Abstract
The biology of human granzymes remains enigmatic in part due to our inability to probe their functions outside of in vitro assays or animal models with divergent granzyme species. We hypothesize that the biology of human granzymes could be better elaborated with a translational imaging technology to reveal the contexts in which granzymes are secreted and biochemically active in vivo. Here, we advance toward this goal by engineering a Granzyme targeting Restricted Interaction Peptide specific to family member B (GRIP B) to measure secreted granzyme B (GZMB) biochemistry with positron emission tomography. A proteolytic cleavage of 64Cu-labeled GRIP B liberates a radiolabeled form of Temporin L, which sequesters the radioisotope by binding to adjacent phospholipid bilayers. Thus, at extended time points postinjection (i.e., hours, not seconds), tissue biodistribution of the radioisotope in vivo reflects relative units of the GZMB activity. As a proof of concept, we show in three syngeneic mouse cancer models that 64Cu-GRIP B detects GZMB from T cells activated with immune checkpoint inhibitors (CPI). Remarkably, the radiotracer detects the proteolysis within tumors but also in lymphoid tissue, where immune cells are activated by a systemic CPI. Control experiments with an uncleavable analogue of 64Cu-GRIP B and tumor imaging studies in germline GZMB knockout mice were applied to show that 64Cu-GRIP B is specific for GZMB proteolysis. Furthermore, we explored a potential noncytotoxic function for GZMB by applying 64Cu-GRIP B to a model of pulmonary inflammation. In summary, we demonstrate that granzyme biochemistry can be assessed in vivo using an imaging modality that can be scaled vertically into human subjects.
Short abstract
A protease-activated chemosensor for in vivo molecular imaging enables the detection of activated immune cells by specifically harnessing granzyme B catalysis.
Introduction
The human granzymes consist of five serine proteases (A, B, H, K, M) that are expressed primarily within the secretory vesicles (i.e., granules) of lymphocytes involved in the host defense, namely, natural killer (NK) and cytotoxic T cells (CTLs).1 In these cell types, granzymes are best understood to be pro-apoptotic effectors against problematic cells, for example, cancer cells or cells infected with pathogens. To impart cytotoxicity, lymphocytes degranulate after docking with a target cell to release granzymes transiently into a pericellular space.2,3 Co-secreted with granzymes are perforin molecules, which form a channel in the plasma membrane of the target cell to facilitate the granzyme transit into the cytoplasm.4,5 Granzyme biochemistry subsequently triggers cell death through several mechanisms, for example, proteolytic activation of caspases or direct DNA damage (granzyme B), and SET-mediated activation of DNA cleavage (granzyme A).6−9
Despite decades of research, granzymes in many ways remain an enigmatic enzyme class. For example, virtually no biological functions are defined for the so-called “orphan” granzymes H, K, and M.10 Moreover, the secretion of granzymes (including A and B) in abundance during conditions unrelated to host defense has suggested noncytotoxic signaling functions.11 These data align with a developing narrative suggesting that dysregulated granzyme secretion and proteolysis may contribute to the pathobiology of chronic human disorders, for example, systemic autoimmune diseases.12 Thus, the canon that granzymes are primarily cytotoxic effectors is being challenged by a more complex biological model in which secreted granzymes can also persist in extracellular space to perform noncytotoxic signaling functions.
More fully elaborating the complex biology of granzymes is challenging in part due to the inadequacy of preclinical models to study their functions.13 In vitro coculture approaches are reductionist models of the more elaborate multicellular conversation occurring in human immunology. In vivo mouse models, for example, germline knockout mice, have revealed roles for granzymes A and B in host defense, but they carry with them the caveat that mice express a larger repertoire of granzymes than humans, which may confound efforts to reveal other granzyme functions due to compensatory effects. Moreover, mouse granzymes are evolutionarily divergent from their human counterparts and bear different protein substrate preferences.14 Thus, any findings in mice by nature will require some corroboration in human cell lines and patients. Collectively, these considerations underscore that an expansion of our knowledge of granzyme biology will ultimately require in vivo technologies that can also be scaled to humans.
In humans, studies of granzymes are generally limited to expression analysis at the protein and mRNA level in biopsied tissue or from biofluids. Missing from the field are technologies that enable measurements of granzyme biochemistry in vivo with a spatiotemporal control. We hypothesized that an imaging tool could address this unmet need were it capable of selectively capturing when and where immune cells present biochemically active granzymes in the extracellular niche.
Although inducible fluorescent imaging probes have a rich history in biomedical research as tools to study protease biology, the limited sensitivity and resolution of fluorescent tomography limits its application in clinically relevant animal models and humans.15,16 Nuclear imaging with positron emission tomography (PET) offers an attractive alternative, as γ-photons are not scattered by tissue and thus produce high-resolution tomographic images of deep tissues. However, developing a protease-activated radiotracer for PET comes with the special design challenge that radioactive decay is continuous and cannot be masked and then conditionally revealed by a proteolytic event. We proposed instead to design an imaging probe in which enzyme catalysis causes probe sequestration in nearby tissues, akin to the biochemical trapping mechanism utilized for metabolic radiotracers like 18F-fluorodeoxyglucose.
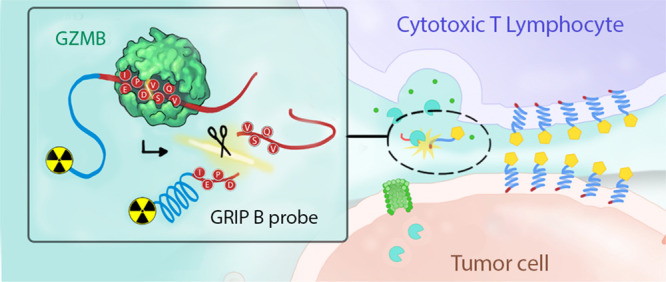
We hypothesized that a “restricted interaction peptide” (RIP) specifically cleaved by an extracellular granzyme could be leveraged to enable in vivo measurements of biochemistry on PET.17 In general, RIPs consist of three domains from N to C terminus, namely, (1) a nontoxic antimicrobial peptide (AMP) coupled to a radioisotope, (2) a specific endoprotease cleavage site, and (3) a peptide “masking” domain that prevents the AMP from adopting its preferred helical conformation that tightly binds phospholipid bilayers (Figure 1A). Upon proteolytic cleavage of the full length, pro-peptide form of the RIP, the radiolabeled AMP is liberated, undergoes a spontaneous conformational shift, and deposits within any nearby membrane.
Figure 1.
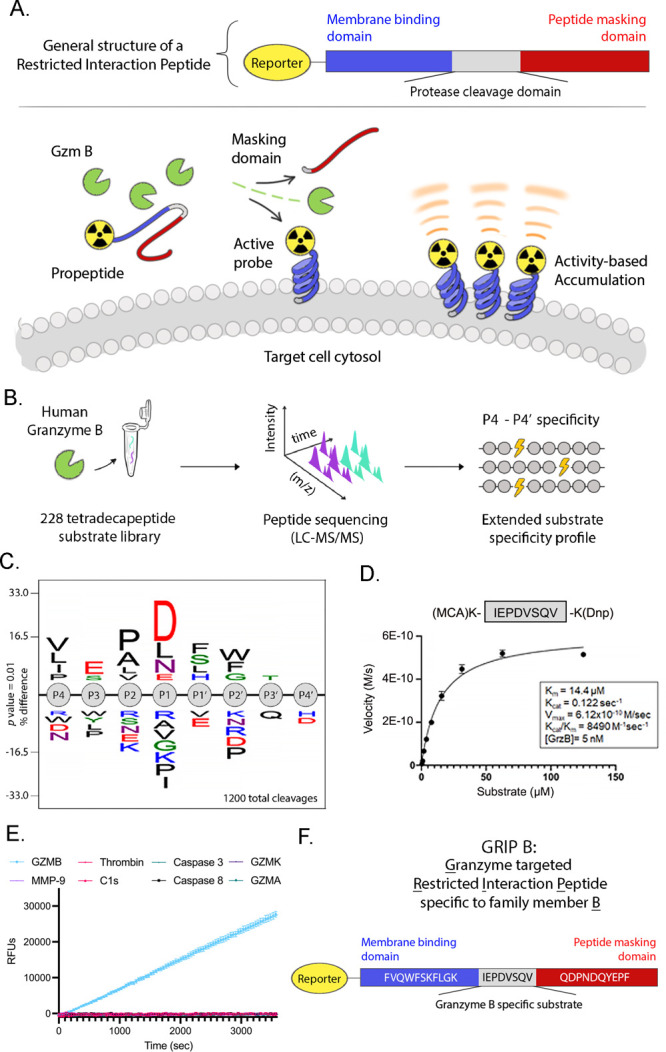
The development and in vitro characterization of GRIP B, a restricted interaction peptide to measure GZMB proteolysis in vivo with imaging. (A) A schematic showing a generalized structure of a restricted interaction peptide and the in vivo mechanism of action. Cleavage of the full-length pro-form by a dedicated endoprotease liberates a tagged (e.g., radiolabeled) antimicrobial peptide, which irreversibly interacts with nearby phospholipid membranes. Thus, the stable accumulation of a peptide at extended time points postinjection (i.e., hours, not seconds) can reflect the relative units of enzyme activity in a region of interest. (B) A schema showing the workflow of the MSP-MS study to identify a GZMB cleavage sequence. Proteolytic products from GZMB activity were produced by incubating the enzyme with a physicochemically diverse library of 228 tetra-decapeptides. Peptide sequencing by LC-MS/MS allowed for the determination of GZMB-generated cleavages. (C) An iceLogo showing the consolidated results of an MSP-MS analysis of the P4–P4′ substrate preferences for human GZMB. (D) A plot showing the Michaelis–Menten kinetics of the human granzyme B proteolysis of the IEPDVSVQ peptide. Coverage of the nonprime and prime sites of GZMB yielded an optimized substrate with an improved catalytic turnover by approximately twofold compared to IEPD alone. (E) A plot showing the IEPDVSVQ peptide is specifically cleaved by GZMB and not by various other relevant proteases. (F) The final amino acid sequence of GRIP B. Thus, the activity-driven accumulation of radioactivity over extended time points postinjection (i.e., hours not seconds) in a region of interest (ROI) reflects the units of enzyme activity for a given protease. The modular design of the RIP technology has also previously allowed for various imaging modalities to be used such as fluorophores, near-infrared dyes, and radiotracers that provide a translational probe that has applicability that ranges from in vivo laboratory applications to quantitative, preclinically relevant animal models via PET. Having established a proof of concept by developing a RIP targeting the protease thrombin, we report herein the design, synthesis, and in vivo assessment of 64Cu-GRIP B, or Granzyme-targeted Restricted Interaction Peptide specific to family member B.
