Abstract
Objective
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the novel coronavirus causing severe respiratory illness (COVID-19). This virus was initially identified in Wuhan city, a populated area of the Hubei province in China, and still remains one of the major global health challenges. RNA interference (RNAi) is a mechanism of post-transcriptional gene silencing that plays a crucial role in innate viral defense mechanisms by inhibiting the virus replication as well as expression of various viral proteins. Dicer, Drosha, Ago2, and DGCR8 are essential components of the RNAi system, which is supposed to be dysregulated in COVID-19 patients. This study aimed to assess the expression level of the mentioned mRNAs in COVID-19patients compared to healthy individuals.
Results
Our findings demonstrated that the expression of Dicer, Drosha, and Ago2 was statistically altered in COVID-19 patients compared to healthy subjects. Ultimately, the RNA interference mechanism as a crucial antiviral defense system was suggested to be dysregulated in COVID-19 patients.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13104-021-05816-0.
Keywords: SARS-CoV-2, RNA interference, Dicer, Drosha, Ago2, DGCR8
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus responsible for severe respiratory disease (COVID-19). The virus was first identified in Wuhan, Hubei, China, in 2019 and spread worldwide. After the Chinese epidemic peak, other countries, especially South Korea, Italy, and Iran saw a significant spread of the disease. [1–3]. SARS‐CoV‐2 categorized in beta-coronaviruses lineage and is closely related to the SARS‐CoV virus [4, 5]. The respiratory system and lungs are most susceptible to damages resulting from the virus that leads to failed functions of the respiratory system, severe acute pulmonary disorders, and consequent mortality [6, 7]. Coronavirus is capable of producing double-stranded RNA (dsRNA) during the infection cycles. Host cell pattern recognition receptors (PRRs) could distinguish viral dsRNA as a foreign pathogen and respond to them by activating several antiviral pathways such as the RNA interference (RNAi) mechanism that is critical for early defense against viral invasion [8]. RNAi is a mechanism of post-transcriptional gene silencing in eukaryote and human cells, that inhibit virus replication and expression of various viral proteins. The RNAi system as an innate immunity pathway is mainly mediated by two non-coding molecules, including microRNA (miRNA) and small interfering RNA (siRNA) [9]. miRNA biogenesis involves Drosha, Dicer and RNAase III proteins, while siRNA processing is exclusively associated with Dicer function. Initially, endogenous or exogenous dsRNAs bounds to form siRNA or miRNA-induced silencing complex (RISC). The most critical component of these complexes is argonaute RISC catalytic component (AGO) proteins, Ago2 in particular. Using a guide RNA strand, the RISC complex could eighter suppress the mRNA translation or degrade the mRNA by Ago slicer activity [10]. Identification of exogenous dsRNAs through PRRs results in the generation of siRNAs (21—25nt in length) mediated by Dicer. One of the siRNA strands bound the RISC complex RISC and cleaves the target mRNA [11]. Similarly, during viral infection, the dsRNA of the viruse is cleaved by Dicer to generate siRNAs, which finally target the complementary viral RNA [12]. The RNAi mechanism could inhibit different types of virus infections such as rotavirus [13], influenza virus [14], and HIV-1[15, 16]. Moreover, siRNA duplexes could effectively reduce the expression of SARS-CoV genes [9]. In addition, viruses are able to encode specific suppressive proteins, which could disrupt the RNAi-mediated host defense. Thus, the balance between host RNAi components and RNAi viral inhibitors could determine disease outcomes [17]. In spite of recent advances in the development of vaccines against SARS-CoV-2, it is still remaining one of the major healthcare challenges worldwide. Thereby, providing detailed insights into the molecular pathogenesis mechanism is highlighted. Dicer, Ago2, DGRC8, and Drosha are the four critical components of RNAi system involved in the innate antiviral defense. To the best of our knowledge, no study has been reported on the RNAi mechanism dysregulation in COVID-19 patients. Therefore, here the expression levels of these genes were evaluated to predict the probable dysregulation of this pathway resulted from COVID-19.
