Abstract
The two-domain actin associated protein coronin interacts with filamentous (F-) actin, facilitating diverse biological processes including cell proliferation, motility, phagocytosis, host-parasite interaction and cargo binding. The conserved N-terminal β-propeller domain is involved in protein: protein interactions, while the C-terminal coiled-coil domain mediates oligomerization, transducing conformational changes. The L. donovani coronin coiled-coil (LdCoroCC) domain exhibited a novel topology and oligomer association with an inherent asymmetry, caused primarily by three a residues of successive heptads. In the T.brucei homolog (TbrCoro), two of these ‘a’ residues are different (Val 493 & 507 replacing LdCoroCC Ile 486 and Met 500 respectively). The elucidated structure possesses a similar topology and assembly while comparative structural analysis shows that the T.brucei coronin coiled-coil domain (TbrCoroCC) too possesses the asymmetry though its magnitude is smaller. Analysis identifies that the asymmetric state is stabilized via cyclic salt bridges formed by Arg 497 and Glu 504. Co-localization studies (LdCoro, TbrCoro and corresponding mutant coiled coil constructs) with actin show that there are subtle differences in their binding patterns, with the double mutant V493I–V507M showing maximal effect. None of the constructs have an effect on F-actin length. Taken together with LdCoroCC, we therefore conclude that the inherent asymmetric structures are essential for kinetoplastids, and are of interest in understanding and exploiting actin dynamics.
Keywords: Trypanosoma brucei, Leishmania, Actin, Coronin, Crystal structure, Oligomerization, In vivo imaging
Graphical abstract
Highlights
-
•
T.brucei coronin coiled-coil domain (TbrCoroCC) is an anti-parallel four-helix bundle as the L.donovani homolog(LdCoroCC).
-
•
Magnitude of asymmetry observed in TbrCoroCC is different from that in LdCoroCC.
-
•
Cyclic salt bridges stabilize the tetramer with contributions from steric interactions.
-
•
LdCoroCC and TbrCoroCC constructs show subtle differences in actin association in vivo.
1. Introduction
Leishmaniasis and trypanosomiasis are parasitic vector borne diseases affecting millions of people in tropical and sub-tropical regions, caused by the kinetoplastid protozoans Leishmania and Trypanosoma species respectively. A characteristic feature of these parasites is the presence of flagella bound to the membrane network, consisting of actin filaments that are vital for the organism. Depletion of this cyto-skeletal protein in T. brucei resulted in subsequent death (Reisler, 1993). Actin exists in two forms: a globular monomeric form knows (G-actin) and as filamentous polymer F-actin. The actin modulation machinery entails filament assembly induced by ATP binding to globular actin (G-actin) forming F-actin, while the release of inorganic phosphate upon hydrolysis results in its dis-assembly, forming ADP-G-actin. These complex arrays of dynamic changes are regulated by actin associated proteins including coronin, cofilin, profilin and the Arp2/3 complex (De Hostos et al., 1993; Ayscough et al., 1998). The widely expressed eukaryotic conserved, F-actin binding protein coronin, first identified in D. discoidium where lack of coronin led to cell migration defects, is a key regulator of actin assembly:dis-assembly dynamics that plays important roles in cell motility, phagocytosis, cytokinesis, immune regulation (Maniak et al., 1995; Hacker et al., 1997; Nagasaki et al., 2001; Bharathi et al., 2004; Morgan and Fernandez, 2008; Xavier et al., 2008). The association of coronin with the Arp2/3 protein complex mediates actin filament branch generation, while the ADP-bound actin filaments are dismantled by coronin and cofilin (Humphries et al., 2002). In Leishmania, S. cerevisiae and D. melanogaster, coronin has been shown to bind with microtubules and crosslink to actin filaments (Goode et al., 1999; Yan et al., 2005) in Toxoplasma it facilitates host cell invasion (Shina et al., 2011; Xavier et al., 2012; Tchang et al., 2013) while in Plasmodium sporozoite mutant coronin exhibits defect in motility (Bane et al., 2016) In Leishmania, the actin filaments are distinct from most organisms, being primarily multi-branched and comparatively short, and overexpression of coronin in L. donovani increases the occurrence of actin filaments (Nayak et al., 2005; Srivastava et al., 2015). The kinetoplastid coronins, are short (Type I, 400–600 residues) and consist of an N-terminal WD40 repeat domain (∼350 residues), and a C-terminal coiled-coil domain (50–80 residue), connected by a variable linker that has unique and conserved regions (de Hostos, 1999; McArdle and Hofmann, 2008; Appleton et al., 2006; Eckert et al., 2011; Nayak et al., 2016). The Coiled coil (CC) domain, made of two or more helices forming a supercoil by mutually burying their hydrophobic regions, are ubiquitous structural motifs, constituting 3–5% of the whole genome, that are primarily involved in homo- and hetero-oligomerization (Wolf et al., 1997; Liu et al., 2006; Rackham et al., 2010). CCs are associated with specific sequence patterns corresponding to the supercoil handedness, with the left-handed supercoil having a heptad repeat abcdefg where hydrophobic residues a, d associate in a knobs into holes pattern, with electrostatic interactions between e, g providing additional stability (O'Shea et al., 1993; Krylov et al., 1994; Monera et al., 1994; Zeng et al., 1997; Kohn et al., 1998; McClain et al., 2001). The CC is dynamic, with changes as subtle as mutation of a single residue leading to drastic consequences, including changes in the oligomer assembly, topology of the helical bundle (Harbury et al., 1993, 1998; Stetefeld et al., 2000; Spoerl et al., 2002; Kammerer et al., 2005; Deng et al., 2008; Kumar et al., 2018).
The L.donovani coronin coiled-coil domain (LdCoroCC), solved earlier in the laboratory showed that the structure, unlike the mammalian homologues (parallel trimer), assembles with a novel topology and oligomeric state (anti-parallel tetramer), with an inherent asymmetry, arising from steric clashes between a residues (Ile 486, Leu493 and Met 500) of three successive heptads (Nayak et al., 2016; Karade et al., 2020). Sequence analysis of the homologous T.brucei coronin coiled-coil domain (TbrCoroCC) indicates that two of the a residues are different, from L.donovani with Val 493 and 507 replacing Ile 486 and Met 500 respectively (Fig. 1A). To understand and to investigate the role of these changes, the structures of T.brucei coronin coiled coil domain (TbrCoroCC) constructs were elucidated and the effects of the constructs along with LdCoro mutants (I486A and I486A-L493A (I + L) M500V) were functionally characterized in vivo.
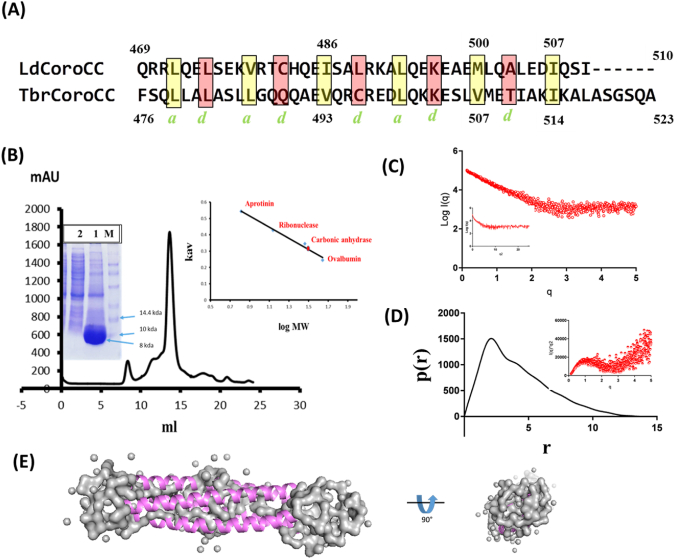
Fig. 1.
