Summary
Background
The SARS-CoV-2 delta (B.1.617.2) variant is highly transmissible and spreading globally, including in populations with high vaccination rates. We aimed to investigate transmission and viral load kinetics in vaccinated and unvaccinated individuals with mild delta variant infection in the community.
Methods
Between Sept 13, 2020, and Sept 15, 2021, 602 community contacts (identified via the UK contract-tracing system) of 471 UK COVID-19 index cases were recruited to the Assessment of Transmission and Contagiousness of COVID-19 in Contacts cohort study and contributed 8145 upper respiratory tract samples from daily sampling for up to 20 days. Household and non-household exposed contacts aged 5 years or older were eligible for recruitment if they could provide informed consent and agree to self-swabbing of the upper respiratory tract. We analysed transmission risk by vaccination status for 231 contacts exposed to 162 epidemiologically linked delta variant-infected index cases. We compared viral load trajectories from fully vaccinated individuals with delta infection (n=29) with unvaccinated individuals with delta (n=16), alpha (B.1.1.7; n=39), and pre-alpha (n=49) infections. Primary outcomes for the epidemiological analysis were to assess the secondary attack rate (SAR) in household contacts stratified by contact vaccination status and the index cases’ vaccination status. Primary outcomes for the viral load kinetics analysis were to detect differences in the peak viral load, viral growth rate, and viral decline rate between participants according to SARS-CoV-2 variant and vaccination status.
Findings
The SAR in household contacts exposed to the delta variant was 25% (95% CI 18–33) for fully vaccinated individuals compared with 38% (24–53) in unvaccinated individuals. The median time between second vaccine dose and study recruitment in fully vaccinated contacts was longer for infected individuals (median 101 days [IQR 74–120]) than for uninfected individuals (64 days [32–97], p=0·001). SAR among household contacts exposed to fully vaccinated index cases was similar to household contacts exposed to unvaccinated index cases (25% [95% CI 15–35] for vaccinated vs 23% [15–31] for unvaccinated). 12 (39%) of 31 infections in fully vaccinated household contacts arose from fully vaccinated epidemiologically linked index cases, further confirmed by genomic and virological analysis in three index case–contact pairs. Although peak viral load did not differ by vaccination status or variant type, it increased modestly with age (difference of 0·39 [95% credible interval –0·03 to 0·79] in peak log10 viral load per mL between those aged 10 years and 50 years). Fully vaccinated individuals with delta variant infection had a faster (posterior probability >0·84) mean rate of viral load decline (0·95 log10 copies per mL per day) than did unvaccinated individuals with pre-alpha (0·69), alpha (0·82), or delta (0·79) variant infections. Within individuals, faster viral load growth was correlated with higher peak viral load (correlation 0·42 [95% credible interval 0·13 to 0·65]) and slower decline (–0·44 [–0·67 to –0·18]).
Interpretation
Vaccination reduces the risk of delta variant infection and accelerates viral clearance. Nonetheless, fully vaccinated individuals with breakthrough infections have peak viral load similar to unvaccinated cases and can efficiently transmit infection in household settings, including to fully vaccinated contacts. Host–virus interactions early in infection may shape the entire viral trajectory.
Funding
National Institute for Health Research.
Introduction
While the primary aim of vaccination is to protect individuals against severe COVID-19 disease and its consequences, the extent to which vaccines reduce onward transmission of SARS-CoV-2 is key to containing the pandemic. This outcome depends on the ability of vaccines to protect against infection and the extent to which vaccination reduces the infectiousness of breakthrough infections.
Research in context.
Evidence before this study
The SARS-CoV-2 delta variant is spreading globally, including in populations with high vaccination coverage. While vaccination remains highly effective at attenuating disease severity and preventing death, vaccine effectiveness against infection is reduced for delta. Determining the extent of transmission from vaccinated delta-infected individuals to their vaccinated contacts is a public health priority. Comparing the upper respiratory tract (URT) viral load kinetics of delta infections with those of other variants gives insight into potential mechanisms for its increased transmissibility. We searched PubMed and medRxiv for articles published between database inception and Sept 20, 2021, using search terms describing "SARS-CoV-2, delta variant, viral load, and transmission". Two studies longitudinally sampled the URT in vaccinated and unvaccinated delta variant-infected individuals to compare viral load kinetics. In a retrospective study of a cohort of hospitalised patients in Singapore, more rapid viral load decline was found in vaccinated individuals than unvaccinated cases. However, the unvaccinated cases in this study had moderate-to-severe infection, which is known to be associated with prolonged shedding. The second study longitudinally sampled professional USA sports players. Again, clearance of delta viral RNA in vaccinated cases was faster than in unvaccinated cases, but only 8% of unvaccinated cases had delta variant infection, complicating interpretation. Lastly, a report of a single-source nosocomial outbreak of a distinct delta sub-lineage in Vietnamese health-care workers plotted viral load kinetics (without comparison with unvaccinated delta infections) and demonstrated transmission between fully vaccinated health-care workers in the nosocomial setting. The findings might therefore not be generalisable beyond the particular setting and distinct viral sub-lineage investigated.
Added value of this study
The majority of SARS-CoV-2 transmission occurs in households, but transmission between fully vaccinated individuals in this setting has not been shown to date. To ascertain secondary transmission with high sensitivity, we longitudinally followed index cases and their contacts (regardless of symptoms) in the community early after exposure to the delta variant of SARS-CoV-2, performing daily quantitative RT-PCR on URT samples for 14–20 days. We found that the secondary attack rate in fully vaccinated household contacts was high at 25%, but this value was lower than that of unvaccinated contacts (38%). Risk of infection increased with time in the 2–3 months since the second dose of vaccine. The proportion of infected contacts was similar regardless of the index cases’ vaccination status. We observed transmission of the delta variant between fully vaccinated index cases and their fully vaccinated contacts in several households, confirmed by whole-genome sequencing. Peak viral load did not differ by vaccination status or variant type but did increase modestly with age. Vaccinated delta cases experienced faster viral load decline than did unvaccinated alpha or delta cases. Across study participants, faster viral load growth was correlated with higher peak viral load and slower decline, suggesting that host–virus interactions early in infection shape the entire viral trajectory. Since our findings are derived from community household contacts in a real-life setting, they are probably generalisable to the general population.
Implications of all the available evidence
Although vaccines remain highly effective at preventing severe disease and deaths from COVID-19, our findings suggest that vaccination is not sufficient to prevent transmission of the delta variant in household settings with prolonged exposures. Our findings highlight the importance of community studies to characterise the epidemiological phenotype of new SARS-CoV-2 variants in increasingly highly vaccinated populations. Continued public health and social measures to curb transmission of the delta variant remain important, even in vaccinated individuals.
