Abstract
Calcium ions trigger many cellular events, including the release of neurotransmitters at the synaptic terminal and excitotoxic cell death. Recently, we have discovered that a transient increase in the level of cytoplasmic Ca2+ triggers the exposure of phosphatidylserine (PS) on the surfaces of necrotic cells in the nematode Caenorhabditis elegans. PS serves as an “eat me” signal that attracts engulfing cells to engulf and degrade necrotic cells. During the above study, we developed a microscopic imaging protocol for real-time monitoring the levels of cytoplasmic Ca2+ and cell surface PS in Caenorhabditis elegans touch neurons. Previously, Ca2+ dynamics was monitored in neurons in Caenorhabditis elegans larvae in time periods ranging from milliseconds to seconds. Methods for monitoring Ca2+ dynamics for a relatively long period of time during embryonic development were not available, let alone for simultaneous monitoring Ca2+ and PS dynamics. The protocol reported here utilizes a deconvolution imaging system with an optimized experimental setting that reduces photo-damage and allows the proper development of embryos during the real-time imaging process. This protocol enables the simultaneous measurement of cytosolic Ca2+ and cell surface PS levels in necrotic touch neurons during embryonic development in a period longer than six hours. Our method provides an easy and sensitive approach to perform long-time Ca2+ and PS recording in living animals, simultaneously or individually. This protocol can be applied to study various cellular and developmental events that involve the dynamic regulation of Ca2+ and/or PS.
Keywords: Calcium ion, Phosphatidylserine, “Eat-me” signal, Time-lapse imaging, Caenorhabditis elegans, Neurons, mec-4, DEG/ENaC channels, Necrosis, Necrotic cells
Background
Cytosolic Ca2+ is an important factor for regulating various cellular events such as gene expression, neurotransmitter release at synaptic terminal sites, muscle contraction, and hormone secretion (Clapham, 2007). Excessive amounts of Ca2+ in the cytoplasm provoke excitotoxic necrosis, often seen in neurodegenerative diseases (Yamashima, 2004; Wojda et al., 2008 ), heart failures ( Nakayama et al., 2007 ), and cancer ( Danese et al., 2017 ). Recently, we have found that a transient increase in Ca2+ level in the cytoplasm of neurons undergoing necrosis is necessary for the exposure of phosphatidylserine (PS), which acts as an “eat me” signal to attract engulfing cells on the surfaces of necrotic cells in the nematode Caenorhabditis elegans ( Furuta et al., 2021 ). In previous studies, in Caenorhabditis elegans larvae, Ca2+ dynamics was monitored in the cell bodies of mechanosensory neurons (touch neurons) with Cameleon, a genetically encoded calcium indicator (GECI) ( Suzuki et al., 2003 ). Different generations of GCaMP, another series of GECIs, were also reported to monitor Ca2+ in the neuronal processes of different sensory neurons in the Caenorhabditis elegans larvae ( Tian et al., 2009 ; Ghosh- Roy et al., 2010 ; Akerboom et al., 2012 ). These Ca2+ recording experiments were performed in Caenorhabditis elegans larvae, and recording time ranged from milliseconds to seconds. However, recording of Ca2+ signal in Caenorhabditis elegans for longer periods of time or during embryonic development has not been reported previously.
We study the necrosis and clearance of Caenorhabditis elegans touch neurons during embryogenesis. In Caenorhabditis elegans, dominant mutations in the DEG/ENaC sodium channel subunit MEC-4 induce the six touch neurons to undergo excitotoxic necrosis (Figure 1A) (Chalfie and Sulston, 1981; Driscoll and Chalfie, 1991). These necrotic neurons expose PS and are subsequently engulfed and digested by neighboring hypodermal cells ( Li et al., 2015 ). Previously, we developed secreted fluorescence PS reporters by tagging MFG-E8, a secreted PS-binding protein, with fluorescent proteins such as mCherry, and expressing each reporter under the control of the dyn-1 promoter (Pdyn-1) ( Li et al., 2015 ). The secreted MFG-E8::mCherry protein associates with PS exposed on cell surfaces and, in this manner, labels the surfaces of necrotic and apoptotic cells (Figure 1B). We established a time-lapse recording protocol that allowed us to monitor, for hours, the dynamics of PS externalization on the surface of necrotic cells in Caenorhabditis elegans embryo ( Li et al., 2015 ).
