Abstract
Livestock farms are generally considered to be the important source of antibiotic resistance genes (ARGs). It is important to explore the spread of ARGs to reduce their harm. This study analyzed 13 resistance genes belonging to 7 types in 68 samples of layer manure including different stages of layer breeding, layer manure fertilizer, and soil from 9 laying hen farms in Guangdong Province. The detection rate of antibiotic resistance genes was extremely high at the layer farm in manure (100%), layer manure fertilizer (100%), and soil (> 95%). The log counts of antibiotic resistance genes in layer manure (3.34–11.83 log copies/g) were significantly higher than those in layer manure fertilizer (3.45–9.80 log copies/g) and soil (0–7.69 log copies/g). In layer manure, ermB was the most abundant antibiotic resistance gene, with a concentration of 3.19 × 109– 6.82 × 1011 copies/g. The average abundances of 5 antibiotic resistance genes were above 1010 copies/g in the descending order ermB, sul2, tetA, sul1, and strB. The relative abundances of ARGs in layer manure samples from different breeding stages ranked as follows: brooding period (BP), late laying period (LL), growing period (GP), early laying period (EL), and peak laying period (PL). There was no significant correlation between the farm scale and the abundance of antibiotic resistance genes. Moreover, the farther away from the layer farm, the lower the abundance of antibiotic resistance genes in the soil. We also found that compost increases the correlation between antibiotic resistance genes, and the antibiotic resistance genes in soil may be directly derived from layer manure fertilizer instead of manure. Therefore, when applying layer manure fertilizer to cultivated land, the risk of antibiotic resistance genes pollution should be acknowledged, and in-depth research should be conducted on how to remove antibiotic resistance genes from layer manure fertilizer to control the spread of antibiotic resistance genes.
Key words: layer manure, soil, compost, antibiotic resistance gene
INTRODUCTION
Intensive breeding of laying hens in confined spaces has led to colonization of poultry by resistant bacteria (Skowron et al., 2016). The use of antibiotics in livestock production may lead to the emergence of antibiotic resistant bacteria and reduce the effectiveness of antibiotics in treating and preventing human bacterial infections (Li et al., 2015; Tiedje et al., 2019). Antibiotics have caused great concern because of their extensive application in agriculture, aquaculture and the medical industry to prevent or treat bacterial infections in humans and animals, which has also caused uncertain threats to global public health. Animal excrement is the main source of residual antibiotics and has become a reservoir of resistant bacteria. There is increasing evidence that antibiotic resistance genes (ARGs) can enter the environment through the discharge of animal manure, causing soil, water, and crop pollution (He et al., 2016). Unlike broiler farms in China, the use of antibiotics is strictly controlled during layer breeding, especially during the laying period. Therefore, there may be differences in antibiotic residues in the feces of laying hens and broilers. For example, the content of chlortetracycline (323 μg/kg), oxytetracycline (147 μg/kg), and doxycycline (303 μg/kg) in layer manure is significantly lower than that in broiler manure (Qiu et al., 2021). The antibiotic concentration is the important factor affecting ARGs (Tong et al., 2020). Previous investigations studied the ARG contents in broiler manure before and after antibiotic use. The results showed that the abundance of certain ARGs in the broiler coop increased even after composting after antibiotics were added to the broiler diet (Subirats et al., 2020). Therefore, antibiotics may lead to some differences in ARGs between laying hens and broilers. In addition, because the use of antibiotics during the incubation and growth periods is allowed for laying hens, most antibiotics are prohibited during the laying period. With growth and development, the abundance of ARGs should be significantly different.
Mainstream farming practices currently include the combined production of plants and animals. Before application of manure to land, composting, as an effective method to treat fecal waste and produce layer manure fertilizer, has become an important link in controlling the development of drug resistance (Cui et al., 2016). Composting is a biological oxidation process that is a harmless way to kill pathogens, and compost products can provide nutrients for crops (Liang et al., 2020). Composting can effectively reduce the concentrations of antibiotics. The antibiotic concentration decreased by more than 4,000-fold along the environmental transmission chain from manure samples from swine farms to aerobic compost, compost-amended agricultural soils, and neighboring agricultural soils (Gao et al., 2020). However, several studies have investigated ARGs have divergent fates during composting. Although compost significantly reduces the abundance of certain resistance genes, it cannot completely eliminate resistance (Cui et al., 2016; Xie et al., 2016; Qian et al., 2017; Zhao et al., 2017). In addition, the abundance of some resistance genes increased after composting (Xia et al., 2019).
Most studies assert that livestock manure is the important source of resistance genes in soil and water bodies and is an ARG reservoir (Zhu et al., 2013). However, manure is not directly applied to farmland but first undergoes harmless treatment processes. Composting is one of the important harmless treatment methods for manure. Therefore, the dynamics of the migration of ARGs from livestock manure to compost products to the surrounding soil provide a reference for better preventing the transfer of resistance genes and bacteria.
The objective of this study was to investigate the distribution characteristics of ARGs from layer manure to receiving environments. We analyzed the distribution of main ARGs in layer manure at different breeding stages, and provided a reference for the application of layer manure. We also investigated ARGs in layer manure from different growth periods and the dynamics of soil resistance genes at different distances from the farm. The study will provide a reference for scientifically and rationally reducing the diffusion of ARGs from layer manure to the ecosystem and evaluating the ecological risks that layer manure fertilizer may cause.
MATERIALS AND METHODS
Laying Hen Farms and Sample Collection
Nine representative laying hen farms with different waste management systems in Guangdong Province were selected for this study (Figure 1A). Manure, layer manure fertilizer and soil samples from laying hen farms at different farm scales were collected. The farm scales included small- and medium-; large-; and collectivization-scales. Detailed information on the laying hen farms is shown in Table S1. Manure samples were collected depending on the breeding periods and manure processing modes of the studied laying hen farms. Manure samples were taken during the brooding period (BP), growing period (GP), early laying period (EL), peak laying period (PL), and late laying period (LL), and different manure treatments included stacked manure (SM) and layer manure fertilizer (OM). Soil samples around the farm included soil from the farm (SI) and soil from farms 100 meters (S100), 300 meters (S300), and 500 meters (S500) away. In total, 37 manure samples and 31 soil samples were collected. These samples were collected from 9 laying hen farms, which were named A to I (S1). Among the 9 laying hen farms, D, G, H, and I employed composting to treat the layer manure. All samples were stored in an ice box, immediately shipped to the laboratory and stored at −80°C.
Figure 1.
