Abstract
Plants produce a variety of metabolites that are essential for plant growth and human health. To fully understand the diversity of metabolites in certain plants, lots of methods have been developed for metabolites detection and data processing. In the data-processing procedure, how to effectively reduce false-positive peaks, analyze large-scale metabolic data, and annotate plant metabolites remains challenging. In this review, we introduce and discuss some prominent methods that could be exploited to solve these problems, including a five-step filtering method for reducing false-positive signals in LC-MS analysis, QPMASS for analyzing ultra-large GC-MS data, and MetDNA for annotating metabolites. The main applications of plant metabolomics in species discrimination, metabolic pathway dissection, population genetic studies, and some other aspects are also highlighted. To further promote the development of plant metabolomics, more effective and integrated methods/platforms for metabolite detection and comprehensive databases for metabolite identification are highly needed. With the improvement of these technologies and the development of genomics and transcriptomics, plant metabolomics will be widely used in many fields.
Key words: plant metabolomics, metabolites, data-processing methods, application
Mass spectrometry-based metabolomics approaches are often used in analysis of complex metabolites in plants. This review discusses the methods on how to effectively reduce false-positive peaks, analyze large-scale datasets, and annotate plant metabolites. LC-MS- and GC-MS-based metabolome analyses have been used for species discrimination, metabolic pathway dissection, and population genetic studies, and plant metabolomics will be widely exploited in many fields of plant sciences.
Introduction
Metabolomics is an important part of systems biology, which aims to study small-molecule metabolites and their changes in organisms, specific tissues, and even single cells (Fiehn, 2002). Plant metabolomics has received much attention as one of the most important parts of metabolomics. It is estimated that there are over 200 000 metabolites in the plant kingdom (Fiehn, 2002; Dixon and Strack, 2003; Fernie and Tohge, 2017), which can be divided into primary metabolites and specialized metabolites (secondary metabolites). The primary metabolites are the type of metabolites mainly involved in plant growth and development, which constitutively accumulated in plant cells (Sulpice and Mckeown, 2015; Fang et al., 2019). While the specialized metabolites play significant roles in plant defense, this kind of metabolites mainly exists in a certain tissue or a given development stage of a plant, resulting in the diversity of plant metabolites (Carreno-Quintero et al., 2013; Fernie and Tohge, 2017; Zaynab et al., 2018; Fang et al., 2019). In addition, the gene function and gene duplication in different plants altered not only the enzymatic characteristics, but also the substrate specificity, making the plant metabolites extremely complex (Fiehn, 2002).
To fully understand the diversity of metabolites in a certain plant, a variety of plant metabolites detecting platforms and approaches have been developed with the advent of the mass spectrometry technology (Zeki et al., 2020). Among them, nuclear magnetic resonance (NMR)- and mass spectrometry (MS)-based methods are the major analytical platforms used in plant metabolomics (Fernie et al., 2004; Zeki et al., 2020). The NMR platform is mainly used to detect metabolites with high abundance (Fernie and Tohge, 2017). While the MS-based methods, including gas chromatography-mass spectrometry (GC-MS), and liquid chromatography-mass spectrometry (LC-MS), are widely used in metabolomics due to their high resolution and sensitivity (Gowda and Djukovic, 2014; Fang et al., 2019; Siddiqui et al., 2020). In GC-MS analysis, metabolites are separated in GC after derivatization, so it is suitable for analyzing volatile metabolites, or metabolites easily to volatilize after derivatization (Fernie et al., 2004). The major metabolites that can be detected in this platform are primary metabolites, such as amino acids, sugars, and organic acids (Figure 1) (Fernie et al., 2004; Fernie and Tohge, 2017). The LC-MS separates compounds in the liquid phase without the requirement of sample pre-treatment, making it the most powerful and comprehensive analytical approach (Fernie and Tohge, 2017). Most secondary metabolites, such as flavonoids, alkaloids, and phenylpropanoids, can be detected in this platform (Figure 1) (Fernie et al., 2004; De Vos et al., 2007; Böttcher et al., 2008). Since different kinds of metabolites are detected in different platforms, it is obvious that a combination of different analytical tools could uncover the diversity of compounds in different plants, even in different tissues (Fernie et al., 2004; t'Kindt et al., 2009; Zeki et al., 2020).
Figure 1.
The data-processing procedures of plant metabolic data.
PCA, principal-component analysis; HCA, hierarchical clustering analysis; PLS, partial least squares analysis; O-PLS, orthogonal to partial least squares analysis.
Except for the analytical platforms, sample preparation steps also have important influences on the metabolites detected (t'Kindt et al., 2009). To save time and cost during sample preparation in a combined metabolomics study, lots of extraction methods, such as two-phase and three-phase methods, were developed (t'Kindt et al., 2009; Zeki et al., 2020). After sample extraction and data acquisition, MS data are analyzed by the following three steps: raw data preprocessing, multivariate statistical analysis, and peak annotation (Figure 1) (Zeki et al., 2020). In many studies, the presence of false-positive signals in LC-MS analysis (Chetnik et al., 2020), lacking software for analyzing ultra-large GC-MS data (Duan et al., 2020), and the absence of a comprehensive database for annotating plant metabolites (Shen et al., 2019), are the major challenges for processing of plant metabolic data. In this review, we briefly introduce several useful strategies and methods to solve these problems effectively. Meanwhile, the application of plant metabolomics, our perspectives on the major problems existed in plant metabolomics, and possible solutions are also discussed.
Reducing false-positive signals in LC-MS analysis of plant metabolomes
Recently, numerous software have been developed for analyzing LC-MS data, including commercial, open-source and online workflows (Wen et al., 2017). The commercial software, such as MassHunter (Agilent Technologies), SIEVE and Compound Discoverer (Thermo Scientific), and Progenesis QI and Markerlynx (Waters), are mainly developed for datasets acquired from the companies’ own instruments, resulting in a limited scope of application for these software (Want and Masson, 2011; Wen et al., 2017; Gorrochategui et al., 2019; Chetnik et al., 2020). While open-source software is widely used in the study of plant metabolomics, to process MS data using these free and open-source software, the acquired raw data should be converted into appropriate formats, such as mzXML, mzML, mzData, and netCDF, using available software (Katajamaa and Oresic, 2007; Want and Masson, 2011; Zeki et al., 2020). Among the developed freely available software for processing LC-MS data (Table 1), the R package XCMS (Smith et al., 2006; Tautenhahn et al., 2012) and MetAlign (Lommen, 2009), are two powerful tools for data preprocessing, including peak filtration, identification, alignment, and quantitation. However, it was time-consuming for them to analyze large-scale datasets. In MZmine, peaks are aligned based on the random sample consensus algorithm, and the data are visualized in multiple visualization modules. To speed up the calculation process, the distributed computing algorithm was also included in this software (Katajamaa et al., 2006; Pluskal et al., 2010). AMDORAP is an easy-to-use tool for detecting more accurate m/z values from raw data. It was estimated that the m/z errors in AMDORAP was within ±3 ppm, while the errors in some other software were over ±100 ppm (Takahashi et al., 2011). MAIT is a programmatic tool that allows for a comprehensive statistical study of LC-MS data. However, the data normalization is not included in MAIT (Fernández-Albert et al., 2014). OpenMS provides a total of 185 tools and ready-to-use workflows for MS data processing, visualization, and quantitation. In OpenMS, a highly flexible and professional software environment is available for users, especially for new users, to reduce potential errors during data analysis (Röst et al., 2016). metaX is a comprehensive workflow for analyzing untargeted metabolomics data, in which data preprocessing, statistics analysis, metabolites identification, pathway annotation, and biomarker selection are all included (Wen et al., 2017). ROIMCR employs the ROIs (the search of regions of interest) in the m/z domain for peak detection and integration without the reduction of peak resolution; and the MCR-ALS method (multivariate curve resolution-alternating least squares) is used to resolve spectra from mixtures without the requirement of peak modeling and alignment. Hence, this is the most useful software to effectively reduce the amount of errors during peak modeling and alignment (Gorrochategui et al., 2019). For the online workflows, MetaboAnalyst is a powerful and comprehensive platform for metabolomics data analysis, including enrichment analysis, pathway analysis, statistical analysis, and so on. However, the original data needs to be converted and aligned by other software to obtain a dataset containing the sample information, such as retention time, m/z values, and intensity (Xia et al., 2009). Another online data visualization and annotation software, MAVEN, is designed for efficient and interactive analysis of LC-MS data using a machine learning-based method to assess peak quality. It can automatically analyze isotope-labeled forms, and graphically map data onto metabolic pathways (Melamud et al., 2010).
Table 1.