Characterization of TbrCoroCC (A) Pairwise sequence alignment of L. donovani and T. brucei coronin coiled-coil domains, with a, d residues highlighted in yellow and red boxes respectively. Residues implicated in the structural asymmetry in LdcoroCC are colored yellow (I486, L493 and M500) (B) Size exclusion chromatography profile of TbrCoroCC shows the protein construct elutes as a tetramer (∼32 kDa) the inset shows molecular weights of standards, while the SDS-PAGE showing the protomer of ∼8 kDa.(C) Experimental scattering curve of TbrCoroCC with the Guinier Region and linear regression (solid line) for Rg evaluation shown in inset (D) Pair distance distribution function p(r) at 10 mg/ml concentration reveals longest dimension (Dmax) of ∼11.5 nm with the unstructured residues from the expression vector possibly accounting for the additional length, in insetKratky plot for the protein TbrCoroCC shows broadness of curve of a protein indicates rod like protein. (E) Different orientations of the low-resolution structure of the protein (envelop), generated by using the in-solution scattering of X-ray intensity data. The TbrCoroCC structure model was fitted well inside the envelope.
2. Methods
2.1. Bacterial strain and growth condition
The E. coli strains DH5α and RPL were grown in Lysogeny Broth (LB) medium at 37 °C and 25 °C.
2.2. Sequence analysis
The coronin gene sequences of L. donoavani and T. brucei homologues were retrieved from GeneDB (http://www.genedb.org) and Uniprot (www.uniprot.org). Sequence alignments of coronin homologues were done by Clustal omega (http://www. ebi.ac.uk/Tools/msa/clustalo/). Coiled coil domain was predicted with MARCOIL web server (http://toolkit.tuebingen.mpg.de/marcoil), and the alignment visualized using ESPript (https://espript.ibcp.fr,) (Robert and Gouet, 2014).
2.3. Cloning, point mutation, protein expression and purification
The T. brucei Coronin coiled-coil domain was amplified using forward and reverse primers GAATTCTCGCAGTTGTTAGCTCTTGCCTCG and CTCGAGGGCAAGGGCCTTTATCTTTG CGAT containing BamHI and EcoRI restriction sites (Bold) designed manually from T.brucei strain (927/4 GUTat10.1) and L.donovani strain (BPK282A1) and the PCR product ligated with T/A vector pTZ57 R/T (Ins TA clone™ PCR cloning kit, Fermentas International Inc.) TA clone digested and sub clone into pET28 (a) (Novagen) expression vector. For in vivo study the digested product ligates and expressed in pLEXSY integrative vector system in Leishmania. We have created single point mutation at V507M using appropriate forward and reverse primers (CTATTGGGGCAGCAACAGGCGGAGATCCAG and TGAAGGATCCTCACGACAACGCTGGA TCT, for double mutant for the V493I–V507M:GACCTGCAGAAGAAGGAGAGCCTGATGATGG and CTTTATCTTTGCGATTGTCTCCATCATAAG respectively using single mutant as template. All the point mutation created by site directed mutagenesis method and confirmed by sequencing.
The genes, cloned in pET28a containing N- and C- terminal His tags were transformed and over-expressed in competent E.coli strain RPL cells and, allowed to grow at 37 °C until OD600 ∼ 0.6 prior to induction with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), harvested after 6–8h at 25 °C by centrifugation at 6500 rpm for 15 min. Cells were re-suspended in the lysis buffer (50 mM TRIS-HCl pH = 7.5, 200 mM NaCl) and subsequently lysed by sonication with a 10s on/10s off pulse for 30 min. On centrifugation at 10,000 rpm for 45 min the cell debris was removed and the supernatant was loaded on an IMAC column (Qiagen, Germany) pre-equilibrated in the buffer, (50 mM TRIS-HCl pH = 7.5, 200 mM Nacl) washed with buffers containing 10 mM and 30 mM imidazole respectively before eluting with 250 mM imidazole in the same buffer. The eluted protein was dialyzed overnight to remove imidazole against a buffer containing 20 mM TRIS-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA and 3 mM β-me, concentrated using 10 kDa cutoff centricon (Amicon).
2.4. Gel filtration analysis
Size exclusion chromatography was carried out using S75 10/300 prepacked columns connected to AKTA pure (GE Healthcare, Cytiva USA).The purified concentrated protein was loaded on pre equilibrated Column. The protein elution volume of 11.6 mL was compared with molecular weight standards (GE catalogue 28403841), the molecular weight and oligomer state estimated using the following standards from GE Healthcare: Ovalbumin (45 kDa), carbonic anhydrase (29 kDa), ribonuclease A (13.7 kDa) and Aprotinin (6.5 kDa). The eluted protein was collected and concentrated for prior to use.
2.5. Western blotting
The proteins from whole Leishmanial cell lysates were separated by SDS-PAGE and blotted to nitrocellulose membrane. Protein bands were detected by incubating with anti His polyclonal primary mouse antibody (Santacruz) for 3 h at room temperature subsequently incubation with horseradish peroxidase (HRP)-conjugated anti-mouse IgG, for 2 h at room temperature. The HRP substrate luminol and H2O2 mixed with 1:1 ratio was spread over the blot which resulted in chemiluminescence and detected in Chemidoc equipment (GE Biosciences).
2.6. Chemical cross linking
Cross linking reactions were set with a 30 mM HEPESE (pH 7.5) and 100 mM NaCl with glutaraldehyde concentrations (0.025 to 0.5%) at 100 μg TbCoroCC. The reactions were stopped with 100 mM Tris (pH = 8) after 30 min of incubation and 20 μg of crosslinked samples were used for SDS page analysis.
2.7. Circular dichroisms spectroscopy
CD spectroscopy assays performed using buffer 20 mM TRIS pH 7.5, 100 mM NaF with protein concentration 5 μM on a J810 ChirascanTM CD spectropolarimeter (Applied Photophysics) (SAIF facility in CSIR-CDRI, Lucknow) calibrated with ammonium (+)-10- camphorsulfonate. The average of four spectra (190–250 nm, scan speed 10 nm/min) of each protein sample was taken. The spectra measured [ϴ]222 as a function of temperature under the same conditions. Spectra were collected in a temperature-controlled quartz cuvette, the successively 10 °C interval in 20–90 °C scanning window at a ramp rate of 2 °C/min.
2.8. Crystallization and data collection
Preliminary crystallization trials were initiated using commercial screens (Crystal Screens 1, 2; PEG ion screen) and around the homologous LdCoroCC conditions with a protein concentration of 10 mg/mL mixed with equal volume of the reservoir solution, equilibrated against 0.5 ml reservoir solution. An initial hit was observed in a Crystal Screen 1 (condition 0.1M HEPES 7.5 and 4.3 M NaCl) in ∼10–14 days and the condition optimized, leading to the growth of diffracting crystals from a hanging drop vapor diffusion where 2 μl of the protein (8 mg/mL) was mixed with equal volume of the precipitate containing 0.1M HEPES pH 7.5 and 3 M NaCl, equilibrated against a 0.5 ml reservoir solution. Crystals were flash frozen with 20% glycerol as a cryo-protectant and data collected (to 2.06–3.02 Å) on the PX-BL21 beamline at INDUS-2, Raja Ramanna Centre for Advances Technologies, Indore, INDIA (TbrCoroCC) or on beamline XRD2, Elettra Sincrotrone, Trieste, Italy (V507M and double mutant).
