Abstract
To investigate the function of histone-lysine N-methyltransferase 2D (KMT2D) on the methylation of H3 lysine 4 (H3K4) in the progression of Ovarian cancer (OV). KMT2D, ESR1 and H3K4me expressions in surgical resected tumors and tumor adjacent tissues of OV from 198 patients were determined using immunohistochemistry (IHC). Human OV cell lines including SKOV3, HO-8910 cells and normal ovarian epithelial cell line IOSE80 were employed for in vitro experiment, and BALB/C female nude mice were used for in vivo study. qRT-PCR and Western blotting were implemented for measuring the KMT2D, ESR1, PTGS2, STAT3, VEGFR2, H3K4me and ELF3 levels. Chromatin immunoprecipitation (ChIP) analysis was used for studying the binding between ESR1 and H3K4me. Edu staining assay was executed to determine cell viability, and colony formation and cell invasion assay. The immunofluorescence method was utilized for the visualization of protein expression and distribution in cells. In this study, KMT2D, ESR1 and H3K4me were found upregulated in OV progression. Mutated H3K4me could inhibit the proliferation, colony formation and invasion ability of OV cells. Mutated H3K4me could also hinder the ESR1 in SKOV3 expressions and HO-8910 cells, which would further mediate PTGS2/STAT3/VEGF pathway. In vivo studies also demonstrated that mutated H3K4me inhibited OV progression via targeting ESR1. All the ChIP-PCR analysis indicated the moderator effect of H3K4me on ESR1. Our findings indicated that ESR1 played an important role in the OV progression. Besides, H3K4me could promote cell proliferation and inhibit apoptosis of OV cells. Meanwhile, it could also targets the ESR1 production to enhance the migration and invasion of OV cells, which was through the activation of ESR1-ELF3-PTGS2-STAT3-VEGF cascade signaling pathway.
Keywords: ovarian cancer, chromatin immunoprecipitation, KMT2D, ESR1, H3K4me
Introduction
Ovarian cancer (OV), was known as one of the cancers which had the highest mortality and mobility, it has been regarded as one of the leading causes of cancer-related deaths in females 1 . Among all the cancers in the United States, 22,530 new cases and 13,980 deaths have been reported to be correlated to OV in the last year 2 . The majority of OV were epithelial ovarian carcinomas (EOCs), which could be further divided into various subtypes according to the tumor cell type while these subtypes might differ in the risk factors, molecular mechanisms or outcome 3 . Although considerable effects have been made in surgery and therapy of OV, the survival rate was not effectively improved and the outcome of patient remained poor, as a consequence of the lacking early specific symptoms and effective biomarkers 4 . Therefore, understanding the mechanisms in OV progression and mining the target for early diagnosis were of vital importance.
Methylation of histones has been widely studied and demonstrated to be participated in multiple genes expression. The protruding tails of histones were consisted of highly dynamic basic amino acids such as lysine and arginine that could be modified by the addition of methyl or other chemical groups 5 . Different methylation states that included mono-, di- and tri-methylation (me1, me2, and me3) had been observed at various lysine residues, such as H3 lysine 4 (H3K4) and H3K27 6 . A previous study showed that H3K4 trimethylation (H3K4me3) contributed to the metastasis of OV 7 . Another publication also demonstrated that chromatin H3K4me3 and H3K27me3 were highly correlated with ovarian tumor cells 8 . However, the potential molecular pathways have not been fully investigated concerning how the H3K4 methylation participated in the OV progression.
Histone-lysine N-methyltransferase 2D (KMT2D) was an essential enzyme in regulating methylation of H3K4 9 . Richard et al. reported that the level of transcription factor estrogen receptor α (ESR1), which was demonstrated to be involved in the OV 2 , was associated with H3K4me1 10 . Moreover, ESR1 would activate prostaglandin G/H synthase 2 (PTGS2) that facilitated migration and invasion of OV cells 11 . Previous studies indicated that forkhead box protein L2 (FOXL2) could decrease the PTGS2 level, while E74-like factor 3 (ELF3) could upregulate PTGS2 level 12,13 . Bioinformatics analysis revealed that PTGS2 might participate in regulating activator of transcription 3 (STAT3), which further regulate the vascular endothelial growth factor (VEGF) expression and promote angiogenesis and increase the risk of tumorigenesis. ELF3 as one of the transcription factor-encoding genes whose expression is significantly higher in long-term ovarian cancer survivors than short-term survivors. Based on the transcriptome analysis, we will hypothesize that ELF3 is a favorable prognostic marker for ovarian cancer and its expression suppresses cancer progression.
In the present study, KMT2D, ESR1 and H3K4me expressions in surgical resected tumors and tumor adjacent tissues of OV were determined using immunohistochemistry. Human OV cell lines including SKOV3, HO-8910 cells and normal ovarian epithelial cell line IOSE80 were employed for in vitro experiment, and BALB/C female nude mice was purchased for in vivo study. qRT-PCR and Western blotting was implemented for measuring the mRNA and protein levels. Edu staining, colony formation and cell invasion assay were implemented for determining cell proliferation and invasion. Immunofluorescence method was utilized for the visualization of protein expression and distribution.
Method
Patients and Tissue Samples
OV samples were obtained from Ovarian Tumor Center, the Affiliated Suzhou Municipal Hospital of Nanjing Medical University. In all, a total of 198 surgical resected tumors and 133 matched tumor adjacent tissues of OV from 198 patients were collected from January 2010 to June 2013 and 2014 to 2017, respectively. Among all the tumor samples, phase I (n = 38), phase II (n = 58), phase III (n = 60), and phase IV (n = 42) were separately collected. The study protocol was performed after approval by the institutional review board of the Affiliated Suzhou Municipal Hospital of Nanjing Medical University, and the study was conducted based on the principles of the Declaration of Helsinki. Informed consents were written by all the donors for their samples offer as per the International Ethical Guidelines.
Online Dataset
The expression of ESR1 in OV stages and survival outcome from TCGA database was analyzed through the UALCAN website (http://ualcan.path.uab.edu/analysis.html).
