Abstract
Alveolar bone remodeling under orthodontic force is achieved by periodontal ligament stem cells (PDLSCs), which are sensitive to mechanical loading. How to regulate functions of PDLSCs is a key issue in bone remodeling during orthodontic tooth movement. This study is aimed at investigating the roles of lncRNA Hedgehog-interacting protein antisense RNA 1 (HHIP-AS1) in the functional regulation of PDLSCs. First, HHIP-AS1 expression was downregulated in PDLSCs under continuous compressive pressure. Then, we found that the alkaline phosphatase activity, in vitro mineralization, and expression levels of bone sialoprotein, osteocalcin, and osterix were increased in PDLSCs by HHIP-AS1. The results of scratch migration and transwell chemotaxis assays revealed that HHIP-AS1 inhibited the migration and chemotaxis abilities of PDLSCs. In addition, the RNA sequencing data showed that 356 mRNAs and 14 lncRNAs were upregulated, including receptor tyrosine kinase-like orphan receptor 2 and nuclear-enriched abundant transcript 1, while 185 mRNAs and 6 lncRNAs were downregulated, including fibroblast growth factor 5 and LINC00973, in HHIP-AS1-depleted PDLSCs. Bioinformatic analysis revealed several biological processes and signaling pathways related to HHIP-AS1 functions, including the PI3K-Akt signaling pathway and JAK-STAT signaling pathway. In conclusion, our findings indicated that HHIP-AS1 was downregulated in PDLSCs under compressive pressure, and it promoted the osteogenic differentiation potential and inhibited the migration and chemotaxis abilities of PDLSCs. Thus, HHIP-AS1 may be a potential target for accelerating tooth movement during orthodontic treatment.
1. Introduction
Orthodontic treatment is a time-consuming process that usually takes 2 to 3 years. Longer treatment cycles are often accompanied by higher risks of complications, such as enamel demineralization, mucosal ulcers, caries, gingival recession, and root resorption [1–7]. It is a common pursuit of orthodontists and patients to accelerate orthodontic tooth movement (OTM) and achieve the treatment goals of health, function, stability, and beauty as quickly as possible. Presently, there are many adjunctive intervention measures to accelerate OTM. These range from surgical interventions to nonsurgical interventions. The surgical interventions include periodontally accelerated osteogenic orthodontics [8], periodontal ligament distraction osteogenesis and dentoalveolar distraction osteogenesis [9], and circumferential supracrestal fiberotomy [10]. The nonsurgical interventions include laser [11], vibration [12], drug [13], ultrasound [14], and electromagnetic therapies [15]. However, the current adjunctive intervention measures are traumatic or cannot accelerate tooth movement completely. Further, none of these measures have been recognized as routine therapies in orthodontic clinics.
During OTM, the force applied to the tooth is transmitted to the alveolar bone through the periodontal ligament (PDL). Research has revealed that compression leads to bone resorption, which is a rate-limiting process, and tension causes bone formation. PDL remodeling, combined with localized resorption and deposition of alveolar bone, enables the tooth to move during orthodontic force application [16, 17]. Furthermore, periodontal ligament stem cells (PDLSCs) with strong self-renewal potential and multiple differentiation potentials can form new bone, cementum, and periodontal ligaments in vitro and in vivo [18, 19]. PDLSCs are regarded as the seed cells for periodontal tissue reconstruction and remodeling. Additionally, PDLSCs are sensitive to mechanical loading, and the effect of orthodontic force on alveolar bone remodeling is achieved by PDLSCs [20]. Positive signals of PDGFRα and nestin in the PDL were gradually enhanced during orthodontic treatment in an OTM rat model and subsequently dropped on the pressure side, indicating that PDLSCs participated in OTM [21]. In vitro investigations demonstrated that the application of different types and magnitudes of force stimuli, including tension and compression, could significantly regulate the osteogenic differentiation of PDLSCs [21–23]. Thus, identifying the key targets and regulatory mechanisms underlying the osteogenic differentiation of PDLSCs can be beneficial for bone remodeling under the orthodontic condition.
Long noncoding RNAs (lncRNAs), which are noncoding RNA molecules longer than 200 nt, participate in many important regulatory processes in the body. The mechanisms underlying the action of lncRNAs mainly involve signals, decoys, guides, and scaffolds [24]. According to existing research, lncRNAs regulate gene expression mainly through transcriptional, posttranscriptional, and epigenetic regulation [25–27]. Moreover, lncRNAs can regulate the osteogenic differentiation of PDLSCs in a variety of ways. Some lncRNAs promote the osteogenic differentiation of PDLSCs, while others inhibit this function; all these lncRNAs play a critical role in bone regeneration and remodeling [28]. For instance, lncRNA TUG1 acted as a “sponge” for miR-222-3p to compete with target genes for miR-222-3p binding sites, which inhibited the negative regulation of miR-222-3p on Smad2/7 and promoted the osteogenic differentiation of PDLSCs [29]. Then, downregulation of lncRNA ANCR could activate the canonical Wnt signaling pathway and promote the osteogenesis of PDLSCs [30]. Additionally, lncRNA Hedgehog-interacting protein antisense RNA 1 (HHIP-AS1) is a newly discovered lncRNA located on human chromosome 4 and is mainly expressed in 18 types of tissues [31]. HHIP-AS1 could promote apoptosis and inhibit proliferation, migration, and chemotaxis of liver cancer cells [31]. In our previous study, we found that HHIP-AS1 expression was downregulated during the osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs) [32]. Furthermore, the expression of HHIP-AS1 in PDLSCs was lower than that in BMSCs [33]. Thus, it was speculated that HHIP-AS1 might be related to the osteogenic differentiation of PDLSCs. However, the role and mechanism of HHIP-AS1 in PDLSCs are still uncertain.
In the present study, we explored the expression of HHIP-AS1 in PDLSCs under compressive pressure and the roles of HHIP-AS1 in the osteogenic differentiation, migration, and chemotaxis of PDLSCs. RNA sequencing (RNA-seq) and bioinformatic analysis were performed to investigate the potential mechanisms of HHIP-AS1. Through this investigation, some original insights which illustrated the potential functions and mechanisms of HHIP-AS1 in PDLSCs were obtained.
2. Materials and Methods
2.1. Cell Cultures
The PDLSCs used in this study were isolated from human impacted third molar teeth. The study was conducted with the permission of the patients and under the guidance of Beijing Stomatological Hospital, Capital Medical University. First, the teeth were sterilized with 75% ethanol and washed with phosphate-buffered saline (PBS); next, the cells were isolated, cultured, and identified, as previously described [34–36]. Briefly, the periodontal ligaments were digested with a solution containing 3 mg/mL collagenase type I (Sigma-Aldrich, St. Louis, MO, USA) and 4 mg/mL dispase (Roche Diagnostics GmbH, Mannheim, Germany) at 37°C for 60 min after they were separated lightly from the middle one-third of the tooth root. Then, single-cell suspensions were successfully obtained using a 70 mm strainer (Nest, Wuxi, China). The cells were grown in MEM Alpha medium (Gibco, Carlsbad, CA, USA) supplemented with 10,000 units/mL penicillin-streptomycin (Gibco), 200 mM L-glutamine (Gibco), and 10% fetal bovine serum (FBS; Gibco) in an incubator at 37°C and under 5% carbon dioxide. PDLSCs from passages 3–5 were used in the following experiments.
