Abstract
Telomeric DNA is maintained within a length range characteristic of an organism or cell type. Significant deviations outside this range are associated with altered telomere function. The yeast telomere-binding protein Rap1p negatively regulates telomere length. Telomere elongation is responsive to both the number of Rap1p molecules bound to a telomere and the Rap1p-centered DNA-protein complex at the extreme telomeric end. Previously, we showed that a specific trinucleotide substitution in the Saccharomyces cerevisiae telomerase gene (TLC1) RNA template abolished the enzymatic activity of telomerase, causing the same cell senescence and telomere shortening phenotypes as a complete tlc1 deletion. Here we analyze effects of six single- and double-base changes within these same three positions. All six mutant telomerases had in vitro enzymatic activity levels similar to the wild-type levels. The base changes predicted from the mutations all disrupted Rap1p binding in vitro to the corresponding duplex DNAs. However, they caused two classes of effects on telomere homeostasis: (i) rapid, RAD52-independent telomere lengthening and poor length regulation, whose severity correlated with the decrease in in vitro Rap1p binding affinity (this is consistent with loss of negative regulation of telomerase action at these telomeres; and (ii) telomere shortening that, depending on the template mutation, either established a new short telomere set length with normal cell growth or was progressive and led to cellular senescence. Hence, disrupting Rap1p binding at the telomeric terminus is not sufficient to deregulate telomere elongation. This provides further evidence that both positive and negative cis-acting regulators of telomerase act at telomeres.
Telomeres, the heterochromatic structures present at the ends of linear chromosomes, serve many functions: they differentiate chromosome ends from broken DNA ends, protecting them from nucleases and recombinases, they help position chromosomes within the nucleus, and they serve as a reservoir of replenishable DNA added de novo to counteract the incomplete replication of chromosome ends by DNA polymerases (reviewed in references 3 and 44). Disruptions in telomere sequence, length, or structure can alter the properties of telomeres, resulting in loss of telomere length regulation, mislocalization of chromosomes within the nucleus, telomere-telomere fusions, and chromosome instability (2, 9, 14, 21, 22, 40).
The length of the telomeric DNA tract, while somewhat variable, is maintained within a range that is characteristic of a given species, cell type, or growth condition (reviewed in reference 13). Telomeric DNA is subject both to lengthening activities, such as recombination and the addition of new DNA by telomerase, and shortening activities, such as incomplete replication and nucleolytic degradation (13). In the yeast Saccharomyces cerevisiae, telomeres typically contain about 300 ± 100 bp of (TG1–3/C1–3A) DNA. However, clonal isolates of S. cerevisiae initially exhibit telomeres within a much tighter size range (35). The full range of telomere lengths is attained only after extended growth in culture and is then maintained for the telomere population. These findings suggested that the lengthening and shortening activities that act on telomeres do so gradually and are closely balanced with each other (35), creating a situation of telomere length homeostasis.
The importance of maintaining telomeres within a given size range becomes evident when cells are grown in the absence of telomerase; telomeres shorten with each round of DNA replication, losing from 4 to 5 bp/generation in yeast to about 50 bp/generation in mammalian cells (4, 37). While no obvious detrimental effects are caused by the initial telomere shortening, eventually the shortening telomeres cease to function to stabilize chromosome ends (15, 27). However, such critically short telomeres still contain substantial tracts of telomeric repeat DNA (15). Furthermore, in both yeast and human cells lacking a functional telomerase, telomeres become critically short earlier (and at a longer length) than in cells containing telomerase (34, 45). These observations suggested that telomerase itself protects short telomeres. Thus, whether a telomere is critically short depends not only on its number of telomeric DNA repeats but also on the status of other components of the telomere, including telomerase.
The proteins that have been implicated in telomere length homeostasis include telomere-associated proteins and proteins involved in DNA replication, recombination, and repair (1, 5, 6, 12). However, little is known about the roles of most of these proteins in regulating telomere length. Mounting evidence suggests that replication of the G-rich strand of telomeres by telomerase is coordinately regulated with replication of the C-rich strand by conventional DNA polymerases (1, 10). In the ciliate Euplotes, inhibition of telomeric C-strand synthesis with aphidicolin results in increased telomerase-mediated G-rich strand synthesis (10). We have previously shown that telomerase is a dimeric (or oligomeric) enzyme complex that remains stably bound to telomeric DNA following polymerization (33). This observation raises the possibility that as the dimeric replicative DNA polymerase complex proceeds toward the end of the chromosome, it may directly contact the dimeric telomerase complex. Such contact may play a role in the observed coordinate regulation of C- and G-strand syntheses (10).
Here we focus on the effects of synthesizing telomere sequences that disrupt binding to the protein Rap1p. This abundant protein binds tightly to duplex telomeric DNA (9) and is the best characterized of the proteins involved in telomere structure and maintenance. In addition to its role at the telomeres, Rap1p binds to promoter elements of many genes to activate transcription and binds to the silent mating type loci, where it acts to repress transcription (36). Certain mutations in Rap1p lead to lengthening of telomeres (21–24, 29, 36). However, elucidating the role of Rap1p at the telomere has been difficult, since mutations that compromise the telomere-specific function of Rap1p may also affect the other essential cellular functions of Rap1p. In contrast, mutations in the template sequence of the RNA subunit of telomerase can be used to synthesize telomeric repeats with altered binding to Rap1p, thereby reducing or altering the binding of Rap1p to the newly synthesized repeats without interfering with its other roles in the cell. In the budding yeast Kluyveromyces lactis, such mutations cause various degrees of telomere lengthening (21, 22, 31).
