Abstract
Root hair formation in Arabidopsis thaliana is a well-established model system for epidermal patterning and morphogenesis in plants. Over the last decades, many underlying regulatory genes and well-established networks have been identified by thorough genetic and molecular analysis. In this study, we used a forward genetic approach to identify genes involved in root hair development in Arabis alpina, a related crucifer species that diverged from A. thaliana approximately 26–40 million years ago. We found all root hair mutant classes known in A. thaliana and identified orthologous regulatory genes by whole-genome or candidate gene sequencing. Our findings indicate that the gene-phenotype relationships regulating root hair development are largely conserved between A. thaliana and A. alpina. Concordantly, a detailed analysis of one mutant with multiple hairs originating from one cell suggested that a mutation in the SUPERCENTIPEDE1 (SCN1) gene is causal for the phenotype and that AaSCN1 is fully functional in A. thaliana. Interestingly, we also found differences in the regulation of root hair differentiation and morphogenesis between the species, and a subset of root hair mutants could not be explained by mutations in orthologs of known genes from A. thaliana. This analysis provides insight into the conservation and divergence of root hair regulation in the Brassicaceae.
Keywords: root hair, Arabis alpina, patterning, morphogenesis, SCN1, R2R3MYB, bHLH
Introduction
Evolutionary studies of development often aim to understand how changes in the function of genes or gene networks result in phenotypic differences. One approach is to compare gene functions in two species that are sufficiently evolutionarily distant to find differences but close enough to identify orthologous genes. In plants, Arabidopsis thaliana serves as an ideal reference system for evolutionary comparisons, due to the rich knowledge of the genetic, molecular, and cell-biological mechanisms underlying various developmental processes. Within the Brassicaceae family, Arabis alpina has been established as an additional genetic model system (Koch et al., 2006; Wang et al., 2009, 2011; Poncet et al., 2010). A. thaliana and A. alpina have an evolutionary distance of 26–40 million years (Koch et al., 2006; Beilstein et al., 2010). The genome of A. alpina is fully sequenced (Willing et al., 2015; Jiao et al., 2017), and orthologous genes can be identified by homology and their relative position on the chromosomes (synteny). The exhaustive search for trichome mutants in A. alpina and their molecular analysis revealed a similar genetic network as described in A. thaliana (Chopra et al., 2014, 2019). However, distinct differences were also found between both Brassicaceae species, in particular for the function of genes involved in trichome patterning (Chopra et al., 2019).
In A. thaliana, root hairs form in single-cell files (H-files), which develop above the cleft of two cortical cells. Non-hair files (N-files) are formed over cortical cells (Dolan et al., 1994; Schiefelbein, 2000). The positional information from the underlying cortex cells is mediated by the leucine-rich repeat receptor-like kinase SCRAMBLED (SCM; Kwak et al., 2005). H-file cells (trichoblasts) and N-file cells (atrichoblasts) differ from early development. Trichoblasts display a higher cell division rate (Berger et al., 1998), reduced cell length (Dolan et al., 1994; Masucci et al., 1996), and denser cytoplasm (Dolan et al., 1994; Galway et al., 1994). In A. thaliana, most trichome patterning genes are also involved in root hair patterning (Schiefelbein, 2003; Schellmann et al., 2007). Proteins encoded by the patterning genes include the R2R3MYB transcription factor WEREWOLF (WER; Lee and Schiefelbein, 1999) and the WD40 protein TRANSPARENT TESTA GLABRA 1 (TTG1; Galway et al., 1994) that form a complex with the bHLH proteins GLABRA3 (GL3; Payne et al., 2000) and ENHANCER OF GLABRA 3 (EGL3; Zhang et al., 2003). The activity of this complex is counteracted by the partially redundant R3MYBs CAPRICE (CPC) and TRIPTYCHON (TRY; Wada et al., 1997; Schellmann et al., 2002; Schiefelbein, 2003) and their activator ENHANCER OF TRY AND CPC 1 (ETC1; Simon et al., 2007). Ultimately, the cooperation of these MYB, bHLH, and WD40 genes activates the expression of GLABRA2 (GL2), which encodes a homeodomain transcription factor that suppresses root hair development in non-root hair cells (Masucci et al., 1996).
Root hair development begins with polarized outgrowth at the basal end of the cell (Molendijk et al., 2001). Initially, RHO-RELATED PROTEIN FROM PLANTS (ROP) localizes to the growth site and remains localized at the very tip of the growing root hair during root hair extension (Molendijk et al., 2001). Tip growth involves the condensation of the endoplasmic reticulum (Ridge et al., 1999), F-actin accumulation (Baluška et al., 2000), and microtubules, which are involved in regulating the localization and size of the bulges (Kost et al., 1999). The second phase of root hair formation begins when hairs are about 40μm long (Dolan et al., 1994). It is characterized by localized growth at the tip of the hair (Carol et al., 2005). This process is organized by polarized cytoplasm in the bulge, which mediates localized secretion and cell wall synthesis (Favery et al., 2001). In summary, the major molecular events during tip growth include a tip-focused calcium influx (Wymer et al., 1997), cytoskeleton re-modelling (Geitmann and Emons, 2000), polarized membrane trafficking (Campanoni and Blatt, 2007), and cell wall synthesis (Favery et al., 2001).
Root hair development in A. alpina is similar to A. thaliana (Chopra et al., 2014, 2019). Early in development two files of morphologically different cells, as judged by their cell length, are formed. However, in contrast to A. thaliana, 30–40 percent of cells in N-file positions also form root hairs (Chopra et al., 2014). The phenotypic analysis of Aattg1 and Aagl3 revealed that both mutants have excessive root hair production (Chopra et al., 2014, 2019). For TTG1, this implies similar functions in A. alpina and A. thaliana. For GL3, however, this finding represents a striking difference to A. thaliana where extra hairs are found in gl3 egl3 double mutants but not in the single mutants (Bernhardt et al., 2003).
In this study, we screened an ethyl methane-sulfonate (EMS)-mutagenized population of A. alpina plants for root hair mutants. We uncovered mutants affecting root hair cell patterning and most steps of root hair development. We identified orthologous genes in A. alpina by taking into consideration not only the sequence similarity but also the relative position of genes on the chromosomes. Candidate genes were sequenced in the mutants to reveal the gene-phenotype relations in A. alpina. These data demonstrate that root hair cell regulation is largely conserved between the two species, but also identify interesting differences.
Materials and Methods
Plant Material and Growth Condition
All A. alpina mutants were isolated from EMS-mutagenized A. alpina Pajares and pep1-1 populations (Wang et al., 2009; Chopra et al., 2014). The A. thaliana scn1-3 mutant was described previously (Carol et al., 2005).
For root hair analysis, seeds were surface sterilized with chlorine gas for 3h. Sterilized seeds were sown on full Murashige-Skoog plates (Murashige and Skoog, 1962) w/o sucrose and stratified at 4°C for 5days. Plants were grown vertically for 7days under long-day conditions (16-h light, 8-h darkness) at 21°C.