Main text
Materials and methods
In silico analysis: Gene Ontology protein–protein interaction (PPI) network analysis
To find the probable molecular roles of the selected genes, gene ontology analysis was carried out by Cytoscape ClueGO plug-in [18]. Further, to predict the correlation and interaction of genes expression levels, PPI network was depicted through string database [19] and Cytoscape string plug-in [20].
Sampling
A case group consisted of 20 COVID-19 patients and 20 healthy individuals recruited from Alzahra Hospital (Isfahan, Iran). Patients were hospitalized in intensive care unit (ICU) with severe symptoms and their status were diagnosed by an infectious specialist. The selected patients had severe symptoms with no history of an underlying disease and autoimmune disorders. The characteristics of patients are mentioned in Additional file 1: Table S1.
RNA extraction
Total RNA was extracted from whole blood using RNA Extraction-Kit (Favor-Prep, Blood/Cultured Cell Total RNA. Taiwan). The quality of RNA was measured at 260/280 nm by a NanoDrop spectrometer (Thermo Scientific, Waltham, MA, USA).
Complementary DNA (cDNA) synthesis
According to the manufacturer's protocol, cDNA synthesis was performed using a BioFACT RT-Kit (Biofact, Korea) and stored at − 20 °C until the next step.
Quantitative real-time PCR (qPCR)
Real-time PCR were performed using SYBR Green master mix (Biofact. Korea) and specific primers conducted on Rotor-Gene 6000 instrument (Corbett Life Science, Mortlake, Australia) as follow: 95 °C for 15 min followed by 40 cycles of 95 °C for 20 s, 60 °C for 30 s and 72 °C for 30 s. The sequences of primers are listed in Additional file 1: Table S2. The data was analyzed using the 2−ΔΔCT method. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) used as internal references gene.
Statistical analysis
To analyze the expression levels of genes between two groups, the unpaired parametric t-test and the nonparametric Mann–Whitney test were used. All results are presented as mean ± SD (standard deviation), P ≤ 0.05 was considered statistically significant, and analysis was done by GraphPad Prism version 8.0.2 (Graph Pad, San Diego, CA, USA).
Results
In silico analysis: gene ontology and PPI network analysis
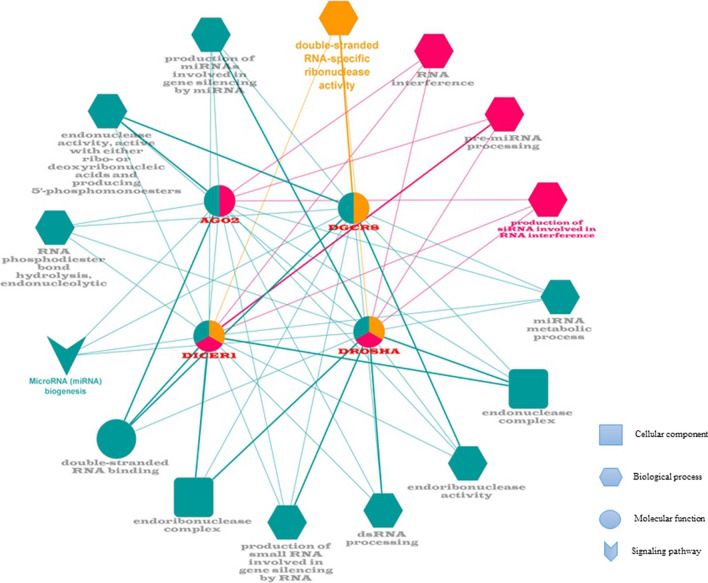
Analyzing gene ontology and signaling pathways indicated that the Dicer, Ago2, Drosha, DGCR8 are involved in RNAi, miRNA biogenesis, and double-strand RNA binding mechanisms (Fig. 1). Moreover, using the string plugin, the PPI network determined high interrelations among proteins. (Fig. 2).
