Abstract
In this study the detection rates of bacterial infection of hip prostheses by culture and nonculture methods were compared for 120 patients with total hip revision surgery. By use of strict anaerobic bacteriological practice during the processing of samples and without enrichment, the incidence of infection by culture of material dislodged from retrieved prostheses after ultrasonication (sonicate) was 22%. Bacteria were observed by immunofluorescence microscopy in 63% of sonicate samples with a monoclonal antibody specific for Propionibacterium acnes and polyclonal antiserum specific for Staphylococcus spp. The bacteria were present either as single cells or in aggregates of up to 300 bacterial cells. These aggregates were not observed without sonication to dislodge the biofilm. Bacteria were observed in all of the culture-positive samples, and in some cases in which only one type of bacterium was identified by culture, both coccoid and coryneform bacteria were observed by immunofluorescence microscopy. Bacteria from skin-flake contamination were readily distinguishable from infecting bacteria by immunofluorescence microscopy. Examination of skin scrapings did not reveal large aggregates of bacteria but did reveal skin cells. These were not observed in the sonicates. Bacterial DNA was detected in 72% of sonicate samples by PCR amplification of a region of the bacterial 16S rRNA gene with universal primers. All of the culture-positive samples were also positive for bacterial DNA. Evidence of high-level infiltration either of neutrophils or of lymphocytes or macrophages into associated tissue was observed in 73% of patients. Our results indicate that the incidence of prosthetic joint infection is grossly underestimated by current culture detection methods. It is therefore imperative that current clinical practice with regard to the detection and subsequent treatment of prosthetic joint infection be reassessed in the light of these results.
Over 50,000 total hip replacement operations are performed annually in the United Kingdom and about 200,000 are performed annually in the United States. The majority of patients undergoing hip replacement experience dramatic relief of pain and restoration of satisfactory hip function (15). A proportion (approximately 20% in Europe [6]) fail, and prosthesis removal and replacement are usually required, with attendant patient trauma and increased medical costs (9). Aseptic mechanical loosening is reported to be the most common cause of prosthetic joint failure (15), after which standard bacteriological culture of specimens from periprosthetic tissue or aspirates is used to detect infection. Estimates of the incidence of infection as a cause of hip failure are usually in the range of 10% (12) to 15% (20), although some studies report an incidence as low as 2% (3) of all revision operations. Failure of the second implant postrevision, however, may be due to infection in up to 40% of patients (10). It has been suggested that the higher rate of infection postrevision could be due to a longer operating time, increased scar tissue formation, or unrecognized infection at the initial revision operation. Our recent study at The Queen’s University of Belfast (37) implicates unrecognized infection as a major cause of prosthetic joint failure. In this study, which was the first to combine sampling by mild ultrasonication to dislodge the bacteria growing within adherent biofilms on the surface of the removed prosthesis with the use of strict anaerobic techniques, we cultured bacteria from 22% of retrieved prostheses (26 of 120 implants). Detection of infections which arise from opportunistic pathogens of the normal microbiota, in particular, the skin, is always problematic, as a clear distinction must be made between infection and potential contamination. This is particularly the case when broth enrichments are used. Our prosthetic joint isolates were obtained by direct plating of the dislodged material onto agar plates and not as a result of broth enrichment. Also, ultrasonication to dislodge the biofilm was required to detect infection by culture. Furthermore, mock processing of a number of autoclaved implants failed to detect any bacteria. It is therefore highly unlikely that the bacteria isolated were present in the sample as a result of exogenous contamination.
Review of the notes for 18 of the 26 patients with culture-positive implants revealed that infection was suspected as the cause of loosening in only 6 patients. Joint fluid, aspirated preoperatively and prior to antibiotic administration, and tissue removed at the time of surgery were processed by the routine clinical laboratory. In only two patients was infection confirmed and were bacteria cultured. In our laboratory, viable bacteria were isolated from only five tissue samples corresponding to prostheses from which bacteria were isolated. This lack of bacteria in cultures of the tissue samples may be due to either the bacteriostatic or the bactericidal effects of cefamandole administered at the time of operation or the host’s defenses. In contrast, bacteria growing within a biofilm on device surfaces have greater resistance to antibiotics (40). Investigation of the antibiotic sensitivity of our isolates indicated that the majority were more than 1,000 times more resistant (minimum bactericidal concentration, >1,024 μg/ml) to cefamandole when growing within in vitro model biofilms than when growing in broth culture (39, 40). These results suggest that the device materials are the major sites colonized by the bacteria in this type of infection and highlight the importance of sampling directly from the prosthesis. In our study the anaerobic bacterium Propionibacterium acnes was isolated either alone or in association with Staphylococcus spp. from 62% of patients. Other published studies report either very low isolation rates for anaerobic bacteria (2) or failure to isolate any (1, 13). This indicates that adherence to strict anaerobic bacteriological practice can increase the detection of prosthetic joint infection and alter our perception of the major causative organisms.
When pathological examination of associated tissue has been carried out, a good correlation between detection of infection by culture and the presence of neutrophilic polymorphonuclear leukocytes in the tissue has been obtained (1, 12, 13, 28). For example, a recent study which involved the detection of infection by both culture and histological assessment of a tissue inflammatory response in five or six tissue specimens reported that 41 of 297 (14%) patients were infected (2). It has been suggested that the observation of neutrophilic infiltration in the associated tissues is a useful means of diagnosing prosthetic joint infection.
Our studies of the inflammatory response in associated tissues showed that in eight of the culture-positive patients neutrophils were not detected, although lymphocytes or macrophages were present. It may be that the lymphocyte or macrophage infiltration is in response to the slow release of bacterial components from a bacterial biofilm growing on the device surface. Histopathological examination of associated tissue samples taken from culture-negative patients showed that 8 had evidence of a high level of neutrophilic infiltration and that a further 36 with low levels or no evidence of neutrophils had large numbers of lymphocytes or macrophages. In total, 87% of our culture-negative patients had evidence either of neutrophilic or of lymphocyte or macrophage infiltration into tissue and may have been infected. These results suggest that even with improved sampling procedures the detection of infection by culture may still be an underestimate of the real incidence of infection. Our study highlights potential inadequacies in current clinical diagnostic methodology (37).
The detection of bacterial rRNA genes as an indicator of the presence of bacteria is an established technique that has been used for the detection of both environmental and medically important bacteria (17, 42). PCR amplification of bacterial 16S rRNA genes has been used successfully to detect bacteria that cause a variety of infections including postoperative endophthalmitis (19, 24), septic arthritis (5), and meningitis (30). In infections such as those of prosthetic joints, however, in which the infectious agents are opportunistic pathogens that may be members of the normal microbiota (e.g., the skin), the potential contamination of samples must be addressed. The direct immunological detection of bacteria in clinical samples can be achieved by the use of monoclonal antibodies (MAbs) and polyclonal antiserum prepared against the bacteria implicated in the clinical infection (32). This technique allows the direct visualization of the bacterial morphology and visual comparison with potential skin-flake contamination.
The aim of the present study was, first, to determine if prosthetic hip infection could be detected reliably by nonculture methods with culture-positive samples. Second, in the light of the inflammatory response in the associated tissues, the aim of the study was to ascertain if nonculturable bacteria were present in culture-negative patient samples. We therefore compared the use of (i) immunolabelling in conjunction with fluorescence microscopy and (ii) PCR amplification of a region of the bacterial 16S rRNA gene for the detection of prosthetic hip infection while taking suitable steps to eliminate and to control for potential contamination by bacteria of the normal microbiota.
MATERIALS AND METHODS
Clinical sample collection and processing.