In this study, we analyzed a cluster of novel mutations made in the template domain of the S. cerevisiae TLC1 gene. We focused on RNA residues that, when incorporated into telomeric DNA, alter the core consensus Rap1p binding site. We show, for the first time in S. cerevisiae, that mutating the telomeric Rap1p binding site can lead to stochastic telomere lengthening, which affects a steadily increasing subpopulation of telomeres until all have been elongated. Telomere lengthening is rapid and RAD52 independent, and it does not correlate with increased core activity of telomerase in vitro, strongly suggesting that rapid elongation results from derepression of the action of telomerase at the telomere. However, for other specific template mutations, disrupting the Rap1p binding site is not sufficient for telomere lengthening, and instead telomeres shorten. These findings provide further evidence that telomere elongation involves both derepression (loss of Rap1p-mediated repression) and activation by one or more cis-acting positive regulators of telomere addition (6, 32, 41), whose functions can be disrupted by mutations in telomeric DNA.
MATERIALS AND METHODS
TLC1 mutant allele construction.
The 212-bp mutant template fragments, generated by two-step overlap PCR (16), were subcloned into pRS313TLC1 at NcoI/HpaI sites. PCR-amplified DNAs were sequenced and shown to contain only the intended mutations.
Strain construction.
YJP116 was generated by disrupting one allele of TLC1 with TRP1 and one allele of RAD52 with LEU2 from the diploid strain YJL537 (gift from Joachim Li). Following transformation with pRS316 containing the wild-type TLC1 gene, including 614 bp of upstream and 222 bp of downstream flanking sequence, the strain was sporulated and Trp+, Leu+, and Ura+ spores were isolated. The resulting strain, strain YJP153, was transformed with pRS313 plasmids containing either TLC1 or the various mutant alleles of TLC1, and loss of the wild-type allele (pRS316-TLC1) was selected on 5-fluoro-orotic acid (5-FOA) medium.
Telomere cloning and sequencing.
Genomic DNA prepared from tlc1-476gCA cells was directly ligated to pBST SK− plasmid DNA that had been digested with EcoRV and XmaI and then further digested with XbaI before being religated and transformed into Escherichia coli Electro-MAX cells. Clones containing telomeric DNA were identified by hybridization to a 32P-labeled telomeric oligonucleotide, and the insert DNA was sequenced. Telomeric DNA was sequenced by the dideoxy-chain termination sequencing method.
Rap1p binding assays.
Competition gel shift assays were carried out as described previously (34).
Southern blotting.
Haploid strains containing the various TLC1 alleles on a pRS313 plasmid were either picked directly from 5-FOA plates lacking histidine and grown overnight in liquid medium lacking histidine (25 generations growth) or restreaked on 5-FOA plates lacking histidine before being grown in liquid medium lacking histidine (35, 45, 55, and 65 generations of growth). Genomic DNA was isolated, digested with XhoI, separated on a 1.2% agarose gel, transferred to Hybond-N+, and hybridized to a 32P-labeled oligonucleotide (5′-TGTGGTGTGTGGGTGTGGTGT-3′) as described previously (34).
Extract preparation and fractionation.
Each of the various strains were grown in 8 liters of liquid medium lacking histidine to an optical density at 600 nm of 0.3. Histidine was then added to a final concentration of 20 mg/liter, and cells were grown further to an optical density at 600 nm of 1.2. Cells were collected by centrifugation, and whole cell extracts were prepared and fractionated on DEAE-agarose as described previously (34).
In vitro telomerase reactions.
The amount of telomerase present in the active DEAE fraction from each of the various strains was assayed by dot blotting, using a 32P-labeled TLC1 gene probe. Telomerase reactions contained equal amounts of telomerase (up to 50% [vol/vol] DEAE fraction), 50 mM Tris-HCl (pH 8), 1 mM spermidine, 1 mM dithiothreitol 50 μM dGTP, 50 μM dATP, 50 μM dCTP, 7.5 μM [α-32P]dTTP (400 Ci/mmol), and 1 μM primer. The DNA primers used are shown in the legend to Fig. 1, and the reactions were carried out as described previously (33).
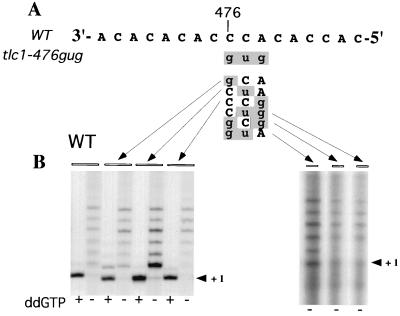
FIG. 1.
Telomerase activity assays using the template mutant enzymes. (A) Sequence of the wild-type (WT) template domain (residues 484 to 468 from the 5′ end) of the TLC1 RNA. (B) Telomerase was partially purified from wild-type cells or the various single- and double-mutant strains and assayed for telomerase activity using a telomeric oligonucleotide in the presence of [32P]dTTP and unlabeled dGTP, dATP, and dCTP (lanes marked ddG−). In lanes marked ddG+, dGTP was replaced with ddGTP. Extension products were purified and separated on a denaturing gel before being exposed to X-ray film. Primers differed only at the 3′ end and matched the sequence of the RNA template from positions 475 through 483 (GTGTGGTGTGTGCA, GTGTGGTGTGTGCG, GTGTGGTGTGTGGA, or GTGTGGTGTGTGGG).
