Abstract
Sixty-six Corynebacterium diphtheriae strains (62 of the gravis biotype and 4 of the mitis biotype) isolated during the Georgian diphtheria epidemic of 1993 to 1998 and 13 non-Georgian C. diphtheriae strains (10 Russian and 3 reference isolates) were characterized by (i) biotyping, (ii) toxigenicity testing with the Elek assay and PCR, (iii) the randomly amplified polymorphic DNA (RAPD) technique, and (iv) pulsed-field gel electrophoresis (PFGE). Fifteen selected strains were ribotyped. Six RAPD types and 15 PFGE patterns were identified among all strains examined, and 12 ribotypes were found among the 15 strains that were ribotyped. The Georgian epidemic apparently was caused by one major clonal group of C. diphtheriae (PFGE type A, ribotype R1), which was identical to the predominant epidemic strain(s) isolated during the concurrent diphtheria epidemic in Russia. A dendrogram based on the PFGE patterns revealed profound differences between the minor (nonpredominant) epidemic strains found in Georgia and Russia. The methodologies for RAPD typing, ribotyping, and PFGE typing of C. diphtheriae strains were improved to enable rapid and convenient molecular typing of the strains. The RAPD technique was adequate for biotype differentiation; however, PFGE and ribotyping were better (and equal to each other) at discriminating between epidemiologically related and unrelated isolates.
After more than 30 years of excellent control, epidemic diphtheria has reemerged in the newly independent states (NIS) of the former Soviet Union (5, 27), including Georgia (9, 24). A diphtheria epidemic began in Moscow in 1990, reputedly among members of a military construction battalion, and spread rapidly throughout the country and the neighboring NIS (7). In Georgia, the epidemic began in 1993 with 28 registered cases, 4 of which were fatal. The number of cases increased to 312 in 1994 and peaked at 429 in 1995 (8, 9). A massive immunization campaign was initiated in the fall of 1995 with the help of the World Health Organization, the United Nations International Children’s Emergency Fund, U.S. Agency for International Development, and other international agencies (29). As a result of this effort, a 33% decline in diphtheria cases was observed from 1995 to 1997 (9). According to National Center for Diseases Control of Georgia data, there were 114 diphtheria cases in Georgia in 1998. At the present time, the epidemic appears to be under control; however, the rate of decline in the number of diphtheria cases in Georgia is only about half that observed in other NIS, emphasizing the need to better characterize the Georgian epidemic and to reevaluate the efficacy of implemented public health measures. In this context, it was of particular interest to characterize molecular epidemiological aspects of the Georgian epidemic and to determine whether the Georgian epidemic Corynebacterium diphtheriae strains were related to the strains responsible for the diphtheria epidemic in Russia. One review of the molecular epidemiology of diphtheria in Russia was recently published by Popovic et al. (20), and the same group reported later (10, 17) on the molecular subtyping of a small number of Georgian C. diphtheriae strains. However, extensive studies on Georgian epidemic-causing strains have not yet been reported, and data about similarities and differences among Georgian and Russian epidemic strains are not available in the peer-reviewed literature.
Several methods are available for the subtyping of C. diphtheriae strains; these include serotyping, phage typing (26), and more modern molecular typing techniques (ribotyping, analysis of DNA restriction fragment patterns, and others) (19, 22). Molecular typing techniques have been shown to be superior (i.e., to provide more specific and discriminating analysis) to serotyping and phage typing for studying the epidemiology of diphtheria outbreaks and for documenting the long-term persistence of C. diphtheriae strains in the population (1, 21), but data pertaining to the superiority of any one molecular typing method over another are insufficient. Ribotyping was reported to be an excellent technique for the typing of C. diphtheriae strains (2). However, it is rather laborious and, in several laboratories, is performed with radiolabeled riboprobes. Pulsed-field gel electrophoresis (PFGE), on the other hand, is generally recognized as the method of choice for typing most bacteria (13) and, while comparably demanding, does not involve the use of radioactive probes. Both methods are labor-intensive and time-consuming (several days are needed before the results can be documented). The randomly amplified polymorphic DNA (RAPD) technique (also called arbitrary primer PCR) is a much more rapid and technically less demanding technique, but its reproducibility has been reported to be of concern (28).
The aims of this study were to (i) identify the major strain(s) causing the diphtheria epidemic in Georgia, (ii) determine the genetic relatedness among the Georgian epidemic strain(s) and the strain(s) isolated during the diphtheria outbreak in Russia, and (iii) evaluate three techniques (ribotyping, PFGE, and RAPD) for the molecular typing of C. diphtheriae strains in order to determine the optimal approach for the molecular epidemiological characterization of diphtheria outbreaks. During the last studies, we also optimized the above methodologies in order to make them more suitable for rapid, easy, and reproducible typing of C. diphtheriae strains.
