Abstract
Background
Lung biopsy tissue samples can be used for infection detection and cancer diagnosis. Metagenomic next-generation sequencing (mNGS) has the potential to further improve diagnosis.
Methods
From July 2018 to May 2020, lung biopsy samples of 133 patients with suspected pulmonary infection or abnormal imaging findings were collected and subjected to clinical microbiological testing, Illumina and Nanopore sequencing to identify pathogens. The neural networks were pretrained by extracting features of human reads from 2,095 metagenomic next-generation sequencing results, and the human reads of lung biopsy samples were entered into the validated pipeline to predict the risk of cancer.
Findings
Based on the pathogen-cancer detection pipeline, the Illumina platform showed 77·6% sensitivity and 97·6% specificity compared to the composite reference standard for infection diagnosis. However, the Nanopore platform showed 34·7% sensitivity and 98·7% specificity. mNGS identified more fungi, which was confirmed by subsequent pathological examination. M. tuberculosis complex was weakly detected. For cancer detection, compared with histology, the Illumina platform showed 83·7% sensitivity and 97·6% specificity, diagnosing an additional 36 cancer patients, of whom half had abnormal imaging findings (pulmonary shadow, space-occupying lesions, or nodules).
Interpretation
For the first time, we have established a pipeline to simultaneously detect pathogens and cancer based on Illumina sequencing of lung biopsy tissue. This pipeline efficiently diagnosed cancer in patients with abnormal imaging findings.
Funding
This work was supported by the National Key Research and Development Program of China and National Natural Science Foundation of China.
Keywords: Lung biopsy tissue, Metagenomic next-generation sequencing, Pulmonary infection, Lung cancer, Genomic instability
Research in context.
Evidence before this study
We searched PubMed with the terms “lung biopsy tissues, mNGS and cancer” for reports published up to July 22, 2021, with no language restrictions. Our search identified three results of relevance to this study, one of which was our previous work. The other two studies both used the Illumina platform. We found no reports describing the diagnostic accuracy of Nanopore sequencing using lung biopsy tissue. We also searched with the terms “mNGS and cancer” and found only one report describing the use of mNGS to identify cryptogenic malignancies in body fluids. The performance of metagenomic next-generation sequencing (mNGS) for diagnosing infection and cancer in lung biopsy tissue remains unclear.
Added value of this study
This study describes the performance of Nanopore sequencing in infectious disease in lung biopsy tissue. Using the Illumina platform, we detected more fungal pathogens than were detected using clinical methods, but the platform had low sensitivity for diagnosing Mycobacterium tuberculosis infection. In addition, based on our pretrained neural networks for cancer prediction, we were able to simultaneously detect pathogens and cancer using the Illumina platform in lung biopsy tissue.
Implications of all the available evidence
Based on our results, we are able to simultaneously detect pathogens and predict the possibility of cancer using the result of Illumina sequencing of lung biopsy tissue in clinical laboratory. In general, the human reads are removed in the bioinformatic pipeline to increase the accuracy of pathogen detection, but our results reveal that human reads can be used to predict the possibility of cancer.
Alt-text: Unlabelled box
1. Introduction
Pulmonary infections, which can be caused by bacteria, fungi, and viruses, can be extremely fatal [1]. Lower respiratory tract infections (LRTIs) include community-acquired pneumonia (CAP), hospital-acquired pneumonia (HAP), and ventilator-associated pneumonia. Reports suggest that these types of pneumonia account for >25% of deaths from pneumonia-associated hospitalisations [2]. Early identification of causative pathogens is crucial for clinical interventions such as the administration of precise antibiotics. However, conventional microbial detection methods can only identify approximately 40% of pathogens [3]. Although polymerase chain reaction (PCR)-based methods do improve the detection sensitivity of pathogens [4], the spectrum of pathogens detected remains narrow.
Clinical metagenomic next-generation sequencing (mNGS) is rapidly transitioning from research laboratories to clinical setting. Because of its culture independency, high throughput, and fast turnaround time (TAT), mNGS has become a promising method for diagnosing infectious disease using the Illumina platform [5], [6], [7]. However, owing to its real-time analysis and long reads for prediction of antibiotic resistance, the Nanopore platform may be more suitable for clinical use [8], [9]. Many studies have focused on the diagnostic performance of bronchoalveolar lavage fluid (BALF) or sputum [10,11] for identifying pulmonary infection, but studies of lung biopsy tissue are rare.
Genome instability, as an important cancer marker [12], has been widely discussed in scientific research, although it has rarely been applied in clinical diagnosis. Additionally, mNGS of body fluids is an emerging approach for identifying occult pathogens in undiagnosed patients, based on genomic instability that allows tumour cells to be distinguished from the pipeline [13]. Generally, clinical mNGS is performed on patients to identify pathogens, and mNGS may yield negative results in patients with underlying malignant neoplasms but no infectious organisms. Several studies have reported that chromosomal instability could be used to identify cancer and as a form of non-invasive prenatal testing [14], [15], [16], so we hypothesised that genome instability analysis may be a useful tool for detecting cancer in lung biopsy tissue, using the human reads of mNGS results.
In this study, we collected 133 lung biopsy samples using computer tomography (CT)-guided puncture or pulmonary wedging and applied Illumina and Nanopore sequencing to explore the diagnostic value of the lung biopsy samples. In addition, we investigated the accuracy of human reads of mNGS in cancer patients.
2. Methods
2.1. Patients and sample collection
The 133 lung biopsy tissues were remnant samples from the clinical laboratory of the Peking University People's Hospital and they were collected between July 2018 and May 2020. Prior to testing, samples were stored at −80°C. All patients were suspected to have pulmonary infection or had abnormal chest imaging results. The inclusion criteria were symptoms such as fever, cough, expectoration, dyspnoea, and abnormal imaging findings such as pulmonary shadows, space-occupying lesions, and other signs of pulmonary infection. Data on the demographic characteristics, clinical laboratory findings, radiography and histology results, clinical treatment, and outcomes of the 133 patients were extracted from the patients’ medical records. The diagnosis of LRTIs was based on microbiological tests, microscopy, and radiography.