One hundred twenty prosthetic hip implants were retrieved from patients undergoing revision hip surgery at Musgrave Park Hospital, Belfast, United Kingdom, during the 14-month period from March 1996 to April 1997. All patients underwent standardized preoperative hygiene procedures. Following skin preparation with Betadine (Seton Healthcare, Oldham, United Kingdom) the incision area was covered with an adhesive plastic drape. All operations were carried out in operating theaters in which a vertical laminar air flow provided a clean environment and all members of the operating team wore disposable impervious drapes. Routine antibiotic prophylaxis consisted of 2 g of cefamandole (Kefadol; Dista Products Ltd., Basingstoke, United Kingdom) given intravenously at the time of anesthetic induction. Further 1-g doses of cefamandole were given 8 and 16 h after surgery. Cefamandole is recommended by the British National Formulary for surgical prophylaxis. It is a broad-spectrum antibiotic active against both gram-positive and gram-negative bacteria. On removal, the surgeon aseptically placed the femoral and acetabular components in separate sterile bags. Tissue in contact with the implants was also removed and placed in sterile bottles. The femoral and acetabular components and the tissue samples were immediately placed in an anaerobic jar for transportation to an anaerobic cabinet (37).
Bacterial isolation and identification.
Samples were handled within the closed atmosphere of an anaerobic cabinet (Don Whitley Mk. III anaerobic cabinet; 80% N2, 10% CO2, and 10% H2; Don Whitley Scientific Ltd., Shipley, United Kingdom). The gas atmosphere of the cabinet was continuously pumped through a solution of 2% glutaraldehyde, and operators wore surgical gloves throughout processing. Bacteria growing within adherent biofilms on the surface of the prostheses were dislodged by mild ultrasonication (5 min, 50 Hz) into Ringer’s solution (25% [vol/vol]) containing cysteine (0.05% [vol/vol]) as a reducing agent (37). This procedure has been shown to have no effect on the viability of pure culture isolates. The use of Ringer’s solution rather than broth as a diluent ensured that the bacteria did not multiply within the sonicate prior to further processing. Determination of total viable bacterial counts was performed as follows: Volumes (0.5 ml) of sonicate were plated onto each of five blood agar (BA) and five anaerobic blood agar (ABA) plates. A known weight of tissue was homogenized (3 min) in Ringer’s solution (25% [vol/vol], 5 ml). Three 0.5-ml volumes of homogenized tissue were plated onto BA and ABA plates. The remaining volume of homogenized tissue was added in equal amounts to tryptone soy broth (TSB) and cooked meat broth (CMB) for enrichment. BA and ABA plates were incubated at 37°C aerobically and anaerobically, respectively, and were examined after 1, 2, 4, and 7 days. Samples were recorded as positive if colonies were observed on a minimum of four of the five plates. The TSB and CMB were incubated at 37°C aerobically and anaerobically, respectively. Both broths were subcultured on days 7 and 14: on BA for aerobic incubation and on ABA for anaerobic incubation. All plates were incubated at 37°C for 48 h. To determine the likelihood that contaminating bacteria were introduced during the sampling procedure, 20 retrieved orthopedic implants were sterilized in a hot-air oven and were processed as described above. In addition, three researchers in our laboratory scraped flakes from the surfaces of their skin into sterile bags containing Ringer’s solution, which were then processed as described above.
Pure bacterial cultures, obtained by picking isolated colonies from the viable count plates, were identified with commercially available kits. Cultures were stored both on Protect Bacterial Preservers (Technical Service Consultants Ltd., Heywood, United Kingdom) at −70°C and on Mueller-Hinton agar slopes at 4°C, which were subcultured at 3-month intervals. The remaining sonicate from each sample was centrifuged (1,000 × g, 20 min), and the concentrated sonicate was stored in 1-ml aliquots at −70°C for further analysis.
Bacterial strains.
The strains used in this study were Bacteroides fragilis NCTC 9343 (National Collection of Type Cultures, Colindale, United Kingdom); Peptostreptococcus magnus NCTC 11804; Peptostreptococcus micros NCTC 11808; Escherichia coli O128, kindly supplied by C. Smyth, Trinity College Dublin, Ireland; and Corynebacterium diphtheriae, Corynebacterium hofmanni, and Corynebacterium xerosis from the culture collection of the Department of Microbiology and Immunobiology, School of Clinical Medicine, The Queen’s University of Belfast. The clinical isolates used in this study (Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus hominis, Staphylococcus capitis, Propionibacterium acnes, and Micrococcus sp.) were all isolated at the Department of Microbiology and Immunobiology, School of Medicine, from retrieved prosthetic hip implants as described above (37).
Immunological detection of bacteria. (i) Production of polyclonal antisera.
A New Zealand White rabbit was immunized with whole cells of S. epidermidis. The rabbit was inoculated subcutaneously at four sites on the back with 0.1 ml of a bacterial suspension of 108 CFU/ml in 0.01 M phosphate-buffered saline (PBS [pH 7.4]; 0.15 M NaCl, 0.0075 M Na2HPO4, 0.0025 M NaH2PO4 · 2H2O). Two further inoculations of bacteria in PBS were made at approximately monthly intervals, and the rabbit was test bled 2 weeks after the final inoculation. The reactivity of the antiserum was then tested by immunofluorescence microscopy (IFM) as described previously (23).
(ii) Production of MAbs.
Four BALB/c mice were immunized with whole cells of two P. acnes strains. The mice were inoculated intraperitoneally with 0.2 ml of a bacterial suspension of 108 CFU/ml in 0.01 M PBS. Further 0.2-ml inoculations were given after 1 month. Serum was tested by immunofluorescence after 3 days, and the mice with the highest titer of antiserum were inoculated again 1 week later. One mouse was killed after 5 days, and the spleen cells from the mouse were fused with P3X 63 Ag8-653 (NS-0/1) mouse myeloma cells by a modification of the method of Galfre and Milstein (16) as described previously (23). Hybridoma cell lines, which produced P. acnes-specific antibodies, were then cloned by limiting dilution (18).
(iii) IFM.
A modification of the IFM procedure described by Lutton et al. (23) was used. Samples of sonicate obtained from prosthetic joints (1 ml) were further centrifuged (10,000 × g; 20 min), and the resulting pellets were resuspended in 100 μl of PBS. Samples (10 μl) were then applied in duplicate to multiwell slides. The slides were air dried and then fixed in 100% methanol for 10 min at −20°C.
The slides were dually labelled for IFM with a combination of MAb QUBPa3 supernatant, which reacted with all P. acnes strains isolated in our previous study, and S. epidermidis polyclonal antiserum, which reacted with all Staphylococcus spp. strains isolated in our previous study (37). The reactivity of MAb QUBPa3 with S. aureus, S. epidermidis, B. fragilis, E. coli, P. magnus, P. micros, C. diphtheriae, C. hofmanni, and C. xerosis was also tested. Similarly, the reactivity of the S. epidermidis polyclonal antiserum with B. fragilis, E. coli, and the gram-positive anaerobic cocci P. magnus and P. micros was tested. For dual labelling, the slides were incubated with undiluted MAb QUBPa3 supernatant, washed, and incubated with rabbit anti-S. epidermidis polyclonal antiserum diluted 1 in 200 in PBS. The slides were again washed and then incubated simultaneously with sheep anti-rabbit fluorescein conjugate (Sigma) and goat anti-mouse rhodamine conjugate (Sigma). After a final wash, all slides were mounted with glycerol-PBS containing an antiphotobleaching agent (Citifluor; Agar Scientific Ltd, Essex, United Kingdom) and were examined with a Leitz fluorescent microscope.
For clinical sonicate samples the detection of bacteria on duplicate wells by immunofluorescence was given a score of between 0 and 3 by using the following criteria: 0, no bacteria; 1, 1 to 10 bacteria/well; 2, 10 to 50 bacteria/well; 3, 50 or more bacteria/well.
To determine whether bacteria detected by immunofluorescence in concentrated sonicate samples could have resulted from skin-flake contamination, skin from researchers working at the Department of Microbiology and Immunobiology, School of Clinical Medicine, was scraped with sterile scalpels into 1-ml volumes of PBS. These were then processed in the same way as the sonicate samples and were examined by dual-labelling IFM.
(iv) CSLM.
Selected wells were examined by confocal scanning laser microscopy (CSLM) with a Leica NCS-NT confocal scanning laser microscope.
Molecular detection of bacteria. (i) PCR sensitivity.