Vaccination was found to be effective in reducing household transmission of the alpha variant (B.1.1.7) by 40–50%,1 and infected, vaccinated individuals had lower viral load in the upper respiratory tract (URT) than infections in unvaccinated individuals,2 which is indicative of reduced infectiousness.3, 4 However, the delta variant (B.1.617.2), which is more transmissible than the alpha variant,5, 6 is now the dominant strain worldwide. After a large outbreak in India, the UK was one of the first countries to report a sharp rise in delta variant infection. Current vaccines remain highly effective at preventing admission to hospital and death from delta infection.7 However, vaccine effectiveness against infection is reduced for delta, compared with alpha,8, 9 and the delta variant continues to cause a high burden of cases even in countries with high vaccination coverage. Data are scarce on the risk of community transmission of delta from vaccinated individuals with mild infections.
Here, we report data from a UK community-based study, the Assessment of Transmission and Contagiousness of COVID-19 in Contacts (ATACCC) study, in which ambulatory close contacts of confirmed COVID-19 cases underwent daily, longitudinal URT sampling, with collection of associated clinical and epidemiological data. We aimed to quantify household transmission of the delta variant and assess the effect of vaccination status on contacts’ risk of infection and index cases’ infectiousness, including (1) households with unvaccinated contacts and index cases and (2) households with fully vaccinated contacts and fully vaccinated index cases. We also compared sequentially sampled URT viral RNA trajectories from individuals with non-severe delta, alpha, and pre-alpha SARS-CoV-2 infections to infer the effects of SARS-CoV-2 variant status—and, for delta infections, vaccination status—on transmission potential.
Methods
Study design and participants
ATACCC is an observational longitudinal cohort study of community contacts of SARS-CoV-2 cases. Contacts of symptomatic PCR-confirmed index cases notified to the UK contact-tracing system (National Health Service Test and Trace) were asked if they would be willing to be contacted by Public Health England to discuss participation in the study. All contacts notified within 5 days of index case symptom onset were selected to be contacted within our recruitment capacity. Household and non-household contacts aged 5 years or older were eligible for recruitment if they could provide written informed consent and agree to self-swabbing of the URT. Further details on URT sampling are given in the appendix (p 13).
The ATACCC study is separated into two study arms, ATACCC1 and ATACCC2, which were designed to capture different waves of the SARS-CoV-2 pandemic. In ATACCC1, which investigated alpha variant and pre-alpha cases in Greater London, only contacts were recruited between Sept 13, 2020, and March 13, 2021. ATACCC1 included a pre-alpha wave (September to November, 2020) and an alpha wave (December, 2020, to March, 2021). In ATACCC2, the study was relaunched specifically to investigate delta variant cases in Greater London and Bolton, and both index cases and contacts were recruited between May 25, and Sept 15, 2021. Early recruitment was focused in West London and Bolton because UK incidence of the delta variant was highest in these areas.10 Based on national and regional surveillance data, community transmission was moderate-to-high throughout most of our recruitment period.
This study was approved by the Health Research Authority. Written informed consent was obtained from all participants before enrolment. Parents and caregivers gave consent for children.
Data collection
Demographic information was collected by the study team on enrolment. The date of exposure for non-household contacts was obtained from Public Health England. COVID-19 vaccination history was determined from the UK National Immunisation Management System, general practitioner records, and self-reporting by study participants. We defined a participant as unvaccinated if they had not received a single dose of a COVID-19 vaccine at least 7 days before enrolment, partially vaccinated if they had received one vaccine dose at least 7 days before study enrolment, and fully vaccinated if they had received two doses of a COVID-19 vaccine at least 7 days before study enrolment. Previous literature was used to determine the 7-day threshold for defining vaccination status.11, 12, 13 We also did sensitivity analyses using a 14-day threshold. The time interval between vaccination and study recruitment was calculated. We used WHO criteria14 to define symptomatic status up to the day of study recruitment. Symptomatic status for incident cases—participants who were PCR-negative at enrolment and subsequently tested positive—was defined from the day of the first PCR-positive result.
Laboratory procedures
SARS-CoV-2 quantitative RT-PCR, conversion of ORF1ab and envelope (E-gene) cycle threshold values to viral genome copies, whole-genome sequencing, and lineage assignments are described in the appendix (pp 13–14).
Outcomes
Primary outcomes for the epidemiological analysis were to assess the secondary attack rate (SAR) in household contacts stratified by contact vaccination status and the index cases’ vaccination status. Primary outcomes for the viral load kinetics analysis were to detect differences in the peak viral load, viral growth rate, and viral decline rate between participants infected with pre-alpha versus alpha versus delta variants and between unvaccinated delta-infected participants and vaccinated delta-infected participants.
We assessed vaccine effectiveness and susceptibility to SARS-CoV-2 infection stratified by time elapsed since receipt of second vaccination as exploratory analyses.
Statistical analysis
To model viral kinetics, we used a simple phenomenological model of viral titre15 during disease pathogenesis. Viral kinetic parameters were estimated on a participant-specific basis using a Bayesian hierarchical model to fit this model to the entire dataset of sequential cycle threshold values measured for all participants. For the 19 participants who were non-household contacts of index cases and had a unique date of exposure, the cycle threshold data were supplemented by a pseudo-absence data point (ie, undetectable virus) on the date of exposure. Test accuracy and model misspecification were modelled with a mixture model by assuming there was a probability p of a test giving an observation drawn from a (normal) error distribution and probability 1 – p of it being drawn from the true distribution.
The hierarchical structure was represented by grouping participants based on the infecting variant and their vaccination status. A single-group model was fitted, which implicitly assumes that viral kinetic parameters vary by individual but not by variant or vaccination status. A four-group model was also explored, where groups 1, 2, 3, and 4 represent pre-alpha, alpha, unvaccinated delta, and fully vaccinated delta, respectively. We fitted a correlation matrix between participant-specific kinetic parameters to allow us to examine whether there is within-group correlation between peak viral titre, viral growth rate, and viral decline rate. Our initial model selection, using leave-one-out cross-validation, selected a four-group hierarchical model with fitted correlation coefficients between individual-level parameters determining peak viral load and viral load growth and decline rates (appendix p 5). However, resulting participant-specific estimates of peak viral load (but not growth and decline rates) showed a marked and significant correlation with age in the exploratory analysis, which motivated examination of models where mean peak viral load could vary with age. The most predictive model overall allowed mean viral load growth and decline rates to vary across the four groups, with mean peak viral load common to all groups but assumed to vary linearly with the logarithm of age (appendix p 5). We present peak viral loads for the reference age of 50 years with 95% credible intervals (95% CrIs). 50 years was chosen as the reference age as it is typical of the ages of the cases in the whole dataset and the choice of reference age made no difference in the model fits or judgment of differences between the groups.