Immunohistochemistry
Immunohistochemistry (IHC) was conducted as per the instructions of manufacturer. Briefly, the constructed 2-µm paraffin wax sections were dewaxed in xylene and rehydrated in grade alcohol and blocked with 5% goat serum. Then, the primary antibody was incubated at 4°C overnight, which were as follows: KMT2D (ab224156, 1:800), ESR1 (ab16660, 1:500), H3K4me (ab185637, 1:800). The quantification of KMT2D, ESR1 and H3K4me was measured by two independent pathologists. The IHC scores were determined by the combination of the percentage of positive staining and the staining intensity 14 . The percentage of positive staining was evaluated as follows: 0, 0%–4%; 1, 5%–24%; 2, 25%–50%; 3, 51%–75%; 4, 76%–100%, while the staining intensity was graded as follows: 0, no staining; 1, weak staining (light yellow); 2, moderate staining (yellow–brown); 3, strong staining (brown). Finally, a score was assigned via the sum of the multiple between positive staining score and corresponding staining intensity score.
Western Blot
For the measurement of protein levels, western blot was performed based on published methods with slight modification 15 . The tissue samples or cells were digested or cracked for easy protein extraction. The collected proteins were preliminarily separated by SDS-PAGE and then applied on a PVDF membrane. Then the membrane was washed with PBS twice and incubated in blocking liquid for 2 h. After the removal of rest liquid, the membrane was probed by antibodies including Anti-KMT2D (ab231239, 1:1000), Anti-ESR1 (ab108398, 1:1000), Anti-H3K4me (ab8580, 1:1000), Anti-ELF3 (ab133621, 1:1000), Anti-PTGS2 (ab15191, 1:1000), Anti-STAT3 (ab68153, 1:1000), Anti-VEGFR2 (ab134191, 1:1000), and Anti-β-actin (ab8226, 1:500) that selected as a control. All the antibodies used were purchased from Abcam Co., Ltd. (Cambridge, USA). The corresponding secondary antibodies were applied together with ECL chromogenic solution. The density of protein bands was examined using an Image analyzer quantitative system.
Chromatin Immunoprecipitation (ChIP)
Cells cultured or collected from tissue samples were preliminarily fixed using 1% formaldehyde solution for 15 min, after which the fixation was terminated by glycine. Cells were washed with PBS and lysed with ChIP lysis buffer (containing 1% Triton X-100 and proteinase inhibitor). Subsequently, DNA gels were performed to ensure that the isolated chromatin was sheared into fragments of 500-800 bp approximately. The magnetic beads were prepared by incubation for 2 h with the H3K4me antibody (lgG was served as a negative control). The complex was washed with PBS and DNA was purified using a column. Finally, qRT-PCR was employed for detecting condensed DNA fraction.
Cell Cultures and Treatment
Human OV cell lines including SKOV3, HO-8910 cells and normal ovarian epithelial cell line IOSE80 were cultured based on the standard protocols. The adenovirus vector for KMT2D interference, the plasmid for point mutation at H3K4 site, as well as the activator and inhibitor of KMT2D were constructed by GenePharma Co., Ltd (Shanghai, China), which were subsequently transfected into SKOV3 and HO-8910 cell lines for 48 h, respectively. The group of non-transfected cells was prepared as model control group, while normal ovarian epithelial cell line IOSE80 was set as normal control group.
Edu Staining Assay
The proliferation of cells was detected with EdU assay (RiboBio, Guangzhou, China) based on the manufacturer’s instructions 16 . The cells from different groups were stained by 50 μmol/L Edu (5-ethynyl-2’-deoxyuridine) for 2 h and then fixed with 4% paraformaldehyde. The cells were reacted with glycine for 5 min, and incubated with 1% Triton X-100 for 10 min, after which Apollo® reaction cocktail was used and incubated with cells for 30 min. Subsequently, cells were incubated with the nuclear marker Hoechst 33342 for DNA staining in a dark room. Finally, the treated samples were mounted in ProLong Gold on glass microscope slides, which were sealed with nail polish and were stored at 4°C.
Colony Formation Assay
Cells (1 × 103 ) were inoculated in culture dishes and cultured for 14 days. After fixation with 4% formaldehyde, 1% crystal violet was added for staining the colonies and then the colonies were counted for quantification.
Cell invasion Assay
Invasion assay was conducted using a 24-well Boyden chambers (Corning, CA, USA) with 8M-inserts coated with fibronectin (Roche, MA, USA) and Matrigel (BD, CA, USA). Cells (1 × 103 ) from each group were inoculated on the upper chamber. The cells on the bottom of the upper chamber were counted for quantification after 24 h.
Immunofluorescence
The immunofluorescence method was utilized for the visualization of protein expression and distribution in cells. Briefly, 4% formaldehyde was obtained for fixing cells, which were then permeated by 0.1% 100X Triton. Cells were subsequently blocked with 10% goat serum for 1 h and incubated with primary antibody Anti-KMT2D (ab231239, 1:1000), Anti-ESR1 (ab108398, 1:1000), Anti-H3K4me (ab8580, 1:1000) at 4°C overnight. The cells were washed with PBS and incubated with fluorescent-labeled secondary antibody correspondingly. The results were observed and examined using a fluorescence microscope.