Results
The Design and Synthesis of GRIP B
To identify an optimal cleavage sequence to install in a RIP targeting GZMB, we performed multiplex substrate profiling using mass spectrometry (MSP-MS) against recombinant human GZMB.18,19 The MSP-MS library contains 228 tetradecameric peptides, which is a physicochemically diverse population of rationally designed substrates with maximum sequence diversity (Figure 1B). On the basis of the observation that most proteases require two optimally positioned amino acids for substrate recognition and cleavage, physicochemical diversity was generated in the peptide library through an incorporation of all neighbor (XY) and near–nearbor (X*Y, X**Y) amino acid pairings. With an incubation of native GZMB at various time points, cleavages were identified by peptide sequencing via liquid chromatography tandem mass spectrometry (LCMS-MS).20 A statistical analysis that considers both cleaved and uncleaved positions in the peptide library was subsequently performed to construct an iceLogo representation of the preferred substrate sequence spanning the granzyme B P4–P4′ sites (Figure 1C and Supporting Information Figure 1).
The iceLogo results suggested that four sequences with conserved sites of P2 = P and P1 = D (i.e., XXPDXXXX) were equally specific and efficient GZMB substrates. Of these, we nominated the sequence IEPDVSQV for two reasons. First, the P4–P1 sequence was previously discovered by our group to be specific to GZMB using an orthogonal approach, namely, a positional scanning synthetic combinatorial library, and we showed this sequence is specifically recognized by GZMB versus other human granzymes.21,22 Second, the IEPD tetrapeptide has been studied in vivo as part of a covalent reversible aldehyde radiotracer targeting GZMB, and the tetrapeptide aldehyde appeared to be effective at labeling GZMB and stable in vivo.23,24
The kinetics of IEPDVSQV cleavage by GZMB was assayed in vitro using a fluorescent quenched peptide substrate, and the incorporation of the P1′-P4′ sequence VSVQ significantly improved the catalytic efficiency (kcat/Km) compared to previously reported values for IEPD alone (∼8000 vs ∼3300 M–1 s–1, see Figure 1D).22 Furthermore, the GZMB-optimized substrate was incubated with various other relevant proteases (Figure 1E). No cleavage was observed with any of these proteases after 1 h compared to significant cleavage by GZMB, highlighting the specificity of the optimized GZMB cleavage sequence. To generate the full-length GRIP B probe, we chose to flank this sequence using Temporin L as the membrane-interacting domain and the PAR1 peptide as the masking domain (Figure 1F).17 Importantly, the full-length GRIP B was efficiently cleaved by a recombinant human GZMB showing that neither Temporin L nor the masking domain interfered with the proteolysis (Supporting Information Figure 2).
In Vitro Mechanism Studies and the Radiosynthesis of 64Cu-GRIP B
We confirmed that the proteolytically cleaved version of GRIP B effectively bound membranes. A N-terminal, 5FAM-tagged version of GRIP B was synthesized and incubated with viable MC38 cells and recombinant human granzyme B or vehicle. Flow cytometry showed that the intact GRIP B had a low interaction with cell membranes, while a preincubation of GRIP B with 20 nM recombinant GZMB, followed by a coincubation with MC38 cells in Hanks’ Balanced Salt Solution (HBSS) at 37 °C for 30 min, resulted in fluorescently labeled cell membranes (Figure 2A). The insertion of the cleaved GRIP B peptide into lipid micelles was further confirmed by measuring the intrinsic tryptophan fluorescence (Supporting Information Figure 3A). Circular dichroism data of full-length GRIP B are indicative of a disordered peptide, showing the efficacy of the masking domain to disrupt the α-helical secondary structure (Supporting Information Figure 3B). Circular dichroism of activated GRIP B confirms that the activated peptide retains the α-helical secondary structure of the cell-penetrating peptide Temporin L upon cleavage of the masking domain (Supporting Information Figure 3C). Lastly, we verified that full-length or proteolytically cleaved GRIP B did not display toxicity toward human red blood cells in vitro (Figure 2B).
Figure 2.
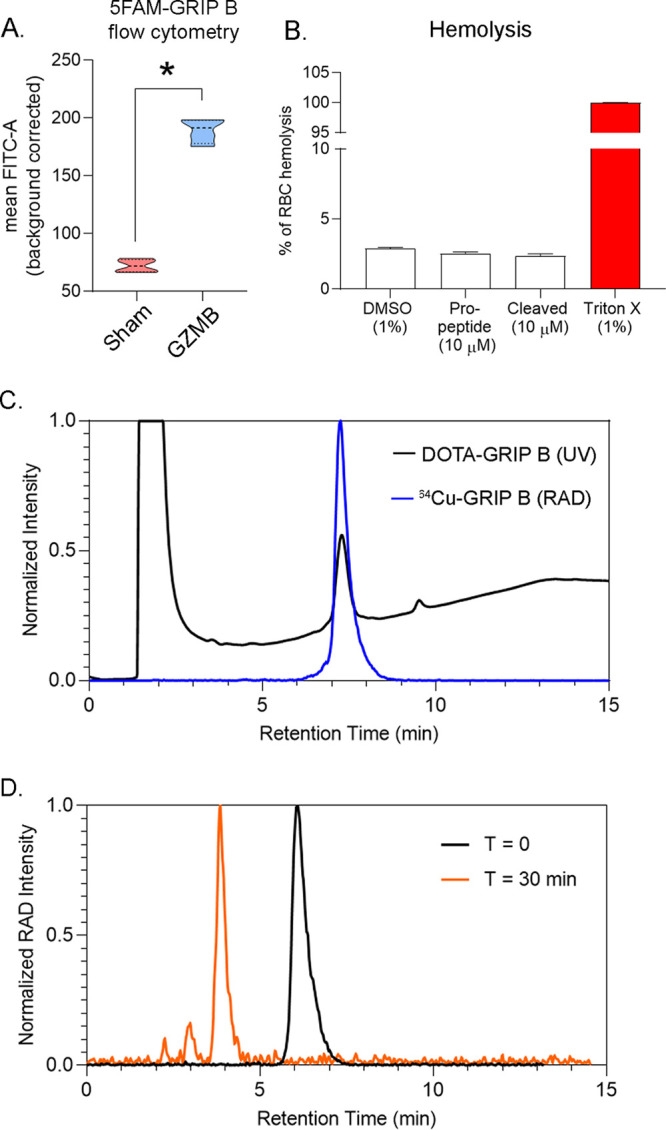
In vitro mechanism of action studies and the synthesis of 64Cu-GRIP B. (A) Mean fluorescence intensity data showing the extent of cell labeling by 5FAM-GRIP B in the presence or absence of GZMB after incubating at 37 °C for 30 min. The data were collected using MC38 cells in triplicate. *P < 0.01. (B) A bar graph representing the extent of red blood cell lysis due to treatment with vehicle (0.1% dimethyl sulfoxide (DMSO)), the full-length GRIP B pro-peptide, and the proteolytically activated truncated peptide. Triton-X is included as a positive control. (C) An HPLC trace showing the overlay of the radioactive trace (blue) with the UV trace of the DOTA-GRIP B precursor. The trace was collected 30 min after the start of the reaction. (D) A radioactive HPLC trace showing the conversion of 64Cu-GRIP B to one major product after a 30 min incubation with 10 nM recombinant human GZMB.