We computed group-level population means and within-sample group means of log peak viral titre, viral growth rate, and viral decline rate. Since posterior estimates of each of these variables are correlated across groups, overlap in the credible intervals of an estimate for one group with that for another group does not necessarily indicate no significant difference between those groups. We, therefore, computed posterior probabilities, pp, that these variables were larger for one group than another. For our model, Bayes factors can be computed as pp/(1–pp). We only report population (group-level) posterior probabilities greater than 0·75 (corresponding to Bayes factors >3) as indicating at least moderate evidence of a difference.
For vaccine effectiveness, we defined the estimated effectiveness at preventing infection, regardless of symptoms, with delta in the household setting as 1 – SAR (fully vaccinated) / SAR (unvaccinated).
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
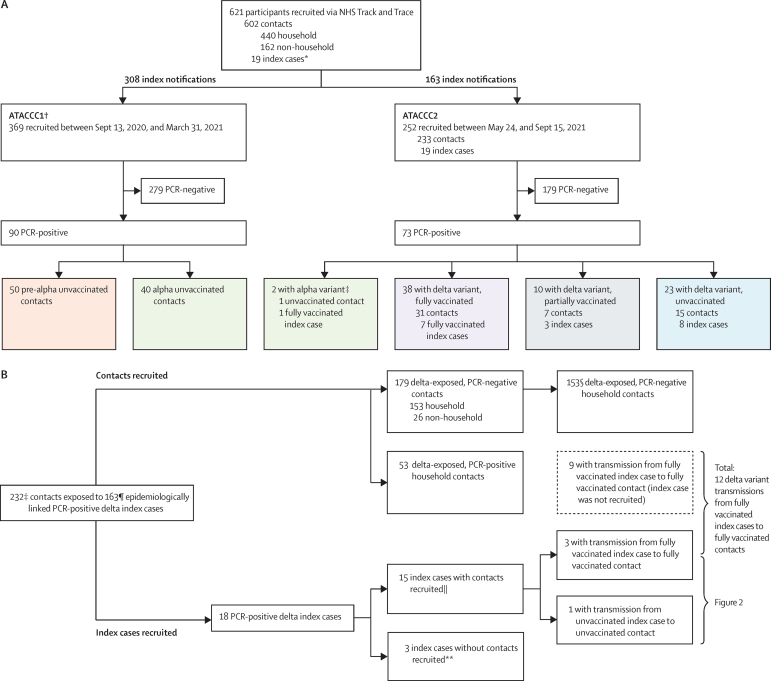
Between Sept 13, 2020, and Sept 15, 2021, 621 community-based participants (602 contacts and 19 index cases) from 471 index notifications were prospectively enrolled in the ATACCC1 and ATACCC2 studies, and contributed 8145 URT samples. Of these, ATACCC1 enrolled 369 contacts (arising from 308 index notifications), and ATACCC2 enrolled 233 contacts (arising from 163 index notifications) and 19 index cases. SARS-CoV-2 RNA was detected in 163 (26%) of the 621 participants. Whole-genome sequencing of PCR-positive cases confirmed that 71 participants had delta variant infection (18 index cases and 53 contacts), 42 had alpha variant infection (one index case and 41 contacts), and 50 had pre-alpha variant infection (all contacts; figure 1A).
Figure 1.
Recruitment, SARS-CoV-2 infection, variant status, and vaccination history for ATACCC study participants
(A) Study recruitment and variant status confirmed by whole-genome sequencing (ATACCC1 and ATACCC2 combined). (B) ATACCC2: delta-exposed contacts included in secondary attack rate calculation (table 1) and transmission assessment (table 2). NHS=National Health Service. *All index cases were from ATACCC2. † All contacts. ‡The two earliest PCR-positive cases from the ATACCC2 cohort (one index case and one contact) were confirmed as having the alpha variant on whole-genome sequencing (recruited on May 28, 2021). This alpha variant-exposed, PCR-positive contact is excluded from figure 1B. §One PCR-negative contact had no vaccination status data available and one PCR-negative contact's index case had no vaccination data available. ¶Vaccination data were available for 138 index cases of 163. ||The contacts of these 15 index cases are included within the 232 total contacts. **These three index cases without contacts are only included in the viral load kinetics analysis (figure 3) and are not included in tables 1 and 2.
Of 163 PCR-positive participants, 89 (55%) were female and 133 (82%) were White. Median age was 36 years (IQR 26–50). Sex, age, ethnicity, body-mass index (BMI) distribution, and the frequency of comorbidities were similar among those with delta, alpha, and pre-alpha infection, and for vaccinated and unvaccinated delta-infected participants, except for age and sex (appendix pp 2–3). There were fewer unvaccinated females than males (p=0·04) and, as expected from the age-prioritisation of the UK vaccine roll-out, unvaccinated participants infected with the delta variant were significantly younger (p<0·001; appendix p 3). Median time between exposure to the index case and study enrolment was 4 days (IQR 4–5). All participants had non-severe ambulatory illness or were asymptomatic. The proportion of asymptomatic cases did not differ among fully vaccinated, partially vaccinated, and unvaccinated delta groups (appendix p 3).
No pre-alpha-infected and only one alpha-infected participant had received a COVID-19 vaccine before study enrolment. Of 71 delta-infected participants (of whom 18 were index cases), 23 (32%) were unvaccinated, ten (14%) were partially vaccinated, and 38 (54%) were fully vaccinated (figure 1A; appendix p 3). Of the 38 fully vaccinated delta-infected participants, 14 had received the BNT162b2 mRNA vaccine (Pfizer–BioNTech), 23 the ChAdOx1 nCoV-19 adenovirus vector vaccine (Oxford–AstraZeneca), and one the CoronaVac inactivated whole-virion vaccine (Sinovac).