To induce osteogenic differentiation, 2.0 × 105 cells were seeded into each well of 6-well plates. Once the cells reached 80% confluency, the medium was changed to an osteogenic-inducing medium that contained 2 mM β-glycerophosphate (Sigma-Aldrich), 100 μM/mL ascorbic acid (Sigma-Aldrich), 10 nM dexamethasone (Sigma-Aldrich), and 1.8 mM KH2PO4 (Sigma-Aldrich) for a period of up to 14 days.
2.2. Plasmid Construction and Viral Infection
The full-length human HHIP-AS1 constructs were created following standard protocols and verified by gene sequencing. The cDNA of HHIP-AS1 was subcloned into the LV5 lentiviral vector (GenePharma Company, Suzhou, China) for overexpression. Subsequently, the LV3 lentiviral vector (GenePharma) was used to construct a short hairpin RNA (shRNA) of HHIP-AS1. The PDLSCs were seeded and cultured in 100 mm dishes overnight and infected with lentiviruses and 6 μg/mL polybrene (Sigma-Aldrich) for 12 h. Then, the transfected PDLSCs were selected by proper antibiotics after 72 h of infection. The target sequences of the shRNAs were as follows: control shRNA (Consh), 5′-TTCTCCGAACGTGTCACGTTTC-3′, and HHIP-AS1 shRNA (HHIP-AS1sh), 5′-GCACCAATGCATCTTGTATGA-3′.
2.3. Real-Time Reverse Transcriptase Polymerase Chain Reaction (Real-Time RT-PCR)
RNA extraction, cDNA synthesis, and real-time RT-PCR were performed as described previously [37]. Total RNA was extracted from PDLSCs using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), after which cDNAs were synthesized by 1 μg aliquots of RNA with oligo (dT) with the use of random primers and reverse transcriptase, according to the manufacturer's protocol (Invitrogen). The cDNA amplifications were conducted using the QuantiTect SYBR Green PCR Kit (Qiagen, Hilden, Germany) and an Icycler iQ Multicolor Real-Time PCR Detection System. All samples were tested in triplicate. The 2−ΔΔCT method was used to determine the expression level of each gene, which was normalized to that of GAPDH. The primers used in this study are listed in Table S1.
2.4. Western Blot Analysis
Total protein was extracted from PDLSCs using RIPA lysis buffer. The Western blot was performed as previously described [35]. Equal amounts of protein (25 μg) were prepared for gel electrophoresis. The samples were electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gels. They were then transferred to polyvinylidene difluoride membranes using a semidry electrophoretic transfer apparatus (Bio-Rad Laboratories, Hercules, CA, USA). The membranes were blocked with 5% nonfat milk for 1 h and subsequently incubated overnight with primary antibodies at 4°C. Then, the membranes were washed three times with Tris-buffered saline with Tween and incubated with horseradish peroxidase-conjugated secondary antibodies (Promega, Madison, WI, USA). After this, bands were visualized using enhanced chemiluminescence detection reagents (Applygen, Beijing, China). GAPDH served as an internal control. We used the following primary antibodies: bone sialoprotein (BSP; Cat No. bs-2668R, Bioss, Beijing, China), osteocalcin (OCN; Cat No. bs-0470R, Bioss), osterix (OSX; Cat No. bs-1110R, Bioss), and GAPDH (Cat No. 60004-1-Ig; Proteintech, Rosemont, IL, USA).
2.5. Continuous Compressive Pressure Assay
First, round glass panes (with a diameter of 30 mm, a thickness of 4 mm, and a density of 2.5 × 103 kg/m3) were customized and sterilized. PDLSCs were seeded at a cell density of 2.0 × 105 and cultured in each well of 6-well plates. Then, we placed a glass pane on the PDLSCs in a single well for 0, 2, 4, and 6 h to subject the PDLSCs to a continuous compressive pressure of 1 g/cm2. The control group was treated with 0 h of pressure.
2.6. Alkaline Phosphatase (ALP) Activity Assay and Alizarin Red Staining
After 5 days of osteogenic induction, ALP activity was quantified using an ALP activity kit (Sigma-Aldrich) according to the manufacturer's protocol. Then, after 14 days of osteogenic induction, the cells were fixed with 70% ethanol and stained with 2% Alizarin Red (Sigma-Aldrich) to assess mineralization. Next, the cells were destained with 10% cetylpyridinium chloride for approximately 30 min to quantify the calcium levels. The OD values were measured at 562 nm absorbance on a multiplate reader and then compared with a standard calcium curve to determine the calcium concentrations.
2.7. Scratch Migration Assay
PDLSCs were seeded into each well of 6-well plates at a 2.0 × 105 cell density. A 1000 μL pipette tip was used to make scratches along the diameters of the cell layers. After washing the wells with PBS, the routine medium was replaced with serum-free medium. Images were captured after 0, 24, and 48 h using a microscope (Olympus) at ×40 magnification. The relative widths of the wounds were calculated by Image-Pro v1.49 (National Institutes of Health).
2.8. Transwell Chemotaxis Assay
Transwell chambers (Corning Costar, MA, USA) with 8 μm pore size membranes were applied. In the upper chambers, 2.0 × 104 cells in 100 μL serum-free medium were distributed, and in the bottom chambers, 600 μL MEM Alpha medium with 10% FBS was contained. After 48 h, the cells in the bottom chambers were fixed using 4% paraformaldehyde and stained using a 0.1% crystal violet staining solution. Finally, the numbers of transferred cells in randomly chosen fields were counted with the assistance of a microscope (Olympus) at ×200 magnification.
2.9. Bioinformatic Analysis of RNA-seq
HHIP-AS1-depleted PDLSCs were used for RNA-seq. Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Subsequently, RNA-seq and bioinformatic analysis were carried out by Guangzhou Epibiotek Co., Ltd. China. RNA libraries were constructed by the Epi™ mini long RNA-seq kit, and the Illumina NovaSeq 6000 was used as the instrument model. The DESeq2 algorithm was applied to filter the differentially expressed genes, following the significant analysis and false discovery rate (FDR) analysis under the following criteria: ∣log2FC | >1 (fold change > 2), FDR < 0.05. Additionally, GO analysis was performed to explain the biological functions of the differentially expressed genes. The GO annotations were downloaded from Gene Ontology (http://www.geneontology.org/), NCBI (http://www.ncbi.nlm.nih.gov/), and UniProt (http://www.uniprot.org/) [38]. Pathway analysis was conducted to identify the significant pathways of the differentially expressed genes in accordance with the KEGG database [39]. Fisher's exact test was used to identify significant GO categories and pathways. Significance was determined by the P value and FDR.
2.10. Statistical Analysis
Data were analyzed by the SPSS version 22 statistical software. Statistical significance, which was set at P ≤ 0.05, was determined by one-way ANOVA or Student's t-test. All experiments were performed independently three times.
