Abstract
The COVID-19 pandemic caused by SARS-CoV-2 requires new treatments both to alleviate the symptoms and to prevent the spread of this disease. Previous studies demonstrated good antiviral and virucidal activity of phospholipase A2s (PLA2s) from snake venoms against viruses from different families but there was no data for coronaviruses. Here we show that PLA2s from snake venoms protect Vero E6 cells against SARS-CoV-2 cytopathic effects. PLA2s showed low cytotoxicity to Vero E6 cells with some activity at micromolar concentrations, but strong antiviral activity at nanomolar concentrations. Dimeric PLA2 from the viper Vipera nikolskii and its subunits manifested especially potent virucidal effects, which were related to their phospholipolytic activity, and inhibited cell–cell fusion mediated by the SARS-CoV-2 spike glycoprotein. Moreover, PLA2s interfered with binding both of an antibody against ACE2 and of the receptor-binding domain of the glycoprotein S to 293T/ACE2 cells. This is the first demonstration of a detrimental effect of PLA2s on β-coronaviruses. Thus, snake PLA2s are promising for the development of antiviral drugs that target the viral envelope, and could also prove to be useful tools to study the interaction of viruses with host cells.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-03985-6.
Keywords: Molecular modelling, Pseudotyped SARS-CoV-2 virus, Receptor binding domain, Replication cycle, Surface plasmon resonance, Time-of-drug-addition assay
Introduction
In December 2019, a rapidly spreading community-acquired pneumonia was discovered in Wuhan (Hubei Province, China) and subsequent studies have shown that this disease is caused by a virus belonging to the coronavirus family, a conclusion confirmed by sequencing the full-length genome from samples (bronchoalveolar lavage fluid) of patients with pneumonia [1]. In February 2020, WHO (https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020) officially named this disease «COVID-19» (coronavirus disease 2019) and the causative virus was designated as SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) by the International Committee on Taxonomy of Viruses [2].
Coronaviruses are enveloped RNA viruses, and form the Coronaviridae family which includes four genera: α-, β-, γ- and δ-coronaviruses [3]. Of the seven known coronaviruses that infect humans, HCoV-229E and HCoV-NL63 belong to α-coronaviruses, while HCoV-HKU1, HCoV-OC43, MERS-CoV, SARS-CoV, and SARS-CoV-2 are β-coronaviruses [4]. Coronaviruses HCoV-229E, HCoV-NL63, HCoV-HKU1 and HCoV-OC43 cause mild disease symptoms similar to the common cold [5]. However, MERS-CoV, SARS-CoV and SARS-CoV-2 cause more severe diseases associated with pneumonia, acute respiratory distress syndrome and «cytokine storms» that can lead to death [6–8]. Cell entry of SARS-CoV-2 occurs as a result of the binding of the spike glycoprotein (glycoprotein S) to angiotensin-converting enzyme (ACE2) expressed on the surface of the host cells [9], the lung tissues being the main target. Intervention at the stage of adsorption/binding or replication of the virus using therapeutic agents can effectively block viral infection [10, 11]. The most common symptom of COVID-19 patients is respiratory distress, and patients admitted to intensive care are usually unable to breathe on their own. In addition, some COVID-19 patients have experienced neurological symptoms including headaches, nausea and vomiting, consistent with the fact that coronaviruses are not always limited to the respiratory tract and can also affect the central nervous system, resulting in neurological complications [12].
COVID-19 was declared a global pandemic in March 2020, by which time it had spread worldwide in only a few months [13]. At present the total number of infected is more than 220 million, and more than 4.5 million have died. Vaccination is currently underway in many countries but is unlikely to be 100% effective, and some medications have been identified which decrease the severity of the disease, but again these do not improve outcomes in all cases; thus, new drugs are urgently needed.
Animal venoms contain a wide range of biologically active compounds of different chemical structures, and have proven to have not only significant pharmacological value but also great potential for drug discovery, with many snake venom components being investigated in preclinical or clinical trials for a variety of therapeutic applications [14–22]. Among snake venom enzymes, phospholipase A2s (PLA2s) are one of the main components [23]. Typically, PLA2s are small proteins with a molecular weight of about 13–15 kDa and they catalyze hydrolysis at the sn2 position of membrane glycerophospholipids to lysophospholipids and free fatty acids [24, 25]. The PLA2 superfamily is currently subdivided into six types, namely secreted PLA2s (sPLA2s), Ca2+-dependent cytosolic cPLA2, Ca2+-independent cytosolic iPLA2, platelet-activating factor acetylhydrolase PAF-AH, lysosomal LPLA2 and adipose-specific AdPLA2 [26]. More than one-third of the members of the PLA2 superfamily belong to sPLA2, which is further subdivided into 10 groups and 18 subgroups. The PLA2s from snake venoms are in groups I and II [27]. Previous studies have shown that PLA2s have antiviral activity against a number of viruses including Dengue and Yellow fever viruses [28], HIV [29] Rous virus [30], adenovirus [31], Newcastle virus [32] and Chikungunya virus [33]. We, therefore, tested several snake venom PLA2s for antiviral activity against SARS-CoV-2. Here we show that all the PLA2s we examined are capable of inhibiting SARS-CoV-2 infection in vitro, although only dimeric PLA2s have high antiviral activity. Additional studies showed that the virucidal and antiviral activities of dimeric PLA2 depend on its catalytic activity. This ability of PLA2 to inactivate the virus suggests a mechanism which has the potential to provide a barrier to the spread of infection.
Materials and methods
Cells and viruses
Vero E6 (ATCC CRL-1586), 293T (ATCC CRL-3216) and 293T/ACE2 (obtained as described [34]) cells were grown in complete DMEM medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS; HyClone, Cytiva, Marlborough, MA, USA), 1 × L-Glutamine (PanEco, Moscow, Russia) and 1 × penicillin–streptomycin (PanEco, Russia). All cell lines tested negative for mycoplasma contamination. SARS-CoV-2 strain hCoV-19/Russia/Moscow_PMVL-4 (GISAID ID: EPI_ISL_470898) was isolated from naso/oropharyngeal swab of COVID-19 patient using the Vero E6 cell line. The stocks of SARS-CoV-2 used in the experiments had undergone six passages. Viral titers were determined as TCID50 in confluent Vero E6 cells in 96-well microtiter plates by endpoint dilutions assay. In some experiments, SARS-CoV-2 strain hCoV-19/Russia/Moscow_PMVL-18 (GISAID ID: EPI_ISL_872633) and SARS-CoV-2 strain hCoV-19/Russia/Moscow_PMVL-20 (GISAID ID: EPI_ISL_872634) were used. Bovine coronavirus (BCV; The State Collection of Viruses of the D.I. Ivanovsky Institute of Virology) was propagated on MDBK cells (ATCC CCL-22). Virus-related work was performed at a biosafety level-3 (BSL-3) facility.
Venoms
The venoms of V. nikolskii and V. ursinii renardi vipers were kindly provided by Siberian Serpentarium Ltd. (Novosibirsk, Russia). Krait B. fasciatus venom (Vinh Son, Vinh Tuong, Vinh Phuc Province, Vietnam) was obtained from the certified farm owned by professional snake breeder Mr. Ha Van Tien. The venom was collected from several tens of snake specimens by farm team members. The certificate of farm has been registered in accordance with the Government's Decree No. 82/2006/ND-CP of August 10, 2006 by the Department of agriculture and rural development of the Socialist Republic of Vietnam and valid until May 31, 2021.
Phospholipases A2
Phospholipase A2 II (BF-PLA2-II, GenBank AAK62361.1) and phospholipase A2 1 (BF-PLA2-1, UniProtKB Q90WA7) were purified from krait B. fasciatus venom as described [35]. Phospholipase A2 Vur-PL2 (UniProtKB F8QN53) was purified from viper V. ursinii renardi venom as described [36]. Dimeric phospholipases HDP-1 and HDP-2 were isolated from vipera V. nikolskii venom and separated into subunits HDP-1P (UniProtKB Q1RP79), HDP-2P (UniProtKB Q1RP78) and HDP-1I (UniProtKB A4VBF0) as described [37].
Modification of HDP-2P
4-Bromophenacyl bromide (Lancaster Synthesis, Morecambe, England) as a 10 mM stock solution in acetone was added to a final concentration of 200 µM to a 20 µM HDP-2P solution in 50 mM Tris HCl buffer, pH 7.5, containing 10 mM Na2SO4. The mixture was incubated for 6 h at room temperature and separated using a Jupiter C18 HPLC column (Phenomenex) and acetonitrile gradient from 20 to 50% in 30 min in the presence of 0.1% trifluoroacetic acid. The phospholipolytic activity was measured (according to [38]) using a synthetic fluorescent substrate 1-palmitoyl-2-(10-pyrenyldecanoyl)-sn-glycero-3-phosphocholine (Molecular Probes, Thermo Fisher Scientific, Waltham, MA, USA) and a Hitachi F-4000 spectrofluorimeter.
Cell viability assay
Cell viability in the presence of the tested PLA2 was assessed using the MTT (Methylthiazolyldiphenyl-tetrazolium bromide) method as previously described [39]. Briefly, Vero E6 cells (2 × 104 cells per well) were incubated with different concentrations of the individual PLA2s in 96-well plates for 48 h, followed by the addition of MTT (Sigma, Merck KGaA, Darmstadt, Germany) according to the manufacturer's instructions. Results are expressed as a percentage of cell viability in comparison to the untreated cell controls. Three experiments were performed with two technical replicates.