Tumor Xenograft in Nude Mice
All animal experiments were conducted in the Affiliated Suzhou Municipal Hospital of Nanjing Medical University. Totally, 62 SPF BALB/C (nu/nu) female nude mice at 6 weeks of age were purchased from SLAC Laboratory Animal Co., Ltd (Shanghai, China) and housed in a clean air laminar flow rack in a sterile laboratory (25°C ± 2°C for constant temperature, 45%–50% for constant humidity). After adapting to feeding for one week, the nude mice were divided into 6 groups (n = 10 in each group): (1) normal group; (2) model control group; (3) KMT2D activator group; (4) KMT2D inhibitor group; (5) KMT2D-shRNA group; (6) H3K4me mutation group. Totally 100 µL of SKOV3 cell suspension (4 × 107/mL) in logarithmic growth period was inoculated subcutaneously in the right armpit of two nude mice, respectively. After 10 days, the transplanted tumor was formed, and the nude mice were sacrificed by CO2 + isoflurane anesthesia. The tumor tissue in the vigorous growth period was cut to about 1.0 mm3 and subcutaneously inoculated into the other groups of nude mice except the normal group under sterile conditions. Meanwhile, the nude mice of KMT2D activator group, KMT2D inhibitor group, KMT2D-shRNA group and H3K4me mutation group were injected with KMT2D protein activator, KMT2D inhibitor, adenovirus interference plasmid of KMT2D gene and point mutation plasmid of H3K4me site by tail vein, respectively. When the size of the transplanted tumor in each group of mice was about 150 mm3 (about 20 days), the mice were sacrificed after anesthesia by CO2 + isoflurane, the mouse ovarian tissue was collected, and the volume of the mouse ovarian tumor tissue was calculated. The relative mRNA expression of ESR1, ELF3, and STAT3 genes was detected by qRT-PCR, and western blotting was conducted to determine the protein levels of KMT2D, ESR1, H3K4me, PTGS2, and VEGFR2 proteins, ChIP-PCR was used to analyze the regulatory characteristics of H3K4me and ESR1 histones.
Real-Time Reverse Transcription Quantitative PCR (qRT-PCR)
After measuring the tumor volume, the mice were sacrificed and dissected to obtain ovarian tumor tissue. RNAiso Plus Kit (Takara, Dalian, China) was obtained for extracting the total RNA within tissues, and Reverse Transcription kit (Thermo Scientific, Waltham, USA) was used to reverse transcribe the RNA into cDNA, and the quantity and purity were evaluated. The reverse transcription product of cDNA was stored at −20°C. The Roche Light Cycler96 real-time fluorescent quantitative PCR system (Roche, Indiana, USA) was used to perform qRT-PCR amplification of c DNA samples under the following conditions: pre-denaturation of 95°C, 30 s, 1 cycle; 40 cycles of 95°C, 5 s and 60°C, 30 s; the last cycle of 95°C, 15 s; 60°C, 30 s; 95°C, 15 s. The specific primers used in qRT-PCR are summarized in Table 1. The level of the target gene was calculated via normalization to the level of internal NADPH using the 2-ΔΔCt method.
Table 1.
PCR primer sequences.
| Gene | Primer-F | Primer-R |
|---|---|---|
| ESR1 | CCCACTCAACAGCGTG TCTC | CGTCGATTATCTGAAT TTGGCCT |
| ELF3 | CATGACCTACGAGAAG CTGAGC | GACTCTGGAGAACCT CTTCCTC |
| STAT3 | TTCACTTGGGTGGA GAAG | CGGACTGGATCTGG GTCT |
Statistical Analysis
All data were expressed in the form of mean ± standard deviation (SD), with each value repeated three times. Student’s t-test of SPSS 20.0 software was used for comparison between the two groups, and Turkey test for One-way ANOVA of SPSS 20.0 software was used for comparison between multiple groups. The P value < 0.05 was regard as a significant difference.
Result
ESR1 Was Upregulated in OV Progression
We preliminarily employed TCGA database to investigate the ESR1 role in OV progression. It was found that ESR1 was significantly upregulated in OV stage II–IV than that in stage I (Fig. 1A). The high ESR1 level was considered to be linked with low survival probability. The collected clinical samples were also grouped as per the pathological feature, FIGO stage. As shown in Fig. 1B, C, the IHC outcome and western blot results presented that ESR1 was barely expressed in adjacent tumor tissues, whereas it was of growing increase as OV progression. KMT2D and H3K4me exhibited the consistent expression with ESR1 that there was a significant difference of expression between the stage I and stage III or stage IV (P < 0.05). The western blot results (Fig. 1C) showed the PTGS2, STAT3 and VEGFR2 expressions were in correlation of ESR1, indicating the existence of underlying regulation relationship. The ChIP-PCR analysis (Fig. 1D) further demonstrated the moderator effect of H3K4me on ESR1. All these findings indicated a promoting role of ESR1 in the OV progression.
Figure 1.
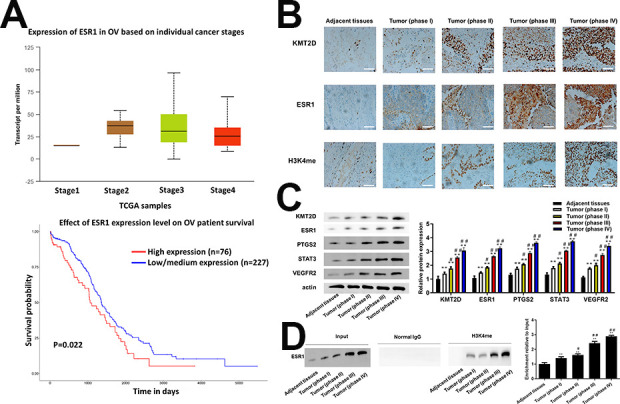
(A) ESR1 expression in OV stage II–IV was higher than stage I and Fig. 1B low survival probability related to high ESR1 level from TCGA (P = 0.022) (B) Immunohistochemical staining images (up) and scores (down) of KMT2D, ESR1 and H3K4me in tumor tissues of phase I (n = 38), phase II (n = 58), phase III (n = 60) and phase IV (n = 42) OV patients and adjacent tissues (including tumor samples and adjacent tissues.). (phase I vs phase II P < 0.05, phase II vs phase III P < 0.001, phase III vs phase IV N.S., two-sided unpaired t-test). (C) The protein expression of KMT2D, ESR1, PTGS2, STAT3, VEGFR2 in tumor tissues of phase I (n = 38), phase II (n = 58), phase III (n = 60) and phase IV (n = 42) OV patients and adjacent tissues (including 198 tumor samples and 133 adjacent tissues). (phase I vs phase II P < 0.05, phase II vs phase III P < 0.001, phase III vs phase IV N.S., two-sided unpaired t-test). (D) The binding of H3K4me and ESR1 was demonstrated using CHIP analysis and the relative expression of ESR1 was calculated via normalized to the Input group. Scale bar: 50 μm. *P < 0.05 vs adjacent tissues, **P < 0.01 vs adjacent tissues, # P < 0.05 vs tumor (phase I), ## P < 0.01 vs tumor (phase I).