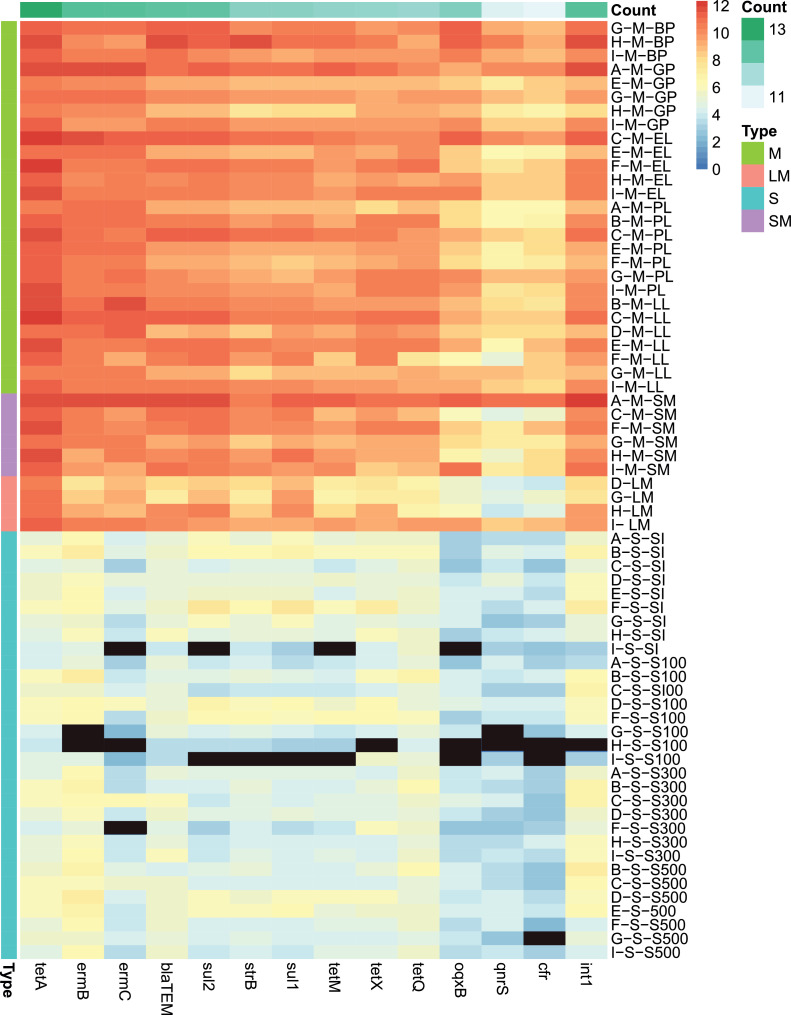
Sample collection and copy numbers of the main ARGs in the sample. (A) Distribution map of nine layer farms (A–I); (B) broad-spectrum quantitative profiles of 13 ARGs and int1 in layer manure, layer manure fertilizer, and soil. The first letter of the sample name represents the farm; the second represents the sample type, manure (M), layer manure fertilizer (LM) or soil (S); the third represents one of the following, brooding period (BP), growing period (GP), early laying period (EL), peak laying period (PL), late laying period (LL), stacked manure (SM), soil at the farm (SI), soil within 100 meters of the farm (S100), soil within 300 meters of the farm (S300), soil within 500 meters of the farm (S500). The absolute ARG concentrations were used for plotting. Black cells indicate the absence of corresponding ARGs in a certain sample. Abbreviation: ARGs, antibiotic resistance genes.
DNA Extraction
DNA of the total microorganisms was extracted from manure and soil samples using the E.Z.N.A. Soil DNA Kit (OMEGA, Norcross, Georgia). The DNA concentration and purity were determined using a NanoDrop 2000 instrument (Thermo Scientific, Waltham, Massachusetts). The extracted DNA was stored in a freezer at −20°C until use.
Quantification of Antibiotic Resistance Genes
Previous studies have shown that tetracycline, sulfonamide, β-lactam, fluoroquinolone, macrolide, chloramphenicol, and streptomycin resistance genes are the most common ARGs in organic waste (Su et al., 2015). Therefore, 13 ARGs were determined, including tetracycline resistance genes (tetA, tetM, tetQ, and tetX), 2 sulfonamide resistance genes (sul1 and sul2), 1 beta-lactam resistance gene (blaTEM), 2 fluoroquinolone resistance genes (oqxB and qnrS), 2 macrolide resistance genes (ermB and ermC), 1 chloramphenicol resistance gene (drfA7), 1 multiple resistance gene (cfr) and one streptomycin resistance gene (strB). Mobile genetic elements (MGEs) can spread resistance genes horizontally in bacteria (Forsberg et al., 2014). int1 is the key MGE for the horizontal transfer of ARGs. All of the ARGs and MGEs were analyzed by PCR and agarose electrophoresis. The detected ARGs and MGEs were analyzed further by qPCR using an iCycler IQ5 thermocycler (Bio-Rad, Hercules, California).
After PCR, the products of all genes were subjected to gel electrophoresis and recovered using a Gel Extraction Kit (OMEGA). The recovered product was ligated into the pMD18-T vector. Standard curves were constructed in which each plasmid was diluted in a 10-fold gradient, and the diluted plasmids were used as templates for qPCR. The qPCR mixture in a volume of 20 μL contained 1 μL of DNA template, 0.5 μL of each primer (ShengGong, China), 10 μL of SYBR qPCR mix (TaKaRa, Japan), and 8 μL of ddH2O. The thermal cycling steps for qPCR amplification were as follows: 95°C for 5 min; followed by 35 cycles of 95°C for 30 s, annealing temperatures (shown in Table S2) for 30 s, 72°C for 30 s; 72°C for 10 min; and a 4°C hold (Yin et al., 2017). Each dilution gradient was repeated 3 times, and 3 negative controls were made at the same time. The lower limit of quantification (LOQ) for each qPCR assay was higher than the result of the negative controls (Table S3). A melting curve of qPCR products was constructed. The squared correlation coefficient (R2) was >0.99, and the amplification efficiency ranged between 85 and 105% for the standard curves, which were used to calculate the copy numbers of ARGs. The absolute abundance of a sample was calculated as follows: absolute abundance (copies/g) = Copy number calculated by the standard curves (copies/μL) × sample DNA elution volume (μL)/sample weight (g). The relative abundance of a sample was calculated as follows: relative abundance (copies/16S rRNA copies) = absolute abundance of ARG / absolute abundance of 16S rRNA.
Statistical Analysis
Spearman's correlation coefficients were calculated using SPSS 19.0 (IBM), and nonparametric tests (Kruskal-Wallis test) was performed (least significant difference test; P < 0.05). For the statistical analysis, the copy numbers of the samples were log transformed as needed to normalize the distributions prior to the analysis of ARGs. Pearson correlation coefficients were determined using R software. After individual node centrality and module separation processes, the constructed network was visualized using Cytoscape.
RESULTS AND DISCUSSION
Distribution Characteristics of ARGs in Layer Manure, Layer Manure Fertilizer, and Soil
In this study, seven classes of ARGs, namely, tetracycline, sulfonamide, β-lactam, quinolone, macrolide, chloramphenicol, and streptomycin resistance genes, were examined because of their widespread contamination of the farm environment. The frequencies of ARG detection in the 3 environmental samples are shown in Table 1. Notably, the detection frequency of the target genes was 100% in layer manure (n = 3 3) and layer manure fertilizer (n = 4). The detection frequency of the target genes was above 90% (n = 31) in soil. Among all samples (n = 68), the highest detection frequency was 100% (tetA, tetQ, and blaTEM), the second was 98.53% (tetX, sul1, strB, and int1), the third was 97.06% (tetM, ermB, sul2, qnrS), and the lowest was 95.59% (ermC, oqxB, and cfr).