Bromodeoxyuridine (BrdU) incorporation assay and flow cytometry
For the BrdU incorporation assay [40], Vero E6 cells were incubated with 100 μg/ml of various PLA2s for 72 h. Then, BrdU was added to the cell culture medium to a final concentration of 10 µM and incubated for 2 h. After BrdU incorporation, cells were permeabilized with 70% ethanol chilled to − 20 °C for 30 min on ice. For the cell cycle assay, cells were treated with 2 N HCl for 30 min to denature chromosome DNA followed by neutralization with 0.1 M Na2B4O7 (pH 8.5) for 30 min. The cells were then stained with fluorescein isothiocyanate (FITC)-anti-BrdU antibody (clone 3D4; Sony Biotechnology, San Jose, CA, USA) and PI (propidium iodide; with RNase A). Data were acquired using a MACSQuant Analyzer 10 Flow Cytometer (Miltenyi Biotec, Bergish Gladbach, Germany), and processed using FlowJo software 10.0.8. (Three Star Inc., Ashland, OR, USA). Dead cells were excluded by forward scatter (FSC) versus side scatter (SSC) dot plot analysis. Doublet cells were excluded by FSC-H versus FSC-A dot plot analysis.
Cytopathic effect (CPE) inhibition assay against SARS-CoV-2
Vero E6 cells were plated in 96-well plates at a density of 2 × 104 cells per well. After 18 h of incubation, 100 TCID50 SARS-CoV-2 were added to the cell monolayer in the absence or presence of various concentrations of PLA2s (tenfold dilutions in a concentration range of 0.001 µg/ml to 100 µg/ml). After 72 h of incubation, differences in cell viability caused by virus-induced cytopathic effects were analyzed using the MTT method as previously described [41, 42]. For this, an MTT stock solution (5 mg/ml in PBS) was added to each well at a final concentration of 0.5 mg/ml. After a 2 h incubation, medium was aspirated from wells and 150 µl of DMSO was added. Absorbance was measured at 590 nm using a SPECTROstar Nano microplate reader (BMG LABTECH GmbH, Ortenberg, Germany). Three independent experiments with triplicate measurements were performed. Data were analyzed using GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA, USA) and the IC50 (concentration of the compound that inhibited 50% of the infection) calculated via nonlinear regression analysis.
Time-of-drug-addition assay
The time-of-drug-addition assay was performed as described in [43]. Briefly, Vero E6 cells (3 × 104 cells/well) were treated with HDP-2 (10 µg/ml) at different stages of virus infection. For “Full-time” treatment, cells were pre-treated with the PLA2 for 1 h prior to virus infection, followed by incubation with a virus for 1 h in the presence of the PLA2. Then cells were washed with PBS, and further cultured with the PLA2-containing medium until the end of the experiment. For “Entry” treatment, the PLA2 were added to the cells for 1 h before virus infection, and maintained during the 1 h viral attachment. Then, the virus-PLA2 mixture was replaced with a fresh culture medium without PLA2 until the end of the experiment. For “Post-entry” experiment, virus was added to the cells to allow infection for 1 h, and then virus-containing supernatant was replaced with PLA2-containing medium until the end of the experiment. The cells were infected with SARS-CoV-2 (multiplicity of infection, MOI = 0.01), and at 18 h after infection the cell culture supernatant of each time point experiment was collected for viral yield measurements using qRT–PCR.
Viral RNA extraction and qRT-PCR
Viral RNA was purified from the cell culture supernatant using the ExtractRNA (Evrogen, Moscow, Russia) according to the manufacturer’s instructions. Real-time one-step qRT–PCR was used for quantitation of SARS-CoV-2 viral load using the «SARS-CoV-2 FRT» RT-PCR kit (Gamaleya National Center of Epidemiology and Microbiology, Russia) with a QuantStudio 5 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA). A standard curve was generated by the determination of copy numbers from serial dilutions (103 -108 copies) of the plasmid.
Pseudovirus assay
To generate SARS-CoV-2 or VSV-G pseudovirus particles, suspension cultures of 293T cells were seeded onto a 10 cm dish and transfected with 10 µg of pLVPG (kindly provided by I.V. Zvyagin, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia) plasmid, 8 µg of pCMV-deltaR8.2 plasmid (Addgene, #12263) and 5 µg of pVAX-1-S-glycoprotein (Evrogen, Moscow, Russia) or pCG1-SARS-2-S-delta 18 UK variant or pCG1-SARS-2-S-delta 18 South Africa variant plasmids (kindly provided by Prof. Stefan Pöhlmann) or pCMV-VSV-G plasmid (Addgene, #8454) using Transporter™ 5 transfection reagent. 72 h after transfection the supernatant was harvested, clarified by centrifugation and passing through a 0.45 µm pore size filter, aliquoted and frozen at – 80 °C.
To evaluate the pseudovirus neutralization activity, HDP-2 was incubated with an equal volume of pseudovirus at 37 °C. Then the mixture was transferred to pre-plated 293T/ACE2 cell monolayers in 96-well plates. After incubation for 72 h, the fluorescence of GFP positive cells was determined using a microplate reader (Hidex Sense Beta Plus, Hidex, Turku, Finland).
Post-entry study
Vero E6 cells were infected with 100TCID50 SARS-CoV-2 PMVL-4 and incubated for 16 h. Then, virus-containing medium was removed and replace with a fresh medium containing different concentrations of HDP-2 and incubated for 48 h. Inhibition of CPE was assessed by MTT assay. In post-entry assay with pseudo-SARS-CoV-2 (lineage B.1.1.7), 293T/ACE2 cells were transduced with pseudo-SARS-CoV-2 for 16 h. Then, different dilutions of HDP-2 were added to the cells. After incubation for 72 h, the fluorescence of GFP was determined using a microplate reader (Hidex Sense Beta Plus, Hidex, Turku, Finland).
Virucidal activity test
The SARS-CoV-2 virus stock (1 × 106 TCID50) was incubated with serial tenfold dilutions of PLA2 (0.1–10 µg/ml) for 1 h at 37 °C. Then, the virus samples treated by PLA2 were diluted below IC50 and titrated onto Vero E6 cells by limiting dilution assay (fivefold dilutions in six replicates) in 96-well plates. A viral stock treated with PBS was used as a control. The plates were incubated at 37 °C (5% CO2) for 72 h. The CPE was scored visually under a microscope and the virus titers were calculated by the Reed and Muench method [44].
Transmission electron microscopy (TEM)
Bovine coronavirus or SARS-CoV-2 was treated with 10 µg/ml HDP-2 for 1 h at 37 °C. Fifty microliters of sample were placed on a glass slide with a 200 mesh copper TEM grid. The grid was then removed, blotted with filter paper and exposed to 1.0% uranyl acetate solution. Excess uranyl acetate was removed, grids were air-dried, and examined under a JEM-2100 Plus electron microscope (JEOL Ltd, Akishima, Japan) at an accelerating voltage of 80 kV.
Cell–cell fusion assay
The inhibitory activity of PLA2 on SARS-CoV-2 S-mediated cell–cell fusion was assessed as previously described [45, 46]. 293T effector cells were transfected with plasmid pUCHR-IRES-GFP (kindly provided by D.V. Mazurov) encoding the GFP and plasmid pVAX-1-S-glycoprotein (Evrogen, Moscow, Russia) encoding a full-length SARS-CoV-2 Spike glycoprotein (293T-GFP-Spike). Vero E6 cells (3 × 104 cells per well), expressing ACE2 receptors on the membrane surface, were used as target cells, and were incubated in 96-well plates for 18 h. Then 104 effector cells (293T-GFP-Spike) per well were added in the presence or absence of PLA2 at various concentrations and incubated at 37 °C for 2 h. The percentage of cell–cell fusion was calculated by counting the fused cells in each well in five random fields using an Olympus (Olympus, Tokyo, Japan) epifluorescence microscope.
ACE2 and RBD binding experiments
Experiments were performed as previously described [47] with minor modifications. Briefly, 293T/ACE2 cells were incubated at 37 °C in PBS (containing 0.5% bovine serum albumin) in the presence of HDP-2P (100 µg/ml) for 20 min. Then, cells were placed on ice and stained with phycoerythrin (PE)-anti-ACE2 antibody (Clone # 535919; R&D Systems, Minneapolis, MN, USA) or isotype-matched control IgG for 1 h. In other experiments, 293T/ACE2 cells were incubated with recombinant RBD protein fused with Fc (kindly provided by R.S. Kalinin, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow, Russia) followed by staining with DyLight 650—conjugated goat anti-human Fc antibody (ThermoFisher Scientific, Waltham, MA, USA). Cells were analyzed with MACSQuant Analyzer 10 Flow Cytometer (Miltenyi Biotec, Bergish Gladbach, Germany).
Molecular modeling
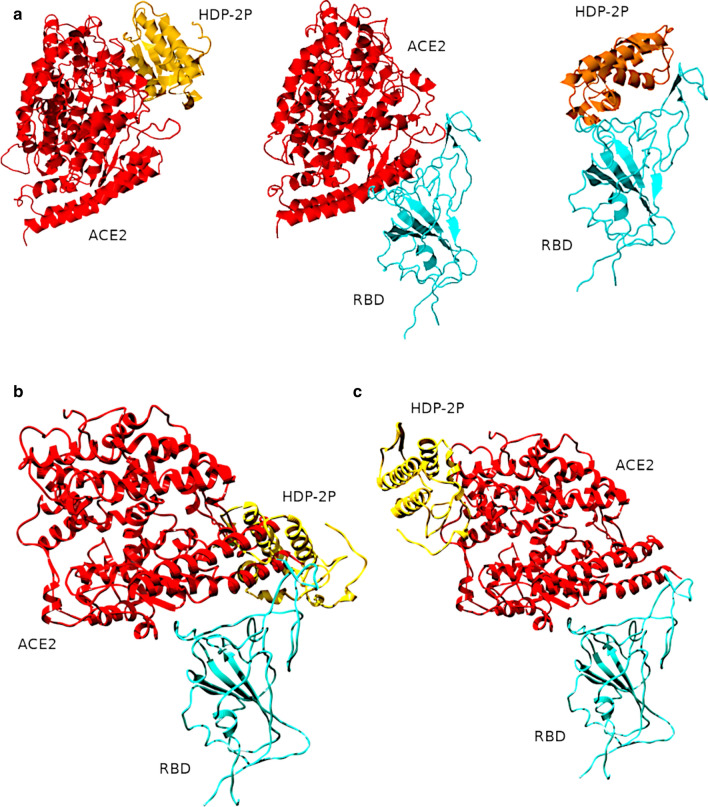
A homology model of the HDP-2P subunit was generated using the Swissmodel Web-server [48] with the X-ray structure (PDB 1jlt) of another snake venom PLA2 vipoxin as a template [49]. Initial docking was performed using the Frodock server [50] with the best-scored positions indicated in Fig. 7A. The data aligned well with the structure of RBD-ACE2 complex (PDB 7A98 [51]) which was used as a positive control, and as a source of individual ACE2 and RBD structures (Fig. 7A central panel). The resulting structures of the complexes of HDP-2P with RBD and ACE2 were submitted for local optimisation docking using the Rosetta-2 Web-server [52]. The resulting optimized structures were visualised using UCSF Chimera [53].