Sensitivity tests were performed with E. coli, B. fragilis, and all the organisms isolated from retrieved orthopedic implants and listed above. Facultative isolates were grown on BA at 37°C for 24 h, and anaerobic isolates were grown on ABA in the anaerobic cabinet for a similar time period. Single colonies of each isolate were inoculated into TSB (10 ml); B. fragilis, however, was inoculated into defined minimal medium (41). Isolates were incubated as described above, and the actively growing cultures were adjusted to an optical density at 540 nm equivalent to 108 CFU/ml and were diluted in 10-fold serial dilutions with PBS. Viable cells were counted as the number of CFU by triplicate plating of diluted samples on either BA or ABA and counting the colonies after incubation at 37°C for 24 h. From 10-fold serial dilutions containing from 108 to 100 CFU/ml, 1-ml volumes were retained for DNA extraction.
(ii) PCR laboratory conditions and control measures.
PCR was carried out under stringent conditions in a hospital diagnostic laboratory accredited by the American Society for Histocompatibility and Immunogenetics. All DNA manipulations pre- and post-PCR were performed in separate designated rooms with separate pipetting devices to avoid contamination of the samples with foreign DNA. Furthermore, as in other studies (4, 27), UV light was used to irradiate all equipment used in the preamplification steps to prevent contaminating DNA from causing false-positive results. Master-mixture water controls and DNA extraction controls were used for every batch of samples processed.
(iii) DNA extraction.
Aliquots (1 ml) of prosthesis sonicate or bacterial culture were centrifuged at 10,000 × g for 15 min. The pellets were resuspended by vortex mixing in 200 μl of cell lysis buffer (10 mM Tris-HCl [pH 8.0], 5 mM EDTA, 0.5% sodium dodecyl sulfate, proteinase K [100 μg/ml]). This reaction took place at 55°C for 3 h, after which the temperature was adjusted to 37°C and the reaction mixture was incubated overnight. The temperature of the samples was increased to 55°C for 1 h before DNA extraction was performed by the addition of equal volumes of saturated phenol-chloroform. After vortex mixing and centrifugation at 10,000 × g for 15 min, the aqueous phase was removed and the DNA was precipitated by the addition of 100% ethanol (2.5 volumes), 3 M sodium acetate (pH 5.2) (0.1 volume), and 2 μl of See DNA (Amersham, Aylesbury, United Kingdom). Following brief vortex mixing and centrifugation at 10,000 × g for 15 min, the supernatant was poured off and the DNA pellet was washed by the addition of 1 ml of 70% ethanol. Following further brief vortex mixing and centrifugation at 10,000 × g for 15 min, any residual ethanol traces were removed by vacuum drying. The extracted DNA was then dissolved in 50 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and was stored at −20°C.
(iv) Oligonucleotide primers.
Oligonucleotide primers were synthesized by Perkin-Elmer-Applied Biosystems, Warrington, United Kingdom. The target DNA sequence was the 16S rRNA gene. Sequence alignment by the clustal method with a weighted residue weight table (34) was carried out with the 16S rRNA genes of the following bacteria: S. capitis, S. epidermidis, Staphylococcus haemolyticus, P. acnes, Micrococcus agilis, B. fragilis, E. coli, and a Peptostreptococcus sp. The primer set selected was D1 (5′-GAG GAA GGT RGG GAY GAC GT) and D2 (5′-AGG CCC GGG AAC GYA TTY ACC G) for amplification of a 216-bp fragment of the 16S rRNA gene. The positions of D1 and D2 are 1199 to 1219 and 1394 to 1415 of the E. coli 16S rRNA positions, respectively (R = AG, Y = CT).
(v) DNA amplification.
The PCR mixture, which was made up to 50 μl in sterile double-distilled water, contained 5 μl of 10× PCR buffer (Perkin-Elmer-Applied Biosystems), 5 μl of MgCl2 (25 mM), each deoxynucleotide triphosphate (Pharmacia Biotech, Milton Keynes, United Kingdom) at a concentration of 200 μM, 20 pM each primer, and 3 U of AmpliTaq polymerase (Perkin-Elmer-Applied Biosystems). Two microliters of lysate containing target DNA was added to the PCR mixture, which was incubated at 96°C for 5 min. PCR was performed for 30 cycles of 1 min at 96°C, 2 min at 55°C, and 1 min at 72°C using a Perkin-Elmer Gene-Amp PCR System 9600 (Perkin-Elmer-Applied Biosystems). The final cycle ended with a 5-min extension at 72°C, and the PCR products were stored in the thermocycler at 15°C until they were collected.
After amplification, 6 μl of the amplified product was run on a 1.5% agarose gel in 1× Tris-borate-EDTA. DNA bands were detected by ethidium bromide staining and were visualized by UV light photography.
RESULTS
Incidence of infection.
Our results indicate that the use of nonculture methods as opposed to culture methods significantly (P < 0.05; chi-square test) increases the level of detection of infected prostheses (Table 1). By including only samples positive by both IFM and 16S rRNA detection and with an inflammatory cell infiltration score (see Table 3) of greater than 1, a conservative estimate for the level of infection in culture-negative samples is 25 of 94 (27%). In contrast, if all samples that were positive by either IFM or 16S rRNA detection are included, the level of infection increases to 69 of 94 (73%) samples.
TABLE 1.
Comparison of the detection rates of prosthetic hip infection by different methods
| Method of detection | No. of samples | No. of positive samples | % Positive samples |
|---|---|---|---|
| Culture of tissue only | 120 | 5 | 4 |
| Culture of tissue and implantsa | 120 | 26 | 22 |
| Immunofluorescence microscopy | 113 | 71 | 63 |
| 16S rRNA gene amplification | 118 | 85 | 72 |
| Inflammatory cell infiltration | 81 | 59b | 73 |
Use of mild ultrasonication to dislodge bacteria growing within adherent biofilms and strict anaerobic procedures.
Inflammatory score greater than 1.
TABLE 3.