Table 1.
Log count of ARGs and int1 in layer manure, layer manure fertilizer, and soil (copies/g).
| Manure |
LM |
Soil |
Total |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | Min | Max | Mean ± std. error | Detection rate/100% | Min | Max | Mean ± std. error | Detection rate/100% | Min | Max | Mean ± std. error | Detection rate/100% | Detection rate/100% |
| tetA | 8.60 | 11.47 | 10.24 ± 0.77 | 100 | 7.27 | 9.53 | 8.36 ± 1.02 | 100 | 3.86 | 7.06 | 5.44 ± 0.79 | 100 | 100 |
| tetM | 7.77 | 10.35 | 9.4 ± 0.59 | 100 | 6.75 | 8.52 | 7.39 ± 0.67 | 100 | 0 | 6.36 | 4.46 ± 1.39 | 93.55 | 97.06 |
| tetQ | 7.20 | 11.31 | 9.69 ± 0.98 | 100 | 6.60 | 8.91 | 7.32 ± 0.93 | 100 | 4.22 | 6.89 | 5.38 ± 0.56 | 100 | 100 |
| tetX | 7.56 | 11.17 | 9.48 ± 0.93 | 100 | 7.45 | 9.40 | 8.28 ± 0.71 | 100 | 0 | 7.27 | 4.98 ± 1.20 | 96.77 | 98.53 |
| ermB | 9.50 | 11.83 | 10.53 ± 0.58 | 100 | 7.46 | 9.17 | 8.36 ± 0.66 | 100 | 0 | 7.26 | 5.87 ± 1.26 | 93.55 | 97.06 |
| ermC | 7.27 | 10.95 | 8.51 ± 0.81 | 100 | 8.67 | 9.67 | 9.2 ± 0.35 | 100 | 0 | 6.39 | 3.67 ± 1.50 | 90.32 | 95.59 |
| sul1 | 8.16 | 11.45 | 10.04 ± 0.85 | 100 | 8.63 | 10.46 | 9.35 ± 0.68 | 100 | 0 | 7.45 | 4.72 ± 1.35 | 96.77 | 98.53 |
| sul2 | 8.70 | 11.54 | 10.36 ± 0.74 | 100 | 7.71 | 10.45 | 8.98 ± 1.02 | 100 | 0 | 7.69 | 4.72 ± 1.37 | 93.55 | 97.06 |
| blaTEM | 6.33 | 11.38 | 9.38 ± 1.22 | 100 | 5.88 | 9.10 | 6.77 ± 1.35 | 100 | 3.41 | 6.14 | 5.15 ± 0.64 | 100 | 100 |
| oqxB | 3.34 | 10.51 | 6.40 ± 1.73 | 100 | 3.92 | 7.07 | 4.83 ± 1.3 | 100 | 0 | 4.91 | 3.41 ± 1.26 | 90.32 | 95.59 |
| qnrS | 5.28 | 11.03 | 8.02 ± 1.34 | 100 | 4.06 | 7.00 | 5.08 ± 1.15 | 100 | 0 | 5.30 | 3.66 ± 0.93 | 93.55 | 97.06 |
| cfr | 5.14 | 9.01 | 7.20 ± 1.08 | 100 | 3.45 | 8.06 | 5.66 ± 1.67 | 100 | 0 | 4.53 | 3.1 ± 1.03 | 90.32 | 95.59 |
| strB | 7.78 | 11.16 | 10.00 ± 0.82 | 100 | 7.44 | 9.58 | 8.52 ± 0.79 | 100 | 0 | 6.39 | 4.82 ± 1.14 | 96.77 | 98.53 |
| int1 | 8.25 | 11.39 | 9.87 ± 0.81 | 100 | 7.84 | 9.80 | 8.86 ± 0.84 | 100 | 0 | 7.20 | 5.36 ± 1.44 | 96.77 | 98.53 |
Abbreviation: ARGs, antibiotic resistance genes.
Although all 13 ARGs were detected in layer manure, layer manure fertilizer, and soil, their abundances were different among the sample types. As shown in Figure 1B, the abundance of detected genes in layer manure was significantly higher than that in layer manure fertilizer and soil (P < 0.05). Lee et al., 2017 and He et al. (2016) previously reported that layer manure could be a main source of ARG contamination in layer farm environments. Based on the measurements within the 3 types of samples, the macrolide resistance gene ermB showed the highest abundances in layer farm environments, in the range of 7.48 × 105–3.40 × 1010 copies/g. Studies in broiler farms also found that the relative abundance of the ermB gene was the highest (Yang et al., 2020), which should create awareness regarding the spread of this resistance gene. The abundance of macrolide resistance genes may be related to the greater use of macrolides during feeding (Zhang et al., 2017). In addition, the absolute abundance of tetA, tetM, tetQ, and tetX was 2.88 × 104–2.95 × 1011 copies/g. Tetracycline resistance genes were the most common types of ARGs. Previous studies reported that tetracycline resistance genes were the predominant ARGs in livestock and poultry manure (Wen et al., 2019). Adding 2 g/L of chlortetracycline to drinking water can increase the abundance of tetracycline resistance genes in chicken feces (Xiong et al., 2018). And studies have shown that 10 ug/L tetracycline can promote the horizontal transfer of tetracycline resistance genes, which shows that even low dose antibiotics can promote the increase of ARGs abundance (Jutkina et al., 2016).
The abundance of sulfa resistance genes (sul2) was 5.25 × 104–3.74 × 1011 copies/g, second only to ermB. From layer manure to organic fertilizer, the levels of the sul2 decreased from 2.29 × 1010 copies/g to 9.55 × 108 copies/g, respectively. Birgit et al. (2016) had similar results in a study in which the levels of the sul2 genes dramatically decreased during composting. The changes in the number of host bacteria may be an explanation for the increase or decrease of ARGs during the composting (Ben et al., 2017). In addition, fluoroquinolone resistance genes oqxB (2.51 × 106 copies/g) and multiple resistance genes cfr (1.58 × 107 copies/g) were the least abundant in layer manure which indicated a smaller risk of transmission of these genes.
