Abstract
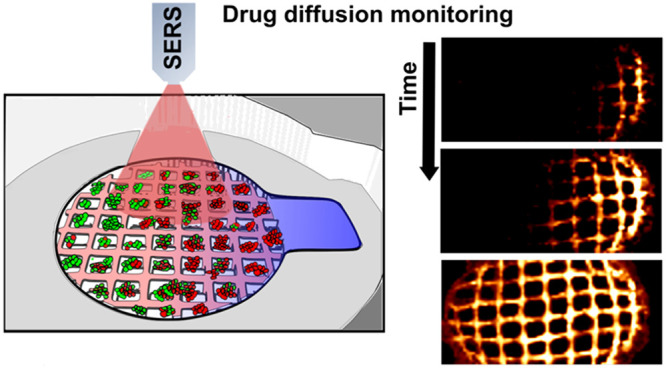
Monitoring dynamic processes in complex cellular environments requires the integration of uniformly distributed detectors within such three-dimensional (3D) networks, to an extent that the sensor could provide real-time information on nearby perturbations in a non-invasive manner. In this context, the development of 3D-printed structures that can function as both sensors and cell culture platforms emerges as a promising strategy, not only for mimicking a specific cell niche but also toward identifying its characteristic physicochemical conditions, such as concentration gradients. We present herein a 3D cancer model that incorporates a hydrogel-based scaffold containing gold nanorods. In addition to sustaining cell growth, the printed nanocomposite inks display the ability to uncover drug diffusion profiles by surface-enhanced Raman scattering, with high spatiotemporal resolution. We additionally demonstrate that the acquired information could pave the way to designing novel strategies for drug discovery in cancer therapy, through correlation of drug diffusion with cell death.
Keywords: surface-enhanced Raman spectroscopy, SERS, nanocomposite scaffold, 3D cell culture, drug diffusion
Introduction
The extracellular environment and its impact on cell fitness is a growing area of research, which has initially focused on mimicking microenvironments by means of tissue-like cell-culture systems in three dimensions (3D).1−3 However, monitoring parameters of interest within these structures, such as spatiotemporal heterogeneity or molecular gradients, is difficult to achieve experimentally. Hence, not only more realistic in vitro models are required but also new imaging technologies should be developed to assess them accurately.4−7
Although 2D cell cultures have allowed the discovery of many biological processes, these systems lack the cell–cell and cell–extracellular matrix (ECM) interactions that ultimately generate specific 3D microenvironments.8,9 Whereas biomolecules can diffuse freely in the extracellular milieu of 2D cell culture models, gradients of soluble molecules are established along tissues by a combination of cellular activity and restricted extracellular diffusion.10,11 Such gradients, along with the 3D internal structure, strongly influence cell responses and phenotypes in solid tissues, including tumors.12,13
During the design of in vitro 3D environments, biological structures can be tailored through the technology used to build them.14−16 Different approaches can be combined to improve the outcome and better mimic the features of a native niche.17,18 Recent experiments involving hydrogels and scaffold-based systems successfully reproduced the 3D physiology of selected human tissues.19 Even though these approaches have revealed new elements in cell biology,20 the incorporated third dimension drastically hampered the efficient capture of dynamic aspects, such as drug or nutrient transport, by optical methods. The ability of optical microscopy to map large extracellular concentration profiles in 3D is limited, especially if other fluorescent labels are present, which may cause signal overlap, e.g., in combination with cell-viability dyes.21,22 Other ordinary sensing techniques involve invasive procedures, which prevent continuous monitoring of gradually evolving processes. Therefore, dynamic parameters are not routinely registered in 3D experiments, thus ignoring their potential effects on cell behavior. The development of alternative analytical methods to rapidly detect gradients in extracellular media is thus required toward a better understanding of cellular niches and their implications for the effectiveness of therapeutic methods.
In this context, surface-enhanced Raman scattering (SERS), a highly sensitive vibrational spectroscopy technique, has emerged as a tool to evaluate changing environments. By enhancing the Raman signal of molecules adsorbed onto plasmonic nanostructures, SERS enables fast, label-free identification of trace analytes upon laser irradiation.23,24 However, the integration of efficient Raman signal enhancers inside cellular environments is still in its infancy, thus hindering the spatiotemporal resolution in SERS bioimaging.25 We have recently reported the fabrication of SERS-active scaffolds from inks containing plasmonic gold nanoparticles,27 as highly efficient platforms for SERS monitoring in 3D. We hypothesized that a similar system could be employed to screen biorelevant compounds in 3D environments while analyzing by SERS the extracellular gradients created upon drug exposure. We selected methylene blue (MB) as a drug candidate because it features a high Raman cross-section and has been used as a photosensitizer agent in antitumor therapy.28−30
We developed a biocompatible gelatin–alginate hydrogel containing gold nanorods (AuNRs), which could be 3D-printed to fabricate cell culture scaffolds.31,32 Alginate provides a strongly anionic character, which facilitates SERS detection of oppositely charged molecules through electrostatic interactions. The resulting fast and sensitive response to MB provided a suitable spatiotemporal resolution. Our results demonstrate real-time monitoring of drug diffusion in 3D cell cultures, through the detection of MB gradients within different environments and upon varying drug administration methodologies.