Summary of software for analyzing plant metabolic data.
| Software | Description | Compatibility | Language | Reference |
|---|---|---|---|---|
| XCMS | data preprocessing, alignment, and quantitation; but it is time-consuming for it to process large-scale datasets | LC-MS, GC-MS | R |
Smith et al. (2006); Tautenhahn et al. (2012) |
| MetAlign | data preprocessing, alignment, and quantitation; but it is time-consuming for it to process large-scale datasets | LC-MS, GC-MS | C | Lommen (2009) |
| Mzmine | distributed computing algorithm-based peak alignment and multiple visualization modules are available for data visualization | LC-MS, GC-MS | Java |
Katajamaa et al. (2006); Pluskal et al. (2010) |
| AMDORAP | accurate m/z detection with the m/z errors within ±3 ppm | LC-MS | R | Takahashi et al. (2011) |
| MAIT | comprehensive statistical analysis tool for LC-MS metabolic data, but the data normalization is not included | LC-MS | R | Fernández-Albert et al. (2014) |
| OpenMS | hundreds of workflows are available for data processing, and a highly flexible and professional software environment is provided for users | LC-MS | C++ | Röst et al. (2016) |
| metaX | a comprehensive workflow for untargeted metabolomics data, including data preprocessing, metabolites identification, pathway annotation, and biomarker selection | LC-MS, GC-MS | R | Wen et al. (2017) |
| ROIMCR | ROI-based peak detection and integration, and an MCR-ALS method is used to resolve peaks from mixture | LC-MS | MATLAB | Gorrochategui et al. (2019) |
| MetaboAnalyst | a powerful platform for metabolomics data analysis, including enrichment analysis, pathway analysis, and statistical analysis; however, the original data need to be converted and aligned by other software | LC-MS, GC-MS, NMR | Java, R | Xia et al. (2009) |
| MAVEN | machine learning-based peak quality assessment, pathway, and isotope-labeling visualization | LC-MS | – | Melamud et al. (2010) |
| apLCMS | a hybrid feature detection approach is used to reduce false-positive and false-negative peaks, but a known-feature database is needed | LC-MS | R | Yu et al. (2009); Yu et al. (2013) |
| MS-FLO | retention time alignment, accurate mass tolerances, peak height similarity, and Pearson’s correlation analysis-based methods to minimize false-positive peaks | LC-MS | Python | DeFelice et al. (2017) |
| rFPF | an EIC profile-based method to remove false-positive features | LC-MS | MATLAB | Ju et al. (2019) |
| Peakonly | precise peak detection using a convolutional neural network-based deep learning method | LC-MS | Python | Melnikov et al. (2020) |
| AMDIS | data deconvolution; without the function of peak alignment | GC-MS | – | Halket et al. (1999); Stein (1999) |
| ChromaTOF | GC-TOF-MS data deconvolution; without published algorithm descriptions | GC-MS | – | – |
| MetaQuant | target metabolome analysis, but an established library is required | GC-MS | Java | Bunk et al. (2006) |
| MET-IDEA | target metabolome analysis, but a list containing m/z and retention time pairs is required | GC-MS | – |
Broeckling et al. (2006); Lei et al. (2012) |
| TagFinder | peak alignment; without the function of baseline correction and peak smooth | GC-MS | Java | Luedemann et al. (2008) |
| MetaboliteDetector | data deconvolution and peak alignment based on a QT4 graphical user interface | GC-MS | C++ | Hiller et al. (2009) |
| ADAP | data deconvolution and peak alignment using a two-phase approach | GC-MS | C++, R |
Jiang et al. (2010); Ni et al. (2012); Ni et al. (2016); Smirnov et al. (2019) |
| MS-DIAL | data deconvolution, peak alignment, and annotation | GC-MS | C | Tsugawa et al. (2015); |
| eRah | peak deconvolution and alignment | GC-MS | R | Domingo-Almenara et al. (2016) |
| IP4M | 62 independent functions for data preprocessing, peak annotation, and pathway enrichment analysis | LC-MS, GC-MS | Java, Perl, R | Liang et al. (2020) |
| autoGCMSDataAnal | TIC peak detection and resolution using raw data; dynamic programming algorithm-based retention time-shift correction | GC-MS | MATLAB | Zhang et al. (2020) |
| QPMASS | large-scale metabolic data analysis (alignment, backfill, and quantitation) | GC-MS | C++ | Duan et al. (2020) |
Although the great ability of the software mentioned above in the processing of LC-MS data, there might also exist some false-positive signals, including background noise, duplicate peaks, and contaminants (no-sample sources), which might originate from sample contamination during sample preparation; column contamination, chemical and solvent noise, retention time drift, and un-optimized analytical separation during metabolites identification; unsuitable settings of alignment parameters and large variations in peak detection across different software during data preprocessing (Sauvage et al., 2008; Want et al., 2010; Yu et al., 2013; Duan et al., 2016; DeFelice et al., 2017; Chetnik et al., 2020). To minimize false-positive signals, it is necessary to be careful with sample preparation as the sample contaminants can co-elute with metabolites. Meanwhile, optimizing instrument conditions and developing an efficient analytical separation method using a series of test samples are needed (Want et al., 2010). Following this, an effective filtering method in the subsequent data preprocessing is critical. The most used data-filtering methods, such as relative standard deviation (RSD), missing value thresholds, mean/median value, correlation approach, and feature clustering across biological samples greatly reduce the total number of detected peaks (Want et al., 2010; Broadhurst et al., 2018; Chong et al., 2019; Alseekh et al., 2021). However, there still remain large numbers of false-positive peaks after applying these methods (Chetnik et al., 2020). To effectively solve this problem, lots of predominant approaches have been developed (Yu et al., 2013; DeFelice et al., 2017; Ju et al., 2019; Kantz et al., 2019; Chetnik et al., 2020; Melnikov et al., 2020). For example, a hybrid feature detection approach was implemented in apLCMS to improve feature detection sensitivity and to reduce the number of spurious peaks. However, a known-feature database is needed in this approach (Yu et al., 2009, 2013). MS-FLO employs retention time alignment, accurate mass tolerances, peak height similarity, and Pearson’s correlation analysis to remove the erroneous peaks. It was estimated that 15.7% of 1481 peaks, which was detected by MZmine2 and MS-DIAL, were removed or marked for users to review (DeFelice et al., 2017). The rFPF method removes the false-positive features using a reproducibility constraint, entropy index, and statistical correlation analysis based on EIC profile. In standard mixture analysis of metabolites, more than 92% of the false-positive peaks were removed after applying this method. In urine sample analysis, nearly 65% of peaks (4660 out of 7182 peaks) were removed (Ju et al., 2019). With the development of computing capability, the deep neural network-based machine learning algorithm has been used in identifying false-positive peaks in untargeted LC-MS data. It was estimated that nearly 90% of false-positive peaks could be removed by this method. However, an appropriate number of training sets is highly needed for this method (Kantz et al., 2019). In a recent study, Chetnik et al. (2020) proposed a machine learning-based method combined with peak quality metrics to filter low-quality peaks in untargeted LC-MS metabolomics data. In the peakonly algorithm, the convolutional neural network-based deep learning method is used to detect true positive peaks in the raw data (Melnikov et al., 2020). Except for these useful methods, a hierarchical five-step filtering approach (Duan et al., 2016) is considered as one of the most efficient methods (Ju et al., 2019), and has been used in some recently published works (Wu et al., 2018, 2020; Li et al., 2019).
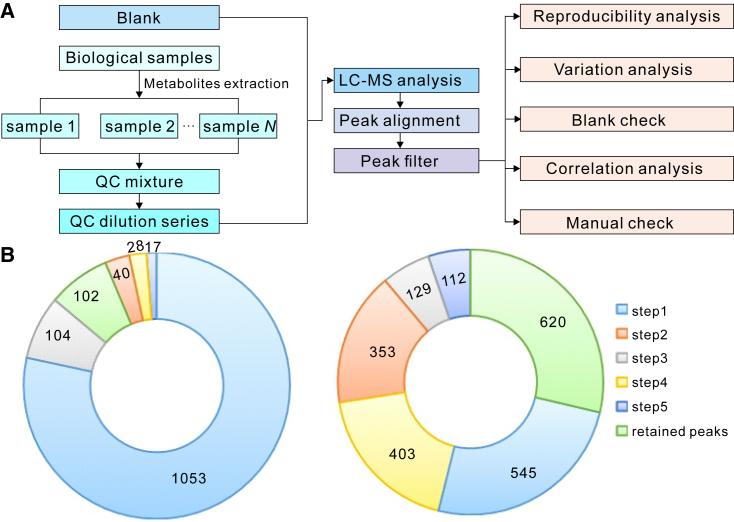
In the five-step filtering method, it is necessary to prepare the blank and quality control (QC) samples, which are the mixture of all the samples to be tested in an experiment. Then a series of QC mixture dilutions are prepared (Figure 2A). All the blank samples, QC mixture, and QC dilution series, are analyzed before the biological samples in LC-MS analysis. After data acquisition and peak alignment, the false-positive signals are filtered in the following steps: step 1, the reproducibility analysis, is used to filter peaks that cannot be detected in 80% of samples. Step 2, the variation analysis, is used to filter peaks with a large variation in the different replicates of the QC mixture (normally, the filtering threshold is RSD > 20%). Step 3, the blank check, is used to filter the contaminative peaks using the peak area ratio of blank to QC mixture. Step 4, the correlation analysis, is used to filter the non-biological peaks using the correlation of the QC dilution series. Step 5, manual check, is used to filter the disqualified peaks by manual inspection (Figure 2A). It was shown that nearly 92.5% of peaks were filtered as false-positive peaks in the validation experiment using artificial samples. Specifically, 1053 out of 1342 total peaks (∼78.5%) were filtered by the first step. In step 2, 40 peaks (∼3%) with RSD > 20% were filtered, and 104 peaks (∼7.7%) with the peak ratio of blank to QC mixture over 1% were eliminated in step 3. Then another 28 peaks (∼2%) were filtered in step 4. Finally, an additional 17 peaks (∼1.3%) were eliminated after a manual check (Figure 2B) (Duan et al., 2016). In the biological samples of rice seeds, 1542 out of 2162 (∼71.3%) peaks were eliminated using the five-step filtering method, including some peaks that were considered as potential biomarkers in traditional metabolomics analysis (Figure 2B). A relative concentration index method is also introduced in this strategy to increase the accuracy of quantitation (Duan et al., 2016). Meanwhile, to achieve more accurate quantitative results in LC-MS analysis, Yu and Huan (2021) proposed an MRC workflow using a series of QC dilutions to achieve the best regression model for correcting the biased signal ratios. The combined utilization of these data-preprocessing, false-positive signals filtering, and the peak quantitation methods will greatly benefit the downstream statistical analysis and biological hypothesis for further study.