(This study was presented in part at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, Calif., 24 to 27 September 1998.)
MATERIALS AND METHODS
Bacterial strains.
A total of 66 C. diphtheriae strains isolated during the diphtheria epidemic of 1993 to 1998 in the Republic of Georgia were characterized in this study. Bacteriological identification of the strains was performed by use of standard microbiological techniques (3) at the National Center for Diseases Control of Georgia (51 strains) and at the Diphtheria Research Laboratory of the Centers for Disease Control and Prevention, Atlanta, Ga. (15 strains). In addition, 10 C. diphtheriae strains isolated during the concurrent diphtheria outbreak in Russia (5 of the gravis biotype and 5 of the mitis biotype) were analyzed. Three control C. diphtheriae strains (NCTC 10648, gravis, tox+; NTCC 10356, belfantii [diphtheria toxin negative]; and ATCC 13812 Park Williams 8, gravis, tox+ [12]) were used as reference strains in our studies.
Biotyping and toxigenicity testing.
Biotyping was performed according to the World Health Organization manual for the laboratory diagnosis of diphtheria (3). Toxigenicity was determined by the Elek immunoprecipitation method (3, 4). The strains also were tested for the presence of the diphtheria toxin gene by PCR amplification (with diphtheria toxin gene-specific primers) as described below.
PCR.
The PCR protocol was performed as described earlier (14) with a RoboCycler 96 thermal cycler (Stratagene, La Jolla, Calif.). The Tox 1 (5′-ATC CAC TTT TAG TGC GAG AAC CTT CGT CA-3′) and Tox 2 (5′-GAA AAC TTT TCT TCG TAC CAC GGG ACT AA-3′) primers were used to amplify a 248-bp fragment of the diphtheria toxin gene (tox) encoding the A subunit of the toxin (14, 18).
RAPD analysis.
The RAPD technique was performed with an RAPD kit (Amersham Pharmacia Biotech, Piscataway, N.J.) containing ready-to-go analysis beads. We examined 10 primers having various G+C contents (including 6 primers supplied with the kit and 4 primers developed at the University of Maryland School of Medicine) and a variety of amplification conditions (various concentrations of template and Mg2+ in the reaction buffer and cycling parameters) in order to determine the optimal conditions for RAPD typing. Primer 3 in the kit (5′-GTA GAC CCG T-3′) was superior to the other primers examined (i.e., it gave distinctive, reproducible patterns having three or more major bands); therefore, all subsequent RAPD typing was performed with this primer. Bacterial DNA for RAPD analysis was derived as previously described (14). The amplification conditions were 95°C for 5 min and then 45 amplification cycles. Each cycle consisted of sequential incubation at 95°C (1 min), 36°C (1 min), and 72°C (2 min). After the cycles, the samples were incubated at 72°C for 5 min and analyzed by electrophoresis with 2% agarose gels in Tris-acetate-EDTA buffer.
PFGE.
PFGE was performed with a CHEF DR-II apparatus (Bio-Rad Laboratories, Hercules, Calif.). We modified the procedure described for PFGE typing of Escherichia coli O157:H7 strains (6) in order to make it suitable for the rapid typing of C. diphtheriae strains. Briefly, bacteria from an overnight blood agar culture were suspended in and washed with 1 ml of 100 mM Tris-HCl buffer (pH 8) containing 100 mM EDTA. The cell suspension was diluted to an optical density at 610 nm of 14 to 15, and proteinase K (2 mg/ml, final concentration) was added to the suspension. An equal volume of 1.2% SeaKem Gold agarose (FMC BioProducts, Rockland, Maine) (in TE buffer (10 mM Tris-HCl, 1 mM EDTA) containing 1% sodium dodecyl sulfate [SDS]) was added, and plugs were cast with a standard casting tray. After the plugs solidified, they were incubated (2 h, 54°C) in 10 ml of 50 mM Tris-HCl buffer (pH 8) containing 50 mM EDTA, 1% sarcosyl, and 0.2 mg of proteinase K/ml and then rinsed in TE buffer (eight or nine changes of buffer, 10 min each). The procedure for making the plugs took 1 day.