RESULTS
In previous studies, we generated and characterized a mutant telomerase RNA (tlc1 allele) bearing a three-nucleotide substitution (tlc1-476gug [lowercase indicates mutated residues]) in the TLC1 template domain (Fig. 1A) (33, 34). This mutant enzyme is completely inactive in vitro. Like yeast strains with tlc1, est2, or est3 gene deletions (18), cells containing only the tlc1-476gug allele show progressive loss of telomeric DNA and cellular senescence (34). Such cells can be rescued by maintenance of telomeres by an alternative, RAD52-dependent telomerase-independent pathway (26). To begin to dissect the effects of the three base changes in the template in the tlc1-476gug allele, we analyzed six S. cerevisiae strains that bear substitutions in the same three nucleotides, 476 to 474, either individually or in pairs. The mutated template bases are shown in Fig. 1A.
We first measured the core enzymatic activity, at saturating primer concentrations (33), of each of the six mutant telomerases in vitro. Telomerase was partially purified from mutant cell extracts and assayed in vitro, using methods and criteria that we developed previously for various wild-type and mutant S. cerevisiae telomerases (7, 33, 34). The enzymatic properties of the six mutant tlc1 telomerases were essentially indistinguishable from wild-type properties (Fig. 1B). While the tlc1-476CuA telomerase showed slightly more activity than the other single-mutant enzymes (when normalized per telomerase RNA present in the active fractions assayed), this increase in activity was within the typical range of variation seen among different wild-type extracts. Since no one- or two-base changes caused a significant loss of activity, we conclude that the ablation of core telomerase activity by the tlc1-476gug triple-base substitution is caused by the concerted effect of all three base changes rather than by any single-base substitution.
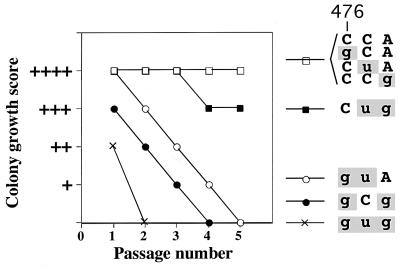
All six tlc1 template mutations were recessive, with the TLC1/tlc1 heterozygotes showing normal telomere length and cell growth (data not shown). This is similar to the results described previously for heterozygotes of five other template mutants of S. cerevisiae telomerase, including the tlc1-476gug mutant (34). Whereas the tlc1-476gug triple-base mutant in the absence of RAD52 gene function showed rapid cellular senescence (within 25 cell divisions [Fig. 2]), each of the single-mutant strains grew normally in both rad52 disrupted (Fig. 2) and RAD52 genetic backgrounds (data not shown). However, each of the three double mutated tlc1 alleles caused a specific and distinguishable effect on cell growth. While tlc1-476Cug allowed for near wild-type growth, both the tlc1-476gCg and tlc1-476guA mutations led to cellular senescence, but reproducibly more slowly than the tlc1-476gug triple mutation (Fig. 2). Together, these data suggest that of the three nucleotides at positions 476 to 474 (Fig. 1A), the wild-type C nucleotide at position 476 plays an especially important role in telomere maintenance, since all three strains that exhibited senescence (tlc1-476gug, -gCg, and -guA) contained a mutation in this position, and the only double-mutant strain that did not senesce (tlc1-476Cug) contained the wild-type C residue in this position.
FIG. 2.
Effects of TLC1 mutations on replicative potential. For each of the tlc1 mutant alleles, residues 476 to 474 are shown; mutated residues are in lowercase and highlighted in gray. Colony growth of strains bearing the indicated tlc1 allele was ranked by approximate colony diameter from + (very small irregularly shaped colonies) to ++++ (indistinguishable from wild type). Colony phenotypes were assessed following one through five restreaks after the loss of a plasmid containing the TLC1 gene.
All of the 476–474 single- and double-base substitutions disrupt Rap1p binding.
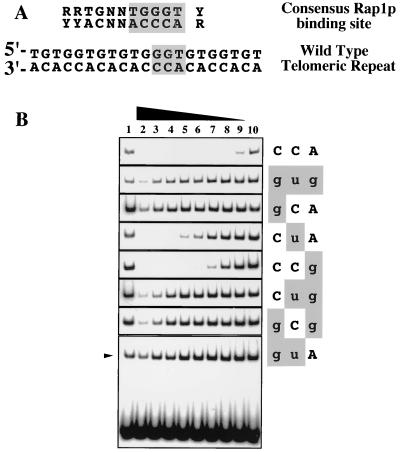
The three nucleotides mutated in telomeric DNA synthesized by tlc1-476gug telomerase lie entirely within the highly conserved core of the consensus binding site for Rap1p, the major telomere-binding protein in S. cerevisiae (Fig. 3A). We therefore assessed the relative affinities of Rap1p for duplex DNA oligonucleotides bearing each of the six 476–474 single or double mutations by gel shift competition assays (Fig. 3B). In this analysis, the affinity of Rap1p for each of the mutated oligonucleotides, relative to the wild-type telomeric sequence, was determined by quantitating the amount of radiolabeled wild-type oligonucleotide bound by Rap1p in the presence of increasing amounts of various unlabeled competitor oligonucleotides. (Rap1p binding affinity was decreased over 300-fold compared to the wild-type level by the 476gtg triple-nucleotide substitution [see also reference 34] and all three double mutations.) The single-nucleotide substitution of 476gCA also decreased Rap1p binding over 300-fold, further emphasizing the importance of position 476 in the telomerase RNA, which templates a DNA nucleotide that lies at the center of the Rap1p core binding site. The remaining two single-base mutations also reduced Rap1p binding affinity compared to the wild-type level, though to lesser extents (14-fold for 476CtA and 4-fold for 476CCg).