Figure 2.
The strategy of a five-step filtering approach for metabolic data.
(A) The flow of the five-step filtering approach.
(B) The validation experiments of the five-step filtering approach using artificial samples (left) and biological samples of rice seed (right). Steps 1 to 5 correspond to the data-filtering procedures in the five-step filtering approach, and the retained peaks are the peaks left after data filtering.
Developing efficient tools for analyzing large-scale GC-MS data
GC-MS is another widely used platform for plant metabolomics analysis (Luo, 2015; Fernie and Tohge, 2017). Compared with LC-MS analysis, lots of fragment ions for certain metabolites are generated in GC-MS analysis, making the processing of GC-MS data more complicated (Duan et al., 2020). Tools initially developed for processing LC-MS data, such as MZmine (Katajamaa et al., 2006; Pluskal et al., 2010), XCMS (Smith et al., 2006; Tautenhahn et al., 2012), and MetAlign (Lommen, 2009), are also used for processing GC-MS data (Robinson et al., 2007; Fernández-Varela et al., 2015). However, it seemed that these tools over-interpreted GC-MS data (Duan et al., 2020). Now, numerous software have been developed for analyzing GC-MS data (Table 1), such as the widely used software AMDIS (Halket et al., 1999; Stein, 1999) and the commercial software ChromaTOF (LECO, St. Joesph, MI, USA). These software were mainly developed for peak deconvolution, and peak alignment cannot be performed in them (Duan et al., 2020). Other software, such as MetaQuant (Bunk et al., 2006) and MET-IDEA (Broeckling et al., 2006; Lei et al., 2012), were mainly developed for target metabolome analysis. For the untargeted GC-MS-based metabolomics analysis, software TagFinder (Luedemann et al., 2008), flagme (Robinson, 2010), APAP (Jiang et al., 2010; Ni et al., 2012, 2016; Smirnov et al., 2019), and MS-DIAL (Tsugawa et al., 2015) show great performance in peak alignment. However, it is time-consuming and compute-intensive for most of them to analyze ultra-large GC-MS data (Duan et al., 2020). To process large-scale GC-MS data accurately and efficiently, the software QPMASS was developed (Duan et al., 2020).
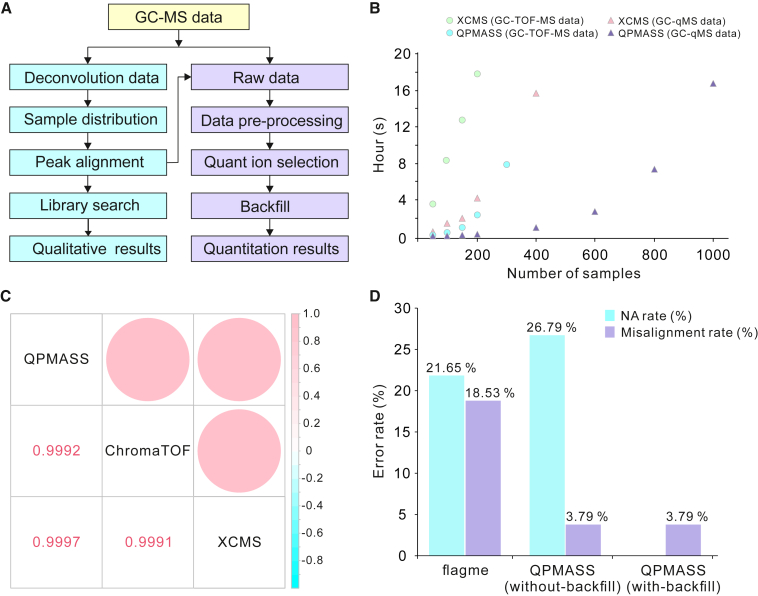
In QPMASS, data from different platforms, including gas chromatography time-of-flight mass spectrometry (GC-TOF-MS) and GC-quadrupole MS (GC-qMS), can be aligned and quantified. The workflow of QPMASS is shown in Figure 3A. To align peaks, the samples are firstly grouped using a furthest-neighbor joining clustering-based hierarchical clustering method (Duan et al., 2020). Then peaks are aligned using a dynamic programming algorithm (Robinson et al., 2007)-based parallel alignment approach to accelerate the speed of alignment. It was found that QPMASS only took 2 h to process 200 GC-TOF-MS data, while the frequently used software XCMS needed 18 h to process the same number of samples. Within 17 h, QPMASS could process 1000 GC-qMS data, while XCMS could only process 400 samples in the same time (Figure 3B). At the same time, a three-parameter strategy was developed in QPMASS to select an optimal quantitative ion (quant ion) for accurate quantitation. In this strategy, the optimal quant ion should not overlap with the adjacent peak, and it should have a better peak shape. If the optimal quant ion cannot be selected using the two aforementioned parameters, the ion with the highest intensity will be used. It has been shown that the quantitation result of QPMASS was significantly correlated with the results from XCMS and ChromaTOF (r2 > 0.99), which are the two major software used for analyzing GC-MS data at present (Figure 3C). To reduce the false-positive and false-negative peaks in the aligned results, peak filtering and backfill methods were also introduced in QPMASS. Using these methods, the NA rate was significantly reduced after backfilling, and the total error rate, including the missing value and misalignment rate, was reduced to 3.79% (Figure 3D) (Duan et al., 2020).
Figure 3.
The performance of QPMASS software.
(A) The workflow of QPMASS.
(B) The processing time of QPMASS and XCMS for different number of GC-TOF-MS (dots) and GC-qMS (triangles) data.
(C) Comparison of quantification performance among QPMASS, XCMS, and ChromaTOF. The legend color and circle size correspond to the correlation of peak areas from different software.
(D) The alignment accuracy of QPMASS. The accuracy of alignment was compared between QPMASS and flagme.
Annotation of metabolites
To annotate metabolites after data preprocessing, lots of databases and strategies for plant metabolites are available now (Table 2), and can be grouped into two major categories: library searching-based and biochemical reaction-based annotation. For the library searching-based annotation, the NIST mass spectral database is one of the most widely used mass spectral reference libraries, and it aims to identify unknown compounds in GC-MS and LC-MS spectra using library searching. In NIST, MS/MS spectra collected from multiple collision energy levels, mass spectra for multiple ion adducts, compound name, formula, CAS number, etc., are all included. METLIN is another widely used database, in which nearly a million of metabolite standards with experimental MS/MS data at multiple energy levels and in positive/negative modes are included. It supports multiple retrieval modes, including simple, advanced, batch, fragment, neutral loss, and MS/MS spectrum match (Smith et al., 2005). BinBase was developed for automatic annotation of GC-TOF/MS data, in which a mass spectral metadata-based filtering algorithm is used to match and generate database objects (Fiehn et al., 2005). MMCD contains over 20 000 metabolites and small molecules of biological interest, and it is compatible for identifying metabolites from both NMR and MS data (Cui et al., 2008). SIRIUS is a powerful tool for metabolite annotation using isotope pattern analysis in its first version. While the fragmentation trees and maximum a posteriori estimation are added in the latter versions to provide a comprehensive assessment of molecular structure using MS/MS data, in the latest version (SIRIUS4), the CSI:FingerID, which combines fragmentation tree computation and machine learning for molecular structure searching, was integrated via a RESTful (representational state transfer) web service (Böcker et al., 2009; Dührkop et al., 2015, 2019). MassBank is a distributed database, in which ESI-MS2 data under different experimental conditions are merged to identify metabolites by mass spectra; and lots of search options are included in this database, such as a quick search by chemical name, mass, and formula; peak search by m/z values and relative intensities (Horai et al., 2010). ReSpect (RIKEN tandem mass spectral database), a plant-specific MS/MS-based database, including records from the literature and MS/MS data from authentic standards (Sawada et al., 2012). Lei et al. (2015) constructed an ultra-high-performance liquid chromatography-tandem mass spectral library of plant natural products, containing thousands of mass spectra with retention time, formula, and structure.