2.9. Data processing, structure determination and refinement
The X-ray diffraction data of TbCorCC were processed using the HKL2000 suite of programs (Otwinowski and Minor 1997; Minor et al., 2006). Indexing the data suggested that the crystal belongs to body centered tetragonal space group I 4 2 2, with unit cell dimensions a = b = 93.3, c = 82.9 Å and the data integrated and scaled to 2.06 Å. Solvent content analyses indicate the asymmetric unit contains two protomers with a solvent content of 50% and Vm of 2.11. The qualities of data were assessed by phenix.Xtriage (Zwart et al., 2005), which confirmed the absence of twinning or diffraction anisotropy. The V493I and double mutant structures were processed using iMosflm & Aimless, from the CCP4 suite of programs (Evans, P. 2014). Data reduction statistics are summarized in Table 1.
Table 1.
Data collection, processing and structural refinement statistics (Values in parenthesis are for highest resolution shell).
| TbrCoroCC |
V493I–V507M |
V507M |
|
|---|---|---|---|
| PDB Id | 7DGX | 7DH4 | 7DHB |
| Resolution range | 24.06–2.06 (2.13–2.06) | 45.61–2.41 (2.496–2.41) | 36.52–3.02 (3.12–3.02) |
| Space group | I 4 2 2 | I 4 2 2 | I 4 2 2 |
|
Unit cell |
a = b = 93.3 c = 82.9 α,β,γ = 90° |
a = b = 93.3 c = 82.9 α,β,γ = 90° |
a = b = 93.3 c = 82.9 α,β,γ = 90° |
| Total reflections | 122,036 (10988) | 94317 (10091) | 6954 (718) |
| Unique reflections | 13712 (1308) | 6752 (681) | 3477 (345) |
| Multiplicity | 8.9 (8.4) | 14 (13.9) | 2.0 (2.0) |
|
Completeness (%) |
99.14 (98.95) |
95.02 (99.12) |
90.65 (95.82) |
| Mean I/sigma(I) | 11.71 (2.22) | 25.8 (6.57) | 7.22 (2.46) |
| Wilson B-factor | 44.9 | 49.21 | 63.15 |
| R-merge | 0.11 (0.61) | 0.053(0.374) | 0.040 (0.19) |
|
R-meas |
0.098(0.451) |
0.013 (0.101) |
0.017 (0.027) |
| CC1/2 | 1 (1) | 1 (0.99) | 1 (0.99) |
| CC∗ | 1 (1) | 1 (0.99) | 1 (1) |
| Reflections used in refinement | 11576 (1136) | 6710 (675) | 3316 (344) |
|
Reflections used for R-free |
556 (64) |
297 (27) |
140 (15) |
| R-work | 0.220 (0.334) | 0.236 (0.265) | 0.223 (0.311) |
| R-free | 0.240 (0.419) | 0.255 (0.434) | 0.264 (0.409) |
| CC(work) | 0.961 (0.776) | 0.966 (0.96) | 0.955 (0.872) |
| CC(free) | 0.971 (0.716) | 0.972(0.875) | 0.975 (0.564) |
|
Number of non-hydrogen atoms |
807 |
752 |
736 |
| macromolecules | 763 | 748 | 731 |
| solvent | 44 | 4 | 5 |
| Protein residues | 99 | 98 | 95 |
|
RMS(bonds) |
0.006 |
0.008 |
0.007 |
| RMS(angles) | 0.77 | 0.97 | 1.07 |
| Ramachandran favored (%) | 100 | 98.94 | 100 |
| Ramachandran allowed (%) | 0 | 1.06 | 1.09 |
| Ramachandran outliers (%) | 0 | 0 | 0 |
| Rotamer outliers (%) | 2.53 | 0 | 1.32 |
| Clashscore | 1.96 | 2.66 | 6.08 |
| Average B-factor | 67.28 | 67.77 | 66.38 |
| macromolecules | 67.33 | 67.83 | 66.42 |
| solvent | 68.01 | 56.82 | 59.80 |
†Rmerge = ƩhklƩi|Ii(hkl)-(I(hkl))|/ƩhklƩi Ii(hkl),where Ii(hkl)is the intensity of the ithobservation of reflection hkl and(I(hkl)) is the average intensity of the i observations a. R = Σ∣Fo∣-∣Fc∣/Σ∣Fo∣.
a. Rfree was calculated using 5% of data excluded from refinement.
The crystal structure was solved by molecular replacement methods using the Phaser program from the phenix suite (Adams et al., 2010) with a canonical dimer of the L. donovani coronin coiled-coil domain structure as the template (PDB ID 5CX2, chains A&B), which gave a unique solution (TFZ 12.5; LLG 196). Refinement of the structure was carried out using the phenix.refine (Afonine et al., 2012)] program from the phenix suite. The refinement strategy was an initial poly alanine rigid body refinement followed by restrained refinements. Electron density maps were computed after each round of refinement, and side chains progressively fitted in COOT (Emsley et al., 2010) and towards the end of the refinement cycles, solvent and other organic groups from the crystallization conditions were added. The crystallographic R-factor and R-free, which were 31.1% and 34.5% after the first rigid body refinement, subsequently converged to 22.6% and 25.7% respectively. The final model contains 99 amino acid residues (52 in chain A and 47 in chain B), 68 solvent molecules.
2.10. Small-angle X-ray scattering (SAXS)
SAXS measurements were done at in-house SAXS instrument SAXSpace (Anton Paar) at Central Drug Research Institute, Lucknow, and samples with 5–15 mg/mL concentration were applied to the system in 30 mM Tris-HCl pH 7.5 and 100 mM NaCl buffer. The protein and buffer frames were selected for processing using Primusqt of ATSAS (Petoukhov et al., 2012). The buffer subtracted protein frames were scaled and averaged. DAMMIF used to generate ab initio models were averaged using DAMAVER (Volkov et al., 2003) and the most typical model was selected. The analogy of models theoretical scattering and experimental scattering superposition was performed with Gasbor, (Svergun et al., 2001) the graph plotted with the help of GraphPad prism 7.0. The processing software was part of the ATSAS package version 3.
2.11. Genetic manipulations and parasite culture
For in vivo study, the amplified gene products were cloned in pLEXSY-hyg2.1integrative vector system (Jena Bisocience). The resulting pLEXSY constructs were linearized by SwaI or SmiI restriction enzyme and approximately 10 μg of purified DNA products were transfected into Leishmania promastigotes by electroporation using Gene pulser (Bio-Rad) as described earlier (Nayak et al., 2005). The transfectants were selected against 10 μg/mL hygromycin and maintained at 26 °C in high glucose DMEM (Gibco) supplemented with 10% heat inactivated fetal bovine serum (Gibco) along with 40 μg/mL gentamycin and 100 μg/mL Penicillin-Streptomycin preparation.