Results and Discussion
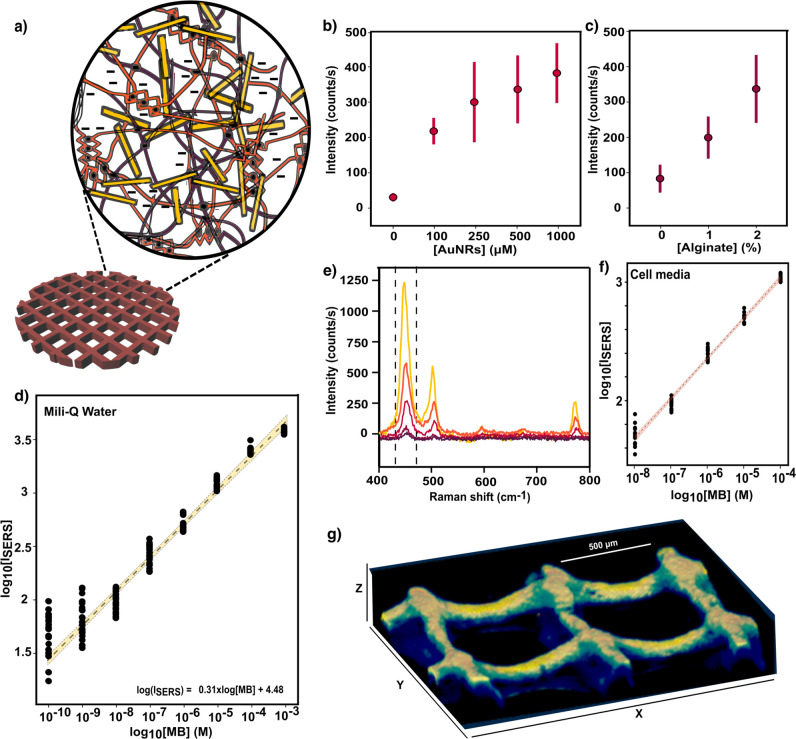
3D-printing strategies are particularly efficient at recreating the tumor niche, due to their ability to control geometric structures within milli-/microscale resolution. Direct printing of hydrogel inks is a common approach toward the design of porous, aqueous environments, which support nutrients and oxygen transport for cell culture.33,34 Consequently, 3D-printing of purposely designed hydrogels renders such scaffolds suitable for cell growth, with a tailored physiological architecture, so that complex in vitro systems can be acurately reconstructed. Among various natural polymers, gelatin-alginate mixtures are characterized by outstanding water retention and cell adhesion facilitated by gelatin-binding domains.35 Since our objective requires SERS-active inks, we incorporated AuNRs within selected mixtures of gelatin–alginate hydrogels (a representative extinction spectrum of the nanocomposite inks is shown in Figure S1). Upon printing, the obtained nanocomposite scaffolds were cross-linked with CaCl2, so divalent cations (Ca2+) bind anionic blocks in different alginate chains, resulting in stable 3D networks containing AuNRs, as schematically depicted in Figure 1a.
Figure 1.
(a) Scheme of a 3D-printed nanocomposite scaffold comprising gelatin and alginate hydrogels, represented by purple and orange fibers, respectively. The homogeneous distribution of gold nanorods (AuNRs) within the polymer matrix is represented by yellow bars. (b) SERS intensity of 1 μM methylene blue (MB) at 450 cm–1 registered by scaffolds (gelatin + 2% alginate) with varying concentrations of AuNRs, from bare scaffolds (no nanoparticles) to 1000 μM colloidal solution. Error bars show standard deviations from 10 different measurements with three different scaffolds (N = 3, n = 10). (c) SERS intensity of 1 μM MB obtained from scaffolds with 500 μM AuNRs and an increasing alginate percentage up to 2%, (N = 3, n = 10). (d) SERS intensity (at 450 cm–1) as a function of MB concentration. The yellow bar is a linear fit in the quantitative detection regions, including a regression line (dotted line) and 95% confidence interval. Each data point corresponds to the signal from 10 spectra collected from three different scaffolds (N = 3, n = 10). (e) SERS spectra of MB at different concentrations (between 10 nM and 10 μM with 10-fold-increase steps) in cell media, DMEM 10% FBS. (f) SERS intensity of MB (the red line is a linear fit with 95% confidence interval) in the presence of cell media components (N = 2, n = 10). An excitation laser line at 785 nm through a 10× objective, with a power of 15.15 mW for 1 s, was used for all measurements. (g) 3D Z-stack reconstruction of SERS intensity at 450 cm–1 upon MB incubation at 10 μM, recorded with a 785 nm excitation laser through a 20× immersion objective in confocal mode, with a power of 7 mW and 10 ms of integration time.
We then evaluated the SERS performance of the printed scaffolds as a function of two key parameters of ink composition: AuNRs concentration and percentage of alginate. MB was incubated with the scaffolds at 1 μM concentration. Increasing AuNR concentrations were found to directly correlate with higher SERS intensities of the MB vibration at 450 cm–1 (C–N–C skeletal bending36). The SERS results in Figure 1b confirm an increase in the number of plasmonic hotspots for Raman signal amplification so that the MB signal was negligible for an ink formulation without nanoparticles. On the other hand, when the percentage of alginate was increased from 0% to 2% at constant AuNR concentration, the registered SERS intensities were significantly affected. As illustrated in Figure 1c, scaffolds containing 2% alginate showed the greatest SERS performance for MB detection. Although a low MB signal could be measured from scaffolds lacking alginate, the effect of this polymer component on signal enhancement was even more intense than that observed at varying AuNR concentrations. We interpret these results in terms of electrostatic interactions between positively charged MB and the anionic alginate-based ink, facilitating analyte penetration through the scaffold. As a consequence, MB molecules are more likely to adsorb on plasmonic nanoparticles, resulting in a higher SERS signal enhancement.37,38 All scaffolds were thus prepared using inks containing 2% alginate and 1 mM AuNRs, thereby achieving a fast, sensitive detection of MB signal; no extensive preincubation times with the analyte were needed, in contrast with previous studies using different analyte–polymer combinations.26,27 SERS spectra collected from varying MB concentrations showed that semiquantitative detection could be achieved in the range from 1 mM down to 0.1 nM (Figure 1d). Within this range, MB concentration could be approximated by the SERS intensity, according to the empirical formula log(ISERS) = 0.31 × log([MB]) + 4.48, where [MB] is the molar concentration of MB and ISERS is the SERS intensity (expressed in counts). Interestingly, the negatively charged hydrogel additionally hinders the interaction of large proteins with AuNRs,39 which in combination with the electrostatic attraction effect would facilitate a reproducible detection of MB in complex cell media (Dulbecco’s modified Eagle medium, DMEM, with 10% of fetal bovine serum, FBS). As shown in Figure 1e, the characteristic peak of MB at 450 cm–1 dominates the SERS spectra, even at low concentrations and when other biomolecules are present at orders of magnitude higher concentrations than that of MB. We propose that our scaffolds can be used for direct, real-time analysis of biological samples with no need for pretreatment separation and concentration steps, which are common requisites for SERS analysis in complex media.40 Although a decreased sensitivity was observed as compared to aqueous samples, semiquantitative measurements could be recorded between 100 μM and 10 nM, thus covering the therapeutic window range for MB, i.e., drug dosage typically used for therapy.41 Therefore, changes in MB concentration would correspond to changes in SERS intensity in a quasi-predictable manner, which was found to follow a slightly different trend: log(ISERS) = 0.33 × log([MB]) + 4.39, as illustrated in Figure 1f.