Fig. 1.
Gene ontology enrichment analysis. microRNA biogenesis was enriched as a signaling pathway in which all four genes are involved. dsRNA binding and endoribonuclease complex were enriched as molecular function and cellular compartment, respectively (p-value ≤ 0.05)
Fig. 2.
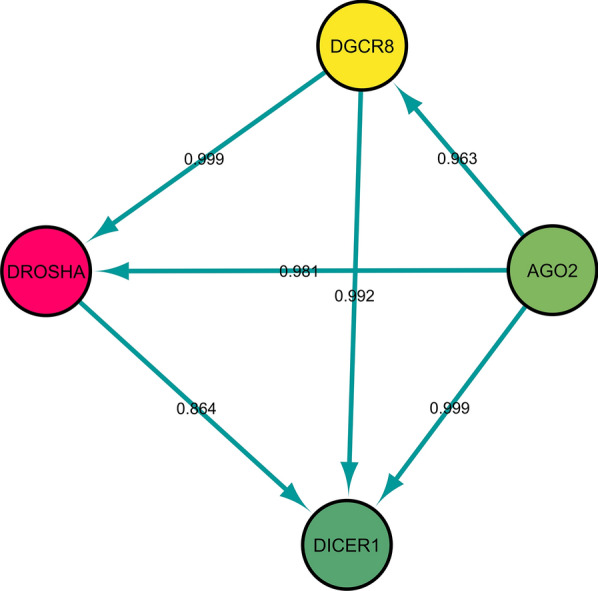
Protein–protein interaction (PPI) network analysis of DICER, Drosha, Ago2, and DGCR8. The edge's labels are based on predicted PPI score
Expression levels of Dicer, Drosha, Ago2, and DGCR8
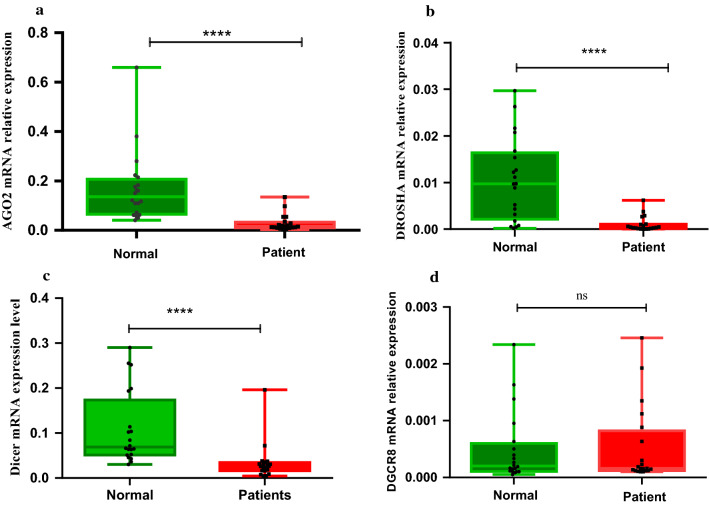
Ago2 and Dicer data were normally distributed; therefore, analyzed by the unpaired parametric t-test. Conversely, Drosha and DGCR8 data were not normally distributed, thus, the non-parametric Mann–Whitney test was applied. It was determined that expression level of Ago2, Dicer, and Drosha were significantly downregulated (Log FC: − 3.46, − 1.92, − 4.23, respectively) in COVID-19 patients compared with control group (Fig. 3 A–C). However, no significant difference of DGCR8 expression (Log FC: − 0.32) was found groups (Fig. 3D). Finally, pearson correlation analysis determined significant interrelation between Ago2 and Drosha expression with Dicer expression level (Additional file 1: Figure S1).
Fig. 3.