To couple GRIP B to a chelator for radiolabeling, the peptide was reacted with 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetratacetic acid mono-N-hydroxysuccinimide ester (DOTA-NHS-ester) on a solid support, which ligated to the amino group on the N-terminal phenylalanine. DOTA-GRIP B was subsequently deprotected, cleaved from the resin, and purified with semiprep high-performance liquid chromatography (HPLC) (Supporting Information Figure 4).
DOTA-GRIP B was next radiolabeled with copper-64, since its half-life (t1/2 ≈ 13 h) would enable studies over a long window of time postinjection to identify the optimal time point for imaging. 64CuCl2 was incubated with DOTA-GRIP B in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer for 30 min at room temperature. The reaction was monitored for completeness via instant thin-layer chromatography and purified using HPLC (Figure 2C). The decay-corrected yield was consistently greater than 95%, with a purity of greater than 99%. The specific activity was ∼0.4 Ci/μmol over three radiosyntheses. An incubation of 64Cu-GRIP B with the recombinant human GZMB showed a conversion within 30 min to one radiolabeled product that comigrated on the HPLC with the cold-cleaved DOTA-peptide fragment (Figure 2D). Lastly, the serum stability was tested in vitro in mouse serum. We observed on HPLC that 64Cu-GRIP B was greater than 98% stable over 4 h at 37 °C (Supporting Information Figure 5).
Immunomodulatory Therapies Induce System-Wide Changes in 64Cu-GRIP B Biodistribution in Mouse Tumor Models
To understand tracer pharmacokinetics and normal tissue biodistribution, we first injected 64Cu-GRIP B intravenously in balb/c mice and conducted a 60 min dynamic PET acquisition (Figure 3A). A region-of-interest analysis showed the probe cleared from a blood pool with t1/2 ≈ 8 min. The dominant mode of clearance was renal, and as with the thrombin RIP probe, the only substantial radiotracer accumulation outside of the kidneys was observed in the liver. We next conducted a biodistribution to evaluate the radiotracer distribution in normal tissues out to 24 h postinjection. The biodistribution data corroborated the imaging findings showing the highest level of tissue-associated activity in the kidney and liver (Supporting Information Table 1).
Figure 3.
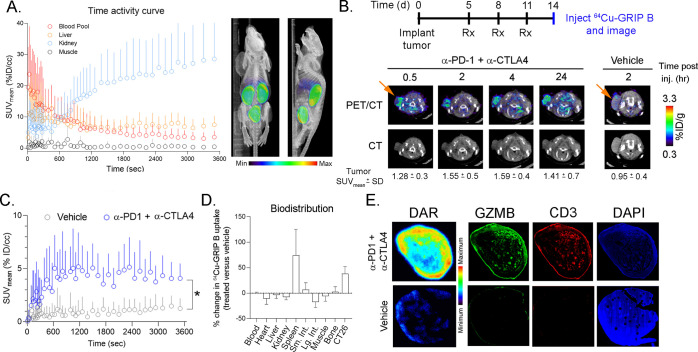
64Cu-GRIP B detects T cell activation in vivo elicited by an immune checkpoint inhibition. (A) A time activity curve showing the renal clearance of 64Cu-GRIP B in a male balb/c mouse bearing a subcutaneous CT26 tumor. (B) Representative transaxial CT and PET/CT images showing the accumulation over time of 64Cu-GRIP B in a CT26 tumor exposed to anti-PD1 and anti-CTLA4 CPI. Also shown is the uptake of 64Cu-GRIP B in a tumor-bearing mouse treated with a vehicle. (C) A time activity curve from a dynamic PET acquisition showing the tumoral uptake of 64Cu-GRIP B in CT26 tumors from mice treated with a vehicle or CPI. *P < 0.05. (D) A plot showing the percentage change in 64Cu-GRIP B uptake per organ in treated vs untreated mice. (E) Digital autoradiography and immunofluorescence showing the colocalization of 64Cu-GRIP B with GZMB and T cells within CT26 tumor slices from mice exposed to a vehicle or CPI.
We evaluated the effect of immunomodulatory therapies on the 64Cu-GRIP B biodistribution in mice bearing subcutaneous CT26 tumors, a mouse colorectal cancer cell line.25 Mice were treated with three intraperitoneal infusions of vehicle or anti-PD1 plus anti-CTLA4 checkpoint inhibitors (CPIs) over 11 d. The radiotracer was injected on day 14, and the tumor uptake was monitored over several time points out to 24 h postinjection on PET. An ROI analysis of static PET/computed tomography (CT) images showed that the 64Cu-GRIP B uptake in the treated tumors steadily rose from 0.5 to 2–4 h postinjection (Figure 3B). Notably, the radioactivity persisted in tumors out to 24 h postinjection, which is consistent with a mechanism of irreversible radiotracer trapping at the tumor. Moreover, the tumoral uptake of 64Cu-GRIP B was significantly higher in the CPI versus vehicle-treated arm at 2 h postinjection. Time activity curves derived from a dynamic PET acquisition showed that the tumoral accumulation of 64Cu-GRIP B in CPI-treated mice was rapid, reaching a level of ∼5% ID/cc within 10 min postinjection (Figure 3C and Supporting Information Table 2). Furthermore, a compartmental modeling showed that k3 ≫ k4 and k4 ≈ 0, which are properties known to be associated with radiotracers like 18F-fluorodeoxyglucose that are irreversibly sequestered in tissues (Supporting Information Figure 6).26 By comparison, the radiotracer uptake in vehicle tumors was significantly lower and did not change over time.
We conducted a biodistribution study at 2 h postinjection to determine relative changes in tracer uptake between tissues in the vehicle and treated groups. These data showed ∼50% induction of radiotracer uptake in the tumors from treated mice compared to control mice (Figure 3D, Supporting Information Figure 7, and Supporting Information Table 2). We also observed a significant increase in the tracer uptake within the spleen, which is consistent with a stimulation of T cells by systemic immune CPIs that we and others have documented.27−29 Digital autoradiography (DAR) showed that 64Cu-GRIP B was significantly higher in the treated versus control tumor. Although the comparatively lower resolution of pseudocolor from DAR limits the ability to rigorously compare the GRIP B localization to the GZMB protein expression on immunofluorescence (IF), qualitatively the regions of the radiotracer binding in tissue appeared to coalign with the expression of GZMB and the T cell marker CD3 (Figure 3E).
Control Studies Attribute the Post-Treatment Changes in 64Cu-GRIP B Biodistribution with GZMB Proteolytic Activity
We tested if the post-treatment changes in the tumoral uptake of 64Cu-GRIP B are driven by the proteolysis of 64Cu-GRIP B. To achieve this, we prepared 64Cu-D-GRIP B, a probe that harbors a d-aspartic acid within the GZMB protease site (IEPdVSQV) to prevent cleavage by GZMB. The probe was functionalized with DOTA and radiolabeled with copper-64 using an approach similar to that for the synthesis of 64Cu-GRIP B (Supporting Information Figure 8). Biodistribution studies showed that the CPI treatment did not cause an increase in the tumoral uptake of 64Cu-D-GRIP B compared to the control (Figure 4A).
Figure 4.