It is highly probable that all but one of the 233 ATACCC2 contacts were exposed to the delta variant because they were recruited when the regional prevalence of delta was at least 90%, and mostly 95–99% (figure 1B).10 Of these, 206 (89%) were household contacts (in 127 households), and 26 (11%) were non-household contacts. Distributions of age, ethnicity, BMI, smoking status, and comorbidities were similar between PCR-positive and PCR-negative contacts (appendix p 4). The median time between second vaccine dose and study recruitment in fully vaccinated contacts with delta variant infection was 74 days (IQR 35–105; range 16–201), and this was significantly longer in PCR-positive contacts than in PCR-negative contacts (101 days [IQR 74–120] vs 64 days [32–97], respectively, p=0·001; appendix p 4). All 53 PCR-positive contacts were exposed in household settings and the SAR for all delta variant-exposed household contacts was 26% (95% CI 20–32). SAR was not significantly higher in unvaccinated (38%, 95% CI 24–53) than fully vaccinated (25%, 18–33) household contacts (table 1). We estimated vaccine effectiveness at preventing infection (regardless of symptoms) with delta in the household setting to be 34% (bootstrap 95% CI –15 to 60). Sensitivity analyses using a 14 day threshold for time since second vaccination to study recruitment to denote fully vaccinated did not materially affect our estimates of vaccine effectiveness or SAR (data not shown). Although precision is restricted by the small sample size, this estimate is broadly consistent with vaccine effectiveness estimates for delta variant infection based on larger datasets.9, 16, 17
Table 1.
SAR in contacts of delta-exposed index cases recruited to the ATACCC2 study
| Total | PCR positive | PCR negative | SAR (95% CI) | p value | |
|---|---|---|---|---|---|
| Contacts | |||||
| All | 231 | 53 | 178 | 23 (18–29) | NA |
| Fully vaccinated | 140 | 31 | 109 | 22 (16–30) | 0·16 |
| Unvaccinated | 44 | 15 | 29 | 34 (22–49) | .. |
| Partially vaccinated | 47 | 7 | 40 | 15 (7–28) | NA |
| Household contacts | |||||
| All | 205 | 53 | 152 | 26 (20–32) | NA |
| Fully vaccinated | 126 | 31 | 95 | 25 (18–33) | 0·17 |
| Unvaccinated | 40 | 15 | 25 | 38 (24–53) | .. |
| Partially vaccinated | 39 | 7 | 32 | 18 (9–33) | NA |
χ2 test was performed to calculate p values for differences in SAR between fully vaccinated and unvaccinated cases. One PCR-negative contact who withdrew from the study without vaccination status information was excluded. NA=not applicable. SAR=secondary attack rate.
The vaccination status of 138 epidemiologically linked index cases of 204 delta variant-exposed household contacts was available (figure 1B, table 2). The SAR in household contacts exposed to fully vaccinated index cases was 25% (95% CI 15–35; 17 of 69), which is similar to the SAR in household contacts exposed to unvaccinated index cases (23% [15–31]; 23 of 100; table 2). The 53 PCR-positive contacts arose from household exposure to 39 PCR-positive index cases. Of these index cases who gave rise to secondary transmission, the proportion who were fully vaccinated (15 [38%] of 39) was similar to the proportion who were unvaccinated (16 [41%] of 39). The median number of days from the index cases’ second vaccination to the day of recruitment for their respective contacts was 73 days (IQR 38–116). Time interval did not differ between index cases who transmitted infection to their contacts and those who did not (94 days [IQR 62–112] and 63 days [35–117], respectively; p=0·43).
Table 2.
Comparison of vaccination status of the 138 epidemiologically linked PCR-positive index cases for 204 delta variant-exposed household contacts
| All household contacts (n=204)* |
Fully vaccinated contacts (n=125) |
Partially vaccinated contacts (n=39) |
Unvaccinated contacts (n=40) |
||||
|---|---|---|---|---|---|---|---|
| PCR positive (n=31) | PCR negative (n=94) | PCR positive (n=7) | PCR negative (n=32) | PCR positive (n=15) | PCR negative (n=25) | ||
| Fully vaccinated index cases (n=50) | 69 | 12 | 31 | 1 | 8 | 4 | 13 |
| Partially vaccinated index cases (n=25) | 35 | 7 | 12 | 3 | 10 | 3 | 0 |
| Unvaccinated index cases (n=63) | 100 | 12 | 51 | 3 | 14 | 8 | 12 |
Non-household exposed contacts (n=24, all PCR negative) were excluded. One PCR-negative household contact who withdrew from the study without vaccination status information was excluded. One PCR-negative household contact who could not be linked to their index case was also excluded.
The rows below show the number of contacts exposed to each category of index case.
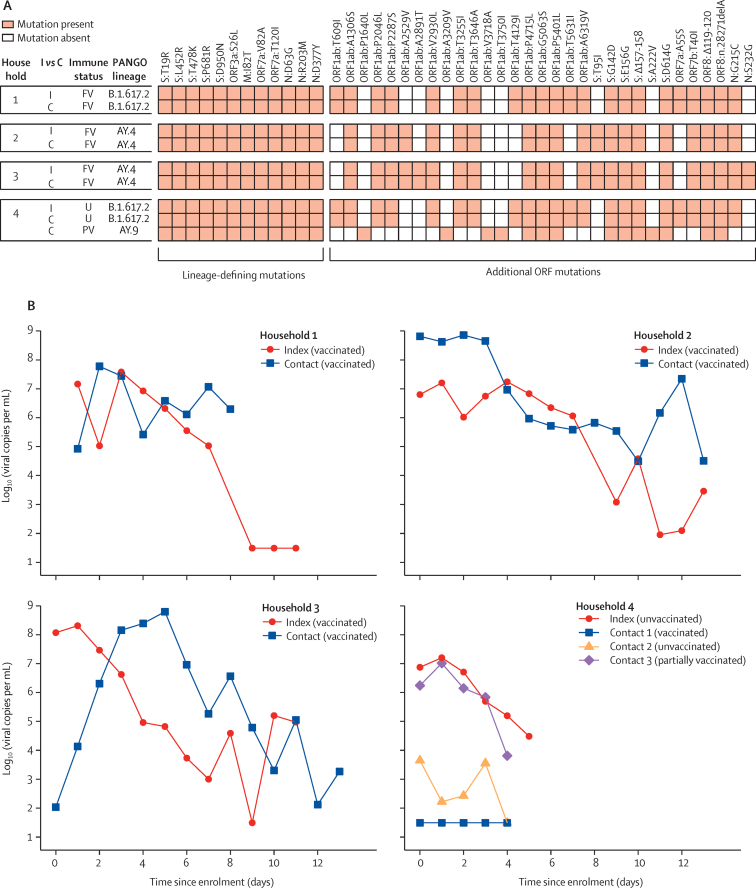
18 of the 163 delta variant-infected index cases that led to contact enrolment were themselves recruited to ATACCC2 and serial URT samples were collected from them, allowing for more detailed virology and genome analyses. For 15 of these, their contacts were also recruited (13 household contacts and two non-household contacts). A corresponding PCR-positive household contact was identified for four of these 15 index cases (figure 1B). Genomic analysis showed that index–contact pairs were infected with the same delta variant sub-lineage in these instances, with one exception (figure 2A). In one household (number 4), an unvaccinated index case transmitted the delta variant to an unvaccinated contact, while another partially vaccinated contact was infected with a different delta sub-lineage (which was probably acquired outside the household). In the other three households (numbers 1–3), fully vaccinated index cases transmitted the delta variant to fully vaccinated household contacts, with high viral load in all cases, and temporal relationships between the viral load kinetics that were consistent with transmission from the index cases to their respective contacts (figure 2B).