Mutated H3K4me Inhibited the Proliferation, Colony Formation and Invasion Ability of OV Cells
To further explore the role of H3K4me and ESR1 in OV progression, we employed two cell lines to construct the H3 overexpression group using KMT2D activator, H3 knockdown group by H3 inhibitor or KMT2D-shRNA, and the mutated H3K4me group.
H3 overexpression promoted while H3 knockdown reduced cell proliferation (Fig. 2, P < 0.05), colony formation (Fig. 3A, P < 0.05), and invasion ability in both SKOV3 and HO-8910 cell lines (Fig. 3B, P < 0.05) as compared with the model control group, and almost recovered to the level in IOSE80 cell lines. Notably, when the H3K4me got mutated, the capability of cell proliferation, colony formation and invasion, though slightly higher than H3 knockdown group, was significantly inhibited compared with model control group.
Figure 2.
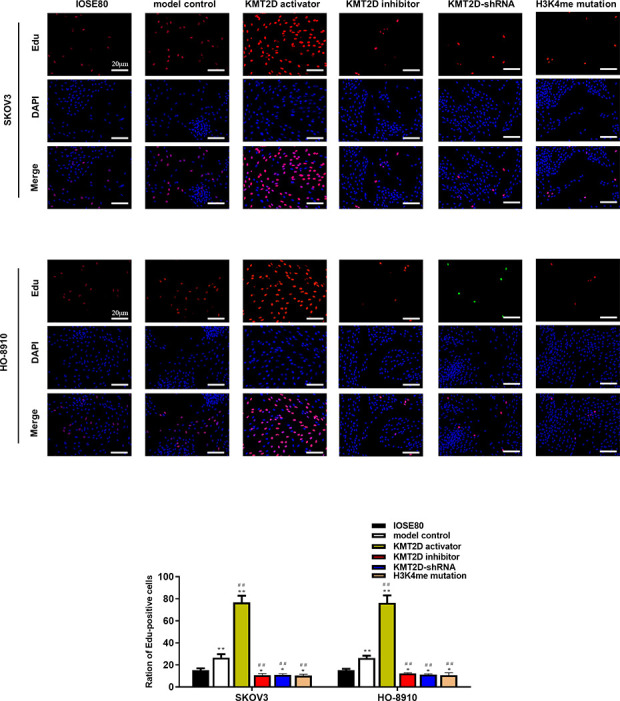
Representative micrographs of EdU and DAPI staining in IOSE80, SKOV3 and HO-8910 cell lines visualized by epifluorescence. Red dots (stained by EdU) represented cells whose DNA were rapidly replicated, and blue dots (stained by DAPI) revealed the distribution of nucleus. Scale bar: 20 μm. *P < 0.05 vs IOSE80, **P < 0.01 vs IOSE80, # P < 0.05 vs model control group, ## P < 0.01 vs model control group.
Figure 3.
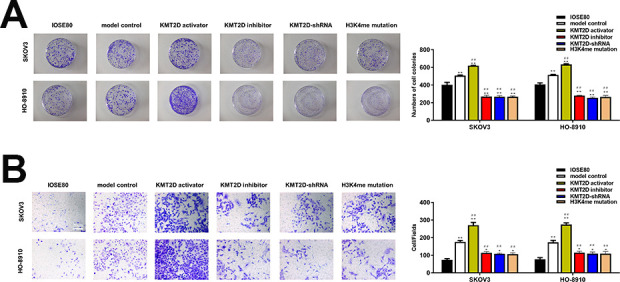
(A) The colony formation of IOSE80, SKOV3, and HO-8910 cell lines. Cells were cultured for 14 days in standard culture dishes and colonies stained with crystal violet were captured and counted. (B) Cell invasion ability of IOSE80, SKOV3 and HO-8910 cell lines detected by transwell assay. *P < 0.05 vs IOSE80, **P < 0.01 vs IOSE80, # P < 0.05 vs model control group, ## P < 0.01 vs model control group.
Mutated H3K4me Hindered the Expression of ESR1
Subsequently, we detected the KMT2D, ESR1, and H3K4me expressions and distributions in each group using immunofluorescence method. As shown in Fig. 4A, KMT2D, ESR1, and H3K4me all located at the nucleus of cells, and evident interaction could be seen among them. Besides, the model control group of SKOV3 cells exhibited higher KMT2D, ESR1, and H3K4me levels than normal control group, which was demonstrated again using western blot method (Fig. 4B). In consistence with immunofluorescence results, the western blot bands suggested the ESR1 level was significantly upregulated in KMT2D activator group than model control group (P < 0.05), while downregulated in H3 inhibitor group and KMT2D-shRNA group (P < 0.05). Importantly, the ESR1 expression was vastly decreased compared with model control group when the H3K4me got mutated, showing H3K4me played a major role in the ESR1 upregulation.
Figure 4.
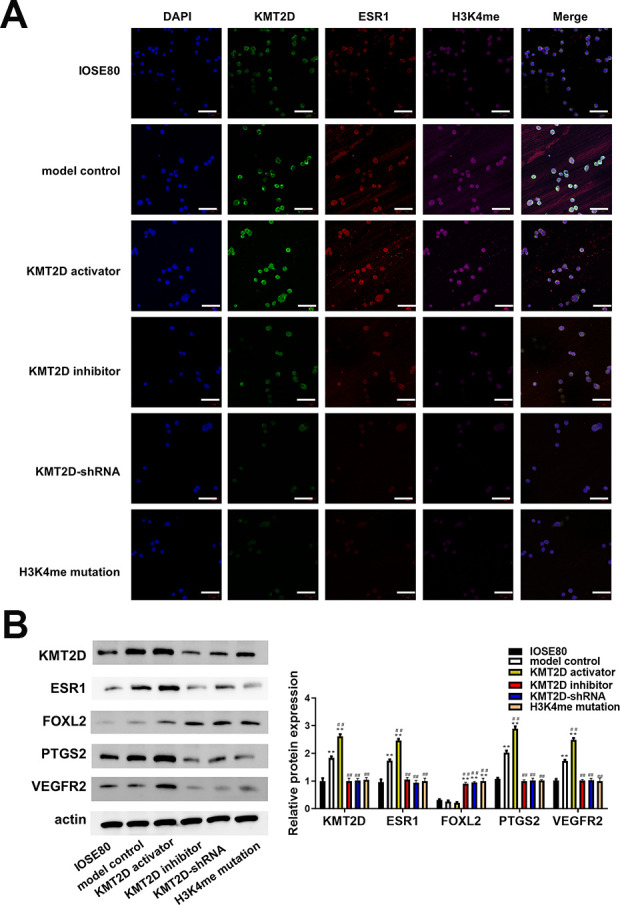
(A) Expression of KMT2D, ESR1, H3K4me in IOSE80 and SKOV3 cell lines were determined using immunofluorescence and located by DAPI staining. (B) Expression of KMT2D, ESR1, FOXL2, PTGS2, and VEGFR2 detected by western blotting. The relative expression was calculated via normalization to the expression of actin. Scale bar: 20 μm. *P < 0.05 vs IOSE80, **P < 0.01 vs IOSE80, # P < 0.05 vs model control group, ## P < 0.01 vs model control group.