Table 2.
Summary of available databases for plant metabolites identification and pathway analysis.
| Database | Compatibility | Link | Description | Reference |
|---|---|---|---|---|
| NIST | LC-MS, GC-MS | https://www.sisweb.com/software/ms/nist.htm | a most widely used mass spectral reference library, in which MS/MS spectra, mass spectra for multiple ion adducts, compound name, formula, CAS number, etc., are all included | – |
| METLIN | LC-MS | http://metlin.scripps.edu | including nearly one million molecular standards with MS/MS data, and supporting multiple retrieval modes | Smith et al. (2005) |
| BinBase | GC-TOF-MS | http://fiehnlab.ucdavis.edu/staff/wohlgemuth/binbase/ | peak filtering and annotation using a mass spectral metadata-based filtering algorithm | Fiehn et al. (2005) |
| MMCD | NMR, LC-MS | http://mmcd.nmrfam.wisc.edu/ | compatible for identifying metabolites from both NMR and MS data | Cui et al. (2008) |
| SIRIUS | LC-MS | https://bio.informatik.uni-jena.de/sirius/ | comprehensive assessment of molecular structure using MS/MS data |
Böcker et al. (2009); Dührkop et al. (2019) |
| MassBank | LC-MS, GC-MS | https://massbank.eu/MassBank/ | a distributed database and ESI-MS2 data, under different experimental conditions, are included | Horai et al. (2010) |
| ReSpect | LC-MS | http://spectra.psc.riken.jp/ | plant-specific MS/MS-based data resource and database | Sawada et al. (2012) |
| CSI:FingerID | LC-MS/MS | https://www.csi-fingerid.uni-jena.de/ | combining fragmentation tree computation and machine learning for molecular structure searching | Dührkop et al. (2015) |
| LC-MS/MS library | LC-MS/MS | http://www.noble.org/apps/Scientific/WebDownloadManager/DownloadArea.aspx | ultra-high-performance liquid chromatography-tandem mass spectral library of plant natural products | Lei et al. (2015) |
| MS2LDA | LC-MS | http://ms2lda.org/ | Mass2Motifs-based method is used to annotate metabolites without the necessary of existing reference spectra; establishing biochemical relationships between molecules | van der Hooft et al. (2016) |
| GNPS | LC-MS | https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp | a natural product and metabolomics analysis platform using molecular networks | Wang et al. (2016) |
| NAP | LC-MS | https://gnps.ucsd.edu/ProteoSAFe/static/gnps-theoretical.jsp | a re-ranking system is used to increase the annotation rates | da Silva et al. (2018) |
| MetDNA | LC-MS | http://metdna.zhulab.cn/ | large-scale and ambiguous identification of metabolites from LC-MS/MS datasets without the need of a standard spectral library | Shen et al. (2019) |
| MMN | LC-MS | / | MicroTom metabolome and transcriptome dataset | Li et al. (2020) |
| KEGG | – | https://www.genome.jp/kegg/ | one of the most complete and widely used databases; containing metabolic pathways from a wide variety of organisms | Ogata et al. (1999) |
| MetaCyc | – | https://metacyc.org/ | experimentally elucidated metabolic pathway database | Caspi et al. (2020) |
| WikiPathways | – | https://www.wikipathways.org | a biological pathway database, including pathways from more than 30 species | Martens et al. (2021) |
| PMN15 | – | https://plantcyc.org/ | genome-wide metabolic pathway databases for 126 plants | Hawkins et al. (2021) |
Although lots of metabolites can be identified by spectral matching using databases mentioned above, it is inconvenient for most of them to annotate the majority of plant metabolites due to lack of standard MS/MS spectra (Shen et al., 2019). To address this problem, an unsupervised method MS2LDA, decomposes fragmentation spectra into co-occurring fragments and losses (Mass2Motifs), has been developed to annotate metabolites without the necessity of existing reference spectra. It has also been used to establish biochemical relationships between molecules (van der Hooft et al., 2016). GNPS (Global Natural Product Social Molecular Networking) is a natural product and metabolomics analysis platform. In this database, metabolites were annotated by MS/MS data-based molecular networking with the notion that the structurally related metabolites have similar mass spectra; therefore, a molecular network is established by the similarity of mass spectra, and the annotation of unknowns can be conducted based on the molecular networks (Wang et al., 2016). An integrated tool in GNPS, called Network Annotation Propagation (NAP), combined the molecular networks with a re-ranking system to increase the annotation rates, which is not only suitable for metabolites in molecular networks with a spectral library match but also useful for those without spectral matches to reference MS/MS data (da Silva et al., 2018). The strategy of MetDNA employs a metabolic reaction network-based recursive algorithm for annotating metabolites by reaction-paired neighbors, without the need for a comprehensive standard spectral library. The reaction pair is defined as the substrate and product in a metabolic reaction, which are linked by the similarity of their MS/MS spectra. Then, these reaction pairs are used as seeds to identify the adjacent metabolites in the metabolic network. The newly identified metabolites can be used as new seeds for recursive analysis. It was estimated that nearly 2000 metabolites could be annotated in an experiment using this method (Shen et al., 2019). Remarkably, most annotation tools/methods might have limitations in annotating isomers at present, since they have same molecular formula and similar mass spectrum, so a combined annotation method, using authenticated standards with the methods mentioned above, is recommended (Alseekh et al., 2021).
Except for the advanced databases or methods for metabolite annotation, numerous databases for metabolic pathway analysis were also developed (Table 2). For example, KEGG (Kyoto Encyclopedia of Genes and Genomes) (https://www.genome.jp/kegg/) is one of the most widely used databases containing numerous metabolic pathways and regulatory pathways (Ogata et al., 1999). MetaCyc (https://metacyc.org/) is a database containing pathways involved in both primary and secondary metabolism, in which nearly 2800 experimentally elucidated pathways from more than 3000 organisms are collected (Caspi et al., 2020). WikiPathways (https://www.wikipathways.org) is a biological pathways database, containing pathways from more than 30 species, including Oryza sativa, Zea mays, and so on (Martens et al., 2021). PMN15 (https://plantcyc.org) is a genome-wide metabolic pathway database for 126 plants (Hawkins et al., 2021).
Applications of plant metabolomics
Recently, with the development of genomics and MS, plant metabolomics has been widely used in multiple aspects, such as species discrimination (Souard et al., 2018), gene function, metabolic pathway analysis (Hirai et al., 2007; Fang et al., 2019), population genetic studies (Gong et al., 2013), and biomarker analysis (Swarbrick et al., 2006; Cañas et al., 2017).
Species discrimination
Metabolic fingerprinting is a promising tool for species discrimination (Duan et al., 2012; Liu et al., 2016; Souard et al., 2018). Mojia Huangqi and Menggu Huangqi are two important Chinese medical herbs, which share great similarities in morphology. To discriminate these two species, AFLP fingerprinting analysis and a GC-TOF/MS-based metabolic fingerprinting method were used. Three candidate AFLP markers, M40E41-5, M33E41-2 (existing in Mojia Huangqi collections), and M38E35-3 (only existing in Menggu Huangqi collections) were identified as candidate DNA markers for differentiation of two Huangqi species in genetic fingerprinting analysis. In GC-TOF/MS analysis, a total of 1193 metabolite features were detected, which could divide Huangqi collections into two distinct clusters (Menggu Huangqi and Mojia Huangqi). Eight metabolites, including malonic acid, xylose, pentonic acid, and other five unknown metabolites, were identified as candidate biomarkers for distinguishing Menggu Huangqi and Mojia Huangqi. After assigning detected metabolites in metabolic pathways and comparing metabolite levels in different species, it was found that some soluble sugars were accumulated in Mojia Huangqi, while some fatty acids, amino acids, and metabolites in polyamine metabolic pathways were decreased. The different metabolite accumulation patterns might relate to the specific distribution region of the two different Huangqi species. The correlation analysis of metabolite features and AFLP markers showed that there were 122 metabolites significantly correlated with 21 AFLP markers, indicating a complex correlation network of metabolic pathways and genetic regulation networks in Huangqi (Duan et al., 2012).