Quantitative real-time PCR analysis of Ago2 (A), Dicer (B), Drosha (C), and DGCR8 (D) in COVID-19 patients and healthy controls. The mRNA expression level of genes was evaluated in the whole blood samples of 20 COVID-19 patients and 20 controls. P values are calculated by parametric t-test for Ago2 and Dicer (p-value < 0.0001) and non-parametric Mann Whitney test for Drosha p-value < 0.0001 andDGCR8 non-significant (ns: p-value 0.213). GAPDH was utilized for an endogenous reference to normalize mRNA expression levels
Discussion
Since the onset of the Severe Acute Respiratory Syndrome (SARS) in 2002–2003 and the Middle East Respiratory Syndrome (MERS) in 2012, many research has been devoted to providing accurate information on the pathogenesis of the disease along with proposing new treatment strategies. Despite outstanding progress in producing effective vaccines, SARS-CoV-2 remains a global health challenge throughout the world. To date, several diagnostic strategies and treatment methods have been investigated, including recovery plasma and serum therapy [21], autophagy and virophagy [22], antiviral drugs [23–26], and nanobiotechnology-based approaches. Due to the necessity developing effective therapeutics based on the virus-pathogenesis mechanism, the RNAi mechanism has the point of intense interest. Several proteins such as host receptors as well as proteins associated with the virus entrance, replication, and survival could be the potential targets for RNAi mechanism [6]. This study investigated the role of the RNAi system as an innate immune response to viral infection in COVID-19 patients. Dicer, Drosha, Ago2, and DGCR8 (critical components of RNAi) were selected for evaluation in COVID-19 patients. Based on the in-silico analysis, these genes are significantly involved in gene silencing processes through miRNA and siRNA (Fig. 1). RNAi is defined as an antiviral defense mechanism that suppresses the expression of viral proteins by targeting mRNAs [10, 27]. Hasan et.al investigated active RNAi systems against SARS-CoV-2 using computational prediction approaches. They reported 24 pre-miRNA hairpins, 131 siRNAs, 12 human miRNA, and 10 siRNA molecules as active therapeutic agents against SARS-CoV-2 [28]. Previous studies have examined the antiviral defense potential of several miRNAs and siRNAs against some viruses including HIV-1 [29], Influenza [30], Zika [31] and Hepatitis C (HCV) [32]. These information introduced some antiviral RNAi therapeutics that could effectively inactivate SARS-CoV-2 [6]. Until a few years ago, siRNAs were thought to be exclusively exogenous, like viral RNA. However, investigations have provided some strong evidence that mammalian cells could produce siRNAs against viral infection. Significantly, the production of endogenous siRNAs in 293 human cells was reported during the response to infection with influenza A virus (IAV) or human enterovirus 71 (HEV71) [33, 34]. Contrary to the antiviral function of RNAi, viruses could be released virus-encoded suppressors of RNAi (VSRs) to escape from the immunity system Some of these suppressors have been identified in cells infected with influenza A virus (non-structural protein 1), human enterovirus 71 (3A protein), and Nodamura virus (B2 protein) [35]. In addition, He et al. found that RNAi (siRNA-RISC) targets replicase 1A region of SARS-CoV, thus inhibiting SARS-CoV infection in-vitro [36]. Given the 80% sequence identity between SARS-CoV and SARS-CoV-2 and high protein homology [37], investigation of SARS-CoV-2 could be effective. We found that the expression levels of Dicer, Drosha, and Ago2 were significantly reduced in patients with COVID-19 compared with healthy individuals. However, the differentially expressed DGCR8 between two groups was not statistically significant. Matskevich et al. investigated the role of RNAi in both Vero and A549 cell lines lacking interferon (IFN) system that were infected with influenza A virus. They concluded that the disfunction of Dicer could increase viral replication and induce apoptosis in infected cells [38]. Moreover, Modai et al. determined that HIV-1 infection could up-regulate the miRNAs and down regulate the level of Dicer1, HRB (HIV-1 Rev-binding protein) and HIV-EP2 (Human Immunodeficiency Virus Type I Enhancer Binding Protein 2), in human Sup-T1 cells. These miRNAs could regulate directly the host genes (Dice1, HRB and HIv-EP2) and HIV-1 interferes with the human immune response against HIV infection [39]. In another study, the significant reduction of Dicer, Drosha, and DGCR8 levels were found in human A-549 cells following Dengue virus (DENV4) infection [40].