3. Results
3.1. HHIP-AS1 Expression Was Downregulated in PDLSCs under Continuous Compressive Pressure
First, we evaluated the change in HHIP-AS1 expression in PDLSCs under compressive force, which imitated the mechanical condition during OTM. Real-time RT-PCR result demonstrated that HHIP-AS1 was downregulated in PDLSCs at 2, 4, and 6 h under continuous compressive pressure compared to that in PDLSCs under no pressure (Figure 1(a)).
Figure 1.
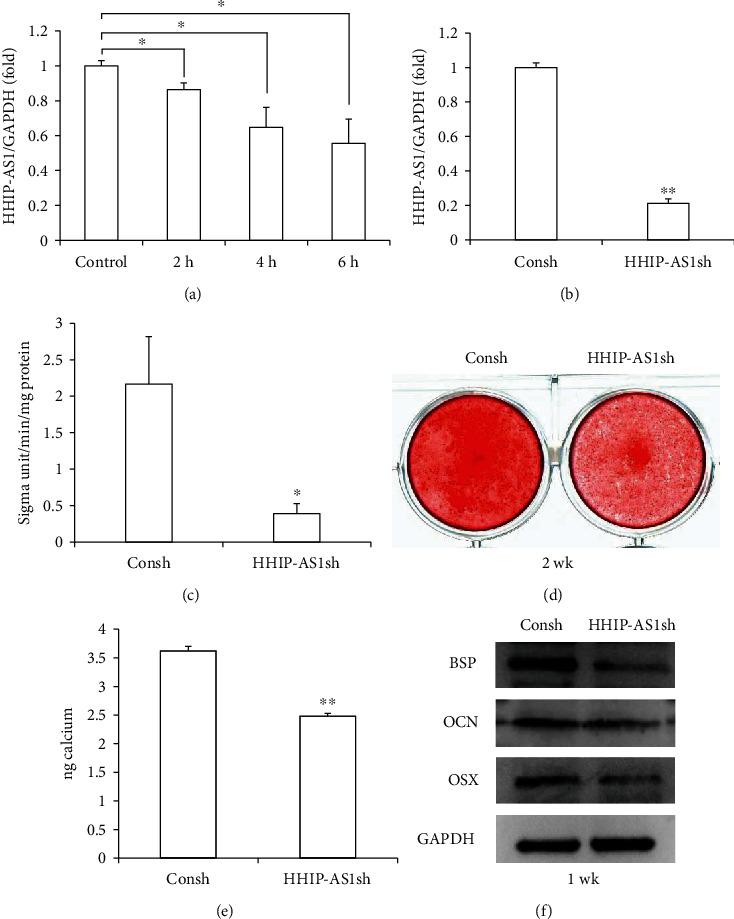
HHIP-AS1 knockdown inhibited the osteogenic differentiation potential of PDLSCs. (a) Real-time RT-PCR result showed the expression level of HHIP-AS1 at 0, 2, 4, and 6 h under continuous compressive pressure. (b) Real-time RT-PCR result showed the knockdown efficiency of HHIP-AS1 in PDLSCs. (c) ALP activity after 5 days of osteogenic induction. (d) Alizarin Red staining result after 2 weeks of osteogenic induction. (e) Calcium quantitative analysis. (f) Western blot results displayed expressions of BSP, OCN, and OSX after 1 week of osteogenic induction. GAPDH was used as internal control in real-time RT-PCR and Western blot. One-way ANOVA or Student's t-test was performed to determine statistical significance. All error bars represent the SD (n = 3). ∗P ≤ 0.05; ∗∗P ≤ 0.01.
3.2. HHIP-AS1 Promoted the Osteogenic Differentiation Potential of PDLSCs
To further investigate the function of HHIP-AS1 in the osteogenic differentiation of PDLSCs, the HHIP-AS1 shRNA lentivirus was used to knock down HHIP-AS1 expression in PDLSCs. The transfected PDLSCs were treated with 2 μg/mL puromycin for 3 days. The real-time RT-PCR result revealed that HHIP-AS1 was efficiently silenced in PDLSCs (Figure 1(b)). Compared with that in the control group (Consh group), the result of the ALP activity assay indicated that HHIP-AS1 knockdown reduced ALP activity after osteogenic induction for 5 days (Figure 1(c)). Additionally, the mineralization of PDLSCs after 2 weeks of induction was inhibited by HHIP-AS1 depletion compared to that in the Consh group. This was tested by the Alizarin Red staining and calcium quantitative measurement (Figures 1(d) and 1(e)). Furthermore, the Western blot experiments revealed the downregulation of osteogenic-related genes, including BSP and OCN, in HHIP-AS1-depleted PDLSCs compared to that in the Consh group after osteogenic induction for 1 week (Figure 1(f)). The Western blot result also showed that the expression of OSX, a key transcription factor, was significantly decreased in HHIP-AS1-depleted PDLSCs (Figure 1(f)).
Subsequently, PDLSCs were transfected with HHIP-AS1 and an empty vector. After selecting the cells with 2 μg/mL puromycin, the overexpression efficiency of HHIP-AS1 was confirmed by real-time RT-PCR (Figure 2(a)). After 5 days of osteogenic induction, ALP activity was enhanced in PDLSCs by the overexpression of HHIP-AS1 compared with that in the control group (Vector group) (Figure 2(b)). Next, the mineralization of PDLSCs was promoted by HHIP-AS1 overexpression compared with that in the Vector group after 2 weeks of induction (Figures 2(c) and 2(d)). Furthermore, the expressions of BSP, OCN, and OSX were significantly upregulated after 1 week of induction in HHIP-AS1-overexpressed PDLSCs (Figure 2(e)).
Figure 2.
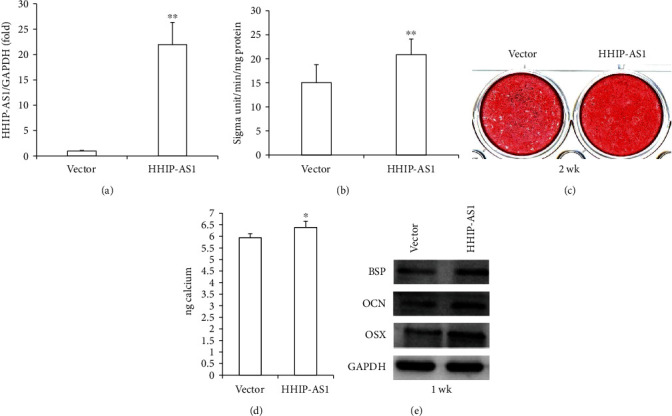
HHIP-AS1 overexpression promoted the osteogenic differentiation potential of PDLSCs. (a) Real-time RT-PCR result showed the overexpression efficiency of HHIP-AS1 in PDLSCs. (b) ALP activity after 5 days of osteogenic induction. (c) Alizarin Red staining result after 2 weeks of osteogenic induction. (d) Calcium quantitative analysis. (e) Western blot results displayed expressions of BSP, OCN, and OSX after 1 week of osteogenic induction. GAPDH was used as internal control in real-time RT-PCR and Western blot. Student's t-test was performed to determine statistical significance. All error bars represent the SD (n = 3). ∗P ≤ 0.05; ∗∗P ≤ 0.01.