2.12. Immunofluorescence microscopy
For immunofluorescence microscopy, log-phase Leishmania promastigotes were adhered on poly-L-lysine coated coverslips for 2 min and then fixed with 4% (w/v) paraformaldehyde in PBS for 15 min. A single wash with 5% (w/v) glycine in PBS was given to quench extra formaldehyde and these fixed cells were permeabilized with 1% Triton X-100 in PBS for 20 min. Blocking was done using 1% BSA in PBS with 0.02% sodium azide for 1 h at room temperature. Primary antibody treatment with anti-actin antibodies (rabbit) (1:500) and anti-His antibodies (mouse) (1:200) was given in same blocking buffer at 4 °C, overnight. Cover slips were washed for about 7–8 times with blocking buffer which was followed by secondary antibody treatment with Alexa Fluor 488 (anti-rabbit) and Alexa Fluor 546 (anti-mice) for 4 h at 4 °C. Cover-slips were then washed again for 7–8 times with blocking buffer and then mounted onto glass slides in mounting media (Calbiochem, Germany) and were stored at 4 °C in the dark until scanning. Confocal images were collected on Leica SP8 confocal microscope using 63X oil plan apochromate lens (1.4 N.A.) at 3X digital zoom. Images were processed and arranged for presentation in Adobe Photoshop (Creative Suit 6).
2.13. Statistical analysis
The results were presented as the pooled data of three independent experiments expressed as mean σ standard deviation (SD). Analysis was done using unpaired t-test with equal SD using PRISM software (Version 6.2).
3. Results
3.1. TbrCoroCC structure determination and analysis
The 47 residue native T. brucei coronin coiled-coil domain (Residues 477–523 of T. brucei coronin, UNIPROT ID Q57W63), and the single and double mutant constructs V507M and V493I–V507M were over-expressed and purified to homogeneity as described in Experimental procedures. The size exclusion profile shows the elution volume corresponds to a tetramer (Fig. 1A and B, Supplementary Fig. S1), further validated by solution X-ray scattering studies (Fig. 1C–E) and glutaraldehyde crosslinking studies (Supplementary Fig. S2). The crystal structure was determined by molecular replacement using a canonical dimer of LdCoroCC (AB dimer of PDB: 5CX2) as a template. The asymmetric unit contains an anti-parallel dimer, with the tetramer generated by the crystallographic two -fold axis. The biological assembly has dimensions of ∼70 × 25.7 × 20.9 Å3 (Fig. 2 A, B). Structures of the single (V507M) and the double mutant (V493I–V507M) constructs were determined using TbrCoroCC as the template and the mutations validated by omit maps (Fig. 2C). Data collection and refinement statistics for the apo and mutant constructs are summarized in Table 1.
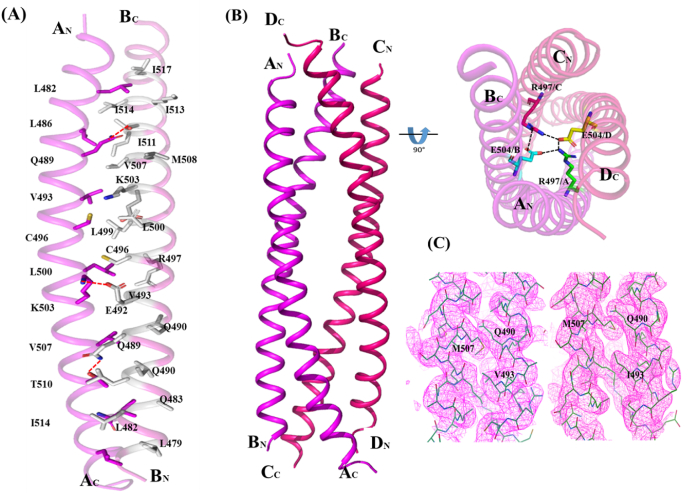
Fig. 2.
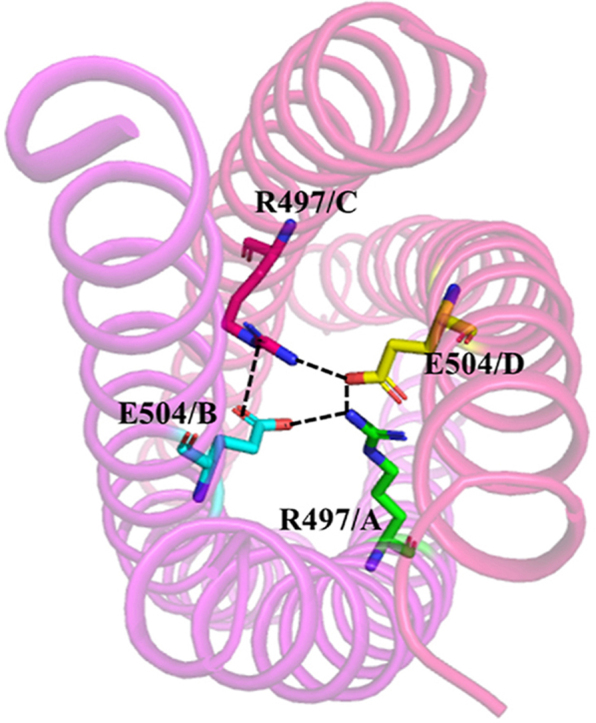
Crystal structure of TbrCoroCC (A) Ribbon representation of TbrCoroCC dimer with residues involved inknobs into holes packing represented as sticks (chain ‘A’- magenta and chain ‘B’- light gray) and hydrogen bonds are shown as dotted lines (B) The biological assembly (tetramer) can be obtained by the application of two fold crystallographic symmetry. The two dimers are colored differently, (magenta and red) and the cyclic salt bridge observed between R497 and E504 are shown as dotted lines in the orthogonal view. (C) Omit map contoured at 1σ ofand V507M (left) and V493I–V507M (right) confirming successful mutations.
The individual protomers A, B are essentially identical in the three TbrCoroCC structures with root mean square displacement (RMSD) values ∼0.5 Å. The protomers associate as a coiled coil between residues 479–514 are stabilized by a:d residues in a knobs-into-holes packing. Significantly, in TbrCoroCC 3 d residues are polar (Gln 489, Lys 503 and Thr 510), and these polar groups provide additional stability: side chains of Gln A 489:Thr B 510 (and B489:A510) form a hydrogen bond (2.8 Å) while Lys 503 forms a salt bridge with Glu 492 of the opposite chain (2.78 Å) (Fig. 2A). The corresponding residues in LdCoroCC are hydrophobic except Lys 496 (equivalent to Lys 503), which forms an ionic interaction with Glu 485 of the partner chain. The salt bridges forming residue Arg 497 and Glu 504 strengthen the stability of structure (Fig. 2B orthogonal view).
Oligomerization analysis using PISA (Krissinel, E., and Henrick, K. 2007) confirms the biological assembly to be a tetramer, consistent with solution experiments. In left-handed coiled-coils with three or more helices, an additional residue, typically e or g is furthermore buried in the interface, referred to as ade/adg packing: core packing analysis of the TbrCoroCC tetramer using the helical analysis programs (Dunin-Horkawicz, S and Lupas, A. N. 2010; Pratap et al., 2013) indicates that TbrCoroCC adopts the ade core packing, similar to LdCoroCC (Fig. 3A and B C &D).The canonical dimer A/B and C/D show identical knobs into holes interactions (Fig 3A) while there is difference at non-canonical dimeric interface between A/D and B/C (Fig. 3B and C): while the B/C dimeric interface extends to the length of the coiled-coil region, helices A and D associate only at either termini (Fig. 3B). As seen in Fig. 3C, the A/D and B/C interfaces are not identical; Residue Ala 518 interacts with residue Ala 483 of the partner chain in the B/C interface, while it interacts with Leu 480 on the A/D interface, suggesting that the interacting helices differ by a turn. A similar asymmetry was observed in the LdCoroCC structure too, though the magnitude was larger (2 turns; Nayak et al., 2016; Karade et al., 2020).