For whole-genome sequencing, plants were grown on plates as described before and the root phenotypes were confirmed in the M3 generation. Next, the seedlings were transferred to soil for three more weeks. Fresh leaf samples of 1-month-old plants were used for DNA extraction.
Sequence and Synteny Analysis
Arabis alpina gene sequences were obtained from the Genomic resources for A. alpina website1 and analyzed with CLC DNA Workbench 5.6.1.
For sequence analysis by Sanger sequencing, primers were designed outside the CDS of a given A. alpina gene to sequence it in the mutants. Library preparation and whole-genome sequencing were carried out by the Cologne Center for Genomics, and the raw sequence reads were deposited into National Center for Biotechnology Information2 under accession number PRJNA745061. Short reads of each mutant sample were aligned to the A. alpina reference genome version 5.1 (Jiao et al., 2017) using Bowtie2 version 2.2.8, which were further processed with SAMtools version 1.4 (Li et al., 2009) to get BAM files. Next, each BAM was provided to SHORE version 0.8 (Ossowski et al., 2008) to call consensus and variants for each mutant sample. SNPs were stringently selected for each mutant with a minimum mutant allele frequency of 0.85 and a coverage of at least 3. Effects of selected mutations on gene integrity were annotated with SHOREmap version 3.0 (Schneeberger et al., 2009; Sun and Schneeberger, 2015).
The EMBL-EBI database (Pfam 32.03; Finn et al., 2016) was used to identify conserved domains.
GBrowse from TAIR 10,4 along with the assembled A. alpina genome, was used to confirm the synteny of the selected genes regarding conserved order and appearance of the neighboring genes.
Plasmids and Stable Plant Transformation
AtSCN1 and AaSCN1 were amplified by PCR, using A. thaliana Col-0 or A. alpina Pajares cDNA as templates, and subsequently cloned into pDONOR201 and/or pDONOR207 using the Gateway system (Thermo Fisher Scientific). For construction of 35S::AtSCN1-YFP and 35S::AaSCN1-YFP, the Gateway destination vector pEXSG-YFP (Feys et al., 2005) was used. For construction of the Aascn1-1 mutant CDS, site-directed mutagenesis was performed on pDONOR207-AaSCN1 and Aascn1-1-YFP was subsequently cloned into pEXSG-YFP (Feys et al., 2005). Plants were transformed using floral dip (Clough and Bent, 1998).
Root Hair and Root Epidermal Cell Measurements
The number of root hairs was determined in a 1mm long section as described before (Grierson and Schiefelbein, 2002). The position of H-files was determined on 7-day-old seedlings with respect to the position of underlying cortex cells.
Staining of Roots
For epidermal cell length analysis, roots were stained with 100mg/ml propidium iodide for 1min and subsequently washed with water. Nitroblue tetrazolium chloride (NBT, Sigma) was used to identify sites of superoxide production (Fryer et al., 2002). Seedlings were covered with NBT solution (0.5mg/ml NBT in 0.1M potassium phosphate pH 7) for 10min.
Microscopical Methods
Stereo microscopy was carried out with the Leica MZ 16F stereo microscope and the LAS AF software (Leica Microsystems, Heidelberg, Germany). Light and fluorescence microscopy were performed with the Leica DMRA2, DMRB, and DM5000B fluorescence microscope (Leica Microsystems, Heidelberg, Germany). Confocal laser scanning microscopy was carried out with the Leica DM5500 CS Microscope and documented with the TCS-SPE imaging systems (Leica Microsystems, Heidelberg, Germany).
Adobe Photoshop CS4 version 14.0.0 and ImageJ (Fabrice Cordelieres, Institut Curie, Orsay, France) were used for image processing.
Statistical Analysis
Data were analyzed with Microsoft Excel 2007. Statistical analysis was carried out with OriginPro 8.5 0G SR0 and Microsoft Excel 2010. Significance was tested using a two-sided t test for normally distributed data and one-way ANOVA for data with non-normal distribution.
Results
Root Hair Development in A. alpina
Root hair development in A. alpina is similar to A. thaliana. Root hairs are initiated at the basal end of root epidermal cells, followed by extensive tip growth (Figures 1A,D; Grierson and Schiefelbein, 2002). However, all cell files in A. alpina can produce root hairs [(Chopra et al., 2014), Figure 1A]. Nevertheless, trichoblasts and atrichoblasts differ in cell length (Chopra et al., 2014), so it is conceivable that A. alpina also produces morphologically distinct H- and N-files in the root epidermis. To substantiate this, root hairs were stained with propidium iodide to identify the H-file position by their position over the cleft of the underlying cortex cells (Figures 1B,C). We measured the length of four consecutive cells in N- and H-files starting immediately below the first cell displaying the formation of a root hair bulge. Cells located close to the root tip show no length difference. More distant cells were about 30% shorter in H-positions as compared to N-positions (Figure 1E), indicating that a morphological difference between these cell types is conserved between the two species.
Figure 1.
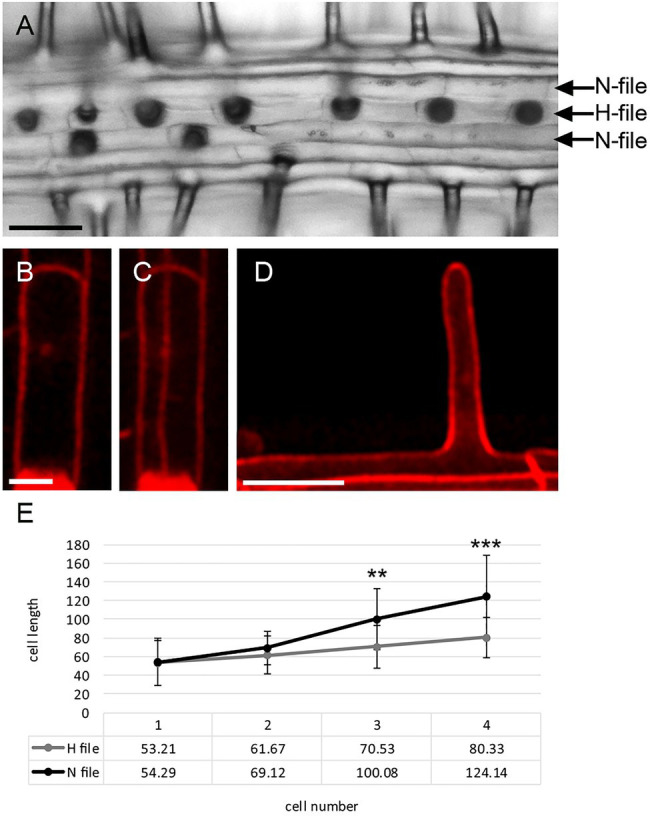
Root hair development in Arabis alpina. (A) Light microscope picture of the root epidermis. The position of cell files relative to the cortex cells is indicated with the H-position defined to be above the cleft between two underlying cells and the N-position above underlying cells. Scale bar: 50mm. Propidium iodide-stained cell in the H-position: (B) the plane of the epidermal cell, (C) the plane of the epidermal cell and of the cortex cells. Scale bar: 25mm. (D) Propidium iodide-stained root hair growing out at the apical end of the root hair cell. Scale bar: 20mm. (E) Quantitative analysis of cell length in N- and H-positions. Cell length is shown for four cells in a cell file with the first cell representing the closest to the tip and the fourth cell being immediately below the first root hair cell showing a bulge. Significance levels were determined at p<0.001 (***), p<0.01 (**).