ESR1 Mediated PTGS2/STAT3/VEGF Pathway In Vitro
The downstream molecules of ESR1 including PTGS2, STAT3 and VEGF were detected by western blot. In agreement of clinical samples, the PTGS2, STAT3 and VEGF expressions were positively regulated by ESR1 (Fig. 4B). Notably, the transcription regulation factor FOXL2 that could inhibit the influence of ESR1 on PTGS2, was hardly expressed in the highly presence of ESR1. Whereas, the FOXL2 level was increased when the KMT2D or ESR1 declined, indicating the underlying modulation between ESR1 and FOXL2.
Mutated H3K4me Inhibited OV Progression Via Targeting ESR1 In Vivo
In order to further reflect the function of ESR1 on the OV progression, in vivo proof experiments were carried out. From the results of OV tumor mass size in Fig. 5A, it could be seen that the tumor size of H3K4me mutation group was significantly reduced compared to the model control group, while the change trend of tumor size of the KMT2D inhibitor and KMT2D-shRNA groups was consistent with it. In contrast, the tumor size of the KMT2D activator group was significantly increased. The results of qRT-PCR analysis (Fig. 5B) showed that the ESR1, ELF3, and STAT3 mRNA expression levels in the model control group and KMT2D activator group were significantly higher than those in the normal group, KMT2D inhibitor group, KTMT2D-shRNA group and H3K4me mutation group. The western blotting results (Fig. 5C) were consistent with the results of our clinical analysis, that was, the.KMT2D, ESR1, H34Kme, PTGS2, and VEGFR2 protein expressions in the model control group and KMT2D activator group were significantly higher than those in the normal group, KMT2D inhibitor group, KMT2D -shRNA group, and H3K4me mutation group. ChIP-PCR analysis (Fig. 5D) verified that the H3K4me protein could bind to the ESR1 gene, it showed that the input DNA expression of the model group and the KMT2D agonist group was significantly increased compared with other groups, and the H3K4me protein expression of these two groups also increased significantly compared with other groups. These results indicated that the combination of H3K4me and ESR1 activated the ESR1-ELF3-PTGS2-STAT3-VEGF signaling pathway in vivo, thereby promoting the development of OV tumors in vivo.
Figure 5.
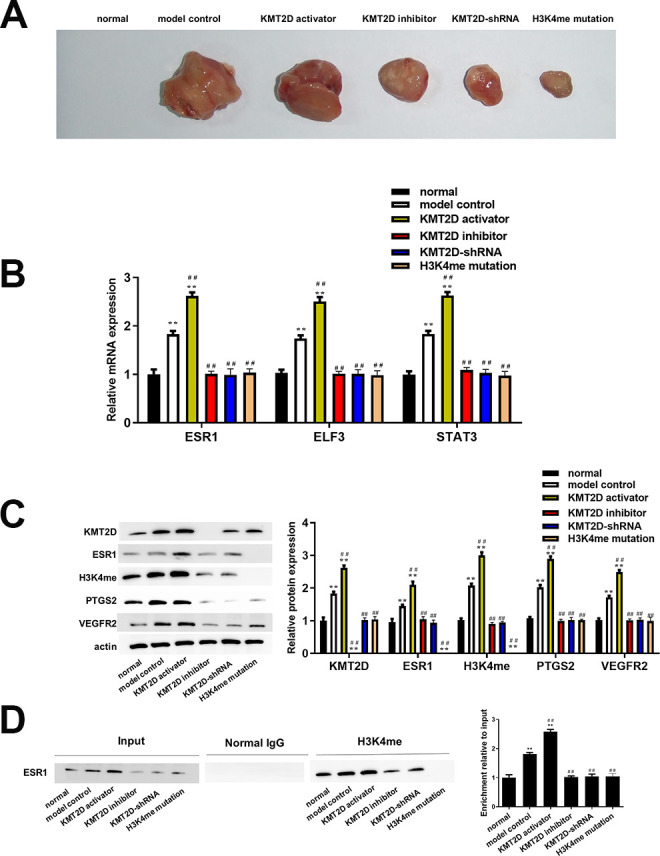
(A) OV tumor size in mice was measured by ruler (mm3). (B) Relative mRNA expression of ESR1, ELF3, and STAT3 determined using qRT-PCR and calculated via normalization to the mRNA level of GAPDH. (C) Expression of KMT2D, ESR1, H4K3me, PTGS2, and VEGFR2 detected by western blotting. The relative expression was calculated via normalization to the expression of actin. (D) The binding of H3K4me and ESR1 was demonstrated using CHIP analysis and the relative expression of ESR1 was calculated via normalized to the Input group. *P < 0.05 vs IOSE80, **P < 0.01 vs IOSE80, # P < 0.05 vs model control group, ## P < 0.01 vs model control group.