Six restriction enzymes (NotI, XbaI, AvrII, EcoRV, SpeI, and SfiI) and a variety of electrophoresis parameters were examined in order to determine the optimal conditions for PFGE typing of the C. diphtheriae strains. In agreement with a previous report (2), SfiI cleavage yielded the optimal number of bands (>10 bands [25]), and we used that enzyme in all subsequent PFGE typing experiments. The plugs were incubated (30 min, room temperature) with restriction enzyme buffer, the DNA in the plugs was digested by incubation (50°C, 5 h) of the plugs with SfiI (New England Biolabs, Beverly, Mass.), and electrophoresis was performed with 1% SeaKem Gold agarose in 0.5× Tris-borate-EDTA buffer. The electrophoresis conditions were as follows. For block 1, the voltage was 183 V, the initial switch time was 8 s, the final switch time was 20 s, and the duration was 20 h; for block 2, the voltage was 183 V, the initial switch time was 5 s, the final switch time was 8 s, and the duration was 20 h. The PFGE patterns were normalized with lambda DNA and low-range PFGE molecular weight markers (Bio-Rad). This modified PFGE protocol, which includes a significantly shorter method for making plugs (1 day instead of the usual 4 days), allowed us to complete the entire procedure (making the plugs and performing the electrophoresis) in approximately 4 days instead of the usual 7 days (2).
Ribotyping.
Bacterial DNA for ribotyping was prepared as described above for PFGE, except that low-melting-point agarose was used instead of SeaKem agarose. Plugs prepared in this manner were preincubated (30 min, room temperature) in restriction enzyme buffer and digested (60°C, 6 h) with BstEII. Preheated (70°C) gel loading buffer (Quality Biologicals, Gaithersburg, Md.) was added to the digested DNA, the mixture was incubated at 70°C for 10 min, and the molten mixture was applied directly to a 1.2% agarose gel. Electrophoresis was performed with Tris-acetate-EDTA buffer (16 h), and the fractionated DNA was transferred to a MagnaGraph nylon membrane (MSI, Westboro, Mass.) by Southern blotting (23). The efficiency of transfer was determined by staining the gel with ethidium bromide (the absence of visible DNA in the gel after blotting indicated a high efficiency of transfer), and the DNA on the membrane was immobilized by UV cross-linking. A radiolabeled cDNA riboprobe was synthesized from a mixture of E. coli 16S and 23S rRNAs (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) by use of a first-strand cDNA synthesis kit (MBI Fermentas, Amherst, N.Y.) and [α-32P]dCTP (Amersham Pharmacia Biotech). The membrane was incubated (65°C, 4 h) with prehybridization buffer (6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 10× Denhardt solution, 0.5% SDS, 100 μg of salmon sperm DNA/ml), and hybridization was performed (65°C, for 16 h) with the riboprobe and fresh prehybridization buffer. After being rigorously washed (65°C, 2× SSC–0.5% SDS, 4 h, two times), the membrane was exposed (3 to 14 days at −80°C) to Biomax MR film (Eastman Kodak Co., Rochester, N.Y.) in a cassette with an enhancing screen, and the film was developed in an automatic film developer.
Analysis of PFGE and ribotyping patterns.
C. diphtheriae isolates were separated into patterns on the basis of two band differences (PFGE) and one band difference (ribotyping) (2, 25), and a dendrogram was constructed based on the PFGE patterns. The patterns were compared by means of the Dice coefficient with a Sun Microsystem (Bio Image, Ann Arbor, Mich.), and clustering of strains was based on the unweighted pair-group method with averages (a tolerance of 3% in the band position was applied). The computer-assisted analysis was performed according to the instructions of the manufacturer.
RESULTS
Biotyping and toxigenicity testing.
Sixty-two of the 66 Georgian C. diphtheriae strains examined in this study were of the gravis biotype, and 4 strains were of the mitis biotype. All strains (including the Russian and reference strains, except for the belfantii strain) were diphtheria toxin positive.
RAPD analysis.
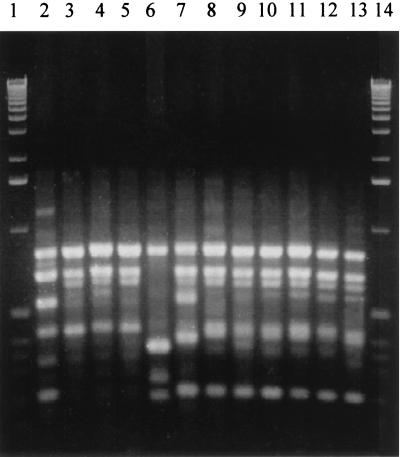
Three RAPD types were identified among the 66 Georgian strains: two RAPD types were found for the 4 mitis strains (AP 2, 3 strains; AP 3, 1 strain), and one RAPD type (AP 1) was identified for all 62 gravis strains (Fig. 1). Among the 10 Russian strains, one RAPD type (AP 1) was found for 5 gravis strains; the remaining 5 Russian strains (mitis) were of the AP 2 (4 strains) and AP 6 (1 strain) RAPD types. The reference strain ATCC 13812 (Park Williams 8) had a typical gravis RAPD type (AP 1); the two remaining reference strains, NCTC 10648 (gravis, tox+) and NCTC 10356 (belfantii), had unique RAPD types (AP 4 and AP 5, respectively). The assay was reproducible: 10 separate analyses of two test strains gave identical results for each strain.