Bacteria detected from culture-negative retrieved prosthetic hip implants by IFM and bacterial 16S rRNA gene detection and associated tissue inflammatory responsea
| Sample | Tissue infiltration by inflammatory cells (score)b
|
Immunofluorescence microscopy result
|
16S rRNA gene detection | Sample | Tissue infiltration by inflammatory cells (score)b:
|
Immunofluorescence microscopy result
|
16S rRNA gene detection | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PMNc | LYMd | MACe | Morphology | Scoref | PMNc | LYMd | MACe | Morphology | Scoref | |||||
| 1 | 1 | 2 | 3 | Cocci | 3 | + | ||||||||
| 2 | 1 | 2 | 3 | Cocci | 3 | + | ||||||||
| 3 | 0 | 2 | 0 | Cocci | 3 | + | ||||||||
| 4 | 0 | 0 | 0 | Cocci | 3 | + | ||||||||
| 5 | 3 | 3 | 2 | Cocci | 2 | + | ||||||||
| 6 | 1 | 3 | 3 | Cocci | 2 | + | ||||||||
| 7 | 0 | 2 | 3 | Cocci | 2 | + | ||||||||
| 8 | 0 | 1 | 1 | Cocci | 2 | + | ||||||||
| 9 | 1 | 2 | 3 | Cocci | 2 | + | ||||||||
| 10 | 0 | 2 | 3 | Cocci | 2 | + | ||||||||
| 11 | 0 | 2 | 2 | Cocci | 1 | + | ||||||||
| 12 | 0 | 1 | 3 | Cocci | 1 | + | ||||||||
| 13 | NSg | NS | NS | Cocci | 2 | + | ||||||||
| 14 | NS | NS | NS | Cocci | 2 | + | ||||||||
| 15 | NS | NS | NS | Cocci | 1 | + | ||||||||
| 16 | 1 | 2 | 3 | Cocci | 3 | − | ||||||||
| 17 | 1 | 2 | 3 | Cocci | 3 | − | ||||||||
| 18 | 1 | 2 | 1 | Cocci | 2 | − | ||||||||
| 19 | NS | NS | NS | Cocci | 3 | − | ||||||||
| 20 | NS | NS | NS | Cocci | 3 | − | ||||||||
| 21 | 2 | 2 | 3 | P. acnes | 2 | + | ||||||||
| 22 | 1 | 2 | 3 | P. acnes | 2 | + | ||||||||
| 23 | 2 | 2 | 3 | P. acnes | 1 | + | ||||||||
| 24 | 2 | 2 | 3 | P. acnes | 1 | + | ||||||||
| 25 | 3 | 2 | 2 | P. acnes | 1 | + | ||||||||
| 26 | 1 | 2 | 3 | P. acnes | 1 | + | ||||||||
| 27 | 1 | 2 | 3 | P. acnes | 1 | + | ||||||||
| 28 | 0 | 1 | 2 | P. acnes | 1 | + | ||||||||
| 29 | 0 | 1 | 2 | P. acnes | 1 | + | ||||||||
| 30 | 1 | 1 | 1 | P. acnes | 1 | + | ||||||||
| 31 | 0 | 1 | 1 | P. acnes | 1 | + | ||||||||
| 32 | 0 | 0 | 1 | P. acnes | 1 | + | ||||||||
| 33 | 0 | 0 | 0 | P. acnes | 1 | + | ||||||||
| 34 | NS | NS | NS | P. acnes | 1 | + | ||||||||
| 35 | NS | NS | NS | P. acnes | 1 | + | ||||||||
| 36 | NS | NS | NS | P. acnes | 1 | + | ||||||||
| 37 | 0 | 0 | 0 | P. acnes | 3 | − | ||||||||
| 38 | 3 | 1 | 1 | P. acnes | 1 | − |
| 39 | NS | NS | NS | P. acnes | 1 | NDh |
| 40 | 1 | 3 | 3 | Cocci and P. acnes | 3 | + |
| 41 | 0 | 0 | 0 | Cocci and P. acnes | 3 | + |
| 42 | 2 | 2 | 3 | Cocci and P. acnes | 2 | + |
| 43 | 1 | 2 | 3 | Cocci and P. acnes | 2 | + |
| 44 | 0 | 2 | 2 | Cocci and P. acnes | 2 | + |
| 45 | 1 | 1 | 2 | Cocci and P. acnes | 1 | + |
| 46 | 0 | 2 | 3 | Cocci and P. acnes | 1 | + |
| 47 | NS | NS | NS | Cocci and P. acnes | 2 | − |
| 48 | 1 | 2 | 3 | 0 | + | |
| 49 | 1 | 1 | 3 | 0 | + | |
| 50 | 1 | 1 | 2 | 0 | + | |
| 51 | 0 | 1 | 1 | 0 | + | |
| 52 | 0 | 0 | 0 | 0 | + | |
| 53 | 0 | 0 | 0 | 0 | + | |
| 54 | 0 | 0 | 0 | 0 | + | |
| 55 | 0 | 1 | 3 | ND | + | |
| 56 | 0 | 1 | 2 | ND | + | |
| 57 | 0 | 1 | 2 | ND | + | |
| 58 | 0 | 0 | 3 | ND | + | |
| 59 | 0 | 0 | 0 | ND | + | |
| 60 | 2 | 3 | 3 | 0 | − | |
| 61 | 1 | 3 | 3 | 0 | − | |
| 62 | 1 | 2 | 3 | 0 | − | |
| 63 | 1 | 2 | 3 | 0 | − | |
| 64 | 1 | 2 | 2 | 0 | − | |
| 65 | 0 | 2 | 3 | 0 | − | |
| 66 | 0 | 2 | 3 | 0 | − | |
| 67 | 1 | 1 | 2 | 0 | − | |
| 68 | 1 | 1 | 1 | 0 | − | |
| 69 | 0 | 1 | 1 | 0 | − | |
| 70 | 0 | 1 | 1 | 0 | − | |
| 71 | 0 | 0 | 1 | 0 | − | |
| 72 | 0 | 0 | 1 | 0 | − | |
| 73 | 0 | 0 | 0 | 0 | − |
A further 21 samples not suitable for histological examination were negative by IFM, of which 10 were positive and 11 were negative by 16S rRNA gene detection.
Number of inflammatory cells per high-power field: 0, no cells; 1, 1 to 10 cells; 2, 10 to 20 cells; 3, 20 or more cells.
PMN, polymorphonuclear leukocyte.
LYM, lymphocyte.
MAC, tissue macrophage.
Number of bacteria per well: 0, no bacteria; 1, 1 to 10 bacteria; 2, 10 to 50 bacteria; 3, 50 or more bacteria.
NS, not suitable for histological examination.
ND, not done due to insufficient sample volume.
Immunological detection of bacteria.
MAb QUBPa3, which reacted with all of the P. acnes strains isolated in our previous study (37), did not cross-react with S. aureus, S. epidermidis, B. fragilis, E. coli, P. magnus, P. micros, C. diphtheriae, C. hofmanni, or C. xerosis by IFM. Similarly, the polyclonal antiserum raised to S. epidermidis, which reacted with all Staphylococcus strains isolated in our previous study (37), did not cross-react with B. fragilis, E. coli, P. magnus, or P. micros.
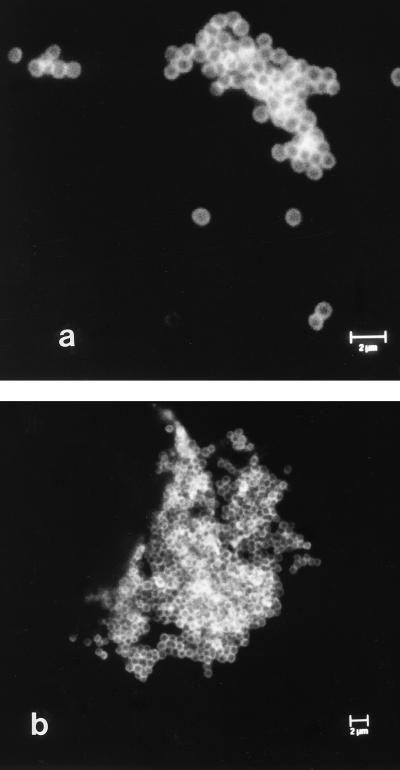
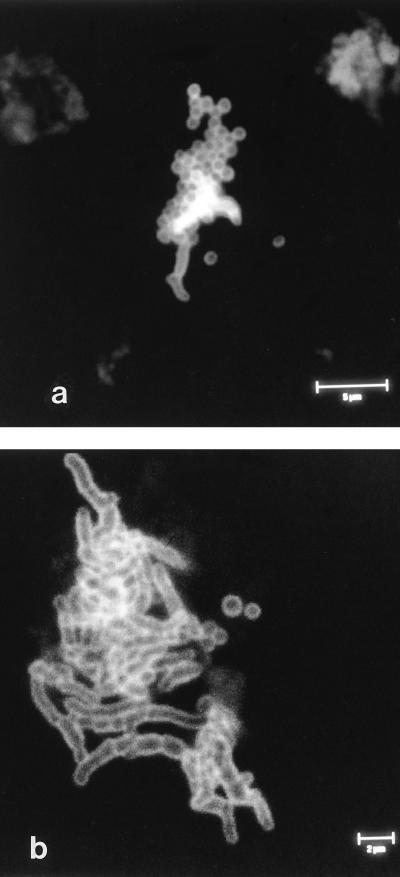
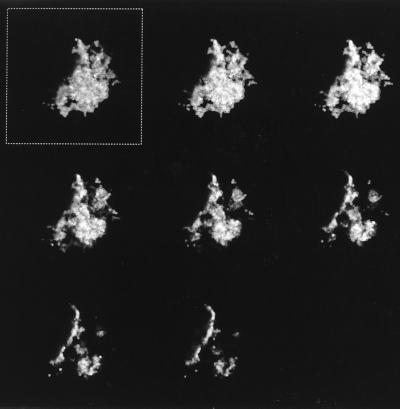
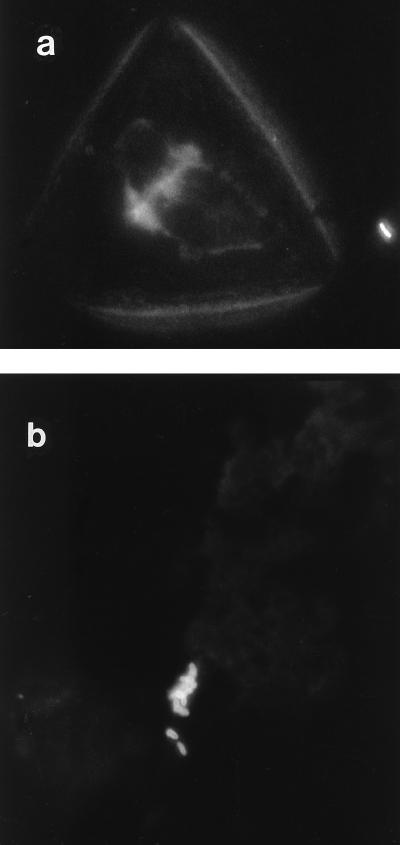
Bacteria were observed singly, in small groups, and in large aggregates by IFM (Fig. 1a and b). In addition, both P. acnes and Staphylococcus spp. were found together in large aggregates in several instances (Fig. 2a and b). The aggregates were between 3 and 4.5 μm in depth, as estimated by CSLM, and consisted of several layers of bacterial cells (Fig. 3). These aggregates were observed only after ultrasonication of the prostheses. When a selected number of prostheses were placed in Ringer’s solution and the solution was removed and processed without ultrasonication, the occasional individual bacterial cell was observed in samples in which large aggregates were observed after ultrasonication. No bacteria were visible by IFM when three researchers at the Department of Microbiology and Immunobiology, School of Clinical Medicine, scraped their skin flakes into the bags containing Ringer’s solution that are normally used for the retrieved prostheses, and the contents of the bags were then processed in the same manner as bags containing prostheses. Concentrated skin scrapings were also applied to microscope slides and were processed for IFM. No large aggregates of bacteria which resembled those observed in the sonicates were detected. IFM revealed sparse small groups of bacteria (up to five or six cells), some isolated bacterial cells, and skin cells (Fig. 4). Skin cells were not observed in sonicates.