Fig. 7.
Molecular models for HDP-2P (gold) complexes with ACE2 (red), and RBD (turquoise) obtained by protein docking. A Structures of HDP-2P complexed with ACE2 (left panel) and S-protein RBD (right panel) determined using Frodock. The central (control) panel shows re-docking of ACE2 to RBD, which results in a structure that resembles published X-ray structures; B A Rosetta-optimized binding interface between HDP-2P and RBD intersects the ACE2-RBD binding surface, which could prevent RBD from interacting with ACE2; C A different Rosetta-optimized binding interface between HDP-2P and ACE2 where HDP-2P docks away from the ACE2-RBD binding surface, implying no direct interplay between viral protein and PLA2
Surface plasmon resonance (SPR)
SPR measurements were performed using a multiparametric SPR instrument NAVI 220A (DBA BioNavis Ltd., Tampere, Finland). Recombinant human ACE2 (Cusabio, Houston, TX, USA) was immobilized on 3D carboxymethyl dextran hydrogel-coated sensor slides. The following protocol was used for ACE2 immobilization:
The sensor was washed with 2 M NaCl solution for 7 min at a flow rate of 30 μl/min;
The sensor was activated by washing with a mixture of 0.05 M N-hydroxysuccinimide and 0.2 M 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (dissolved in 5 mM 2-(N-morpholino)ethanesulfonic acid buffer) for 7 min at a flow rate of 30 μl/min;
ACE2 (0.35 mg/ml) dissolved in 5 mM acetic acid, pH 4 was applied for 25 min at a flow rate of 10 μl/min;
The sensor was washed with water for 40 min to complete the coupling reaction;
Ethanolamine (pH 9) was applied for 6 min at a flow rate of 30 μl/min to quench the reaction.
Protein immobilization was confirmed by comparing the baseline level at the beginning and at the end of the procedure. Measurements were carried out simultaneously in two channels, one of which was used as a control. HDP-2P (0.5 mg/ml) dissolved in PBS pH 7.5 was applied for 20 min at a flow rate of 10 μl/min. RBD (0.1 mg/ml, kindly provided by V.Yu. Kost, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Moscow) dissolved in PBS pH 7.5 was applied for 25 min at a flow rate of 10 μl/min.
Statistical analysis
Data were analyzed using Student’s t test or one-way ANOVA with a Tukey’s post hoc test for multiple comparisons. P < 0.05 was considered statistically significant. All analyses were performed with GraphPad Prism version 6.
Results
Secretory PLA2s from snake venoms
We studied eight samples of snake venom PLA2s and their subunits. Two PLA2s were from the venom of the krait Bungarus fasciatus: BF-PLA2-II (phospholipase A2 II; GenBank AAK62361.1), and BF-PLA2-1 (basic phospholipase A2 1; UniProtKB Q90WA7), both of which belong to group IA. All other PLA2s tested in this work belong to group IIA: Vur-PL2 (UniProtKB F8QN53) was from the viper V. ursinii renardi. HDP-1 and HDP-2 were from the viper V. nikolskii; these are dimers composed of the enzymatically active subunits HDP-1P (UniProtKB Q1RP79) and HDP-2P (UniProtKB Q1RP78), respectively, combined with the enzymatically inactive HDP-1I (UniProtKB A4VBF0) for both dimers. HDP-2 was separated into HDP-2P and HDP-1I for further studies using reverse phase chromatography. All these PLA2s are Asp-49 enzymes with the exception of HDP-1I which contains Gln residue instead of Asp at position 49. An inactive version of HDP-2P was also created by treating it with 4-bromophenacyl bromide, which selectively alkylates the His residue in the active site. This modified analogue was purified by reversed-phase HPLC (yield of the target derivative = 45%) and analyzed by mass spectrometry, which revealed the increase in mass by 198 Da, the molecular weight of one 4-bromophenacyl residue. Testing the enzyme activity of the modified HDP-2P (called HDP-2P inact) showed that phospholipolytic activity decreased by about 2200-fold (Figure S1).
Snake venom PLA2s possess high antiviral activity against SARS-CoV-2
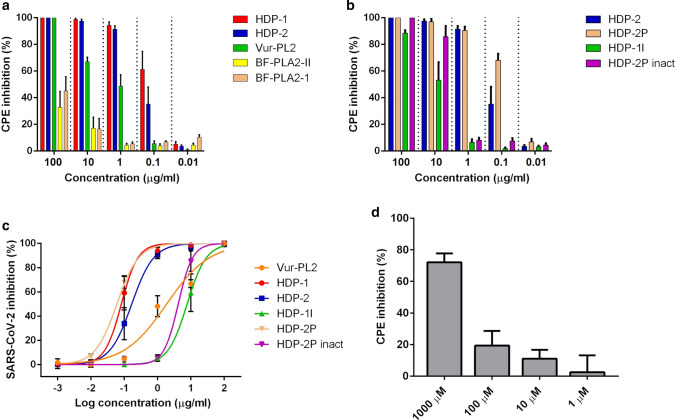
The cytopathic effects (CPE) of SARS-CoV-2 on Vero E6 cells were used to assay the antiviral activity of five snake venom PLA2s, two of which, HDP-1 and HDP-2, were dimeric, and the antiviral activity of two subunits (HDP-1I and HDP-2P) isolated from HDP-2 was also examined. The Vero E6 cell line is widely used in virology, as viruses produce obvious CPE. The variants of SARS-CoV-2 used in this work belong to lineage B.1.1, which is the main lineage in Russia and is also widely spread worldwide. Our data show SARS-CoV-2 infection results in several morphological changes including cell rounding, detachment and cytolysis (Figure S2). We found that all five tested PLA2s showed antiviral activity as morphological changes induced by the virus were prevented by PLA2s (Figure S2). The two dimeric PLA2s HDP-1 and HDP-2 showed the strongest antiviral activity, inhibiting ~ 50% of the CPE at a concentration of 0.1 µg/ml (Fig. 1a, b, Table 1). Vur-PL2 was less potent, inhibiting 50% of the CPE at 1 µg/ml, while BF-PLA2-II and BF-PLA2-1 showed the lowest level of activity, inhibiting less than 50% of the CPE at 100 µg/ml (Fig. 1a).
Fig. 1.
Snake venom PLA2s possess high inhibitory activity against the cytopathic effect of SARS-CoV-2 on the Vero E6 cells. Vero E6 cells were infected with SARS-CoV-2 at 100 TCID50 (50% tissue culture infectious dose) in the presence of PLA2s at the indicated concentrations for 72 h. CPE inhibition was then measured by colorimetric assay with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). n = 3. a Influence of PLA2s from different venoms on CPE of SARS-CoV-2. b Influence of HDP-2 and its subunits on CPE of SARS-CoV-2. c Dose–response curves for determination of half-maximum inhibitory concentration (IC50) values. Nonlinear regression was used to fit experimental points. d Arachidonic acid (AA) weakly inhibits the CPE of SARS-CoV-2 on Vero E6 cells. Vero E6 cells were infected with SARS-CoV-2 at 100 TCID50 in the presence of different concentrations of the AA for 72 h. CPE inhibition was then measured by colorimetric MTT assay. Results are representative of at least five biologically independent samples and shown as mean ± SD
Table 1.
Half-maximum inhibitory concentration (IC50) values for antiviral effects of PLA2s against SARS-CoV-2
| PLA2 | IC50, µg/ml | 95% CIa, µg/ml |
|---|---|---|
| Vur-PL2 | 1.88 | 1.27–2.79 |
| HDP-1 | 0.08 | 0.058–0.12 |
| HDP-2 | 0.17 | 0.12–0.24 |
| HDP-1I | 7.71 | 5.28–11.27 |
| HDP-2P | 0.06 | 0.05–0.07 |
| HDP-2P inact | 4.17 | 3.01–5.77 |
aCI confidence interval
Next, we studied the antiviral activity of two isolated subunits of HDP-2: the catalytically active subunit HDP-2P showed twofold higher antiviral activity than the parent HDP-2, while the enzymatically inactive subunit HDP-1I had ~ 100 fold lower activity (Fig. 1b, c). This suggests that antiviral activity may be associated with phospholipolytic activity. To determine the contribution of the phospholipase activity of the HDP-2P subunit to the inhibition of SARS-CoV-2, the enzymatic activity of this subunit was inhibited by chemical modification of the active site (by producing HDP-2P inact): after such modification the antiviral activity decreased 70-fold (Fig. 1b, Table 1). As PLA2s catalyze hydrolysis at the sn2 position of membrane glycerophospholipids to lysophospholipids and free fatty acids, mostly arachidonic acid (AA), we also tested the influence of AA on the CPE. AA did inhibit the CPE, but only at high (mM) concentration (Fig. 1d).