FIG. 1.
RAPD patterns of C. diphtheriae strains. Lanes 1 and 14, molecular weight standards (1-kb ladder); lanes 2 through 5, strains of the mitis biotype, including RAPD types AP 3 (lane 2) and AP 2 (lanes 3, 4, and 5); lane 6, NCTC 10356 (negative control strain, AP 5); lane 7, NCTC 10648 (positive control strain, AP 4); lanes 8 through 13, strains of the gravis biotype, including RAPD type AP 1 Georgian strains (lanes 8, 9, and 10), Russian strains (lanes 11 and 12), and ATCC 13812 Park Williams 8 (lane 13).
PFGE.
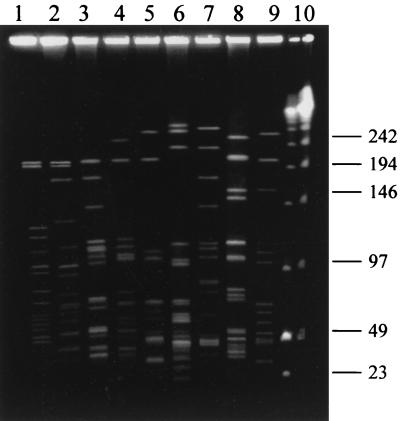
A total of 15 PFGE types were identified in the entire strain collection. Of those, seven types were identified among the 66 Georgian C. diphtheriae strains. The 62 gravis strains classified as a single type (AP 1) by RAPD typing were further differentiated by PFGE into five PFGE types (A, B, C, D, and G). Type A predominated (54 strains), followed by B (4 strains), D (2 strains), and G and C (1 strain each). The four Georgian mitis strains were classified as PFGE type E (three strains, RAPD type AP 2) and type F (one strain, RAPD type AP 3). The 10 Russian C. diphtheriae strains were grouped into six PFGE types (A, H, I, J, K, and L), of which one (containing 4 of the 5 Russian gravis strains) was the same as the predominant Georgian PFGE type (type A). Each of the three reference strains had a unique PFGE pattern (Fig. 2).
FIG. 2.
PFGE patterns of SfiI-digested DNA of C. diphtheriae strains. Lanes 1 through 4 and 6, strains of the mitis biotype, including PFGE types F (lane 1), E (lane 2), H (lane 3), L (lane 4), and J (lane 6); lanes 5 and 7 through 9, strains of the gravis biotype, including PFGE types I (lane 5), A (lane 7), M (lane 8, ATCC 13812), and N (lane 9, NCTC 10648); lane 10, combination of λ ladder and low-range PFGE size markers (kilobases).
Ribotyping.
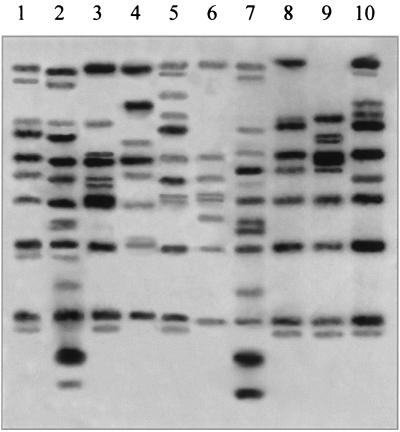
Fifteen strains (including at least one randomly chosen strain from each of the 15 PFGE types, except for G, K, and L) also were analyzed by ribotyping (Fig. 3). Six ribotype patterns (R 1, R 2, R 3, R 4, R 5, and R 6) were identified among the Georgian C. diphtheriae strains in this small collection, and an additional three patterns were found among the four Russian strains analyzed. In addition, the positive and negative control strains and the reference strain ATCC 13812 had unique ribotypes (R 11, R 12, and R 10, respectively). There was a strong correlation between the PFGE types and the ribotypes: all strains of one PFGE type were grouped in a single ribotype and vice versa. For example, all C. diphtheriae strains of PFGE type A belonged to a single ribotype (R 1), and all strains of ribotype R 2 belonged to a single PFGE type (B). Two predominant Russian epidemic strains reported (2) to be indistinguishable by PFGE but to have a minor difference in ribotyping patterns (G 1 and G 4) were indistinguishable by both PFGE typing and ribotyping in this study, and they were classified as PFGE type A, ribotype R 1. The correlation among biotypes, PFGE types, RAPD types, and ribotypes is shown in Table 1.