There are no reports of Ago2 expression in human cells. However, defective fly Ago-2, in Drosophila melanogaster, is susceptible to infection with Drosophila C virus and cricket paralysis virus [41]. Demirci et al. are predicted that about 30 viral mature miRNA-like sequences could target 1,367 human genes, affecting their functions at different cellular processes. Furthermore, some human miRNAs could target SARS-CoV-2 genes such as S, M, N, E proteins and open reading frames. Ultimately, these valuable insights on miRNAs mechanism could promise further advances in developing novel therapeutic agents for SARS-CoV-2 infections [42]. A recent in silico targeted SARS and COVID-19 genomes using human miRNAs. SARS and COVID-19 targeting miRNAs, 848 and 873 respectively, were found in which 315 and 290 miRNAs are exclusive for COVID-19 and SARS, respectively. The results also identified 19 miRNA targeting isolates out of 29 COVID-19 isolates [43]. In addition, Arisan et al. suggested seven key miRNAs linked with viral pathogenicity and host responses that could be used as effective therapeutic approaches in the treatment of COVID-19 and its pathological consequences [44]. Overall, dysregulation of Dicer, Drosha, Ago2 could play an important role in promoting viral infection. An explanation for the reported dysregulation in the three mRNAs mentioned is that SARS-COV-2 may defeat key components of the RNAi system to escape antiviral defense resulting in uncontrolled replication and transcription. On the other hand, given the high average age of patients admitted to the ICU and the clinical manifestations of the disease, it could be suggested that the RNAi mechanism is unable to defend against viral infection and the virus rapidly disables the system. Finally, correlation analysis demonstrated a significant relation between Ago2 expression to Dicer and Drosha to Dicer (Additional file 1: Figure S1). These significant interactions may indicate the interrelation of RNAi components, in which dysregulation of one factor could exacerbate the effects of the virus infection and consequently lead to the malfunction of the RNAi mechanism. As predicted in Fig. 2, these proteins may be strongly correlated based on high scores. However, according to the results of correlation analysis, only two interrelations (Ago2 with Dicer, and Drosha with Dicer) were confirmed.
Conclusion
No vaccine or drug has been developed against COVID-19 with 100 efficiencies. In this regard, development of novel therapeutic agents considering the role of RNAi mechanism is of great importance. In conclusion, we observed the dysregulation of key RNAi components, Dicer, Ago2, and Drosha, as crucial antiviral defense factors in RNAi system in COVID-19 patients. This finding could provide insights to further identification of SARS-CoV-2 pathogenesis for COVID-19 treatment based on the RNAi system.
Limitations
Difficulty of sampling and small size of population.
Supplementary Information
Additional file 1: Table S1. Sex and age of the enrolled patients in this study. Table S2. Real-time primer sequences. Figure S1. Correlation analysis graphs. The interrelation of (A) Ago2 with Dicer (p-value ≤0.002; r: 0.7430; 95% confidence interval; 0.4477 to 0.8922; R squared: 0.5521) and (B) Drosha with Dicer (p-value ≤ 0.0034; r: 0.6223; 95% confidence interval: 0.2481 to 0.8349; R squared: 0.3872) was determined with the regression line. The expression values were calculated as -log relative expression.
Acknowledgements
The authors would like to appreciate the Alzahra University Hospital staff for support of this work.