Resistance Gene Abundance in Layer Farms of Different Scales
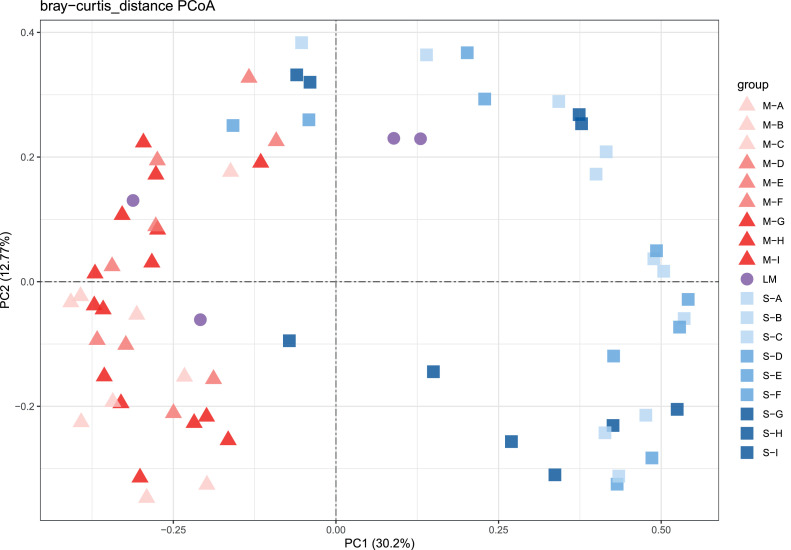
Few studies in the past have focused on the relationship between ARGs and the scale of farms. Therefore, we analyzed the correlation between layer farms of different scales and ARGs. Previous studies have found that ARGs were mainly subjected to impacts from environmental factors and microbial communities (Cao et al., 2020). The samples from manure and soil are concentrated on the left and right of the figure, respectively, while the samples from layer manure fertilizer are mainly concentrated in the middle (Figure 2). In addition, the samples of the same scale were not clustered together. Principal coordinate analysis (PCoA) demonstrated that the difference in the abundance of ARGs between samples mainly depended on whether they originated from manure, layer manure fertilizer or soil, and there was no significant correlation with the scale of the layer farm. The above results indicate that the farm scale did not affect the abundance of ARGs.
Figure 2.
Principal coordinate analysis (PCoA) showing the clustering of communities based on b-diversity. The nodes are shaped according to the types of substances, including layer manure (triangle), layer manure fertilizer (round), and soil (square). The color from light to dark indicates the scale of the farm from small to large, respectively.
Abundance of ARGs and int1 in Different Breeding Stages
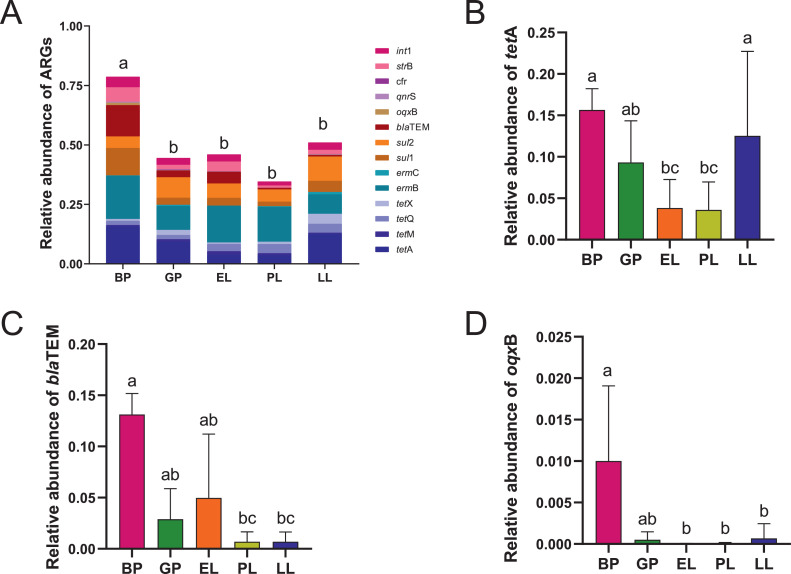
Ma et al. (2016) detected 4 tetracycline ARGs in 20-day-old layer manure, and as many as 16 ARGs in 80-day-old layer manure. To study the dynamics of ARGs over the breeding cycle of laying hens, we collected layer manure from different periods and analyzed the relative abundance of the main ARGs (Figure 3A). The comparison of ARG profiles indicated that the abundance of ARGs in different breeding periods was quite different. The relative abundance of ARGs in the manure of layers at different breeding stages from high to low was as follows: the brooding period, the late laying period, the early laying period, the growing period, and the peak laying period. This may be related to the heavy use of antibiotics during the brooding period (Rivera-Gomis et al., 2021). The relative abundance during the brooding period was significantly higher than that during the other breeding stages (P < 0.05). The reason for this phenomenon is that the relative abundance of tetA (Figure 3B), blaTEM (Figure 3C), and oqxB (Figure 3D) in the brooding period was significantly higher than peak laying period (P < 0.05). The decrease in the abundance of tetA, blaTEM, and oqxB may be related to the prohibition of antibiotics during the laying period, which reduces the risk of residual resistance genes in eggs. The relative abundance of ermB varied from 1.22 × 10−1–2.34 × 10−1 during the integral breeding period. Moreover, a study also found the macrolide resistance gene (erm-ARG) ermB in the waste of intensive chicken farms with high abundance (Mu et al., 2015). The relative abundance of the sulfonamide resistance gene sul2 (9.13 × 10−2–1.54 × 10−1) was always high during the breeding process. Previous studies have shown that sul1 and sul2 are very abundant sulfonamide resistance genes in manure and soil to which manure is applied (Zhao et al., 2017). Therefore, ermB and sul2 consistently maintained a high abundance without a significant decrease, which indicates that they pose a high risk during the laying period and can exist more stably without selection pressure. However, this study is based solely on a snapshot of various samples collected at a single time in point, so the conclusion drawn is limited to one observation.
Figure 3.
The relative abundance of ARGs and int1 in different breeding stages. (A) Changes in ARGs and int1 in different breeding stages. (B–D) Changes in tetA, sul1 and blaTEM in different breeding stages. Brooding period (BP), growing period (GP), early laying period (EL), peak laying period (PL), late laying period (LL). Different letters indicate significant differences between the means (Kruskal-Wallis test; P < 0.05) of treatments. Abbreviation: ARGs, antibiotic resistance genes.
The Relative Abundance of ARGs and int1 in LM and SM
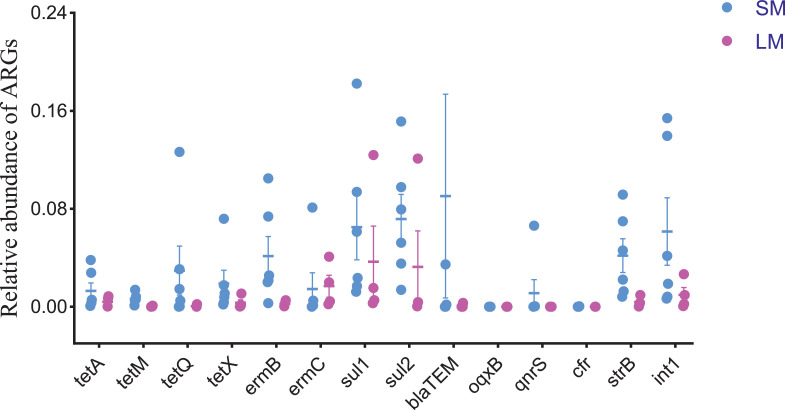
Composting is widely used to treat and reutilize animal manures for ARG and pathogen reduction (Guo et al., 2020; Peng et al., 2020). Stacked manure is manure that has not been composted, just piled manure together for several months. The distributions of detected ARGs in stacked manure and layer manure fertilizer are shown in Figure 4. It can be seen that there was no significant difference in ARGs between stacked manure and layer manure fertilizer (P > 0.05), but the value between the upper and lower quartiles in stacked manure was notably higher than that in layer manure fertilizer. OqxB and cfr had the lowest relative abundances in stacked manure and layer manure fertilizer. The relative abundances of tetM, tetQ oqxB, and cfr were less than 1.4 × 10−2. Composting resulted in better ARG reduction efficiency in layer manure fertilizer than did manure stacking. These results suggested that specific compost treatment components can effectively decrease the concentration of ARGs in layer farms. Current studies demonstrate the effective elimination of ARG residues, and the absolute abundance of TRGs, MLSBRGs, β-LRGs, and PRGs significantly declined after composting (Zhang et al., 2016; Cheng et al., 2019). In layer manure fertilizer, the relative abundances of sul1 and sul2 were higher than those of other ARGs. The results indicate that composting had lower removal efficiency for sulfonamide resistance genes than for other ARGs. Holman et al. (2016) found similar results and showed that sul1 was more persistent than most tetracycline and macrolide resistance genes during cattle manure composting. The removal efficiencies of tetX, ermC, and int1 from compost were also limited. Many studies have reported that composting may not completely remove ARGs, so compost products are still considered an important reservoir for ARGs (Su et al., 2015; Qian et al., 2016). In general, composting is a vital link for controlling the dissemination of antibiotic resistance, however, close attention should still be given to those genes that are not easily removed.