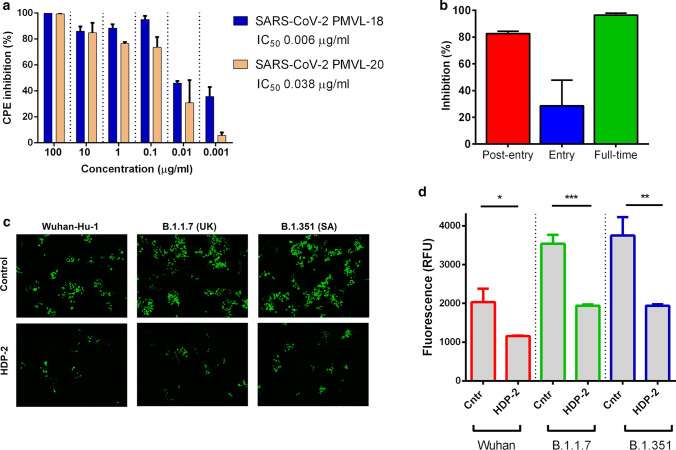
A number of genetically distinct variants of SARS-CoV-2 have appeared in recent months, some of which are associated with an increased rate of spread of the virus [54–57]. To determine whether PLA2s exhibit antiviral activity against these viruses, HDP-2 was tested using two SARS-CoV-2 isolates containing deletions and mutation in glycoprotein S. HDP-2 showed potent antiviral activity with IC50 values of 0.006 and 0.038 μg/ml for hCoV-19/Russia/Moscow_PMVL-18 and hCoV-19/Russia/Moscow_PMVL-20 SARS-CoV-2 strains, respectively (Fig. 2a).
Fig. 2.
PLA2s exhibit antiviral activity against viruses containing mutations and interfere with different stages of the SARS-CoV-2 replication cycle. a CPE inhibition assay. Vero E6 cells were infected with SARS-CoV-2 strain PMVL-19 or PMVL-20 at 100 TCID50 in the presence of different concentrations of HDP-2 for 72 h. CPE inhibition was then measured by colorimetric assay with MTT. b Time-of-drug-addition assay. Vero E6 cells were infected with SARS-CoV-2 at a multiplicity of infection (MOI) of 0.01, and virus yield in the infected cell supernatants was quantified by qRT-PCR 18 h after infection. c 293T/ACE2 were infected with different pseudo-SARS-CoV-2-GFP either in the presence of vehicle (PBS, Control) or HDP-2 (10 µg/ml). Representative fluorescent microscopy images of 293T/ACE2 cells infected with the pseudo-SARS-CoV-2 and treated with HDP-2. HDP-2 treatment led to a decrease in the entry of pseudoviruses, which was manifested in a decrease in the number of GFP-positive cells compared to the control. Scale bars, 100 µm. d Infectivity of pseudo-SARS-CoV-2 particles on 293T/ACE2 cells was quantified by measuring GFP fluorescence. Wuhan, B.1.1.7 and B.1.351 are the Wuhan reference strain, the lineage B.1.1.7 (United Kingdom) and the lineage B.1.351 (South Africa), respectively. Significant difference was determined using a Student’s t test: *p˂0.05; **p˂0.01; ***p˂0.001. All results are shown as mean ± SD of n = 3 or 5 biologically independent samples. RFU relative fluorescence units
To investigate which stage(s) of the SARS-CoV-2 replication cycle were interrupted by PLA2, we treated virus-infected Vero E6 cells with PLA2 (10 µg/ml) at different time points (a time-of-drug-addition assay), and measured the viral load using quantitative real-time PCR (qRT-PCR) (Fig. 2b). The assay showed that HDP-2 efficiently inhibited SARS-CoV-2 infection at stages “full-time” and “post-entry”, while smaller inhibition (~ 40%) was observed at stage “entry”. We next determined whether HDP-2 can block SARS-CoV-2 pseudo-virions entering the cell through binding to ACE2 (Fig. 2c, d). For this, the pseudo-SARS-CoV-2 (either the Wuhan reference strain or the lineage B.1.1.7 (United Kingdom) and B.1.351 (South Africa) spike protein) was incubated with HDP-2, after which the mixture was added to 293T cells expressing the human ACE2 receptor (293T/ACE2 cells). After 72 h of infection, green fluorescent cell (GFP cells) counts were performed. At 10 µg/ml, HDP-2 showed ~ 50% inhibition of pseudo-SARS-CoV-2 entry into 293T/ACE2 cells (Fig. 2d), indicating that PLA2 can interfere with SARS-CoV-2 attachment to the cellular surface. Lentivirus pseudotyped with vesicular stomatitis virus G glycoprotein (VSV-G) was used as a control, and we found HDP-2 also blocked the entry of pseudo-VSV-G into 293T/ACE2 cells (Figure S3). To analyze HDP-2 effects at the stage «post-entry» in more detail, Vero E6 cells were infected with SARS-CoV-2 for 24 h, after which various dilutions of HDP-2 were added to the cells. It was found that HDP-2 blocked the CPE in virus-infected cells at concentrations as low as 0.1–0.01 µg/ml, which indicated its high activity at the «post-entry» stage (Figure S4A). For further analysis of “post-entry” mechanisms, we used pseudotyped SARS-CoV-2 virus. Treatment of cells with HDP-2 at a concentration of 100 μg/ml for 72 h after transduction with a pseudovirus revealed an ~ twofold decrease in the expression compared to the control (Figure S4B). Taken together, these results suggest that HDP-2 inhibits «post-entry» stages of virus infection prior to/at translation.
Snake venom PLA2s exhibit low cytotoxic activity against Vero E6 cells
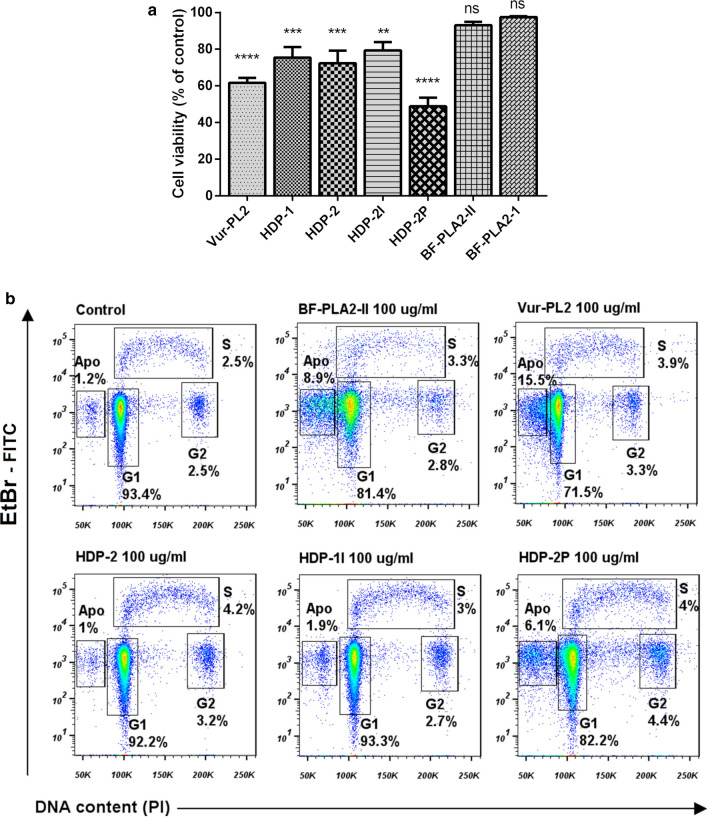
To evaluate direct effects of PLA2s on Vero E6 cells, the cytotoxicity of PLA2s was studied. PLA2s slightly decreased cell viability, but did not produce morphological changes in the cell monolayer (Figure S5). The highest, albeit moderate, cytotoxicity was manifested by Vur-PL2 and HDP-2P which at 100 μg/ml reduced cell viability on average by 38% and 51%, respectively (Fig. 3a). Only HDP-2P at the maximal concentration used (100 μg/ml) had pronounced cytotoxicity with a change in cell morphology.
Fig. 3.
Snake venom PLA2s exhibit low cytotoxicity to Vero E6 cells. a Cytotoxicity data for all tested PLA2s at 100 µg/ml. Data are mean ± SD from three independent experiments with two measurements. The data are presented as the mean ± SD. One-way ANOVA with Tukey post hoc test was applied to assess statistically significant differences: **p˂0.01; ***p˂0.001; ****p˂0.0001; ns p > 0.05. b Flow cytometry analysis of the cell cycle. Vero E6 cells treated with different PLA2s at 100 µg/ml. After 72 h of incubation, cells were labeled with bromodeoxyuridine (BrdU), treated with HCl, stained with FITC-labeled anti-BrdU antibody and propidium iodide (PI), and analyzed by flow cytometry. Vero E6 treated with PBS were used as a control. Numbers show percentages of cells in S phase (upper) and G1 or G2 phase (lower) in each dot plot. Apo apoptotic cells
The effect of PLA2 on cell proliferation was also examined using cell cycle analysis. For this, the maximum concentration (100 µg/ml) of each PLA2 was added to Vero E6 cells, which were then incubated for 72 h (Fig. 3b). Analysis of the treated cells by flow cytometry showed that the majority of these cells were in G1 phase, with a small percentage in the early S and G2 phases. The cell cycle profiles for cells treated with PLA2s did not broadly differ from the profile for untreated cells, except that BF-PLA2-II, Vur-PL2 and HDP-2P treatment resulted in small increases in the percentage of apoptotic cells.
Dimeric PLA2 HDP-2 and its subunits HDP-2P and HDP-1I possess potent virucidal activity and inactivate coronavirus
To elucidate whether the PLA2s exert their antiviral activity through phospholipolytic effects on the cell membrane or on the virus, we analyzed the virucidal activity of the HDP-2 and HDP-2P (which manifested the highest antiviral activity), as well as of HDP-1I and HDP-2P inact. To probe a direct inhibitory effect on viral particles, SARS-CoV-2 was treated with various concentrations (0.1–10 µg/ml) of PLA2 or buffer, and the results were evaluated using visual scoring of the CPE on Vero E6 cells (Fig. 4). Complete suppression of the infectivity of SARS-CoV-2 was observed when the viral stock was treated with HDP-2P even at 0.1 µg/ml, at which concentration HDP-2, HDP1I and HDP-2P inact showed significant residual viral infectivity. HDP-2P inact also showed residual infectivity at 1 µg/ml, supporting our hypothesis that HDP-2 can only act efficiently when it retains phospholipolytic activity.
Fig. 4.