FIG. 3.
Ribotyping patterns of C. diphtheriae strains. Lanes 1, 2, 5, 7, 8, and 10, strains of the gravis biotype, including ribotypes R 1 (lane 1), R 4 (lane 2), R 8 (lane 5), R 3 (lane 7), R 10 (lane 8, ATCC 13812), and R 11 (lane 10, NCTC 10648); lanes 3, 4, and 6, strains of the mitis biotype, including ribotypes R 6 (lane 3), R 5 (lane 4), and R 9 (lane 6); lane 9, negative control strain NCTC 10356 (ribotype R 12).
TABLE 1.
Correlation among the biotypes, PFGE types, ribotypes, and RAPD types of the C. diphtheriae strains
| Strainsa | Biotype | PFGE type (location in Fig. 2) | Ribotype (location in Fig. 3) | RAPD type (location in Fig. 1) |
|---|---|---|---|---|
| 54/4 | Gravis | A (lane 7) | R 1 (lane 1) | AP 1 (lanes 8–13) |
| 4/0 | Gravis | B (NS)b | R 2 (NS) | AP 1 (lanes 8–13) |
| 1/0 | Gravis | C (NS) | R 3 (lane 7) | AP 1 (lanes 8–13) |
| 2/0 | Gravis | D (NS) | R 4 (lane 2) | AP 1 (lanes 8–13) |
| 3/0 | Mitis | E (lane 2) | R 5 (lane 4) | AP 2 (lanes 3–5) |
| 1/0 | Mitis | F (lane 1) | R 6 (lane 3) | AP 3 (lane 2) |
| 1/0 | Gravis | G (NS) | NDc (ND) | AP 1 (lanes 8–13) |
| 0/2 | Mitis | H (lane 3) | R 7 (NS) | AP 2 (lanes 3–5) |
| 0/1 | Gravis | I (lane 5) | R 8 (lane 5) | AP 1 (lanes 8–13) |
| 0/1 | Mitis | J (lane 6) | R 9 (lane 6) | AP 6 (NS) |
| 0/1 | Mitis | K (NS) | ND (ND) | AP 2 (lanes 3–5) |
| 0/1 | Mitis | L (lane 4) | ND (ND) | AP 2 (lanes 3–5) |
| ATCC 13812 | Gravis | M (lane 8) | R 10 (lane 8) | AP 1 (lanes 8–13) |
| NCTC 10648 | Gravis | N (lane 9) | R 11 (lane 7) | AP 4 (lane 7) |
| NCTC 10356 | Belfantii | O (NS) | R 12 (lane 10) | AP 5 (lane 6) |
Number of Georgian strains/number of Russian strains, unless otherwise indicated.
NS, not shown.
ND, not done.
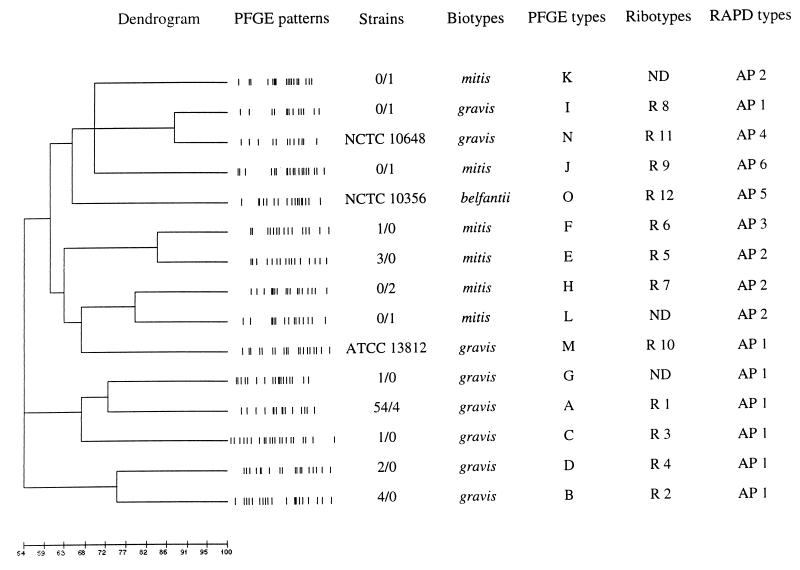
Dendrogram.
A dendrogram based on the patterns obtained by PFGE typing is shown in Fig. 4. There was a 100% relatedness between the major epidemic strains isolated in Georgia and Russia, and relatedness was lower (<75%) among the minor (nonpredominant) Georgian and Russian epidemic strains. Cluster analysis based on 11 of the 12 available ribotyping patterns (all except for R 7) revealed that the percentage of genetic relatedness among strains identified by this approach was identical to that seen in the dendrogram based on the PFGE patterns (data not shown).