FIG. 1.
Examples of confocal laser scanning micrographs of bacteria in material removed by ultrasonication (sonicate) from culture-negative hip prostheses to illustrate coccoid cells present singly and in small groups (a) and in a large aggregate (b). Bacteria were labelled with anti-Staphylococcus spp. polyclonal antiserum and an anti-P. acnes-specific MAb, followed by labelling with suitable fluorescently conjugated secondary antibodies.
FIG. 2.
Confocal laser scanning micrographs of sonicates obtained from culture-negative hip prostheses to illustrate a large number of coccoid cells associated with a smaller number of coryneform cells (a) and a large number of coryneform cells associated with a smaller number of coccoid cells (b) labelled as described in the legend to Fig. 1.
FIG. 3.
Confocal laser scanning micrograph illustrating the depth of the dislodged biofilm aggregate of coccoid cells shown in Fig. 1b. The series of images were captured at 0.5-μm intervals from the top (image in the box) to the bottom (images displayed left to right) of the aggregate. The depth of this aggregate was estimated to be 3.5 μm.
FIG. 4.
Immunofluorescence micrographs of material scraped from skin illustrate an isolated bacterial cell and a skin cell (a) and a small group of bacteria (b). Immunolabelling was carried out as described in the legend to Fig. 1.
All 24 clinical sonicate samples which were positive by culture were also positive by IFM (Table 2). In 11 of the samples the same bacteria that had been isolated by culture were detected by IFM. A further 11 samples that were positive by culture for either a Staphylococcus sp. or P. acnes were positive for both organisms by IFM.
TABLE 2.
Bacteria detected from culture-positive retrieved prosthetic hip implants by culture, IFM, and bacterial 16S rRNA gene detection and associated tissue inflammatory response
| Sample no. | Bacterium(a) isolated | Tissue infiltration by inflammatory cells (score)a
|
IFM result
|
16S rRNA gene detection | |||
|---|---|---|---|---|---|---|---|
| PMNb | LYMc | MACd | Morphology | Scoree | |||
| 1 | S. epidermidis | 1 | 1 | 2 | Cocci | 3 | + |
| 2 | S. epidermidis | 2 | 1 | 3 | Cocci | 3 | + |
| 3 | S. aureus | 0 | 1 | 0 | Cocci | 2 | + |
| 4 | S. capitis | 0 | 0 | 1 | Cocci | 1 | + |
| 5 | S. epidermidis | 0 | 1 | 2 | Cocci and P. acnes | 2 | + |
| 6 | S. hominis | 0 | 1 | 2 | Cocci and P. acnes | 1 | + |
| 7 | S. epidermidis | 1 | 1 | 3 | P. acnes | 2 | + |
| 8 | S. epidermidis and S. hominis | 0 | 2 | 3 | Cocci | 2 | + |
| 9 | S. epidermidis and S. hominis | 1 | 2 | 3 | Cocci | 2 | + |
| 10 | S. epidermidis and S. capitis | 2 | 2 | 2 | Cocci and P. acnes | 1 | + |
| 11 | P. acnes | 2 | 2 | 3 | P. acnes | 1 | + |
| 12 | P. acnes | 1 | 1 | 1 | P. acnes | 1 | + |
| 13 | P. acnes | NSf | NS | NS | P. acnes | 3 | + |
| 14 | P. acnes | 0 | 2 | 3 | Cocci and P. acnes | 2 | + |
| 15 | P. acnes | 0 | 1 | 2 | Cocci and P. acnes | 2 | + |
| 16 | P. acnes | 2 | 2 | 3 | Cocci and P. acnes | 1 | + |
| 17 | P. acnes | NS | NS | NS | Cocci and P. acnes | 3 | + |
| 18 | P. acnes | NS | NS | NS | Cocci and P. acnes | 3 | + |
| 19 | P. acnes | NS | NS | NS | Cocci and P. acnes | 1 | + |
| 20 | P. acnes | NS | NS | NS | Cocci and P. acnes | 1 | + |
| 21 | P. acnes | NS | NS | NS | Cocci and P. acnes | 1 | + |
| 22 | P. acnes | 0 | 1 | 2 | NDg | ND | + |
| 23 | S. haemolyticus and P. acnes | 1 | 1 | 1 | Cocci and P. acnes | 1 | + |
| 24 | Micrococcus sp. and P. acnes | 1 | 2 | 3 | Cocci and P. acnes | 1 | + |
| 25 | S. epidermidis and P. acnes | NS | NS | NS | Cocci | 1 | + |
Number of inflammatory cells per high-power field: 0, no cells; 1, 1 to 10 cells; 2, 10 to 20 cells; 3, 20 or more cells.
PMN, polymorphonuclear leukocyte.
LYM, lymphocyte.
MAC, tissue macrophage.
Number of bacteria per well: 0, no bacteria; 1, 1 to 10 bacteria; 2, 10 to 50 bacteria; 3, 50 or more bacteria.
NS, not suitable for histological examination.
ND, not done due to insufficient sample volume.
Bacteria were also detected by IFM in 47 of the 89 (53%) culture-negative samples examined. Staphylococcus spp. and P. acnes were detected alone in 20 and 19 samples, respectively, and both organisms were detected in a further 8 samples (Table 3). Staphylococcus spp. were found in greater quantity than P. acnes, with 17 of the 20 Staphylococcus-positive samples having IFM scores of 2 or 3, which corresponds to between 10 and greater than 50 bacteria per field of view. In comparison, only 3 of 19 P. acnes-positive samples had a similar score.
Molecular detection of bacteria.
The lower limits of detection of S. aureus, S. epidermidis, S. hominis, S. capitis, P. acnes, Micrococcus spp., E. coli, and B. fragilis bacterial cells by PCR amplification of a region of the bacterial 16S rRNA gene were examined. Results indicated that, by our protocol, P. acnes and Micrococcus spp. were detected only at concentrations greater than or equal to 105 CFU/ml (data not shown). The remaining six organisms could be detected if they were present in numbers greater than or equal to 104 CFU/ml (data not shown).
Bacterial DNA was amplified from sonicate samples of all 25 implants which were culture positive (Table 2). Bacterial DNA was also amplified from sonicate samples of 60 of the 93 (65%) culture-negative implants examined (Table 3).
Comparison of tissue pathology and immunological and molecular detection.