Figure 4.
The relative abundance of ARGs and int1 in LM and SM. Stacked manure (SM, n = 6), layer manure fertilizer (LM, n = 4). Abbreviation: ARGs, antibiotic resistance genes.
The Relative Abundance of ARGs and int1 in Soil
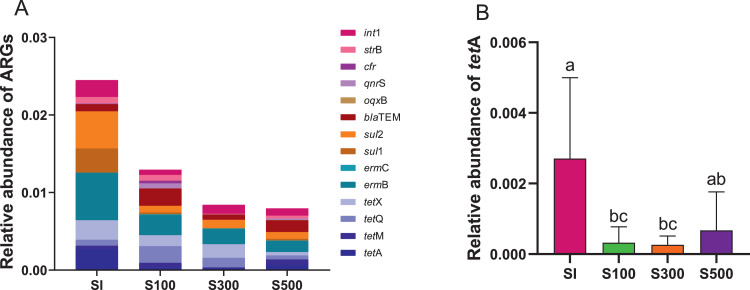
Poultry is currently regarded as a reservoir from which multidrug resistance can be readily transferred to the surrounding ecosystem (Dandachi et al., 2020). ARGs in animal manures enter the soil through manure application, which leads to significant increases in ARGs in soil (Chen et al., 2019a,b). In addition to being directly affected by layer manure fertilizer, the soil is also affected by groundwater and rain due to the proximity to the layer farm (Nnadozie and Odume, 2019; Zhu et al., 2021), so the distance to the layer farm may affect the abundance of ARGs in the soil. Therefore, we analyzed the distribution of ARGs in the soil at different distances from the farm, as shown in Figure 5A. The relative concentration of total ARGs in SI was 2.51 × 10−2, which was higher than that in surrounding soil. However, the difference between the groups was not significant. We found that the abundance of tetA in the soil outside the laying hen farm was lower than that in the farm, especially the S100 and S300 were significantly lower than the SI. (Figure 5B). This indicated that the risk of tetA transmission outside the farm was lower than that on the farm. Moreover, the 2 ARGs, blaTEM and ermB, had high abundances both inside and outside the layer farms and did not decrease significantly with increasing distance. Therefore, these 2 resistance genes may be more likely to spread in the soil off-site, posing an ARG contamination risk.
Figure 5.
The abundance of ARGs and int1 in the soil at different distances from the farm. (A) The relative abundance of ARGs and int1 in soil. (B) The difference in the abundance of tetA in the soil. Soil on the farm (SI), soil within 100 meters of the farm (S100), soil within 300 meters of the farm (S300), soil within 500 meters of the farm (S500). Different letters indicate significant differences between the means (Kruskal-Wallis test; P < 0.05) of treatments. Abbreviation: ARGs, antibiotic resistance genes.
The relative abundance of the total ARGs gradually decreased with distance from the farm, but the difference was not significant. This indicates that the relative abundance of ARGs in the soil is mainly affected by layer manure fertilizer, but has a low correlation with the distance from layer farms.
Correlation Analysis of ARGs in Different Environments
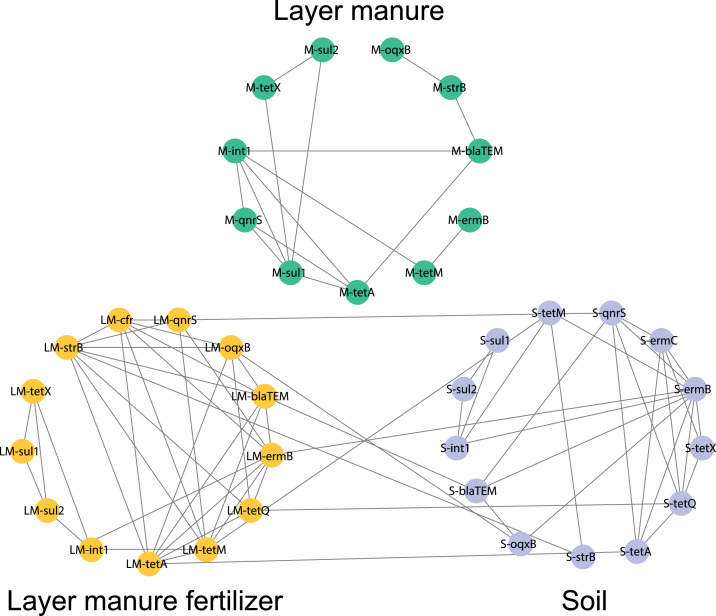
Layer manure, which is processed into organic fertilizer or directly applied to the soil, contains a large number of ARGs. Therefore, it is fundamental to research the spread of ARGs in different environments. The soil samples collected in this study were all applied with layer manure fertilizer instead of directly applying layer manure. Layer manure fertilizer is a harmless product from layer manure after aerobic composting. In the present study, significant correlations were found among various ARGs in particular environments. Nevertheless, the correlation of ARGs in different samples varied greatly (Figure 6). In the 3 environmental samples, ermB was significantly correlated with tetM (P < 0.05). According to a study by Jia et al. (2017), tetM and ermB can be acquired by the same microorganism. Therefore, tetM and ermB may be carried by the same microorganism to contaminate the environment (Shen et al., 2008).
Figure 6.
Network analysis of different ARGs (relative abundance) based on Pearson's correlation coefficients (P < 0.05, R > 0.50) in layer manure, layer manure fertilizer and soil. The nodes are colored according to the type of environment, including layer manure (green), layer manure fertilizer (yellow), and soil (purple). Abbreviation: ARGs, antibiotic resistance genes.