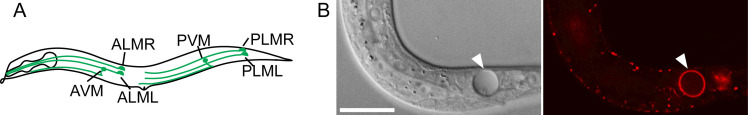
Figure 1. Caenorhabditis elegans six touch neurons and PS exposed on the surface of a necrotic cell.
(A) Diagram of a hermaphrodite indicating the positions and names of the six touch neurons. (B) A necrotic cell caused by mec-4(e1611) mutation in Differential Interference Contrast (DIC) image (left) and exposed PS visualized by MFG-E8::mCherry reporter (right). The scale bar is 15 µm.
To investigate the effect of cytoplasmic Ca2+ in triggering PS exposure on the surfaces of necrotic neurons, we constructed an in vivo Ca2+ reporter CGaMP5 that was expressed under the control of promoter Pmec-7, a promoter that is specifically active in touch neurons ( Furuta et al., 2021 ). We generated a strain that co-expressed this Ca2+ reporter and the PS reporter MFG-E8::mCherry, and established a time-lapse recording protocol that allowed us to simultaneously monitor the level of Ca2+ in the cytoplasm of two touch neurons (PLML and PLMR) that undergo necrosis and the level of PS on the outer surfaces of these neurons during embryogenesis. This protocol, together with genetic and pharmacological approaches, enabled us to discover that a transient increase in the level of cytoplasmic Ca2+ triggers the exposure of PS on the surfaces of touch neurons during necrosis and that the endoplasmic reticulin (ER) contributes to the PS exposure by releasing Ca2+ to the cytoplasm ( Furuta et al., 2021 ).
Monitoring cytoplasmic Ca2+ and cell surface PS in necrotic cells simultaneously is challenging because long-time imaging of embryos tends to cause photo-damage that impairs proper embryonic development. To overcome this problem, we operated our protocol using the DeltaVision Elite Deconvolution Imaging System from GE Healthcare, Inc., and optimized our experimental settings to minimize photo-damage. Our protocol uses the DeltaVision Elite Deconvolution Imaging System or an equivalent imaging system, the Caenorhabditis elegans strain carrying the Ca2+ and PS reporters, and requires the routine laboratory setup for propagating Caenorhabditis elegans. This protocol can also be adapted to monitor cytoplasmic Ca2+ or PS exposure individually. Our protocol is thus applicable to other Ca2+- and/or PS exposure-related cellular and developmental events. Furthermore, if the Pmec-7 promoter is replaced by other promoters, this protocol can be used to monitor cytosolic Ca2+ in cell types other than the touch neurons.
Materials and Reagents
-
Reporters
The Ca2+ reporter construct: Pmec-7GCaMP5G
-
The PS reporter: Pdyn-1mfg-e8::mCherry
Note: The reporter constructs are available upon request (Furuta et al., 2021).
-
The Caenorhabditis elegans strain that carries both reporters
Strain name: ZH3052
Genotype: enIs92[Pmec-7GCaMP5G , Pdyn-1mfg-e8::mCherry, punc-76(+)]V unc-76(e911)V; mec-4(e1611)X .
Note: This strain is available upon request (Furuta et al., 2021).