All 25 of the culture-positive samples were also positive by IFM and 16S rRNA gene amplification. Tissue pathology results were available for 18 of these patients, and in 8 of these patients neutrophilic infiltration was not observed. Lymphocytes or macrophages were, however, observed in all 18 patients (Table 2). A giant cell-foreign body reaction was also noted in three of the culture-positive tissue samples.
Of the 94 culture-negative samples, 38 (40%) were positive by both IFM and 16S rRNA gene amplification. Tissue pathology was available for 32 of the 38 samples, and there was evidence of inflammatory cell infiltration in 29 samples. Two of the three samples in which tissue infiltration was not observed had IFM scores of 3, equivalent to 50 or more bacteria per well (Table 3). Neutrophilic infiltration was not observed in 12 of the 29 samples, although lymphocytes or macrophages were evident. A further eight samples were negative by 16S rRNA gene amplification but positive by immunolabelling, and one of these eight samples was also negative for inflammatory cell infiltration.
Seventeen samples were positive by 16S gene amplification but negative by IFM. Tissue pathology was available for only seven of these samples, and cell infiltration was observed in four samples. Tissue pathology results were available for 15 samples which were negative by both IFM and 16S rRNA gene detection. There was evidence of a giant cell-foreign body reaction in one of these, and the inflammatory cell score was zero for only one sample. Tissue pathology was not available for the remaining 12 culture-, IFM-, and 16S rRNA gene amplification-negative samples.
DISCUSSION
This study is the first to compare immunological and DNA detection methods with culture for the detection of bacterial infection of retrieved prosthetic hip joints. Bacteria were detected by immunolabelling and fluorescence microscopy with a P. acnes-specific MAb and polyclonal antiserum specific for Staphylococcus spp. only after the retrieved prostheses were subjected to mild ultrasonic treatment to dislodge the bacteria growing within a biofilm on the prosthetic joint surface. By this method bacteria were detected in all of the samples which were positive by culture and in 53% of the culture-negative samples. Bacteria, either coccoid or coryneform in morphology, were clearly visible by IFM. These were recorded as containing either P. acnes or Staphylococcus spp. due to the lack of cross-reactivity of the antibodies with other coccoid bacteria which are implicated in prosthetic implant infection, such as P. magnus (14) and other commensal and pathogenic coryneform bacteria. Therefore, the total number of samples positive by immunodetection was 71 of 113 (63%).
Previously, we have observed bacterial biofilms on retrieved prostheses by scanning electron microscopy (37); however, processing for scanning electron microscopy precludes examination of the biofilm by other methods. The use of CSLM in the present study showed that bacteria dislodged from implants by ultrasonication could be found in aggregates that varied in depth from 3 to 4.5 μm and that consisted of several layers of bacterial cells. It therefore seems that infecting bacteria growing within adherent biofilms are removed in large aggregates by the ultrasonication procedure. As the diluent used throughout was Ringer’s salt solution, no enrichment was involved. Samples were also stored at −70°C; therefore, these aggregates could not have arisen as a result of multiplication of contaminating bacteria. It is worth noting that as these aggregates may contain over 300 bacteria, our estimates of the number of bacteria infecting an implant (37), calculated by total viable colony count per implant, are likely to be a gross underestimate. Each bacterial aggregate will give rise to only 1 CFU, and therefore, the total number of bacteria colonizing the implant will be considerably more than the number of CFU counted. Examination for potential skin-flake contamination, generated by scraping and rubbing the skin surfaces of laboratory personnel, showed that bacteria present as a result of this type of contamination should have been easily distinguishable by IFM from bacterial aggregates dislodged from the prosthesis biofilm. Skin cells were not observed in the sonicates. Examination of skin scrapings revealed few bacteria and no large aggregates. The estimated density of P. acnes ranges from 102 to 105 per cm2 of skin (26), and propionibacteria account for approximately half of the total skin microbiota (35). It would therefore take gross skin contamination for this to be visible by immunofluorescence microscopy. Contamination of the prostheses by the skin of the patient, surgical operating staff, or laboratory personnel should have easily been detectable. Furthermore, if prostheses were placed in diluent which was then aspirated, without sonication, bacterial aggregates were not detected in the diluent. If prostheses were placed in fresh diluent and then sonicated, bacteria could be observed in some samples. Sonication was therefore necessary to remove the adherent bacteria, which again makes it highly unlikely that the bacteria detected arose from skin contaminants. The contamination of clinical samples by the normal skin microbiota is a potential problem in clinical diagnostic procedures involving an enrichment step in which small numbers of contaminants have the opportunity to multiply along with infecting microbes. We have clearly shown that when no enrichment step is used and samples are processed in Ringer’s solution rather than broth diluent, it is possible to distinguish between infecting and contaminating microbes. This is probably largely due to quantitative differences; if there are any contaminating bacteria they are few in number compared to the number of infecting microbes.
Bacterial 16S rRNA was detected in all culture-positive samples by use of two broad-range oligonucleotide primers that had been designed in order to amplify by PCR rRNA genes from a wide spectrum of bacteria implicated in prosthetic hip infection. Bacterial 16S rRNA genes (termed PCR positive) were detected in all of the culture-positive samples and in a further 65% of the culture-negative samples. In total, 85 of 118 (72%) samples were positive by PCR. Culture-negative, PCR-positive samples are unlikely to represent samples with false-positive results as suitable stringent control procedures were carried out to ensure that such samples were not positive due to the presence of contaminating bacterial DNA (4, 27). The lower limit of detection was also estimated to be approximately 104 CFU/ml. Samples were therefore unlikely to be positive as a result of exogenous contamination. Various methods could have been used to increase the efficiency of the PCR, including the use of nested PCR; however, as these should also have increased the chance of detecting contaminants, they were not used. In a comparison of culture and 16S rRNA gene detection methods for detection of total knee arthroplasty infection, a similar level of infection was reported. Evidence of bacterial infection on the basis of PCR amplification of synovial fluid aspirates was reported for 32 of 50 specimens (64%) (25).
Although the data for the tissue pathology are incomplete, both the rRNA gene detection and IFM results related well to the presence of inflammatory cells in implant-associated tissue; in only three instances was the inflammatory cell score zero. In two of these the IFM score was high, which suggests that the negative tissue pathology result may have been due to inadequate tissue sampling (1).
In some instances, although the tissue pathology score was high, bacteria were only evident in low numbers by IFM. It may be that some of the bacteria are strongly adherent to the device and sonication is insufficient to remove them, an observation we have made with in vitro models of biofilms (38). Also, in some cases the bacteria may predominantly colonize bone rather than the prosthesis. Alternatively, any infecting bacteria may not have reacted with our sera. We are addressing these issues. The lack of observation of neutrophilic infiltration in eight of the culture-positive samples, in which lymphocytes or macrophages were present, suggests that neutrophil infiltration should perhaps not be used as the sole indicator of infection.
It is likely that prosthetic joint infection may be quite variable in nature, depending on the type of infecting bacteria, whether the infection is polymicrobial, and the length of time that the implant has been infected. When bacteria are prevalent in the surrounding tissues, the infection may be typified by high levels of neutrophil infiltration in tissue; however, when the majority of bacteria are growing on the device surface within a biofilm, the infection may be typified by lymphocyte and macrophage infiltration. Within the tissues, cells of the immune system will come into contact with living whole bacterial cells, whereas it is likely that the major immunostimulants of biofilm bacteria will be secreted or released products. An immunoglobulin response to polysaccharides, thought to be mainly teichoic acids, released by S. epidermidis is detectable in the serum of patients with prosthetic joint infection (21). Teichoic acids are well recognized as belonging to the group of immunomodulatory molecules which may be present in or released from the bacterial cell envelope (31). Whether the bacteria involved in these infections, other than S. aureus, are able to produce other immunomodulatory molecules, such as superantigen toxins, is unknown. The immunostimulatory properties of P. acnes are well documented and comparable to those of bacteria such as Mycobacterium tuberculosis and Bordetella pertussis (11). Therefore, low-grade chronic infection with P. acnes could be sufficient to stimulate the observed inflammatory cell infiltration. A lack of a neutrophilic inflammatory response is characteristic of diseases such as typhoid and tuberculosis, which are caused by bacteria which grow intracellularly (33). Interestingly, there are reports in the literature that P. acnes can also grow intracellularly (11). It should also be noted that the presence of giant cells without evidence of neutrophils, although indicative of reaction to the device materials (28), may not necessarily rule out bacterial infection of the device.