This study was approved by the Peking University People's Hospital Institutional Review Board (No. 2019PHB010-01). All samples were obtained with the patient's consent.
2.2. DNA extraction, library preparation, and sequencing
We performed nucleic acid extraction using 400 μL of ground lung biopsy tissue and 100 μL of ATL buffer solution (1·5 mL buffer ATL and 10 μL reagent DX; Qiagen) in a pathogen lysis tube L (Qiagen), at a frequency of 30 Hz for 10 min (TissuerLyser II; Qiagen). DNA was extracted from the supernatant using the QIAamp DNA mini kit (Qiagen), as described in the manufacturer's protocol. Sterile deionised water was extracted as the negative parallel control (NTC). The concentrations of DNA were measured by Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). Illumina sequencing libraries were prepared using NEBNext Ultra II DNA Library Prep Kit (New England BioLabs Inc.) and sequenced using the Novaseq 6000 System (150-bp paired-end reads; Illumina). Approximately 20 million reads were generated for each sample. Nanopore sequencing libraries were prepared according to the manufacturer's instructions for the Rapid Barcoding Kit SQK-RPB004 (DNA concentration <20 ng/μL; Oxford Nanopore) and SQK-RBK004 (DNA concentration >20 ng/μL; Oxford Nanopore). Up to five barcoded samples per flow cell were loaded on the Nanopore instrument (GridION X5, Oxford Nanopore) for sequencing. Approximately 0·8 G of data were generated for each sample. Amplification of 7 and 17 samples failed according to the library procedure using the Illumina and Nanopore platforms, respectively. The analysis pipeline of the Illumina platform has been described in our previous work [5], and the taxonomy was based on Centrifuge for Nanopore sequencing.
All species detected by the Illumina and Nanopore platforms were looked up in PubMed to determine whether the organisms cause pneumonia. After removing normal flora or colonising bacteria for Illumina sequencing, the positive pathogenic microorganisms were defined as those with a ratio of unique reads per million (RPM) above 10, and the RPM ratio = RPMsample/RPM (no template control [NTC]) or RPM ratio = RPMsample if the organism was not detected in the parallel NTC [7]. For Nanopore sequencing, unique reads >3 for bacteria and unique reads >1 for fungi were considered positive for pathogenic microorganism identification [11]. M. tuberculosis was considered positive when more than one read was detected. The cut-off values for bacteria (except for M. tuberculosis) and fungi to determine the LRTI and non-LRTI group were based on receiver-operating characteristic (ROC) curves (Supplementary Table S1).
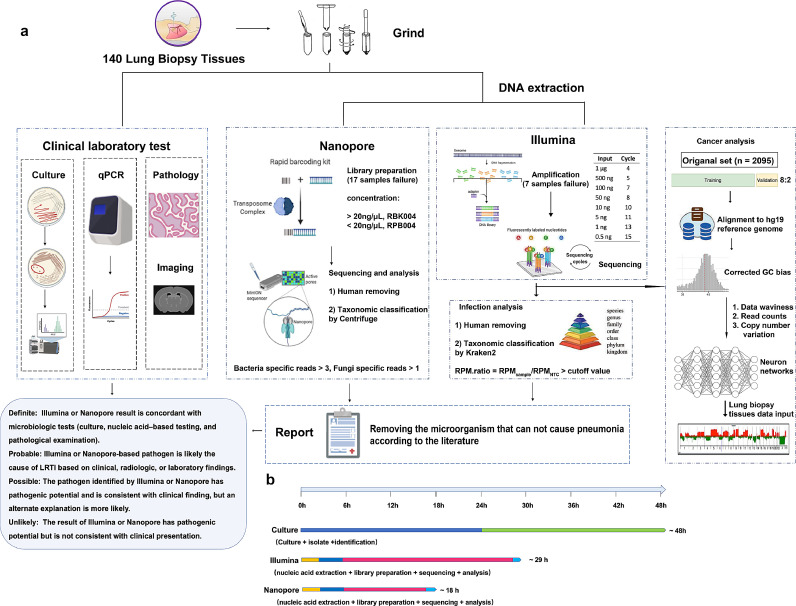
The results were divided into four categories (define, probable, possible, and unlikely) based on our laboratory rules (Fig. 1a). Detailed information on each patient was shown in Supplementary Table S2 and Supplementary Table S3. The composite reference standard included the results from all microbiological tests (including culture), pathological examinations, and clinical adjudications. Definite and probable results were considered positive for clinical diagnosis, and possible and unlikely results were considered negative for clinical diagnosis.
Fig. 1.
Schematic workflow of this study. (a) Lung biopsy samples analysis from 133 patients. Several microbiological tests were performed on these samples, including BALF culture, qPCR, ELISA, GeneXpert MTB/RIF, and histology. DNA was extracted and prepared for Illumina sequencing (seven samples showed library failure), Nanopore sequencing (17 samples showed library failure), and final analysis. The pathogen results of Illumina and Nanopore sequencing were adjudicated based on the literature from PubMed and their clinical condition. Three neural networks were pretrained by extracting features of human reads from 2,095 metagenomic next-generation sequencing results. The Illumina sequencing results were mapped to the human reference and then applied to the model. (b) The TAT of different methods. Abbreviations: BALF, bronchoalveolar lavage fluid; ELISA, enzyme-linked immunosorbent assay; qPCR, quantitative polymerase chain reaction; TAT, turnaround time.