Abbreviations
- DENV4
Dengue virus
- dsRNA
Double-stranded RNA
- HEV71
Human enterovirus 71
- IAV
Influenza A virus
- MERS
Middle East respiratory syndrome
- miRNA
MicroRNA
- PPI
Protein–protein interaction
- PRRs
Pattern recognition receptors
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- SARS
Severe acute respiratory syndrome
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- siRNA
Small interfering RNA
- VSRs
Virus-encoded suppressors of RNAi
Authors’ contributions
SRM designed the study and wrote the manuscript. MSS gathered the data and wrote the manuscript. FK, SF, SS, and FAB performed the experiment. ZTE and AA gathered the data. MK, KS, RS, MH analyzed manuscript data. HL edited and substantively revised the manuscript for important intellectual content. MS edited and supervised the study. All authors read and approved the final manuscript.
Funding
This study was supported by Medical Genetics Research Center of Genome, Isfahan university of medical sciences, Isfahan, Iran (Grant number 199134).
Availability of data and materials
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Declarations
Ethics approval and consent to participate
The informed written consent was obtained from the patient's parent or legal guardian. This study was approved by the Ethics Committee of Isfahan University of Medical Sciences, Isfahan, Iran, with ethics code IR.MUI.MED.REC.1399.439.
Consent for publication
Not Applicable.
Competing interests
The author declares no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mansoor Salehi, Email: M_salehi@med.mui.ac.ir.
Hajie Lotfi, Email: lotfi.hajie@yahoo.com.
References
- 1.Hsu LY, Chia PY, Vasoo SJSMJ. A midpoint perspective on the COVID-19 pandemic. Singap Med J. 2020;61(7):381–383. doi: 10.11622/smedj.2020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prem K, Liu Y, Russell TW, Kucharski AJ, Eggo RM, Davies N, et al. The effect of control strategies to reduce social mixing on outcomes of the COVID-19 epidemic in Wuhan, China: a modelling study. Lancet Public Health. 2020;5(5):e261–e270. doi: 10.1016/S2468-2667(20)30073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fauci AS, Lane HC, Redfield RR. Covid-19—navigating the uncharted. Mass Med Soc 2020. [DOI] [PMC free article] [PubMed]
- 4.Velavan TP, Meyer CGJTM, Health i. The COVID‐19 epidemic. 2020;25(3):278. [DOI] [PMC free article] [PubMed]
- 5.Kumar S, Nyodu R, Maurya VK, Saxena SK. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Coronavirus Disease 2019 (COVID-19) Springer; 2020. pp. 23–31. [Google Scholar]
- 6.Uludağ H, Parent K, Aliabadi HM, Haddadi A. Prospects for RNAi therapy of COVID-19. Front Bioeng Biotechnol. 2020;8:916. doi: 10.3389/fbioe.2020.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheervalilou R, Shirvaliloo M, Dadashzadeh N, Shirvalilou S, Shahraki O, Pilehvar-Soltanahmadi Y, et al. COVID-19 under spotlight: a close look at the origin, transmission, diagnosis, and treatment of the 2019-nCoV disease. J Cell Physiol. 2020;235(12):8873–8924. doi: 10.1002/jcp.29735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Renner DM, Comar CE, Whelan JN, Reyes HM, Cardenas-Diaz FL, et al. SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial derived cells and cardiomyocytes. bioRxiv. 2020;12:4. doi: 10.1073/pnas.2022643118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmadi A, Hekmatnezhad H. The sound of getting rid of coronavirus by RNA interference technology: RNAi against COVID-19. J Curr Biomed Rep. 2020;1(2):45–47. doi: 10.52547/JCBioR.1.2.45. [DOI] [Google Scholar]