FIG. 4.
Dendrogram portraying the genetic diversity of the Georgian and Russian epidemic C. diphtheriae strains. Representative PFGE patterns of SfiI-digested DNA of C. diphtheriae strains are shown. Data for strains are number of Georgian strains/number of Russian strains, unless otherwise indicated.
DISCUSSION
This paper is the first peer-reviewed publication to report on the molecular epidemiology of the 1990s diphtheria epidemic in Georgia and extends previously available information (15) on the molecular characterization of diphtheria strains isolated during an epidemic in an NIS besides Russia. In our study, the Georgian epidemic appeared to be linked to one major clonal group of C. diphtheriae (PFGE type A, ribotype R 1), which was identical to the predominant epidemic strain(s) isolated during the concurrent outbreak in Russia (ribotypes G1 and/or G4 [2]). This finding supports the idea that the Georgian epidemic was caused by the introduction of a major epidemic diphtheria strain from Russia, as has been hypothesized previously based on epidemiological data (8, 9). On the other hand, the minor epidemic strains found in Georgia and Russia were genetically heterogeneous (Fig. 4). Since the diphtheria toxin gene is known to be carried by a lysogenic bacteriophage and can integrate into the DNA of nontoxigenic strains, it is possible that several genetically diverse, nontoxigenic Georgian strains acquired the toxin gene from a newly introduced toxigenic strain of C. diphtheriae and became able to cause diphtheria. In addition, only a limited number of minor C. diphtheriae epidemic strains from Russia were analyzed in our study, limiting our ability to comprehensively evaluate similarities and differences among minor epidemic strains found in these two countries. More detailed characterization of the toxin genes in predominant Russian and Georgian epidemic strains and molecular typing of C. diphtheriae strains from Russian regions bordering Georgia will need to be done in order to address these possibilities.
All strains but one (including the Russian and reference strains but excluding the belfantii strain) were found to produce diphtheria toxin and to carry the toxin-encoding gene when examined by the Elek immunoprecipitation method and by PCR analysis, respectively. An excellent correlation has been reported (11, 16) between the results of the Elek test and PCR analysis that detects a specific fragment of the diphtheria toxin gene, and our observations are in agreement with that finding.
The diphtheria strains in our collection were first typed by the RAPD technique, and we found that this technique was useful for the preliminary rapid typing of C. diphtheriae strains. The assay was fast (ca. 5 h to the point at which results could be documented), and its reproducibility was excellent when major bands were compared. In addition, the RAPD technique differentiated well between strains of the gravis and mitis biotypes: 68 of the 69 Georgian and non-Georgian gravis strains characterized in this study had the same RAPD pattern (AP 1), and 1 gravis strain (the positive control strain NCTC 10648) had a slightly different (one major band difference) pattern (AP 4) (Fig. 1). Also, none of the nine mitis strains had the same RAPD patterns as the gravis strains. These results suggest that the RAPD technique can be used as a rapid method for the preliminary biotyping of C. diphtheriae strains or as an additional approach for biotype determination, especially when the results of traditional biotyping are not clear-cut. However, some epidemiologically unrelated strains were not differentiated by this technique, indicating that more discriminating typing methods must be used for a comprehensive molecular epidemiological evaluation of diphtheria outbreaks and epidemics.
PFGE and ribotyping are believed to possess good discriminating power for typing the majority of bacterial species (13). In our study, we found that PFGE and ribotyping were superior to the RAPD technique in discriminating between epidemiologically related and unrelated strains of C. diphtheriae. For example, PFGE and ribotyping of strain ATCC 13812 (PFGE type M, ribotype R 10) showed that it was distinct from the epidemiologically unrelated predominant Georgian epidemic strains (PFGE type A, ribotype R 1), whereas these strains were identified as having the same type (AP 1) by RAPD typing. Moreover, dendrogram analysis indicated that there was less than 55% genetic relatedness between strain ATCC 13812 and the predominant Georgian epidemic strains (Fig. 4).