Dissection of metabolic pathways
Metabolite profiling combined with gain-of function and loss-of-function analysis seem to be powerful tools for plant functional genomics analysis (Fiehn et al., 2000; Fernie et al., 2004). Tanshinone diterpenoids produced from Salvia miltiorrhiza have various pharmacological activities. To thoroughly investigate the tanshinone metabolism pathway, the role of diterpene synthases (diTPSs), which are involved in diterpenoid biosynthesis, were analyzed in S. miltiorrhiza. It was found that there were five CPSs (SmCPS1 to SmCPS5) and two KSs (SmKSL1 and SmKSL2) in the genome of an inbred line bh2-7. Among them, SmCPS1 and SmKSL1 led to the biosynthesis of tanshinones in roots; SmCPS2 and SmKSL1 controlled the biosynthesis of tanshinones in the aerial; SmCPS4 and SmKSL2 could catalyze the formation of ent-13-epi-manoyl oxide in floral sepals; SmCPS5 and SmKSL2 were confirmed to be involved in gibberellin biosynthesis, while there was no product observed when combining SmCPS3 with either SmKSL1 or SmKSL2. To further explore the roles of SmCPSs in diterpenoid biosynthesis in S. miltiorrhiza, RNAi gene silencing was carried out. After analyzing metabolites in the roots of wild-type and SmCPS1-RNAi plants, it was found that 39 metabolites obtained from LC-QTOF/MS and 19 metabolites from GC-QqQ/MS analysis were significantly decreased in SmCPS1-RNAi lines, while another four metabolites (from GC-QqQ/MS analysis) were increased. Most of these differential compounds (21 out of 58 decreased metabolites, and 3 out of 4 increased metabolites) detected in the SmCPS1-RNAi plants were diterpenoids. Among 21 annotated metabolites with reduced level in the roots of SmCPS1-RNAi plants, 2 of them were rearranged abietane diterpenoids, a further 19 metabolites were tanshinones or plausible biosynthetic intermediates. These 19 features could further be divided into 5 main groups according to their structures, indicating a complex tanshinone metabolism network (Cui et al., 2015). In a further study, CYP71Ds (CYP71D373, CYP71D411, CYP71D464, and CYP71D375) were identified as candidate enzymes for tanshinone biosynthesis using a combination of methods for metabolic analysis of RNAi lines, genome sequencing, and biochemical analysis. Among them, CYP71D373 and CYP71D375 were required for heterocyclization to form the D-ring of tanshinones, and CYP71D411 was a C20 hydroxylase (Ma et al., 2021). In Catharanthus roseus, the iridoid synthase was identified as the enzyme involved in the iridoid biosynthetic pathway by biochemical assays, gene silencing, untargeted LC-MS analysis, co-expression analysis, and localization studies (Geu-Flores et al., 2012). For an un-sequenced species, the comprehensive analysis of metabolome, environmental, and physical data provide the information of plant adaption to the environment and pointing to genome specialization (Meijón et al., 2016).
Population genetic studies
The variation of metabolite abundance in genetic population is mainly influenced by genetic factors (Toubiana et al., 2012; Alseekh et al., 2015). Numerous loci or genes underlying variation of metabolites levels and agronomic traits have been identified by integrating metabolic profiling and genetic analysis (Luo, 2015; Fernie and Tohge, 2017). In Arabidopsis, 4213 QTLs were identified for metabolites that were detected in the 160 recombinant inbred lines (Ler × Cvi) (Keurentjes et al., 2006). Gong et al. (2013) detected 2800 mQTLs (metabolic quantitative trait loci) for 900 metabolites in a rice population with 241 recombinant inbred lines. Then 24 candidate genes were identified for numerous mQTLs based on chemical structure and pathway analysis. In rice, lots of loci were detected for the majority of metabolites using GWASs (genome-wide association studies) in a natural population. Then 36 genes were identified as candidate genes for the variation of metabolites related to the physiological and nutritional characteristics, in which the functions of five genes were verified by biochemical analysis (Chen et al., 2014). In maize, four genes involved in the primary metabolism were functionally characterized by the combination of metabolic profiling, GWAS, and qGWAS analysis (quantitative genome-wide association study, which links the expression levels of genes with the levels of the identified metabolites) (Wen et al., 2018). Two glycosyltransferase genes OsUGT706D1 (flavone 7-O-glucosyltransferase) and OsUGT707A2 (flavone 5-O-glucosyltransferase) have been proved to be not only involved in the synthesis or metabolism of flavonoids in rice but also related to UV tolerance after metabolic profiling, mGWAS, and biochemical analysis (Peng et al., 2017). To systematically study the effect of metabolites on tomato flavor, the flavor-related metabolites, such as sugars, volatile compounds, and organic acids, were detected among 398 tomato varieties. In addition, the flavors of different tomato varieties were evaluated in consumer panels. The correlation analysis showed that there were 33 metabolites significantly related to consumer preferences, and 37 metabolites obviously related to tomato flavor. Then whole-genome sequencing and GWAS were conducted to identify genetic loci for these flavor chemicals. The results showed that the contents of glucose and fructose were significantly associated with SNPs on chromosomes 9 and 11, and both loci were located in a domestication and improvement sweep (the large S. lycopersicum with lower nucleotide diversity), indicating that the sweetness (negatively correlated with fruit size)-related loci were lost during selection of fruit size. At the same time, lots of loci for carotenoid-derived volatiles, methyl salicylate, and guaiacol were also identified. The comprehensive analysis of metabolites and the correlated loci provided the understanding of the flavor deficiencies in modern tomato varieties and information for molecular breeding (Tieman et al., 2017).
Other applications
Plants produce a variety of secondary metabolites, and most of them can be used as allelochemicals (chemical barriers) for defense against herbivores, as well as for sexual communication (Nishida, 2014). The combination of plant metabolomics with ecological studies, that is chemical-ecology and eco-metabolomics, are necessary for dissecting the biochemical basis of numerous ecological interactions (Peters et al., 2018). At the same time, most plant secondary metabolites are the major source of drugs, so the development of plant metabolomics engineering will have significant influence on the discovery of new natural products as drugs (Li and Vederas, 2009). In addition, plant metabolomics has been widely used in identifying biomarkers for biological processes or traits of interest, which will greatly benefit the rapid phenotypic screening and crop breeding. For example, metabolites, such as leucine, isoleucine, aspartate, and glutamate, are correlated with the leaf age and the leaf senescence in Arabidopsis. The different ratio of glycine/serine can even be observed before any senescence symptoms in the rosettes, indicating it could be used as predictive indicator for plant senescence traits (Diaz et al., 2005). The metabolites related to disease resistance in barley were also identified by analyzing metabolites among different susceptibility and resistance barley varieties (Swarbrick et al., 2006). Chlorogenates (reversibly biosynthesized from quinic acid and shikimate) appear to be important biomarkers for selecting larger kernels in maize (Cañas et al., 2017).
Perspectives
With the development of genomics, transcriptomics, and MS, metabolomics developed rapidly in recent years, and has been widely applied in the study of pharmacology, toxicology, and disease diagnosis (Bar et al., 2020; Suceveanu et al., 2020), as well as in gene function analysis, and in improvement of crop yield and quality (Keurentjes et al., 2006; Schauer et al., 2006, 2008; Zhu et al., 2018). Normally, hundreds or even thousands of biological samples need to be analyzed in these studies. To fully understand the biological significance contained in these datasets, numerous research platforms, and data preprocessing, multivariate analysis, and metabolic pathway analysis methods or databases have been developed. Despite the prominent advantages of these methods and databases, there are still some problems currently existing in plant metabolomics, such as lack of rapid detection methods for trace substances, effective method for sample preparation, complete database for plant metabolites, and so on. Hence, it is necessary to develop a more broad-spectrum, in situ, and real-time detection method for plant samples based on existing technologies, such as mass spectrometry imaging technology (DESI-MSI). At the same time, an integrated platform is also needed for rapid and efficient analysis of the ultra-large MS data and annotation metabolites from different platforms. With the improvement of these technologies, and the combination of genomics and transcriptomics, plant metabolomics will play more important roles in gene function analysis, crop quality analysis, and many other studies in the future.
Funding
This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27010202), the National Natural Science Foundation of China (31920103003), and the National Key Research and Development Program of China (2016YFD0100904).
Acknowledgments
No conflict of interest declared.
