zhou Q.H., Qin Y.M. Phytosphinganine affects plasmodesmata permeability via facilitating PDLP5-stimulated callose accumulation in Arabidopsis. Mol. Plant. 2020;13:128–143. doi: 10.1016/j.molp.2019.10.013. [DOI] [PubMed] [Google Scholar]
- Liu N.J., Wang N., Bao J.J., Zhu H.X., Wang L.J., Chen X.Y. Lipidomic analysis reveals the importance of GIPCs in Arabidopsis leaf extracellular vesicles. Mol. Plant. 2020;13:1523–1532. doi: 10.1016/j.molp.2020.07.016. [DOI] [PubMed] [Google Scholar]
- Luttgeharm K.D., Kimberlin A.N., Cahoon E.B. Plant sphingolipid metabolism and function. Subcell. Biochem. 2016;86:249–286. doi: 10.1007/978-3-319-25979-6_11. [DOI] [PubMed] [Google Scholar]
- Magnin-Robert M., Le B.D., Markham J., Dorey S., Clément C., Baillieul F., Dhondt-Cordelier S. Modifications of sphingolipid content affect tolerance to hemibiotrophic and necrotrophic pathogens by modulating plant defense responses in Arabidopsis. Plant Physiol. 2015;169:2255–2274. doi: 10.1104/pp.15.01126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makiyama T., Nakamura H., Nagasaka N., Yamashita H., Honda T., Yamaguchi N., Nishida A., Murayama T. Trafficking of acetyl-C16-ceramide-NBD with long-term stability and no cytotoxicity into the golgi complex. Traffic. 2015;16:476–492. doi: 10.1111/tra.12265. [DOI] [PubMed] [Google Scholar]
- Mandon E.C., Ehses I., Rother J., van Echten G., Sandhoff K. Subcellular localization and membrane topology of serine palmitoyltransferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver. J. Biol. Chem. 1992;267:11144–11148. [PubMed] [Google Scholar]
- Mao C., Obeid L.M. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim. Biophys. Acta. 2008;1781:424–434. doi: 10.1016/j.bbalip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham J.E., Li J., Cahoon E.B., Jaworski J.G. Separation and identification of major plant sphingolipid classes from leaves. J. Biol. Chem. 2006;281:22684–22694. doi: 10.1074/jbc.M604050200. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Jaworski J.G. Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007;21:1304–1314. doi: 10.1002/rcm.2962. [DOI] [PubMed] [Google Scholar]
- Markham J.E., Molino D., Gissot L., Bellec Y., Hématy K., Marion J., Belcram K., Palauqui J.C., Satiat-Jeunemaître B., Faure J.D. Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell. 2011;23:2362–2378. doi: 10.1105/tpc.110.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham J.E., Lynch D.V., Napier J.A., Dunn T.M., Cahoon E.B. Plant sphingolipids: function follows form. Curr. Opin. Plant Biol. 2013;16:350–357. doi: 10.1016/j.pbi.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Mauri L., Sonnino S., Prinetti A. Chemical and physicochemical properties of gangliosides. Methods Mol. Biol. 2018;1804:1–17. doi: 10.1007/978-1-4939-8552-4_1. [DOI] [PubMed] [Google Scholar]
- Melser S., Batailler B., Peypelut M., Poujol C., Bellec Y., Wattelet-Boyer V., Maneta-Peyret L., Faure J.D., Moreau P. Glucosylceramide biosynthesis is involved in Golgi morphology and protein secretion in plant cells. Traffic. 2010;11:479–490. doi: 10.1111/j.1600-0854.2009.01030.x. [DOI] [PubMed] [Google Scholar]