Dimeric PLA2 HDP-2 and its subunits HDP-2P and HDP-1I possess potent virucidal activity; they inactivate coronaviruses and disrupt their membranes. a Virucidal activity: SARS-CoV-2 (1 × 106 TCID50) was incubated with serial tenfold dilutions of PLA2s for 1 h at 37 °C. The treated viruses were used to infect Vero E6 cells. Virus infectivity was calculated by the reduction of the virus titer after treatment with PLA2 compared to untreated virus. The data are presented as mean ± SD of three independent experiments and analyzed by one-way ANOVA with a Tukey post hoc test: all PLA2 concentrations were significantly different to control, p < 0.0001. TCID50 50% tissue culture infectious dose. Cntr control. b, c Transmission electron microscopy (TEM) demonstrates that HDP-2 disrupts the integrity of the viral envelope and alters viral morphology. Untreated bovine coronavirus BCV (b) or SARS-CoV-2 (c) (Control) were treated with 10 μg/ml of HDP-2 for 1 h at 37 °C. Two different representative images are shown for both control and treated virions. In c, the arrow shows the accumulation of contrasting material inside the virion. Viral particles exposed to HDP-2 show a disrupted envelope and, as a consequence, altered morphology of viral units. In b, scale bar = 50 nm
The mechanisms of action of HDP-2 against bovine coronavirus (BCV; a prototypical SARS-CoV-2 coronavirus) and SARS-CoV-2 itself were evaluated using transmission electron microscopy. The viral envelope of the untreated BCV was intact and the virions had a predominantly spherical morphology, while exposure to 10 μg/ml HDP-2 resulted in the destruction of the envelope and gross distortion of the virus ultrastructure, revealing a significant change in the morphology (Fig. 4b). The treated virus suspension contained two forms of virions:virions with partially destroyed membranes and defective morphology, or virions with completely destroyed membranes (deformed, disintegrated particles). Similar effects of HDP-2 on the morphology of SARS-CoV-2 were observed (Fig. 4c) as virions treated with HDP-2 revealed many empty virus shells which were permeable to negative stain (Fig. 4c). These damaged virions are likely caused by lipid hydrolysis in the viral membrane.
Snake venom PLA2s inhibit cell–cell fusion mediated by SARS-CoV-2 glycoprotein S
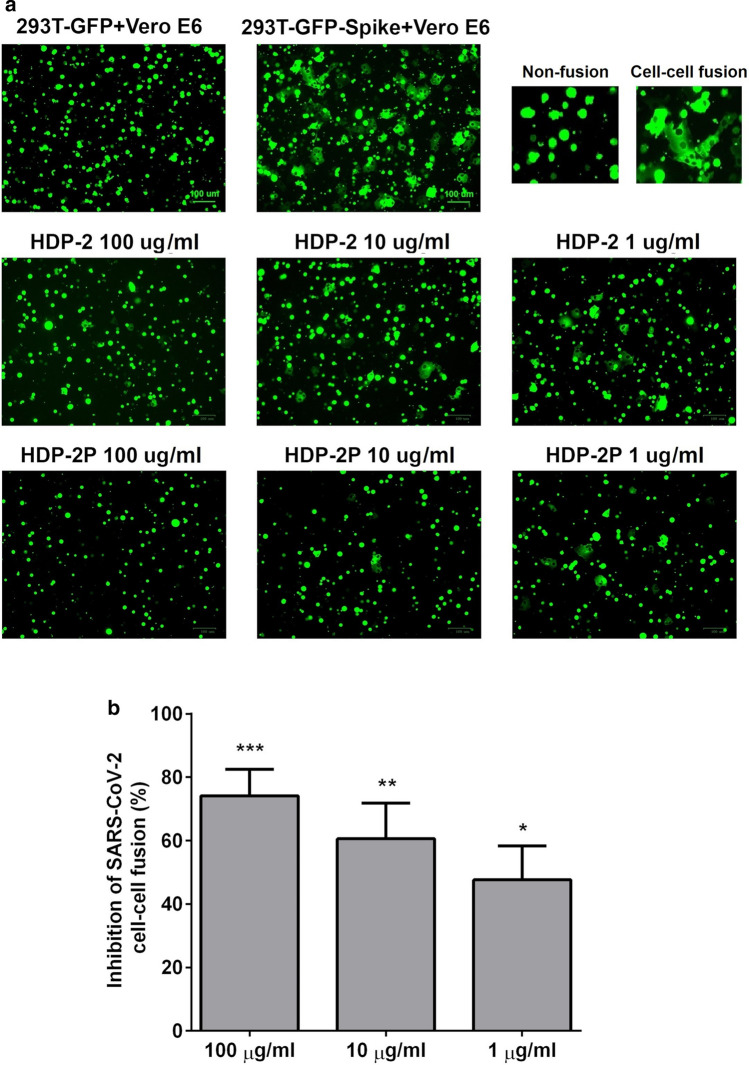
To study cell fusion mediated by glycoprotein S interaction with the ACE2 receptor, we used 293T cells expressing green fluorescent protein (GFP) and SARS-CoV-2 glycoprotein S (293T-GFP-Spike), and Vero E6 cells expressing ACE2. After co-cultivation of the effector 293T-GFP-Spike cells and target Vero E6 cells in the presence of the five snake venom PLA2s, the number of fused cells was counted using a fluorescence microscope. The data showed that HDP-1 and HDP-2 inhibited glycoprotein S mediated cell–cell fusion by ~ 50% at 1 µg/ml and 70% at 100 µg/ml (Fig. 5A, B). HDP-2P at 100 µg/ml completely blocked the cell–cell fusion (Fig. 5A), although Vur-PL2 and BF-PLA2-II at 100 µg/ml showed no significant inhibition (Figure S6).
Fig. 5.
Snake venom PLA2s inhibit cell–cell fusion mediated by SARS-CoV-2 glycoprotein S. A Images of SARS-CoV-2 glycoprotein S mediated cell–cell fusion after a 2 h incubation at different concentrations of PLA2s. Scale bar = 100 µm. B Percent inhibition of cell–cell fusion by HDP-2. Cell–cell fusion was calculated relative to the number of fused cells in wells untreated with HDP-2. Experiments were repeated twice, and the data are expressed as means ± SD. One-way ANOVA with a Tukey post hoc test was applied to assess statistically significant differences: *p˂0.05; **p˂0.01; ***p˂0.001
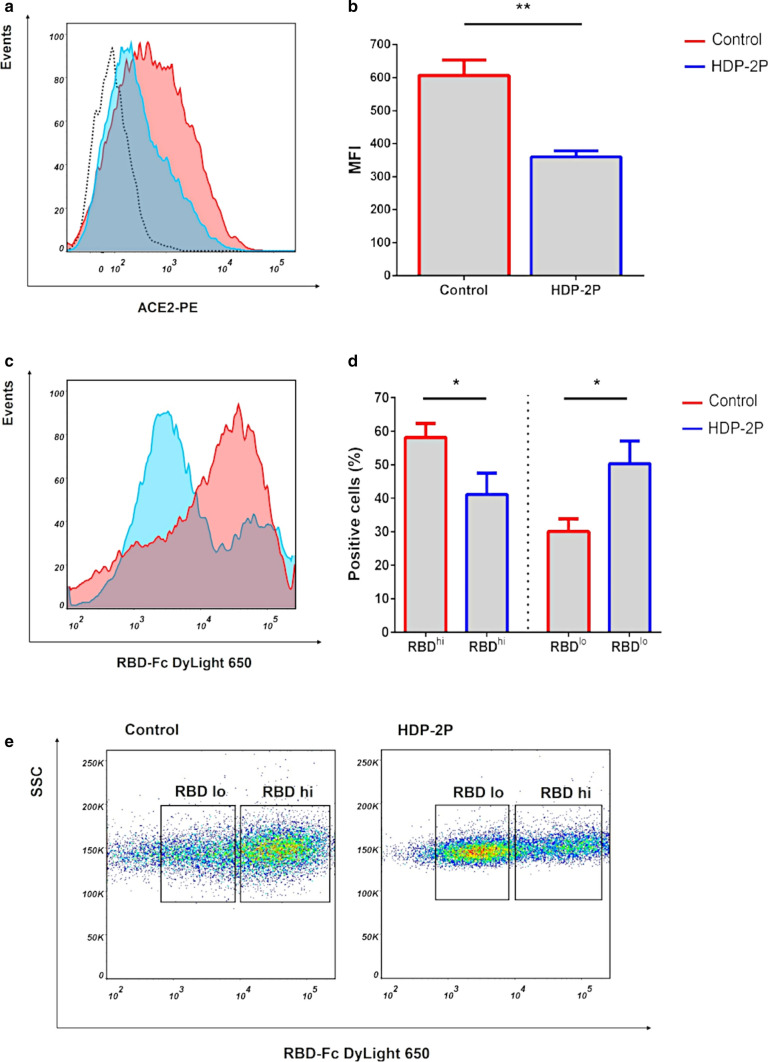
HDP-2P reduces the binding of anti-ACE2 antibody and receptor-binding domain (RBD) of glycoprotein S to ACE2 receptor
As the catalytic subunit HDP-2P (at 100 µg/ml) completely inhibits glycoprotein S mediated cell–cell fusion and suppresses SARS-CoV-2 replication, an interaction of HDP-2P with the ACE2 receptor, the major receptor for SARS-CoV-2, is a reasonable hypothesis. To test this, we analyzed the effect of this PLA2 on the binding of an ACE2-specific antibody to 293T/ACE2 cells (Fig. 6a, b). Binding of this antibody to ACE2 decreased twofold in the presence of HDP-2P, as measured by flow cytometry analysis, which suggests that HDP-2P may bind to ACE2 receptors. To test this hypothesis, we analyzed the HDP-2P inhibitory effect on the binding of the SARS-CoV-2 receptor binding domain protein (RBD) to the ACE2 receptor on 293T/ACE2 cells using flow cytometry (Fig. 6c, d). The staining of the cells revealed two populations which differed in the fluorescence intensity possessing a high (RBDhi) and a low (RBDlo) fluorescence (Fig. 6e). While most cells in the control were RBDhi, in the cell population treated with HDP-2P there was an increase in the percentage of RBDlo cells and a decrease in RBDhi cells. Thus, the treatment with HDP-2P decreased the number of RBD bound by the cell, indicating HDP-2P-mediated inhibition of RBD protein binding to the ACE2 receptor. Using real-time biomolecular interaction analysis with surface plasmon resonance (SPR), we further studied the interaction of HDP-2P with ACE2 protein. The results demonstrated that HDP-2P could bind to the ACE2 (Figure S7 A) and a sensorgram showed this occurred within 3 min, i.e. it was a very fast interaction. The complex formed was stable as it did not dissociate before washing out the sensor with buffer. As a control, the binding of SARS-CoV-2 RBD to ACE2 was also examined (Figure S7 B). RBD interaction with ACE2 proceeded slowly, taking about 20 min to reach equilibrium, and the complex formed began to dissociate immediately after reaching equilibrium. These data indicate that HDP-2P binds to ACE2 and this binding is stronger than that of SARS-CoV-2 RBD.