Discussion
OV is a deadly gynecological malignant tumor, and the mortality rate ranks first among various gynecological tumors. More and more studies have shown that histone modification, especially histone methylation modification, is closely related to the OV occurrence and development 17 –19 . In this work, we found that the expression of ESR1 showed a significant positive correlation with the OV development through clinical organization and data analysis. In vitro cell experiment results show that the H3K4me mutation can significantly inhibit the proliferation, colony formation and invasion ability of OV cells. At the same time, the PTGS2/STAT3/VEGF pathway is activated by significantly increasing the expression of ESR1. Besides, we further confirmed that H3K4me could target ESR1 to activate the PTGS2/STAT3/VEGF pathway, thereby affecting the OV progression.
The ovary is the main part of estrogen synthesis. During the process of cancer formation, estrogen regulates the main physiological activities of ovarian epithelium through its specific hormone receptor subtypes, including regulating balance, hyperplasia, and abnormal differentiation. However, the role of estrogen receptors and the mechanism that induces cancer are currently unclear. Our results showed that ESR1 expression was significantly increased in OV tissues, and the KMT2D and H3K4me expressions were also significantly increased. It was reported that the increased KMT2D activity would promote the recruitment of ESR1 by chromatin helix, thus promoting the cancer development 20 , which suggested that KMT2D may have the function of activating ESR1, and ESR1 may play an important role in the process of OV. Other studies had also obtained similar results 21,22 . It is speculated that ESR1 may activate relevant cancer signaling pathways and lead to the proliferation of OV cells. Therefore, the high ESR1 in OV expression can be used as a feature of metastatic changes in OV. Although the cause of cancer promotion of ESR1 is not known, many investigations had shown that the regulatory effect of ESR1 may be related to the amount of estrogen. After the formation of estrogen, ESR1 overexpression was shown in the urogenital system, that was, it was exposed to estrogen, and finally promoted the formation of oncogenes. Our results indicated that ESR1 could significantly promote the OV progress.
H3K4 methylation modification is usually associated with gene transcription activation and plays an important role in regulating gene expression 23,24 . There are few reports on solid tumors with abnormal H3K4me1 expression, as well as OV. The H3K4me1 expression was significantly increased in colorectal cancer cells, and KMT2C induced the deposition of H3K4me1, suggesting that KMT2C made an important contribution to the expression of H3K4me1 25 . Similarly, a strong relationship between H3K4me1 and KMT2D has been reported in many literatures 26 –28 . On the question of H3K4me1, we observed consistent KMT2D and H3K4me expressions in both cell and animal models, and the mutation of H3K4me caused a significant decrease in the KMT2D and ESR1 expressions in the OV cells, a possible explanation for this might be that KMT2D may accelerate the H3K4me1 deposition, leading to an accelerated process in OV. Apart from H3K4me1, H3K4me2 may also participate in the OV process, some observations and experimental data further support the concept of H3K4me2 as an OV-promoting gene 29 –31 . Consistent with our results, our results showed that the H3K4me expression gradually increased as OV progresses. At present, the mechanism and path of H3K4me2 action in tumor cells are not yet clear, but several in vivo and in vitro studies have shown that changing the H3K4me2 expression may have potential significance for the OV treatment. As mentioned in the literature review, there was a close relationship between H3K4me3 and oncogene transcription efficiency 32 . H3K4me3 is highly aggregated in the 5’ region of transcriptionally active genes, the higher H3K4me3 expression, the slower apoptosis of tumor cells 33 . Abnormal H3K4me3 expression can also cause epigenetic changes, which can regulate tumor-related protein expression and tumor-related functions (proliferation, etc.) 34 . Therefore, abnormal H3K4me3 expression has become a mechanism responsible for tumor-related changes in the epigenome 35 . In vivo experiments showed that the H3K4me mutant caused a significant reduction in OV tumor volume, this also accorded with the observations of other studies 36 –38 . Based on the results of this study, we speculated that measuring changes in H3K4me expression may provide important clues for assessing the OV patients’ prognosis. Although the methylation of three H3K4 modes is related to the transcriptional activation of cancer, the overall change in gene expression in cells may depend on the complex interactions between induced target genes 39,40 . Therefore, we cannot rule out the possibility that one or several H3K4 methylation patterns synergistically promote cancer development.
ETS transcription factor 3 (ELF3) belongs to the ETS transcription factor family. Studies had found that it was closely related to malignant phenotypes such as tumor growth, invasion and metastasis 41 . Prostaglandin-endoperoxide synthase 2 (PTGS2, also known as Cox2) is an enzyme responsible for catalyzing the formation of pathological prostaglandin E2 (PGE2), it is involved in tumor growth, angiogenesis, and infiltration 42 . Signal transducer and activator of transcription 3 (STAT3) is the convergent point of various oncogenic signaling pathways, which can regulate the transcriptional activity of tumor cell proliferation, invasion, migration and other related target genes. Vascular endothelial growth factor (VEGF) is a highly specific vascular growth factor, which can significantly increase the integrin expression level on endothelial cells and malignant tumor cells, and contribute to the migration, adhesion and infiltration of malignant tumor cells 43 . The current study found that the PTGS2, STAT3, and VEGFR2 expressions in OV tissue increased significantly, and were related to the ESR1expression. ELF3 could be used as a gene marker for lymph node metastasis of colorectal cancer 44 . Bioinformatics analysis showed that the STAT3 expression could be regulated by PTGS2, the PTGS2 expression could be active by ELF3, and the PTGS2 was overexpressed in most OV tissues 45 . The PTGS2 expression was significantly negatively correlated with the survival time of OV 46 . The specific mechanism may be that PTGS2 promoted the OV process by accelerating endothelial cell proliferation, division, and regulating p53 and/or bcl-2 signaling pathways. The results of this study showed that the PTGS2 expression was significantly negatively correlated with the cancer stage and OV tumor size, suggesting that PTGS2 may be related to the OV progression and participate in tumor invasion and metastasis. Therefore, PTGS2 detection helps to reflect the biological behavior and clinical staging of OV. It is well known that STAT3 and VEGF signaling pathways played an important role in tumor progression 47,48 . These findings supported our results that STAT3 and VEGFR2 were highly expressed in OV tissues.