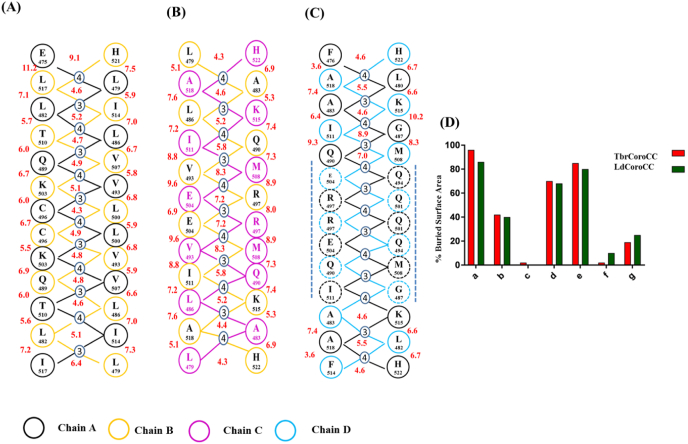
Fig. 3.
Knobs into hole packing and PISA Buried surface area calculation. The ladder representation of the knobs into holes packing patterns in the interfaces of TbrCoroCC, with the corresponding inter-Cβ distances for the (A) A/B interface, (B) B/C interface and (C) A/D interface. Residues of the ladder not associating in knobs into holes manner are shown as dotted circles(D) Bar diagram representation of the buried surface area contribution of residues of the heptad, the residues at position a, d and e are more buried compared to other positions. The red color is used to represent TbrCoroCC and dark green color for LdCoroCC.
3.2. Structural asymmetry and comparison of TbrCoroCC structure with LdCoroCC
Structural superposition of the TbrCoroCC tetramer with LdCoroCC, shows that the structures are similar, the protomers of TbrCoroCC was superposed with LdCoroCC, the structures superpose with a rms deviation of 0.75 A°, while the dimers/tetramers superposing with an overall r.m.s deviation of 1.2–2.2 Å with similar inter-Cβ distances in the A/B, and B/C interfaces but not in the A/D interface, with the D helix is shifted by ∼ a turn, smaller than the 2 turns observed in LdCoroCC (Fig. 3A–C, Supplementary Table 1. Mutation of LdCoroCC Ile 486 to alanine, resulted in a symmetric AD/BC interface, but as a consequence, the canonical dimer interface (AB/CD) become asymmetric (Karade et al., 2020). The structural superposition and sequence analysis shows, that there is one heptad shift in case of T.brucei.
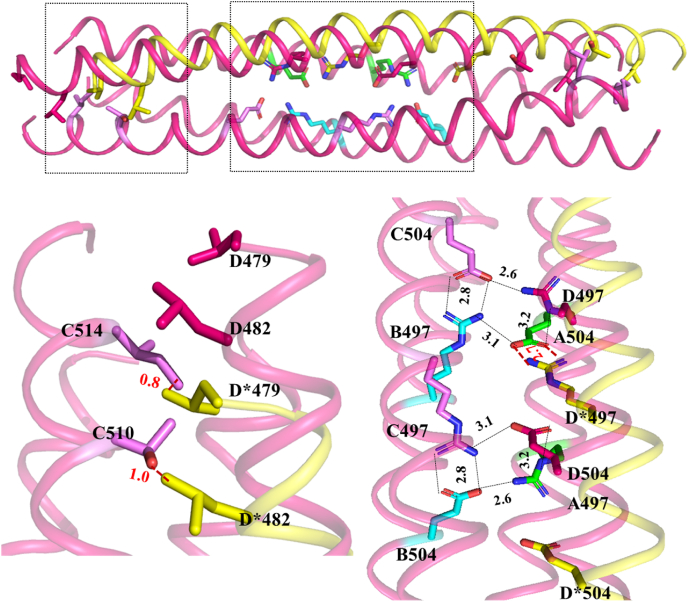
To further investigate and identify the cause of asymmetry in TbrCoroCC, the symmetric tetramer was modeled, wherein the D helix is replaced by its symmetry equivalent D∗, generated by imposing symmetry at the A/D and B/C interfaces. This was generated by superposing helix A on helix B and using this transformation matrix on helix C to generate the symmetric hypothetical helix D∗. Visual analysis of the symmetric tetramer ABCD∗ reveals that there are steric clashes occurring between helices D∗ and C, at residues 479 and 482 with 510 and 514 with side chain atoms coming to distances of ∼0.8–1.0Å (Fig. 4). The steric clashes persist, even with alternate rotamer configurations. Significantly, the cyclic salt bridges observed between Arg. 497 and Glu 504 (Fig. 2B) forms only two ionic contacts in the symmetric interface, unlike the eight interactions that stabilize the native structure (Fig. 4). In the elucidated structures, Glu 504 of chain C interacts with Arg 497 of B & D chain with distances 2.8 and 2.6 Å respectively, while Arg B 497 interacts with Glu A 504 (distance 3.1Å) which also interacts with δ-N atom of Arg D 497 (3.2 Å; Fig 4). A similar salt bridge network is also observed in the LdCoroCC structure, with equivalent residues Arg 490 and Glu 497 involved in ionic interactions among three of the protomers, while in the fourth, the side chain points to the solvent (Nayak et al., 2016; Karade et al., 2020).
Fig. 4.
Hypothetical symmetric TbrCoroCC four-helix bundle – Cartoon representation showing the TbrCoroCC structure in magenta, with the symmetric D∗ helix shown in yellow with the insets showing an enlarged view of the regions of difference. The steric clashes occur at either end of the helical bundles, though only one is shown for clarity. The break of salt-bridge interactions between Arg 497 and Glu 504.
3.2.1. Sub-cellular distribution and actin coronin co-localization
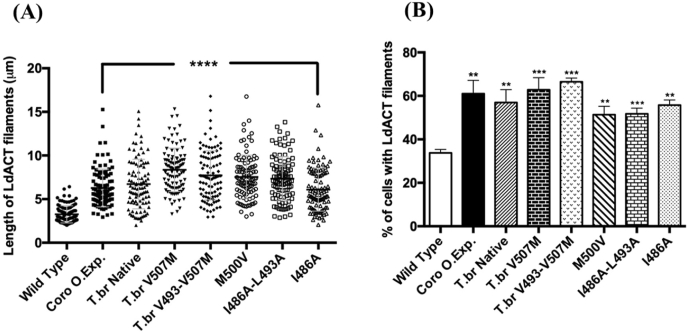
Wild-type and mutant (L.donovani (I486A, I486A-L493A (I + L), M500V) and T.brucei (V507M & V493I–V507M) coronins were over-expressed in L. donovani promastigotes (Fig. 5). LdAct filaments were analyzed by immunofluorescence microscopy for the number of cells containing LdAct filaments and their lengths. The over-expressed mutant constructs have significantly longer LdAct filaments in comparison to wild type cells. Also, while LdAct filaments were in about 30% wild type cells, the corresponding values for the mutant constructs were ∼60%. Interestingly, Leishmania cells overexpressing point mutated versions of T.brucei coronin have more actin filaments when compared to cells expressing native T.brucei coronin. A similar pattern however was not observed for cells expressing point mutated variants of L.donovani coronin, and here the occurrence of LdAct filaments was lesser than to the cells over-expressing native LdCoroCC (Fig. 6, Fig. 7).
Fig. 5.