2.3. An artificial intelligence method for prediction of cancer risk from the mNGS dataset
Datasets for training were collected from human reads of mNGS containing 2095 samples. Of the patients from whom the 2095 samples were obtained, 755 (36·0%) were female and 1340 (64·0%) were male. Of the female patients, 359 (47·5%) had cancer, and of the male patients, 641 (47·8%) had cancer. Of the patients, 22·6% were aged under 40 years, 34·1% were aged 40–60 years, 36·7% were aged 60–80 years, and 6·6% were aged 80 years or older. The samples were split into training and validation datasets in a ratio of 8:2. After controlling for quality in FASTQ data, cleaned DNA sequences were aligned to the human hg19 (GRCh37) reference genome. During NGS, guanine-cytosine (GC) content bias may occur in the priming, size selection, and probability of sequencing errors, which might account for abnormal conditions in the analysis. Thus, we corrected for GC bias from the mapped read counts data using LOESS regression. Subsequently, features, such as the waviness (standard deviation of the read fold change of each bin) of data and normalized read counts, were noted after the mapping step, and we obtained information on the copy number variation from the calling method, which was rebuilt on the base of XHMM [17] and Canoes [18] (i.e., two read counts based on the copy number variation calling methods). Then, all the features were normalized and fitted to dozens of predefined neuron networks. They were constructed using three classical neural network structures (full connect, convolutional neural network, and long short-term memory [an artificial recurrent neural network architecture usually used in the field of deep learning]) and different in hyper-parameters, which is suitable for large-scale-fold changed data of read counts. Twenty models were selected so that the models were complementary. Each model was used to predict the possibility of cancer in the sample (0 or 1). The predicted score was calculated using the total number of positive results from the 20 models.
The raw data of the chromosome copy variation and the picture of each patient were deposited in, or linked to, Zenodo (https://doi.org/10.5281/zenodo.5079188). Metagenomic sequencing data with the human reads removed were also deposited as National Center for Biotechnology Information (NCBI) sequence read archive (SRA) under Bioproject PRJNA744354.
2.4. Real-time quantitative polymerase chain reaction
Quantitative PCR (qPCR) detection of Mycobacterium tuberculosis was performed using MeltPro Mycobacterium tuberculosis Test Kit (Zeesan), according to the manufacturer's instructions and measured using an ABI QuantStudio 5 system (Thermo Fisher Scientific). The primer, probe, and PCR-mix were provided by the manufacturer.
2.5. Statistical analysis
Student's t-test and the chi-square test were used to assess the statistical significance of differences in continuous and categorical data, respectively. Statistical significance was set at P<0·05. SPSS software (version25·0; IBM Corporation, Armonk, NY, USA) was used for the statistical analysis.
2.6. Role of the funding source
The funders did not play any role in the study design, data collection, management, analysis, interpretation, review, approval of the manuscript, or the decision to submit the manuscript for publication.
3. Results
3.1. Patient characteristics
In this study, 133 patients were divided into an LRTI group (n=49) and a non-LRTI group (n=84), according to the diagnostic criteria for CAP [19], HAP [20] and suspected infection, based on a clinical specialist's opinion. The prevalence of malignancy was significantly higher in the non-LRTI group than in the LRTI group (41·7% and 16·3%, P=0·003, chi-square test), while the prevalence of haematological disease was significantly higher in the LRTI group than in the non-LRTI group (14·3% and 3·6%, P=0·024, chi-square test). The incidence of antibiotic use was significantly higher in the LRTI group than in the non-LRTI group (P<0·001, chi-square test). Patients with LRTIs tended to have slightly longer stays in hospital than their non-LRTI counterparts (Table 1). Clinical laboratory test results showed that white blood cell counts, C-reactive protein levels, and procalcitonin levels were elevated in the LRTI group than in the non-LRTI group (Table 1).
Table 1.
The characteristics of patients in LRTIs and non-LRTIs group.
| LRTIs (n=49) | Non-LRTIs (n=84) | P | |
|---|---|---|---|
| Age, mean (range), years | 52·60 (20-77) | 62·05 (33-91) | 0·000 |
| Sex, male, n (%) | 30 (61·22) | 47 (55·95) | 0·552 |
| Infection types | |||
| CAP | 25 (51·02) | 0 | NA |
| HAP | 4 (8·16) | 0 | NA |
| TB | 14 (28·57) | 0 | NA |
| ILD | 2 (4·08) | 0 | NA |
| AECOPD | 1 (2·04) | 0 | NA |
| Obstructive Pneumonia | 1 (2·04) | 0 | NA |
| Lung abscess | 2 (4·08) | 0 | NA |
| History of aspiration, n (%) | 1 (2·04) | 0 | NA |
| Any comorbidity, n (%) | 31 (63·27) | 57 (67·86) | 0·589 |
| Diabetes | 7 (14·29) | 11 (13·10) | 0·846 |
| Hypertension | 10 (20·41) | 26 (30·95) | 0·187 |
| Cardiovascular disease | 5 (10·20) | 9 (10·71) | 0·926 |
| COPD | 0 | 6 (7·14) | NA |
| Malignancy | 8 (16·33) | 35 (41·67) | 0·003 |
| Chronic liver disease | 2 (4·08) | 5 (5·95) | 0·641 |
| Haematological disease | 7 (14·29) | 3 (3·57) | 0·024 |
| Renal disease | 3 (6·12) | 0 | NA |
| Hospital, mean (range), days | 14·50 (3-60) | 12·71 (2-30) | 0·270 |
| Antibiotic use, n (%) | 34 (69·39) | 17 (20·23) | 0·000 |
| White blood cell count, 109/L | 9·37 (1·18-29·8) | 8·97 (3·89-30·28) | 0·640 |
| <4 | 4 (8·16) | 1 (1·20) | 0·021 |
| 4–10 | 26 (53·06) | 61 (73·49) | |
| >10 | 19 (38·78) | 21 (25·30) | |
| Percentage of neutrophils | |||
| 40–75% | 28 (57·14) | 57 (68·67) | 0·181 |
| >75% | 21 (42·86) | 26 (31·33) | |
| Percentage of lymphocytes | |||
| <20% | 28 (57·14) | 45 (54·22) | 0·744 |
| 20–50% | 21 (42·86) | 38 (45·78) | |
| CRP, mg/L | 41·35 (0·31-182·58) | 20·44 (0·43-103·98) | 0·066 |
| <10 mg/L | 12/22 (54·55) | 22/43 (51·16) | 0·244 |
| 10–50 mg/L | 4/22 (18·18) | 15/43 (34·88) | |
| >50 mg/L | 6/22 (27·27) | 6/43 (13·95) | |
| Procalcitonin, ng/mL | 0·93 (0·02-11·12) | 0·80 (0·02-15·46) | 0·917 |
| <0·5 ng/mL | 14/18 (77·78) | 29/32 (90·63) | 0·209 |
| >0·5 ng/mL | 4/18 (22·22) | 3/32 (9·37) |
Abbreviations: LRTIs, lower respiratory tract infections; CAP, community-acquired pneumonia; HAP, hospital-acquired pneumonia; TB, pulmonary tuberculosis; ILD, interstitial pneumonia; AECOPD, acute exacerbation of chronic obstructive pulmonary disease; COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; NA, not applicable; Data were presented as n (%) or means (range); Significance was determined by Student's t-test for the comparation of age, others were determined by chi-square test.