Isolation of Root Hair Mutants in A. alpina
We used a forward genetic approach to define different functional steps during root hair development in A. alpina. Two EMS screens were performed in independent populations: one in Pajares (Wang et al., 2009), representing 4,205M1 plants, and the other in the pep1-1 background (Wang et al., 2009; Albani et al., 2012; Bergonzi et al., 2013; Zhou et al., 2021), representing 6,800M1 individuals. Seeds of five M1 plants were pooled, and 50 M2 seedlings from each pool were screened for root hair phenotypes. Forty-five root hair pattern and root hair morphology mutant phenotypes were confirmed in the M3 generation (Table 1). We found two types of patterning mutants: Seven mutants showed more root hairs (Figure 2B), and six had fewer or no root hairs (Figure 2C) compared to the wild type (Figures 2A and 3A). The morphology mutants were divided into six classes. Three lines produced only small bulges (Figure 3B), suggesting that cell elongation is disturbed. In six mutants, hairs initiated from a swelling of the epidermal cell (Figure 3C). Three lines showed a bursting phenotype (Figure 3D). Typically, these root hairs ruptured at the tip soon after initiation. One line produced multiple sites of growth from each epidermal cell (Figure 3E). Sixteen mutants exhibited short hairs (Figure 3F), and three mutants produced branched (Figure 3G) and wavy root hairs (Figure 3H). A similar range of phenotypes has been previously described in A. thaliana (Grierson and Schiefelbein, 2002).
Table 1.
Classes of root hair mutants in A. alpina.
| Root hair phenotype | Pajares | pep1-1 | Total | |
|---|---|---|---|---|
| Patterning | More hairs | 2 | 5 | 7 |
| Fewer hairs | 3 | 3 | 6 | |
| Morphology | Only bulge | 1 | 2 | 3 |
| Swollen hairs | 1 | 5 | 6 | |
| Bursting hairs | 1 | 2 | 3 | |
| Multiple hairs | 1 | 0 | 1 | |
| Short hairs | 2 | 14 | 16 | |
| Branched/wavy hairs | 1 | 2 | 3 | |
| Total | 12 | 33 | 45 |
Figure 2.

Root hair patterning phenotypes in A. alpina. Light microscopic images of (A) wild type, (B) increased root hair number (line 1,264), (C) no root hairs (line 1,101). Scale bar: 100mm.
Figure 3.
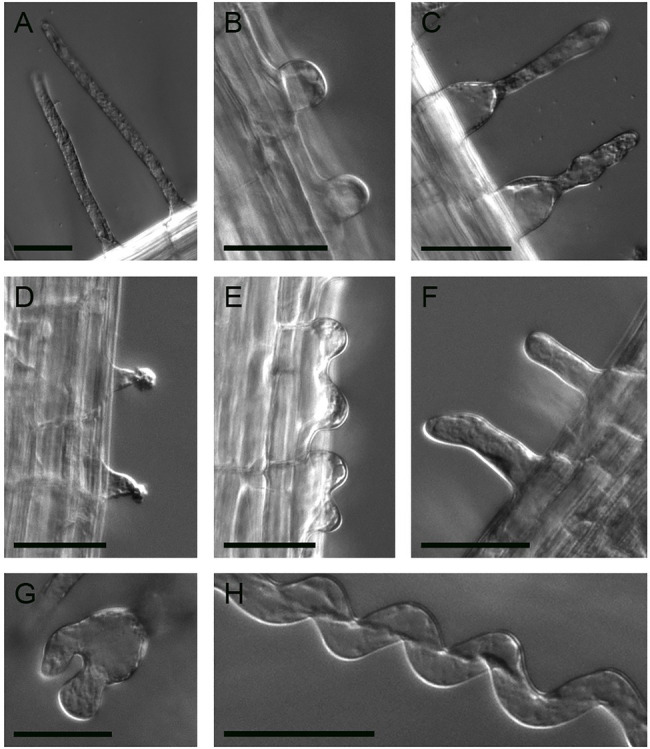
Root hair morphogenesis phenotypes. (A) Wild-type root hair. (B) Root hair growth stops after bulge formation (line 366). (C) Root hairs are swollen at the base (line 1,221). (D) Bursting root hairs (line 1,185). (E) Multiple outgrowths from each epidermal cell (line 1,384). (F) Short root hairs (line 1,091). (G) Branched root hairs (line 216). (H) Wavy root hairs (line 216). Scale bar: 50μm.
Identification of Root Hair Genes in A. alpina
A. alpina and A. thaliana are both members of the Brassicaceae and therefore relatively closely related (Koch et al., 2006; Beilstein et al., 2010). Hence, we reasoned that most mutant phenotypes found in the A. alpina root hair screen are probably caused by mutations in orthologs of genes shown to affect root hair development in A. thaliana. In a first step, we selected genes known to be involved in root hair development of A. thaliana (Supplementary Table S1). We also included genes involved in trichome development, because of genetic overlap between the two processes, and pollen tube genes, since pollen tubes show tip growth similar to root hairs. In total, we considered 136 A. thaliana genes. In a next step, we identified 283 corresponding paralogs or orthologs based on the annotated A. alpina genome (Willing et al., 2015; Supplementary Table S1). This way, we constructed a list of candidate A. alpina genes that might be impaired in function in our set of root hair mutants.
Identification of Mutant-Specific Alleles
We then sequenced candidate genes in selected root hair mutants, as previously described for trichome genes in A. alpina (Chopra et al., 2014, 2019). However, the high number of candidate genes rendered gene by gene sequencing inefficient. Therefore, we performed whole-genome sequencing of the genomic DNA from different mutants, aiming for an average of 15X fold coverage. Each SNP identified in the genes of interest is listed in Supplementary Table S2 along with the position in the chromosome, the sequence of the reference allele and the mutant allele, the location of exchange regarding the gene model, mutant allele coverage, and mutant allele frequency. Annotated SNPs, stringently selected for each mutant, with a minimum mutant allele frequency of 0.85 and minimum mutant allele coverage of 3, were chosen for further analysis. Next, we correlated SNPs to the phenotype of the respective mutants. As summarized in Supplementary Table S3, 26 mutant-specific alleles were identified. We considered stop codons as mutant-specific alleles, while other mutations were only considered when leading to a non-synonymous amino acid exchange.