Fig. 6.
HDP-2P reduces the binding of an anti-ACE2 antibody and RBD of glycoprotein S to ACE2 receptor at 293T/ACE2 cells. a, b Inhibition of anti-ACE2 antibody binding to 293T/ACE2 cells by HDP-2P. 293T/ACE2 cells were incubated with (100 µg/ml) or without (control) HDP-2P for 30 min and stained using human phycoerythrin (PE) conjugated anti-mouse ACE2 antibody. MFI mean fluorescence intensity. c, d Cells were incubated with PBS (control) or HDP-2P. The RBD protein fused with human Fc was then added for 1 h. After washing, the binding of RBD was detected using a DyLight 650-conjugated secondary anti-human Fc antibody. e Representative RBD binding profile in control and HDP-2P-treated cells. The cell populations binding high (RBDhi) and low (RBDlo) amount of RBD are shown. The percentage of positive cells was determined using flow cytometry analysis. Significant difference was determined using a Student’s t test: *p˂0.05, **p˂0.01. Results are mean ± SD and are representative of at least three independent determinations
Molecular modeling of interactions of HDP-2P with ACE2 and RBD of SARS-CoV-2
To clarify possible molecular interactions of HDP-2P with ACE2 and RBD of SARS-CoV-2, we performed molecular modelling using Frodock and Rosetta 2 docking servers. Both ACE2 and RBD were predicted to be potential docking partners of HDP-2P with scoring function values better than the control ACE2-RBD complex. The Rosetta algorithm scores were − 578.669 and − 250.419 for ACE2 and RBD interaction with HDP-2P, respectively, vs. − 248.931 for the control complex. This suggests a putative mechanism of snake venom PLA2s action on SARS-CoV-2. The predicted complex of HDP-2P with RBD has a phospholipase site near the structurally disordered RBD loop which, according to X-ray structures, interacts with ACE2. A partial overlap (Fig. 7B) between HDP-2P and ACE2 binding interfaces on RBD could explain some of PLA2s direct effects on viral attachment to cells. Not all structural data, however, are consistent with this hypothesis: one predicted complex where the interaction of HDP-2P at the opening of ACE2 catalytic cleft was scored as the most energy favourable among all those considered here, showed that the binding interfaces of HDP-2P and RBD with ACE2 do not intersect (Fig. 7C), thus, suggesting that HDP-2P binding to ACE2 may exert only indirect effects on viral attachment.
Discussion
Current efforts to combat the COVID-19 pandemic are primarily focused on hygiene, quarantine of infected people, social distancing and the development and use of vaccines [58, 59]. Nevertheless, despite considerable success with the latter, many hundreds of thousands of people are still being infected, and, both now and in the future (due to the rapid viral mutation rate) patients are in need of effective therapeutic interventions. Russia belongs to the countries with the highest number of confirmed cases of COVID-19, making it a potential source of new virus variants. The vast majority of variants found in Russia originated from Europe and are of haplotype B; the variants of SARS-CoV-2 used in this study belong to lineage B.1.1. We selected this because at the beginning of this work, this lineage was the main lineage in Russia [60] and was also widespread in the United Kingdom (33%), United States of America (13%), Japan (9%), Germany (4%), and Turkey (3%); indeed more than forty thousand sequences in GISAID are assigned to this lineage. In addition, several virus variants of this lineage are referred to as wild-type strains and some VOCs including B.1.1.7 were derived from this lineage.
We show here that a promising starting point for a novel COVID-19 antiviral agent are snake venom PLA2s, as a number of these have antiviral activity against SARS-CoV-2. The most potent PLA2 in our study was the dimeric phospholipase HDP-2, and our work has revealed the possible mechanism by which this enzyme can inhibit infection. Our data have shown that HDP-2 must retain enzyme activity to have antiviral activity, and treatment of SARS-CoV-2 and BCV with HDP-2 causes not only a change in the morphology of virions but significant destruction of the viral membrane. These data suggest that HDP-2 cleaves the membrane lipids. This is consistent with previous studies which have shown that crotoxin, a dimeric PLA2 isolated from Crotalus durissus terrificus venom, and its enzymatically active subunit PLA2-CB, inactivated Dengue Virus Type 2 (Flaviviridae), Rocio virus (Flaviviridae), Oropouche virus (Peribunyaviridae) and Mayaro virus (Togaviridae) by cleaving the envelope glycerophospholipids that originate from the membranes of host cells, and were inactive against non-enveloped Coxsackie B5 (Picornaviridae) and encephalomyocarditis virus (Picornaviridae) or viruses budding through the plasma membrane [28, 61]. A recent study of PLA2s isolated from the venom of Bothrops asper pit viper showed the high virucidal activity against Flaviviridae strains for an enzymatically active PLA2 Mt-I, while a natural enzymatically-inactive PLA2 homolog Mt-II and inactivated Mt-I possessed greatly diminished activity [62]. These data supported the role of catalysis in the viral inhibition mechanism. In addition, several studies support the idea that PLA2s belonging to groups V and X inhibit adenoviral infection by hydrolysis of the plasma membrane of host cells [31]. Not all enveloped viruses, however, are susceptible to PLA2, and this is likely to be because the composition of phospholipids in the endoplasmic reticulum membrane differs from those in the plasma membrane, resulting in different compositions of phospholipids in the envelopes of different viruses [63, 64]. Thus, depending on the origin, and, therefore, the composition of the envelope, PLA2s can differently affect the viruses and exert different virucidal activities. Interestingly, it was recently shown that HDP-2 induced an aggregation and stacking of lipid bilayers [65].
The products of PLA2 activity may also play an important role in their activity; for example lysophosphatidylcholine, from PLA2 mediated hydrolysis of phosphatidylcholine, has been shown to inhibit membrane fusion caused by influenza (Orthomyxoviridae), simian immunodeficiency (Retroviridae), Sendai (Paramyxoviridae), and rabies (Rhabdoviridae) viruses blocking viral entry into host cells [66, 67]. Another product of membrane lipid hydrolysis by PLA2s is AA, the inhibitory effect of which on virus replication has been demonstrated for MERS-CoV [68]. We found that AA inhibited CPE of SARS-CoV-2, albeit at a high concentration. Thus, these data show that AA released upon membrane lipid hydrolysis has the potential to contribute to the inhibitory action of PLA2s.
Investigation of the stage(s) of the replication cycle affected by PLA2 showed that stages “full-time” and “post-entry” were efficiently inhibited by HDP-2 and stage “entry” was less influenced (although it should be noted that a few snake venom PLA2s can be internalized and exert effects inside cells [69, 70]). Moderate inhibition of post‑entry steps of Chikungunya virus infection was observed for phospholipase A2CB from the venom of Crotalus durissus terrificus [33]. Using pseudotyped virus labelled with GFP we observed that in a “post-entry” experiment, HDP-2 inhibited the expression of GFP. These data suggest that transcription, integration, and/or translation are affected by PLA2. However as HDP-2 is active against SARS-CoV-2 at a much higher concentrations, the later stages of the replication cycle which include assembly, budding and release may also be damaged by PLA2s perhaps because these stages proceed with the participation of lipids from the host cell which can be degraded by PLA2.
The envelopes of coronaviruses contain a spike protein (glycoprotein S) which forms a “crown”, and mediates viral entry into cells. Glycoprotein S is composed of two domains: S1, which binds to the ACE2 receptor on the surface of host cells, and S2 which mediates fusion of the viral envelope with the cell membrane. Following binding to ACE2, target cell proteases activate glycoprotein S, cleaving the site between S1 and S2, and rendering the latter now able to mediate viral fusion and penetration [71, 72]. We found that HDP-1 and HDP-2 are able to block SARS-CoV-2 glycoprotein S mediated cell–cell fusion, perhaps acting in a similar manner to peptide fusion inhibitors [45]. It is also possible that PLA2s may inhibit SARS-CoV-2 binding to ACE2, and our data showing that HDP-2P reduced the binding of anti-ACE2 and RBD to 293T/ACE2 cells support this hypothesis. In addition, we demonstrated a direct HDP-2P-ACE2 interaction using SPR. Further support comes from molecular modelling of HDP-2P interactions with ACE2 and RBD of SARS-CoV-2. Docking studies showed that HDP-2P can form fairly stable complexes both with ACE2 and RBD, and, as SARS-CoV-2 utilizes ACE2 for cell adhesion and entering, HDP-2P could interrupt viral adhesion directly by interacting with the ACE2-binding site on RBD. Alternatively, the interaction of HDP-2P with ACE2 may influence viral adhesion indirectly by altering ACE2 conformation. Such competitive effects have been previously observed with a number of peptides from scorpion venom, which are active against retroviruses such as HIV/SIV through their ability to bind to the HIV glycoprotein gp120 due to molecular mimicry of the CD4+ receptor. As a result, they block gp120-CD4 interactions, which are important for the initiation of conformational changes in the viral envelope that trigger virus entry into the cells [73].