3.3. HHIP-AS1 Inhibited the Migration and Chemotaxis Abilities of PDLSCs
Next, we explored the effect of HHIP-AS1 on cell migration and chemotaxis. The greater migration ability of HHIP-AS1-depleted PDLSCs than that of the Consh group at 24 and 48 h was confirmed by the scratch migration assay and quantitative analysis (Figures 3(a) and 3(b)). Furthermore, the results of the transwell chemotaxis assay and quantitative analysis also revealed a stronger chemotaxis ability of HHIP-AS1-depleted PDLSCs than that of the Consh group at 48 h (Figures 3(c) and 3(d)).
Figure 3.

HHIP-AS1 knockdown promoted the migration and chemotaxis abilities of PDLSCs. The results of the (a) scratch migration assay and (b) quantitative analysis at 24 and 48 h (scale bars: 1 mm). The results of the (c) transwell chemotaxis assay and (d) quantitative analysis at 48 h (scale bars: 200 μm). Student's t-test was performed to determine statistical significance. All error bars represent the SD (n = 9). ∗∗P ≤ 0.01.
In contrast, the scratch migration assay and quantitative analysis results showed that HHIP-AS1 overexpression inhibited the migration ability of PDLSCs at 24 and 48 h (Figures 4(a) and 4(b)). Meanwhile, the results of the transwell chemotaxis assay and quantitative analysis revealed that HHIP-AS1 overexpression suppressed the chemotaxis ability at 48 h (Figures 4(c) and 4(d)).
Figure 4.

HHIP-AS1 overexpression inhibited the migration and chemotaxis abilities of PDLSCs. The results of the (a) scratch migration assay and (b) quantitative analysis at 24 and 48 h (scale bars: 1 mm). The results of the (c) transwell chemotaxis assay and (d) quantitative analysis at 48 h (scale bars: 200 μm). Student's t-test was performed to determine statistical significance. All error bars represent the SD (n = 9). ∗∗P ≤ 0.01.
3.4. The Profiling of Differentially Expressed mRNAs and lncRNAs in HHIP-AS1-Depleted PDLSCs
The differentially expressed mRNAs and lncRNAs in HHIP-AS1-depleted PDLSCs were identified by RNA-seq. In total, 541 differentially expressed protein-coding genes were detected (∣log2FC | >1, FDR < 0.05); among these, 356 were upregulated and 185 were downregulated in HHIP-AS1-depleted PDLSCs compared to that in the Consh group (Table S2). Additionally, 20 differentially expressed lncRNAs were also detected (∣log2FC | >1, FDR < 0.05); among these, 14 were upregulated and 6 were downregulated in HHIP-AS1-depleted PDLSCs compared to that in the Consh group (Table S3).
To ensure the reliability of the RNA-seq data, three differentially expressed mRNAs (receptor tyrosine kinase-like orphan receptor 2 (ROR2), C-X-C motif chemokine ligand 12 (CXCL12), and fibroblast growth factor 5 (FGF5)) and two differentially expressed lncRNAs (nuclear-enriched abundant transcript 1 (NEAT1) and LINC00973) were randomly chosen for expression detection by real-time RT-PCR. Real-time RT-PCR results confirmed that HHIP-AS1 knockdown resulted in the upregulation of ROR2 (Figure 5(a)), CXCL12 (Figure 5(b)), and NEAT1 (Figure 5(d)) and downregulation of FGF5 (Figure 5(c)) and LINC00973 (Figure 5(e)) in PDLSCs. Furthermore, we repeated the continuous compressive pressure assay and found that when PDLSCs were stimulated by continuous compressive pressure, ROR2 and CXCL12 were both significantly upregulated and FGF5 was significantly downregulated at 2 h under pressure (Figure S1).
Figure 5.
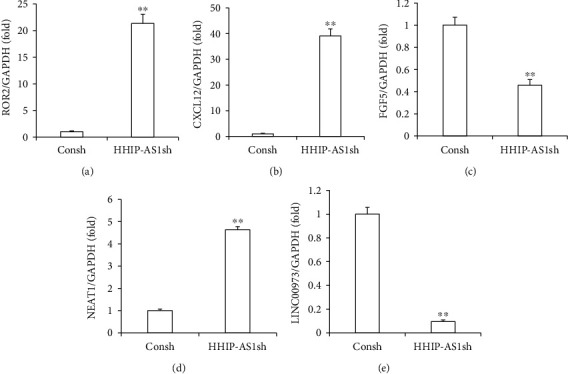
HHIP-AS1 knockdown resulted in upregulation of ROR2, CXCL12, and NEAT1 and downregulation of FGF5 and LINC00973 in PDLSCs. Real-time RT-PCR results showed the expressions of (a) ROR2, (b) CXCL12, (c) FGF5, (d) NEAT1, and (e) LINC00973. GAPDH was used as internal control. Student's t-test was performed to determine statistical significance. All error bars represent the SD (n = 3). ∗∗P ≤ 0.01.
3.5. Bioinformatic Analysis of RNA-seq Data
First, GO analysis was conducted from three aspects: biological process, molecular function, and cellular component (Figure S2). For biological processes, we obtained 535 upregulated and 360 downregulated GO functions (P < 0.05) (Table S4). The relevance between gene expression and biological processes was represented by the negative logarithm of the P value (−LgP). The bubble diagram clearly illustrated the top 20 enriched GO terms (Figure 6(a)). Among these enriched terms, some important upregulated GO functions that might be related to osteogenic differentiation included extracellular matrix (ECM) organization, cell adhesion, and skeletal system development, while some important downregulated GO functions that might be related to osteogenic differentiation included DNA methylation on cytosine, regulation of cell proliferation, and cell chemotaxis (Table S4).
Figure 6.