FIG. 3.
Effects of TLC1 mutations on Rap1p binding in vitro. (A) Sequence of the conserved (8) consensus Rap1p binding site, which includes and extends the duplex GGTGT sequence that is seen to be bound directly in a Rap1p-DNA cocrystal structure (20). The Rap1 binding consensus sequence is aligned with a typical wild-type telomeric repeat, used as the labeled probe for gel shift analyses. (B) Gel shift analyses using 32P-labeled wild-type telomeric repeat oligonucleotides, incubated with S100 whole cell extracts in the absence (lanes 1 and 10) or presence of 3,000, 1,000, 300, 100, 30, 10, 3, or 1 molar excess of unlabeled competitor oligonucleotides (lanes 2 through 9, respectively) and separated on a native gel. For all TLC1 alleles other than tlc1-476guA, only the shifted bands are shown.
Each of the 476–474 single- and double-base substitutions has a distinctive effect on telomere length.
It was shown previously that in the yeast K. lactis, three telomeric sequence mutations that disrupt Rap1p binding resulted in immediate telomere lengthening and loss of length regulation (31). In these three mutants, the severity of the lengthening phenotype correlated with the degree of loss of Rap1 binding affinity in vitro (21). However, it was not shown whether such lengthening resulted from increased telomeric recombination, increased telomerase activity, or both. We therefore assessed telomere lengths in the six strains described here. We deleted the RAD52 gene, thereby preventing any recombination-mediated telomere maintenance. Three distinct types of telomere changes were observed: telomere lengthening and loss of length regulation, stable maintenance of telomeres at a shortened mean length, and progressive telomere shortening (Fig. 4).
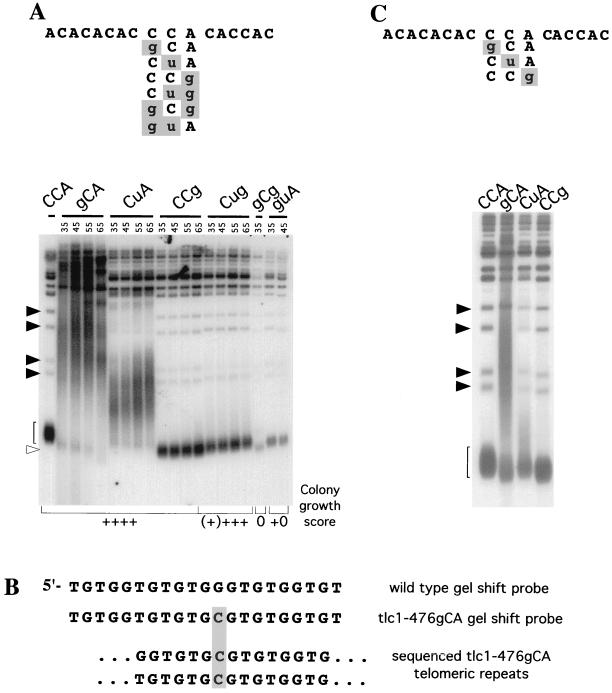
FIG. 4.
Effects of TLC1 mutations on telomere length. (A and C) Southern analysis of XhoI-digested genomic DNA hybridized to a wild-type telomeric repeat oligonucleotide. Genomic DNA was isolated from strains after 35, 45, 55, and 65 (A) or 25 (C) generations of growth in the presence of the indicated TLC1 allele. Filled arrowheads indicate non-Y′-containing telomeres; the open arrowhead and brackets indicate Y′-containing telomeres. (B) DNA sequences of one strand of the synthetic duplex DNA oligonucleotides used in the gel shift experiment shown in Fig. 3, aligned with the same strand of the tlc1-476gCA mutant gel shift probe. Sequence of two telomeric repeats subcloned from tlc1-476gCA telomeres are shown aligned below.
In the single-base tlc1-476gCA and tlc1-476CuA mutant strains, telomeres lengthened immediately. The mean telomere length in tlc1-476gCA cells was approximately 10 times greater than the wild-type length (∼3 kbp, compared to ∼300 bp for wild-type telomeres [Fig. 4A, gCA lanes]). The telomeres in the tlc1-476CuA mutant were also heterogeneous and an average of ∼1 kbp longer than wild-type telomeres (Fig. 4A, CuA lanes). Thus, the severity of overall loss of length control and loss of regulation correlated with the loss of in vitro binding affinity to Rap1p (Fig. 3B and Table 1), consistent with a role of Rap1p as a negative regulator of telomere addition. The results with these two template mutants were therefore consistent with those seen previously with K. lactis mutant telomeric repeats (31). In both of these lengthening mutants, telomere elongation was observed on chromosome ends containing subtelomeric Y′ elements as well as ends lacking Y′ elements (Fig. 4A).
TABLE 1.