Root Hair Patterning Mutants
The analysis of the seven mutants with ectopic root hair production revealed one line with a mutation in the AaTTG1 and six lines with mutations in the AaGL3 genes (Supplementary Table S3), with similar phenotypes as described previously (Chopra et al., 2014, 2019). None of the mutants with an ectopic root hair phenotype exhibited mutations in AaWER, although the Arabidopsis wer mutant shows this phenotype. Six mutants exhibited fewer root hairs as compared to the wild type. None of these showed mutations in any of the selected candidate genes compiled in Supplementary Table S1.
Root Hair Morphology Mutants
Among the 38 root hair morphology mutants (Figure 4), we identified 12 genes displaying mutations in our collection of candidate genes (Supplementary Figure S1; Supplementary Table S3).
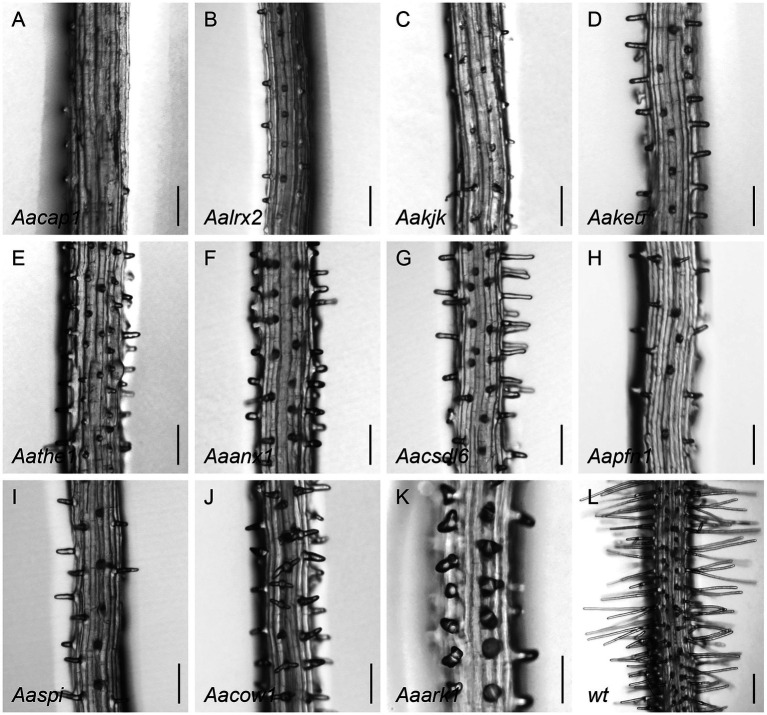
Figure 4.
Root hair phenotypes of identified A. alpina mutants in (A) [CA2+]CYT-ASSOCIATED PROTEIN KINASE 1 (CAP1), (B) LEUCINE-RICH REPEAT/EXTENSIN 2 (LRX2), (C) KOJAK (KJK), (D) KEULE (KEU), (E) THESEUS 1 (THE1), (F) ANXUR 1 (ANX1), (G) CELLULOSE SYNTHASE-LIKE D6 (CSLD6), (H) PROFILIN 1 (PFN1), (I) SPIRRIG (SPI), (J) CAN OF WORMS1 (COW1), (K) ARMADILLO REPEAT-CONTAINING KINESIN 1 (ARK1), and of the (L) wild type. Scale bar: 250μm.
Sequencing of three mutants showing only small bulges confirmed allele-specific changes in two of them. We found an E to K exchange at position 465 in the [CA2+]CYT-ASSOCIATED PROTEIN KINASE 1 (CAP1) gene (Figure 4A; Supplementary Figure S1A). CAP1 is a receptor-like kinase which maintains cytoplasmic Ca2+ gradients and is important for root hair growth in A. thaliana. The Atcap1 mutant displays shorter, slightly malformed hairs (Bai et al., 2014). In the Aacap1 line, the mutant phenotype appeared slightly stronger as compared to the mutant in A. thaliana, as only small bulges without any outgrowth were formed. The second small bulge mutant showed a P to T exchange at position 588 in LEUCINE-RICH REPEAT/EXTENSIN 2 (LRX2; Figure 4B; Supplementary Figure S1B). In A. thaliana, LRX2 and its paralog LRX1 act together during root hair elongation such that Atlrx1 single mutants and Atlrx1 Atlrx2 double mutants but not Atlrx2 mutants exhibit aborted, swollen, or branched root hairs (Baumberger et al., 2003).
Sequencing of the six mutants with swollen root hairs revealed no relevant mutations in any of the selected candidate genes.
All three mutant lines showing bursting root hairs carried mutations in the KOJAK (KJK) gene (Figure 4C; Supplementary Figure S1C). The phenotype is similar to that of A. thaliana kjk mutants (Favery et al., 2001), indicating a similar function of KJK in both species. KJK is a cellulose synthase-like protein required for the synthesis of non-cellulose cell wall polysaccharides. In one line, we found a premature STOP codon at a similar position [W (843) to STOP] as in Atkjk-3 (Favery et al., 2001) and two other alleles presented G (248) to R and T (854) to M substitutions in conserved regions, which are expected to cause severe defects in protein function.
Analysis of 16 short root hair mutants revealed mutations in eight known root hair genes. In one line, we found a mutation in the KEULE (KEU) gene (Figure 4D; Supplementary Figure S1D), which encodes a Sec1 protein required for cytokinesis (Grierson and Schiefelbein, 2002). The mutation leads to a G (487) to R exchange within the Sec1 domain. Unlike in A. thaliana (Grierson and Schiefelbein, 2002), we did not observe swollen root hairs in the Aakeu mutant. In addition, we found mutant lines with SNPs in THESEUS 1 (THE1) and ANXUR 1 (ANX1; Hématy et al., 2007; Boisson-Dernier et al., 2009; Miyazaki et al., 2009). Both encode receptor-like tyrosine kinase proteins (Cheung and Wu, 2011). Aathe1 has a G (614) to R exchange in the tyrosine kinase domain and Aaanx1 a D (813) to N exchange after the tyrosine kinase domain (Figures 4E,F; Supplementary Figures S1E,F). The respective A. thaliana mutants appear to have a stronger phenotype as the root hairs are not only shorter but may collapse or burst (Duan et al., 2010; Cheung and Wu, 2011). We also identified one short root hair line with an E (289) to K exchange in the CELLULOSE SYNTHASE-LIKE D6 (CSLD6) protein (Figure 4G; Supplementary Figure S1G). The members of the CSLD family, including KJK (CSLD3), are known to be essential for the structural integrity of cell walls of tip-growing cells (Bernal et al., 2008). It is therefore conceivable that AaCSLD6 plays a role in root hair development in A. alpina. In one of the short root hair mutants, we found a G (64) to E substitution in PROFILIN1 (PFN1; Figure 4H; Supplementary Figure S1H). PFN is an actin-binding protein involved in the organization of the cytoskeleton which plays a general role in cell elongation in A. thaliana (Ramachandran et al., 2000). Similar as in Aapfn1, A. thaliana 35S::antisense PFN 1 lines displayed shorter root hairs (Ramachandran et al., 2000). Two short root hair mutants had STOP codons in SPIRRIG (SPI) at positions 2,107 and 1976, similar to Atspi-4.2 (Saedler et al., 2009; Figure 4I; Supplementary Figure S1I). Four spi alleles in A. alpina with similar phenotypes have been previously reported (Chopra et al., 2019; Stephan et al., 2021). In four short root hair mutants, we found meaningful SNPs in the CAN OF WORMS 1 (COW1) gene (Grierson et al., 1997; Böhme et al., 2004; Figure 4J; Supplementary Figure S1J). cow1 hairs in A. thaliana are shorter and wider than wild type and occasionally produce two hairs (Grierson et al., 1997; Böhme et al., 2004). Detailed analysis of the four identified Aacow1 alleles revealed the same range of phenotypes. Mutations in AaCOW1 included one premature STOP codon at position 505, two splicing site mutations [G (1849) to A and G (1679) to A], resulting in STOP codons at different positions similar to Atcow1 alleles (Grierson et al., 1997; Böhme et al., 2004). In one allele, we found a P (293) to L substitution in the conserved CRAL-TRIO lipid binding domain (Supplementary Figure S1).