-
Consumables and Reagents
Microscope slides (Premiere, catalog number: 9101)
Coverslips (22 × 22 mm) (Fisher Scientific, catalog number: 12-542B)
DeltaVision immersion oil N = 1.516 (Cytiva, catalog number: 29162940)
Handmade worm pick, which is a platinum wire (Alfa Aesar, catalog number: 10287) mounted on a Pasteur pipette (Fisher Scientific, catalog number: 13-678-20B) (Figure 2)
High vacuum grease (Fisher Scientific, catalog number: 14-635)
Agarose (Fisher Scientific, catalog number: BP160-500)
KH2PO4 (MilliporeSigma, catalog number: PX1565)
Na2HPO4 (MilliporeSigma, catalog number: 567550)
NaCl (Fisher Scientific, catalog number: S671)
NH4Cl (Fisher Scientific, catalog number: S671-3)
4% agarose solution (see Recipes)
M9 buffer (see Recipes)
Figure 2. A handmade worm pick.
Equipment
Stereomicroscope (Nikon, catalog number: SMZ645) or any stereomicroscope from other manufacturers for handling Caenorhabditis elegans.
The DeltaVision Elite Deconvolution Imaging System (GE Healthcare, Inc.), including AP1 F1-Delta Vision microscope (Olympus) equipped with 20×, 63×, and 100× Uplan Apo objectives, excitation/emission filter sets, fluorescent light source, DIC microscopy accessories, motorized stage (X, Y, and Z-axis), and a Coolsnap HQ2 digital camera (Photometrics) (Figure 3). For fluorescent imaging, two sets of fluorescence filters (Chroma Inc.) are used, including the GFP filter (excitation wavelength 475/28 nm; emission wavelength 525/50 nm) and the mCherry filter (excitation wavelength 575/25 nm; emission wavelength 632/60 nm). The DeltaVision microscope is kept in a room where the temperature is maintained at 20°C.
A computer for image processing and analysis
Figure 3. DeltaVision deconvolution microscope.

Software
SoftWoRx 5.5 Software (for the deconvolution and processing of images) (GE Healthcare, Inc.)
Procedure
-
Mounting Animals on an Agar Pad
Melt the 4% agarose solution in a microwave oven.
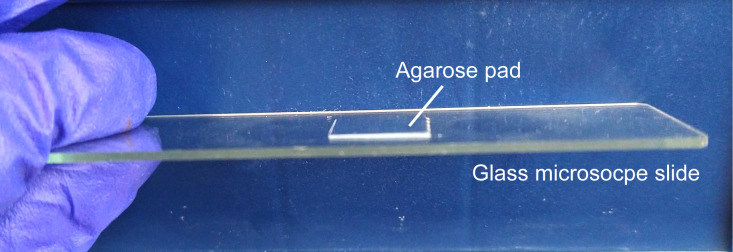
Dispense 300 µl of the agarose solution on a glass microscope slide, flatten the drop immediately by placing another glass slide on the top. After agarose is solidified, gently separate the two glass slides by sliding one against the other. The thickness of agarose is approximately 0.2 mm (Figure 4).
Place 3 µl of M9 buffer at the center of the agar pad. The M9 buffer is a buffer for collecting/washing worms.
Under the stereotype microscope, transfer embryos with a worm pick from a plate to the drop of M9 buffer. Try not to transfer more embryos than necessary (see Note 1).
Gently squeeze a thin line of high vacuum grease around the agarose pad. Gently place a coverslip over the drop of liquid. Avoid air bubbles. Vacuum grease prevents the drying of the agarose pad. A completed slide is shown in Figure 5.
-
Time-lapse recording of PLM touch neurons in Caenorhabditis elegans embryos
Set up microscope parameters. For differential interference contrast (DIC) images, the exposure time to white light is usually 0.2 s with a 10% neutral density filter. The neutral density filter partially blocks the light, and the percentage of the filter represents how much light travels through. For recording the signal of the Ca2+ reporter GCaMP5G, the exposure time for the GFP channel is 0.1 s with a 5% neutral density filter. For recording the signal of the PS reporter MFG-E8::mCherry, the exposure time for the mCherry channel is also 0.1 s with a 5% neutral density filter.