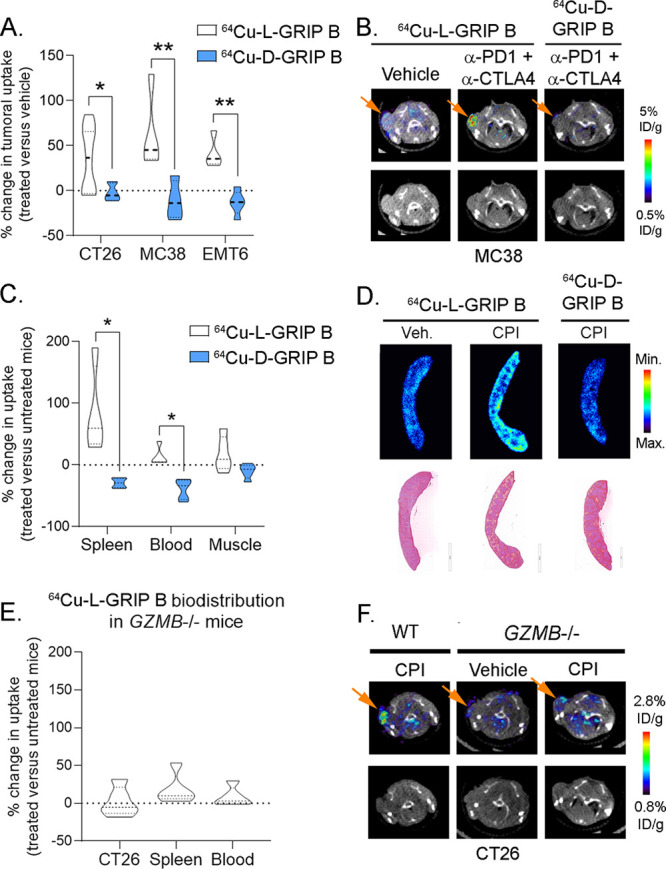
64Cu-GRIP B biodistribution in vivo is dependent on the GZMB proteolytic activity. (A) A bar graph summarizing the post-treatment effects on the tumoral uptake of 64Cu-GRIP B (or 64Cu-L-GRIP B) and 64Cu-D-GRIP B, an uncleavable negative control tracer bearing d-amino acids in the GZMB cut site. Three cohorts of mice bearing subcutaneous CT26, MC38, or EMT6 mice were studied. CT26 and MC38 were implanted in male balb/c and C57Bl6 mice, and EMT6 was implanted in female Balb/c mice. *P < 0.05, **P < 0.01. (B) Representative transaxial PET/CT and CT images from the MC38 cohort showing the tumoral uptake of 64Cu-L-GRIP B and 64Cu-D-GRIP B in mice treated with a vehicle or CPI. (C) A bar graph showing the post-treatment effects on the splenic uptake of 64Cu-GRIP B and 64Cu-D-GRIP B in mice treated with a vehicle vs CPI. These data were taken from the CT26 cohort, and similar trends were observed in the other mouse cohorts. *P < 0.01. (D) Autoradiography and H&E showing the relative intensity of 64Cu-L-GRIP B and 64Cu-D-GRIP B uptake in spleen sections. (E) A bar graph summarizing the post-treatment effects on the tumoral and splenic uptake of 64Cu-GRIP B in germline GZMB–/– treated with a vehicle or CPI. The GZMB–/– mice were inoculated with CT26 tumors for this study. (F) Representative transaxial PET/CT and CT images from the CT26 cohort showing the tumoral uptake of 64Cu-L-GRIP B in C57Bl6 mice treated with CPI or germline GZMB–/– mice treated with vehicle or CPI.
We compared the biodistribution of 64Cu-GRIP B or 64Cu-D-GRIP B in mice bearing MC38 (mouse colorectal cancer) or EMT6 (mouse mammary breast carcinoma) xenografts. The mice were treated with vehicle or anti-PD1 and anti-CTLA4 CPIs following the schema used for the CT26 cohort. Biodistribution data showed a significant increase in the tumoral uptake of 64Cu-L-GRIP B in both cohorts, while 64Cu-D-GRIP B was not induced in tumors compared to controls, as expected (Figure 4A and Supporting Information Tables 3–7). Moreover, the absolute levels of the 64Cu-D-GRIP B uptake in treated tumors were low and comparable with the baseline uptake of 64Cu-L-GRIP B in untreated tumors (Figure 4B). The basal 64Cu-D-GRIP B uptake in the spleen was also low and unaffected by a treatment with immune checkpoint inhibitors (Figure 4C,D).
To confirm that GZMB is responsible for post-treatment changes in the 64Cu-GRIP B biodistribution, we inoculated germline homozygous GZMB knockout mice30 with CT26 tumors and assessed the relative biodistribution of 64Cu-L-GRIP B in mice after a treatment with a vehicle or CPIs. We observed no significant post-treatment changes in the radiotracer uptake among tumors and spleen exposed with CPI compared to vehicle (Figure 4E,F, Supporting Information Table 8).
Post-Treatment Changes in the Tumoral Uptake of 64Cu-GRIP B Correlate with Tumor Volume Changes
Tumors enriched with comparatively higher levels of GZMB activity could be expected to more significantly debulk compared to GZMB-poor tumors. Thus, we next asked if post-treatment changes in the tumoral uptake of 64Cu-GRIP B correlated with antitumor effects. The tumoral uptake of 64Cu-GRIP B at day 11 was significantly anticorrelated with the percent change in tumor volume at day 11 compared to day 0 (Figure 5A). 64Cu-GRIP B tumor-to-blood ratios at day 11 also significantly correlated with the percent change in tumor volume. In contrast, neither the tumoral uptake nor the tumor-to-blood ratio of 64Cu-GRIP B correlated with percent changes in the tumor volume in the GZMB knockout mouse background (Figure 5B).
Figure 5.
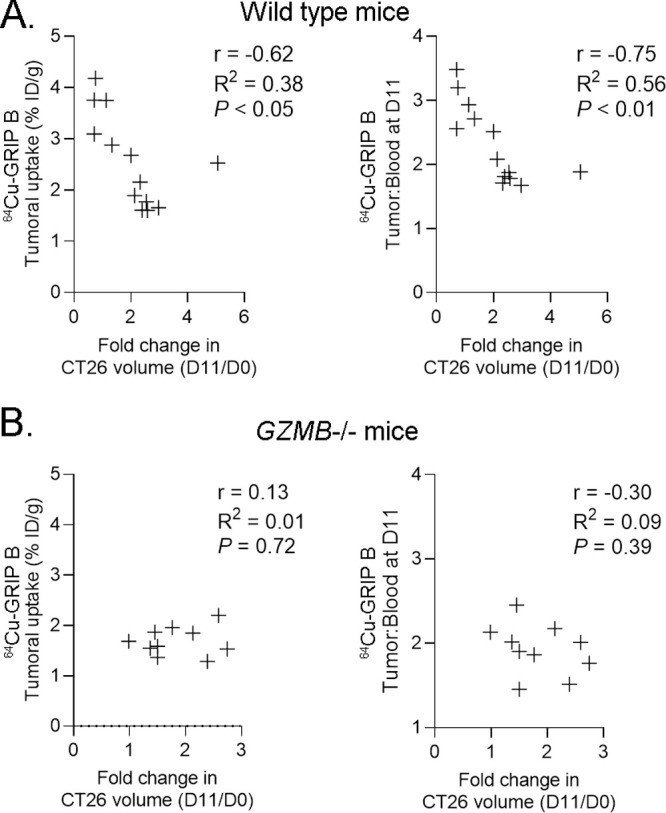
Post-treatment changes in the tumoral uptake of 64Cu-GRIP B correlate with the magnitude of volumetric tumor response to CPI in wild-type mice but not in GZMB–/– mice. (A) Scatter plots showing the correlation between the fold change in the tumor volume from day 11 to day 0 and the 64Cu-GRIP B tumoral uptake (left) or tumor-to-blood ratio (right). The data were collected from two cohorts of wild-type mice bearing CT26 tumors. (B) Scatter plots showing the correlation between fold change in tumor volume from day 11 to day 0 and the 64Cu-GRIP B tumoral uptake (left) or tumor-to-blood ratio (right). The data were collected from two cohorts of GZMB–/– mice bearing CT26 tumors.
Exploratory Studies with 64Cu-GRIP B PET Suggests a Role for Secreted GZMB in Pulmonary Inflammation
Though not well-defined, noncytotoxic functions for secreted GZMB have been proposed in several physiological processes, including inflammation.31−33 To test if 64Cu-GRIP B can localize potentially pathogenic reservoirs of secreted GZMB due to inflammation, we next performed 64Cu-GRIP B PET/CT on wild-type mice that had received intratracheal instillations of lipopolysaccharide (LPS). PET/CT was performed 4 d after the instillation, a time point at which a T cell recruitment to the lungs has occurred.34,35 An ROI analysis showed a significantly higher radiotracer accumulation in the lungs of mice treated with low (0.1 mg/kg) and high (3 mg/kg) doses of LPS compared to a vehicle (Figure 6A,B). Autoradiography and immunofluorescence of the lungs showed a visually higher radiotracer binding in the LPS-treated lung. Consistent with the autoradiography, the overall levels of GZMB, CD3, Ly6G, and NKp46/NCR1 staining were higher in the lung parenchyma of LPS versus vehicle-treated mice (Figure 6C and Supporting Information Figure 9). We observed regions in the lung that were positive for GZMB but lacked evidence of 64Cu-GRIP B binding. We hypothesize these regions harbor high levels of GZMB positive immune cells that are not actively secreting GZMB. Since 3 mg/kg LPS can trigger a system-wide T cell activation, we further examined a radiotracer uptake in a larger panel of mouse organs. Ex vivo biodistribution studies showed that 64Cu-GRIP B was significantly higher in numerous tissues in the LPS-treated versus vehicle mice at either dose, including lymphoid organs like the spleen and thymus (Figure 6D and Supporting Information Table 9). Remarkably, the system-wide impact on T cell activation due to the intratracheal instillation of LPS was also visually obvious by a comparison of maximum intensity projections between treatment arms (Figure 6E).