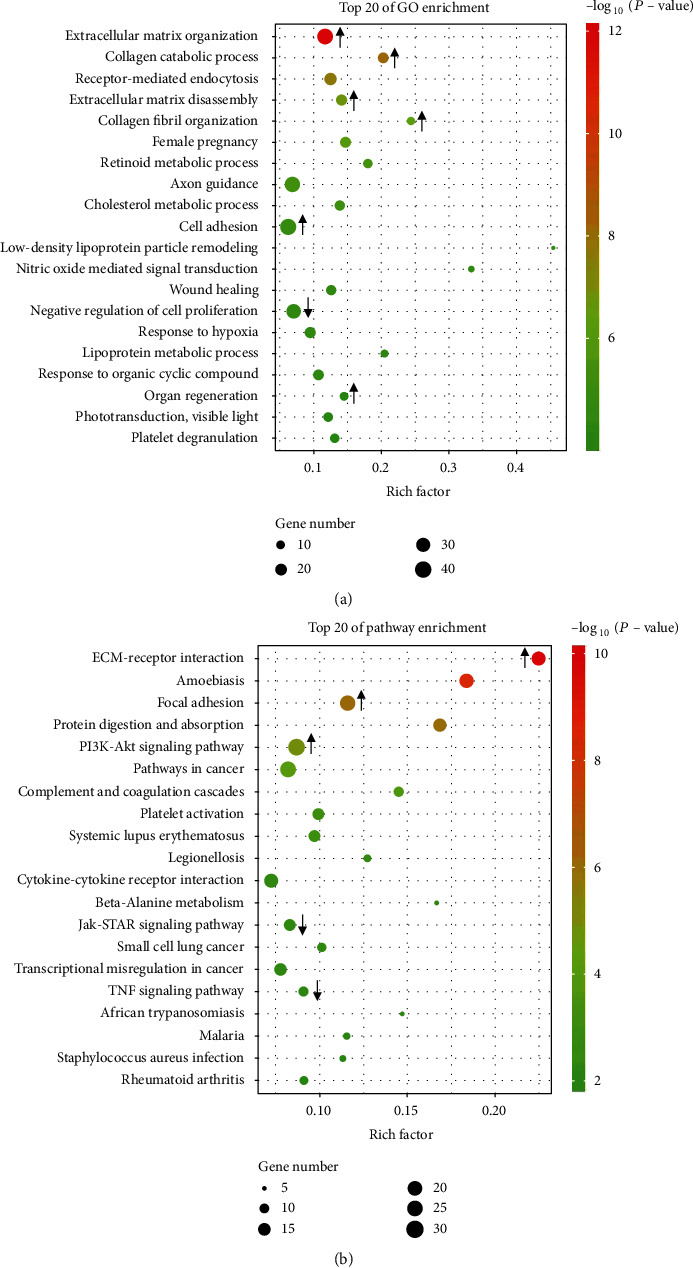
GO and pathway analysis of RNA-seq data. (a) Bubble diagram of the top 20 enriched GO terms of the differentially expressed genes in HHIP-AS1-depleted PDLSCs. (b) Bubble diagram of the top 20 enriched pathway terms of the differentially expressed genes in HHIP-AS1-depleted PDLSCs. The arrows mark the changes of key GO terms and signaling pathways.
Next, the significantly altered pathways that connected the differentially expressed genes with biological functions were identified by KEGG analysis. Twenty-four upregulated pathways and 20 downregulated pathways were detected, which may play critical roles during HHIP-AS1 regulation (P < 0.05) (Table S5, Figure S3). The top 20 enriched pathways were depicted as the bubble diagram (Figure 6(b)). Among these signaling pathways, the upregulated pathways that correlated to osteogenic differentiation included the PI3K-Akt signaling pathway, calcium signaling pathway, and Hedgehog signaling pathway. Additionally, the downregulated pathways that correlated to osteogenic differentiation were the TNF signaling pathway, JAK-STAT signaling pathway, and cell cycle (Table S5).
4. Discussion
To assist alveolar bone remodeling during OTM, it is necessary to investigate the regulation and related mechanisms of PDLSCs under mechanical stimuli. In the present study, PDLSCs were subjected to continuous compressive pressure at 1 g/cm2 for 2, 4, and 6 h to imitate the compressive condition during OTM. We found that HHIP-AS1 was downregulated in PDLSCs under compressive pressure. A study showed that after the application of compressive force at 1 g/cm2 for 12 and 24 h, PDLSCs suffered an elongated morphology, and the osteogenic marker type I collagen (Col-I) was suppressed in PDLSCs. However, the morphology and Col-I expression were recovered after force withdrawal [40]. Thus, the compressive pressure caused the downregulation of HHIP-AS1, which might further lead to the inhibition of osteogenic differentiation. To verify this hypothesis, the function of HHIP-AS1 in the osteogenic differentiation of PDLSCs was explored. Initially, we found that HHIP-AS1 knockdown inhibited ALP activity, an early-stage marker of osteogenic differentiation, and in vitro mineralization, a late-stage marker of osteogenic differentiation. We further revealed that HHIP-AS1 overexpression promoted ALP activity and in vitro mineralization. Moreover, HHIP-AS1 also upregulated the expressions of osteogenic differentiation markers, including BSP and OCN, and the transcription factor, OSX, in PDLSCs. These results suggested that HHIP-AS1 enhanced the osteogenic differentiation of PDLSCs, and downregulation of HHIP-AS1 led to the inhibition of osteogenic differentiation.
In addition to the mineralization ability, we further investigated the roles of HHIP-AS1 in the migration and chemotaxis abilities of PDLSCs. The results of the scratch migration assay, transwell chemotaxis assay, and quantitative analysis illustrated that HHIP-AS1 knockdown promoted the migration and chemotaxis abilities of PDLSCs and that HHIP-AS1 overexpression inhibited the migration and chemotaxis abilities. This was consistent with HHIP-AS1 function in the previous research on liver cancer cells [31]. Moreover, it has been reported that the application of an external force can disturb the balance between the synthesis and degradation of periodontal tissues, and the activated remodeling of PDL and alveolar bone leads to tooth movement. On the resorption side, the PDL and alveolar bone are degraded to provide space for tooth movement [17]. Taken together, our findings indicated that on the compressive side, downregulation of HHIP-AS1 led to the reduction of bone formation and enhanced migration and chemotaxis abilities of PDLSCs, which promoted the movement of PDLSCs and thereby accelerated tooth movement. Further investigation will be carried on in an animal model to validate these findings.