We found that PFGE and ribotyping had identical discriminating power; i.e., the number of PFGE types was the same as the number of ribotypes, and all strains grouped in a PFGE type were grouped in a ribotype and vice versa. However, ribotyping has been reported (2) to be able to differentiate some C. diphtheriae strains having the same PFGE type. For example, two strains having the same PFGE type (A) were found to have distinct ribotypes (G1 and G4) in the above-referenced study. In our study, however, the same two strains were found to have a single ribotype (R 1). Several reasons may account for this discrepancy. First, the different conditions used for PFGE and ribotyping in the two studies may have contributed to the observed differences. This explanation seems likely because only minor, not reproducible from run to run, variations were observed (2) in the ribotyping patterns of the two strains. Second, a difference in three or more bands was used to distinguish PFGE types in the above-referenced study, whereas we used a difference in just two bands. Therefore, it is possible that some strains identified as having distinct PFGE types in our study were identified as being of the same PFGE type in the other study. Third, we ribotyped only a limited number of strains having the same PFGE type and therefore may not have included some strains having a single PFGE type that could be further differentiated by ribotyping. The discrepancy could also be due to genetic changes occurring in bacterial strains stored for a prolonged period of time and subcultured regularly; inversion, deletion, acquisition, or loss of mobile genetic elements (prophages, transposons, and so forth) may result in slight changes in PFGE and/or ribotyping patterns.
Our observation that PFGE and ribotyping have comparably good discriminatory power suggests that they could be useful, in combination or alone, for the molecular characterization of C. diphtheriae strains. The method to be used can be determined by the equipment available and the expertise of the laboratory staff. However, our modification of the PFGE procedure (which includes a significantly shorter protocol for making plugs) allows us to perform PFGE in a much shorter time than does the previously developed PFGE protocol or ribotyping and supports the use of PFGE when time is of the essence and the necessary equipment is available. Ribotyping with automated ribotyping equipment may be an attractive alternative but, at the present time, the equipment and supplies are prohibitively expensive for most laboratories. Therefore, we did not evaluate the use of an automated riboprinter for typing of the C. diphtheriae strains in this study.
ACKNOWLEDGMENTS
We thank Tanja Popovic and Izabella Mazurova for providing some of the C. diphtheriae strains and Levan Baidoshvili, Tinatin Kartvelishvili, Eka Zhorzholiani, and Naira Jamaspishvili for help in collecting epidemiological data about the diphtheria outbreak in Georgia. We thank Arnold Kreger for helpful discussions and editorial comments.
M.K. and P.I. were supported by an International Training and Research in Emerging Infectious Diseases grant from the Fogarty International Center, National Institutes of Health.
REFERENCES
- 1.Coyle M B, Groman N B, Russell J Q, Harnisch J P, Robin M, Holmes K K. The molecular epidemiology of three biotypes of Corynebacterium diphtheriae in the Seattle outbreak, 1972–1982. J Infect Dis. 1989;4:670–679. doi: 10.1093/infdis/159.4.670. [DOI] [PubMed] [Google Scholar]
- 2.De Zoysa A, Efstratiou A, George R C, Jahkola M, Vuopio-Varkila J, Deshevoi S, Tseneva G, Rikushin Y. Molecular epidemiology of Corynebacterium diphtheriae from northwestern Russia and surrounding countries studied by using ribotyping and pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:1080–1083. doi: 10.1128/jcm.33.5.1080-1083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Efstratiou A, Maple P A. WHO manual for the laboratory diagnosis of diphtheria. Reference no. ICP-EPI 038 (C). Geneva, Switzerland: World Health Organization; 1994. [Google Scholar]
- 4.Engler K H, Glushkevich T, Mazurova I, George R C, Efstratiou A. A modified Elek test for detection of toxigenic corynebacteria in the diagnostic laboratory. J Clin Microbiol. 1997;35:495–498. doi: 10.1128/jcm.35.2.495-498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galazka A M, Robertson S E, Oblapenko G P. Resurgence of diphtheria. Eur J Epidemiol. 1995;1:95–105. doi: 10.1007/BF01719954. [DOI] [PubMed] [Google Scholar]
- 6.Gautom R K. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol. 1997;35:2977–2980. doi: 10.1128/jcm.35.11.2977-2980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardy I R B, Dittmann S, Sutter R W. Current situation and control strategies for resurgence of diphtheria in newly independent states of the former Soviet Union. Lancet. 1996;347:1739–1744. doi: 10.1016/s0140-6736(96)90811-9. [DOI] [PubMed] [Google Scholar]