Figure 6.
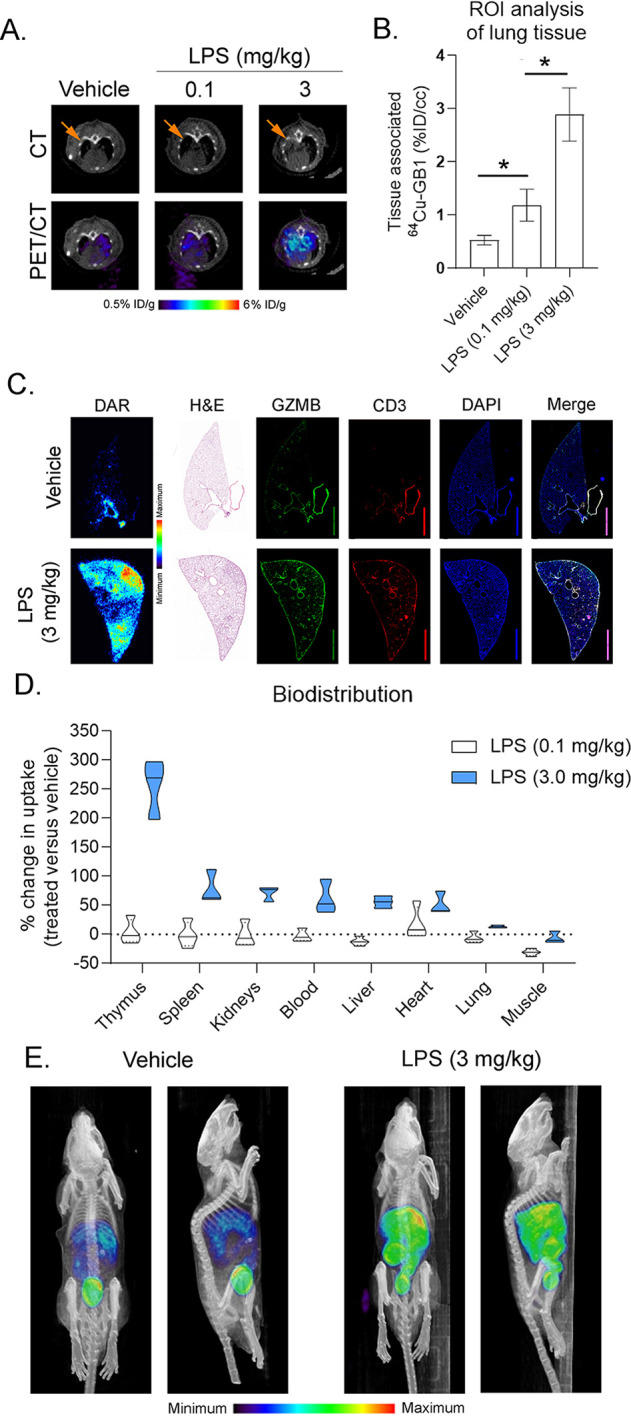
64Cu-GRIP B PET detects secreted GZMB elicited by an endotoxin-mediated inflammatory response. (A) Representative 64Cu-GRIP B PET/CT studies showing a higher radiotracer accumulation in the lungs of mice treated with 0.1 or 3.0 mg/kg LPS compared to mice that received a sham. (B) ROI analysis of the right lung lobe shows a significantly higher radiotracer uptake in LPS-treated vs sham-treated mice (n = 3 per arm). *P < 0.01. (C) Autoradiography, immunofluorescence, and H&E of the right lung lobe shows a higher tracer accumulation in the treated lung as well as a higher GZMB and CD3 staining. (D) A bar graph showing the percent change in the radiotracer uptake per organ between the LPS- vs sham-treated mice (n = 4 per arm). All changes were determined to be statistically significant, P < 0.05. (E) Representative maximum intensity projections showing the systemwide changes in the tracer biodistribution due to a treatment with 3.0 mg/kg LPS.
Discussion
In this report, we show that granzyme biochemistry can be detected in vivo with nuclear imaging. This was achieved by a development of 64Cu-GRIP B, a peptide-based chemosensor whose biodistribution was engineered to be controlled by the proteolytic activity of secreted GZMB. Crucial to the design of GRIP B was the discovery of a highly efficient P4–P4′ substrate sequence for GZMB that gates the activation of the probe. Subsequently, in vitro and in vivo studies demonstrated that GZMB cleaves GRIP B and that changes in the tissue uptake of 64Cu-GRIP B are driven by immune cell activation and the secretion of GZMB. Applying 64Cu-GRIP B in vivo identified in real time multiple organs with elevated GZMB, a feature that we expect will enable a more holistic mapping of complex human immunology through quantitative imaging. Moreover, the exploratory study that applied 64Cu-GRIP B to an inflammatory model in which the secreted GZMB has an unknown significance shows how this imaging technology may be used to begin a probing of noncytotoxic functions for this enzyme class.
These imaging data come at an exciting time, as ongoing work from other laboratories have shown that active site-directed inhibitors of GZMB can also be rendered into Ga-68 or F-18 radiopharmaceuticals designed to detect GZMB in vivo.23,24,36 As with 64Cu-GRIP B, the tumoral uptake of these active site inhibitors is enhanced by a systemic CPI treatment, and the magnitude of post-treatment changes in the tumoral uptake is anticorrelated with volumetric tumor responses. The development of 64Cu-GRIP B complements these innovations by enabling a readout of enzyme turnover to pair with the measurement of biochemically active GZMB molecules provided by the peptide aldehydes. The unique mechanism of GRIP B may also enable an imaging at extended time points postinjection (i.e., >4 h), which could improve the signal-to-background and/or reveal occult tumors through a catalytic amplification of probe retention.
Our data suggest that 64Cu-GRIP B may have a role in monitoring an early treatment response to immunotherapies. The documentation of early tumor responses to CPIs is currently a challenge clinically with standard of care 18F fluorodeoxyglucose (FDG) PET and CT. In the case of PET, the immunoresponsive tumor newly perfused with metabolically active cytotoxic T cells tends to consume the same or more 18F FDG than it did in its pretreatment state, which mimics the imaging findings of an unresponsive, progressing tumor.37 Pseudoprogression is also observed on CT in up to 10% of all patients,38 presumably due to the transient increase in cellularity within the immunoresponsive tumors. Custom algorithms (e.g., PERCIMT, iRECIST) are under development to better interpret 18F FDG PET/CT post-immunotherapies, but the challenges with 18F FDG PET/CT have rightly motivated a swell of new experimental radiotracers to more clearly distinguish responsive from nonresponsive tumors.39−42 The detection of a system-wide T-cell activation in normal tissues also implies that 64Cu-GRIP B might be useful for the identification of patients at risk for developing immune-related adverse events. Direct detection of extracellular GZMB proteolysis may have advantages over biomarkers that measure immune cell markers (e.g., CD4, CD8) but do not distinguish between indolent or activated T cells.43 We are currently translating 64Cu-GRIP B into patients to study these applications.
Lastly, these data provide an important confirmation that the RIP platform can be engineered to image functionally discrete proteases beyond thrombin. The versatility of the RIP platform to conjugate with chemically discrete payloads may also permit radiolabeling with other diagnostic (e.g., Zr-89, F-18) or therapeutic (e.g., Lu-177, Ac-225) radioisotopes. The granzyme family has divergent, highly specific substrate specificities from which we expect the RIP technology’s activity-based accumulation benefits. Moving forward, we aim to expand this platform to enable a preclinical imaging with other modalities (e.g., fluorescence tomography) and develop imaging probes to detect the other granzyme species.
Materials and Methods
General Methods
All reagents were purchased from commercial sources and used without further purification. 64Cu-hydrochloride acid was purchased from University of Wisconsin Madison. Recombinant human GZMB was purchased from Sigma-Aldrich. The mouse cancer cell lines CT26 and EMT6were purchased from ATCC. MC38 was purchased from Kerafast. Antimouse PD-1 (CD279) (BE0146) and antimouse CTLA-4 (CD152) (BE0164) were purchased from Bio X Cell; Anti granzyme B (ab4059) was purchased from Abcam; anti-CD3 (MCA1477) was purchased from Bio-Rad; AF488 anti-Rabbit (A21206), AF546 anti-Mouse (A111081), and AF633 anti-Mouse (A21052) secondary antibodies were purchased from Invitrogen. AF2225 anti-mouse NKp46/NCR1 was purchased from R&D system. BE0075-1 anti-mouse Ly6G was purchased from Bio X Cell. AF488 anti-RAT (A48269) and AF647 anti-goat (A21446) were purchased from Invitrogen. 4′,6-Diamidino-2-phenylindole (DAPI) (D1306) was purchased from Life Technologies Corporation. Antibodies for immunofluorescence. All cell lines were cultured according to manufacturer’s instructions. Active, human MMP-9 was purchased from Millipore Sigma. Human, activated Complement C 1s protease was purchased from Millipore Sigma. Thrombin from human plasma was purchased from Sigma-Aldrich. Active, human placenta cathepsin B was purchased from Sigma-Aldrich. Active, human recombinant caspase-8 was purchased from Enzo Lifesciences. Active, human recombinant caspase-3 was purchased from Enzo Lifesciences. Recombinant human granzyme A was purchased from R&D Systems. Recombinant human granzyme K was purchased from Enzo Lifesciences.