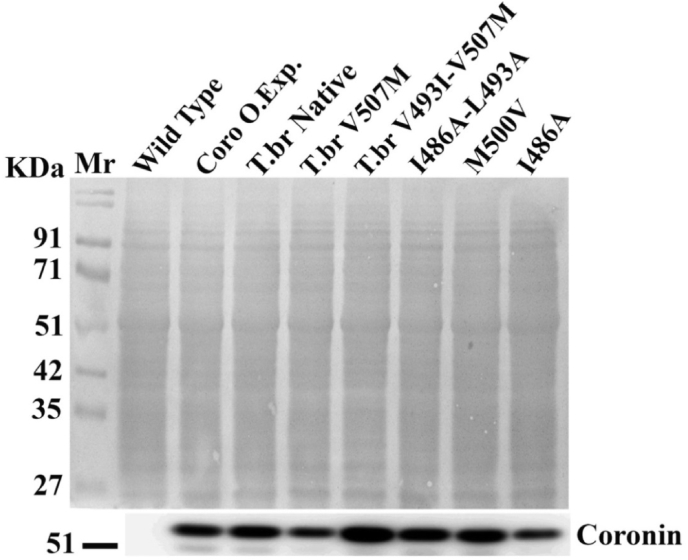
Full length Coronin expression: Expression of LdCoro, TbrCoro and respective point mutated variants in L. donovani cells as analyzed by western blotting using anti-His6 antibodies; Coomassie stained PVDF membrane along with Western blot, Mr, molecular weight marker.
Fig. 6.
Quantitative analysis of LdACT filament length in the Leishmania(A) cells showing a marked increase in the average length of LdACT filaments in cells overexpressing mutated coronin variants as compared with wild type cells; length of more than 100 filaments was measured from three independently coronin prepared samples. Statistical significance was drawn through unpaired “t” test ∗∗∗P < 0.0001. (B) Quantitative analysis of the number of cells containing LdACT filaments in native LdCoro, native TbrCoro and mutant coronin overexpressing cells compared with wild type cells. Only the cells that contained >2 μm long LdACT filaments were considered; the data shown are representative of at least three independent experiments (>1000 cells). The values shown are mean ± standard deviation.
Fig. 7.
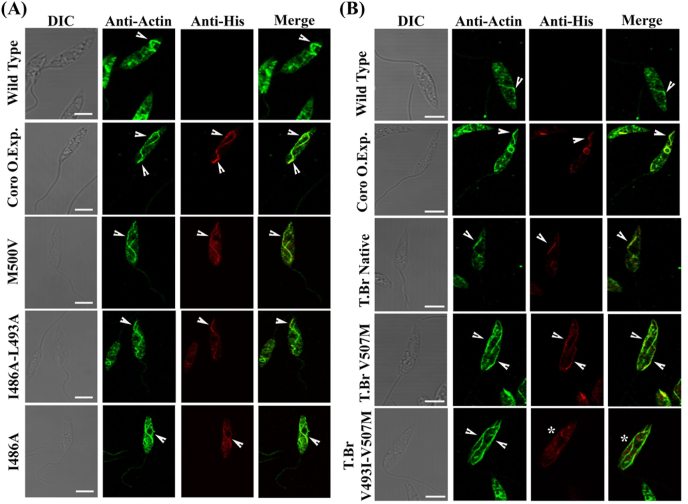
Immunofluorescence confocal images:(A) showing actin filaments in wild type, LdCoro overexpressing cells and LdCoro point mutant variants overexpressing cells. Labeling of LdACT was done by using anti-LdACT antibodies. Arrowheads mark filamentous structures of L. donovani actin (LdACT). (B) TbrCoro overexpressing cells and TbrCoro point mutant variants overexpressing cells. Labelling of LdACT was done by using anti-LdACT antibodies. Arrow heads mark filamentous structures of L. donovani actin (LdACT) Bar, 5 μm.
The point mutation variants of LdCoro viz. M500V, I486A + L493A and I486A showed similar co-localization pattern as native LdCoro when expressed in Leishmania cells. Intriguingly, the L.donovani cells expressing native Tbr Coro showed similar interaction of coronin and LdAct filaments despite of distinct structure of TbrCoroCC from LdCoroCC. Also the point mutation constructs TbrCoroV493I–V507M and TbrCoro V507M resulted in similar increase in length and occurrence of LdAct filaments when expressed in Leishmania cells though the co-localization was reduced (7A, B) with no co-localization observed in the double mutant (Fig. 7 B bottom right).
4. Discussion
The coiled coil domain of T. brucei coronin showed distinct differences in a, d residues compared to the L. donovani homolog, whose structure possesses a distinct asymmetry. Structural characterization of TbrCoroCC shows that the biological molecule is a tetramer, a dimer of dimers, as the homologous LdCoroCC. The oligomer association was additionally confirmed by solution scattering studies and glutaraldehyde cross-linking experiments. Comparative analysis of the TbrCoroCC with LdCoroCC shows that the structures are similar, with an r.m.s deviation of ∼1.2–2.2 Å, with both having an ade core packing, though small differences are observed in the individual amino acid contributions as well as super-helical parameters.
In the T. brucei coronin CC crystal structures, the asymmetric unit contains a dimer with the tetramer generated by the crystallographic two fold axis. The structure analysis shows that asymmetry causing residues in TbrCoroCC are distinct from LdCoroCC with residues Leu 479, Leu 482, Thr 510 and Ile 514 having steric clashes and reduced polar interactions between Arg 497 and Glu 504 in the symmetric model. In the symmetric LdCoroCC structurealso the equivalent residues have steric hindrance, though this region is not in the CC.
Leishmania coronin (LdCoro) upon over-expression increases the length of Leishmania actin (LdAct) and therefore number of cells showing the filamentous structures of LdAct also increases (Nayak et al., 2005). It has further been shown that LdAct filaments interact with the unique region of the linker domain Ldcoronin, though neither the unique domain nor the β-propellor WD40 domain alone is able to promote LdAct filament formation (Srivastava et al., 2015). This raises a question whether β-propeller domain has an allosteric effect on the quaternary structure of LdCoro due to which LdAct filament length increases only after full length LdCoro overexpression. Notably, T. brucei coronin (TbCoro), which has 60% sequence similarity with LdCoro, was also able to show patterned co-localization with LdAct filament and render effects on the filament length, typical to LdCoro. The mutant TbCoroCC structures, generated on the basis of steric hindrance causing residues of LdCoroCC,are ∼ identical to the native TbCoroCC structure, and structural analysis indeed shows that the residues involved in the asymmetry are distinct from LdCoroCC. Therefore, it was obvious to see their LdAct interaction and effects on filament length. Nevertheless, as an exception, the mutant TbrCoro V493I–V507M did not show patterned co-localization with LdAct filaments, though it affected an increase in LdAct filament length. Although it is difficult to explain the reason for this drastic change in interaction behavior of TbrCoro V493I–V507M, could be further examined, by generating mutants that disrupt the oligomer assembly, either in LdCoroCC or TbrCoroCC, by the alteration of salt bridge/apolar interactions that stabilize them. Consequently, our perception that interaction of LdCoro with LdAct filaments is an important characteristic which promotes LdAct filament formation is now commuting. It appears that coronin (TbCoro or LdCoro) overexpression has a moonlighting effect on LdAct filament length. Although, it is a testable hypothesis, it provides a clue on our previous observations on no increase in LdAct filament lengths upon overexpression of either beta-propeller or unique + coiled coil domains alone (Srivastava et al., 2015). The observed effects of overexpressing TbCorocc constructs on LdAct makes us speculate on whether a similar effect would be observed in T. brucei actin filaments with the LdCoroCC mutant constructs. However, there are reports that the actin filaments in both these kinetoplastids are distinct: trypanosoma do not contain actin filaments as observed in Leishmania, rather displaying a diffused distribution of actin throughout the cells in the insect stage and only in the endocytic pathway in the bloodstream form (Garcia-Salcedo et al., 2004; Sahasrabuddhe et al., 2004; Nayak et al., 2005); hence, a direct comparison might not be feasible. There could be a strong possibility that beta-propeller is a known protein-protein interaction scaffold, recruits an unknown LdAct interacting partner that actually promotes LdAct filament formation. As the re-introduction of the hypothesized steric hindrance causing residues of LdCoroCC in TbCoroCC shows no significant structural changes, the analysis implies that the salt bridge forming residues Arg497 and Glu504 might be the major contributor for asymmetry in TbrCoroCC. Based on the observed asymmetries in LdCoroCC and TbCoroCC, it is also tempting to hypothesize the asymmetry in LdCoroCC and TbrCoroCC are probably essential for the functionality of kinetoplastid coronins.