- 8.Imnadze P, Tsereteli Z, Baidoshvili L, Katsitadze G. Abstracts of the International Conference on Emerging Infectious Diseases. Atlanta, Ga: Centers for Disease Control and Prevention; 1998. Diphtheria epidemic in Georgia: current situation, abstr. P-14.12; p. 120. [Google Scholar]
- 9.Khetsuriani, N., P. Imnadze, and N. Dekanosidze. Diphtheria epidemic in Georgia, 1993–1997. J. Infect. Dis., in press. [DOI] [PubMed]
- 10.Kobaidze K, Quick L, Nakao H, Popovic T. Program and abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Molecular characterization of C. diphtheriae from the Republic of Georgia, abstr. C-347; p. 181. [Google Scholar]
- 11.Kobaidze, K., H. Nakao, L. Quick, and T. Popovic. Direct PCR for detection of toxigenic Corynebacterium diphtheriae from the Republic of Georgia after a prolonged storage. J. Infect. Dis., in press. [DOI] [PubMed]
- 12.Lampidis T, Barksdale L. Park-Williams number 8 strain of Corynebacterium diphtheriae. J Bacteriol. 1971;105:77–85. doi: 10.1128/jb.105.1.77-85.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maslow J N, Mulligan M E, Arbeit R D. Molecular epidemiology: application of contemporary techniques to the typing of microorganisms. Clin Infect Dis. 1993;17:153–164. doi: 10.1093/clinids/17.2.153. [DOI] [PubMed] [Google Scholar]
- 14.Mikhailovich V M, Melnikov V G, Mazurova I K, Wachsmuth I K, Wenger J D, Wharton M, Nakao H, Popovic T. Application of PCR for detection of toxigenic Corynebacterium diphtheriae strains isolated during the Russian diphtheria epidemic, 1990 through 1994. J Clin Microbiol. 1995;33:3061–3063. doi: 10.1128/jcm.33.11.3061-3063.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakao H, Pruckler J M, Mazurova I K, Narvskaia O V, Glushkevich T, Marijevski V F, Kravetz A N, Fields B S, Wachsmuth I K, Popovic T. Heterogeneity of diphtheria toxin gene, tox, and its regulatory element, dtxR, in Corynebacterium diphtheriae strains causing epidemic diphtheria in Russia and Ukraine. J Clin Microbiol. 1996;34:1711–1716. doi: 10.1128/jcm.34.7.1711-1716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakao H, Popovic T. Development of a direct PCR assay for detection of the diphtheria toxin gene. J Clin Microbiol. 1997;35:1651–1655. doi: 10.1128/jcm.35.7.1651-1655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakao H, Kim C, Kobaidze K, Popovic T. Program and abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. A new molecular subtyping assay for Corynobacterium diphtheriae: comparison with standard ribotyping, abstr. C-43; p. 128. [Google Scholar]
- 18.Pallen M J, Hay A J, Puckey L H, Efstratiou A. Polymerase chain reaction for screening clinical isolates of corynebacteria for the production of diphtheria toxin. J Clin Pathol. 1994;47:353–356. doi: 10.1136/jcp.47.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pappenheimer A M, Murphy J R. Studies on molecular epidemiology of diphtheria. Lancet. 1983;ii:923–926. doi: 10.1016/s0140-6736(83)90449-x. [DOI] [PubMed] [Google Scholar]
- 20.Popovic T, Kombarova S Y, Reeves M W, Nakao H, Mazurova I K, Wharton M, Wachsmuth I K, Wenger J D. Molecular epidemiology of diphtheria in Russia, 1985–1994. J Infect Dis. 1996;174:1064–1072. doi: 10.1093/infdis/174.5.1064. [DOI] [PubMed] [Google Scholar]
- 21.Popovic T, Kim C, Reiss J, Reeves M, Nakao H, Golaz A. Use of molecular subtyping to document long-term persistence of Corynebacterium diphtheriae in South Dakota. J Clin Microbiol. 1999;37:1092–1099. doi: 10.1128/jcm.37.4.1092-1099.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rappuoli R, Perugini M, Falsen E. Molecular epidemiology of the 1984–1986 diphtheria outbreak in Sweden. N Engl J Med. 1988;318:12–14. doi: 10.1056/NEJM198801073180103. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 9.34–9.46. [Google Scholar]
- 24.Sasse A, Malfait P, Padron T, Erikashvili M, Freixa E, Moren A. Outbreak of diphtheria in Republic of Georgia. Lancet. 1994;343:1358–1359. doi: 10.1016/s0140-6736(94)92491-0. [DOI] [PubMed] [Google Scholar]
- 25.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toshach S, Valentine A, Sigurdson S. Bacteriophage typing of Corynebacterium diphtheriae. J Infect Dis. 1977;136:655–660. doi: 10.1093/infdis/136.5.655. [DOI] [PubMed] [Google Scholar]
- 27.Vitek C R, Wharton M. Diphtheria in the former Soviet Union: reemergence of a pandemic disease. Emerg Infect Dis. 1998;4:539–549. doi: 10.3201/eid0404.980404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization. WHO/UNICEF strategy for diphtheria control in the newly independent states. Copenhagen, Denmark: Regional Office for Europe, World Health Organization; 1995. [Google Scholar]