3.2. Detection performances of the Illumina and Nanopore platforms in lung biopsy samples
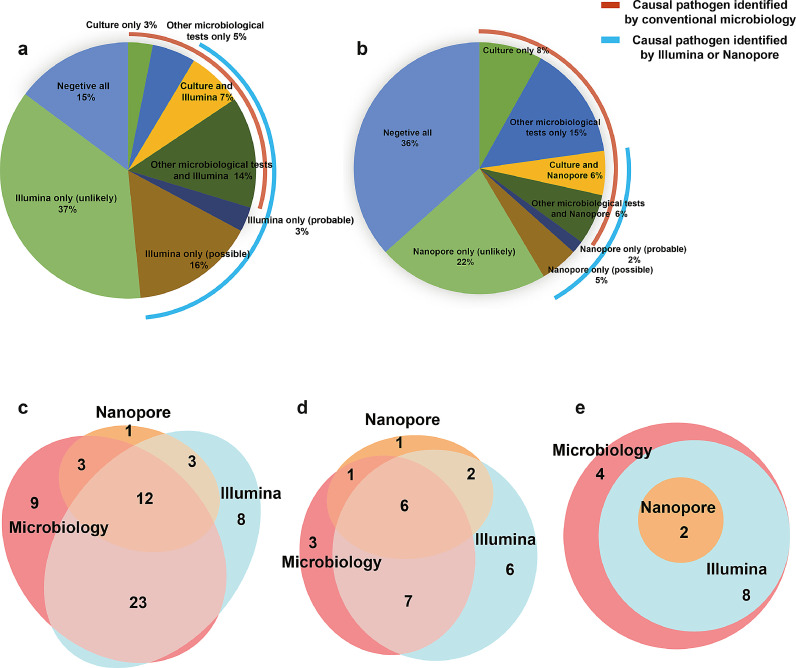
Our workflow for lung biopsy samples using conventional clinical microbiological tests, Illumina sequencing, and Nanopore sequencing was shown in Fig. 1a. From sample retrieval to result analysis, the TAT of the Illumina platform was longer than that of the Nanopore platform and shorter than that of the culture (Fig. 1b). Of the enrolled 133 samples, only 18 (13·5%) were culture positive, and the positive result was elevated to 43 (32·3%) samples with the addition of other microbiological tests (Fig. 2a, b). The Illumina and Nanopore platforms detected 40 (30·1%) and 18 (14·6%) samples, respectively, with definite or probable pathogens (Fig. 2a, b). Overall, compared with lung biopsy tissue culture results, the Illumina platform had 83·3% clinical sensitivity and 79·1% clinical specificity. In addition, there were 25 patients with positive other microbiological tests, indicating a clinical sensitivity of 74·4% and specificity of 95·6% compared with all microbiological testing. Compared with the composite reference standard, the Illumina platform showed 77·6% clinical sensitivity and 97·6% clinical specificity (Table 2, Supplementary Table S4).
Fig. 2.
Performance of Illumina and Nanopore sequencing. The proportion of samples with pathogens identified by the Illumina (a) and Nanopore (b) platforms. The different detection efficiency of all microorganisms (c), fungi (d), and Mycobacterium (e) in microbiological tests (BALF culture, qPCR, ELISA, GeneXpert-TB, antibody), and the Illumina and Nanopore platforms. Abbreviations: BALF, bronchoalveolar lavage fluid; ELISA, enzyme-linked immunosorbent assay; qPCR, quantitative polymerase chain reaction.
Table 2.
The performance of Illumina relative to culture, all microbiological testing, and composite reference standard (n=133).
| Illumina positive | Illumina negative | Agreement % | |
|---|---|---|---|
| Positive by culture (n=18) | 15 | 3 | 83·33% |
| Negative by culture (n=115) | 24 | 91 | 79·13% |
| Positive by all microbiological testing (n=43) | 32 | 11 | 74·42% |
| Negative by all microbiological testing (n=90) | 4 | 86 | 95·56% |
| Positive by composite reference standard (n=49) | 38 | 11 | 77·55% |
| Negative by composite reference standard (n=84) | 2 | 82 | 97·62% |
Compared with the culture results, after the removal of the samples involving library failure, the Nanopore platform showed a sensitivity and specificity of 38·9% and 91·4%, respectively. The sensitivity declined to 33·3% and the specificity increased to 97·5% when other microbiological test results were considered. Compared with the composite reference standard, the Nanopore platform showed 34·7% clinical sensitivity and 98·7% clinical specificity (Table 3, Supplementary Table S5).
Table 3.