- 10.Jeang K-T. RNAi in the regulation of mammalian viral infections. BMC Biol. 2012;10(1):1–6. doi: 10.1186/1741-7007-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laganà A, Veneziano D, Russo F, Pulvirenti A, Giugno R, Croce CM, et al. Computational design of artificial RNA molecules for gene regulation. RNA Bioinformatics. Springer; 2015. pp. 393–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawlings RA, Krishnan V, Walter NG. Viral RNAi suppressor reversibly binds siRNA to outcompete Dicer and RISC via multiple turnover. J Mol Biol. 2011;408(2):262–276. doi: 10.1016/j.jmb.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Déctor MA, Romero P, López S, Arias CF. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 2002;3(12):1175–1180. doi: 10.1093/embo-reports/kvf234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Q, McManus MT, Nguyen T, Shen C-H, Sharp PA, Eisen HN, et al. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci. 2003;100(5):2718–2723. doi: 10.1073/pnas.0437841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coburn GA, Cullen BR. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J Virol. 2002;76(18):9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez MA, Clotet B, Esté JA. RNA interference of HIV replication. Trends Immunol 2002; 23(12):559–61. [DOI] [PubMed]
- 17.Karjee S, Mukherjee SK. RNAi suppressor: the hidden weapon of SARS-CoV. J Biosci. 2020;45(1):1–6. doi: 10.1007/s12038-020-00071-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(suppl_1):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res. 2018;18(2):623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sheervalilou R, Shirvaliloo M, Sargazi S, Bahari S, Saravani R, Shahraki J, et al. Convalescent blood: current perspective on the efficacy of a legacy approach in COVID-19 treatment. Blood Purif. 2021 doi: 10.1159/000513164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sargazi S, Sheervalilou R, Rokni M, Shirvaliloo M, Shahraki O, Rezaei N. The role of autophagy in controlling SARS-CoV-2 infection: an overview on virophagy-mediated molecular drug targets. Cell Biol Int. 2021;45(8):1599–1612. doi: 10.1002/cbin.11609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bimonte S, Crispo A, Amore A, Celentano E, Cuomo A, Cascella M. Potential antiviral drugs for SARS-Cov-2 treatment: preclinical findings and ongoing clinical research. In Vivo. 2020;34(3 suppl):1597–1602. doi: 10.21873/invivo.11949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko W-C, Rolain J-M, Lee N-Y, Chen P-L, Huang C-T, Lee P-I, et al. Arguments in favour of remdesivir for treating SARS-CoV-2 infections. Int J Antimicrob Agents. 2020;55(4):105933. doi: 10.1016/j.ijantimicag.2020.105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheervalilou R, Shirvaliloo M, Sargazi S, Shirvalilou S, Shahraki O, Pilehvar-Soltanahmadi Y, et al. Application of nanobiotechnology for early diagnosis of SARS-CoV-2 infection in the COVID-19 pandemic. Appl Microbiol Biotechnol. 2021;105(7):2615–2624. doi: 10.1007/s00253-021-11197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaushik AK, Dhau JS, Gohel H, Mishra YK, Kateb B, Kim N-Y, et al. Electrochemical SARS-CoV-2 sensing at point-of-care and artificial intelligence for intelligent COVID-19 management. ACS Appl Bio Mater. 2020;3(11):7306–7325. doi: 10.1021/acsabm.0c01004. [DOI] [PubMed] [Google Scholar]
- 27.Fischer LT, James QY. RNAi, a new therapeutic strategy against viral infection. Cell Res. 2004;14(6):460–466. doi: 10.1038/sj.cr.7290248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasan M, Ashik AI, Chowdhury MB, Tasnim AT, Nishat ZS, Hossain T, et al. Computational prediction of potential siRNA and human miRNA sequences to silence orf1ab associated genes for future therapeutics against SARS-CoV-2. Inform Med Unlocked. 2021;24:100569. doi: 10.1016/j.imu.2021.100569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanghvi VR, Steel LF. RNA silencing as a cellular defense against HIV-1 infection: progress and issues. FASEB J. 2012;26(10):3937–3945. doi: 10.1096/fj.12-210765. [DOI] [PubMed] [Google Scholar]