- Merrill A.H. J,r., Stokes T.H., Momin A., Park H., Portz B.J., Kelly S., Wang E., Sullards M.C., Wang M.D. Sphingolipidomics: a valuable tool for understanding the roles of sphingolipids in biology and disease. J. Lipid Res. 2009;50:97–102. doi: 10.1194/jlr.R800073-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson L.V., Zäuner S., Markham J.E., Haslam R.P., Desikan R., Mugford S., Albrecht S., Warnecke D., Sperling P., Heinz E. Functional characterization of a higher plant sphingolipid Delta4-desaturase: defining the role of sphingosine and sphingosine-1-phosphate in Arabidopsis. Plant Physiol. 2009;149:487–498. doi: 10.1104/pp.108.129411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina J.G., Okada Y., Wansadhipathi-Kannangara N.K., Pratt S., Shams-Eldin H., Schwarz R.T., Steel P.G., Fawcett T., Denny P.W. Functional analyses of differentially expressed isoforms of the Arabidopsis inositol phosphorylceramide synthase. Plant Mol. Biol. 2010;73:399–407. doi: 10.1007/s11103-010-9626-3. [DOI] [PubMed] [Google Scholar]
- Mortimer J.C., Yu X., Albrecht S., Sicilia F., Huichalaf M., Ampuero D., Michaelson L.V., Murphy A.M., Matsunaga T., Kurz S. Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell. 2013;25:1881–1894. doi: 10.1105/tpc.113.111500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- Nagano M., Ishikawa T., Ogawa Y., Iwabuchi M., Nakasone A., Shimamoto K., Uchimiya H., Kawai-Yamada M. Arabidopsis Bax inhibitor-1 promotes sphingolipid synthesis during cold stress by interacting with ceramide-modifying enzymes. Planta. 2014;240:77–89. doi: 10.1007/s00425-014-2065-7. [DOI] [PubMed] [Google Scholar]
- Nagano M., Takahara K., Fujimoto M., Tsutsumi N., Uchimiya H., Kawai-Yamada M. Arabidopsis sphingolipid fatty acid 2-hydroxylases (AtFAH1 and AtFAH2) are functionally differentiated in fatty acid 2-hydroxylation and stress responses. Plant Physiol. 2012;159:1138–1148. doi: 10.1104/pp.112.199547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N., Kato M., Takahashi Y., Shimazaki K., Tamura K., Tokuji Y., Kihara A., Imai H. Degradation of long-chain base 1-phosphate (LCBP) in Arabidopsis: functional characterization of LCBP phosphatase involved in the dehydration stress response. J. Plant Res. 2012;125:439–449. doi: 10.1007/s10265-011-0451-9. [DOI] [PubMed] [Google Scholar]
- Ng C.K., Carr K., McAinsh M.R., Powell B., Hetherington A.M. Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature. 2001;410:596–599. doi: 10.1038/35069092. [DOI] [PubMed] [Google Scholar]
- Ngo T.N., Nguyen N.D.P., Nguyen N.T.L., Pham N.K.T., Phan N.M., Bui T.D., Dang V.S., Tran C.L., Mai D.T., Nguyen T.P. Markhasphingolipid A, new phytosphingolipid from the leaves of Markhamia stipulata var. Canaense V.S. Dang. Nat. Prod. Res. 2020;34:1820–1826. doi: 10.1080/14786419.2018.1561686. [DOI] [PubMed] [Google Scholar]
- Nishiura H., Tamura K., Morimoto Y., Imai H. Characterization of sphingolipid long-chain base kinase in Arabidopsis thaliana. Biochem. Soc. Trans. 2000;28:747–748. [PubMed] [Google Scholar]
- Norberg P., Månsson J.E., Liljenberg C. Characterization of glucosylceramide from plasma membranes of plant root cells. Biochim. Biophys. Acta. 1991;1066:257–260. doi: 10.1016/0005-2736(91)90195-e. [DOI] [PubMed] [Google Scholar]