The performance of Nanopore relative to culture, all microbiological testing, and composite reference standard (n=123).
| ONT positive | ONT negative | Agreement % | |
|---|---|---|---|
| Positive by culture (n=18) | 7 | 11 | 38·89% |
| Negative by culture (n=105) | 9 | 96 | 91·43% |
| Positive by all microbiological testing (n=42) | 14 | 28 | 33·33% |
| Negative by all microbiological testing (n=81) | 2 | 79 | 97·53% |
| Positive by composite reference standard (n=49) | 17 | 32 | 34·69% |
| Negative by composite reference standard (n=74) | 1 | 73 | 98·65% |
3.3. Detailed information of samples with conflicting results
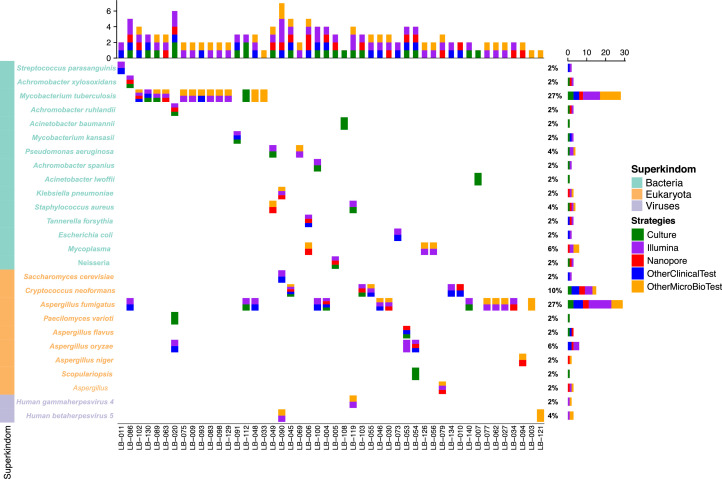
Twenty pathogens were cultured from the 133 lung biopsy samples, and two samples were co-infected. Eight samples were infected with fungi (Aspergillus flavus [n=1], Scopulariopsis sp. [n=1], Paecilomyces varioti [n=1], Cryptococcus neoformans [n=2] and Aspergillus fumigatus [n=3]). Four patients were infected with acid-fast bacilli and the remaining eight samples (40%) were infected with other bacteria. Other microbiological tests showed positive results for 25 other pathogens. Three were identified in BALF culture, seven were galactomannan positive, one was antibody positive, and 14 were detected using PCR (Fig. 3, Supplementary Table S2). Comparison of the results of the clinical microbiological tests with the Illumina and Nanopore platforms, showed that nine pathogens could identify only in microbiological tests and the Illumina platform detected additional eleven pathogens; however, Nanopore sequencing only detected four additional pathogens and three samples were positive on both the Illumina and Nanopore platforms, but negative on the microbiological test results. Consistent results were obtained for the remaining 38 pathogens with the clinical microbiological tests and the Illumina and Nanopore platforms (Fig. 2c).
Fig. 3.
Five strategies for pathogen detection. The different conditions of pathogen species detection in culture, other microbiological tests (BALF culture, qPCR, ELISA, GeneXpert MTB/RIF, antibody testing), other clinical tests (histology, clinical condition), and the Illumina and Nanopore platforms. Abbreviations: BALF, bronchoalveolar lavage fluid; ELISA, enzyme-linked immunosorbent assay; qPCR, quantitative polymerase chain reaction.
In terms of the detection of fungi, the culture results were not consistent with the Illumina or Nanopore sequencing results in two samples, and one sample that was Aspergillus fumigatus-positive in the BALF sample, but negative in the lung biopsy sample. Seven pathogens were detected using the microbiological test and with Illumina sequencing, one pathogen was detected in both the clinical samples and with Nanopore sequencing, and six pathogens could be detected with all the methods (Fig. 2d). Compared to the microbiological tests, Illumina and Nanopore sequencing identified nine additional fungi, and this result was consistent with that of the histology (Supplementary Table S6).
In terms of Mycobacterium detection, the clinical microbiological tests detected 14 pathogens; however, Illumina and Nanopore sequencing identified ten and two pathogens, respectively, and no unique Mycobacterium were identified in other samples using these two platforms (Fig. 2e). Only one sample was positive for acid-fast bacilli on culture and Mycobacterium kansasii was identified using the Illumina platform. In terms of the additional positive results in the microbiological test, one sample was cultured with acid-fast positive bacillus, three samples were positive using the quantitative PCR (qPCR), but we did not detect any bacteria in the Mycobacterium genus using either the Illumina or the Nanopore platforms (Supplementary Table S7).
3.4. Cancer detection based on the Illumina sequencing results
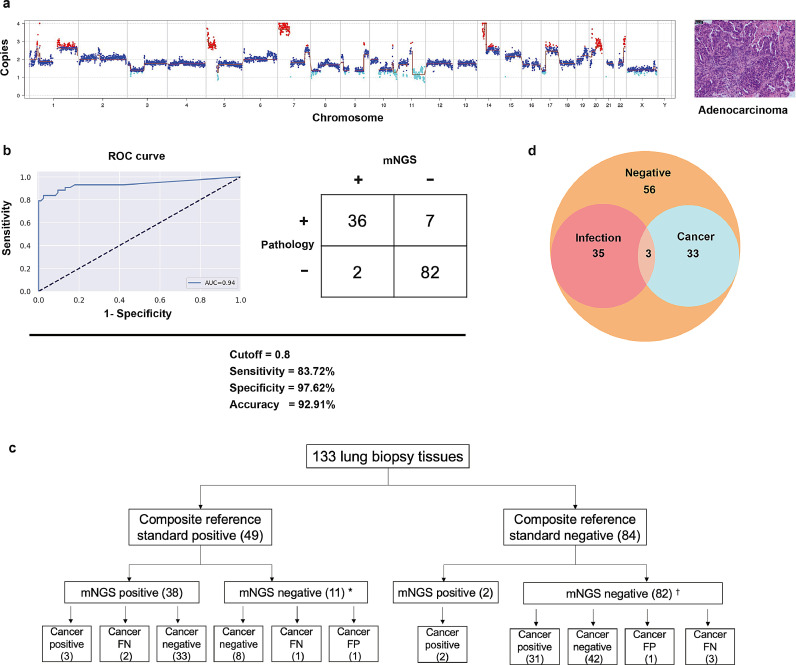
For diagnosing infections, we could accurately detect infection in 30·1% (40) of the samples on the basis of the Illumina sequencing results. Except for the consistent pathogen detection results, the diagnosis of the remaining patients was unclear. Most of these patients had suspected infections or abnormal imaging findings, and there may have been alternate causes for this, including malignant tumours. We then applied the human reads from mNGS to map the reference human database to explore chromosomal deletions or duplications with our predefined neural networks. Except for the six samples with inadequate human reads, genome instability was assessed in the remaining 127 lung biopsy samples to determine the presence of cancer in the patients. As shown in Fig. 4a, a large number of gains and losses were identified in adenocarcinoma patient, which was confirmed by pathological examination. Out of the 127 patients, 43 were confirmed to have malignancy on histology, and the cut-off values were established on the basis of the ROC curve (area under the curve =0·94; Fig. 4b). Compared with the results of the pathological examination, the mNGS had clinical sensitivity of 83·7%, specificity of 97·6%, and 92·9% accuracy (Fig. 4b). In both clinical and mNGS negative infection diagnosis patients, we found 31 patients with malignancy. Among patients who tested positive for a pathogen, we identified five cancer patients (Fig. 4c). In summary, the Illumina platform simultaneously detected 38 patients with infection and 36 patients with cancer, which was confirmed by clinical results (Fig. 4d). We identified 18 cancer patients with an initial diagnosis of pulmonary shadow, a space-occupying lesion, or nodules (Supplementary Table S9).