Summary of tlc1 mutant phenotypesa
| TLC1 allele | Template sequenceb | Fold reduction in Rap1p binding | Telomere length (bp) | Telomerase activity | Senescence |
|---|---|---|---|---|---|
| TLC1 | ACCCA | 1 | 350 | + | No |
| tlc1-478gug | gugCA | >300c | 150c | +c | No |
| tlc1-476gug | ACgug | >300c | *c | −c | Yes |
| tlc1-476gCA | ACgCA | >300 | 3,000 | + | No |
| tlc1-476CuA | ACCuA | 14 | 800 | + | No |
| tlc1-476CCg | ACCCg | 4 | 250 | + | No |
| tlc1-476Cug | ACCug | >300 | 250 | + | No |
| tlc1-476gCg | ACgCg | >300 | *200 | + | Slow |
| tlc1-476guA | ACguA | >300 | *200 | + | Slow |
Various mutant alleles of TLC1 are shown, along with the positions of the mutated residues (lowercase) in the TLC1 RNA and the effects that each mutation has on Rap1p binding in vitro, on telomere length in vivo, and on telomerase activity in vitro. An asterisk indicates that telomere length was not stably maintained by the mutant enzyme; in the case of the two senescent double mutants (gCg and guA), the telomere length at which the strain senesced is also given.
From positions 478 to 474.
Data reported previously (34).
In addition, the 476gCA telomeres were heterogeneous in size and included telomeric DNA that was both larger and smaller than the wild-type telomere DNA. The continuous smear of telomeric hybridization signal extending below the shortest wild-type telomeric band (bracket in the Southern blot in Fig. 4A) indicated that degradation of telomeric DNA had occurred. In addition, a contribution to the shortening of telomeric DNA fragments through the previously described mechanism of telomeric rapid deletion events cannot be excluded (24). These results, together with comparable results obtained with K. lactis telomerase RNA template mutant strains (21, 31, 38), are consistent with Rap1p functioning to stabilize telomeric DNA from degradation, and possibly telomere rapid deletion, as well as overelongation in vivo.
Interestingly, as described further below, a distinguishable subpopulation of telomeres in the 476gCA mutant was short but length regulated, showing progressive shortening as the cells were passaged (Fig. 4A). Surprisingly, despite the presence of long newly added telomeric DNA tracts lacking Rap1p binding sites, tlc1-476gCA colonies grew well and looked normal at the level of gross colony morphology. To rule out the possibility that the tlc1-476gCA telomerase has an increased frequency of misincorporation in vivo, which might have allowed it to reconstitute a Rap1p binding site, we sequenced telomeric DNA subcloned out of this strain. As found previously for five other template mutations of S. cerevisiae telomerase (34), the telomeric repeats synthesized in vivo matched the sequence predicted from precise copying of the mutated region of the telomerase RNA template (Fig. 4B). The Rap1 core consensus binding sites in the sequenced repeats, and in the duplex oligonucleotides used in the gel shift analysis, were identical. Hence we conclude that the tlc1-476gCA mutant telomerase faithfully copies its mutated RNA template in vivo, predicting that the telomeric DNA synthesized by this enzyme is poorly or not bound by Rap1p in vivo.
The two remaining long-term viable strains, the single-base tlc1-476CCg mutant and the doubly mutated tlc1-476Cug strain, had telomeres that were stably maintained at a length ∼100 bp shorter than wild-type telomeres (Fig. 4A, CCg and Cug lanes). Even upon long autoradiographic exposures, no degradation like that in the gCA lanes was detectable. These two mutants behaved much like other tlc1 template mutants that we have analyzed, which also produced short and stable telomeres (34). In these mutants, most of the telomeric DNA consisted of wild-type repeats and telomeres contained at most only one or two distally added mutant repeats (34).
The lack of RAD52 function in the tlc1-476CCg and tlc1-476Cug cells argues that telomeres were maintained in these strains by the enzymatically active telomerase (Fig. 1B) rather than the alternative RAD52-dependent mechanism (26). Thus, although these mutations disrupted in vitro Rap1p binding to various degrees (4-fold for 476CCg and >300-fold for 476Cug), they did not cause the telomere lengthening expected from the disruption of binding of a negative regulator of telomere homeostasis. These results indicate that a reduction in telomeric Rap1p is not inevitably associated with telomere lengthening.
Cellular senescence occurred in both of the double-mutant strains (tlc1-476guA and tlc1-476gCg) that underwent progressive telomere shortening (Fig. 4A, gCg and guA lanes). The gradual loss of viability of these strains (Fig. 2) indicated that their telomeres progressively became functionally destabilized. This phenotype is typical of rad52 cells lacking telomerase activity (37). Notably, however, as shown in Fig. 1B, the telomerase in both these mutant strains was enzymatically active.
Telomere elongation by mutant tlc1-476gCA telomerase was rapid and stochastic.