Multiplex Substrate Profiling by Mass Spectrometry
Human GZMB (100 nM) was incubated with a library containing 228 synthetic tetra-decapeptides (500 nM). Aliquots (10 μL) were removed at three time intervals and subsequently quenched with 10 μL of 8 M guanidinium hydrochloride. Aliquots were then flash frozen until all time points were taken. Prior to the mass spectrometry, samples were desalted using C18 tips (Rainin). Aliquots were then analyzed by LC-MS/MS sequencing using a Quadrupole Orbitrap mass spectrometer (LTQ Orbitrap XL) coupled to a 10 000 psi nanoACQUITY Ultra Performance Liquid Chromatography (UPLC) System (Waters) for a peptide separation by reverse-phase liquid chromatography (RPLC). Peptides were separated over a Thermo ES901 C18 column (75 μm inner diameter, 50 cm length) coupled to an EASY-Spray ion source and eluted by the application of a flow rate of 300 nL/min with a 65 min linear gradient from 2–50% in Buffer B (acetonitrile, 0.5% formic acid). Survey scans were recorded over a 325–1500 m/z range and up to the three most-intense precursor ions (MS1 features of charge ≥2) were selected for a higher-energy collisional dissociation (HCD) at a resolution of 30 000 at m/z 200 for MS/MS[CB2]. Data were acquired using Xcalibur software and processed as previously described.18,44 Briefly, raw mass spectrometry data were processed to generate peak lists using MSConvert. Peak lists were then searched in Protein Prospector ver. 6.2.2 against a proprietary database containing the sequences from the 228 tetra-decapeptide library.45 Searches used a mass accuracy tolerance of 20 ppm for precursor ions and 30 ppm for fragment ions. Variable modifications included a N-terminal pyroglutamate conversion from glutamine or glutamate and an oxidation of tryptophan, proline, and tyrosine. Searches were subsequently processed using the MSP-xtractor software (http://www.craiklab.ucsf.edu/extractor.html), which extracts the peptide cleavage site and spectral counts of the corresponding cleavage products. Spectral counts were used for the relative quantification of peptide cleavage products. Human GZMB samples were processed as three biological replicates per time point, and a nonenzyme control was used for each replicate to remove unspecific cleavages from the data analysis.
Fmoc-Solid-Phase Peptide Synthesis
A quenched fluorogenic peptide synthesized of the sequence NH2-K(MCA)IEPDVSQVK(DNP)-COOH (MCA = 7-methoxycoumarin-4-acetic acid, DNP = 2-dinitrophenyl) was synthesized by an Fmoc solid-phase synthesis on a Biotage SyroII peptide synthesizer at ambient temperature. The synthesis scale was at 12.5 μM using a preloaded lysine DNP Wang resin, where the DNP quencher was linked to the epsilon nitrogen of the lysine. Coupling reactions were performed with 4.9 equiv of O-(1H-6-chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluoro-phosphate (HCTU), 5 equiv of Fmoc-amino acid–OH, and 20 equiv of N-methylmorpholine (NMM) in 500 μL of N,N-dimethylformamide (DMF) for 8 min while being shaken. Each amino acid position was double-coupled, and subsequent Fmoc deprotection was performed with 500 μL of 40% 4-methylpiperadine in DMF for 10 min followed by six washes with 500 μL of DMF for 3 min. The final amino acid coupling contained the fluorophore, lysine (MCA), where MCA was linked to the epsilon nitrogen of the lysine. Peptides were cleaved from Wang’s resin with 500 μL of solution composed of 95% trifluoroacetic acid, 2.5% water, and 2.5% triisopropylsilane for 1 h while being shaken. The crude peptide product was then precipitated in 30 mL of cold 1:1 diethyl ether/hexanes and then solubilized in a 1:1:1 mixture of DMSO/water/acetonitrile. The solubilized crude was purified by high-performance liquid chromatography (HPLC) using an Agilent Pursuit 5 C18 column (5 mm bead size, 150 × 21.2 mm) on an Agilent PrepStar 218 series preparative HPLC. The mobile phases A and B were water + 0.1% trifluoroacetic acid (TFA) and acetonitrile + 0.1% TFA, respectively. The purified peptide product had the solvent removed under a reduced atmosphere and was solubilized into a DMSO stock with a final concentration of 10 mM. The purity was confirmed by liquid chromatography–mass spectrometry, and the stock was stored at −20 °C. Fluorescently labeled 5FAM-GRIP B was purchased from CPC Scientific at 95–98% purity.
Synthesis of DOTA-GRIP B
DOTA-GRIP B (Dota-hexanoic acid-FVQWFSKFLGKIEPDVSQVQDPNDQYEPF-COOH) was synthesized first by the use of standard solid-phase peptide synthesis conditions as outlined above. The resin-bound peptide with N-terminal hexanoic acid was triply coupled with 2 equiv of dota-NHS, 5 equiv of HCTU, and 20 equiv of N,N-diisopropylethylamine (DIPEA) for 12 h. The DOTA-GRIP B probe was then cleaved, purified, and analyzed as described for the fluorogenic peptide.
In Vitro Kinetics
Kinetic measurements were performed in Corning black 384-well flat-bottom plates and read on a BioTek H4 multimode plate reader. The proteolysis of the quenched fluorogenic peptide (NH2-K(MCA)IEPDVSQVK(DNP)-COOH) by GZMB was performed at a final enzyme concentration of 5 nM in phosphate-buffered saline (PBS). Michaelis–Menten kinetics were performed in triplicate at 37 °C, and activity was monitored for 1 h. The Vo value was calculated at 1–30 min in RFU/s. Initial velocities were then converted to molar per second using a standard curve of cleaved substrate. Specificity experiments were done with all final enzyme concentrations of 5 nM and final quenched fluorogenic peptide concentration of 60 μM. The specificity assay buffer was PBS with 1 mM dithiothreitol (DTT). Kinetics were performed in quadruplicate at 37 °C, and the activity was monitored for 1 h.
Intrinsic Tryptophan Fluorescence Spectroscopy Measuring Lipid Insertion
The fluorescence of the tryptophan within the full-length and activated GRIP B was monitored in the presence or absence of lipid micelles on a BioTek H4 multimode plate reader. Sodium dodecyl sulfate (SDS) was solubilized as a 5 mg mL–1 stock. Full-length and activated GZMB-RIP were solubilized in PBS to a final concentration of 0.01 mg mL–1 with a final peptide/lipid molar ratio of 1:40. Tryptophan emission spectra of the peptide/lipid suspension were acquired with an excitation wavelength of 295 nm and by scanning from 310 to 450 nm. The bandwidth was 5 nm for both excitation and emission. The spectra of the peptides in PBS in the absence of SDS lipids were acquired at the same concentration of 0.01 mg mL–1.
Circular Dichroism of Activated GRIP B
Spectra were acquired on a Jasco J-810 spectrometer with samples maintained at 310 K. Spectra were recorded from 250 to 200 nm using a spectral bandwidth of 1 nm and a scan rate of 100 nm min–1. The buffer was 20 mM Na2HPO4. Final peptide concentrations were 0.1 mg/mL, and SDS lipid concentrations were kept at a minimum of a 1:40 peptide/lipid molar ratio. Spectra were treated using Jasco spectra analysis software, where a spectrum of the peptide-free suspension was subtracted and means-movement smoothing with a convolution width of five points was applied.
Toxicity Assay Measuring Hemolysis of Human Erythrocytes
Blood from healthy anonymous donors was harvested from Trima Leukoreduction chambers (Vitalant). Erythrocytes were isolated from the anonymous blood samples. Full-length GRIP B and activated GRIP B were measured for their hemolytic activity on healthy human erythrocytes in triplicate. Aliquots of human erythrocytes were suspended in PBS (pH 7.4) and incubated with serial dilutions of both peptides, which were initially solubilized in DMSO. DMSO and 1% Triton X-100 were incubated in parallel as negative and positive controls, respectively. The incubation was for 1 h at 37 °C. After the incubation, the samples were centrifuged for 5 min at 2000g, after which the supernatant was collected. The supernatant was measured for the release of hemoglobin by the erythrocytes using a BioTek H4 multimode plate reader, monitoring the optical density of the supernatant at a wavelength of 540 nm.