- 30.Peng S, Wang J, Wei S, Li C, Zhou K, Hu J, et al. Endogenous cellular microRNAs mediate antiviral defense against influenza A virus. Molecular Therapy-Nucleic Acids. 2018;10:361–375. doi: 10.1016/j.omtn.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giulietti M, Righetti A, Cianfruglia L, Šabanović B, Armeni T, Principato G, et al. To accelerate the Zika beat: candidate design for RNA interference-based therapy. Virus Res. 2018;255:133–140. doi: 10.1016/j.virusres.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Basavappa M, Lu J, Dong S, Cronkite DA, Prior JT, et al. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat Microbiol. 2016;2(3):1–9. doi: 10.1038/nmicrobiol.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu Y, Xu Y, Zhang Y, Zhou H, Deng Y-Q, Li X-F, et al. Human virus-derived small RNAs can confer antiviral immunity in mammals. Immunity. 2017;46(6):992–1004.e5. doi: 10.1016/j.immuni.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Ding S-W, Han Q, Wang J, Li W-X. Antiviral RNA interference in mammals. Curr Opin Immunol. 2018;54:109–114. doi: 10.1016/j.coi.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He M-L, Zheng B, Peng Y, Peiris JS, Poon LL, Yuen KY, et al. Inhibition of SARS-associated coronavirus infection and replication by RNA interference. JAMA. 2003;290(20):2665–2666. doi: 10.1001/jama.290.20.2665. [DOI] [PubMed] [Google Scholar]
- 37.Donia A, Bokhari H. RNA interference as a promising treatment against SARS-CoV-2. Int Microbiol. 2020:1–2. [DOI] [PMC free article] [PubMed]
- 38.Matskevich AA, Moelling K. Dicer is involved in protection against influenza A virus infection. J Gen Virol. 2007;88(10):2627–2635. doi: 10.1099/vir.0.83103-0. [DOI] [PubMed] [Google Scholar]
- 39.Modai S, Farberov L, Herzig E, Isakov O, Hizi A, Shomron N. HIV-1 infection increases microRNAs that inhibit Dicer1, HRB and HIV-EP2, thereby reducing viral replication. PLoS ONE. 2019;14(1):e0211111. doi: 10.1371/journal.pone.0211111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casseb SMM, Simith DDB, Melo KFLD, Mendonca M, Santos A, Carvalho VL, et al. Drosha, DGCR8, and Dicer mRNAs are down-regulated in human cells infected with dengue virus 4, and play a role in viral pathogenesis. Genet Mol Res. 2016 doi: 10.4238/gmr.15027891. [DOI] [PubMed] [Google Scholar]
- 41.Van Rij RP, Saleh M-C, Berry B, Foo C, Houk A, Antoniewski C, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20(21):2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saçar Demirci MD, Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8:e9369. doi: 10.7717/peerj.9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fulzele S, Sahay B, Yusufu I, Lee TJ, Sharma A, Kolhe R, et al. COVID-19 virulence in aged patients might be impacted by the host cellular microRNAs abundance/profile. Aging Dis. 2020;11(3):509–522. doi: 10.14336/AD.2020.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arisan ED, Dart A, Grant GH, Arisan S, Cuhadaroglu S, Lange S, et al. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases identify 7 Key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12(6):614. doi: 10.3390/v12060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sex and age of the enrolled patients in this study. Table S2. Real-time primer sequences. Figure S1. Correlation analysis graphs. The interrelation of (A) Ago2 with Dicer (p-value ≤0.002; r: 0.7430; 95% confidence interval; 0.4477 to 0.8922; R squared: 0.5521) and (B) Drosha with Dicer (p-value ≤ 0.0034; r: 0.6223; 95% confidence interval: 0.2481 to 0.8349; R squared: 0.3872) was determined with the regression line. The expression values were calculated as -log relative expression.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.