The detection of bacteria in sonicates from patients with culture-negative implants indicates that these implants were colonized by bacteria which were not isolated by the microbiological techniques used. This may be because the implants were infected with viable but nonculturable bacteria. Interestingly, in some instances only one type of bacterium was detected by culture, but both coccoid and coryneform bacteria were observed by IFM. The possibility that the bacteria were nonculturable as a result of the cefamandole administered intravenously at the time of operation must be addressed, although it is generally considered that bacteria growing within an adherent biofilm on the implant surface are protected from antibiotic therapy (36). We examined the antibiotic sensitivities of the bacteria that we isolated from prostheses. The majority were approximately 1,000 times more resistant (minimum bactericidal concentration, >1,024 μg/ml) to cefamandole when they were growing within in vitro model biofilms than when they were growing in broth culture (39, 40). This indicates that it is unlikely that the cefamandole rendered the bacteria unculturable. Naylor et al. (29) and Costerton et al. (7) have previously proposed that alterations of bacterial metabolism may be responsible for the enhanced antibiotic resistance of bacteria growing within a biofilm.
The detection of bacterial DNA in culture-negative clinical samples by PCR amplification despite prolonged antibiotic therapy resulted in several researchers concluding that antibiotic administration did not eradicate the infecting bacteria but rendered them nonculturable. For example, Canvin et al. (5) described a case of septic arthritis in a patient with rheumatoid arthritis and prosthetic knee joints. Cultures of synovial fluid from the patient’s knees were initially positive for S. aureus but rapidly became sterile after 1 week of antibiotic therapy. However, synovial fluid samples taken until 10 weeks of therapy showed the persistence of S. aureus by the presence of specific staphylococcal DNA by PCR, leading the investigators to conclude that either nonviable debris persisted after infection or that organisms were still viable but rendered nonculturable by antibiotic administration. Similarly, Ni et al. (30) used PCR to detect meningococcal DNA in 54 cerebrospinal fluid samples for which antibiotic administration had prevented isolation by conventional culture techniques. In the present study, the observation of whole bacterial cells by IFM indicates that our samples do not only contain bacterial DNA debris. Another possible explanation for the lack of viability is that the bacteria from the implants are sensitive to mild ultrasonication. Although pure cultures of laboratory-grown bacteria are not killed by this level of ultrasound (37), bacteria which have been growing in vivo and subsequently stressed by removal from the patient and transportation to the laboratory for processing may have greater sensitivity.
It is also possible that viable but nonculturable bacteria are so highly adapted to the environment of the in vivo biofilm that the conditions required for their continued growth, and therefore successful isolation, are not met by the growth media and isolation procedures used. In effect, growth within the in vivo biofilm renders the bacteria more fastidious with respect to growth requirements. One possible explanation for the difficulty in the isolation of these bacteria could be the dilution of bacterial signaling molecules, of which a critical concentration may be necessary to trigger growth of the bacteria. Quorum sensing, involving, for example, N-acyl homoserine lactones in gram-negative bacteria (8) and peptide pheromone in gram-positive bacteria (22), is well characterized. Whether prolonged growth in this environment also results in irreversible genotypic changes remains an open question. Bacterial pathogens subjected to the highly variable challenge of the human immune system are well known for genetically based reversible variation (31); however, bacteria growing for long periods in what may be a relatively static environment, largely protected from the immune system, may, in the long term, become less adaptable.
Advantages and disadvantages are associated with the use of both 16S rRNA gene amplification and immunolabelling to detect infection. Benefits of both methods include rapid detection of infection when culture techniques have proved ineffective and the ability to detect bacteria in clinical samples in the presence of antimicrobial drugs. The major advantage of rRNA gene detection is that the use of universal broad-range primers allows recognition of any bacteria that may be present and polymicrobial infections can be detected as it is not necessary to predict which bacteria may be present. In contrast, for immunological detection of infection to be effective, the MAbs and polyclonal antiserum must be prepared against the bacteria thought to cause the infection, and other bacteria will therefore not be detected. This could explain why more culture-negative samples were positive for infection by rRNA gene detection than by immunolabelling in the present study. As well as being a disadvantage, this requirement can also be an advantage as the identity of the infecting bacteria can be ascertained immunologically. Also, direct visualization of the bacterial morphology makes it very unlikely that a false-positive diagnosis will be made or, indeed, that contaminating bacteria will be mistaken for infecting bacteria. In contrast, bacterial 16S rRNA gene amplification alone does not identify the infecting bacteria. For definitive identification of infecting bacteria, the amplification products could be directly sequenced or, in the case of mixed infections, first cloned into E. coli plasmid vectors and then sequenced.
In conclusion, this study implicates unrecognized infection as a potential major cause of prosthetic hip failure. IFM allows a rapid quantitative and qualitative assessment of infected prostheses and distinguishes the bacteria from the infected prostheses from bacteria that may result from skin contamination. The IFM results indicate that 63% of retrieved hip prostheses may be colonized with bacteria. 16S rRNA genes were detected from 72% of retrieved prostheses. We are investigating the nature of the bacteria detectable by PCR amplification but not by IFM. The follow-up of these patients with respect to the successes of the second prostheses is a critical part of our study; however, the treatment of patients with primary prostheses must be addressed in the short term, given the long-term nature of this type of infection. Unfortunately, neither of the nonculture detection methods allows the determination of antimicrobial susceptibility. This limitation can be partly overcome, however, by using current knowledge of prosthetic hip infection as a guide to the expected pathogens and their antimicrobial susceptibilities, in particular when they are growing within biofilms. The improved detection of infection could then be coupled with appropriate antibiotic therapy for a longer postoperative period, and this should improve the clinical outcomes for patients undergoing revision hip surgery.
ACKNOWLEDGMENTS
M. M. Tunney and the work were funded by the Arthritis Research Campaign of the United Kingdom (project grant P0522); G. Ramage was funded by a Department of Education for Northern Ireland Studentship, and D. Hanna was funded by a European Social Fund Grant.
We thank the operating theater staff, Musgrave Park Hospital, Belfast, and we thank A. Maule, School of Biology and Biochemistry, The Queen’s University of Belfast, for assistance with the use of the confocal scanning laser microscope.