Flow Cytometry with 5FAM-GRIP B
MC38 cells (2 × 105/well) were seeded into a 12-well plate and incubated at 37 °C for 48 h. 5FAM-GRIP B (200 nM) and GZMB (20 nM) were dissolved in HBSS and incubated at 37 °C for 2 h. The 200 nM RIP with/without 50 nm GZMB in HBSS (300 μL) was added into the well with the cells, followed by an incubation at 37 °C for 30 min. The probe solution was removed, and cells were washed with PBS four to five times. Trypsin (100 μL) was added, followed by 3 min of incubation at 37 °C. PBS was added into the wells, and all cells were collected and washed with PBS for one time before being further diluted with PBS (300 μL) and passed through a cell strainer. Experiments were performed on a BD FACSCanto II Cell Analyzer. Data were analyzed by the use of FlowJo and Prism 8.0.
Radiosynthesis and In Vitro Characterization of 64Cu-GRIP B
Into a 1.5 mL reaction vial was added 5 mCi of 64Cu-chloride (aqueous), and the pH was adjusted to 7.0 with Na2CO3 (2 M). A solution of DOTA-GRIP B (50 μg in 20 μL of DMSO) and 0.1 M NH4OAC buffer (200 μL) was added into this reaction vial. The reaction mixture was incubated at 50 °C for 30 min. The reaction progress was monitored by analytical HPLC equipped with an Agilent Pursuit analytical column (C18, 200 Å, 4.6 mm × 10 cm, 5 μm) or Phenomenex Luna analytical column (C18, 100 Å, 4.6 mm × 250 cm, 10 μm) (70:30 MeOH/H2O to 95:5 MeOH/H2O over 10 min). The crude reaction was purified using a C18 Sep-Pak cartridge and eluted with a small volume of CH3CN. The CH3CN was then removed at 50 °C under vacuum and a gentle stream of N2(g) to afford neat 64Cu-GRIP B. The chelation efficacy is usually greater than 90% based on the HPLC. A formulation comprising 10% DMSO, 10% tween 80, and 80% saline was adopted for further mice studies. The cleavage of 64Cu-GRIP B by granzyme B was verified in vitro by an addition of the radiotracer (∼200 μCi) to the recombinant granzyme B (10 nM) in 500 uL of PBS. The vial was then incubated at 37 °C. Rad-HPLC was used to monitor the cleavage of the radiotracer at the dedicated time points.
Animal Studies
All animal experiments were approved by the Institutional Animal Care and Use Committee at UCSF. Four to six week old male or female balb/c mice and C57BL6/J mice were purchased from Jackson Laboratory and housed with free access to the water and food. All mice were inoculated with 5 × 106 CT26, MC38, or EMT6 cells in a mixture of media and Matrigel (Corning) (v/v 1:1) subcutaneously into the left shoulder. Antimouse PD-1 (CD279) (BE0146) and antimouse CTLA-4 (CD152) (BE0164) were purchased from Bio X Cell and stored at 4 °C during the treatment studies. Mice bearing subcutaneous tumors received antimouse CTLA-4 (200 ug) or/and antimouse PD-1 (200 ug) and as a combination therapy or PBS as the vehicle on days 5, 8, and 11 following the tumor inoculation. Mice were weighed, and the tumor volume was measured with calipers on the same day of the treatment. On day 14, all mice were used for PET/CT or BioD studies.
GZMB Homozygous Knockout Mice
Germline homozygous GZMB knockout cluster mice were created with a homozygous null mutation in the GZMB gene. GZMB knockout cluster mice were generously donated by Dr. T. Ley, originating from ref (30).
Small Animal PET/CT
64Cu-L-GRIP B or 64Cu-D-GRIP B (∼100 μCi/mouse) in 100–150 μL of 10% DMSO and 10% Tween 80 in saline was injected via tail vein. After a period of uptake time, mice were anesthetized with isoflurane (∼2%) and imaged with a microPET/CT scanner (Inveon, Siemens). For a static imaging, mice were scanned for 30 min for a PET data acquisition and 10 min for a CT data acquisition. For the dynamic acquisitions, the mice were anesthetized, positioned on the scanner bed, and injected intravenously with a radiotracer. The dynamic acquisition was performed for 60 min followed by a 10 min CT acquisition.
List-mode PET data were histogrammed to generate sinograms that were reconstructed using a 2D ordered subsets expectation maximization algorithm provided by the scanner manufacturer. An attenuation correction was applied using the coregistered CT data that were acquired immediately following the PET data acquisition. The CT data were acquired using the following setting: 220° angular coverage with 120 steps, X-ray tube operating at 80 kVp and 0.5 mA with each angular step exposure time set as 175 ms. All reconstructed three-dimensional (3D) PET volume image voxels were calibrated to becqerel per milliliter using a precalibrated quantification factor. AMIDE software was used for the reconstruction of PET/CT data and an image analysis.
Biodistribution Studies
At dedicated time points post-radiotracer injection, mice were euthanized with CO2(g) asphyxiation, and the blood was collected by a direct cardiac puncture. Tissues were harvested, weighed, and counted on a γ-counter (Hidex). The amount of radioactivity in the tissues was determined by comparison with a standard of known activity. The samples were decay-corrected and expressed as the percentage of the injected dose/weight of the harvested tissues (%ID/g).
Digital Autoradiography
Tumors or designated tissue were flash frozen in an optimal cutting temperature (OCT) in dry ice. The tissues were sectioned with a microtome (Leica) into slices with 10–20 um thickness and directly mounted on glass slides (VWR). A GE Storage Phosphor Screen was exposed by such slides with radioactive tissue. After 10 half-lives of copper-64, the screen was developed on a phosphorimager (Typhoon 9400). The images were further analyzed by using Fiji software.
Histology
Hematoxylin and eosin (H&E) staining and immunofluorescence staining were performed by the Pathology core facility at UCSF and Acepix Biosciences. For IF studies, tumor samples were soaked in acetone at −20 °C for 20 min, followed by a soak in MeOH at 4 °C for 10 min. Antigen retrieval was conducted with citrate buffer 10 mM pH = 6, and samples were blocked with a universal blocking buffer plus 5% goat and donkey serum. The primary antibodies, namely, anti-GZMB (ab4059, Abcam) (1:50), anti NKp46/NCR1(1:1000, R&D system), anti Ly6G (1:5000, Bio X Cell), and anti-CD3 (MCA1477, Bio-Rad) (1:100), were added into samples and incubated at 4 °C overnight. Such primary antibodies were detected by AF488 anti-Rabbit (A21206, Invitrogen) (1:200), AF546 anti-Mouse (A111081, Invitrogen) (1:200), AF488 anti-Rat (A48269, Invitrogen) (1:500), AF647 anti-goat (A21446, Invitrogen) (1:500), and AF633 anti-Mouse (A21052, Invitrogen) (1:200) secondary antibodies by an incubation with samples. DAPI Nucleic Acid Stain (D1306, Life Technologies Corporation) was used to stain the nucleus by being incubating with samples (10 min at room temperature). Immunofluorescence results were performed by the Gladstone Institutes’ Histology & Light Microscopy Core. Images of whole sections were acquired on a VERSA automated slide scanner (Leica Biosystems), equipped with an Andor Zyla 5.5 sCMOS camera (Andor Technologies). Individual images were created with the ImageScope software (Aperio Technologies).
Statistics
All statistical analyses were performed by using PRISM v8.0 or ORIGIN software. A statistically significant difference was determined by an unpaired, two-tailed Student’s t test. Changes only at the 95% confidence level (P < 0.05) were regarded as statistically significant.
Acknowledgments
The authors acknowledge Professors Y. Seo, H. VanBrocklin, Mr. J. Blecha, and R. Tang (UCSF) for technical assistance and Professor T. Ley (WUSTL) for contributing GZMB knockout breeding pairs. M.J.E. was supported by an American Cancer Society Research Scholar Grant (130635-RSG-17-005-01-CCE) and the National Institute Biomedical Imaging and Bioengineering (R01EB025207). M.J.E., L.F., and C.S.C. were supported by the National Cancer Institute (R01CA258297) and a Prostate Cancer Foundation Challenge Grant. M.J.E, D.M.W., and C.S.C. were supported by the National Institute of Allergy and Infectious Diseases (R01AI161027). L.F. was supported by the National Cancer Institute (R01CA223484). C.S.C. was supported by the National Cancer Institute (P41CA196276). Small animal PET/CT studies were performed on the instrument supported by National Institutes of Health, Grant No. S10RR023051. Research from UCSF reported in this publication was supported in part by the National Cancer Institute of the National Institutes of Health under Award No. P30CA082103. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank C. Mathy from Dr. T. Kortemme’s lab and Dr. R. Newberry from Dr. W. Degrado’s lab for their assistance with circular dichroism.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.1c00529.