Sequencing of mutants with wavy and branched root hairs revealed STOP codons at positions 979 and 899 in the ARMADILLO REPEAT-CONTAINING KINESIN 1 (ARK1) gene (Yang et al., 2007; Yoo and Blancaflor, 2013; Figure 4K; Supplementary Figure S1K). ARK1 controls microtubule organization during root hair tip growth. In Atark1 mutants, fragmentation and random orientation of microtubules results in wavy/spiral and branched root hairs (Yang et al., 2007; Yoo and Blancaflor, 2013; Rishmawi et al., 2014).
The protein sequence analysis of all identified morphology genes showed a high similarity between A. alpina and A. thaliana (Supplementary Table S4), and synteny is also maintained for these genes between both species (Supplementary Figure S2).
Multiple Hairs: Analysis of AaSCN1 in A. alpina
One of the first mutants identified showed multiple bulges that are initiated from each epidermal cell and multiple growing axes initiated from each bulge (Figure 5). This mutant was shown to have a STOP codon in the SCN1 gene (Carol et al., 2005; Supplementary Figure S1). We decided to analyze this mutant in more detail in parallel to the systematic analysis of the other mutants. SCN1 encodes a RhoGTPase GDP dissociation inhibitor (RhoGDI), which is involved in localizing the RHD2/AtrbohC NADPH oxidase to the tip of root hairs (Carol et al., 2005). The RHD2/AtrbohC NADPH oxidase is required for ROS production, which in turn is essential for root hair elongation (Foreman et al., 2003). The AaSCN1 protein shows 84 percent similarity to AtSCN1 (Supplementary Table S4), and synteny is also maintained between the two species (Supplementary Figure S2).
Figure 5.
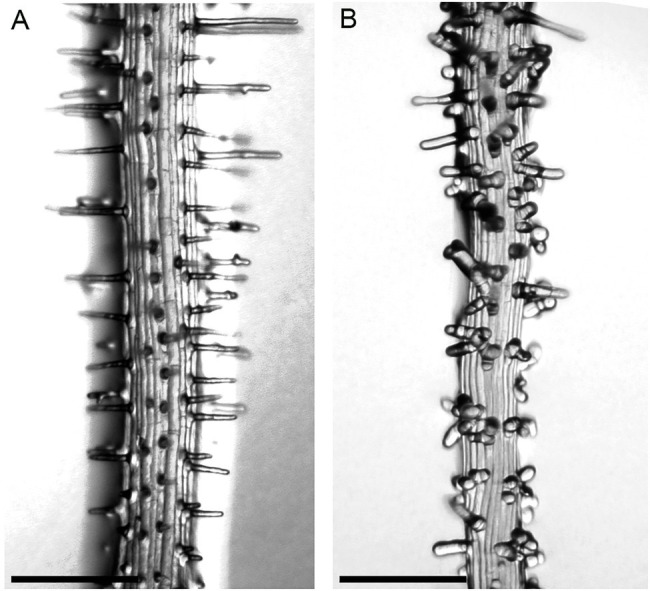
Root hair phenotype of the Aascn1 mutant. (A) Wild type. (B) Aascn1 exhibits multiple bulges initiated from each epidermal cell and multiple growing axes initiated from each bulge. Scale bar: 250μm.
To monitor the spatial production of ROS, we used Nitrobluetetrazolium (NBT). NBT is reduced to a blue formazan precipitate in the presence of ROS (Fryer et al., 2002). NBT staining in the root hairs of wild type showed blue staining at one single point of the hair tip in the wild type (Figure 6A). In the Aascn1-1 mutant, the blue color was not observed in a focal point but in a broader area (Figure 6B). Next, we analyzed the localization of the SCN1 protein from A. alpina and A. thaliana. The SCN1 CDS was fused to YFP and expressed in Col-0 plants under control of the 35S promoter. Two lines for AtSCN1-YFP and two lines for AaSCN1-YFP were analyzed. Both A. alpina and A. thaliana SCN1-YFP were detected at the tip of growing hairs (Figures 6C,D). However, AaSCN1-YFP was less restricted to the tip than its A. thaliana ortholog.
Figure 6.
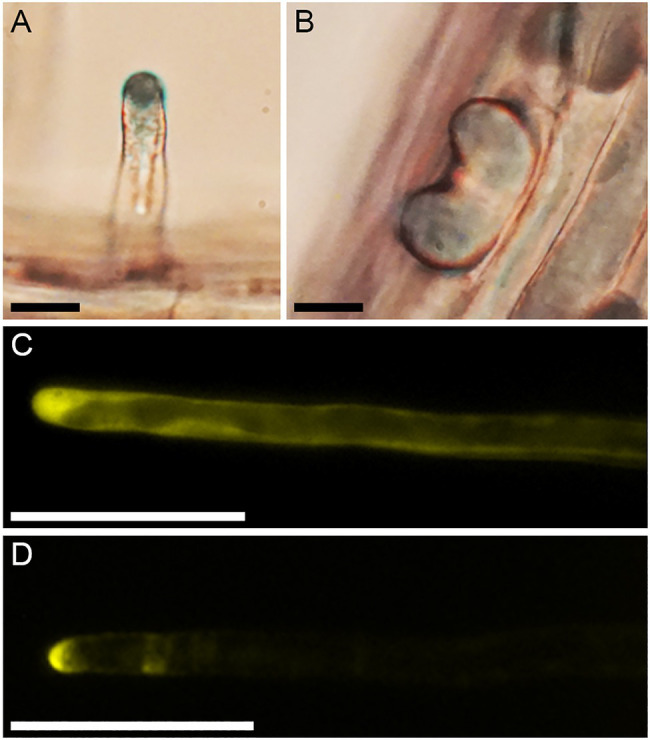
ROS and SCN1 localization in root hairs. NBT staining of (A) wild type A. alpina and (B) Aascn1-1. Localization of the SCN1 protein from (C) A. alpina and (D) Arabidopsis thaliana in A. thaliana Col-0 background. Scale bar: 10μm (A, B) and 50μm (C, D).