In layer manure, there was a weak correlation among ARGs, but after composting, the correlation among ARGs was enhanced in layer manure fertilizer. In organic matter, StrB had a positive correlation with 9 other ARGs. tetA, tetQ, tetM, ermB, oqxB, and blaTEM in layer manure fertilizer showed a significant positive correlation with each other (P < 0.05). The ARGs in layer manure showed a lower correlation than those in layer manure fertilizer and soil. It is worth noting that tetA had a significant positive correlation between layer manure fertilizer and soil (P < 0.01), and tetQ, tetM, ermB and blaTEM showed similar results (P < 0.05). This result highlights the possibility of ARGs spreading from layer manure fertilizer to soil. The results from a recent study showed that compost application to land could lead to enhanced contamination levels of ARGs in compost-amended soils (Xu et al., 2015; Ben et al., 2017). Many studies have indicated that livestock manure is an important source of ARGs in the environment (Cerqueira et al., 2019; Liao et al., 2019). However, from our investigation, the high abundance of ARGs in manure does not mean a high risk of ARG transmission because composting has become an efficient method to decompose manure waste and produce layer manure fertilizer. Notably, compared with the ARGs in the layer manure, the ARGs in the layer manure fertilizer had a significant correlation with the ARGs in the soil. This result means that we need to pay more attention to the content of ARGs and new pollutants in compost products.
ARGs and MGEs have often been linked to or located in integrons (Cao et al., 2020). In this study, int1 was significantly correlated with ermB, sul2, and tetM in layer manure fertilizer and soil (P < 0.01), whereas these factors were weakly correlated in layer manure. Studies found that tetM exists in conjugative plasmids of various gram-positive and gram-negative bacteria (Ojo et al., 2006; Pachulec and Does, 2010; Shaskolskiy et al., 2018). During swine manure composting, int1 was significantly correlated with tetM and ermB (Qian et al., 2019). Previous studies have shown that integrons can integrate exogenous ARG cassettes, which promote multidrug resistance (Rapa and Labbate, 2013). In addition, ARG cassettes are usually located in plasmids or transposons, making them easy to disseminate in the environment (Partridge et al., 2018). These findings suggest that the promotion of horizontal gene transfer by int1 may be an important contributor to dissemination in different media. Huddleston, (2018) suggested that int1 participates in Horizontal gene transfer (HGT) processes and induces bacteria to acquire ARGs (e.g., sulI, dfrA1, tetA, and floR) in the environment. Furthermore, compared with manure, ARGs in layer manure fertilizer are more related, which may be caused by the existence of a common host for multiple ARGs. The correlation of ARGs in soil was also significantly higher than that in manure. The ARGs in soil may mainly come from layer manure fertilizer, which also confirms our speculation. This result illustrates that composting may lead to the multidrug resistance of microorganisms and aggravate the harmful effects of resistant bacteria. Eventually, animal husbandry and even humans themselves may be in crisis, so we need to pay more attention.
CONCLUSIONS
This study investigated the distribution of ARG in layer manure, layer manure fertilizer, and soil. The number of ARGs in layer manure during the incubation period was significantly higher than that in other periods, and the scale of the layer farm had no significant effect on the abundance of ARGs. We found that the correlation of ARGs in layer manure fertilizer was significantly higher than that of ARGs in layer manure, which may be due to the emergence of multidrug resistant bacteria. Furthermore, the ARGs in the soil and layer manure fertilizer show a significant positive correlation, indicating that the ARGs in the soil may come from layer manure fertilizer instead of directly from layer manure.
Acknowledgments
ACKNOWLEDGMENTS
This work was supported by the Natural Science Foundation of Guangdong Province [grant numbers 2020A151501925]; WENS Foodstuff Group Co., Ltd, quantification of odor-causing substances in manure fermentation and optimization of their removal methods [grant numbers YQ20200608FCXY079]; the earmarked fund for Modern Agro-industry Technology Research System [grant numbers CARS-40]; and the Laboratory of Lingnan Modern Agriculture Project [grant numbers NZ2021027].
DISCLOSURES
We declare that no conflict of interest exits in this manuscript, and manuscript is approved by all authors for publication. This manuscript is original and has not been published in whole or in part previously.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2021.101485.
Appendix. Supplementary materials
REFERENCES
- Ben W., Wang J., Pan X., Qiang Z. Dissemination of antibiotic resistance genes and their potential removal by on-farm treatment processes in nine swine feedlots in shandong province, China. Chemosphere. 2017;167:262–268. doi: 10.1016/j.chemosphere.2016.10.013. [DOI] [PubMed] [Google Scholar]
- Birgit W., Guo-Chun D., Robert K., Kornelia S. Full-scale mesophilic biogas plants using manure as c-source: bacterial community shifts along the process cause changes in the abundance of resistance genes and mobile genetic elements. Fems. Microbiol. Ecol. 2016:fiv163. doi: 10.1093/femsec/fiv163. [DOI] [PubMed] [Google Scholar]
- Cao R., Ben W., Qiang Z., Zhang J. Removal of antibiotic resistance genes in pig manure composting influenced by inoculation of compound microbial agents. Bioresour. Technol. 2020;317 doi: 10.1016/j.biortech.2020.123966. [DOI] [PubMed] [Google Scholar]
- Cerqueira F., Matamoros V., Bayona J.M., Berendonk T.U., Piña B. Antibiotic resistance gene distribution in agricultural fields and crops. A soil-to-food analysis. Environ. Res. 2019;177 doi: 10.1016/j.envres.2019.108608. [DOI] [PubMed] [Google Scholar]
- Chen C., Pankow C.A., Oh M., Heath L.S., Zhang L., Du P., Xia K., Pruden A. Effect of antibiotic use and composting on antibiotic resistance gene abundance and resistome risks of soils receiving manure-derived amendments. Environ. Int. 2019;128:233–243. doi: 10.1016/j.envint.2019.04.043. [DOI] [PubMed] [Google Scholar]
- Chen Z., Zhang W., Yang L., Stedtfeld R.D., Peng A., Gu C., Boyd S.A., Li H. Antibiotic resistance genes and bacterial communities in cornfield and pasture soils receiving swine and dairy manures. Environ. Pollut. 2019;248:947–957. doi: 10.1016/j.envpol.2019.02.093. [DOI] [PubMed] [Google Scholar]