We show in Figure 1g the result of scanning the SERS signal of 10 μM MB (typical concentration for anticancer therapy) in cell media.29,41 A confocal Raman microscope with a 20× water immersion objective and scanning steps of 13.3, 10, and 20 μm in X, Y, and Z, respectively, were employed for precise MB 3D screening throughout a total volume of (2.0 × 1.5 × 0.3) mm3. The signal recorded along the XY-plane in Figure S2 shows constant intensity values, accounting for a homogeneous distribution of AuNRs at this scale. On the contrary, a SERS intensity decay was registered at deeper imaging planes, due to laser spot broadening, decrease of collection efficiency, and other factors (see also Figure S3). SERS images with higher spatial resolution could be obtained using a 63× water immersion objective, allowing us to visualize the local AuNR distribution along the scaffold (Figure S4). However, this strategy restricted MB monitoring to much smaller (μm-sized) areas, since longer measurement times were required to obtain high spatial resolution at large scales.
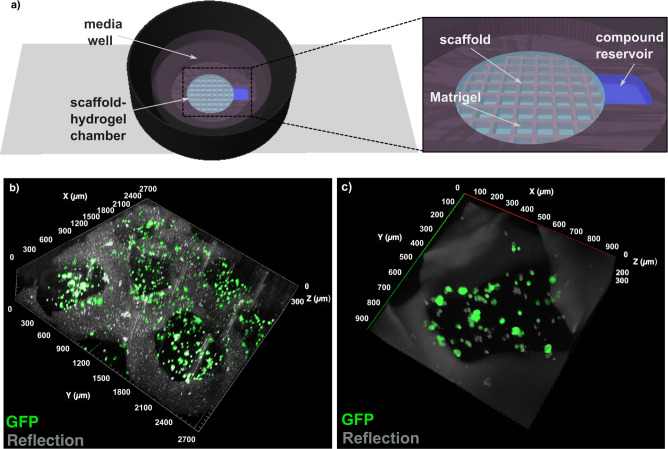
Aiming at the integration of the nanocomposite scaffolds into 3D cell environments, we characterized the rheological behavior of the hydrogels to be used as inks for 3D printing, followed by the biocompatibility and mechanical stability of the scaffolds for extended periods of time (more than 1 week in vitro). The results (Figure S5) demonstrate a shear-thinning behavior of the inks within the usual shear rate range for 3D printing, thus enhancing their printability. By use of these inks, grid-like scaffolds with holes of around 700 μm were obtained, according to scanning electron microscopy (SEM) images (Figure S5g). Regarding the stability of the printed scaffolds, maximum swelling was recorded after 7 days of incubation in complete media (cDMEM) and hydrolytic degradation was noticeable after 14 days of incubation (Figure S5f). MCF-7 cancer cells, which exhibit features of mammary epithelium, were selected as the biological model owing to their well-described behavior in 3D cell cultures and their sensitivity to MB therapy.29,42 Using a tailored cell culture device was essential toward successfully implementing a homogeneous cell distribution within 3D-printed plasmonic scaffolds. The system (Figure 2a) comprised a central chamber where the supporting scaffolds could be fixed while using a commercial extracellular matrix (Matrigel) to uniformly sustain the cells (Figure 2b). A lateral reservoir was incorporated into the device (Figure 2a, inset) to challenge the 3D cell environment with reproducible MB gradients. Meanwhile, the media well, located above the scaffold/hydrogel chamber, provides the necessary nutrients to the 3D culture and supports the use of dip-in immersion objectives during SERS measurements (dimensions indicated in Figure S6). Following this approach, MCF-7 cells were efficiently seeded in the device and remained viable for several days, forming tumor-like cell aggregates after 3–4 days in vitro, throughout the entire 3D extracellular matrix (see confocal microscopy images in Figures 2c and S7).
Figure 2.
(a) Scheme of a custom-made device to integrate nanocomposite scaffolds within a 3D tumor cell environment, comprising MCF-7 cells and Matrigel. The presence of a lateral reservoir (see inset) allows for controlled MB delivery. (b) 3D reconstruction of confocal images of growing MCF-7 cells embedded in Matrigel within the supporting scaffold. Dimensions XYZ = 2700 μm × 2700 μm × 300 μm. (c) Higher magnification image showing cancer cell clusters formed under these conditions. Dimensions XYZ = 900 μm × 900 μm × 300 μm.