Align the DIC light path carefully for optimal DIC effect according to the manufacturer's instruction.
Under the 63× or 100× objectives, find embryos and mark the positions by using the “point marking” and “position visiting” functions of the software. It is recommended not to mark too many embryos at the same time (see Note 2). Carefully choose the embryos that are younger than the 2-fold stage (460 min post the 1st embryonic cell division).
Set up the positions for the serial z-section recording. The serial z-section recording is performed from the ventral surfaces and proceeding inwards. The setting of 10-15 z-sections at 0.5 μm/section is sufficient to cover the necessary region.
Set up the time-lapse recording parameters. We found no striking Ca2+ or PS signal prior to the 560 min post-1st embryonic cell division. The starting point of time-lapse recording is set at 560 min-post the 1st embryonic cell division, which is 100 min after an embryo reaches the 2-fold stage (Figure 6). Recording of the Caenorhabditis elegans tail, where the touch neurons PLML and PLMR reside, in the strain ZH3052, is set in the 2.5-min interval for the first 2 h of recording and in the 5-min interval for the next 4 h.
Start recording DIC, GFP, and mCherry images once the embryos reach 560 min post-1st embryonic cell division, which is 100 min post the 2-fold stage.
Keep observing the images during the recording process. Adjust the starting focal plane during the interval of recording if any change of focal place takes place. Abort recording if an embryo slows down in motion or stops its development due to photo-damage. Examples of time-lapse images of cytoplasmic Ca2+ and cell surface PS in the necrotic PLML touch neurons are shown in Figure 7.
Figure 4. Trim the flattened agar pad into an approximately 12 × 12 mm square.
Figure 5. A completed slide.
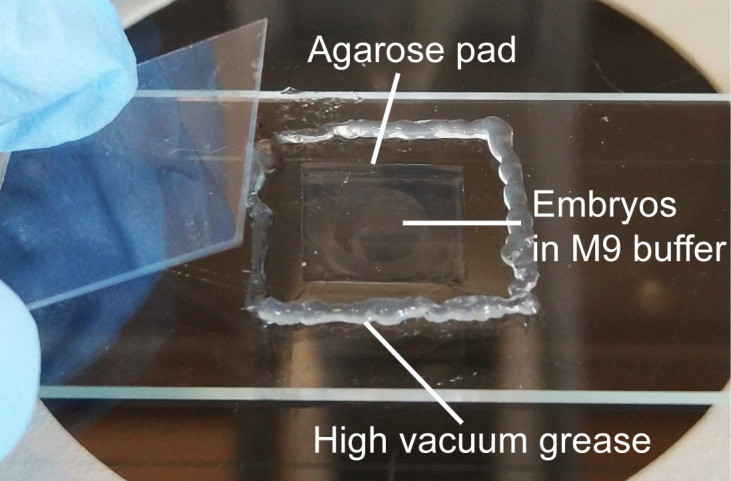
Figure 6. DIC image of a 2-fold stage Caenorhabditis elegans embryo.
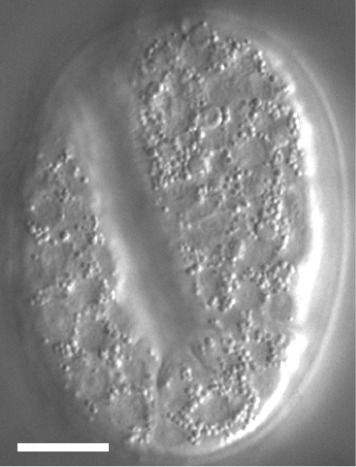
The scale bar is 10 µm.