- Cheng D., Feng Y., Liu Y., Xue J., Li Z. Dynamics of oxytetracycline, sulfamerazine, and ciprofloxacin and related antibiotic resistance genes during swine manure composting. J. Environ. Manage. 2019;230:102–109. doi: 10.1016/j.jenvman.2018.09.074. [DOI] [PubMed] [Google Scholar]
- Cui E., Wu Y., Zuo Y., Chen H. Effect of different biochars on antibiotic resistance genes and bacterial community during chicken manure composting. Bioresour. Technol. 2016;203:11–17. doi: 10.1016/j.biortech.2015.12.030. [DOI] [PubMed] [Google Scholar]
- Dandachi I., Fayad E., Sleiman A., Daoud Z., Rolain J.M. Dissemination of multidrug-resistant and mcr-1 gram-negative Bacilli in broilers, farm workers, and the surrounding environment in Lebanon. Microb. Drug. Resist. 2020;26:368–377. doi: 10.1089/mdr.2019.0137. [DOI] [PubMed] [Google Scholar]
- Forsberg K.J., Patel S., Gibson M.K., Lauber C.L., Knight R., Fierer N., Dantas G. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509:612. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Gao S., Bates C., Zeng Y., Yang Y. The microbial network property as a bio-indicator of antibiotic transmission in the environment. Sci. Total Environ. 2020;758 doi: 10.1016/j.scitotenv.2020.143712. [DOI] [PubMed] [Google Scholar]
- Guo H., Gu J., Wang X., Nasir M., Yu J., Lei L., Wang Q. Elucidating the effect of microbial inoculum and ferric chloride as additives on the removal of antibiotic resistance genes from chicken manure during aerobic composting. Bioresour. Technol. 2020;309 doi: 10.1016/j.biortech.2020.122802. [DOI] [PubMed] [Google Scholar]
- He L.Y., Ying G.G., Liu Y.S., Su H.C., Chen J., Liu S.S., Zhao J.L. Discharge of swine wastes risks water quality and food safety: antibiotics and antibiotic resistance genes from swine sources to the receiving environments. Environ. Int. 2016;92-93:210–219. doi: 10.1016/j.envint.2016.03.023. [DOI] [PubMed] [Google Scholar]
- Holman D.B., Xiying H., Edward T., Eun Y.H., Alexander T.W., Zhi Z. Effect of co-composting cattle manure with construction and demolition waste on the archaeal, bacterial, and fungal microbiota, and on antimicrobial resistance determinants. Plos. One. 2016;11 doi: 10.1371/journal.pone.0157539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J.R. Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect. Drug. Resist. 2018;7:167–176. doi: 10.2147/IDR.S48820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B., Raphenya A.R., Alcock B., Waglechner N., Guo P., Tsang K.K., Lago B.A., Dave B.M., Pereira S., Sharma A.N., Doshi S., Courtot M., Lo R., Williams L.E., Frye J.G., Elsayegh T., Sardar D., Westman E.L., Pawlowski A.C., Johnson T.A., Brinkman F.S., Wright G.D., McArthur A.G. Card 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucl. Acids. Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jutkina J., Rutgersson C., Flach C.F., Joakim Larsson D.G. An assay for determining minimal concentrations of antibiotics that drive horizontal transfer of resistance. Sci. Total Environ. 2016;548-549:131–138. doi: 10.1016/j.scitotenv.2016.01.044. [DOI] [PubMed] [Google Scholar]
- Lee J., Shin S.G., Jang H.M., Kim Y.B., Lee J., Kim Y.M. Characterization of antibiotic resistance genes in representative organic solid wastes: food waste-recycling wastewater, manure, and sewage sludge. Sci. Total Environ. 2017;579:1692–1698. doi: 10.1016/j.scitotenv.2016.11.187. [DOI] [PubMed] [Google Scholar]
- Li Q., Wang Y., Zou Y.D., Liao X.D., Liang J.B., Xin W., Wu Y.B. Co-addition of manure increases the dissipation rates of tylosin A and the numbers of resistance genes in laboratory incubation experiments. Sci. Total Environ. 2015;527-528:126–134. doi: 10.1016/j.scitotenv.2015.04.117. [DOI] [PubMed] [Google Scholar]
- Liao H., Zhao Q., Cui P., Chen Z., Yu Z., Geisen S., Friman V.P., Zhou S. Efficient reduction of antibiotic residues and associated resistance genes in tylosin antibiotic fermentation waste using hyperthermophilic composting. Environ. Int. 2019;133 doi: 10.1016/j.envint.2019.105203. [DOI] [PubMed] [Google Scholar]
- Liang J., Jin Y., Mi J., Zhu T., Wu Y. Adding compound microbial agent twice promoted doxycycline degradation with low risk on spreading tetracycline resistance genes in the composting of laying-hen manure. Environ. Pollut. 2020;265 doi: 10.1016/j.envpol.2020.114202. [DOI] [PubMed] [Google Scholar]
- Ma L., Xia Y., Li B., Yang Y., Li L., Tiedje J.M., Zhang T. Metagenomic assembly reveals hosts of antibiotic resistance genes and the shared resistome in pig, chicken, and human feces. Environ. Sci. Technol. 2016;50:420–427. doi: 10.1021/acs.est.5b03522. [DOI] [PubMed] [Google Scholar]
- Mu Q., Li J., Sun Y., Mao D., Wang Q., Luo Y. Occurrence of sulfonamide-, tetracycline-, plasmid-mediated quinolone- and macrolide-resistance genes in livestock feedlots in northern china. Environ. Sci. Pollut. Res. Int. 2015;22:6932–6940. doi: 10.1007/s11356-014-3905-5. [DOI] [PubMed] [Google Scholar]
- Nnadozie C.F., Odume O.N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 2019;254 doi: 10.1016/j.envpol.2019.113067. [DOI] [PubMed] [Google Scholar]
- Ojo K.K., Ruehlen N.L., Close N.S., Luis H., Bernardo M., Leitao J., Roberts M.C. The presence of a conjugative Gram-positive Tn2009 in Gram-negative commensal bacteria. J. Antimicrob. Chemother. 2006;57:1065–1069. doi: 10.1093/jac/dkl094. [DOI] [PubMed] [Google Scholar]
- Pachulec E., Does C.V.D. Conjugative plasmids of neisseria gonorrhoeae. Plos. One. 2010;5:e9962. doi: 10.1371/journal.pone.0009962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge S.R., Kwong S.M., Neville F., Jensen S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018;31 doi: 10.1128/CMR.00088-17. e00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Gu J., Wang X., Wang Q., Sun W., Hu T., Guo H., Ma J., Bao J. Insight into the fate of antibiotic resistance genes and bacterial community in co-composting green tea residues with swine manure. J. Environ. Manage. 2020;266 doi: 10.1016/j.jenvman.2020.110581. [DOI] [PubMed] [Google Scholar]
- Qian X., Gu J., Sun W., Wang X.J., Stedfeld R. Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 2017;344:716–722. doi: 10.1016/j.jhazmat.2017.11.020. [DOI] [PubMed] [Google Scholar]
- Qian X., Sun W., Gu J., Wang X.J., Sun J.J., Yin Y.N., Duan M.L. Variable effects of oxytetracycline on antibiotic resistance gene abundance and the bacterial community during aerobic composting of cow manure. J. Hazard. Mater. 2016;315:61–69. doi: 10.1016/j.jhazmat.2016.05.002. [DOI] [PubMed] [Google Scholar]