To confirm that the phenotype observed in the Aascn1-1 mutant is caused by the mutation in AaSCN1, we initially tried rescue experiments in the A. alpina Pajares scn1-1 mutant but failed to isolate transgenic lines. We therefore performed rescue experiments in A. thaliana. We reasoned that the Atscn1-3 mutant (Carol et al., 2005) may be rescued by wild-type SCN1 from A. alpina but not by the Aascn1-1 mutant protein. Transgenic plants, expressing A. thaliana and A. alpina SCN1 wild-type versions, rescued the root hair defect of Atscn1-3 in T1 and T2 generations (Figure 7). This indicates that the AaSCN1 protein is fully functional in A. thaliana. Transgenic lines, expressing the Aascn1-1 mutant version, showed a root hair defective phenotype similar to Atscn1-3 (Figure 7). This indicates that the Aascn1-1 mutation renders the protein defective and suggests that the mutation in AaSCN1 causes the root hair branching phenotype.
Figure 7.

Root hairs of Atscn1-3 line overexpressing SCN1 versions. Wild-type AaSCN1 CDS expression in (A) YFP channel and (B) transmission channel. Mutant Aascn1-1 CDS expression in (C) YFP channel and (D) transmission channel. Scale bar: 75μm.
Discussion
This study used a forward genetic approach to identify genes involved in root hair morphogenesis and patterning in A. alpina. Our phenotypic comparison revealed a similar range of phenotypes as previously described for A. thaliana (Grierson and Schiefelbein, 2002). By comparing genes known from A. thaliana and those involved in the closely related process of trichome development in A. alpina, we uncovered genes affecting most steps of root hair development. Overall, 26 mutant-specific alleles were identified through candidate-driven gene selection and sequencing and whole-genome sequencing including re-sequencing of the respective mutated genes.
Importantly, we correlate mutant-specific alleles with phenotypes. In cases where we found several alleles correlating with the same phenotype, the evidence that the mutations are causal for the phenotype is particularly strong. In cases where we found only one allele, additional proof by rescue experiments is necessary. In these cases, we consider it reasonable to assume that relevant mutations in genes that lead to the same phenotype in A. alpina and A. thaliana reflect a similar function of these genes in both species.
Our sequence analysis of candidate genes in the respective mutants revealed on average about two mutants for each considered gene. As observed in other screens (Pollock and Larkin, 2004), the distribution of alleles is highly asymmetric, such that we found six gl3 alleles and only one allele for many other genes. However, the average allele frequency of two alleles for each gene suggests that we have identified a representative set of mutants (Chopra et al., 2019).
Differences and Similarities in Root Hair Patterning Between A. alpina and A. thaliana
In A. thaliana, the R2R3MYB transcription factor WER (Lee and Schiefelbein, 1999) and the WD40 protein TTG1 (Galway et al., 1994) form a complex with the bHLH proteins GL3/EGL3 (Payne et al., 2000; Zhang et al., 2003) that activates the expression of GL2, which in turn suppresses root hair development in non-root hair cells (Masucci et al., 1996).
As expected, various mutants with ectopic root hairs showed relevant mutations in AaTTG1 and AaGL3. Similar as had been shown for trichomes (Chopra et al., 2019), Aagl3 mutants showed a strong phenotype and none of the mutants was defective in AaEGL3. We found no mutations in AaWER. This is particularly interesting as we have previously found no gl1 mutants in glabrous trichome mutants (Chopra et al., 2019). While this may be due to the mutant screen not having been fully saturated, it is also possible that the MYB genes act redundantly in A. alpina, such that single gene mutations do not lead to a phenotype.
Surprisingly, all six mutants displaying a reduced number of root hairs had no mutations in any of the known A. thaliana genes in which mutations lead to fewer root hairs.
Differences and Similarities in Root Hair Morphology Between A. alpina and A. thaliana
The analysis of the root hair morphology mutants derived from our screen revealed that various genes, including ARK1, SPI, COW, PFN1, CAP1, and KJK, have similar functions as described in A. thaliana. In addition, we found one short root hair mutant with a relevant amino acid substitution in another AaKJK family member, the AaCSLD6 gene. Although the corresponding mutant was not yet described in A. thaliana, our finding suggests that CSLD6 is involved in root hair morphogenesis.
We also found possible differences in the gene-phenotype relationships. While in A. thaliana only lrx1 and the lrx1 lrx2 double mutants show a root hair phenotype (Baumberger et al., 2003), we found that a relevant amino acid exchange in the AaLRX2 gene correlates with a short root phenotype. This suggests that the relative importance of the AaLRX1 and AaLRX2 genes is different in A. alpina and A. thaliana. Also, unlike in A. thaliana (Grierson and Schiefelbein, 2002), we observed short but not swollen root hairs in the Aakeu mutant. This suggests that mutations in the same gene lead to slightly different phenotypes in the two species. Similarly, the1 and anx1 mutants in A. thaliana exhibit collapsed, burst, and short root hairs (Duan et al., 2010), while the corresponding A. alpina mutants showed short but intact root hairs.
Truncation of AaSCN1 Leads to a Multiple Root Hair Phenotype
Our attempt to analyze the function of the AaSCN1 gene was challenging because we failed to generate transgenic lines in A. alpina to prove that the mutant phenotype is indeed caused by the mutation in the gene. However, interspecies experiments using A. thaliana provided evidence that the mutation in the AaSCN1 gene renders it non-functional, suggesting that the scn1 mutant phenotypes are very similar in the two species. This includes not only the morphological phenotype but also the ROS distribution in the mutants of both species (Carol et al., 2005). Moreover, we showed that YFP fused SCN1 from A. thaliana and A. alpina localized to the tip of growing root hairs.
Perspective
In this work, we discovered interesting differences in the gene-to-phenotype relationship between A. alpina and A. thaliana and their further analysis may help to understand the evolution of the underlying gene regulatory networks. Moreover, we found root hair mutants without relevant SNPs in any of the known A. thaliana root hair genes. In principle, it is possible that in these cases, SNPs in the promoter regions cause transcriptional changes of the candidate genes. However, as this was rarely observed in EMS screens before, it is conceivable that the mutant phenotypes can be explained by genetic redundancies or mechanistic differences in root hair formation in A. alpina. In either case, the identification of the corresponding genes will likely reveal new players important for root hair development.
Data Availability Statement
The data sets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA745061.