Figure 2.
Virological, epidemiological, and genomic evidence for transmission of the SARS-CoV-2 delta variant (B.1.617.2) in households
(A) Genomic analysis of the four households with lineage-defining mutations for delta18 and additional mutations within ORFs displayed to give insight into whether strains from individuals within the household are closely related. Lineages AY.4 and AY.9 are sub-lineages of delta. (B) Viral trajectories and vaccination status of the four index cases infected with the delta variant for whom infection was detected in their epidemiologically linked household contacts. All individuals had non-severe disease. Each plot shows an index case and their household contacts. Undetectable viral load measurements are plotted at the limit of detection (101·49). C=contact. I=index case. FV=fully vaccinated. ORF=open reading frame. PV=partially vaccinated. U=unvaccinated.
Inclusion criteria for the modelling analysis selected 133 participant's viral load RNA trajectories from 163 PCR-positive participants (49 with the pre-alpha variant, 39 alpha, and 45 delta; appendix p 14). Of the 45 delta cases, 29 were fully vaccinated and 16 were unvaccinated; partially vaccinated cases were excluded. Of the 133 included cases, 29 (22%) were incident (ie, PCR negative at enrolment converting to PCR positive subsequently) and 104 (78%) were prevalent (ie, already PCR positive at enrolment). 15 of the prevalent cases had a clearly resolvable peak viral load. Figure 3 shows modelled viral RNA (ORF1ab) trajectories together with the viral RNA copy numbers measured for individual participants. The E-gene equivalent is shown in the appendix (p 2). Estimates derived from E-gene cycle threshold value data (appendix pp 5, 7, 9, 11) were similar to those for ORF1ab.
Figure 3.
ORF1ab viral load trajectories from 14 days before to 28 days after peak for 133 participants infected with pre-alpha or alpha variants (uncaccinated), or the delta variant (vaccinated and unvaccinated) variants
Black circles are measured values, with the first datapoint for each participant being taken to the day of enrolment. Plots are rooted on the day of peak viral load for each participant, denoted as day 0 on the x-axis. Curves show the model posterior median estimate, with a 95% credible interval shading. 133 infected participants, comprising 114 contacts and 19 index cases. *Index cases.
Although viral kinetics appear visually similar for all four groups of cases, we found quantitative differences in estimated viral growth rates and decline rates (Table 3, Table 4). Population (group-level) estimates of mean viral load decline rates based on ORF1ab cycle threshold value data varied in the range of 0·69–0·95 log10 units per mL per daxes 4; appendix p 10), indicating that a typical 10-day period was required for viral load to decline from peak to undetectable. A faster decline was seen in the alpha (pp=0·93), unvaccinated delta (pp=0·79), and fully vaccinated delta (pp=0·99) groups than in the pre-alpha group. The mean viral load decline rate of the fully vaccinated delta group was also faster than those of the alpha group (pp=0·84) and the unvaccinated delta group (pp=0·85). The differences in decline rates translate into a difference of about 3 days in the mean duration of the decline phase between the pre-alpha and delta vaccinated groups.
Table 3.
Estimates of VL growth rates for pre-alpha, alpha, and delta (unvaccinated and fully vaccinated) cases, derived from ORF1ab cycle threshold data
| VL growth rate (95% CrI), log10units per day | Posterior probability estimate is less than pre-alpha | Posterior probability estimate is less than alpha | Posterior probability estimate is less than delta (unvaccinated) | Posterior probability estimate is less than delta (fully vaccinated) | |
|---|---|---|---|---|---|
| Pre-alpha (n=49) | 3·24 (1·78–6·14) | .. | 0·44 | 0·27 | 0·21 |
| Alpha (n=39) | 3·13 (1·76–5·94) | 0·56 | .. | 0·32 | 0·25 |
| Delta, unvaccinated (n=16) | 2·81 (1·47–5·47) | 0·73 | 0·68 | .. | 0·44 |
| Delta, fully vaccinated (n=29) | 2·69 (1·51–5·17) | 0·79 | 0·75 | 0·56 | .. |
VL growth rates are shown as within-sample posterior mean estimates. Remaining columns show population (group-level) posterior probabilities that the estimate on that row is less than an estimate for a different group. Posterior probabilities are derived from 20 000 posterior samples and have sampling errors of <0·01. VL=viral load. CrI=credible interval.
Table 4.
Estimates of VL decline rates for pre-alpha, alpha, and delta (unvaccinated and fully vaccinated) cases, derived from ORF1ab cycle threshold data
| VL decline rate (95% CrI), log10units per day | Posterior probability estimate is larger than pre-alpha | Posterior probability estimate is larger than alpha | Posterior probability estimate is larger than delta (unvaccinated) | Posterior probability estimate is larger than delta (fully vaccinated) | |
|---|---|---|---|---|---|
| Pre-alpha (n=49) | 0·69 (0·58–0·81) | .. | 0·07 | 0·21 | 0·01 |
| Alpha (n=39) | 0·82 (0·67–1·01) | 0·93 | .. | 0·60 | 0·16 |
| Delta, unvaccinated (n=16) | 0·79 (0·59–1·04) | 0·79 | 0·40 | .. | 0·15 |
| Delta, fully vaccinated (n=29) | 0·95 (0·76–1·18) | 0·99 | 0·84 | 0·85 | .. |
VL decline rates are shown as within-sample posterior mean estimates. Remaining columns show population (group-level) posterior probabilities that the estimate on that row is less than an estimate for a different group. Posterior probabilities are derived from 20 000 posterior samples and have sampling errors of <0·01. VL=viral load. CrI=credible interval.
Viral load growth rates were substantially faster than decline rates, varying in the range of 2·69–3·24 log10 units per mL per day between groups, indicating that a typical 3-day period was required for viral load to grow from undetectable to peak. Our power to infer differences in growth rates between groups was more restricted than for viral decline, but there was moderate evidence (pp=0·79) that growth rates were lower for those in the vaccinated delta group than in the pre-alpha group.