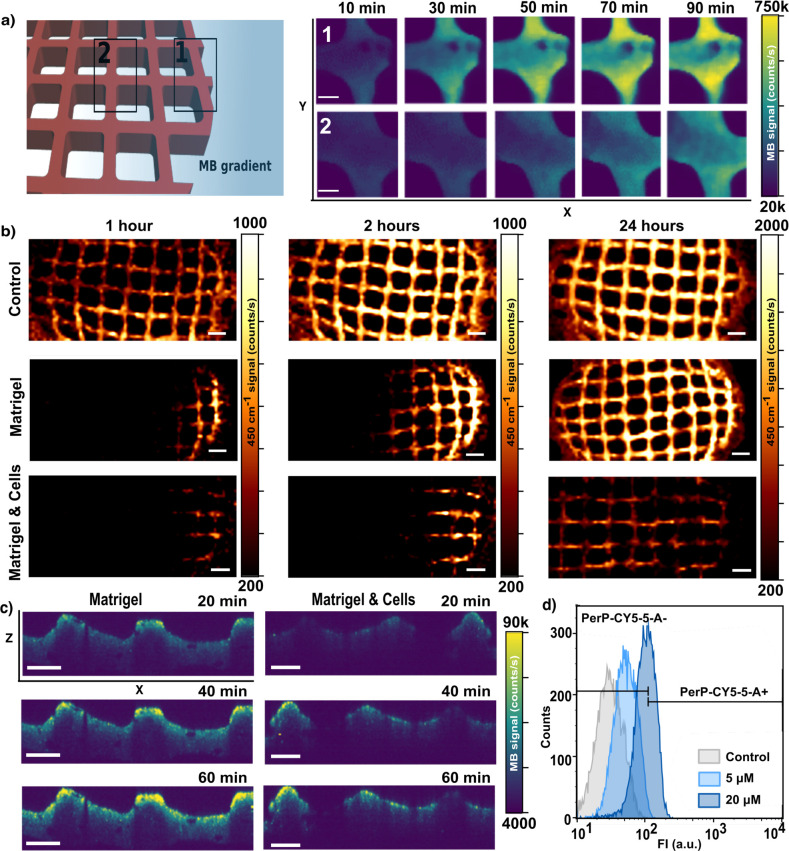
As a first example of application, we monitored drug delivery using poly lactic-co-glycolic acid (PLGA) capsules containing MB. On account of the well-known spontaneous PLGA degradation in cell environments,43,44 encapsulated MB was gradually released into the medium, where it could be detected by SERS. As shown in Figure S8, we first confirmed that drug monitoring could be performed over time, within 3D-printed nanocomposite scaffolds. Subsequently, we assessed the potential of the system to visualize MB diffusion. In these experiments, scaffolds were placed within our homemade devices, followed by addition of MB from the lateral reservoir (Figure 2a). As illustrated in Figure 3a, the localized drug delivery created a MB gradient that could be imaged by SERS. We initially selected two neighboring areas in the plasmonic scaffold, separated by 1 mm from each other, to register SERS intensity upon MB administration (10 μM). The area located closer to the compound reservoir (labeled as 1) showed a rapid response to MB (fewer than 30 min), whereas the farthest area (labeled as 2) reached a comparable level with an 1 h delay. These results, in addition to those presented in Figure S9, demonstrate a suitable spatiotemporal resolution in SERS imaging, which may account for MB gradients along the scaffold, in three dimensions. We then analyzed the impact of the ECM, in this case Matrigel, and of the presence of cells, on MB diffusion. To this end, we performed a similar experiment (localized drug delivery from the compound reservoir), but incorporating Matrigel, either with or without embedded MCF-7 cells. Upon MB administration, we scanned sufficiently large areas (8 mm × 3 mm) of the scaffold at different incubation times (1, 2, and 24 h). The corresponding maps (Figure 3b) confirm the formation of MB gradients (at different conditions and times) with meaningful and varying SERS profiles. In contrast to the control conditions, significantly restricted MB diffusion occurred in the presence of both Matrigel and MCF-7 cells. We observed (Figure S10) that hindered diffusion strongly affected MB distribution at early time points (1 and 2 h) whereas a similar equilibrium situation was reached for all experimental conditions within 24 h. Similar effects are shown in Figure 3c, in which the administration of MB was performed from the top of the scaffold chamber so as to establish a gradient along the Z-axis. Using the confocal microscopy mode, we observed that the presence of MCF-7 not only hindered MB diffusion but also originated a more heterogeneous distribution along the XZ-plane (control condition shown in Figure S11). Besides, the total SERS intensity at 24 h was consistently lower in experiments with cells (Figure S10b,c). To clarify these results, we investigated whether MCF-7 cells were effectively taking up MB from the extracellular milieu, thereby affecting its diffusion. In this context, MB could also serve as a fluorophore probe with emission around 690 nm so that cells uptaking MB would display higher emission intensities at this wavelength. We interrogated MCF-7 cells upon Matrigel depolymerization and subsequent cell retrieval, via flow cytometry (Figure 3d), which confirmed intracellular accumulation of MB in a dose-dependent manner. We thus interpret the creation of different diffusion profiles as the result of three main factors: drug diffusivity, extracellular matrix permeability, and intracellular accumulation of MB.45,46
Figure 3.
(a) Schematic view of the experimental setup to monitor MB diffusion by SERS. Two neighboring areas of the plasmonic scaffold, separated from each other by 1 mm in X, were selected to register the SERS intensity at successive times upon MB administration. Maps were recorded with a 785 nm laser through a 20× objective with laser power of 15 mW, 10 ms integration time, and a step size of 16 μm. Scale bars: 200 μm. (b) MB diffusion patterns along the X-axis in pristine nanocomposite scaffolds, scaffolds embedded in Matrigel, and Matrigel loaded with high cell density (2 × 106 cells/mL). The SERS signal (at 450 cm–1) was acquired at indicated times (1, 2, and 24 h) after MB delivery from the lateral reservoir. An excitation laser at 785 nm through a 10× objective, with a power of 15.15 mW for 0.1 s, was used for all measurements. Scale bars: 600 μm. (c) SERS profile maps (XZ plane) of the nanocomposite scaffold within Matrigel and cell-containing Matrigel, after MB administration from the top media well. Confocal SERS mappings were recorded with a 785 nm excitation laser through a 20× water immersion objective, 7 mW power, 50 ms integration time, and a step size of 10 μm. Scale bars: 200 μm. (d) Flow cytometry analysis of the uptake of MB by cells retrieved from the 3D culture. The fluorescence from MB increased in a dose-dependent manner. The gating indicates positive and negative cell populations after applying the PerCP-Cy5.5 emission filter (670/40).