Fig. 4.
Genome instability analysis of patients with cancer that was identified using Illumina sequencing. (a) Genome instability data and histology of a patient with a lung adenocarcinoma. (b) The ROC curve and contingency table comparing histology to predict the score based on the Illumina sequencing results. (c) Flowchart of clinical evaluation of 133 samples. (d) The positive detection that is consistent with clinical findings. *One sample had inadequate data; †five samples had inadequate data. Abbreviations: CNV, copy number variation; FN, false negative; FP, false positive; ROC, receiver-operating characteristic.
4. Discussion
In this retrospective study, we investigated utility of the Illumina and Nanopore platforms for detecting infectious pathogens in lung biopsy samples and we established a method to simultaneously detect pathogens and cancer using these samples.
Lung biopsy samples are rarely used for detecting pathogens in the clinical setting because performing a lung biopsy is challenging. In recent studies, bronchoalveolar lavage fluid or sputum have been used to diagnose pulmonary infections, and only few studies have explored the use of lung biopsy tissue in detecting pulmonary infections [5], [10], [11]. Although some studies have confirmed that diagnosing infections is easier in BALF samples than in lung biopsy samples due to complications present in the lung biopsy samples such as malignancy, pulmonary nodules, or pulmonary shadows [21]. In this study, we demonstrated for the first time that infection and solid tumour can be detected simultaneously in lung biopsy samples using mNGS; therefore, lung biopsies could be used for diagnosing pulmonary diseases.
Since the introduction of sequencing techniques in clinical samples [22], mNGS has been widely used for pathogen detection owing to its high throughput capacity and fast TAT [5], [6], [7]. We compared mNGS and culture results in lung biopsy samples in a previous study [23], and found a sensitivity and specificity of 100% and 76·5%, respectively, for bacteria, and 57·1% and 61·5%, respectively, for fungi. In this study, the Illumina sequencing showed 83·3%, 74·4%, and 77·6% agreement with culture, microbiological tests, and the composite reference standard results, respectively, a level of agreement similar to that of lung biopsy reported in our previous study [23].
The Oxford Nanopore Technologies platform is an attractive method for diagnosing infectious diseases because of its rapid TAT [24]. Therefore, we tested the diagnostic performance of the Nanopore platform in this study. However, the agreement between culture results and Nanopore sequencing results was inferior to that of the clinical findings. The possible reason is that there were small number of microbe reads in too many samples, even microbe reads of zero in some samples (Supplementary Table S8), and the microbe read levels were lower than that of other studies [10], [25]. Because the samples were collected after grinding for clinical culture testing or other PCR-based methods, and stored at –80°C, it was difficult to remove the human genome in the wet experiments. The proportion of human genome was much larger after PCR amplification during library preparation. The sensitivity of the Nanopore platform in our study was much lower than that reported by Gu et al [11]. This may have been partly due to the use of different sample types in the two studies. Gu et al. detected cfDNA in body fluid, and the concentration of pathogens and positive rate in lung biopsy tissue may be lower than in body fluid. Another possible reason for the lower sensitivity in our study is the higher host background noise in lung biopsy tissue. Additionally, Gu et al. minimised the host background noise by centrifugation and DNA extraction, but we did not remove the human genome because they were frozen. Charalampous et al. [10] showed that the Nanopore platform is more efficient in wet experiments when the human genome is removed from the samples.
The incidence of pulmonary fungal infections has been rising in recent years, but the early diagnosis of pulmonary fungal infections is difficult due to non-specific clinical manifestations at the early stages. Similar to the findings of our previous work and other studies [23], [26], the mNGS did improve the diagnosis of pulmonary fungal infections in this study, and we detected inconsistent results between culture and the Illumina or Nanopore sequencing in two samples. We also identified four Aspergillus spp. isolates and one Saccharomyces cerevisiae isolate that were not detected by microbiological tests and which were confirmed by pathological examination. The Illumina and Nanopore sequencing results that identified Cryptococcus neoformans were confirmed by pathological examination (Supplementary Table S6). Although mNGS improved the sensitivity in detecting fungal infections, combining other clinical methods such as galactomannan testing or pathological examination is useful to combat the risk of contamination during the experiment.
In 14 patients with Mycobacterium infection (13 M. tuberculosis, one nontuberculous mycobacteria) based on clinical microbiological test results, only 10 and two patients were identified using the Illumina and Nanopore platforms, respectively (Supplementary Table S7). Three samples with acid-fast positive bacilli were detected only in the pathological examination, but these were negative in all clinical microbiological tests and Illumina and Nanopore sequencing, and negative results were confirmed by qPCR. For Mycobacterium infection, a positive result that is only supported by pathological examination results should be re-checked using another clinical microbiological tests or assessment of clinical symptoms. As for the lower sensitivity of Mycobacterium detection in mNGS, it is possible that the extraction of DNA from M. tuberculosis was suboptimal. Zhou et al. [27] illustrated that the diagnostic ability of mNGS for M. tuberculosis was similar to that of Xpert and higher than that of conventional methods. However, Chen et al. [28] reported that the sensitivity of mNGS was superior to those of conventional culture methods and Xpert. These inconsistent reports could be due to low read ratio of M. tuberculosis and existent bias in different studies [29]. To improve the sensitivity to M. tuberculosis in mNGS, we could extend the lysis time or enriched the DNA by hybridisation [30].