REFERENCES
- 1.Athanasou N A, Pandey R, DeSteiger R, Crook D, McLardy Smith P. Diagnosis of infection by frozen section during revision arthroplasty. J Bone Jt Surg Br Vol. 1995;77:28–33. [PubMed] [Google Scholar]
- 2.Atkins B L, Athanasou N A, Deeks J J, Crook D W M, Simpson H, Peto T E A, McLardy-Smith P, Berendt A R The Osiris Collaborative Study Group. Prospective evaluation of criteria for microbiological diagnosis of prosthetic-joint infection at revision arthroplasty. J Clin Microbiol. 1998;36:2932–2939. doi: 10.1128/jcm.36.10.2932-2939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrack R L, Harris W H. The value of aspiration of the hip joint before revision total hip arthroplasty. J Bone Jt Surg Am Vol. 1993;75:66–76. doi: 10.2106/00004623-199301000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Brakstad O G, Aasbakk K, Maeland J A. Detection of Staphylococcus aureus by polymerase chain-reaction amplification of the NUC gene. J Clin Microbiol. 1992;30:1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canvin J M G, Goutcher S C, Hagig M, Gemmell C G, Sturrock R D. Persistence of Staphylococcus aureus as detected by polymerase chain reaction in the synovial fluid of a patient with septic arthritis. Br J Rheumatol. 1997;36:203–206. doi: 10.1093/rheumatology/36.2.203. [DOI] [PubMed] [Google Scholar]
- 6.Christel P, Dijan P. Recent advances in adult hip joint surgery. Curr Opin Rheum. 1994;6:161–171. doi: 10.1097/00002281-199403000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Costerton J W, Lewandowski Z, DeBeer D, Caldwell D, Korber D, James G. Biofilms, the customized microniche. J Bacteriol. 1994;176:2137–2142. doi: 10.1128/jb.176.8.2137-2142.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 9.Dreghorn C R, Hamblen D L. Revision arthroplasty: a high price to pay. Br Med J. 1989;298:648–649. doi: 10.1136/bmj.298.6674.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupont J A. Significance of operative cultures in total hip arthroplasty. Clin Orthop. 1986;211:122–127. [PubMed] [Google Scholar]
- 11.Eady E A, Ingham E. Propionibacterium acnes—friend or foe? Rev Med Microbiol. 1994;5:163–173. [Google Scholar]
- 12.Fehring T K, McAlister J A. Frozen histologic section as a guide to sepsis in revision joint arthroplasty. Clin Orthop Rel Res. 1994;304:229–237. [PubMed] [Google Scholar]
- 13.Feldman D S, Lonner J H, Desai P, Zuckerman J D. The role of intraoperative frozen sections in revision total joint arthroplasty. J Bone Jt Surg Am Vol. 1995;77:1807–1813. doi: 10.2106/00004623-199512000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Felten A, Desplaces N, Nizard R, Sedel L, Lagrange P. Infections osteoarticulaires A Peptostreptococcus magnus apres chirurgie orthopedique. Pathol Biol. 1998;46:442–448. [PubMed] [Google Scholar]
- 15.Fitzgerald R H. Total hip arthroplasty sepsis. Orthop Clin N Am. 1992;23:259–264. [PubMed] [Google Scholar]
- 16.Galfre G, Milstein C. Preparation of monoclonal antibodies: strategies and procedures. Methods Enzymol. 1981;73:1–46. doi: 10.1016/0076-6879(81)73054-4. [DOI] [PubMed] [Google Scholar]
- 17.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 19.Hykin P G, Tobal K, McIntyre G, Matheson M M, Towler H M A, Lightman S L. The diagnosis of delayed post-operative endophthalmitis by polymerase chain reaction of bacterial DNA in vitreous samples. J Med Microbiol. 1994;40:408–415. doi: 10.1099/00222615-40-6-408. [DOI] [PubMed] [Google Scholar]
- 20.Lachiewicz P F, Rogers G D, Thomason H C. Aspiration of the hip joint before revision total hip arthroplasty. J Bone Jt Surg Am Vol. 1996;78:749–754. doi: 10.2106/00004623-199605000-00015. [DOI] [PubMed] [Google Scholar]
- 21.Lambert P A, VanMaurik A, Parvatham S, Akhtar Z, Fraise A P, Krikler S J. Potential of exocellular carbohydrate antigens of Staphylococcus epidermidis in the serodiagnosis of orthopaedic prosthetic infection. J Med Microbiol. 1996;44:355–361. doi: 10.1099/00222615-44-5-355. [DOI] [PubMed] [Google Scholar]
- 22.Lazazera B A, Grossman A D. The ins and outs of peptide signalling. Trends Microbiol. 1998;6:288–293. doi: 10.1016/s0966-842x(98)01313-4. [DOI] [PubMed] [Google Scholar]
- 23.Lutton D A, Patrick S, Crockard A D, Stewart L D, Larkin M J, Dermott E, McNeill T A. Flow cytometric analysis of within-strain variation in polysaccharide expression by Bacteroides fragilis by use of murine monoclonal antibodies. J Med Microbiol. 1991;35:229–237. doi: 10.1099/00222615-35-4-229. [DOI] [PubMed] [Google Scholar]
- 24.Madhavan H N, Therese K L, Anand A R. Diagnostic value of polymerase chain reaction (PCR) in bacterial and Propionibacterium acnes endophthalmitis. Investig Ophthalmol Vis Sci. 1997;38:S1104. [Google Scholar]
- 25.Mariani B D, Martin D S, Levine M J, Booth R E, Tuan R S. Polymerase chain reaction detection of bacterial infection in total knee arthroplasty. Clin Orthop Rel Res. 1996;331:11–22. doi: 10.1097/00003086-199610000-00003. [DOI] [PubMed] [Google Scholar]
- 26.McGinley K J, Webster G F, Leyden J L. Regional variation of cutaneous propionibacteria. Appl Environ Microbiol. 1978;35:62–66. doi: 10.1128/aem.35.1.62-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meier A, Persing D H, Finken M, Bottger E C. Elimination of contaminating DNA within polymerase chain reaction reagents: implications for a general approach to detection of uncultured pathogens. J Clin Microbiol. 1993;31:646–652. doi: 10.1128/jcm.31.3.646-652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirra J M, Marder R A, Amstutz H C. The pathology of failed total joint arthroplasty. Clin Orthop. 1982;170:175–183. [PubMed] [Google Scholar]
- 29.Naylor P T, Myrvik Q N, Gristina A. Antibiotic-resistance of biomaterial-adherent coagulase-negative and coagulase-positive staphylococci. Clin Orthop Rel Res. 1990;261:126–133. [PubMed] [Google Scholar]
- 30.Ni H, Knight A I, Cartwright K, Palmer W H, McFadden J. Polymerase chain reaction for diagnosis of meningococcal meningitis. Lancet. 1992;340:1432–1434. doi: 10.1016/0140-6736(92)92622-m. [DOI] [PubMed] [Google Scholar]
- 31.Patrick S, Larkin M J. Immunological and molecular aspects of bacterial virulence. Chichester, United Kingdom: John Wiley & Sons; 1995. [Google Scholar]
- 32.Patrick S, Stewart L D, Damani N, Wilson K G, Lutton D A, Larkin M J, Poxton I, Brown R. Immunological detection of Bacteroides fragilis in clinical samples. J Med Microbiol. 1995;43:99–109. doi: 10.1099/00222615-43-2-99. [DOI] [PubMed] [Google Scholar]
- 33.Robbins S L, Cotran R S, Kumar V. Pathologic basis of disease. 5th ed. Philadelphia, Pa: The W. B. Sanders Company; 1997. [Google Scholar]
- 34.Ruimy R, Breittmayer V, Elbaze P. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA-specific DNA. Int J Syst Bacteriol. 1994;44:416–426. doi: 10.1099/00207713-44-3-416. [DOI] [PubMed] [Google Scholar]
- 35.Tancrede C. Role of human microflora in health and disease. Eur J Clin Microbiol Infect Dis. 1992;11:1012–1015. doi: 10.1007/BF01967791. [DOI] [PubMed] [Google Scholar]
- 36.Tunney M M, Gorman S P, Patrick S. Infection associated with medical devices. Rev Med Microbiol. 1996;7:195–205. [Google Scholar]
- 37.Tunney M M, Patrick S, Gorman S P, Nixon J R, Anderson N, Davis R I, Hanna D, Ramage G. Improved detection of infection in hip replacements: a currently underestimated problem. J Bone Jt Surg Br Vol. 1998;80:568–572. doi: 10.1302/0301-620x.80b4.8473. [DOI] [PubMed] [Google Scholar]
- 38.Tunney, M. M., G. Ramage, and S. Patrick. 1998. Unpublished data.
- 39.Tunney M M, Ramage G, Patrick S, Nixon J R, Murphy P G, Gorman S P. Antimicrobial susceptibility of bacteria isolated from orthopedic implants following revision hip surgery. Antimicrob Agents Chemother. 1998;42:3002–3005. doi: 10.1128/aac.42.11.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tunney M M, Ramage G, Patrick S, Nixon J R, Murphy P G, Gorman S P. Improved antibiotic therapy for elimination and prevention of prosthetic hip infection. J Pharm Pharmacol. 1998;50:40. [Google Scholar]
- 41.Van Tassell R L, Wilkins T D. Isolation of auxotrophs of Bacteroides fragilis. Can J Microbiol. 1978;24:1619–1621. doi: 10.1139/m78-260. [DOI] [PubMed] [Google Scholar]
- 42.Wilson M J, Weightmann A J, Wade W G. Applications of molecular ecology in the characterisation of uncultured microorganisms associated with human disease. Rev Med Microbiol. 1997;8:91–101. [Google Scholar]