Two aspects of the telomere lengthening seen in tlc1-476gCA single-mutant cells were particularly noteworthy. First, the extent of telomere elongation/deregulation appeared complete within the first 35 generations of growth in the presence of the mutant enzyme. To obtain a minimal estimate of the rate of telomeric DNA elongation in this strain, genomic DNA was isolated from colonies picked directly from the 5-FOA counterselection plate. After only ∼25 generations of growth in the absence of the wild-type enzyme (the first point at which DNA could be analyzed), the subpopulation of elongated telomeres had reached a new average length of approximately 3 kbp longer than wild type (Fig. 4B). Thus, a conservative minimal estimate of the elongation rate is 120 bp/cell division (Fig. 4B, compare strains CCA and gCA). Given that telomeres in wild-type cells increase and decrease in length only very gradually, by no more than 3 to 5 bp/cell division (35, 37), this rate represents minimally a 25-fold increase over the normal rate of net telomeric DNA addition in vivo. Second, while the majority of the telomeric molecules were elongated in the gCA mutant, a subpopulation was not elongated, and instead these telomeres were gradually shortened as the cells continued to divide (Fig. 4A and B). This population of short telomeres eventually disappeared, possibly because they were converted to longer telomeres and/or because they led to cessation of cell division, and hence cells containing these telomeres became greatly underrepresented in the cell culture.
DISCUSSION
Mutations in the template domain of the telomerase RNA subunit have been invaluable tools for elucidating the enzymatic properties of telomerase. Such mutations are also proving useful in elucidating telomere structure-function relationships. We showed previously that substitution of a specific three-base sequence located within the telomerase RNA template completely destroys telomerase activity and function (33, 34). In the present study, we created a series of one- and two-nucleotide substitutions in this three-base sequence. None significantly altered core in vitro telomerase activity, but each had a different effect on telomere maintenance and long-term cell viability (Table 1). Thus, we can begin to dissect away effects caused by lack of telomerase activity from effects caused by changes in the telomeric sequence synthesized. Notably, 476gCA telomerase, which caused significant telomere lengthening in vivo, and 476CCg telomerase, which maintained shortened but stable telomeres, both had wild-type-like activity in vitro. In addition, the telomerase of the tlc1-476Cug double mutant, which grew well, was somewhat less active in vitro than the telomerase of the tlc1-476guA mutant, yet the latter cells senesced. Hence, the telomere lengthening and shortening and cellular senescence phenotypes caused by these mutations likely result from altered regulation of the action of telomerase at telomeres or the altered telomeric DNA sequences, rather than significantly altered intrinsic core enzymatic activity.
The base changes expected to be copied from each of the six mutated telomerase RNA templates into telomeric DNA all reduced, to various degrees, in vitro binding by the DNA sequence-specific binding protein Rap1p. Hence, when transferred to the telomere, we predict that these base changes could correspondingly disrupt, at the distal ends of the telomeres, the telomeric DNA-protein complex centered on Rap1p. In the two mutants that showed telomere elongation, as with three previously reported telomerase RNA template mutants of K. lactis (21, 31), the predicted reduction in binding by Rap1p at the terminal telomeric repeats correlated with the degree of loss of length regulation. However, loss of Rap1 binding affinity clearly is not sufficient to cause elongation in all cases, since telomeres in the other four S. cerevisiae mutants described here were shorter than wild type.
Telomere shortening could also potentially be indicative of increased negative regulation by Rap1p at telomeres. For example, other effects of the base changes in the telomeric repeats on Rap1p binding could include a change in the bending of the Rap1p-bound mutant repeat, changing the overall properties of the higher-order telomeric DNA-protein complex. However, for the shortening effects to be entirely Rap1p mediated, all such base changes would have had to have caused a gain of function in Rap1p length regulation. While possible, this does not seem likely, given that all four of the shortening mutations described in this study, as well as two different template mutations that were analyzed previously that disrupt the Rap1p binding site and cause shortening (34), would have had a similar gain-of-function effect. It therefore seems likely that impairment of at least one other function besides Rap1 binding affinity contributes to the telomere shortening observed in these mutants.
This work provides further evidence that telomere addition is subject to at least two levels of control: repression by Rap1p and activation by another factor(s). Such other possible factors include Est1p and/or Cdc13p, both of which bind single-stranded telomeric DNA in vitro with DNA sequence specificity and are required for telomere addition in vivo. Additionally, a specific DNA sequence or structure at the telomeric 3′ end, which may be altered by the four shortening mutations, could be required for telomere elongation by telomerase. We have shown previously that telomerase, in addition to the base pairing between the RNA template domain and telomeric DNA, shows sequence-specific interactions with DNA in vitro (33, 34). Thus, the interaction of the telomeric DNA with telomerase enzyme, comprised minimally of Est2p and the TCL1 RNA (18), could also potentially be altered in vivo by the changes in the telomeric sequence caused by the tlc1 template mutations. Finally, it has been proposed that the Rap1p-centered higher-order DNA-protein complex of the telomere helps to maintain telomere length homeostasis by limiting the elongation of long telomeres and stimulating the elongation of short telomeres (21–25, 29, 31). If so, loss of Rap1p binding would certainly exacerbate the phenotypes seen here.
We propose that the rapid and stochastic telomere elongation catalyzed in vivo by 476gCA telomerase results from altered regulation of telomerase activity at the telomere. The behavior of 476gCA telomeres also strongly suggests that on any given telomere molecule in the population, telomere overelongation/deregulation initiates stochastically. We therefore propose that in tlc1-476gCA cells, as the initial population of telomeres becomes shortened under the direction of the tlc1-476gCA allele, at each cell division a subset of these telomeres loses regulation. Once overelongation/deregulation initiates on any one telomere, it rapidly progresses to the fully elongated/deregulated state and apparently does not regain length control. Thus, the whole population of telomeres eventually switches over to the deregulated state. A similar phenomenon of conversion of the telomere population to a deregulated state was seen previously with K. lactis cells mutated both in the telomerase RNA template and in the C terminus of Rap1p. However, in that case no telomere shortening was observed prior to the loss of length regulation (21).