We estimated mean peak viral load for 50-year-old adults to be 8·14 (95% CrI 7·95 to 8·32) log10 copies per mL, but peak viral load did not differ by variant or vaccination status. However, we estimated that peak viral load increases with age (pp=0·96 that the slope of peak viral load with log[age] was >0), with an estimated slope of 0·24 (95% CrI –0·02 to 0·49) log10 copies per mL per unit change in log(age). This estimate translates to a difference of 0·39 (–0·03 to 0·79) in mean peak log10 copies per mL between those aged 10 years and 50 years.
Within-group individual participant estimates of viral load growth rate were positively correlated with peak viral load, with a correlation coefficient estimate of 0·42 (95% CrI 0·13 to 0·65; appendix p 8). Hence, individuals with faster viral load growth tend to have higher peak viral load. The decline rate of viral load was also negatively correlated with viral load growth rate, with a correlation coefficient estimate of –0·44 (95% CrI –0·67 to –0·18), illustrating that individuals with faster viral load growth tend to experience slower viral load decline.
Discussion
Households are the site of most SARS-CoV-2 transmission globally.19 In our cohort of densely sampled household contacts exposed to the delta variant, SAR was 38% in unvaccinated contacts and 25% in fully vaccinated contacts. This finding is consistent with the known protective effect of COVID-19 vaccination against infection.8, 9 Notwithstanding, these findings indicate continued risk of infection in household contacts despite vaccination. Our estimate of SAR is higher than that reported in fully vaccinated household contacts exposed before the emergence of the delta variant.1, 20, 21 The time interval between vaccination and study recruitment was significantly higher in fully vaccinated PCR-positive contacts than fully vaccinated PCR-negative contacts, suggesting that susceptibility to infection increases with time as soon as 2–3 months after vaccination—consistent with waning protective immunity. This potentially important observation is consistent with recent large-scale data and requires further investigation.17 Household SAR for delta infection, regardless of vaccination status, was 26% (95% CI 20–32), which is higher than estimates of UK national surveillance data (10·8% [10·7–10·9]).10 However, we sampled contacts daily, regardless of symptomatology, to actively identify infection with high sensitivity. By contrast, symptom-based, single-timepoint surveillance testing probably underestimates the true SAR, and potentially also overestimates vaccine effectiveness against infection.
We identified similar SAR (25%) in household contacts exposed to fully vaccinated index cases as in those exposed to unvaccinated index cases (23%). This finding indicates that breakthrough infections in fully vaccinated people can efficiently transmit infection in the household setting. We identified 12 household transmission events between fully vaccinated index case–contact pairs; for three of these, genomic sequencing confirmed that the index case and contact were infected by the same delta variant sub-lineage, thus substantiating epidemiological data and temporal relationships of viral load kinetics to provide definitive evidence for secondary transmission. To our knowledge, one other study has reported that transmission of the delta variant between fully vaccinated people was a point-source nosocomial outbreak—a single health-care worker with a particular delta variant sub-lineage in Vietnam.22
Daily longitudinal sampling of cases from early (median 4 days) after exposure for up to 20 days allowed us to generate high-resolution trajectories of URT viral load over the course of infection. To date, two studies have sequentially sampled community cases of mild SARS-CoV-2 infection, and these were from highly specific population groups identified through asymptomatic screening programmes (eg, for university staff and students23 and for professional athletes24).
Our most predictive model of viral load kinetics estimated mean peak log10 viral load per mL of 8·14 (95% CrI 7·95–8·32) for adults aged 50 years, which is very similar to the estimate from a 2021 study using routine surveillance data.25 We found no evidence of variation in peak viral load by variant or vaccination status, but we report some evidence of modest but significant (pp=0·95) increases in peak viral load with age. Previous studies of viral load in children and adults4, 25, 26 have not used such dense sequential sampling of viral load and have, therefore, been restricted in their power to resolve age-related differences; the largest such study25 reported a similar difference between children and adults to the one we estimated. We found the rate of viral load decline was faster for vaccinated individuals with delta infection than all other groups, and was faster for individuals in the alpha and unvaccinated delta groups than those with pre-alpha infection.
For all variant vaccination groups, the variation between participants seen in viral load kinetic parameter estimates was substantially larger than the variation in mean parameters estimated between groups. The modest scale of differences in viral kinetics between fully vaccinated and unvaccinated individuals with delta infection might explain the relatively high rates of transmission seen from vaccinated delta index cases in our study. We found no evidence of lower SARs from fully vaccinated delta index cases than from unvaccinated ones. However, given that index cases were identified through routine symptomatic surveillance, there might have been a selection bias towards identifying untypically symptomatic vaccine breakthrough index cases.
The differences in viral kinetics we found between the pre-alpha, alpha, and delta variant groups suggest some incremental, but potentially adaptive, changes in viral dynamics associated with the evolution of SARS-CoV-2 towards more rapid viral clearance. Our study provides the first evidence that, within each variant or vaccination group, viral growth rate is positively correlated with peak viral load, but is negatively correlated with viral decline rate. This finding suggests that individual infections during which viral replication is initially fastest generate the highest peak viral load and see the slowest viral clearance, with the latter not just being due to the higher peak. Mechanistically, these data suggest that the host and viral factors determining the initial growth rate of SARS-CoV-2 have a fundamental effect on the trajectory throughout infection, with faster replication being more difficult (in terms of both peak viral load and the subsequent decline of viral load) for the immune response to control. Analysis of sequentially sampled immune markers during infection might give insight into the immune correlates of these early differences in infection kinetics. It is also possible that individuals with the fastest viral load growth and highest peaks contribute disproportionately to community transmission, a hypothesis that should be tested in future studies.
Several population-level, single-timepoint sampling studies using routinely available data have found no major differences in cycle threshold values between vaccinated and unvaccinated individuals with delta variant infection.10, 27, 28 However, as the timepoint of sampling in the viral trajectory is unknown, this restricts the interpretation of such results. Two other studies longitudinally sampled vaccinated and unvaccinated individuals with delta variant infection.23, 29 A retrospective cohort of hospitalised patients in Singapore29 also described a faster rate of viral decline in vaccinated versus unvaccinated individuals with delta variant, reporting somewhat larger differences in decline rates than we estimated here. However, this disparity might be accounted for by the higher severity of illness in unvaccinated individuals in the Singaporean study (almost two-thirds having pneumonia, one-third requiring COVID-19 treatment, and a fifth needing oxygen) than in our study, given that longer viral shedding has been reported in patients with more severe illness.30 A longitudinal sampling study in the USA reported that pre-alpha, alpha, and delta variant infections had similar viral trajectories.24 The study also compared trajectories in vaccinated and unvaccinated individuals, reporting similar proliferation phases and peak cycle threshold values, but more rapid clearance of virus in vaccinated individuals. However, this study in the USA stratified by vaccination status and variant separately, rather than jointly, meaning vaccinated individuals with delta infection were being compared with, predominantly, unvaccinated individuals with pre-alpha and alpha infection. Moreover, sampling was done as part of a professional sports player occupational health screening programme, making the results not necessarily representative of typical community infections.