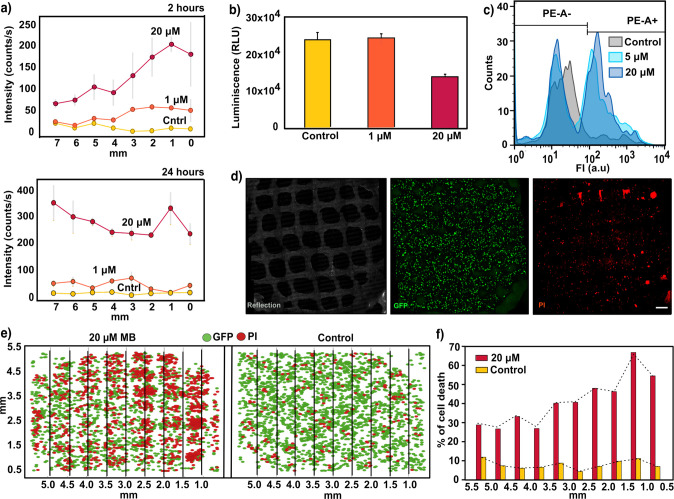
We finally sought to monitor the SERS signal at different MB concentrations over time, correlating with the cell viability resulting from treatments with the selected MB doses. It should be noted that the use of MB as a drug against tumors requires a drug activation step by illumination at an appropriate wavelength (≈640 nm).47 Upon exposure to light, the photosensitizer (MB molecules) would start generating reactive oxygen species (ROS), eventually inducing cell death. In the experiments discussed so far, MB was applied without light activation (dark conditions) so that no significant cytotoxic activity was observed (Figure S12). In this case, we monitored both the distribution of MB and its cytotoxicity by challenging 3D cell cultures with increasing doses of photoactivated MB solution (control, 1 μM, and 20 μM). This means that free MB was illuminated by a red light lamp prior to administration from the lateral reservoir of the cell culture device. Subsequently, drug diffusion was imaged through the SERS fingerprint of MB at two different time points (2 and 24 h), followed by evaluation of cell viability at the end of the experiment (24 h). The patterns of MB distribution through the scaffold at selected times are plotted in Figure 4a. As expected, the amount of MB was found to dictate both the recorded SERS intensities and the number of dying cells. For example, whereas at 1 μM MB no SERS signal was detected at areas far away (more than 5 mm) from the lateral reservoir during the first 2 h, a high SERS intensity could be readily registered when 20 μM MB was used. Quantification of cell viability by CellTiter-Glo test (Figure 4b) and flow cytometry (Figure 4c) after recovery of cellular aggregates from Matrigel demonstrated the dose-dependent cytotoxic effect of MB on the entire cell population and for single-cell measurements. Likewise, live confocal imaging (Figure 4d) was conducted by following dead cells labeled with propidium iodide (PI). Images of the entire culture volume around the scaffold were captured to examine the spatiotemporal distribution of affected cells and to correlate drug gradients with the spatial distribution of dead cells in 3D.
Figure 4.
(a) SERS intensity at increasing doses of MB (control, 1 μM, and 20 μM), registered by scaffolds at different points from the reservoir after 2 h (upper panel) and 24 h (lower panel). Gray lines represent the standard deviation from six spectra (N = 2 samples, n = 3 spectra). An excitation laser at 785 nm through a 10× objective, with a power of 15.15 mW for 1 s, was used for all measurements. (b) Luminescence output 24 h after MB administration monitored with CellTiter-Glo 3D cell viability assay. Error bars indicate the standard derivation of multiple wells measured from the same experiment. (c) Flow cytometry analysis representing quantified single cells labeled with propodium iodide (PI) after treatment with 1 μM and 20 μM MB. The gating indicates the positive and negative cell population after applying a PE emission filter (585/42). (d) Maximum intensity projection (XY) images of a representative live confocal image. 3D cells growing in Matrigel and within the nanocomposite scaffold were labeled with PI for visualizing cytotoxic effects 24 h after dispensing 1 μM MB. Images correspond to a 400 μm thick Z-stack. Scale bar: 500 μm. (e) Two-dimensional representation of the Z-stack projection of 3D live confocal images. Analysis was performed after a 2 h MB treatment to visualize the cytotoxicity gradient. Lines indicate columns into which the images were divided to study the distribution of PI (red) and GFP signals (green). (f) Automated quantification of cell death percentage for control and 20 μM samples, segmented by columns. Dotted line indicates the profile of cell death with the distance.
Imaging of cytotoxicity gradients was attempted 24 h after the addition of photoactivated MB. However, homogeneous cell death across the 3D culture was observed at this time by confocal microscopy imaging (Figure S13). This result is in agreement with SERS results for drug distribution over 24 h (Figures 3b and 4a), which show uniform drug distribution throughout the scaffold. Therefore, we developed an alternative approach to enhance the cell death gradient, based on our previous MB transport studies. The new strategy consisted of replacing cell media at 2 h after MB administration from the lateral reservoir, thereby rinsing extracellular MB from the 3D culture. By subsequent irradiation with the red lamp, we photoactivated primarily MB that had been uptaken by cells, as well as any remaining MB present in the area closest to the reservoir. Automated quantification of fluorescence images after 24 h (Figure 4e) revealed a well-defined gradient of dying cells along the X-axis (Figure 4f), in contrast to results obtained under control conditions or without extracellular drug removal (see Figure S14 for statistical analysis).