It has been proposed that normally, two mutually reinforcing telomeric components contribute to length control: a proper regulatory end structure that is disrupted upon incorporation of mutant residues into the terminal telomeric repeat(s); and a higher-order structure, destabilized by telomere shortening, that normally acts to limit telomerase access to telomeres (21, 22, 29, 31, 38). In addition, the specific importance of the terminal few repeats has been demonstrated by mutating them in K. lactis (21, 22, 38). Such telomerase RNA template mutations cause deregulated telomere lengthening, either immediately following expression of the mutant enzyme, like that we report here for the S. cerevisiae tlc1-476gCA and tlc1-476CuA mutants, or only after hundreds of generations of growth (31). Loss of the telomeric cap structure also apparently allows telomere degradation and telomere-telomere fusions (30, 40, 45). While the two immediate-telomere-lengthening mutations described in K. lactis decreased Rap1p binding affinity in vitro, the two delayed-lengthening mutations that have been described in K. lactis did not, suggesting that at least two types of regulatory mechanisms are involved in telomere homeostasis (21, 31). It was proposed that mutations causing immediate telomere lengthening do so by altering the cap structure at the extreme terminus of the telomere, leading to either an increased rate of telomere addition by telomerase, as we have demonstrated in the present work with S. cerevisiae telomerase, or increased telomeric recombination (22, 38). On the other hand, mutations that require hundreds of generations to manifest a telomere lengthening phenotype may do so by causing a more global alteration in telomere structure. This requires a more extensive replacement of wild-type repeat tracts by mutant telomeric repeats throughout the length of the telomere, and the phenotypic delay has been proposed to occur because these mutant repeats only gradually infiltrate throughout the body of the telomeres (31).
Both tlc1-476gCA RAD52 and tlc1-476gCA rad52 cells, which contain long telomeres with extremely low affinity for Rap1p, showed an apparently normal FACS profile (C. D. Smith, unpublished observation). Furthermore, these cells had only slightly abnormal cytology, with minimally increased numbers of the large, unbudded, multinucleated cells typically associated with a failure to properly segregate chromosomes (38; J. C. Prescott, E. H. Blackburn, and C. D. Smith, unpublished observation). The long tracts of mutant telomeric repeats were added directly onto a preexisting tract of wild-type repeats, suggesting that an internal tract of wild-type telomeric repeats may be sufficient to maintain at least some telomere functions, although not telomere length regulation. Alternatively, the presence of an internal tract of wild-type telomeric DNA may facilitate Rap1p binding to the mutant repeats, thus maintaining a functional elongated telomere.
Several different telomere phenotypes are caused by various mutations affecting Rap1p and/or its telomeric DNA binding sites, which include lengthening, shortening, and mislocalization in the nucleus, suggests that Rap1p has multiple roles in telomere maintenance. Mutations in the C-terminal domain of Rap1p, which normally mediates interactions with Sir and Rif proteins, causes telomere lengthening and, in some situations, cellular phenotypes indicative of loss of capping (21–23). Human telomeres contain two distinct telomere-binding proteins, TRF1 and TRF2 (39, 40). It is possible that the roles carried out by Rap1p in yeasts are divided between TRF1 and TRF2 in mammals. As with yeast telomerase RNA template mutations that cause telomere elongation concomitant with disruption of Rap1 binding (21, 31; this work), disrupting the DNA binding domain of TRF1 causes telomere lengthening (39), suggesting that telomere addition in human cells is also responsive to TRF1 occupancy of the telomere. Mutations that disrupt the binding of TRF2 to telomeres cause telomere-telomere fusions and apoptosis, suggesting that TRF2 protects the telomeric terminus (19, 40). This function of TRF2 may be analogous to the role of Rap1p in the formation of a telomeric cap structure. Thus, the data suggest that Rap1p serves both a TRF1-homologous role in regulating telomere length homeostasis and a TRF2-homologous role in protecting the end of the chromosome from being recognized as damaged DNA.
Telomeres generally contain single-stranded 3′ overhangs during at least part of the cell cycle (28, 43). Telomere addition requires that this 3′ terminus, which is itself synthesized by telomerase, be made accessible to telomerase. It is possible that the wild-type telomeric repeats present when the mutant template telomerase first replaces wild-type telomerase cannot base pair with the mutant template in a manner that allows productive synthesis by telomerase. The necessity for such template position-specific base pairing has been demonstrated for Tetrahymena telomerase (42). In Oxytricha nova, the 3′ overhang is bound both in vivo and in vitro by a heterodimeric protein complex in which nearly every nucleotide is stacked with an aromatic amino acid (11, 17). Two S. cerevisiae proteins, Est1p and Cdc13p, bind single-stranded telomeric TG1–3 repeat DNA in vitro (32, 41) and could possibly be counterparts of the Oxytricha heterodimer. While neither Est1p nor Cdc13p is required for core enzymatic activity of telomerase in vitro, both are required for telomerase action in vivo, suggesting that they may modulate the interaction between telomerase and the telomeric 3′ end (7, 18, 25). Hence, the mutations described here that disrupt Rap1p binding, yet do not cause telomere lengthening, may exert their effects by disrupting sequence-specific binding by Estp1p, Cdc13p, and/or some other positive regulator of telomerase action. The DNA sequence requirements for Est1p and Cdc13p binding are not sufficiently understood to predict what effects the mutations analyzed here would have on their binding.