Moreover, to identify the mechanism of HHIP-AS1 action, RNA-seq and subsequent bioinformatic analysis were carried out. RNA-seq identified 541 differentially expressed mRNAs and 20 differentially expressed lncRNAs in HHIP-AS1-depleted PDLSCs. Among these, 356 mRNAs and 14 lncRNAs were upregulated, including ROR2 and NEAT1, while 185 mRNAs and 6 lncRNAs were downregulated, including FGF5 and LINC00973. Furthermore, the expression of ROR2, CXCL12, FGF5, NEAT1, and LINC00973, as tested by real-time RT-PCR, confirmed the RNA-seq results. ROR2, belonging to a family of receptor tyrosine kinases, could initiate the commitment of mesenchymal stem cells (MSCs) to the osteoblastic lineage and promote differentiation at both early and late stages of osteoblastogenesis. Using a mouse calvaria ex vivo organ culture model, it was demonstrated that these effects of ROR2 resulted in increased bone formation [41]. However, this was not consistent with our findings that HHIP-AS1 knockdown promoted the expression of ROR2 and inhibited the osteogenic differentiation. Meanwhile, CXCL12, a chemokine, influenced the migration, growth, survival, and differentiation of MSCs. The CXCL12/CXCR4 axis played an important role in the development and maintenance of the skeletal system through the recruitment of multipotent MSCs for bone regeneration [42]. Hwang et al. [43] treated the critical size defects of mouse calvaria with CXCL12 and bone morphogenetic protein 2 (BMP2). The highest degree of new bone regeneration was shown by the strong osteogenic differentiation and enhanced cell migration effects. In our study, although HHIP-AS1 depletion caused the upregulation of CXCL12, which promoted migration and chemotaxis, the suppression of osteogenesis was not consistent with the function of CXCL12. Then, NEAT1, a recently identified lncRNA, promoted the osteogenic differentiation of BMSCs by regulating the miR-29b-3p/BMP1 axis; this indicated its potential role in bone metabolism was contrary to our results [44]. Interestingly, we found that ROR2 and CXCL12 were both significantly upregulated in PDLSCs stimulated by continuous compressive pressure. Sometimes, the gene plays different roles in different cells or tissues under different conditions. ROR2, CXCL12, and NEAT1 might play a different role in PDLSCs during OTM. Further studies will be carried on to test this hypothesis. Additionally, the expression of FGF5 was reduced in HHIP-AS1-depleted PDLSCs and it was also significantly downregulated in PDLSCs stimulated by continuous compressive pressure. A study revealed that FGF5 facilitated cell proliferation through extracellular signal-regulated kinase 1/2 activation and enhanced the osteogenic differentiation of tonsil-derived MSCs [45]. Thus, we speculated that FGF5 played an important role in mediating the HHIP-AS1 function. Furthermore, the expressions of secreted phosphoprotein 1 (SPP1) and spondin 1 (SPON1) were upregulated in HHIP-AS1-depleted PDLSCs. SPP1 was demonstrated to be a factor for triggering an osteopenic state caused by the mechanical load during tooth movement [46, 47]. Meanwhile, SPON1 was upregulated under compression and was confirmed as a negative regulator of bone mass [48, 49]. These results indicated that SPP1 and SPON1 might also be crucial downstream genes of HHIP-AS1 that could have significant impacts on the regulation of PDLSCs and should be explored in further investigations.
Furthermore, various biological processes, ranging from the development of individuals to the occurrence of diseases, have been shown to be affected by lncRNAs [50, 51]. To clarify the underlying mechanism of HHIP-AS1 in PDLSCs, we performed GO analysis to explore the biological processes in which differentially expressed mRNAs are involved. The results showed that ECM organization, cell adhesion, skeletal system development, DNA methylation on cytosine, regulation of cell proliferation, and cell chemotaxis were the top GO terms and were related to HHIP-AS1 regulation in PDLSCs. Moreover, pathway analysis revealed that the PI3K/Akt signaling pathway, calcium signaling pathway, and Hedgehog signaling pathway were upregulated, while the TNF signaling pathway, JAK-STAT signaling pathway, and cell cycle were downregulated in HHIP-AS1-depleted PDLSCs, which were also related to HHIP-AS1 regulation in PDLSCs. Intensive studies have elucidated that many critical signaling pathways participate in regulating the osteogenic differentiation of MSCs, and multiple signaling pathways usually interact with each other to form a complex regulatory network. In our study, HHIP-AS1 depletion suppressed the osteogenic differentiation potential and upregulated the PI3K/Akt signaling pathway. Similarly, miR-let-7c-5p inhibited the expression of HMGA2 and PI3K/Akt signaling pathway and promoted the osteogenic differentiation of dental pulp stem cells in the lipopolysaccharide-mediated inflammatory environment [52]. Also, HHIP-AS1 depletion led to the downregulation of the JAK-STAT signaling pathway. A recent study showed that BMSCs promoted the healing of rabbit tibial fractures through the JAK-STAT signaling pathway [53]. These results indicated that the PI3K/Akt and JAK-STAT signaling pathways were key pathways for HHIP-AS1 regulation in PDLSCs. In general, HHIP-AS1 had a significant effect on the biological functions of PDLSCs by regulating downstream genes and related pathways. However, the underlying molecular mechanisms remain to be further explored.
5. Conclusions
In conclusion, we found here that HHIP-AS1 was downregulated in PDLSCs under compressive pressure. Additionally, HHIP-AS1 promoted the osteogenic differentiation potential and inhibited the migration and chemotaxis abilities of PDLSCs. Bioinformatic analysis of RNA-seq data revealed candidate downstream genes of HHIP-AS1 and key signaling pathways related to HHIP-AS1 regulation. Overall, our findings provide a potential target as well as a theoretical basis for the functional modification of PDLSCs and bone remodeling under the mechanical condition of orthodontic treatment.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81970964 and 81771108), CAMS Innovation Fund for Medical Sciences (2019-I2M-5-031 to Z.P.F.), and Key Program of Jiangxi Provincial Science and Technology Department of China (20171BBH80012 to J.G.).
Contributor Information
Zhipeng Fan, Email: zpfan@ccmu.edu.cn.
Jie Guo, Email: kqgjsdu@163.com.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Supplementary Materials
Table S1: primer sequences used in the real-time RT-PCR.
Table S2: the differentially expressed mRNAs in HHIP-AS1-depleted PDLSCs.
Table S3: the differentially expressed lncRNAs in HHIP-AS1-depleted PDLSCs.
Figure S1: ROR2 and CXCL12 were upregulated while FGF5 was downregulated under compressive pressure. Real-time RT-PCR results showed expressions of ROR2 (a), CXCL12 (b), and FGF5 (c) at 2 h under continuous compressive pressure. GAPDH was used as internal control. Student's t-test was performed to determine statistical significance. All error bars represent the SD (n = 3). ∗P ≤ 0.05; ∗∗P ≤ 0.01.
Figure S2: significant GO analysis of differentially expressed genes from aspects of biological process, molecular function, and cellular component. (a) Upregulated GO terms. (b) Downregulated GO terms.
Table S4: the significant upregulated and downregulated GO functions in HHIP-AS1-depleted PDLSCs.
Table S5: the significant upregulated and downregulated pathways in HHIP-AS1-depleted PDLSCs.
Figure S3: significant pathway analysis of differentially expressed genes in HHIP-AS1-depleted PDLSCs. (a) Upregulated pathways. (b) Downregulated pathways.