Our study has limitations. First, we recruited only contacts of symptomatic index cases as our study recruitment is derived from routine contact-tracing notifications. Second, index cases were defined as the first household member to have a PCR-positive swab, but we cannot exclude the possibility that another household member might already have been infected and transmitted to the index case. Third, recording of viral load trajectories is subject to left censoring, where the growth phase in prevalent contacts (already PCR-positive at enrolment) was missed for a proportion of participants. However, we captured 29 incident cases and 15 additional cases on the upslope of the viral trajectory, providing valuable, informative data on viral growth rates and peak viral load in a subset of participants. Fourth, owing to the age-stratified rollout of the UK vaccination programme, the age of the unvaccinated, delta variant-infected participants was lower than that of vaccinated participants. Thus, age might be a confounding factor in our results and, as discussed, peak viral load was associated with age. However, it is unlikely that the higher SAR observed in the unvaccinated contacts would have been driven by younger age rather than the absence of vaccination and, to our knowledge, there is no published evidence showing increased susceptibility to SARS-CoV-2 infection with decreasing age.31 Finally, although we did not perform viral culture here—which is a better proxy for infectiousness than RT-PCR—two other studies27, 32 have shown cultivable virus from around two-thirds of vaccinated individuals infected with the delta variant, consistent with our conclusions that vaccinated individuals still have the potential to infect others, particularly early after infection when viral loads are high and most transmission is thought to occur.30
Our findings help to explain how and why the delta variant is being transmitted so effectively in populations with high vaccine coverage. Although current vaccines remain effective at preventing severe disease and deaths from COVID-19, our findings suggest that vaccination alone is not sufficient to prevent all transmission of the delta variant in the household setting, where exposure is close and prolonged. Increasing population immunity via booster programmes and vaccination of teenagers will help to increase the currently limited effect of vaccination on transmission, but our analysis suggests that direct protection of individuals at risk of severe outcomes, via vaccination and non-pharmacological interventions, will remain central to containing the burden of disease caused by the delta variant.
This online publication has been corrected. The corrected version first appeared at thelancet.com/infection on November 2, 2021
Data sharing
An anonymised, de-identified version of the dataset can be made available upon request to allow all results to be reproduced. Modelling code will also be made publicly available on the GitHub repository.
Declaration of interests
NMF reports grants from UK Medical Research Council, UK National Institute of Health Research, UK Research and Innovation, Community Jameel, Janssen Pharmaceuticals, the Bill & Melinda Gates Foundation, and Gavi, the Vaccine Alliance; consulting fees from the World Bank; payment or honoraria from the Wellcome Trust; travel expenses from WHO; advisory board participation for Takeda; and is a senior editor of the eLife journal. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
This work is supported by the National Institute for Health Research (NIHR200927), a Department of Health and Social Care COVID-19 Fighting Fund award, and the NIHR Health Protection Research Units (HPRUs) in Respiratory Infections and in Modelling and Health Economics. NMF acknowledges funding from the MRC Centre for Global Infectious Disease Analysis and the Jameel Institute. PSF and MAC are supported by the UK Dementia Research Institute. JD is supported by the NIHR HPRU in Emerging and Zoonotic Infections. MGW is supported by the NIHR HPRU in Healthcare Associated Infections and Antimicrobial Resistance. GPT is supported by the Imperial NIHR Biomedical Research Centre. We thank all the participants who were involved in the study, Public Health England staff for facilitating recruitment into the study, the staff of the Virus Reference Department for performing PCR and sequencing assays, and the Immunisations Department for assisting with analysis of vaccination data. We also thank Kristel Timcang, Mohammed Essoussi, Holly Grey, Guilia Miserocchi, Harriet Catchpole, Charlotte Williams, Niamh Nichols, Jessica Russell, Sean Nevin, Lulu Wang, Berenice Di Biase, Alice Panes, Esther Barrow, and Lauren Edmunds for their involvement in logistics, conducting data entry, or quality control; and the Molecular Diagnostics Unit at Imperial College London, in particular Lucy Mosscrop, Carolina Rosadas de Oliveira, and Patricia Watber, for performing RNA extraction, quantitative RT-PCR, and preparing samples for sequencing.
Contributors
AS, JD, MZ, NMF, WB, and ALal conceptualised the study. AS, SH, JD, KJM, AK, JLB, MGW, ND-F, RV, RK, JF, CT, AVK, JC, VQ, EC, JSN, SH, EM, TP, HH, CL, JS, SB, JP, CA, SA, and NMF were responsible for data curation and investigation. AS, SH, KJM, JLB, AC, NMF, and ALal did the formal data analysis. MAC, AB, DJ, SM, JE, PSF, SD, and ALac did the laboratory work. RV, RK, JF, CT, AVK, JC, VQ, EC, JSN, SH, EM, and SE oversaw the project. AS, SH, JD, KJM, JLB, NMF, and ALal accessed and verified the data. JD, MZ, and ALal acquired funding. NMF sourced and oversaw the software. AS and ALal wrote the initial draft of the manuscript. AS, JD, GPT, MZ, NMF, SH, and ALal reviewed and edited the manuscript. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
The ATACCC Study Investigators
Anjna Badhan, Simon Dustan, Chitra Tejpal, Anjeli V Ketkar, Janakan Sam Narean, Sarah Hammett, Eimear McDermott, Timesh Pillay, Hamish Houston, Constanta Luca, Jada Samuel, Samuel Bremang, Samuel Evetts, John Poh, Charlotte Anderson, David Jackson, Shahjahan Miah, Joanna Ellis, and Angie Lackenby.
Contributor Information
Ajit Lalvani, Email: a.lalvani@imperial.ac.uk.