Overall, our results indicated that ESR1 played an important role in the OV progression. Besides, H3K4me could promote cell proliferation and inhibit apoptosis of OV cells. Meanwhile, it could also target the production of ESR1 to enhance the migration and invasion of OV cells, which was related to the activation of the ESR1-ELF3-PTGS2-STAT3-VEGF cascade signaling pathway.
Conclusion
ESR1 played an important role in the OV progression. Besides, H3K4me could promote cell proliferation and inhibit OV apoptosis. Meanwhile, it could also target the ESR1 production to enhance the migration and invasion of OV cells, which was through the activation of ESR1-ELF3-PTGS2-STAT3-VEGF cascade signaling pathway.
Acknowledgments
We would like to acknowledge everyone for their helpful contributions on this paper.
Ethical Approval: Ethical approval was given by the Affiliated Suzhou Municipal Hospital of Nanjing Medical University, Suzhou, China.
Statement of Human and Animal Rights: All procedures in this study were conducted in accordance with the Affiliated Suzhou Municipal Hospital of Nanjing Medical University, Suzhou, China.
Statement of Informed Consent: Written informed consent was obtained from the patients for their anonymized information to be published in this article.
Authors’ contributions: Each author has made an important scientific contribution to the study and has assisted with the drafting or revising of the manuscript.
Availability of data and materials: The data are free access to available upon request.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Qing Lv  https://orcid.org/0000-0003-2968-5237
https://orcid.org/0000-0003-2968-5237
References
- 1. Mihanfar A, Fattahi A, Nejabati HR. MicroRNA-mediated drug resistance in ovarian cancer. J Cell Physiol. 2019;234(4):3180–3191. [DOI] [PubMed] [Google Scholar]
- 2. Wang K, Zhu G, Bao S, Chen S. Long non-coding RNA LINC00511 mediates the effects of ESR1 on proliferation and invasion of ovarian cancer through miR-424-5p and miR-370-5p. Cancer Manag Res. 2019;11:10807–10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones BA, Varambally S, Arend RC. Histone methyltransferase ezh2: a therapeutic target for ovarian cancer. Mol Cancer Ther. 2018;17(3):591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nezhat FR, Apostol R, Nezhat C, Pejovic T. New insights in the pathophysiology of ovarian cancer and implications for screening and prevention. Am J Obstet Gynecol. 2015;213(3):262–267. [DOI] [PubMed] [Google Scholar]
- 5. Leszinski G, Gezer U, Siegele B, Stoetzer O, Holdenrieder S. Relevance of histone marks H3K9me3 and H4K20me3 in cancer. Anticancer Res. 2012;32(5):2199–2205. [PubMed] [Google Scholar]
- 6. Zhao S, Zhong Y, Fu X, Wang Y, Ye P, Cai J, Liu Y, Sun J, Mei Z, Jiang Y, Liu J. H3K4 methylation regulates LPS-induced proinflammatory cytokine expression and release in macrophages. Shock. 2019;51(3):401–406. [DOI] [PubMed] [Google Scholar]
- 7. Lyu T, Jiang Y, Jia N, Che X, Li Q, Yu Y, Hua K, Bast RC, Jr, Feng W. SMYD3 promotes implant metastasis of ovarian cancer via H3K4 trimethylation of integrin promoters. Int J Cancer. 2020;146(6):1553–1567. [DOI] [PubMed] [Google Scholar]
- 8. Chapman-Rothe N, Curry E, Zeller C, Liber D, Stronach E, Gabra H, Ghaem-Maghami S, Brown R. Chromatin H3K27me3/H3K4me3 histone marks define gene sets in high-grade serous ovarian cancer that distinguish malignant, tumour-sustaining and chemo-resistant ovarian tumour cells. Oncogene. 2013;32(38):4586–4592. [DOI] [PubMed] [Google Scholar]
- 9. Froimchuk E, Jang Y, Ge K. Histone H3 lysine 4 methyltransferase KMT2D. Gene. 2017;627:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cowper-Sal lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, Moore JH, Lupien M. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet. 2012;44(11):1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nagaraja AS, Dorniak PL, Sadaoui NC, Kang Y, Lin T, Armaiz-Pena G, Wu SY, Rupaimoole R, Allen JK, Gharpure KM, Pradeep S, et al. Sustained adrenergic signaling leads to increased metastasis in ovarian cancer via increased PGE2 synthesis. Oncogene. 2016;35(18):2390–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yenuganti VR, Vanselow J. Oleic acid induces down-regulation of the granulosa cell identity marker FOXL2, and up-regulation of the Sertoli cell marker SOX9 in bovine granulosa cells. Reprod Biol Endocrinol. 2017;15(1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wondimu EB, Culley KL, Quinn J, Chang J, Dragomir C, Plumb D, Goldring M, Otero-Adran M. The E74-like factor 3 (ELF3) is a central mediator of cartilage degradation in a surgically-induced osteoarthritis model in mice. Osteoarthr Cart. 2015;23:A288. [Google Scholar]
- 14. Li Q, Qin T, Bi Z, Hong H, Ding L, Chen J, Wu W, Lin X, Fu W, Zheng F, Yao Y, et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat Commun. 2020;11(1):1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peng PH, Chieh-Yu Lai J, Hsu KW, Wu KJ. Hypoxia-induced lncRNA RP11-390F4.3 promotes epithelial-mesenchymal transition (EMT) and metastasis through upregulating EMT regulators. Cancer Lett. 2020;483:35–45. [DOI] [PubMed] [Google Scholar]
- 16. Gao Q, Gu Y, Jiang Y, Fan L, Wei Z, Jin H, Yang X, Wang L, Li X, Tai S, Yang B, et al. Long non-coding RNA Gm2199 rescues liver injury and promotes hepatocyte proliferation through the upregulation of ERK1/2. Cell Death Dis. 2018;9(6):602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Borghese B, Zondervan KT, Abrao MS, Chapron C, Vaiman D. Recent insights on the genetics and epigenetics of endometriosis. Clin Genet. 2017;91(2):254–264. [DOI] [PubMed] [Google Scholar]