Several studies have shown that cytomegalovirus (CMV) and Epstein-Barr virus (EBV) can be detected in the blood of healthy donors [31], [32], which makes it difficult for the doctors to decide whether to use antiviral drugs for treatment. In our study, we identified three patients with positive CMV or EBV results that were confirmed by the clinical test (LB-090, LB-119, LB-121). All three patients were immunosuppressed with haematological diseases and prolonged severe pneumonia, and all three patients died. Reactivation of CMV or EBV is common after haematopoietic stem cell transplantation, and is associated with a poor prognosis [33], [34]. We recommend that if EBV or CMV are detected, the results should be interpreted based on the sample type and the clinical symptoms.
Previous studies have also reported chromosomal instability (chromosomal duplication or deletion) in patients with non-small cell lung cancer and adenocarcinoma [35], [36]. Generally, human genome data are removed during bioinformatic analysis for infection diagnosis in the mNGS pipeline. However, this is the first study to show that residual human genome data from mNGS could detect chromosomal changes in patients with malignancy, improve the accuracy of diagnosis and the TAT of the pathological examination (approximately 48 h), and reduce the testing cost (¥ 600 for Illumina sequencing, ¥ 1000 for Nanopore sequencing, and ¥ 1500 for histological examination). Compared with the histology results, there were two samples that were false positive, and the histology result of one patient was negative but accompanied by aplastic anaemia (LB-121). We also identified pulmonary sclerosing pneumocytoma and a benign tumour with a positive predictive score (LB-097). In patients with an initial diagnosis of pulmonary shadow, a space-occupying lesion, or nodules, we also detected cancer in 18 patients and this was consistent with the histology results (Supplementary Table S9). We assessed the seven samples with positive histology results incorrectly, and the predicted score of three samples was zero and the predicted scores of the remaining four samples were lower than that of a confirmed positive result (Supplementary Table S9). Therefore, the cut-off value should be adjusted based on the results of a larger cohort study in the future.
In addition, we analysed the human reads of the Nanopore sequencing results for cancer prediction. However, there were too few human reads for cancer prediction (Supplementary Table S8). Our model was based on the read counts; however, there are some advantages to using the read length rather than the read counts for Nanopore sequencing. A low human reads count could influence the coverage of the reference genome, leading to more instability of the genome. Thus, in the future, the model needs to be adjusted for use with Nanopore results.
There are some limitations to our study. First, human genome was not removed in this pipeline. Therefore, inadequate microbe reads were generated in the Nanopore platform. Second, we installed the real-time analysis process that was unsuccessful in the server and cluster of our clinical laboratory, and therefore, the TAT of Nanopore sequencing was longer. Third, the pathogens identified by mNGS were not validated by qPCR, and RNA viruses were excluded. Due to the small sample size, the cut-offs for viruses were difficult to determine. In the future, the cut-off for bacteria, fungi, and cancer need to be adjusted based on the results of larger cohort studies. Finally, we did not identify the type or stage of lung cancer, but we speculate that this could be determined if more samples were included in our model.
In summary, we have reported successful simultaneous detection of pathogens and cancer for the first time in lung biopsy samples using mNGS based on the Illumina sequencing results. This pipeline efficiently diagnosed cancer in patients with abnormal imaging findings such as pulmonary shadows, space-occupying lesions, or nodules. However, we recommend the removal of human genome in wet experiments for Nanopore sequencing to improve the accuracy in detecting pathogens; but, this study also shows that human data should not be ignored when using bioinformatics pipelines, as it can detect genome instability.
Contributors
HW conceived, designed, and supervised the study. YG and HL acquired the data. YG, HC, ZL, WD, and JW analysed and interpreted the data. YG, CJ, YY and SS conducted the clinical work associated with the study. ZL provided the technical support. YG, ZL and WD verified the underlying data. YG wrote the draft, and HW revised it. All authors read and approved the final version of the manuscript. The corresponding author attests that all listed authors meet the authorship criteria and that no others meeting the criteria have been omitted.
Data sharing statement
The raw data of this study are available from the corresponding author upon reasonable request.
Declaration of Competing Interest
YG, HL, HC, YY, LJ, SS, CJ and HW declare that they have no conflict of interest. ZL is affiliated with Simcere Diagnostics Co., Ltd. WD and JW are affiliated with MatriDx Biotechnology Co., Ltd.
Acknowledgements
This study was supported by National Key Research and Development Program of China (2018YFE0102100) and National Natural Science Foundation of China (81625014).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103639.