ATACCC Study Investigators:
Anjna Badhan, Simon Dustan, Chitra Tejpal, Anjeli V Ketkar, Janakan Sam Narean, Sarah Hammett, Eimear McDermott, Timesh Pillay, Hamish Houston, Constanta Luca, Jada Samuel, Samuel Bremang, Samuel Evetts, John Poh, Charlotte Anderson, David Jackson, Shahjahan Miah, Joanna Ellis, and Angie Lackenby
Supplementary Material
References
- 1.Harris RJ, Hall JA, Zaidi A, Andrews NJ, Dunbar JK, Dabrera G. Effect of vaccination on household transmission of SARS-CoV-2 in England. N Engl J Med. 2021;385:759–760. doi: 10.1056/NEJMc2107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine-Tiefenbrun M, Yelin I, Katz R, et al. Initial report of decreased SARS-CoV-2 viral load after inoculation with the BNT162b2 vaccine. Nat Med. 2021;27:790–792. doi: 10.1038/s41591-021-01316-7. [DOI] [PubMed] [Google Scholar]
- 3.Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyngse FP, Mølbak K, Træholt Franck K, et al. Association between SARS-CoV-2 transmissibility, viral load, and age in households. medRxiv. 2021 doi: 10.1101/2021.02.28.21252608. published online June 4. (preprint). [DOI] [Google Scholar]
- 5.Liu Y, Rocklöv J. The reproductive number of the delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J Travel Med. 2021 doi: 10.1093/jtm/taab124. published online Aug 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Challen R, Dyson L, Overton CE, et al. Early epidemiological signatures of novel SARS-CoV-2 variants: establishment of B.1.617.2 in England. medRxiv. 2021 doi: 10.1101/2021.06.05.21258365. published online June 7. (preprint). [DOI] [Google Scholar]
- 7.Sheikh A, McMenamin J, Taylor B, Robertson C. SARS-CoV-2 delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397:2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 (delta) variant. N Engl J Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seppälä E, Veneti L, Starrfelt J, et al. Vaccine effectiveness against infection with the delta (B.1.617.2) variant, Norway, April to August 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.35.2100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Public Health England SARS-CoV-2 variants of concern and variants under investigation in England Technical briefing 20. Aug 6, 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1009243/Technical_Briefing_20.pdf
- 11.Dagan N, Barda N, Kepten E, et al. BNT162b2 mRNA COVID-19 vaccine in a nationwide mass vaccination setting. N Engl J Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung H, He S, Nasreen S, et al. Effectiveness of BNT162b2 and mRNA-1273 COVID-19 vaccines against symptomatic SARS-CoV-2 infection and severe COVID-19 outcomes in Ontario, Canada: test negative design study. BMJ. 2021;374 doi: 10.1136/bmj.n1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO WHO COVID-19 case definition. 2020. https://www.who.int/publications/i/item/WHO-2019-nCoV-Surveillance_Case_Definition-2020.2
- 15.Ferguson NM, Kien DT, Clapham H, et al. Modeling the impact on virus transmission of wolbachia-mediated blocking of dengue virus infection of Aedes aegypti. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.3010370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott P, Haw D, Wang H, et al. REACT-1 round 13 final report: exponential growth, high prevalence of SARS-CoV-2 and vaccine effectiveness associated with delta variant in England during May to July 2021. https://spiral.imperial.ac.uk/bitstream/10044/1/90800/2/react1_r13_final_preprint_final.pdf [DOI] [PMC free article] [PubMed]
- 17.Pouwels K, Pritchard E, Matthews P, et al. Impact of delta on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. https://www.ndm.ox.ac.uk/files/coronavirus/covid-19-infection-survey/finalfinalcombinedve20210816.pdf [DOI] [PMC free article] [PubMed]
- 18.O'Toole Á, Hill V, Pybus OG, et al. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2 with grinch. Wellcome Open Res. 2021;6:121. doi: 10.12688/wellcomeopenres.16661.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson HA, Mousa A, Dighe A, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) setting-specific transmission rates: a systematic review and meta-analysis. Clin Infect Dis. 2021;73:e754–e764. doi: 10.1093/cid/ciab100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.House T, Pellis L, Pritchard E, McLean AR, Walker AS. Total effect analysis of vaccination on household transmission in the Office for National Statistics COVID-19 infection survey. arXiv. 2021 http://arxiv.org/abs/2107.06545 published online July 14. (preprint). [Google Scholar]
- 21.de Gier B, Andeweg S, Joosten R, et al. Vaccine effectiveness against SARS-CoV-2 transmission and infections among household and other close contacts of confirmed cases, the Netherlands, February to May 2021. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.31.2100640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chau NVV, Ngoc NM, Nguyet LA, et al. An observational study of breakthrough SARS-CoV-2 Delta variant infections among vaccinated healthcare workers in Vietnam 2021. E Clin Med. 2021;41 doi: 10.1016/j.eclinm.2021.101143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ke R, Martinez PP, Smith RL, et al. Daily sampling of early SARS-CoV-2 infection reveals substantial heterogeneity in infectiousness. medRxiv. 2021 doi: 10.1101/2021.07.12.21260208. published online July 12. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kissler SM, Fauver JR, Mack C, et al. Densely sampled viral trajectories suggest longer duration of acute infection with B.1.1.7 variant relative to non-B.1.1.7 SARS-CoV-2. medRxiv. 2021 doi: 10.1101/2021.02.16.21251535. published online Aug 25. (preprint). [DOI] [Google Scholar]
- 25.Jones TC, Biele G, Mühlemann B, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021;373 doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madera S, Crawford E, Langelier C, et al. Nasopharyngeal SARS-CoV-2 viral loads in young children do not differ significantly from those in older children and adults. Sci Rep. 2021;11 doi: 10.1038/s41598-021-81934-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riemersma KK, Grogan BE, Kita-Yarbro A, et al. Shedding of infectious SARS-CoV-2 despite vaccination when the delta variant is prevalent—Wisconsin, July 2021. Version 4. medRxiv. 2021 doi: 10.1101/2021.07.31.21261387. published online Aug 24. (preprint). [DOI] [Google Scholar]
- 28.Brown CM, Vostok J, Johnson H, et al. Outbreak of SARS-CoV-2 infections, including COVID-19 vaccine breakthrough infections, associated with large public gatherings–Barnstable County, Massachusetts, July 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1059–1062. doi: 10.15585/mmwr.mm7031e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chia PY, Ong S, Chiew CJ, et al. Virological and serological kinetics of SARS-CoV-2 delta variant vaccine-breakthrough infections: a multi-center cohort study. medRxiv. 2021 doi: 10.1101/2021.07.28.21261295. published online July 31. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viner RM, Mytton OT, Bonell C, et al. Susceptibility to SARS-CoV-2 infection among children and adolescents compared with adults: a systematic review and meta-analysis. JAMA Pediatr. 2021;175:143–156. doi: 10.1001/jamapediatrics.2020.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shamier MC, Tostmann A, Bogers S, et al. Virological characteristics of SARS-CoV-2 vaccine breakthrough infections in health care workers. medRxiv. 2021 doi: 10.1101/2021.08.20.21262158. published online Aug 21. (preprint). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
An anonymised, de-identified version of the dataset can be made available upon request to allow all results to be reproduced. Modelling code will also be made publicly available on the GitHub repository.