Conclusions
In summary, the present study provides compelling evidence supporting the use of plasmonic scaffolds as suitable SERS platforms for drug transport monitoring. By integrating 3D-printed composite scaffolds within a customized cell culture device, we could achieve a closer recreation of different compound gradients. The same system allowed us to monitor the impact of drug diffusion and to better understand its cytotoxic effect on a 3D cell culture model. The selection of MB as a proof-of-concept drug candidate allowed us to demonstrate that label-free SERS studies can be performed in 3D, under various conditions, which may enable the calculation of diffusion coefficients in future studies. The real-time images obtained regarding these processes evidenced the value of this system to study the extracellular environment in vitro, thereby deciphering the biochemical and biophysical factors involved in drug transport, which might harbor therapeutic relevance.
Acknowledgments
J.P. acknowledges an FPU fellowship from the Spanish Ministry of Science, Innovation and Universities. L.M.L.-M. acknowledges funding from the European Research Council (Grants ERC AdG 787510, 4DbioSERS) and the Maria de Maeztu Units of Excellence Program from the Spanish State Research Agency (Grant MDM-2017-0720). A.C. was funded by MICINN (Grant PID2019-108787RB-I00 (FEDER/EU)) and the European Research Council (ERC Consolidator Grant 819242).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.1c03070.
Experimental details and additional images on SERS and mechanical characterization, MB diffusion and cytotoxicity (PDF)
Author Contributions
∞ J.P. and B.M.-M. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Barcellos-Hoff M. H.; Lyden D.; Wang T. C. The Evolution of the Cancer Niche during Multistage Carcinogenesis. Nat. Rev. Cancer 2013, 13, 511–518. 10.1038/nrc3536. [DOI] [PubMed] [Google Scholar]
- Rodrigues J.; Heinrich M. A.; Teixeira L. M.; Prakash J. 3D In Vitro Model (R)Evolution: Unveiling Tumor–Stroma Interactions. Trends in Cancer 2021, 7, 249–264. 10.1016/j.trecan.2020.10.009. [DOI] [PubMed] [Google Scholar]
- Jordahl S.; Solorio L.; Neale D. B.; McDermott S.; Jordahl J. H.; Fox A.; Dunlay C.; Xiao A.; Brown M.; Wicha M.; Luker G. D.; Lahann J. Engineered Fibrillar Fibronectin Networks as Three-Dimensional Tissue Scaffolds. Adv. Mater. 2019, 31, 1904580. 10.1002/adma.201904580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos V.; Pleitez M. A.; Aime S.; Brindle K. M. Emerging Technologies to Image Tissue Metabolism. Cell Metab. 2019, 29, 518–538. 10.1016/j.cmet.2018.09.004. [DOI] [PubMed] [Google Scholar]
- Vidavsky N.; Kunitake J. A. M. R.; Diaz-Rubio M. E.; Chiou A. E.; Loh H. C.; Zhang S.; Masic A.; Fischbach C.; Estroff L. A. Mapping and Profiling Lipid Distribution in a 3D Model of Breast Cancer Progression. ACS Cent. Sci. 2019, 5, 768–780. 10.1021/acscentsci.8b00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert S.; Gutbrod M. S.; Liebsch G.; Meier R.; Meinert C.; Hutmacher D. W. Automated 3D Microphysiometry Facilitates High-Content and Highly Reproducible Oxygen Measurements within 3D Cell Culture Models. ACS Sens. 2021, 6, 1248–1260. 10.1021/acssensors.0c02551. [DOI] [PubMed] [Google Scholar]
- Cheng J. X.; Xie X. S. Vibrational Spectroscopic Imaging of Living Systems: An Emerging Platform for Biology and Medicine. Science 2015, 350, aaa8870. 10.1126/science.aaa8870. [DOI] [PubMed] [Google Scholar]
- Ma C.; Witkowski M. T.; Harris J.; Dolgalev I.; Sreeram S.; Qian W.; Tong J.; Chen X.; Aifantis I.; Chen W. Leukemia-on-a-Chip: Dissecting the Chemoresistance Mechanisms in B Cell Acute Lymphoblastic Leukemia Bone Marrow Niche. Sci. Adv. 2020, 6, eaba5536. 10.1126/sciadv.aba5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holle A. W.; Young J. L.; Van Vliet K. J.; Kamm R. D.; Discher D.; Janmey P.; Spatz J. P.; Saif T. Cell-Extracellular Matrix Mechanobiology: Forceful Tools and Emerging Needs for Basic and Translational Research. Nano Lett. 2018, 18, 1–8. 10.1021/acs.nanolett.7b04982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C.; Deforet M.; Akkari L.; Thompson C. B.; Joyce J. A.; Xavier J. B. Metabolic Origins of Spatial Organization in the Tumor Microenvironment. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 2934–2939. 10.1073/pnas.1700600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker L. H.; Hamm G. R.; Sargeant R. J. E.; Goodwin R. J. A.; Mackay C. L.; Campbell C. J.; Clarke D. J. Untargeted Metabolite Mapping in 3D Cell Culture Models Using High Spectral Resolution FT-ICR Mass Spectrometry Imaging. Anal. Chem. 2019, 91, 9522–9529. 10.1021/acs.analchem.9b00661. [DOI] [PubMed] [Google Scholar]