- Qian X., Gu J., Sun W., Wang X., Li H. Effects of passivators on antibiotic resistance genes and related mechanisms during composting of copper-enriched pig manure. Sci. Total Environ. 2019;674:383–391. doi: 10.1016/j.scitotenv.2019.04.197. [DOI] [PubMed] [Google Scholar]
- Qiu T., Wu D., Zhang L., Zou D., Sun Y., Gao M., Wang X. A comparison of antibiotics, antibiotic resistance genes, and bacterial community in broiler and layer manure following composting. Environ. Sci. Pollut. Res. Int. 2021;28:14707–14719. doi: 10.1007/s11356-020-11469-6. [DOI] [PubMed] [Google Scholar]
- Rapa R.A., Labbate M. The function of integron-associated gene cassettes in vibrio species: the tip of the iceberg. Front. Microbiol. 2013;4:385. doi: 10.3389/fmicb.2013.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Gomis J., Marín P., Otal J., Galecio J.S., Martínez-Conesa C., Cubero M.J. Resistance patterns to C and D antibiotic categories for veterinary use of Campylobacter spp., Escherichia coli and Enterococcus spp. commensal isolates from laying hen farms in Spain during 2018. Prev. Vet. Med. 2021;186 doi: 10.1016/j.prevetmed.2020.105222. [DOI] [PubMed] [Google Scholar]
- Shaskolskiy B., Dementieva E., Leinsoo A., Petrova N., Chestkov A., Kubanov A., Deryabin D., Gryadunov D. Tetracycline resistance of Neisseria gonorrhoeae in Russia, 2015-2017. Infect. Genet. Evol. 2018;63:236–242. doi: 10.1016/j.meegid.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Shen X., Yang H., Yu S., Yao K., Wang Y., Yuan L., Yang Y. Macrolide-resistance mechanisms in Streptococcus pneumoniae isolates from Chinese children in association with genes of tetM and integrase of conjugative transposons 1545. Microb. Drug. Resist. 2008;14:155–161. doi: 10.1089/mdr.2008.0773. [DOI] [PubMed] [Google Scholar]
- Skowron K., Jeleńska A., Paluszak Z., Szala B. Prevalence and distribution of VRE (vancomycin resistant enterococci) and VSE (vancomycin susceptible enterococci) strains in the breeding environment. Ann. Agric. Environ. Med. 2016;23:231–236. doi: 10.5604/12321966.1203882. [DOI] [PubMed] [Google Scholar]
- Su J.Q., Wei B., Ou-Yang W.Y., Huang F.Y., Zhao Y., Xu H.J., Zhu Y.G. Antibiotic resistome and its association with bacterial communities during sewage sludge composting. Environ. Sci. Technol. 2015;49:7356–7363. doi: 10.1021/acs.est.5b01012. [DOI] [PubMed] [Google Scholar]
- Subirats J., Murray R., Scott A., Lau H.F., Topp E. Composting of chicken litter from commercial broiler farms reduces the abundance of viable enteric bacteria, firmicutes, and selected antibiotic resistance genes. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141113. [DOI] [PubMed] [Google Scholar]
- Tiedje J.M., Wang F., Manaia C.M., Virta M., Sheng H., Liping M.A., Zhang T., Topp E. Antibiotic resistance genes in the human-impacted environment: a one health perspective. Pedosphere. 2019;29:273–282. [Google Scholar]
- Tong L., Qin L., Guan C., Wilson M.E., Li X., Cheng D., Ma J., Liu H., Gong F. Antibiotic resistance gene profiling in response to antibiotic usage and environmental factors in the surface water and groundwater of Honghu Lake, China. Environ. Sci. Pollut. Res. Int. 2020;27:31995–32005. doi: 10.1007/s11356-020-09487-5. [DOI] [PubMed] [Google Scholar]
- Yang Y., Xie X., Tang M., Liu J., Tuo H., Gu J., Tang Y., Lei C., Wang H., Zhang A. Exploring the profile of antimicrobial resistance genes harboring by bacteriophage in chicken feces. Sci. Total. Environ. 2020;700 doi: 10.1016/j.scitotenv.2019.134446. [DOI] [PubMed] [Google Scholar]
- Wen X., Mi J., Wang Y., Ma B., Zou Y., Liao X., Liang J.B., Wu Y.B. Occurrence and contamination profiles of antibiotic resistance genes from swine manure to receiving environments in guangdong province southern china. Ecotox. Environ. Safe. 2019;173:96–102. doi: 10.1016/j.ecoenv.2019.02.023. [DOI] [PubMed] [Google Scholar]
- Xia H., Chen J., Chen X., Kui H., Wu Y. Effects of tetracycline residuals on humification, microbial profile and antibiotic resistance genes during vermicomposting of dewatered sludge. Environ. Pollut. 2019;252:1068–1077. doi: 10.1016/j.envpol.2019.06.048. [DOI] [PubMed] [Google Scholar]
- Xie W.Y., Yang X.P., Li Q., Wu L.H., Shen Q.R., Zhao F.J. Changes in antibiotic concentrations and antibiotic resistome during commercial composting of animal manures. Environ. Pollut. 2016;219:182–190. doi: 10.1016/j.envpol.2016.10.044. [DOI] [PubMed] [Google Scholar]
- Xiong W., Wang Y., Sun Y., Ma L., Zeng Q., Jiang X., Li A., Zeng A., Zhang T. Antibiotic-mediated changes in the fecal microbiome of broiler chickens define the incidence of antibiotic resistance genes. Microbiome. 2018;6:34. doi: 10.1186/s40168-018-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Xu Y., Wang H., Guo C., Qiu H., He Y., Zhang Y., Li X., Meng W. Occurrence of antibiotics and antibiotic resistance genes in a sewage treatment plant and its effluent-receiving river. Chemosphere. 2015;119:1379–1385. doi: 10.1016/j.chemosphere.2014.02.040. [DOI] [PubMed] [Google Scholar]
- Yin Y., Gu J., Wang X., Song W., Zhang K., Sun W., Zhang X., Zhang Y., Li H. Effects of copper addition on copper resistance, antibiotic resistance genes, and intl1 during swine manure composting. Front. Microbiol. 2017;8:344. doi: 10.3389/fmicb.2017.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Sui Q., Tong J., Buhe C., Wang R., Chen M., Wei Y. Sludge bio-drying: effective to reduce both antibiotic resistance genes and mobile genetic elements. Water. Res. 2016;106:62–70. doi: 10.1016/j.watres.2016.09.055. [DOI] [PubMed] [Google Scholar]
- Zhang Y.J., Hu H.W., Gou M., Wang J.T., Chen D., He J.Z. Temporal succession of soil antibiotic resistance genes following application of swine, cattle and poultry manures spiked with or without antibiotics. Environ. Pollut. 2017;231:1621–1632. doi: 10.1016/j.envpol.2017.09.074. [DOI] [PubMed] [Google Scholar]
- Zhao X., Wang J., Zhu L., Ge W., Wang J. Environmental analysis of typical antibiotic-resistant bacteria and ARGs in farmland soil chronically fertilized with chicken manure. Sci. Total Environ. 2017;593-594:10–17. doi: 10.1016/j.scitotenv.2017.03.062. [DOI] [PubMed] [Google Scholar]
- Zhu Y.G., Johnson T.A., Su J.Q., Qiao M., Guo G.X., Stedtfeld R.D., Hashsham S.A., Tiedje J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U. S. A. 2013;110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Wang X., Yang T., Su J., Qin Y., Wang S., Gillings M., Wang C., Ju F., Lan B., Liu C., Li H., Long X.E., Wang X., Jetten M.S.M., Wang Z., Zhu Y.G. Air pollution could drive global dissemination of antibiotic resistance genes. ISME J. 2021;15:270–281. doi: 10.1038/s41396-020-00780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.