In summary, we demonstrate here for the first time the antiviral activity of snake PLA2s against SARS-CoV-2, suggesting that these enzymes are active against the Coronaviridae family as they are against many other virus families. Dimeric phospholipase HDP-2 and its catalytic subunit showed virucidal activity in the nanomolar range, activity that was significantly decreased by inhibition of phospholipase enzymatic activity, indicating the effect is most likely due to the cleavage of phospholipids on the virus envelope, which can lead to the destruction of the lipid bilayer and destabilization of surface glycoproteins in the virus. Additionally, PLA2s may prevent virus entry into the cell by inhibiting its binding to ACE2. These data highlight the potential of PLA2s as a natural product that could prove to be fruitful as the starting point for the development of antiviral drugs.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Mr. V.G. Zhukhovitsky for his help in experiments with electron microscopy and Mr. V. Yu. Kost for providing receptor binding domain of S-glycoprotein.
Author contributions
Conceptualization, AES, SCRL, YNU; Methodology, MAS; Investigation, AES, SDG, DSK, VAM, AVO, MAN, SDG; Resources VGS; Data Curation, VAG; Writing—Original Draft, AES; Writing—Review and Editing, SCRL, YNU, VIT; Supervision, YNU, VIT. All authors read and approved the final manuscript.
Funding
This work was supported by the Russian Foundation for Basic Research (RFBR) Grant No. 20-04-60277. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Availability of data and materials
Data are available on request from the corresponding author.
Code availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable. No animals or humans were involved in this study. The venoms used in this work were obtained from commercial sources as indicated in Materials and Methods.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Andrei E. Siniavin, Email: andreysi93@ya.ru
Yuri N. Utkin, Email: utkin@ibch.ru
References
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acids Res. 2018;46:D708–D717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hulse JD. Human coronaviruses: the deadly seven. ACTA Sci Microbiol. 2020;3:86–89. doi: 10.31080/ASMI.2020.03.0612. [DOI] [Google Scholar]
- 5.Chiu SS, Chan KH, Chu KW, Kwan SW, Guan Y, Poon LLM, Peiris JSM. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, Qiu Z, Liu J, Fan T, Liu C, Tian P, Wang Y, Ni Z, Zhang S, Luo J, Liu D, Li W. Analysis of the clinical characteristics of 77 COVID-19 deaths. Sci Rep. 2020;10:16384. doi: 10.1038/s41598-020-73136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Souza WM, Buss LF, Candido DDS, Carrera JP, Li S, Zarebski AE, Pereira RHM, Prete CA, Jr, de Souza-Santos AA, Parag KV, Belotti MCTD, Vincenti-Gonzalez MF, Messina J, da Silva Sales FC, Andrade PDS, Nascimento VH, Ghilardi F, Abade L, Gutierrez B, Kraemer MUG, Braga CKV, Aguiar RS, Alexander N, Mayaud P, Brady OJ, Marcilio I, Gouveia N, Li G, Tami A, de Oliveira SB, Porto VBG, Ganem F, de Almeida WAF, Fantinato FFST, Macário EM, de Oliveira WK, Nogueira ML, Pybus OG, Wu CH, Croda J, Sabino EC, Faria NR. Epidemiological and clinical characteristics of the COVID-19 epidemic in Brazil. Nat Hum Behav. 2020;4:856–865. doi: 10.1038/s41562-020-0928-4. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Petitjean SJL, Koehler M, Zhang Q, Dumitru AC, Chen W, Derclaye S, Vincent SP, Soumillion P, Alsteens D. Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor. Nat Commun. 2020;11:1–10. doi: 10.1038/s41467-020-18319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, Leist SR, Schäfer A, Dinnon KH, 3rd, Stevens LJ, Chappell JD, Lu X, Hughes TM, George AS, Hill CS, Montgomery SA, Brown AJ, Bluemling GR, Natchus MG, Saindane M, Kolykhalov AA, Painter G, Harcourt J, Tamin A, Thornburg NJ, Swanstrom R, Denison MR, Baric RS. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020;12:eabb5883. doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen KY. Coronaviruses-drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khatoon F, Prasad K, Kumar V. Neurological manifestations of COVID-19: available evidences and a new paradigm. J Neurovirol. 2020;26:619–630. doi: 10.1007/s13365-020-00895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoehl S, Rabenau H, Berger A, Kortenbusch M, Cinatl J, Bojkova D, Behrens P, Böddinghaus B, Götsch U, Naujoks F, Neumann P, Schork J, Tiarks-Jungk P, Walczok A, Eickmann M, Vehreschild MJGT, Kann G, Wolf T, Gottschalk R, Ciesek S. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. N Engl J Med. 2020;382:1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schöni R. The use of snake venom-derived compounds for new functional diagnostic test kits in the field of haemostasis. Pathophysiol Haemost Thromb. 2005;34:234–240. doi: 10.1159/000092430. [DOI] [PubMed] [Google Scholar]
- 15.Rivero JVR, de Castro FOF, Stival AS, Magalhães MR, Carmo Filho JR, Pfrimer IAH. Mechanisms of virus resistance and antiviral activity of snake venoms. J Venom Anim Toxins Incl Trop Dis. 2011;17:387–393. doi: 10.1590/S1678-91992011000400005. [DOI] [Google Scholar]
- 16.Koh CY, Kini RM. From snake venom toxins to therapeutics—cardiovascular examples. Toxicon. 2012;59:497–506. doi: 10.1016/j.toxicon.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 17.Calderon LA, Sobrinho JC, Zaqueo KD, de Moura AA, Grabner AN, Mazzi MV, Marcussi S, Nomizo A, Fernandes CF, Zuliani JP, Carvalho BM, da Silva SL, Stábeli RG, Soares AM. Antitumoral activity of snake venom proteins: new trends in cancer therapy. Biomed Res Int. 2014;2014:203639. doi: 10.1155/2014/203639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.da Mata ÉCG, Mourão CBF, Rangel M, Schwartz EF. Antiviral activity of animal venom peptides and related compounds. J Venom Anim Toxins Incl Trop Dis. 2017;23:3. doi: 10.1186/s40409-016-0089-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu JF, Pereira CM, Bittar C, Batista MN, Campos GRF, da Silva S, Cintra ACO, Zothner C, Harris M, Sampaio SV, Aquino VH, Rahal P, Jardim ACG. Multiple effects of toxins isolated from Crotalus durissus terrificus on the hepatitis C virus life cycle. PLoS ONE. 2017;12:e0187857. doi: 10.1371/journal.pone.0187857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Aziz TMA, Soares AG, Stockand JD. Snake venoms in drug discovery: valuable therapeutic tools for life saving. Toxins (Basel) 2019;11:564. doi: 10.3390/toxins11100564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh A, Roy R, Nandi M, Mukhopadhyay A. Scorpion venom-toxins that aid in drug development: a review. Int J Pept Res Ther. 2019;25:27–37. doi: 10.1007/s10989-018-9721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herzig V, Cristofori-Armstrong B, Israel MR, Nixon SA, Vetter I, King GF. Animal toxins—Nature’s evolutionary-refined toolkit for basic research and drug discovery. Biochem Pharmacol. 2020;181:114096. doi: 10.1016/j.bcp.2020.114096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutiérrez JM, Lomonte B. Phospholipases A2: unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon. 2013;62:27–39. doi: 10.1016/j.toxicon.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Kini RM. Snake venom phospholipase A2 enzymes in cell biology. In: Rochat H, Martin-Eauclaire MF, editors. Animal toxins methods and tools in biosciences and medicine. Basel: Birkhäuser; 2000. pp. 304–318. [Google Scholar]
- 25.Berg OG, Gelb MH, Tsai MD, Jain MK. Interfacial enzymology: the secreted phospholipase A2-paradigm. Chem Rev. 2001;101:2613–2653. doi: 10.1021/cr990139w. [DOI] [PubMed] [Google Scholar]
- 26.Dennis EA, Cao J, Hsu YH, Magrioti V, Kokotos G. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem Rev. 2011;111:6130–6185. doi: 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Paula R, Castro H, Rodrigues C, Melo P, Fuly A. Structural and pharmacological features of phospholipases A2 from snake venoms. Protein Pept Lett. 2009;16:899–907. doi: 10.2174/092986609788923365. [DOI] [PubMed] [Google Scholar]
- 28.Muller VDM, Russo RR, Oliveira Cintra AC, Sartim MA, De Melo A-P, Figueiredo LTM, Sampaio SV, Aquino VH. Crotoxin and phospholipases A 2 from Crotalus durissus terrificus showed antiviral activity against dengue and yellow fever viruses. Toxicon. 2012;59:507–515. doi: 10.1016/j.toxicon.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 29.Kim J-O, Chakrabarti BK, Guha-Niyogi A, Louder MK, Mascola JR, Ganesh L, Nabel GJ. Lysis of human immunodeficiency virus type 1 by a specific secreted human phospholipase A2. J Virol. 2007;81:1444–1450. doi: 10.1128/JVI.01790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drayton HA. Inactivation of Rous virus by phospholipase A. Nature. 1961;192:896. doi: 10.1038/192896a0. [DOI] [PubMed] [Google Scholar]
- 31.Mitsuishi M, Masuda S, Kudo I, Murakami M. Group V and X secretory phospholipase A2 prevents adenoviral infection in mammalian cells. Biochem J. 2006;393:97–106. doi: 10.1042/BJ20050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohn A, Klibansky C. Studies on the inactivation of cell-fusing property of newcastle disease virus by phospholipase A. Virology. 1967;31:385–388. doi: 10.1016/0042-6822(67)90183-3. [DOI] [PubMed] [Google Scholar]