- Lyssiotis C. A.; Kimmelman A. C. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017, 27, 863–875. 10.1016/j.tcb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D.; Weinberg R. A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Pampaloni F.; Reynaud E. G.; Stelzer E. H. K. The Third Dimension Bridges the Gap between Cell Culture and Live Tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- Jeong S. Y.; Lee J. H.; Shin Y.; Chung S.; Kuh H. J. Co-Culture of Tumor Spheroids and Fibroblasts in a Collagen Matrix-Incorporated Microfluidic Chip Mimics Reciprocal Activation in Solid Tumor Microenvironment. PLoS One 2016, 11, e0159013. 10.1371/journal.pone.0159013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H. G.; Jeong Y. H.; Kim Y.; Choi Y. J.; Moon H. E.; Park S. H.; Kang K. S.; Bae M.; Jang J.; Youn H.; Paek S. H.; Cho D. W. A Bioprinted Human-Glioblastoma-on-a-Chip for the Identification of Patient-Specific Responses to Chemoradiotherapy. Nat. Biomed. Eng. 2019, 3, 509–519. 10.1038/s41551-019-0363-x. [DOI] [PubMed] [Google Scholar]
- Brancato V.; Oliveira J. M.; Correlo V. M.; Reis R. L.; Kundu S. C. Could 3D Models of Cancer Enhance Drug Screening?. Biomaterials 2020, 232, 119744. 10.1016/j.biomaterials.2019.119744. [DOI] [PubMed] [Google Scholar]
- Pati F.; Jang J.; Ha D. H.; Won Kim S.; Rhie J. W.; Shim J. H.; Kim D. H.; Cho D. W. Printing Three-Dimensional Tissue Analogues with Decellularized Extracellular Matrix Bioink. Nat. Commun. 2014, 5, 3935. 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijal G.; Li W. A Versatile 3D Tissue Matrix Scaffold System for Tumor Modeling and Drug Screening. Sci. Adv. 2017, 3, e1700764. 10.1126/sciadv.1700764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L.; Mooney D. J. Biomaterials and Emerging Anticancer Therapeutics: Engineering the Microenvironment. Nat. Rev. Cancer 2016, 16, 56–66. 10.1038/nrc.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T.; Nagano T. Fluorescent Probes for Sensing and Imaging. Nat. Methods 2011, 8, 642–645. 10.1038/nmeth.1663. [DOI] [PubMed] [Google Scholar]
- Hong G.; Antaris A. L.; Dai H. Near-Infrared Fluorophores for Biomedical Imaging. Nat. Biomed. Eng. 2017, 1, 0010. 10.1038/s41551-016-0010. [DOI] [Google Scholar]
- Kim N.; Thomas M. R.; Bergholt M. S.; Pence I. J.; Seong H.; Charchar P.; Todorova N.; Nagelkerke A.; Belessiotis-Richards A.; Payne D. J.; Gelmi A.; Yarovsky I.; Stevens M. M. Surface Enhanced Raman Scattering Artificial Nose for High Dimensionality Fingerprinting. Nat. Commun. 2020, 11, 207. 10.1038/s41467-019-13615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer J.; Jimenez De Aberasturi D.; Aizpurua J.; Alvarez-Puebla R. A.; Auguié B.; Baumberg J. J.; Bazan G. C.; Bell S. E. J.; Boisen A.; Brolo A. G.; Choo J.; Cialla-May D.; Deckert V.; Fabris L.; Faulds K.; García De Abajo F. J.; Goodacre R.; Graham D.; Haes A. J.; Haynes C. L.; Huck C.; Itoh T.; Käll M.; Kneipp J.; Kotov N. A.; Kuang H.; Le Ru E. C.; Lee H. K.; Li J. F.; Ling X. Y.; Maier S. A.; Mayerhöfer T.; Moskovits M.; Murakoshi K.; Nam J. M.; Nie S.; Ozaki Y.; Pastoriza-Santos I.; Perez-Juste J.; Popp J.; Pucci A.; Reich S.; Ren B.; Schatz G. C.; Shegai T.; Schlücker S.; Tay L. L.; Thomas K. G.; Tian Z. Q.; Van Duyne R. P.; Vo-Dinh T.; Wang Y.; Willets K. A.; Xu C.; Xu H.; Xu Y.; Yamamoto Y. S.; Zhao B.; Liz-Marzán L. M. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plou J.; García I.; Charconnet M.; Astobiza I.; García-Astrain C.; Matricardi C.; Mihi A.; Carracedo A.; Liz-Marzán L. M. Multiplex SERS Detection of Metabolic Alterations in Tumor Extracellular Media. Adv. Funct. Mater. 2020, 30, 1910335. 10.1002/adfm.201910335. [DOI] [Google Scholar]
- Bodelón G.; Montes-García V.; Costas C.; Pérez-Juste I.; Pérez-Juste J.; Pastoriza-Santos I.; Liz-Marzán L. M. Imaging Bacterial Interspecies Chemical Interactions by Surface-Enhanced Raman Scattering. ACS Nano 2017, 11, 4631–4640. 10.1021/acsnano.7b00258. [DOI] [PubMed] [Google Scholar]
- García-Astrain C.; Lenzi E.; Jimenez de Aberasturi D.; Henriksen-Lacey M.; Binelli M. R.; Liz-Marzán L. M. 3D-Printed Biocompatible Scaffolds with Built-In Nanoplasmonic Sensors. Adv. Funct. Mater. 2020, 30, 2005407. 10.1002/adfm.202005407. [DOI] [Google Scholar]
- Srichan C.; Ekpanyapong M.; Horprathum M.; Eiamchai P.; Nuntawong N.; Phokharatkul D.; Danvirutai P.; Bohez E.; Wisitsoraat A.; Tuantranont A. Highly-Sensitive Surface-Enhanced Raman Spectroscopy (SERS)-Based Chemical Sensor Using 3D Graphene Foam Decorated with Silver Nanoparticles as SERS Substrate. Sci. Rep. 2016, 6, 23733. 10.1038/srep23733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos A. F.; Terra L. F.; Wailemann R. A. M.; Oliveira T. C.; de Morais Gomes V.; Mineiro M. F.; Meotti F. C.; Bruni-Cardoso A.; Baptista M. S.; Labriola L. Methylene Blue Photodynamic Therapy Induces Selective and Massive Cell Death in Human Breast Cancer Cells. BMC Cancer 2017, 17, 194. 10.1186/s12885-017-3179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P. T.; Lin C. L.; Lin C. W.; Chang N. C.; Tsai W. B.; Yu J. Methylene-Blue-Encapsulated Liposomes as Photodynamic Therapy Nano Agents for Breast Cancer Cells. Nanomaterials 2019, 9, 14. 10.3390/nano9010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastoriza-Santos I.; Kinnear C.; Pérez-Juste J.; Mulvaney P.; Liz-Marzán L. M. Plasmonic Polymer Nanocomposites. Nat. Rev. Mater. 2018, 3, 375–391. 10.1038/s41578-018-0050-7. [DOI] [Google Scholar]