Data availability
The coordinates have been deposited in the Protein Data Bank (PDB ID: 7DGX, 7DH4 & 7DHB) and SASBDB Id SASDK46.
CRediT authorship contribution statement
Pankaj Singh Parihar: Conceptualization, Investigation, Validation, Formal analysis, Writing – original draft. Aastha Singh: Investigation, Writing – original draft. Sharanbasappa Shrimant Karade: Conceptualization, Formal analysis, Writing – review & editing. Amogh Anant Sahasrabuddhe: Conceptualization, Methodology, Formal analysis, Validation, Writing – review & editing, Supervision, Project administration. J. Venkatesh Pratap: Conceptualization, Methodology, Validation, Formal analysis, Writing – review & editing, Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank Dr Hemlata, Division of Parasitology, CDRI for providing T.brucei culture for genomic DNA isolation. The authors also thank Dr Ravi Makde, Dr Biplab Ghosh & Dr Ashwani Kumar, Scientists in charge of Beamline-BL21, RRCAT, Indore, and Dr. Babu A. Manjasetty at XRD2, Elettra beamline for their assistance in data collection. PSP and AS thank the Council for Scientific & Industrial Research (CSIR) India for their fellowships. The authors also thank the Department of Science & Technology (DST), Government of India, for providing access to the Elettra Beamline. The manuscript bears CDRI communication number 10301.
Glossary
- TbrCoroCC
T.brucei coiled coil domain
- LdCoroCC
L.donovani coiled coil domain
- LdCoro
L.donovani full length Coronin
- TbrCoro
T.brucei full length Coronin
- Arp2/3
Actin-related protein 2/3 complex
- CC
Coiled Coil
- LdAct
L.donovani actin
- ADP
Adenosine diphosphate
- ATP
Adenosine triphosphate
Handling editor: Glaucius Oliva
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2021.10.002.
Contributor Information
Amogh Anant Sahasrabuddhe, Email: amogh_sahasrabuddhe@cdri.res.in.
J. Venkatesh Pratap, Email: jvpratap@cdri.res.in.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66(2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix. refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68(4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton B.A., Wu P., Wiesmann C. The crystal structure of murine coronin-1: a regulator of actin cytoskeletal dynamics in lymphocytes. Structure. 2006;14(1):87–96. doi: 10.1016/j.str.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Ayscough K.R. In vivo functions of actin-binding proteins. Curr. Opin. Cell Biol. 1998;10(1):102–111. doi: 10.1016/s0955-0674(98)80092-6. [DOI] [PubMed] [Google Scholar]
- Bane K.S., Lepper S., Kehrer J., Sattler J.M., Singer M., Reinig M., Frischknecht F. The actin filament-binding protein coronin regulates motility in Plasmodium sporozoites. PLoS Pathog. 2016;12(7) doi: 10.1371/journal.ppat.1005710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathi V., Pallavi S.K., Bajpai R., Emerald B.S., Shashidhara L.S. Genetic characterization of the Drosophila homologue of coronin. J. Cell Sci. 2004;117(10):1911–1922. doi: 10.1242/jcs.01034. [DOI] [PubMed] [Google Scholar]
- de Hostos E.L. The coronin family of actin-associated proteins. Trends Cell Biol. 1999;(9):345–350. doi: 10.1016/s0962-8924(99)01620-7. [DOI] [PubMed] [Google Scholar]
- De Hostos E.L., Rehfueß C., Bradtke B., Waddell D.R., Albrecht R., Murphy J., Gerisch G. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J. Cell Biol. 1993;120(1):163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Zheng Q., Liu J., Cheng C.S., Kallenbach N.R., Lu M. Self-assembly of coiled-coil tetramers in the 1.40 Å structure of a leucine-zipper mutant. Protein Sci. 2007;16(2):323–328. doi: 10.1110/ps.062590807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunin-Horkawicz S., Lupas A.N. Measuring the conformational space of square four-helical bundles with the program samCC. J. Struct. Biol. 2010;170(2):226–235. doi: 10.1016/j.jsb.2010.01.023. [DOI] [PubMed] [Google Scholar]
- Eckert C., Hammesfahr B., Kollmar M. A holistic phylogeny of the coronin gene family reveals an ancient origin of the tandem-coronin, defines a new subfamily, and predicts protein function. BMC Evol. Biol. 2011;11(1):1–7. doi: 10.1186/1471-2148-11-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66(4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Salcedo, Perez-Morga D., Gijon P., Dilbeck V., Pays E., Nolan D.P. A differential role for actin during the lifecycle of. T. brucei. EMBO J. 2004;23(4):780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode B.L., Wong J.J., Butty A.C., Peter M., McCormack A.L., Yates J.R. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 1999;144(1):83–98. doi: 10.1083/jcb.144.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker U., Albrecht R., Maniak M. Fluid-phase uptake by macropinocytosis in Dictyostelium. J. Cell Sci. 1997;110(2):105–112. doi: 10.1242/jcs.110.2.105. [DOI] [PubMed] [Google Scholar]
- Harbury P.B., Plecs J.J., Tidor B., Alber T., Kim P.S. High-resolution protein design with backbone freedom. Science. 1998;282(5393):1462–1467. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- Harbury P.B., Zhang T., Kim P.S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Humphries C.L., Balcer H.I., D'Agostino J.L., Winsor B., Drubin D.G., Barnes G., Goode B.L. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J. Cell Biol. 2002;159(6):993–1004. doi: 10.1083/jcb.200206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer R.A., Kostrewa D., Progias P., Honnappa S., Avila D., Lustig A., Winkler F.K., Pieters J., Steinmetz M.O. A conserved trimerization motif controls the topology of short coiled coils. Proc. Natl. Acad. Sci. Unit. States Am. 2005;(39):13891–13896. doi: 10.1073/pnas.0502390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karade S.S., Ansari A., Srivastava V.K., Nayak A.R., Pratap J.V. Molecular and structural analysis of a mechanical transition of helices in the L. donovani coronin coiled-coil domain. Int. J. Biol. Macromol. 2020;143:785–796. doi: 10.1016/j.ijbiomac.2019.09.138. [DOI] [PubMed] [Google Scholar]
- Kohn W.D., Kay C.M., Hodges R.S. Orientation, positional, additivity, and oligomerization-state effects of interhelical ion pairs in alpha-helical coiled-coils. J. Mol. Biol. 1998;283:993–1012. doi: 10.1006/jmbi.1998.2125. [DOI] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Krylov D., Mikhailenko I., Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g interhelical interactions. EMBO J. 1994;13:2849–2861. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Ramappa R., Pamidimukkala K., Rao C.D., Suguna K. New tetrameric forms of the rotavirus NSP4 with antiparallel helices. Arch. Virol. 2018;163(6):1531–1547. doi: 10.1007/s00705-018-3753-6. [DOI] [PubMed] [Google Scholar]