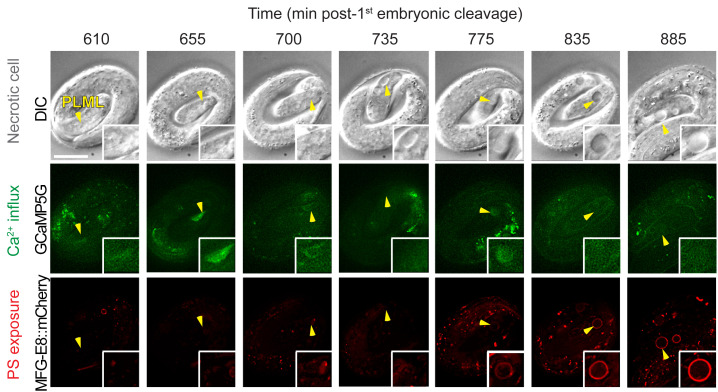
Figure 7. Example of images of cytoplasmic Ca2+ and cell surface PS in necrotic PLML touch neuron caused by the mec-4(e1611) mutation.
The top row shows DIC images depicting morphological changes of the PLML touch neuron undergoing necrosis (marked by arrowhead). The middle row shows cytoplasmic Ca2+ signal in the PLML neuron. The bottom row shows cell surface PS in the PLML neuron. Recording was started at 100 min after the embryo reached the 2-fold stage. The scale bar is 10 µm. Images are adapted from Furuta et al. (2021) .
Data analysis
Images are deconvolved using the SoftWoRx software. For the quantification of Ca2+ signal intensity, the cell bodies of the PLML and PLMR neurons are outlined by polygons, and the total signal intensity (Cax) is obtained. The total signal of Ca2+ in the neighboring region of the same size is also measured by moving the same polygon to the neighboring region in the tail (CaBackground). The relative Ca2+ intensity is calculated as CaR = Cax/CaBackground. To maintain the quality of quantification of Ca2+ dynamics, images out of focus or obscure images that are results of the fast-moving embryos should be avoided (see Note 3). For measuring Ca2+ dynamics, we excluded the embryos with fewer than 10 time points with images of good quality. For quantification of PS intensity, the cell surface area containing the MFG-E8::mCherry signal is outlined by two closed polygons. The total signal intensity, as well as the area size of each polygon, is recorded. The unit PS signal intensity (UPS) of the “donut shape” area between the two polygons is calculated as follows: UPS = (PSexternalpolygon−PSinternalpolygon)/(Areaexternalpolygon−Areainternalpolygon). The background mCherry signal (PSbackground) of a “circular” area nearby and the area size (Areabackground) are obtained, and the unit intensity is calculated as UPSbackground = PSbackground/Areabackground. The relative PS signal intensity RPS = UPS/UPSbackground. The value 1 indicates that there is no enrichment of signal comparing to the non-specific background signal.
Notes
For time-lapse imaging, it is strongly recommended to place up to 10 embryos on one slide to reduce oxygen consumption by embryos during the recording period. Additionally, transfer the embryos at the stage close to a developmental stage of choice [e.g., if you want to start recording from the 2-fold stage, transfer 1.5-fold stage embryos (Figure 8)].
To capture images in 2.5 or 5 min intervals, and to reduce the fluorescent light damage, it is recommended to record no more than 6 embryos in one experiment.
Embryos are constantly moving inside the eggshell during recording, and thus PLM neurons go out of focus from time to time. Furthermore, due to the fast-moving of embryos, some images are blurred. The number of good images captured largely depends on the embryo movement.
Figure 8. A DIC image of a 1.5-fold stage Caenorhabditis elegans embryo.
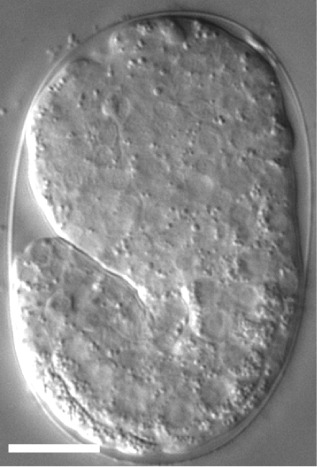
The scale bar is 10 µm.