Published: September 4, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
References
- Alseekh S., Aharoni A., Brotman Y., Contrepois K., D'Auria J., Ewald J., Ewald J.C., Fraser P.D., Giavalisco P., Hall R.D. Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nat. Methods. 2021;18:747–756. doi: 10.1038/s41592-021-01197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseekh S., Tohge T., Wendenberg R., Scossa F., Omranian N., Li J., Kleessen S., Giavalisco P., Pleban T., Mueller-Roeber B. Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell. 2015;27:485–512. doi: 10.1105/tpc.114.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar N., Korem T., Weissbrod O., Zeevi D., Rothschild D., Leviatan S., Kosower N., Lotan-Pompan M., Weinberger A., Le Roy C.I. A reference map of potential determinants for the human serum metabolome. Nature. 2020;588:135–140. doi: 10.1038/s41586-020-2896-2. [DOI] [PubMed] [Google Scholar]
- Böttcher C., von Roepenack-Lahaye E., Schmidt J., Schmotz C., Neumann S., Scheel D., Clemens S. Metabolome analysis of biosynthetic mutants reveals a diversity of metabolic changes and allows identification of a large number of new compounds in Arabidopsis. Plant Physiol. 2008;147:2107–2120. doi: 10.1104/pp.108.117754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcker S., Letzel M.C., Lipták Z., Pervukhin A. SIRIUS: decomposing isotope patterns for metabolite identification. Bioinformatics. 2009;25:218–224. doi: 10.1093/bioinformatics/btn603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhurst D., Goodacre R., Reinke S.N., Kuligowski J., Wilson I.D., Lewis M.R., Dunn W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics. 2018;14:72. doi: 10.1007/s11306-018-1367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckling C.D., Reddy I.R., Duran A.L., Zhao X.C., Sumner L.W. MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal. Chem. 2006;78:4334–4341. doi: 10.1021/ac0521596. [DOI] [PubMed] [Google Scholar]
- Bunk B., Kucklick M., Jonas R., Münch R., Schobert M., Jahn D., Hiller K. MetaQuant: a tool for the automatic quantification of GC/MS-based metabolome data. Bioinformatics. 2006;22:2962–2965. doi: 10.1093/bioinformatics/btl526. [DOI] [PubMed] [Google Scholar]
- Cañas R.A., Yesbergenova-Cuny Z., Simons M., Chardon F., Armengaud P., Quillere I., Cukier C., Gibon Y., Limami A.M., Nicolas S. Exploiting the genetic diversity of maize using a combined metabolomic, enzyme activity profiling, and metabolic modeling approach to link leaf physiology to kernel yield. Plant Cell. 2017;29:919–943. doi: 10.1105/tpc.16.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno-Quintero N., Bouwmeester H.J., Keurentjes J.J. Genetic analysis of metabolome-phenotype interactions: from model to crop species. Trends Genet. 2013;29:41–50. doi: 10.1016/j.tig.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Caspi R., Billington R., Keseler I.M., Kothari A., Krummenacker M., Midford P.E., Ong W.K., Paley S., Subhraveti P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes—a 2019 update. Nucleic Acids Res. 2020;48:D445–D453. doi: 10.1093/nar/gkz862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Gao Y.Q., Xie W.B., Gong L., Lu K., Wang W.S., Li Y., Liu X.Q., Zhang H.Y., Dong H.X. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 2014;46:714–721. doi: 10.1038/ng.3007. [DOI] [PubMed] [Google Scholar]
- Chetnik K., Petrick L., Pandey G. MetaClean: a machine learning-based classifier for reduced false positive peak detection in untargeted LC-MS metabolomics data. Metabolomics. 2020;16:117. doi: 10.1007/s11306-020-01738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Wishart D.S., Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinformatics. 2019;68:e86. doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- Cui G.H., Duan L.X., Jin B.L., Qian J., Xue Z.Y., Shen G.A., Snyder J.H., Song J.Y., Chen S.L., Huang L.Q. Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza. Plant Physiol. 2015;169:1607–1618. doi: 10.1104/pp.15.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Lewis I.A., Hegeman A.D., Anderson M.E., Li J., Schulte C.F., Westler W.M., Eghbalnia H.R., Sussman M.R., Markley J.L. Metabolite identification via theMmadison Metabolomics Consortium Database. Nat. Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- da Silva R.R., Wang M.X., Nothias L.F., van der Hooft J.J.J., Caraballo-Rodríguez A.M., Fox E., Balunas M.J., Klassen J.L., Lopes N.P., Dorrestein P.C. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol. 2018;14:e1006089. doi: 10.1371/journal.pcbi.1006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos R.C.H., Moco S., Lommen A., Keurentjes J.J.B., Bino R.J., Hall R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007;2:778–791. doi: 10.1038/nprot.2007.95. [DOI] [PubMed] [Google Scholar]
- DeFelice B.C., Mehta S.S., Samra S., Cajka T., Wancewicz B., Fahrmann J.F., Fiehn O. Mass Spectral Feature List Optimizer (MS-FLO): a tool to minimize false positive peak reports in untargeted liquid chromatography-mass spectroscopy (LC-MS) data processing. Anal. Chem. 2017;89:3250–3255. doi: 10.1021/acs.analchem.6b04372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C., Purdy S., Christ A., Morot-Gaudry J.F., Wingler A., Masclaux-Daubresse C.L. Characterization of markers to determine the extent and variability of leaf senescence in Arabidopsis. A metabolic profiling approach. Plant Physiol. 2005;138:898–908. doi: 10.1104/pp.105.060764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R.A., Strack D. Phytochemistry meets genome analysis, and beyond. Phytochemistry. 2003;62:815–816. doi: 10.1016/s0031-9422(02)00712-4. [DOI] [PubMed] [Google Scholar]
- Domingo-Almenara X., Brezmes J., Vinaixa M., Samino S., Ramirez N., Ramon-Krauel M., Lerin C., Diaz M., Ibanez L., Correig X. eRah: a computational tool integrating spectral deconvolution and alignment with quantification and identification of metabolites in GC/MS-based metabolomics. Anal. Chem. 2016;88:9821–9829. doi: 10.1021/acs.analchem.6b02927. [DOI] [PubMed] [Google Scholar]
- Duan L.X., Chen T.L., Li M., Chen M., Zhou Y.Q., Cui G.H., Zhao A.H., Jia W., Huang L.Q., Qi X.Q. Use of the metabolomics approach to characterize Chinese medicinal material Huangqi. Mol. Plant. 2012;5:376–386. doi: 10.1093/mp/ssr093. [DOI] [PubMed] [Google Scholar]
- Duan L.X., Ma A.M., Meng X.B., Shen G.A., Qi X.Q. QPMASS: a parallel peak alignment and quantification software for the analysis of large-scale gas chromatography-mass spectrometry (GC-MS)-based metabolomics datasets. J. Chromatogr. A. 2020;1620:460999. doi: 10.1016/j.chroma.2020.460999. [DOI] [PubMed] [Google Scholar]
- Duan L.X., Molnár I., Snyder J.H., Shen G.A., Qi X.Q. Discrimination and quantification of true biological signals in metabolomics analysis based on liquid chromatography-mass spectrometry. Mol. Plant. 2016;9:1217–1220. doi: 10.1016/j.molp.2016.05.009. [DOI] [PubMed] [Google Scholar]
- Dührkop K., Fleischauer M., Ludwig M., Aksenov A.A., Melnik A.V., Meusel M., Dorrestein P.C., Rousu J., Böcker S. Sirius 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods. 2019;16:299–302. doi: 10.1038/s41592-019-0344-8. [DOI] [PubMed] [Google Scholar]
- Dührkop K., Shen H.B., Meusel M., Rousu J., Böcker S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. U S A. 2015;112:12580–12585. doi: 10.1073/pnas.1509788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang C.Y., Fernie A.R., Luo J. Exploring the diversity of plant metabolism. Trends Plant Sci. 2019;24:83–98. doi: 10.1016/j.tplants.2018.09.006. [DOI] [PubMed] [Google Scholar]
- Fernández-Albert F., Llorach R., Andres-Lacueva C., Perera A. An R package to analyse LC/MS metabolomic data: MAIT (Metabolite Automatic Identification Toolkit) Bioinformatics. 2014;30:1937–1939. doi: 10.1093/bioinformatics/btu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Varela R., Tomasi G., Christensen J.H. An untargeted gas chromatography mass spectrometry metabolomics platform for marine polychaetes. J. Chromatogr. A. 2015;1384:133–141. doi: 10.1016/j.chroma.2015.01.025. [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Tohge T. The genetics of plant metabolism. Annu. Rev. Genet. 2017;51:287–310. doi: 10.1146/annurev-genet-120116-024640. [DOI] [PubMed] [Google Scholar]
- Fernie A.R., Trethewey R.N., Krotzky A.J., Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004;5:763–769. doi: 10.1038/nrm1451. [DOI] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- Fiehn O., Kopka J., Dormann P., Altmann T., Trethewey R.N., Willmitzer L. Metabolite profiling for plant functional genomics. Nat. Biotech. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Fiehn O., Wohlgemuth G., Scholz M. Setup and annotation of metabolomic experiments by integrating biological and mass spectrometric metadata. Mol. Cell. Biol. 2005;3615:224–239. [Google Scholar]