- Paladino S., Sarnataro D., Tivodar S., Zurzolo C. Oligomerization is a specific requirement for apical sorting of glycosyl-phosphatidylinositol-anchored proteins but not for non-raft-associated apical proteins. Traffic. 2007;8:251–258. doi: 10.1111/j.1600-0854.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- Panzenboeck L., Troppmair N., Schlachter S., Koellensperger G., Hartler J., Rampler E. Chasing the major sphingolipids on earth: automated annotation of plant glycosyl inositol phospho ceramides by glycolipidomics. Metabolites. 2020;10:375. doi: 10.3390/metabo10090375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pata M.O., Wu B.X., Bielawsk i J., Xiong T.C., Hannun Y.A., Ng C.K. Molecular cloning and characterization of OsCDase, a ceramidase enzyme from rice. Plant J. 2008;55:1000–1009. doi: 10.1111/j.1365-313X.2008.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer M., Stegmann M., Mueller M.J., Waller F. Pseudomonas syringae infection triggers de novo synthesis of phytosphingosine from sphinganine in Arabidopsis thaliana. FEBS Lett. 2010;584:4053–4056. doi: 10.1016/j.febslet.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Pike L.J. Rafts defined: a report on the Keystone Symposium on lipid rafts and cell function. J. Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- Puli M.R., Rajsheel P., Aswani V., Agurla S., Kuchitsu K., Raghavendra A.S. Stomatal closure induced by phytosphingosine-1-phosphate and sphingosine-1-phosphate depends on nitric oxide and pH of guard cells in Pisum sativum. Planta. 2016;244:831–841. doi: 10.1007/s00425-016-2545-z. [DOI] [PubMed] [Google Scholar]
- Qin X., Zhang R.X., Ge S., Zhou T., Liang Y.K. Sphingosine kinase AtSPHK1 functions in fumonisin B1-triggered cell death in Arabidopsis. Plant Physiol. Biochem. 2017;119:70–80. doi: 10.1016/j.plaphy.2017.08.008. [DOI] [PubMed] [Google Scholar]
- Rennie E.A., Ebert B., Miles G.P., Cahoon R.E., Christiansen K.M., Stonebloom S., Khatab H., Twell D., Petzold C.J., Adams P.D. Identification of a sphingolipid α-glucuronosyltransferase that is essential for pollen function in Arabidopsis. Plant Cell. 2014;26:3314–3325. doi: 10.1105/tpc.114.129171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboni L., Giussani P., Viani P. Sphingolipid transport. Adv. Exp. Med. Biol. 2010;688:24–45. doi: 10.1007/978-1-4419-6741-1_2. [DOI] [PubMed] [Google Scholar]
- Saucedo-García M., Guevara-García A., González-Solís A., Cruz-García F., Vázquez-Santana S., Markham J.E., Lozano-Rosas M.G., Dietrich C.R., Ramos-Vega M., Cahoon E.B. MPK6, sphinganine and the LCB2a gene from serine palmitoyltransferase are required in the signaling pathway that mediates cell death induced by long chain bases in Arabidopsis. New Phytol. 2011;191:943–957. doi: 10.1111/j.1469-8137.2011.03727.x. [DOI] [PubMed] [Google Scholar]
- Schulze H., Sandhoff K. Sphingolipids and lysosomal pathologies. Biochim. Biophys. Acta. 2014;1841:799–810. doi: 10.1016/j.bbalip.2013.10.015. [DOI] [PubMed] [Google Scholar]
- Sheets E.D., Lee G.M., Simson R., Jacobson K. Transient confinement of a glycosylphosphatidylinositol-anchored protein in the plasma membrane. Biochemistry. 1997;36:12449–12458. doi: 10.1021/bi9710939. [DOI] [PubMed] [Google Scholar]
- Shi L., Bielawski J., Mu J., Dong H., Teng C., Zhang J., Yang X., Tomishige N., Hanada K., Hannun Y.A. Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 2007;17:1030–1040. doi: 10.1038/cr.2007.100. [DOI] [PubMed] [Google Scholar]