Author Contributions
MH and MM conceived and designed the analysis. MM and DC collected the data. MM, DC, AS, HS, and LS performed the analysis. MH, LS, and MM wrote the paper. KS, MA, and GC provided access to crucial research components. AS, KS, MA, and GC provided revisions to scientific content of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft grant HU 497 (MH), the SFB680 (MH), and an International Max Planck Research School fellowship (MM).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Sabine Lohmer for her excellent technical assistance.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.767772/full#supplementary-material
References
- Albani M. C., Castaings L., Wötzel S., Mateos J. L., Wunder J., Wang R., et al. (2012). PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genet. 8:1003130. doi: 10.1371/journal.pgen.1003130, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Ma X., Zhang G., Song S., Zhou Y., Gao L., et al. (2014). A receptor-like kinase mediates ammonium homeostasis and is important for the polar growth of root hairs in Arabidopsis. Plant Cell 26, 1497–1511. doi: 10.1105/tpc.114.124586, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluška F., Salaj J., Mathur J., Braun M., Jasper F., Šamaj J., et al. (2000). Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 227, 618–632. doi: 10.1006/dbio.2000.9908, PMID: [DOI] [PubMed] [Google Scholar]
- Baumberger N., Steiner M., Ryser U., Keller B., Ringli C. (2003). Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J. 35, 71–81. doi: 10.1046/j.1365-313X.2003.01784.x, PMID: [DOI] [PubMed] [Google Scholar]
- Beilstein M. A., Nagalingum N. S., Clements M. D., Manchester S. R., Mathews S. (2010). Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 107, 18724–18728. doi: 10.1073/pnas.0909766107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F., Hung C.-Y., Dolan L., Schiefelbein J. (1998). Control of cell division in the root epidermis of Arabidopsis thaliana. Dev. Biol. 194, 235–245. doi: 10.1006/dbio.1997.8813, PMID: [DOI] [PubMed] [Google Scholar]
- Bergonzi S., Albani M. C., Van Themaat E. V. L., Nordström K. J. V., Wang R., Schneeberger K., et al. (2013). Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340, 1094–1097. doi: 10.1126/science.1234116, PMID: [DOI] [PubMed] [Google Scholar]
- Bernal A. J., Yoo C. M., Mutwil M., Jensen J. K., Hou G., Blaukopf C., et al. (2008). Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing arabidopsis cells. Plant Physiol. 148, 1238–1253. doi: 10.1104/pp.108.121939, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt C., Lee M. M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J., et al. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, 6431–6439. doi: 10.1242/dev.00880 [DOI] [PubMed] [Google Scholar]
- Böhme K., Li Y., Charlot F., Grierson C., Marrocco K., Okada K., et al. (2004). The Arabidopsis COW1 gene encodes a phosphatidylinositol transfer protein essential for root hair tip growth. Plant J. 40, 686–698. doi: 10.1111/j.1365-313X.2004.02245.x, PMID: [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Roy S., Kritsas K., Grobei M. A., Jaciubek M., Schroeder J. I., et al. (2009). Disruption of the pollen-expressed FERONIA homologs ANXUR1 and ANXUR2 triggers pollen tube discharge. Development 136, 3279–3288. doi: 10.1242/dev.040071, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanoni P., Blatt M. R. (2007). Membrane trafficking and polar growth in root hairs and pollen tubes. J. Exp. Bot. 58, 65, –74. doi: 10.1093/jxb/erl059 [DOI] [PubMed] [Google Scholar]
- Carol R. J., Takeda S., Linstead P., Durrant M. C., Kakesova H., Derbyshire P., et al. (2005). A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438, 1013–1016. doi: 10.1038/nature04198, PMID: [DOI] [PubMed] [Google Scholar]
- Cheung A. Y., Wu H. M. (2011). THESEUS 1, FERONIA and relatives: A family of cell wall-sensing receptor kinases? Curr. Opin. Plant Biol. 14, 632–641. doi: 10.1016/j.pbi.2011.09.001, PMID: [DOI] [PubMed] [Google Scholar]
- Chopra D., Mapar M., Stephan L., Albani M. C., Deneer A., Coupland G., et al. (2019). Genetic and molecular analysis of trichome development in Arabis alpina. Proc. Natl. Acad. Sci. U. S. A. 116, 12078–12083. doi: 10.1073/pnas.1819440116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra D., Wolff H., Span J., Schellmann S., Coupland G., Albani M. C., et al. (2014). Analysis of TTG1 function in Arabis alpina. BMC Plant Biol. 14, 16. doi: 10.1186/1471-2229-14-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S., Bent A. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x, PMID: [DOI] [PubMed] [Google Scholar]
- Dolan L., Duckett C. M., Grierson C., Linstead P., Schneider K., Lawson E., et al. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development. 120, 2465–2474. doi: 10.1242/dev.120.9.2465 [DOI] [Google Scholar]
- Duan Q., Kita D., Li C., Cheung A. Y., Wu H. M. (2010). FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. U. S. A. 107, 17821–17826. doi: 10.1073/pnas.1005366107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B., Ryan E., Foreman J., Linstead P., Boudonck K., Steer M., et al. (2001). KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev. 15, 79–89. doi: 10.1101/gad.188801, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B. J., Wiermer M., Bhat R. A., Moisan L. J., Medina-Escobar N., Neu C., et al. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17, 2601–2613. doi: 10.1105/tpc.105.033910, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J. H. F., Mylona P., Miedema H., Angel Torres M., et al. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. doi: 10.1038/nature01485, PMID: [DOI] [PubMed] [Google Scholar]
- Fryer M. J., Oxborough K., Mullineaux P. M., Baker N. R. (2002). Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 53, 1249–1254. doi: 10.1093/jxb/53.372.1249 [DOI] [PubMed] [Google Scholar]
- Galway M. E., Masucci J. D., Lloyd A. M., Walbot V., Davis R. W., Schiefelbein J. W. (1994). The TTG gene is required to specify epidermal cell fate andcell patterning in the arabidopsis root. Dev. Biol. 166, 740–754. doi: 10.1006/dbio.1994.1352, PMID: [DOI] [PubMed] [Google Scholar]
- Geitmann A., Emons A. M. C. (2000). The cytoskeleton in plant and fungal cell tip growth. J. Microsc. 198, 218–245. doi: 10.1046/j.1365-2818.2000.00702.x, PMID: [DOI] [PubMed] [Google Scholar]
- Grierson C. S., Roberts K., Feldmann K. A., Dolan L. (1997). The COW1 locus of Arabidopsis acts after RHD2, and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol. 115, 981–990. doi: 10.1104/pp.115.3.981, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C., Schiefelbein J. (2002). Root hairs. Arab. B. 1:e0060. doi: 10.1199/tab.0060, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K., Sado P. E., Van Tuinen A., Rochange S., Desnos T., Balzergue S., et al. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17, 922–931. doi: 10.1016/j.cub.2007.05.018, PMID: [DOI] [PubMed] [Google Scholar]