- Geu-Flores F., Sherden N.H., Courdavault V., Burlat V., Glenn W.S., Wu C., Nims E., Cui Y.H., O'Connor S.E. An alternative route to cyclic terpenes by reductive cyclization in iridoid biosynthesis. Nature. 2012;492:138–142. doi: 10.1038/nature11692. [DOI] [PubMed] [Google Scholar]
- Gong L., Chen W., Gao Y.Q., Liu X.Q., Zhang H.Y., Xu C.G., Yu S.B., Zhang Q.F., Luo J. Genetic analysis of the metabolome exemplified using a rice population. Proc. Nalt. Acad. Sci. U S A. 2013;110:20320–20325. doi: 10.1073/pnas.1319681110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrochategui E., Jaumot J., Tauler R. ROIMCR: a powerful analysis strategy for LC-MS metabolomic datasets. BMC Bioinformatics. 2019;20:256. doi: 10.1186/s12859-019-2848-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda G.A.N., Djukovic D. Overview of mass spectrometry-based metabolomics: opportunities and challenges. Methods Mol. Biol. 2014;1198:3–12. doi: 10.1007/978-1-4939-1258-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halket J.M., Przyborowska A., Stein S.E., Mallard W.G., Down S., Chalmers R.A. Deconvolution gas chromatography mass spectrometry of urinary organic acids-potential for pattern recognition and automated identification of metabolic disorders. Rapid Commun. Mass Spectrom. 1999;13:279–284. doi: 10.1002/(SICI)1097-0231(19990228)13:4<279::AID-RCM478>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Hawkins C., Ginzburg D., Zhao K.M., Dwyer W., Xue B., Xu A., Rice S., Cole B., Paley S., Karp P. Plant metabolic network 15: a resource of genome-wide metabolism databases for 126 plants and algae. J. Integr. Plant Biol. 2021 doi: 10.1111/jipb.13163. [DOI] [PubMed] [Google Scholar]
- Hiller K., Hangebrauk J., Jager C., Spura J., Schreiber K., Schomburg D. MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal. Chem. 2009;81:3429–3439. doi: 10.1021/ac802689c. [DOI] [PubMed] [Google Scholar]
- Hirai M.Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. U S A. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai H., Arita M., Kanaya S., Nihei Y., Ikeda T., Suwa K., Ojima Y., Tanaka K., Tanaka S., Aoshima K. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- Jiang W.X., Qiu Y.P., Ni Y., Su M.M., Jia W., Du X.X. An automated data analysis pipeline for GC-TOF-MS metabonomics studies. J. Proteome Res. 2010;9:5974–5981. doi: 10.1021/pr1007703. [DOI] [PubMed] [Google Scholar]
- Ju R., Liu X.Y., Zheng F.J., Zhao X.J., Lu X., Zeng Z.D., Lin X.H., Xu G.W. Removal of false positive features to generate authentic peak table for high-resolution mass spectrometry-based metabolomics study. Anal. Chim. Acta. 2019;1067:79–87. doi: 10.1016/j.aca.2019.04.011. [DOI] [PubMed] [Google Scholar]
- Kantz E.D., Tiwari S., Watrous J.D., Cheng S., Jain M. Deep neural networks for classification of LC-MS spectral peaks. Anal. Chem. 2019;91:12407–12413. doi: 10.1021/acs.analchem.9b02983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katajamaa M., Oresic M. Data processing for mass spectrometry-based metabolomics. J. Chromatogr. A. 2007;1158:318–328. doi: 10.1016/j.chroma.2007.04.021. [DOI] [PubMed] [Google Scholar]
- Katajamaa M., Miettinen J., Oresic M. MZmine: toolbox for processing and visualization of mass spectrometry based molecular profile data. Bioinformatics. 2006;22:634–636. doi: 10.1093/bioinformatics/btk039. [DOI] [PubMed] [Google Scholar]
- Keurentjes J.J.B., Fu J.Y., de Vos C.H.R., Lommen A., Hall R.D., Bino R.J., van der Plas L.H.W., Jansen R.C., Vreugdenhil D., Koornneef M. The genetics of plant metabolism. Nat. Genet. 2006;38:842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- Lei Z.T., Jing L., Qiu F., Zhang H., Huhman D., Zhou Z.Q., Sumner L.W. Construction of an ultrahigh pressure liquid chromatography-tandem mass spectral library of plant natural products and comparative spectral analyses. Anal. Chem. 2015;87:7373–7381. doi: 10.1021/acs.analchem.5b01559. [DOI] [PubMed] [Google Scholar]
- Lei Z.T., Li H.Q., Chang J.N., Zhao P.X., Sumner L.W. MET-IDEA version 2.06; improved efficiency and additional functions for mass spectrometry-based metabolomics data. Metabolomics. 2012;8:S105–S110. [Google Scholar]
- Li J.W.H., Vederas J.C. Drug discovery and natural products: end of an era or an endless frontier? Science. 2009;325:161–165. doi: 10.1126/science.1168243. [DOI] [PubMed] [Google Scholar]
- Li S.Z., Zeng S.L., Wu Y., Zheng G.D., Chu C., Yin Q., Chen B.Z., Li P., Lu X., Liu E.H. Cultivar differentiation of Citri Reticulatae Pericarpium by a combination of hierarchical three-step filtering metabolomics analysis, DNA barcoding and electronic nose. Anal. Chim. Acta. 2019;1056:62–69. doi: 10.1016/j.aca.2019.01.004. [DOI] [PubMed] [Google Scholar]
- Li Y., Chen Y., Zhou L., You S.J., Deng H., Chen Y., Alseekh S., Yuan Y., Fu R., Zhang Z.X. Microtom metabolic network: rewiring tomato metabolic regulatory network throughout the growth cycle. Mol. Plant. 2020;13:1203–1218. doi: 10.1016/j.molp.2020.06.005. [DOI] [PubMed] [Google Scholar]
- Liang D.D., Liu Q., Zhou K.J., Jia W., Xie G.X., Chen T.L. IP4M: an integrated platform for mass spectrometry-based metabolomics data mining. BMC Bioinformatics. 2020;21:444. doi: 10.1186/s12859-020-03786-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Bai X., Yang F.Q., Zhang X.J., Hu Y.J., Li P., Wan J.B. Discriminating from species of curcumae radix (yujin) by a UHPLC/Q-TOFMS-based metabolomics approach. Chin. Med. 2016;11:21. doi: 10.1186/s13020-016-0095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen A. MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Anal. Chem. 2009;81:3079–3086. doi: 10.1021/ac900036d. [DOI] [PubMed] [Google Scholar]
- Luedemann A., Strassburg K., Erban A., Kopka J. TagFinder for the quantitative analysis of gas chromatography-mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics. 2008;24:732–737. doi: 10.1093/bioinformatics/btn023. [DOI] [PubMed] [Google Scholar]
- Luo J. Metabolite-based genome-wide association studies in plants. Curr. Opin. Plant Biol. 2015;24:31–38. doi: 10.1016/j.pbi.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Ma Y., Cui G.H., Chen T., Ma X.H., Wang R.S., Jin B.L., Yang J., Kang L.P., Tang J.F., Lai C.J.S. Expansion within the CYP71D subfamily drives the heterocyclization of tanshinones synthesis in Salvia miltiorrhiza. Nat. Commun. 2021;12:685. doi: 10.1038/s41467-021-20959-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens M., Ammar A., Riutta A., Waagmeester A., Slenter D.N., Hanspers K., Miller R.A., Digles D., Lopes E.N., Ehrhart F. WikiPathways: connecting communities. Nucleic Acids Res. 2021;49:D613–D621. doi: 10.1093/nar/gkaa1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijó M., Feito I., Oravec M., Delatorre C., Weckwerth W., Majada J., Valledor L. Exploring natural variation of Pinus pinaster Aiton using metabolomics: is it possible to identify the region of origin of a pine from its metabolites? Mol. Ecol. 2016;25:959–976. doi: 10.1111/mec.13525. [DOI] [PubMed] [Google Scholar]
- Melamud E., Vastag L., Rabinowitz J.D. Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A.D., Tsentalovich Y.P., Yanshole V.V. Deep learning for the precise peak detection in high-resolution LC-MS data. Anal. Chem. 2020;92:588–592. doi: 10.1021/acs.analchem.9b04811. [DOI] [PubMed] [Google Scholar]
- Ni Y., Qiu Y.P., Jiang W.X., Suttlemyre K., Su M.M., Zhang W.C., Jia W., Du X.X. ADAP-GC 2.0: deconvolution of coeluting metabolites from GC-TOF-MS data for metabolomics studies. Anal. Chem. 2012;84:6619–6629. doi: 10.1021/ac300898h. [DOI] [PubMed] [Google Scholar]
- Ni Y., Su M.M., Qiu Y.P., Jia W., Du X.X. ADAP-GC 3.0: improved peak detection and deconvolution of co-eluting metabolites from GC/TOF-MS data for metabolomics studies. Anal. Chem. 2016;88:8802–8811. doi: 10.1021/acs.analchem.6b02222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida R. Chemical ecology of insect-plant interactions: ecological significance of plant secondary metabolites. Biosci. Biotechnol. Biochem. 2014;78:1–13. doi: 10.1080/09168451.2014.877836. [DOI] [PubMed] [Google Scholar]
- Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M., Shahzad R., Gul A., Subthain H., Shen S.Q., Lei L., Zheng Z.G., Zhou J.J., Lu D.D., Wang S.C. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 2017;8:1975. doi: 10.1038/s41467-017-02168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K., Worrich A., Weinhold A., Alka O., Balcke G., Birkemeyer C., Bruelheide H., Calf O.W., Dietz S., Duhrkop K. Current challenges in plant eco-metabolomics. Int. J. Mol. Sci. 2018;19:1385. doi: 10.3390/ijms19051385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskal T., Castillo S., Villar-Briones A., Oresic M. MZmine2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D. 2010. flagme: Analysis of Metabolomics GC/MS Data, R package version 1.14.0.https://bioconductor.org/packages/release/bioc/html/flagme.html [Google Scholar]