References
- 1.Fisher M. A., Wenger R. M., Hans M. G. Pretreatment characteristics associated with orthodontic treatment duration. American Journal of Orthodontics and Dentofacial Orthopedics . 2010;137(2):178–186. doi: 10.1016/j.ajodo.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 2.Mavreas D., Athanasiou A. E. Factors affecting the duration of orthodontic treatment: a systematic review. European Journal of Orthodontics . 2008;30(4):386–395. doi: 10.1093/ejo/cjn018. [DOI] [PubMed] [Google Scholar]
- 3.Julien K. C., Buschang P. H., Campbell P. M. Prevalence of white spot lesion formation during orthodontic treatment. The Angle Orthodontist . 2013;83(4):641–647. doi: 10.2319/071712-584.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baricevic M., Mravak-Stipetic M., Majstorovic M., Baranovic M., Baricevic D., Loncar B. Oral mucosal lesions during orthodontic treatment. International Journal of Paediatric Dentistry . 2011;21(2):96–102. doi: 10.1111/j.1365-263X.2010.01078.x. [DOI] [PubMed] [Google Scholar]
- 5.Pinto A. S., Alves L. S., Maltz M., Susin C., Zenkner J. E. A. Does the duration of fixed orthodontic treatment affect caries activity among adolescents and young adults? Caries Research . 2018;52(6):463–467. doi: 10.1159/000488209. [DOI] [PubMed] [Google Scholar]
- 6.Boke F., Gazioglu C., Akkaya S., Akkaya M. Relationship between orthodontic treatment and gingival health: a retrospective study. Eur J Dent . 2014;8(3):373–380. doi: 10.4103/1305-7456.137651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roscoe M. G., Meira J. B. C., Cattaneo P. M. Association of orthodontic force system and root resorption: a systematic review. American Journal of Orthodontics and Dentofacial Orthopedics . 2015;147(5):610–626. doi: 10.1016/j.ajodo.2014.12.026. [DOI] [PubMed] [Google Scholar]
- 8.Amit G., Jps K., Pankaj B., Suchinder S., Parul B. Periodontally accelerated osteogenic orthodontics (PAOO) - a review. Journal of Clinical and Experimental Dentistry . 2012;4(5):e292–e296. doi: 10.4317/jced.50822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kharkar V. R., Kotrashetti S. M., Kulkarni P. Comparative evaluation of dento-alveolar distraction and periodontal distraction assisted rapid retraction of the maxillary canine: a pilot study. International Journal of Oral and Maxillofacial Surgery . 2010;39(11):1074–1079. doi: 10.1016/j.ijom.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Taner T. U., Haydar B., Kavuklu I., Korkmaz A. Short-term effects of fiberotomy on relapse of anterior crowding. American Journal of Orthodontics and Dentofacial Orthopedics . 2000;118(6):617–623. doi: 10.1067/mod.2000.110637. [DOI] [PubMed] [Google Scholar]
- 11.Qamruddin I., Alam M. K., Mahroof V., Fida M., Khamis M. F., Husein A. Effects of low-level laser irradiation on the rate of orthodontic tooth movement and associated pain with self-ligating brackets. American Journal of Orthodontics and Dentofacial Orthopedics . 2017;152(5):622–630. doi: 10.1016/j.ajodo.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 12.Jing D., Xiao J., Li X., Li Y., Zhao Z. The effectiveness of vibrational stimulus to accelerate orthodontic tooth movement: a systematic review. BMC Oral Health . 2017;17(1):p. 143. doi: 10.1186/s12903-017-0437-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F., Li G., Hu H., Liu R., Chen J., Zou S. Effect of parathyroid hormone on experimental tooth movement in rats. American Journal of Orthodontics and Dentofacial Orthopedics . 2013;144(4):523–532. doi: 10.1016/j.ajodo.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Alazzawi M. M. J., Husein A., Alam M. K., et al. Effect of low level laser and low intensity pulsed ultrasound therapy on bone remodeling during orthodontic tooth movement in rats. Progress in Orthodontics . 2018;19(1):p. 10. doi: 10.1186/s40510-018-0208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dogru M., Akpolat V., Dogru A. G., Karadede B., Akkurt A., Karadede M. I. Examination of extremely low frequency electromagnetic fields on orthodontic tooth movement in rats. Biotechnology and Biotechnological Equipment . 2014;28(1):118–122. doi: 10.1080/13102818.2014.901669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan V., Davidovitch Z. Cellular, molecular, and tissue-level reactions to orthodontic force. American Journal of Orthodontics and Dentofacial Orthopedics . 2006;129(4):469.e1–469.e32. doi: 10.1016/j.ajodo.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Henneman S., Von den Hoff J. W., Maltha J. C. Mechanobiology of tooth movement. European Journal of Orthodontics . 2008;30(3):299–306. doi: 10.1093/ejo/cjn020. [DOI] [PubMed] [Google Scholar]
- 18.Seo B.-M., Miura M., Gronthos S., et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. The Lancet . 2004;364(9429):149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Zheng Y., Ding G., et al. Periodontal ligament stem cell-mediated treatment for periodontitis in miniature swine. Stem Cells . 2008;26(4):1065–1073. doi: 10.1634/stemcells.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H., Yang R., Zhou Y.-h. Mechanobiology of periodontal ligament stem cells in orthodontic tooth movement. Stem Cells International . 2018;2018:7. doi: 10.1155/2018/6531216.6531216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L., Liu W., Zhao J., et al. Mechanical stress regulates osteogenic differentiation and RANKL/OPG ratio in periodontal ligament stem cells by the Wnt/β-catenin pathway. Biochimica et Biophysica Acta . 2016;1860(10):2211–2219. doi: 10.1016/j.bbagen.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Tang N., Zhao Z., Zhang L., et al. Up-regulated osteogenic transcription factors during early response of human periodontal ligament stem cells to cyclic tensile strain. Archives of Medical Science . 2012;8(3):422–430. doi: 10.5114/aoms.2012.28810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen T., Qiu L., Chang H., et al. Cyclic tension promotes osteogenic differentiation in human periodontal ligament stem cells. International Journal of Clinical and Experimental Pathology . 2014;7(11):7872–7880. [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K. C., Chang H. Y. Molecular mechanisms of long noncoding RNAs. Molecular Cell . 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitz S. U., Grote P., Herrmann B. G. Mechanisms of long noncoding RNA function in development and disease. Cellular and Molecular Life Sciences . 2016;73(13):2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Q., Hao Q., Prasanth K. V. Nuclear long noncoding RNAs: key regulators of gene expression. Trends in Genetics . 2018;34(2):142–157. doi: 10.1016/j.tig.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ju C., Liu R., Zhang Y. W., et al. Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomedicine & Pharmacotherapy . 2019;115:p. 108912. doi: 10.1016/j.biopha.2019.108912. [DOI] [PubMed] [Google Scholar]
- 28.Qiu W., Wu B. L., Fang F. C. Overview of noncoding RNAs involved in the osteogenic differentiation of periodontal ligament stem cells. World J Stem Cells . 2020;12(4):251–265. doi: 10.4252/wjsc.v12.i4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu D., Yin L., Sun D., et al. Long noncoding RNA TUG1 promotes osteogenic differentiation of human periodontal ligament stem cell through sponging microRNA-222-3p to negatively regulate Smad2/7. Archives of Oral Biology . 2020;117:p. 104814. doi: 10.1016/j.archoralbio.2020.104814. [DOI] [PubMed] [Google Scholar]