It was noted previously that a short conserved core sequence within the otherwise highly variable telomeric repeat units of yeasts coincides with the Rap1 binding core (8). Here we have reported widely varying effects on telomeres caused by small mutations within this core sequence in S. cerevisiae TLC1 RNA. We propose that the striking evolutionary conservation of this core sequence in the telomeric DNA of yeasts is imposed because this sequence has to serve as a binding site not only for Rap1p but also for Est1p, Cdc13p, and possibly other factors including the telomerase RNP itself.
ACKNOWLEDGMENTS
This work was supported by grant GM26259 from the National Institutes of Health to E.H.B. J.C.P. was supported by a Special Fellow award from the Leukemia Society of America.
We thank the members of the Blackburn laboratory for helpful ideas and discussions and specifically Shivani Nautiyal and Chris D. Smith for critical reading of the manuscript. Plasmid pBRΔHS LEU2, used to disrupt RAD52, was a gift from Dennis Livingston.
REFERENCES
- 1.Adams A K, Holm C. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4614–4620. doi: 10.1128/mcb.16.9.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ancelin K, Brun C, Gilson E. Role of the telomeric DNA-binding protein TRF2 in the stability of human chromosome ends. Bioessays. 1998;20:879–883. doi: 10.1002/(SICI)1521-1878(199811)20:11<879::AID-BIES2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn E H. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 4.Blasco M A, Lee H W, Hande M P, Samper E, Lansdorp P M, DePinho R A, Greider C W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 5.Boulton S J, Jackson S P. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourns B D, Alexander M K, Smith A M, Zakian V A. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol Cell Biol. 1998;18:5600–5608. doi: 10.1128/mcb.18.9.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohn M, Blackburn E H. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 8.Cohn M, McEachern M J, Blackburn E H. Telomeric sequence diversity within the genus Saccharomyces. Curr Genet. 1998;33:83–91. doi: 10.1007/s002940050312. [DOI] [PubMed] [Google Scholar]
- 9.Conrad M N, Wright J H, Wolf A J, Zakian V A. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 10.Fan X, Price C M. Coordinate regulation of G− and C strand length during new telomere synthesis. Mol Biol Cell. 1997;8:2145–2155. doi: 10.1091/mbc.8.11.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottschling D E, Zakian V A. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- 12.Gravel S, Larrivee M, Labrecque P, Wellinger R J. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 13.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 14.Greider C W. Telomeres and senescence: the history, the experiment, the future. Curr Biol. 1998;8:R178–R181. doi: 10.1016/s0960-9822(98)70105-8. [DOI] [PubMed] [Google Scholar]
- 15.Hande M P, Samper E, Lansdorp P, Blasco M A. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J Cell Biol. 1999;144:589–601. doi: 10.1083/jcb.144.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.Horvath M P, Schweiker V L, Bevilacqua J M, Ruggles J A, Schultz S C. Crystal structure of the Oxytricha nova telomere end binking protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 18.Hughes T R, Morris D K, Salinger A, Walcott N, Nugent C I, Lundblad V. The role of the EST genes in yeast telomere replication. Ciba Found Symp. 1997;211:41–47. doi: 10.1002/9780470515433.ch4. [DOI] [PubMed] [Google Scholar]
- 19.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 20.König P, Rhodes D. Recognition of telomeric DNA. Trends Biochem Sci. 1997;22:43–47. doi: 10.1016/s0968-0004(97)01008-6. [DOI] [PubMed] [Google Scholar]
- 21.Krauskopf A, Blackburn E H. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature. 1996;383:354–357. doi: 10.1038/383354a0. [DOI] [PubMed] [Google Scholar]
- 22.Krauskopf A, Blackburn E H. Rap1 protein regulates telomere turnover in yeast. Proc Natl Acad Sci USA. 1998;95:12486–12491. doi: 10.1073/pnas.95.21.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyrion G, Boakye K A, Lustig A J. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5159–5173. doi: 10.1128/mcb.12.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Lustig A J. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 25.Lingner J, Cech T R, Hughes T R, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundblad V, Blackburn E H. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 27.Lundblad V, Szostak J W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 28.Makarov V L, Hirose Y, Langmore J P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 29.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 30.McEachern M J, Blackburn E H. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 31.McEachern M J, Blackburn E H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 32.Nugent C I, Hughes T R, Lue N F, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 33.Prescott J, Blackburn E H. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prescott J, Blackburn E H. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 1997;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- 35.Shampay J, Blackburn E H. Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1988;85:534–538. doi: 10.1073/pnas.85.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 37.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 38.Smith C D, Blackburn E H. Uncapping and deregulation of telomeres lead to detrimental cellular consequences in yeast. J Cell Biol. 1999;145:203–214. doi: 10.1083/jcb.145.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 40.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 41.Virta-Pearlman V, Morris D K, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Gilley D, Blackburn E H. A novel specificity for the primer-template pairing requirement in Tetrahymena telomerase. EMBO J. 1998;17:1152–1160. doi: 10.1093/emboj/17.4.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wellinger R J, Wolf A J, Zakian V A. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 44.Zakian V A. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 45.Zhu J, Wang H, Bishop J M, Blackburn E H. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc Natl Acad Sci USA. 1999;96:3723–3728. doi: 10.1073/pnas.96.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]