- Zhu K.; Shin S. R.; van Kempen T.; Li Y. C.; Ponraj V.; Nasajpour A.; Mandla S.; Hu N.; Liu X.; Leijten J.; Lin Y. D.; Hussain M. A.; Zhang Y. S.; Tamayol A.; Khademhosseini A. Gold Nanocomposite Bioink for Printing 3D Cardiac Constructs. Adv. Funct. Mater. 2017, 27, 1605352. 10.1002/adfm.201605352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erben A.; Hörning M.; Hartmann B.; Becke T.; Eisler S. A.; Southan A.; Cranz S.; Hayden O.; Kneidinger N.; Königshoff M.; Lindner M.; Tovar G. E. M.; Burgstaller G.; Clausen-Schaumann H.; Sudhop S.; Heymann M. Precision 3D-Printed Cell Scaffolds Mimicking Native Tissue Composition and Mechanics. Adv. Healthcare Mater. 2020, 9, 2000918. 10.1002/adhm.202000918. [DOI] [PubMed] [Google Scholar]
- Tibbitt M. W.; Anseth K. S. Hydrogels as Extracellular Matrix Mimics for 3D Cell Culture. Biotechnol. Bioeng. 2009, 103, 655–663. 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afewerki S.; Sheikhi A.; Kannan S.; Ahadian S.; Khademhosseini A. Gelatin-Polysaccharide Composite Scaffolds for 3D Cell Culture and Tissue Engineering: Towards Natural Therapeutics. Bioeng. Transl. Med. 2019, 4, 96–115. 10.1002/btm2.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu S.; Nithiyanantham U. In situ Formation of Curcumin Stabilized Shape-selective Ag Nanostructures in Aqueous Solution and their Pronounced SERS aActivity. RSC Adv. 2013, 3, 25278. 10.1039/c3ra44471f. [DOI] [Google Scholar]
- Shin Y.; Jeon I.; You Y.; Song G.; Lee T. K.; Oh J.; Son C.; Baek D.; Kim D.; Cho H.; Hwang H.; Kim T.; Kwak S. K.; Kim J.; Lee J. Facile Microfluidic Fabrication of 3D Hydrogel SERS Substrate with High Reusability and Reproducibility via Programmable Maskless Flow Microlithography. Adv. Opt. Mater. 2020, 8, 2001586. 10.1002/adom.202001586. [DOI] [Google Scholar]
- Kim D. J.; Park S. G.; Kim D. H.; Kim S. H. SERS-Active-Charged Microgels for Size- and Charge-Selective Molecular Analysis of Complex Biological Samples. Small 2018, 14, 1802520. 10.1002/smll.201802520. [DOI] [PubMed] [Google Scholar]
- Lieleg O.; Ribbeck K. Biological hydrogels as selective diffusion barriers. Trends Cell Biol. 2011, 21, 543–551. 10.1016/j.tcb.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont E.; De Bleye C.; Rademaker G.; Coïc L.; Horne J.; Sacré P. Y.; Peulen O.; Hubert P.; Ziemons E. Development of a Prototype Device for near Real-Time Surface-Enhanced Raman Scattering Monitoring of Biological Samples. Talanta 2021, 224, 121866. 10.1016/j.talanta.2020.121866. [DOI] [PubMed] [Google Scholar]
- Orth K.; Beck G.; Genze F.; Rück A. Methylene Blue Mediated Photodynamic Therapy in Experimental Colorectal Tumors in Mice. J. Photochem. Photobiol., B 2000, 57, 186–192. 10.1016/S1011-1344(00)00105-6. [DOI] [PubMed] [Google Scholar]
- Vantangoli M. M.; Madnick S. J.; Huse S. M.; Weston P.; Boekelheide K. MCF-7 Human Breast Cancer Cells Form Differentiated Microtissues in Scaffold-Free Hydrogels. PLoS One 2015, 10, e0135426. 10.1371/journal.pone.0135426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X.; Li T.; Xie X.; Feng Y.; Chen Z.; Yang H.; Wu C.; Deng S.; Liu Y. PLGA-Based Drug Delivery Systems for Remotely Triggered Cancer Therapeutic and Diagnostic Applications. Front. Bioeng. Biotechnol. 2020, 8, 381. 10.3389/fbioe.2020.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghitman J.; Biru E. I.; Stan R.; Iovu H. Review of Hybrid PLGA Nanoparticles: Future of Smart Drug Delivery and Theranostics Medicine. Mater. Des. 2020, 193, 108805. 10.1016/j.matdes.2020.108805. [DOI] [Google Scholar]
- Dewhirst M. W.; Secomb T. W. Transport of Drugs from Blood Vessels to Tumour Tissue. Nat. Rev. Cancer 2017, 17, 738–750. 10.1038/nrc.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B.; Qu C.; Park K.; Konieczny S. F.; Korc M. Recapitulation of Complex Transport and Action of Drugs at the Tumor Microenvironment Using Tumor-Microenvironment-on-Chip. Cancer Lett. 2016, 380, 319–329. 10.1016/j.canlet.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosowski E. M.; de Souza B. T. L.; Mito M. S.; Constantin R. P.; Mantovanelli G. C.; Mewes J. M.; Bizerra P. F. V.; da Costa Menezes P. V. M.; Gilglioni E. H.; Utsunomiya K. S.; Marchiosi R.; dos Santos W. D.; Filho O. F.; Caetano W.; de Souza Pereira P. C.; Goncalves R. S.; Constantin J.; Ishii-Iwamoto E. L.; Constantin R. P. The Photodynamic and Direct Actions of Methylene Blue on Mitochondrial Energy Metabolism: A Balance of the Useful and Harmful Effects of This Photosensitizer. Free Radical Biol. Med. 2020, 153, 34–53. 10.1016/j.freeradbiomed.2020.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.