- 30.Jia Q., Jiang W., Ni L. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Archives of Oral Biology . 2015;60(2):234–241. doi: 10.1016/j.archoralbio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Bo C., Li X., He L., Zhang S., Li N., An Y. A novel long noncoding RNA HHIP-AS1 suppresses hepatocellular carcinoma progression through stabilizing HHIP mRNA. Biochemical and Biophysical Research Communications . 2019;520(2):333–340. doi: 10.1016/j.bbrc.2019.09.137. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W., Dong R., Diao S., du J., Fan Z., Wang F. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Research & Therapy . 2017;8(1):p. 30. doi: 10.1186/s13287-017-0485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong R., Du J., Wang L., et al. Comparison of long noncoding RNA and mRNA expression profiles in mesenchymal stem cells derived from human periodontal ligament and bone marrow. BioMed Research International . 2014;2014:12. doi: 10.1155/2014/317853.317853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu D., Wang Y., Jia Z., et al. Demethylation of IGFBP5 by histone demethylase KDM6B promotes mesenchymal stem cell-mediated periodontal tissue regeneration by enhancing osteogenic differentiation and anti-inflammation potentials. Stem Cells . 2015;33(8):2523–2536. doi: 10.1002/stem.2018. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Jia Z., Diao S., et al. IGFBP5 enhances osteogenic differentiation potential of periodontal ligament stem cells and Wharton's jelly umbilical cord stem cells, via the JNK and MEK/Erk signalling pathways. Cell Proliferation . 2016;49(5):618–627. doi: 10.1111/cpr.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu S., Long J., Yu J., et al. Analysis of differentiation potentials and gene expression profiles of mesenchymal stem cells derived from periodontal ligament and Wharton’s jelly of the umbilical cord. Cells, Tissues, Organs . 2013;197(3):209–223. doi: 10.1159/000343740. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Zhang C., Yang H., Han X., Fan Z., Hou B. Depletion of PRDM9 enhances proliferation, migration and chemotaxis potentials in human periodontal ligament stem cells. Connective Tissue Research . 2020;61(5):498–508. doi: 10.1080/03008207.2019.1620224. [DOI] [PubMed] [Google Scholar]
- 38.Ashburner M., Ball C. A., Blake J. A., et al. Gene ontology: tool for the unification of biology. Nature Genetics . 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Draghici S., Khatri P., Tarca A. L., et al. A systems biology approach for pathway level analysis. Genome Research . 2007;17(10):1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng L., Yang R., Liu D., et al. PDL progenitor-mediated PDL recovery contributes to orthodontic relapse. Journal of Dental Research . 2016;95(9):1049–1056. doi: 10.1177/0022034516648604. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y., Bhat R. A., Seestaller-Wehr L. M., et al. The orphan receptor tyrosine kinase Ror2 promotes osteoblast differentiation and enhances ex vivo bone formation. Molecular Endocrinology . 2007;21(2):376–387. doi: 10.1210/me.2006-0342. [DOI] [PubMed] [Google Scholar]
- 42.Gilbert W., Bragg R., Elmansi A. M., et al. Stromal cell-derived factor-1 (CXCL12) and its role in bone and muscle biology. Cytokine . 2019;123:p. 154783. doi: 10.1016/j.cyto.2019.154783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang H. D., Lee J. T., Koh J. T., Jung H. M., Lee H. J., Kwon T. G. Sequential treatment with SDF-1 and BMP-2 potentiates bone formation in calvarial defects. Tissue Engineering. Part A . 2015;21(13-14):2125–2135. doi: 10.1089/ten.tea.2014.0571. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y., Chen B., Li D., Zhou X., Chen Z. LncRNA NEAT1/miR-29b-3p/BMP1 axis promotes osteogenic differentiation in human bone marrow-derived mesenchymal stem cells. Pathology, Research and Practice . 2019;215(3):525–531. doi: 10.1016/j.prp.2018.12.034. [DOI] [PubMed] [Google Scholar]
- 45.Park G. C., Song J. S., Park H. Y., et al. Role of fibroblast growth factor-5 on the proliferation of human tonsil-derived mesenchymal stem cells. Stem Cells and Development . 2016;25(15):1149–1160. doi: 10.1089/scd.2016.0061. [DOI] [PubMed] [Google Scholar]
- 46.Terai K., Takano-Yamamoto T., Ohba Y., et al. Role of osteopontin in bone remodeling caused by mechanical stress. Journal of Bone and Mineral Research . 1999;14(6):839–849. doi: 10.1359/jbmr.1999.14.6.839. [DOI] [PubMed] [Google Scholar]
- 47.Choi E. K., Lee J. H., Baek S. H., Kim S. J. Gene expression profile altered by orthodontic tooth movement during healing of surgical alveolar defect. American Journal of Orthodontics and Dentofacial Orthopedics . 2017;151(6):1107–1115. doi: 10.1016/j.ajodo.2016.10.039. [DOI] [PubMed] [Google Scholar]
- 48.Li Y., Li M., Tan L., et al. Analysis of time-course gene expression profiles of a periodontal ligament tissue model under compression. Archives of Oral Biology . 2013;58(5):511–522. doi: 10.1016/j.archoralbio.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Palmer G. D., Attur M. G., Yang Q., et al. F-spondin deficient mice have a high bone mass phenotype. PLoS One . 2014;9(5, article e98388) doi: 10.1371/journal.pone.0098388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bouckenheimer J., Assou S., Riquier S., et al. Long non-coding RNAs in human early embryonic development and their potential in ART. Human Reproduction Update . 2016;23(1):19–40. doi: 10.1093/humupd/dmw035. [DOI] [PubMed] [Google Scholar]
- 51.Arun G., Diermeier S. D., Spector D. L. Therapeutic targeting of long non-coding RNAs in cancer. Trends in Molecular Medicine . 2018;24(3):257–277. doi: 10.1016/j.molmed.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan H., Zhao H., Wang J., et al. MicroRNA let-7c-5p promotes osteogenic differentiation of dental pulp stem cells by inhibiting lipopolysaccharide-induced inflammation via HMGA2/PI3K/Akt signal blockade. Clinical and Experimental Pharmacology & Physiology . 2019;46(4):389–397. doi: 10.1111/1440-1681.13059. [DOI] [PubMed] [Google Scholar]
- 53.Wang P., Zhang Z. Bone marrow-derived mesenchymal stem cells promote healing of rabbit tibial fractures via JAK-STAT signaling pathway. Experimental and Therapeutic Medicine . 2020;19(3):2310–2316. doi: 10.3892/etm.2020.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: primer sequences used in the real-time RT-PCR.
Table S2: the differentially expressed mRNAs in HHIP-AS1-depleted PDLSCs.
Table S3: the differentially expressed lncRNAs in HHIP-AS1-depleted PDLSCs.
Figure S1: ROR2 and CXCL12 were upregulated while FGF5 was downregulated under compressive pressure. Real-time RT-PCR results showed expressions of ROR2 (a), CXCL12 (b), and FGF5 (c) at 2 h under continuous compressive pressure. GAPDH was used as internal control. Student's t-test was performed to determine statistical significance. All error bars represent the SD (n = 3). ∗P ≤ 0.05; ∗∗P ≤ 0.01.
Figure S2: significant GO analysis of differentially expressed genes from aspects of biological process, molecular function, and cellular component. (a) Upregulated GO terms. (b) Downregulated GO terms.
Table S4: the significant upregulated and downregulated GO functions in HHIP-AS1-depleted PDLSCs.
Table S5: the significant upregulated and downregulated pathways in HHIP-AS1-depleted PDLSCs.
Figure S3: significant pathway analysis of differentially expressed genes in HHIP-AS1-depleted PDLSCs. (a) Upregulated pathways. (b) Downregulated pathways.
Data Availability Statement
The data used to support the findings of this study are included within the article.