- Liu J., Zheng Q., Deng Y., Cheng C.S., Kallenbach N.R., Lu M. A seven-helix coiled coil. Proc. Natl. Acad. Sci. Unit. States Am. 2006;(42):15457–15462. doi: 10.1073/pnas.0604871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniak M., Rauchenberger R., Albrecht R., Murphy J., Gerisch G. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein Tag. Cell. 1995;83(6):915–924. doi: 10.1016/0092-8674(95)90207-4. [DOI] [PubMed] [Google Scholar]
- McArdle B., Hofmann A. In: The Coronin Family of Proteins. Clemen C.S., Eichinger L., Rybakin V., editors. Vol. 48. Springer; New York, NY: 2008. Coronin structure and implications. (Sub-cellular Biochemistry). [Google Scholar]
- McClain D.L., Binfet J.P., Oakley M.G. Evaluation of the energetic contribution of interhelical Coulombic interactions for coiled coil helix orientation specificity. J. Mol. Biol. 2001;313:371–383. doi: 10.1006/jmbi.2001.5044. [DOI] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta. Crystallogr D. Biol. Crystallogr. 2006;62(Aug):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Monera O.D., Kay C.M., Hodges R.S. Electrostatic interactions control the parallel and antiparallel orientation of alpha-helical chains in two-stranded alpha-helical coiled-coils. Biochemistry. 1994;33:3862–3871. doi: 10.1021/bi00179a010. [DOI] [PubMed] [Google Scholar]
- Morgan R.O., Fernandez M.P. The Coronin Family of Proteins. Springer; New York, NY: 2008. Molecular phylogeny and evolution of the coronin gene family; pp. 41–55. [DOI] [PubMed] [Google Scholar]
- Nagasaki A., Hibi M., Asano Y., Uyeda T.Q. Genetic approaches to dissect the mechanisms of two distinct pathways of cell cycle-coupled cytokinesis in Dictyostelium. Cell Struct. Funct. 2001;26(6):585–591. doi: 10.1247/csf.26.585. [DOI] [PubMed] [Google Scholar]
- Nayak A.R., Karade S.S., Srivastava V.K., Rana A.K., Gupta C.M., Sahasrabuddhe A.A., Pratap J.V. Structure of Leishmania donovani coronin coiled coil domain reveals an antiparallel 4 helix bundle with inherent asymmetry. J. Struct. Biol. 2016;195(1):129–138. doi: 10.1016/j.jsb.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Nayak R.C., Sahasrabuddhe A.A., Bajpai V.K., Gupta C.M. A novel homologue of coronin colocalizes with actin in filament-like structures in Leishmania. Mol. Biochem. Parasitol. 2005;143(2):152–164. doi: 10.1016/j.molbiopara.2005.06.001. [DOI] [PubMed] [Google Scholar]
- O'Shea E.K., Lumb K.J., Kim P.S. Peptide ‘Velcro’: design of a heterodimeric coiled coil. Curr. Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Vol. 276. Academic press; 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. (Methods in Enzymology). [DOI] [PubMed] [Google Scholar]
- Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45(2):342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J.V., Luisi B.F, Calladine C.R. Geometric principles in packing of helices. Philos. Trans A. Maths. Phys. Eng. Sci. 2013;371(1993) doi: 10.1098/rsta.2012.0369. [DOI] [PubMed] [Google Scholar]
- Rackham O.J., Madera M., Armstrong C.T., Vincent T.L., Woolfson D.N., Gough J. The evolution and structure prediction of coiled coils across all genomes. J. Mol. Biol. 2010;(3):480–493. doi: 10.1016/j.jmb.2010.08.032. [DOI] [PubMed] [Google Scholar]
- Reisler E. Actin molecular structure and function. Curr. Opin. Cell Biol. 1993;5(1):41–47. doi: 10.1016/s0955-0674(05)80006-7. [DOI] [PubMed] [Google Scholar]
- Shina M.C., Müller-Taubenberger A., Ünal C., Schleicher M., Steinert M., Eichinger L., Müller R., Blau-Wasser R., Glöckner G., Noegel A.A. Redundant and unique roles of coronin proteins in Dictyostelium. Cell. Mol. Life Sci. 2011;68(2):303–313. doi: 10.1007/s00018-010-0455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids. Res. 2014;42(W1):W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasrabuddhe A.A., Bajpai V.K., Gupta C.M. A novel form of actin in Leishmania : molecular characterization, subcellular localization and association with subpellicular microtubules. Mol. Biochem. Parasitol. 2004;134(1):105–114. doi: 10.1016/j.molbiopara.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Spoerl Z., Stumpf M., Noegel A.A., Hasse A. Oligomerization, F-actin interaction, and membrane association of the ubiquitous mammalian coronin 3 are mediated by its carboxyl terminus. J. Biol. Chem. 2002;277(50):48858–48867. doi: 10.1074/jbc.M205136200. [DOI] [PubMed] [Google Scholar]
- Srivastava R., Prasadareddy Kajuluri L., Pathak N., Gupta C.M., Sahasrabuddhe A.A. Oligomerization of coronin: implication on actin filament length in Leishmania. Cytoskeleton. 2015;72(12):621–632. doi: 10.1002/cm.21269. [DOI] [PubMed] [Google Scholar]
- Stetefeld J., Jenny M., Schulthess T., Landwehr R., Engel J., Kammerer R.A. Crystal structure of a naturally occurring parallel right-handed coiled coil tetramer. Nat. Struct. Biol. 2000;7(9):772–776. doi: 10.1038/79006. [DOI] [PubMed] [Google Scholar]
- Svergun D.I., Petoukhov M.V., Koch M.H. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80(6):2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchang V.S., Mekker A., Siegmund K., Karrer U., Pieters J. Diverging role for coronin 1 in antiviral CD4+ and CD8+ T cell responses. Mol. Immunol. 2013;56(4):683–692. doi: 10.1016/j.molimm.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Volkov V.V., Svergun D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36(3):860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf E., Kim P.S., Berger B. MultiCoil: a program for predicting two-and three-stranded coiled coils. Protein Sci. 1997;(6):1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier C.P., Eichinger L., Fernandez M.P., Morgan R.O., Clemen C.S. The Coronin Family of Proteins. Springer; New York, NY: 2008. Evolutionary and functional diversity of coronin proteins; pp. 98–109. [DOI] [PubMed] [Google Scholar]
- Xavier C.P., Rastetter R.H., Blömacher M., Stumpf M., Himmel M., Morgan R.O., Fernandez M.P., Wang C., Osman A., Miyata Y., Gjerset R.A. Phosphorylation of CRN2 by CK2 regulates F-actin and Arp2/3 interaction and inhibits cell migration. Sci. Rep. 2012;2:241. doi: 10.1038/srep00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Collins R.F., Grinstein S., Trimble W.S. Coronin-1 function is required for phagosome formation. Mol. Biol. Cell. 2005;16(7):3077–3087. doi: 10.1091/mbc.E04-11-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Zhu H., Lashuel H.A., Hu J.C. Oligomerization properties of GCN4 leucine zipper e and g position mutants. Protein Sci. 1997;6:2218–2226. doi: 10.1002/pro.5560061016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart P, Gross-Kunstleve R.W., Adams P.D. Characterization of X-ray Datasets. CCP4 Newsletts. 2005;42 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates have been deposited in the Protein Data Bank (PDB ID: 7DGX, 7DH4 & 7DHB) and SASBDB Id SASDK46.