Appendix. Supplementary materials
References
- 1.Collaborators GBDCoD Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the global burden of disease study. Lancet. 2017;390(10100):1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corrado RE, Lee D, Lucero DE, Varma JK, Vora NM. Burden of adult community-acquired, health-care-associated, hospital-acquired, and ventilator-associated pneumonia: New York city, 2010 to 2014. Chest. 2017;152(5):930–942. doi: 10.1016/j.chest.2017.04.162. [DOI] [PubMed] [Google Scholar]
- 3.Jain S, Self WH, Wunderink RG. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373(5):415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gadsby NJ, Russell CD, McHugh MP. Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin Infect Dis. 2016;62(7):817–823. doi: 10.1093/cid/civ1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Yin Y, Gao H. Clinical utility of in-house metagenomic next-generation sequencing for the diagnosis of lower respiratory tract infections and analysis of the host immune response. Clin Infect Dis. 2020;71(Suppl 4):S416–SS26. doi: 10.1093/cid/ciaa1516. [DOI] [PubMed] [Google Scholar]
- 6.Blauwkamp TA, Thair S, Rosen MJ. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. doi: 10.1038/s41564-018-0349-6. [DOI] [PubMed] [Google Scholar]
- 7.Wilson MR, Sample HA, Zorn KC. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380(24):2327–2340. doi: 10.1056/NEJMoa1803396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet. 2019;20(6):341–355. doi: 10.1038/s41576-019-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leggett RM, Alcon-Giner C, Heavens D. Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat Microbiol. 2020;5(3):430–442. doi: 10.1038/s41564-019-0626-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charalampous T, Kay GL, Richardson H. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat Biotechnol. 2019;37(7):783–792. doi: 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- 11.Gu W, Deng X, Lee M. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat Med. 2021;27(1):115–124. doi: 10.1038/s41591-020-1105-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Gu W, Talevich E, Hsu E. Detection of cryptogenic malignancies from metagenomic whole genome sequencing of body fluids. Genome Med. 2021;13(1):98. doi: 10.1186/s13073-021-00912-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watkins TBK, Lim EL, Petkovic M. Pervasive chromosomal instability and karyotype order in tumour evolution. Nature. 2020;587(7832):126–132. doi: 10.1038/s41586-020-2698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolhaqueiro ACF, Ponsioen B, Bakker B. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat Genet. 2019;51(5):824–834. doi: 10.1038/s41588-019-0399-6. [DOI] [PubMed] [Google Scholar]
- 16.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105(42):16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fromer M, Moran JL, Chambert K. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91(4):597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Backenroth D, Homsy J, Murillo LR. CANOES: detecting rare copy number variants from whole exome sequencing data. Nucleic Acids Res. 2014;42(12):e97. doi: 10.1093/nar/gku345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metlay JP, Waterer GW, Long AC. Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American thoracic society and infectious diseases society of America. Am J Respir Crit Care Med. 2019;200(7):e45–e67. doi: 10.1164/rccm.201908-1581ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalil AC, Metersky ML, Klompas M. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the infectious diseases society of America and the American thoracic society. Clin Infect Dis. 2016;63(5):e61–e111. doi: 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chellapandian D, Lehrnbecher T, Phillips B. Bronchoalveolar lavage and lung biopsy in patients with cancer and hematopoietic stem-cell transplantation recipients: a systematic review and meta-analysis. J Clin Oncol. 2015;33(5):501–509. doi: 10.1200/JCO.2014.58.0480. [DOI] [PubMed] [Google Scholar]
- 22.Wilson MR, Naccache SN, Samayoa E. Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl J Med. 2014;370(25):2408–2417. doi: 10.1056/NEJMoa1401268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Gao H, Meng H. Detection of pulmonary infectious pathogens from lung biopsy tissues by metagenomic next-generation sequencing. Front Cell Infect Microbiol. 2018;8:205. doi: 10.3389/fcimb.2018.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petersen LM, Martin IW, Moschetti WE, Kershaw CM, Tsongalis GJ. Third-generation sequencing in the clinical laboratory: exploring the advantages and challenges of nanopore sequencing. J Clin Microbiol. 2019;58(1):e01315–e01319. doi: 10.1128/JCM.01315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanderson ND, Street TL, Foster D. Real-time analysis of nanopore-based metagenomic sequencing from infected orthopaedic devices. BMC Genomics. 2018;19(1):714. doi: 10.1186/s12864-018-5094-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian YY, Wang HY, Zhou Y. Improving pulmonary infection diagnosis with metagenomic next generation sequencing. Front Cell Infect Microbiol. 2020;10 doi: 10.3389/fcimb.2020.567615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, Wu H, Ruan Q. Clinical evaluation of diagnosis efficacy of active mycobacterium tuberculosis complex infection via metagenomic next-generation sequencing of direct clinical samples. Front Cell Infect Microbiol. 2019;9:351. doi: 10.3389/fcimb.2019.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen P, Sun W, He Y. Comparison of metagenomic next-generation sequencing technology, culture and GeneXpert MTB/RIF assay in the diagnosis of tuberculosis. J Thorac Dis. 2020;12(8):4014–4024. doi: 10.21037/jtd-20-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pankhurst LJ, Del Ojo Elias C, Votintseva AA. Rapid, comprehensive, and affordable mycobacterial diagnosis with whole-genome sequencing: a prospective study. Lancet Respir Med. 2016;4(1):49–58. doi: 10.1016/S2213-2600(15)00466-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckert SE, Chan JZ, Houniet D, The Pathseek C, Breuer J, Speight G. Enrichment by hybridisation of long DNA fragments for Nanopore sequencing. Microb Genom. 2016;2(9) doi: 10.1099/mgen.0.000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu M, Gao J, Li S, Zeng M, Wu J, Luo M. Metagenomic analysis and identification of emerging pathogens in blood from healthy donors. Sci Rep. 2020;10(1):15809. doi: 10.1038/s41598-020-72808-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smatti MK, Yassine HM, AbuOdeh R. Prevalence and molecular profiling of Epstein Barr virus (EBV) among healthy blood donors from different nationalities in Qatar. PLoS One. 2017;12(12) doi: 10.1371/journal.pone.0189033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Kong J, Chang YJ. Patients with refractory cytomegalovirus (CMV) infection following allogeneic haematopoietic stem cell transplantation are at high risk for CMV disease and non-relapse mortality. Clin Microbiol Infect. 2015;21(12):1121 e9-15. doi: 10.1016/j.cmi.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Styczynski J, Gil L, Tridello G. Response to rituximab-based therapy and risk factor analysis in Epstein Barr Virus-related lymphoproliferative disorder after hematopoietic stem cell transplant in children and adults: a study from the infectious diseases working party of the European group for blood and marrow transplantation. Clin Infect Dis. 2013;57(6):794–802. doi: 10.1093/cid/cit391. [DOI] [PubMed] [Google Scholar]
- 35.Jamal-Hanjani M, Wilson GA, McGranahan N. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 36.Gillette MA, Satpathy S, Cao S. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell. 2020;182(1):200-25 e35. doi: 10.1016/j.cell.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.