- Robinson M.D., De Souza D.P., Keen W.W., Saunders E.C., McConville M.J., Speed T.P., Likic V.A. A dynamic programming approach for the alignment of signal peaks in multiple gas chromatography-mass spectrometry experiments. BMC Bioinformatics. 2007;8:419. doi: 10.1186/1471-2105-8-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röst H.L., Sachsenberg T., Aiche S., Bielow C., Weisser H., Aicheler F., Andreotti S., Ehrlich H.C., Gutenbrunner P., Kenar E. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat. Methods. 2016;13:741–748. doi: 10.1038/nmeth.3959. [DOI] [PubMed] [Google Scholar]
- Sauvage F.L., Gaulier J.M., Lachatre G., Marquet P. Pitfalls and prevention strategies for liquid chromatography-tandem mass spectrometry in the selected reaction-monitoring mode for drug analysis. Clin. Chem. 2008;54:1519–1527. doi: 10.1373/clinchem.2008.105478. [DOI] [PubMed] [Google Scholar]
- Sawada Y., Nakabayashi R., Yamada Y., Suzuki M., Sato M., Sakata A., Akiyama K., Sakurai T., Matsuda F., Aoki T. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: a plant-specific MS/MS-based data resource and database. Phytochemistry. 2012;82:38–45. doi: 10.1016/j.phytochem.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Schauer N., Semel Y., Balbo I., Steinfath M., Repsilber D., Selbig J., Pleban T., Zamir D., Fernie A.R. Mode of inheritance of primary metabolic traits in tomato. Plant Cell. 2008;20:509–523. doi: 10.1105/tpc.107.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N., Semel Y., Roessner U., Gur A., Balbo I., Carrari F., Pleban T., Perez-Melis A., Bruedigam C., Kopka J. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 2006;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- Shen X.T., Wang R.H., Xiong X., Yin Y.D., Cai Y.P., Ma Z.J., Liu N., Zhu Z.J. Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics. Nat. Commun. 2019;10:1516. doi: 10.1038/s41467-019-09550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui M.A., Pandey S., Azim A., Sinha N., Siddiqui M.H. Metabolomics: an emerging potential approach to decipher critical illnesses. Biophys. Chem. 2020;267:106462. doi: 10.1016/j.bpc.2020.106462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A., Qiu Y.P., Jia W., Walker D.I., Jones D.P., Du X.X. ADAP-GC 4.0: application of clustering-assisted multivariate curve resolution to spectral deconvolution of gas chromatography-mass spectrometry metabolomics data. Anal. Chem. 2019;91:9069–9077. doi: 10.1021/acs.analchem.9b01424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.A., O'Maille G., Want E.J., Qin C., Trauger S.A., Brandon T.R., Custodio D.E., Abagyan R., Siuzdak G. Metlin—a metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Smith C.A., Want E.J., O'Maille G., Abagyan R., Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Souard F., Delporte C., Stoffelen P., Thevenot E.A., Noret N., Dauvergne B., Kauffmann J.M., Van Antwerpen P., Stevigny C. Metabolomics fingerprint of coffee species determined by untargeted-profiling study using LC-HRMS. Food Chem. 2018;245:603–612. doi: 10.1016/j.foodchem.2017.10.022. [DOI] [PubMed] [Google Scholar]
- Stein S.E. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Am. Soc. Mass Spectrom. 1999;10:770–781. [Google Scholar]
- Suceveanu A.I., Mazilu L., Katsiki N., Parepa I., Voinea F., Pantea-Stoian A., Rizzo M., Botea F., Herlea V., Serban D. NLRP3 inflammasome biomarker—could be the new tool for improved cardiometabolic syndrome outcome. Metabolites. 2020;10:448. doi: 10.3390/metabo10110448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R., Mckeown P.C. Moving toward a comprehensive map of central plant metabolism. Annu. Rev. Plant Biol. 2015;66:187–210. doi: 10.1146/annurev-arplant-043014-114720. [DOI] [PubMed] [Google Scholar]
- Swarbrick P.J., Schulze-Lefert P., Scholes J.D. Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ. 2006;29:1061–1076. doi: 10.1111/j.1365-3040.2005.01472.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H., Morimoto T., Ogasawara N., Kanaya S. AMDORAP: non-targeted metabolic profiling based on high-resolution LC-MS. BMC Bioinformatics. 2011;12:259. doi: 10.1186/1471-2105-12-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R., Patti G.J., Rinehart D., Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal. Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D., Zhu G.T., Resende M.F.R., Lin T., Taylor M., Zhang B., Ikeda H., Liu Z.Y., Fisher J., Zemach I. A chemical genetic roadmap to improved tomato flavor. Science. 2017;355:391–394. doi: 10.1126/science.aal1556. [DOI] [PubMed] [Google Scholar]
- t'Kindt R., Morreel K., Deforce D., Boerjan W., Van Bocxlaer J. Joint GC-MS and LC-MS platforms for comprehensive plant metabolomics: repeatability and sample pre-treatment. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2009;877:3572–3580. doi: 10.1016/j.jchromb.2009.08.041. [DOI] [PubMed] [Google Scholar]
- Toubiana D., Semel Y., Tohge T., Beleggia R., Cattivelli L., Rosental L., Nikoloski Z., Zamir D., Fernie A.R., Fait A. Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genet. 2012;8:e1002612. doi: 10.1371/journal.pgen.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., Kanazawa M., VanderGheynst J., Fiehn O., Arita M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 2015;12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hooft J.J.J., Wandy J., Barrett M.P., Burgess K.E.V., Rogers S. Topic modeling for untargeted substructure exploration in metabolomics. Proc. Natl. Acad. Sci. U S A. 2016;113:13738–13743. doi: 10.1073/pnas.1608041113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.X., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C.A., Luzzatto-Knaan T. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Want E., Masson P. Processing and analysis of GC/LC-MS-based metabolomics data. Methods Mol. Biol. 2011;708:277–298. doi: 10.1007/978-1-61737-985-7_17. [DOI] [PubMed] [Google Scholar]
- Want E.J., Wilson I.D., Gika H., Theodoridis G., Plumb R.S., Shockcor J., Holmes E., Nicholson J.K. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010;5:1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- Wen B., Mei Z.L., Zeng C.W., Liu S.Q. metaX: a flexible and comprehensive software for processing metabolomics data. BMC Bioinformatics. 2017;18:183. doi: 10.1186/s12859-017-1579-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W.W., Jin M., Li K., Liu H.J., Xiao Y.J., Zhao M.C., Alseekh S., Li W.Q., Lima F.D.E., Brotman Y. An integrated multi-layered analysis of the metabolic networks of different tissues uncovers key genetic components of primary metabolism in maize. Plant J. 2018;93:1116–1128. doi: 10.1111/tpj.13835. [DOI] [PubMed] [Google Scholar]
- Wu S.W., Fan Z., Xiao Y.L. Comprehensive relative quantitative metabolomics analysis of lycopodium alkaloids in different tissues of Huperzia serrata. Synth. Syst. Biotechnol. 2018;3:44–55. doi: 10.1016/j.synbio.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.T., Li X.T., Wang W., Shan Y.H., Wang C.T., Zhu M.L., La Q., Zhong Y., Xu Y., Nan P. Integrated metabolomics and transcriptomics study of traditional herb Astragalus membranaceus Bge. var. mongolicus (Bge.) Hsiao reveals global metabolic profile and novel phytochemical ingredients. BMC Genomics. 2020;21:697. doi: 10.1186/s12864-020-07005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.G., Psychogios N., Young N., Wishart D.S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37:W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.X., Huan T. Patterned signal ratio biases in mass spectrometry-based quantitative metabolomics. Anal. Chem. 2021;93:2254–2262. doi: 10.1021/acs.analchem.0c04113. [DOI] [PubMed] [Google Scholar]
- Yu T.W., Park Y., Johnson J.M., Jones D.P. apLCMS—adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T.W., Park Y., Li S.Z., Jones D.P. Hybrid feature detection and information accumulation using high-resolution LC-MS metabolomics data. J. Proteome Res. 2013;12:1419–1427. doi: 10.1021/pr301053d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaynab M., Fatima M., Abbas S., Sharif Y., Umair M., Zafar M.H., Bahadar K. Role of secondary metabolites in plant defense against pathogens. Microb. Pathog. 2018;124:198–202. doi: 10.1016/j.micpath.2018.08.034. [DOI] [PubMed] [Google Scholar]
- Zeki Ö.C., Eylem C.C., Recber T., Kir S., Nemutlu E. Integration of GC-MS and LC-MS for untargeted metabolomics profiling. J. Pharm. Biomed. Anal. 2020;190:113509. doi: 10.1016/j.jpba.2020.113509. [DOI] [PubMed] [Google Scholar]
- Zhang Y.Y., Zhang Q., Zhang Y.M., Wang W.W., Zhang L., Yu Y.J., Bai C.C., Guo J.Z., Fu H.Y., She Y.B. A comprehensive automatic data analysis strategy for gas chromatography-mass spectrometry based untargeted metabolomics. J. Chromatogr. A. 2020;1616:460787. doi: 10.1016/j.chroma.2019.460787. [DOI] [PubMed] [Google Scholar]
- Zhu G.T., Wang S.C., Huang Z.J., Zhang S.B., Liao Q.G., Zhang C.Z., Lin T., Qin M., Peng M., Yang C.K. Rewiring of the fruit metabolome in tomato breeding. Cell. 2018;172:249–261. doi: 10.1016/j.cell.2017.12.019. [DOI] [PubMed] [Google Scholar]