Recipes
-
4% agarose solution
Place 2.0 g agarose in 50 ml deionized water and microwave until agarose is completely melted. After usage, the solidified solution can be stored at room temperature and melted in a microwave oven again.
-
M9 buffer (1 L)
Dissolve 3.0 g KH2PO4 (22.0 mM), 5.8 g Na2HPO4 (40.9 mM), 0.5 g NaCl (8.6 mM), and 1.0 g NH4Cl (18.7mM) in deionized water. Adjust volume to 1 L with deionized water. Autoclave.
Acknowledgments
This work is supported by NIH R01GM104279. This protocol was adapted from Furuta et al. (2021).
Competing interests
The authors have declared that no competing interests exist.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Akerboom J., Chen T. W., Wardill T. J., Tian L., Marvin J. S., Mutlu S., Calderon N. C., Esposti F., Borghuis B. G., Sun X. R., et al.(2012). Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci 32(40): 13819-13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chalfie M. and Sulston J.(1981). Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans . Dev Biol 82(2): 358-70. [DOI] [PubMed] [Google Scholar]
- 3. Clapham D. E.(2007). Calcium signaling. Cell 131(6): 1047-1058. [DOI] [PubMed] [Google Scholar]
- 4. Danese A., Patergnani S., Bonora M., Wieckowski M. R., Previati M., Giorgi C. and Pinton P.(2017). Calcium regulates cell death in cancer: Roles of the mitochondria and mitochondria-associated membranes(MAMs). Biochim Biophys Acta Bioenerg 1858(8): 615-627. [DOI] [PubMed] [Google Scholar]
- 5. Driscoll M. and Chalfie M.(1991). The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration . Nature 349(6310): 588-93. [DOI] [PubMed] [Google Scholar]
- 6. Furuta Y., Pena-Ramos O., Li Z., Chiao L. and Zhou Z.(2021). Calcium ions trigger the exposure of phosphatidylserine on the surface of necrotic cells. PLoS Genet. 17(2): e1009066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghosh-Roy A., Wu Z., Goncharov A., Jin Y. and Chisholm A. D.(2010). Calcium and cyclic AMP promote axonal regeneration in Caenorhabditis elegans and require DLK-1 kinase . J Neurosci 30(9): 3175-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Z., Venegas V., Nagaoka Y., Morino E., Raghavan P,. Audhya A., Nakanishi Y. and Zhou Z.(2015). Necrotic Cells Actively Attract Phagocytes through the Collaborative Action of Two Distinct PS-Exposure Mechanisms. PLoS Genet 11(6): e1005285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakayama H., Chen X., Baines C. P., Klevitsky R., Zhang X., Zhang H., Jaleel N., Chua B. H., Hewett T. E., Robbins J., et al.(2007). Ca2+- and mitochondrialdependent cardiomyocyte necrosis as a primary mediator of heart failure . J Clin Invest 117: 2431-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suzuki H., Kerr R., Bianchi L., Frøkjaer-Jensen C., Slone D., Xue J., Gerstbrein B., Driscoll M. and Schafer W. R.(2003). In vivo imaging of C. elegans mechanosensory neurons demonstrates a specific role for the MEC-4 channel in the process of gentle touch sensation . Neuron 39(6): 1005-17. [DOI] [PubMed] [Google Scholar]
- 11. Tian L., Hires S. A., Mao T., Huber D., Chiappe M. E., Chalasani S. H., Petreanu L., Akerboom J., McKinney S. A., Schreiter E. R., et al.(2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods 6(12): 875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wojda U., Salinska E. and Kuznicki J.(2008). Calcium ions in neuronal degeneration. IUBMB Life 60(9): 575-590. [DOI] [PubMed] [Google Scholar]
- 13. Yamashima T.(2004). Ca2+-dependent proteases in ischemic neuronal death: a conserved’calpain-cathepsin cascade’ from nematodes to primates . Cell Calcium 36(3-4): 285-293. [DOI] [PubMed] [Google Scholar]