- 18. Chung VY, Tan TZ, Tan M, Wong MK, Kuay KT, Yang Z, Ye J, Muller J, Koh CM, Guccione E, Thiery JP, et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci Rep. 2016;6:19943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuang Y, Lu F, Guo J, Xu H, Wang Q, Xu C, Zeng L, Yi S. Histone demethylase KDM2B upregulates histone methyltransferase EZH2 expression and contributes to the progression of ovarian cancer in vitro and in vivo. Onco Targets Ther. 2017;10:3131–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toska E, Osmanbeyoglu HU, Castel P, Chan C, Hendrickson RC, Elkabets M, Dickler MN, Scaltriti M, Leslie CS, Armstrong SA, Baselga J. PI3 K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science. 2017;355(6331):1324–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang JH, Shi YF, Chen XD, Qi WJ. The influence of aquaporin-1 and microvessel density on ovarian carcinogenesis and ascites formation. Int J Gynecol Cancer. 2006;16(Suppl 1):400–405. [DOI] [PubMed] [Google Scholar]
- 22. Li AJ, Baldwin RL, Karlan BY. Estrogen and progesterone receptor subtype expression in normal and malignant ovarian epithelial cell cultures. Am J Obstet Gynecol. 2003;189(1):22–27. [DOI] [PubMed] [Google Scholar]
- 23. Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49(5):825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pfaffenbach KT, Lee AS. The critical role of GRP78 in physiologic and pathologic stress. Curr Opin Cell Biol. 2011;23(2):150–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larsson C, Cordeddu L, Siggens L, Pandzic T, Kundu S, He L, Ali MA, Pristovšek N, Hartman K, Ekwall K, Sjöblom T. Restoration of KMT2C/MLL3 in human colorectal cancer cells reinforces genome-wide H3K4me1 profiles and influences cell growth and gene expression. Clin Epigenetics. 2020;12(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MDM, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sze CC, Shilatifard A. MLL3/MLL4/COMPASS family on epigenetic regulation of enhancer function and cancer. Cold Spring Harb Perspect Med. 2016;6(11):a026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol. 2013;33(23):4745–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gray SG. Epigenetic treatment of neurological disease. Epigenomics. 2011;3(4):431–450. [DOI] [PubMed] [Google Scholar]
- 30. McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23(2):260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12(2):142–148. [DOI] [PubMed] [Google Scholar]
- 32. Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339(2):240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131(1):58–69. [DOI] [PubMed] [Google Scholar]
- 34. Low D, Mizoguchi A, Mizoguchi E. DNA methylation in inflammatory bowel disease and beyond. World J Gastroenterol. 2013;19(32):5238–5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13(7):497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ke XS, Qu Y, Rostad K, Li WC, Lin B, Halvorsen OJ, Haukaas SA, Jonassen I, Petersen K, Goldfinger N, Rotter V, et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One. 2009;4(3):e4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Giakountis A, Moulos P, Sarris ME, Hatzis P, Talianidis I. Smyd3-associated regulatory pathways in cancer. Semin Cancer Biol. 2017;42:70–80. [DOI] [PubMed] [Google Scholar]
- 38. Zhang E, He X, Zhang C, Su J, Lu X, Si X, Chen J, Yin D, Han L, De W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19(1):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tajima K, Yae T, Javaid S, Tam O, Comaills V, Morris R, Wittner BS, Liu M, Engstrom A, Takahashi F, Black JC, et al. SETD1A modulates cell cycle progression through a miRNA network that regulates p53 target genes. Nat Commun. 2015;6:8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sahlberg SH, Spiegelberg D, Glimelius B, Stenerlöw B, Nestor M. Evaluation of cancer stem cell markers CD133, CD44, CD24: association with AKT isoforms and radiation resistance in colon cancer cells. PLoS One. 2014;9(4):e94621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Erdogan E, Klee EW, Thompson EA, Fields AP. Meta-analysis of oncogenic protein kinase Ciota signaling in lung adenocarcinoma. Clin Cancer Res. 2009;15(5):1527–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ristimäki A, Nieminen O, Saukkonen K, Hotakainen K, Nordling S, Haglund C. Expression of cyclooxygenase-2 in human transitional cell carcinoma of the urinary bladder. Am J Pathol. 2001;158(3):849–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tiper IV, Temkin SM, Spiegel S, Goldblum SE, Giuntoli RL, 2nd, Oelke M, Schneck JP, Webb TJ. VEGF potentiates GD3-mediated immunosuppression by human ovarian cancer cells. Clin Cancer Res. 2016;22(16):4249–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakarai C, Osawa K, Matsubara N, Ikeuchi H, Yamano T, Okamura S, Kamoshida S, Tsutou A, Takahashi J, Ejiri K, Hirota S, et al. Significance of ELF3 mRNA expression for detection of lymph node metastases of colorectal cancer. Anticancer Res. 2012;32(9):3753–3758. [PubMed] [Google Scholar]
- 45. Li S, Miner K, Fannin R, Carl Barrett J, Davis BJ. Cyclooxygenase-1 and 2 in normal and malignant human ovarian epithelium. Gynecol Oncol. 2004;92(2):622–627. [DOI] [PubMed] [Google Scholar]
- 46. Ali-Fehmi R, Che M, Khalifeh I, Malone JM, Morris R, Lawrence WD, Munkarah AR. The effect of cyclooxygenase-2 expression on tumor vascularity in advanced stage ovarian serous carcinoma. Cancer. 2003;98(7):1423–1429. [DOI] [PubMed] [Google Scholar]
- 47. Jiang C, Masood M, Rasul A, Wei W, Wang Y, Ali M, Mustaqeem M, Li J, Li X. Altholactone inhibits NF-κB and STAT3 activation and induces reactive oxygen species-mediated apoptosis in prostate cancer DU145 cells. Molecules 2017;22(2):240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. González-Palomares B, Martín PJ, de Las Casas ML, de Castro SV, Fernández SR, Lázaro MV, De la Orden García V, Aragon JA. Vascular endothelial growth factor (VEGF) polymorphisms and serum VEGF levels in women with epithelial ovarian cancer, benign tumors, and healthy ovaries. Int J Gynecol Cancer. 2017;27(6):1088–1095. [DOI] [PubMed] [Google Scholar]