- Simon-Plas F., Perraki A., Bayer E., Gerbeau-Pissot P., Mongrand S. An update on plant membrane rafts. Curr. Opin. Plant Biol. 2011;14:642–649. doi: 10.1016/j.pbi.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Simons K., van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Siskind L.J. Mitochondrial ceramide and the induction of apoptosis. J. Bioenerg. Biomembr. 2005;37:143–153. doi: 10.1007/s10863-005-6567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D.C., Lord J.M., Roberts L.M., Johannes L. Glycosphingolipids as toxin receptors. Semin. Cell Dev. Biol. 2004;15:397–408. doi: 10.1016/j.semcdb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Sperling P., Franke S., Lüthje S., Heinz E. Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol. Biochem. 2005;43:1031–1038. doi: 10.1016/j.plaphy.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Sperling P., Heinz E. Plant sphingolipids: structural diversity, biosynthesis, first genes and functions. Biochim. Biophys. Acta. 2003;1632:1–15. doi: 10.1016/s1388-1981(03)00033-7. [DOI] [PubMed] [Google Scholar]
- Sperling P., Ternes P., Moll H., Franke S., Zähringer U., Heinz E. Functional characterization of sphingolipid C4-hydroxylase genes from Arabidopsis thaliana. FEBS Lett. 2001;494:90–94. doi: 10.1016/s0014-5793(01)02332-8. [DOI] [PubMed] [Google Scholar]
- Subei A.M., Cohen J.A. Sphingosine 1-phosphate receptor modulators in multiple sclerosis. CNS Drugs. 2015;29:565–575. doi: 10.1007/s40263-015-0261-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner W., Malinsky J., Opekarová M. In plant and animal cells, detergent-resistant membranes do not define functional membrane rafts. Plant Cell. 2011;23:1191–1193. doi: 10.1105/tpc.111.086249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazona P., Feussner K., Feussner I. An enhanced plant lipidomics method based on multiplexed liquid chromatography-mass spectrometry reveals additional insights into cold- and drought-induced membrane remodeling. Plant J. 2015;84:621–633. doi: 10.1111/tpj.13013. [DOI] [PubMed] [Google Scholar]
- Ternes P., Feussner K., Werner S., Lerche J., Iven T., Heilmann I., Riezman H., Feussner I. Disruption of the ceramide synthase LOH1 causes spontaneous cell death in Arabidopsis thaliana. New Phytol. 2011;192:841–854. doi: 10.1111/j.1469-8137.2011.03852.x. [DOI] [PubMed] [Google Scholar]
- Thudichum J.L.W. Baillière, Tindall and Cox; London: 1884. A Treatise on the Chemical Constitution of the Brain. [Google Scholar]
- Tian S., Muneeruddin K., Choi M.Y., Tao L., Bhuiyan R.H., Ohmi Y., Furukawa K., Furukawa K., Boland S., Shaffer S.A. Genome-wide CRISPR screens for Shiga toxins and ricin reveal Golgi proteins critical for glycosylation. PLoS Biol. 2018;16:e2006951. doi: 10.1371/journal.pbio.2006951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellström H., Hellgren L.I., Wieslander A., Sandelius A.S. Lipid asymmetry in plant plasma membranes: phosphate deficiency-induced phospholipid replacement is restricted to the cytosolic leaflet. FASEB J. 2010;24:1128–1138. doi: 10.1096/fj.09-139410. [DOI] [PubMed] [Google Scholar]
- van Helvoort A., Giudici M.L., Thielemans M., van Meer G. Transport of sphingomyelin to the cell surface is inhibited by brefeldin A and in mitosis, where C6-NBD-sphingomyelin is translocated across the plasma membrane by a multidrug transporter activity. J. Cell Sci. 1997;110:75–83. doi: 10.1242/jcs.110.1.75. [DOI] [PubMed] [Google Scholar]
- van Weely S., van den Berg M., Barranger J.A., Sa Miranda M.C., Tager J.M., Aerts J.M. Role of pH in determining the cell-type-specific residual activity of glucocerebrosidase in type 1 Gaucher disease. J. Clin. Invest. 1993;91:1167–1175. doi: 10.1172/JCI116276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoek B., Haas R., Wrage K., Linscheid M., Heinz E. Lipids and enzymatic-activities in vacuolar membranes isolated via protoplasts from oat primary leaves. Z. Naturforsch. 1983;38:770–777. [Google Scholar]