- 33.Santos IA, Shimizu JF, de Oliveira DM, Martins DOS, Cardoso-Sousa L, Cintra ACO, Aquino VH, Sampaio SV, Nicolau-Junior N, Sabino-Silva R, Merits A, Harris M, Jardim ACG. Chikungunya virus entry is strongly inhibited by phospholipase A2 isolated from the venom of Crotalus durissus terrificus. Sci Rep. 2021;11:8717. doi: 10.1038/s41598-021-88039-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruglova N, Siniavin A, Gushchin V, Mazurov D. Different neutralization sensitivity of SARS-CoV-2 cell-to-cell and cell-free modes of infection to convalescent sera. Viruses. 2021;13:1133. doi: 10.3390/v13061133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran TV, Siniavin AE, Hoang AN, Le MTT, Pham CD, Phung TV, Nguyen KC, Ziganshin RH, Tsetlin VI, Weng CF, Utkin YN. Phospholipase A2 from krait Bungarus fasciatus venom induces human cancer cell death in vitro. PeerJ. 2019;7:e8055. doi: 10.7717/peerj.8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai IH, Wang YM, Cheng AC, Starkov V, Osipov A, Nikitin I, Makarova Y, Ziganshin R, Utkin Y. cDNA cloning, structural, and functional analyses of venom phospholipases A2 and a Kunitz-type protease inhibitor from steppe viper Vipera ursinii renardi. Toxicon. 2011;57:332–341. doi: 10.1016/j.toxicon.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Ramazanova AS, Zavada LL, Starkov VG, Kovyazina IV, Subbotina TF, Kostyukhina EE, Dementieva IN, Ovchinnikova TV, Utkin YN. Heterodimeric neurotoxic phospholipases A2-The first proteins from venom of recently established species Vipera nikolskii: Implication of venom composition in viper systematics. Toxicon. 2008;51:524–537. doi: 10.1016/j.toxicon.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Radvanyi F, Jordan L, Russo-Marie F, Bon C. A sensitive and continuous fluorometric assay for phospholipase A2 using pyrene-labeled phospholipids in the presence of serum albumin. Anal Biochem. 1989;177:103–109. doi: 10.1016/0003-2697(89)90022-5. [DOI] [PubMed] [Google Scholar]
- 39.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 40.Luo Y, Deng X, Cheng F, Li Y, Qiu J. SMC1-mediated intra-S-phase arrest facilitates bocavirus DNA replication. J Virol. 2013;87:4017–4032. doi: 10.1128/JVI.03396-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Runfeng L, Yunlong H, Jicheng H, Weiqi P, Qinhai M, Yongxia S, Chufang L, Jin Z, Zhenhua J, Haiming J, Kui Z, Shuxiang H, Jun D, Xiaobo L, Xiaotao H, Lin W, Nanshan Z, Zifeng Y. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol Res. 2020;156:104761. doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Meyer S, Bojkova D, Cinatl J, Van Damme E, Buyck C, Van Loock M, Woodfall B, Ciesek S. Lack of antiviral activity of darunavir against SARS-CoV-2. Int J Infect Dis. 2020;97:7–10. doi: 10.1016/j.ijid.2020.05.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang G, Yang ML, Duan ZL, Liu FL, Jin L, Long CB, Zhang M, Tang XP, Xu L, Li YC, Kamau PM, Yang L, Liu HQ, Xu JW, Chen JK, Zheng YT, Peng XZ, Lai R. Dalbavancin binds ACE2 to block its interaction with SARS-CoV-2 spike protein and is effective in inhibiting SARS-CoV-2 infection in animal models. Cell Res. 2021;31:17–24. doi: 10.1038/s41422-020-00450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoins. Am J Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 45.Xia S, Yan L, Xu W, Agrawal AS, Algaissi A, Tseng CK, Wang Q, Du L, Tan W, Wilson IA, Jiang S, Yang B, Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, Qi F, Bao L, Du L, Liu S, Qin C, Sun F, Shi Z, Zhu Y, Jiang S, Lu L. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fenard D, Lambeau G, Maurin T, Lefebvre JC, Doglio A. A peptide derived from bee venom-secreted phospholipase A2 inhibits replication of T-cell tropic HIV-1 strains via interaction with the CXCR4 chemokine receptor. Mol Pharmacol. 2001;60:341–347. doi: 10.1124/mol.60.2.341. [DOI] [PubMed] [Google Scholar]
- 48.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banumathi S, Rajashankar KR, Nötzel C, Aleksiev B, Singh TP, Genov N, Betzel C. Structure of the neurotoxic complex vipoxin at 1.4 A resolution. Acta Crystallogr D Biol Crystallogr. 2001;57:1552–1559. doi: 10.1107/s0907444901013543. [DOI] [PubMed] [Google Scholar]
- 50.Ramírez-Aportela E, López-Blanco JR, Chacón P. FRODOCK 2.0: fast protein-protein docking server. Bioinformatics. 2016;32:2386–2388. doi: 10.1093/bioinformatics/btw141. [DOI] [PubMed] [Google Scholar]
- 51.Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leman JK, Weitzner BD, Lewis SM, et al. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat Methods. 2020;17:665–680. doi: 10.1038/s41592-020-0848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield COVID-19 Genomics Group. McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kemp SA, Datir RP, Collier DA, Ferreira IATM, Carabelli A, Harvey W, Robertson DL, Gupta RK. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion ∆H69/V70. bioRxiv. 2020 doi: 10.1101/2020.12.14.422555. [DOI] [Google Scholar]
- 56.Liu Z, Zheng H, Lin H, Li M, Yuan R, Peng J, Xiong Q, Sun J, Li B, Wu J, Yi L, Peng X, Zhang H, Zhang W, Hulswit RJG, Loman N, Rambaut A, Ke C, Bowden TA, Pybus OG, Lu J. Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J Virol. 2020;94:e00790–e820. doi: 10.1128/JVI.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 58.Kruse RL. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Research. 2020;9:72. doi: 10.12688/f1000research.22211.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar M, Al Khodor S. Pathophysiology and treatment strategies for COVID-19. J Transl Med. 2020;18:353. doi: 10.1186/s12967-020-02520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gushchin VA, Dolzhikova IV, Shchetinin AM, Odintsova AS, Siniavin AE, Nikiforova MA, Pochtovyi AA, Shidlovskaya EV, Kuznetsova NA, Burgasova OA, Kolobukhina LV, Iliukhina AA, Kovyrshina AV, Botikov AG, Kuzina AV, Grousova DM, Tukhvatulin AI, Shcheblyakov DV, Zubkova OV, Karpova OV, Voronina OL, Ryzhova NN, Aksenova EI, Kunda MS, Lioznov DA, Danilenko DM, Komissarov AB, Tkachuck AP, Logunov DY, Gintsburg AL. Neutralizing activity of sera from Sputnik V-vaccinated people against variants of concern (VOC: B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.617.3) and Moscow endemic SARS-CoV-2 variants. Vaccines. 2021;9:779. doi: 10.3390/vaccines9070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muller VD, Soares RO, dos Santos NN, Jr, Trabuco AC, Cintra AC, Figueiredo LT, Caliri A, Sampaio SV, Aquino VH. Phospholipase A2 isolated from the venom of Crotalus durissus terrificus inactivates dengue virus and other enveloped viruses by disrupting the viral envelope. PLoS ONE. 2014;9:e112351. doi: 10.1371/journal.pone.0112351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brenes H, Loría GD, Lomonte B. Potent virucidal activity against Flaviviridae of a group IIA phospholipase A2 isolated from the venom of Bothrops asper. Biologicals. 2020;63:48–52. doi: 10.1016/j.biologicals.2019.12.002.40. [DOI] [PubMed] [Google Scholar]
- 63.Lenard J. Viral membranes. In: Mahy BWJ, Van Regenmortel MHV, editors. Encyclopedia of virology. 3. Academic Press; 2008. pp. 308–314. [Google Scholar]
- 64.Lorizate M, Kräusslich HG. Role of lipids in virus replication. Cold Spring Harb Perspect Biol. 2011;3:1–20. doi: 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alekseeva AS, Tretiakova DS, Chernikov VP, Utkin YN, Molotkovsky JG, Vodovozova EL, Boldyrev IA. Heterodimeric V. nikolskii phospholipases A2 induce aggregation of the lipid bilayer. Toxicon. 2017;133:169–179. doi: 10.1016/j.toxicon.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 66.Gunther-Ausborn S, Praetor A, Stegmann T. Inhibition of influenza-induced membrane fusion by lysophosphatidylcholine. J Biol Chem. 1995;270:29279–29285. doi: 10.1074/jbc.270.49.29279. [DOI] [PubMed] [Google Scholar]
- 67.Günther-Ausborn S, Stegmann T. How lysophosphatidylcholine inhibits cell-cell fusion mediated by the envelope glycoprotein of human immunodeficiency virus. Virology. 1997;235:201–208. doi: 10.1006/viro.1997.8699. [DOI] [PubMed] [Google Scholar]
- 68.Yan B, Chu H, Yang D, Sze KH, Lai PM, Yuan S, Shuai H, Wang Y, Kao RY, Chan JF, Yuen KY. Characterization of the lipidomic profile of human coronavirus-infected cells: implications for lipid metabolism remodeling upon coronavirus replication. Viruses. 2019;11:73. doi: 10.3390/v11010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Praznikar ZJ, Kovacic L, Rowan EG, Romih R, Rusmini P, Poletti A, Krizaj I, Pungercar J. A presynaptically toxic secreted phospholipase A2 is internalized into motoneuron-like cells where it is rapidly translocated into the cytosol. Biochim Biophys Acta. 2008;1783:1129–1139. doi: 10.1016/j.bbamcr.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 70.da Lomeo RS, Gonçalves AP, da Silva CN, de Paula AT, Costa Santos DO, Fortes-Dias CL, Gomes DA, de Lima ME. Crotoxin from Crotalus durissus terrificus snake venom induces the release of glutamate from cerebrocortical synaptosomes via N and P/Q calcium channels. Toxicon. 2014;85:5–16. doi: 10.1016/j.toxicon.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 71.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hoffmann M, Kleine-Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li C, Dowd CS, Zhang W, Chaiken IM. Phage randomization in a charybdotoxin scaffold leads to CD4-mimetic recognition motifs that bind HIV-1 envelope through non-aromatic sequences. J Pept Res. 2001;57:507–518. doi: 10.1046/j.1397-002x.2001.00876.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on request from the corresponding author.
Not applicable.