- Jiao W. B., Accinelli G. G., Hartwig B., Kiefer C., Baker D., Severing E., et al. (2017). Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res. 27, 778–786. doi: 10.1101/gr.213652.116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M. A., Kiefer C., Ehrich D., Vogel J., Brochmann C., Mummenhoff K. (2006). Three times out of Asia minor: the phylogeography of Arabis alpina L. (Brassicaceae). Mol. Ecol. 15, 825–839. doi: 10.1111/j.1365-294X.2005.02848.x, PMID: [DOI] [PubMed] [Google Scholar]
- Kost B., Mathur J., Chua N.-H. (1999). Cytoskeleton in plant development. Curr. Opin. Plant Biol. 2, 462–470. doi: 10.1016/S1369-5266(99)00024-2, PMID: [DOI] [PubMed] [Google Scholar]
- Kwak S. H., Shen R., Schiefelbein J. (2005). Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307, 1111–1113. doi: 10.1126/science.1105373, PMID: [DOI] [PubMed] [Google Scholar]
- Lee M. M., Schiefelbein J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. doi: 10.1016/S0092-8674(00)81536-6, PMID: [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci J. D., Rerie W. G., Foreman D. R., Zhang M., Galway M. E., Marks M. D., et al. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122, 1253–1260. doi: 10.1242/dev.122.4.1253, PMID: [DOI] [PubMed] [Google Scholar]
- Miyazaki S., Murata T., Sakurai-Ozato N., Kubo M., Demura T., Fukuda H., et al. (2009). ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr. Biol. 19, 1327–1331. doi: 10.1016/j.cub.2009.06.064, PMID: [DOI] [PubMed] [Google Scholar]
- Molendijk A. J., Bischoff F., Rajendrakumar C. S. V., Friml J., Braun M., Gilroy S., et al. (2001). Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 20, 2779–2788. doi: 10.1093/emboj/20.11.2779, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R. M., Lanz C., Warthmann N., Weigel D. (2008). Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18, 2024–2033. doi: 10.1101/gr.080200.108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C. T., Zhang F., Lloyd A. M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction With GL1 and TTG1. Genetics 156, 1349–1362. doi: 10.1093/genetics/156.3.1349, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock D. D., Larkin J. C. (2004). Estimating the degree of saturation in mutant screens. Genetics 168, 489–502. doi: 10.1534/genetics.103.024430, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet B. N., Herrmann D., Gugerli F., Taberlet P., Holderegger R., Gielly L., et al. (2010). Tracking genes of ecological relevance using a genome scan in two independent regional population samples of Arabis alpina. Mol. Ecol. 19, 2896–2907. doi: 10.1111/j.1365-294X.2010.04696.x, PMID: [DOI] [PubMed] [Google Scholar]
- Ramachandran S., Christensen H. E. M., Ishimaru Y., Dong C. H., Chao-Ming W., Cleary A. L., et al. (2000). Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 124, 1637–1647. doi: 10.1104/pp.124.4.1637, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge R. W., Uozumi Y., Plazinski J., Hurley U. A., Williamson R. E. (1999). Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol. 40, 1253–1261. doi: 10.1093/oxfordjournals.pcp.a029513, PMID: [DOI] [PubMed] [Google Scholar]
- Rishmawi L., Sun H., Schneeberger K., Hülskamp M., Schrader A. (2014). Rapid identification of a natural knockout allele of armadillo repeat-containing kinesin1 that causes root hair branching by mapping-by-sequencing. Plant Physiol. 166, 1280–1287. doi: 10.1104/pp.114.244046, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler R., Jakoby M., Marin B., Galiana-Jaime E., Hülskamp M. (2009). The cell morphogenesis gene SPIRRIG in Arabidopsis encodes a WD/BEACH domain protein. Plant J. 59, 612–621. doi: 10.1111/j.1365-313X.2009.03900.x, PMID: [DOI] [PubMed] [Google Scholar]
- Schellmann S., Hülskamp M., Uhrig J. (2007). Epidermal pattern formation in the root and shoot of Arobidopsis. Biochem. Soc. Trans., 146–148. doi: 10.1042/BST0350146 [DOI] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beerman A., et al. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21, 5036–5046. doi: 10.1093/emboj/cdf524, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. W. (2000). Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 124, 1525–1531. doi: 10.1104/pp.124.4.1525, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. (2003). Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6, 74–78. doi: 10.1016/S136952660200002X, PMID: [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A. H., Nielsen K. L., et al. (2009). SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods 6, 550–551. doi: 10.1038/nmeth0809-550, PMID: [DOI] [PubMed] [Google Scholar]
- Simon M., Lee M. M., Lin Y., Gish L., Schiefelbein J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311, 566–578. doi: 10.1016/j.ydbio.2007.09.001, PMID: [DOI] [PubMed] [Google Scholar]
- Stephan L., Jakoby M., Hülskamp M. (2021). Evolutionary comparison of the developmental/physiological phenotype and the molecular behavior of SPIRRIG Between Arabidopsis thaliana and Arabis alpina. Front. Plant Sci. 11:2115. doi: 10.3389/fpls.2020.596065, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Schneeberger K. (2015). Shoremap v3.0: fast and accurate identifi cation of causal mutations from forward genetic screens. Methods Mol. Biol. 1284, 381–395. doi: 10.1007/978-1-4939-2444-8_19 [DOI] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog. CPC. Science. 277, 1113–1116. doi: 10.1126/science.277.5329.1113, PMID: [DOI] [PubMed] [Google Scholar]
- Wang R., Albani M., Vincent C., Bergonzi S., Luan M., Bai Y., et al. (2011). Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell 23, 1307–1321. doi: 10.1105/tpc.111.083451, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Farrona S., Vincent C., Joecker A., Schoof H., Turck F., et al. (2009). PEP1 regulates perennial flowering in Arabis alpina. Nature 459, 423–427. doi: 10.1038/nature07988, PMID: [DOI] [PubMed] [Google Scholar]
- Willing E.-M., Rawat V., Mandáková T., Maumus F., James G. V., Nordström K. J. V., et al. (2015). Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat. Plants 1:14023. doi: 10.1038/nplants.2014.23, PMID: [DOI] [PubMed] [Google Scholar]
- Wymer C. L., Bibikova T. N., Gilroy S. (1997). Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 12, 427–439. doi: 10.1046/j.1365-313X.1997.12020427.x, PMID: [DOI] [PubMed] [Google Scholar]
- Yang G., Gao P., Zhang H., Huang S., Zheng Z. L. (2007). A mutation in MRH2 kinesin enhances the root hair tip growth defect caused by constitutively activated ROP2 small GTPase in Arabidopsis. PLoS One 2. doi: 10.1371/journal.pone.0001074 [Epub ahead of print], PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C. M., Blancaflor E. B. (2013). Overlapping and divergent signaling pathways for ARK1 and AGD1 in the control of root hair polarity in Arabidopsis thaliana. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00528 [Epub ahead of print], PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C. T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. doi: 10.1242/dev.00681, PMID: [DOI] [PubMed] [Google Scholar]
- Zhou Y., Gan X., Viñegra de la Torre N., Neumann U., Albani M. (2021). Beyond flowering time: diverse roles of an APETALA2-like transcription factor in shoot architecture and perennial traits. New Phytol. 229, 444–459. doi: 10.1111/nph.16839, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA745061.