- Vu H.S., Shiva S., Roth M.R., Tamura P., Zheng L., Li M., Sarowar S., Honey S., McEllhiney D., Hinkes P. Lipid changes after leaf wounding in Arabidopsis thaliana: expanded lipidomic data form the basis for lipid co-occurrence analysis. Plant J. 2014;80:728–743. doi: 10.1111/tpj.12659. [DOI] [PubMed] [Google Scholar]
- Wang Y.M., Wu A.Z., Zhong Y., Peng G.T., Fan Q., Zhu C.C., Zhang C.X. New glycosylsphingolipids from Psychotria serpens L. Nat. Prod. Res. 2020;34:2095–2100. doi: 10.1080/14786419.2019.1574789. [DOI] [PubMed] [Google Scholar]
- Wang L., Liu C., Liu Y., Luo M. Fumonisin B1-induced changes in cotton fiber elongation revealed by sphingolipidomics and proteomics. Biomolecules. 2020;10:1258. doi: 10.3390/biom10091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke D., Heinz E. Recently discovered functions of glucosylceramides in plants and fungi. Cell Mol. Life Sci. 2003;60:919–941. doi: 10.1007/s00018-003-2243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattelet-Boyer V., Brocard L., Jonsson K. Enrichment of hydroxylated C24- and C26-acyl-chain sphingolipids mediates PIN2 apical sorting at trans-Golgi network subdomains. Nat Commun. 2016;7:12788. doi: 10.1038/ncomms12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G., Viitanen L., Alm C., Mattjus P., Salminen T.A., Edqvist J. Identification of a glycosphingolipid transfer protein GLTP1 in Arabidopsis thaliana. FEBS J. 2008;275:3421–3437. doi: 10.1111/j.1742-4658.2008.06498.x. [DOI] [PubMed] [Google Scholar]
- Wu J.X., Li J., Liu Z., Yin J., Chang Z.Y., Rong C., Wu J.L., Bi F.C., Yao N. The Arabidopsis ceramidase AtACER functions in disease resistance and salt tolerance. Plant J. 2015;81:767–780. doi: 10.1111/tpj.12769. [DOI] [PubMed] [Google Scholar]
- Yan D., Yadav S.R., Paterlini A., Nicolas W.J., Petit J.D., Brocard L., Belevich I., Grison M.S., Vaten A., Karami L. Sphingolipid biosynthesis modulates plasmodesmal ultrastructure and phloem unloading. Nat. Plants. 2019;5:604–615. doi: 10.1038/s41477-019-0429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Li M., Phillips A., Li L., Ali U., Li Q., Lu S., Hong Y., Wang X., Guo L. Nonspecific phospholipase C4 hydrolyzes phosphosphingolipids and sustains plant root growth during phosphate deficiency. Plant Cell. 2021;33:766–780. doi: 10.1093/plcell/koaa054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachowski A. Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem. J. 1993;294(Pt 1):1–14. doi: 10.1042/bj2940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zienkiewicz A., Gömann J., König S., Herrfurth C., Liu Y.T., Meldau D., Feussner I. Disruption of Arabidopsis neutral ceramidases 1 and 2 results in specific sphingolipid imbalances triggering different phytohormone-dependent plant cell death programmes. New Phytol. 2020;226:170–188. doi: 10.1111/nph.16336. [DOI] [PubMed] [Google Scholar]
- Zhu G., Yin N., Luo Q., Liu J., Chen X., Liu L., Wu J. Enhancement of sphingolipid synthesis improves osmotic tolerance of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2020;86:e02911–e02919. doi: 10.1128/AEM.02911-19. [DOI] [PMC free article] [PubMed] [Google Scholar]