-
1.
Heatmap of Granzyme B specificity (PDF)
-
2.
GRIP B cleavage by granzyme B HPLC traces
-
3.
Intrinsic tryptophan fluorescence membrane insertion and circular dichroism
-
4.
Structure of DOTA-GRIP B
-
5.
Stability of 64Cu-GRIP B in mouse serum
-
6.
Compartmental analysis of tumoral uptake of 64Cu-GRIP B
-
7.
Coronal 64Cu-GRIP B PET/CT images of mice bearing CT26 tumors and treated with vehicle or checkpoint inhibitors
-
8.
Analytical HPLC chromatogram showing synthesis of 64Cu-D-GRIP B
-
9.
Immunofluorescence data of LPS versus vehicle treated lung tissue
-
10.
Table summary of SUVmean values
-
11.
Tables of biodistribution data
Author Contributions
∇ (N.Z. and C.B.) Co-first authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Voskoboinik I.; Whisstock J. C.; Trapani J. A. Perforin and granzymes: function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. 10.1038/nri3839. [DOI] [PubMed] [Google Scholar]
- Ritter A. T.; Angus K. L.; Griffiths G. M. The role of the cytoskeleton at the immunological synapse. Immunol. Rev. 2013, 256, 107–117. 10.1111/imr.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannelli J. R.; Sullivan J. A.; Mandell G. L.; Engelhard V. H. Reorientation and fusion of cytotoxic T lymphocyte granules after interaction with target cells as determined by high resolution cinemicrography. J. Immunol 1986, 136, 377–382. [PubMed] [Google Scholar]
- Bykovskaja S. N.; Rytenko A. N.; Rauschenbach M. O.; Bykovsky A. F. Ultrastructural alteration of cytolytic T lymphocytes following their interaction with target cells. II. Morphogenesis of secretory granules and intracellular vacuoles. Cell. Immunol. 1978, 40, 175–185. 10.1016/0008-8749(78)90325-8. [DOI] [PubMed] [Google Scholar]
- Bykovskaja S. N.; Rytenko A. N.; Rauschenbach M. O.; Bykovsky A. F. Ultrastructural alteration of cytolytic T lymphocytes following their interaction with target cells. I. Hypertrophy and change of orientation of the Golgi apparatus. Cell. Immunol. 1978, 40, 164–174. 10.1016/0008-8749(78)90324-6. [DOI] [PubMed] [Google Scholar]
- Shi L.; Kam C. M.; Powers J. C.; Aebersold R.; Greenberg A. H. Purification of three cytotoxic lymphocyte granule serine proteases that induce apoptosis through distinct substrate and target cell interactions. J. Exp. Med. 1992, 176, 1521–1529. 10.1084/jem.176.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H.; Henkart P. A. Cytotoxic lymphocyte granzymes trigger a target cell internal disintegration pathway leading to cytolysis and DNA breakdown. J. Immunol 1994, 152, 1057–1063. [PubMed] [Google Scholar]
- Henkart P. A. Lymphocyte-mediated cytotoxicity: two pathways and multiple effector molecules. Immunity 1994, 1, 343–346. 10.1016/1074-7613(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Sutton V. R.; Trapani J. A. Proteases in lymphocyte killer function: redundancy, polymorphism and questions remaining. Biol. Chem. 2010, 391, 873–879. 10.1515/bc.2010.088. [DOI] [PubMed] [Google Scholar]
- Grossman W. J.; et al. The orphan granzymes of humans and mice. Curr. Opin. Immunol. 2003, 15, 544–552. 10.1016/S0952-7915(03)00099-2. [DOI] [PubMed] [Google Scholar]
- van Daalen K. R.; Reijneveld J. F.; Bovenschen N. Modulation of Inflammation by Extracellular Granzyme A. Front. Immunol. 2020, 11, 931. 10.3389/fimmu.2020.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah E.; Rosen A. Granzyme B cleavage of autoantigens in autoimmunity. Cell Death Differ. 2010, 17, 624–632. 10.1038/cdd.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granville D. J. Granzymes in disease: bench to bedside. Cell Death Differ. 2010, 17, 565–566. 10.1038/cdd.2009.218. [DOI] [PubMed] [Google Scholar]
- Kaiserman D.; et al. The major human and mouse granzymes are structurally and functionally divergent. J. Cell Biol. 2006, 175, 619–630. 10.1083/jcb.200606073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Hong H.; Zhang Y.; Cai W. Molecular Imaging of Proteases in Cancer. Cancer Growth Metastasis 2009, 2, 13–27. 10.4137/CGM.S2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A. Granzyme B nanoreporter for early monitoring of tumor response to immunotherapy. Sci. Adv. 2020, 6, eabc2777. 10.1126/sciadv.abc2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page M. J.; et al. Non-invasive imaging and cellular tracking of pulmonary emboli by near-infrared fluorescence and positron-emission tomography. Nat. Commun. 2015, 6, 8448. 10.1038/ncomms9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue A. J.; et al. Global identification of peptidase specificity by multiplex substrate profiling. Nat. Methods 2012, 9, 1095–1100. 10.1038/nmeth.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapek J. D. Jr.; et al. Quantitative Multiplex Substrate Profiling of Peptidases by Mass Spectrometry. Mol. Cell Proteomics 2019, 18, 968–981. 10.1074/mcp.TIR118.001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivry S. L.; et al. Global substrate specificity profiling of post-translational modifying enzymes. Protein Sci. 2018, 27, 584–594. 10.1002/pro.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S.; Craik C. S. Selective chemical functional probes of granzymes A and B reveal granzyme B is a major effector of natural killer cell-mediated lysis of target cells. Chem. Biol. 2005, 12, 567–577. 10.1016/j.chembiol.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Ruggles S. W.; Fletterick R. J.; Craik C. S. Characterization of structural determinants of granzyme B reveals potent mediators of extended substrate specificity. J. Biol. Chem. 2004, 279, 30751–30759. 10.1074/jbc.M400949200. [DOI] [PubMed] [Google Scholar]
- Larimer B. M.; et al. Granzyme B PET Imaging as a Predictive Biomarker of Immunotherapy Response. Cancer Res. 2017, 77, 2318–2327. 10.1158/0008-5472.CAN-16-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer B. M.; et al. The Effectiveness of Checkpoint Inhibitor Combinations and Administration Timing Can Be Measured by Granzyme B PET Imaging. Clin. Cancer Res. 2019, 25, 1196–1205. 10.1158/1078-0432.CCR-18-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M. G.; et al. Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J. Immunother. 2013, 36, 477–489. 10.1097/01.cji.0000436722.46675.4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Qi J. Direct estimation of kinetic parametric images for dynamic PET. Theranostics 2013, 3, 802–815. 10.7150/thno.5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai C. S.; et al. Tumor-conditional anti-CTLA4 uncouples antitumor efficacy from immunotherapy-related toxicity. J. Clin. Invest. 2019, 129, 349–363. 10.1172/JCI123391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise W.; et al. CTLA-4 suppresses the pathogenicity of self antigen-specific T cells by cell-intrinsic and cell-extrinsic mechanisms. Nat. Immunol. 2010, 11, 129–135. 10.1038/ni.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer J. R.; et al. Management of Immune-Related Adverse Events in Patients Treated With Immune Checkpoint Inhibitor Therapy: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2018, 36, 1714–1768. 10.1200/JCO.2017.77.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusel J. W.; Wesselschmidt R. L.; Shresta S.; Russell J. H.; Ley T. J. Cytotoxic lymphocytes require granzyme B for the rapid induction of DNA fragmentation and apoptosis in allogeneic target cells. Cell 1994, 76 (6), 977–87. 10.1016/0092-8674(94)90376-X. [DOI] [PubMed] [Google Scholar]
- Hiebert P. R.; Granville D. J. Granzyme B in injury, inflammation, and repair. Trends Mol. Med. 2012, 18, 732–741. 10.1016/j.molmed.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Cullen S. P.; Brunet M.; Martin S. J. Granzymes in cancer and immunity. Cell Death Differ. 2010, 17, 616–623. 10.1038/cdd.2009.206. [DOI] [PubMed] [Google Scholar]
- Hirota J. A.; et al. Granzyme B deficiency exacerbates lung inflammation in mice after acute lung injury. Am. J. Respir. Cell Mol. Biol. 2013, 49, 453–462. 10.1165/rcmb.2012-0512OC. [DOI] [PubMed] [Google Scholar]
- Su X.; Looney M.; Robriquet L.; Fang X.; Matthay M. A. Direct visual instillation as a method for efficient delivery of fluid into the distal airspaces of anesthetized mice. Exp. Lung Res. 2004, 30, 479–493. 10.1080/01902140490476382. [DOI] [PubMed] [Google Scholar]
- D’Alessio F. R.; et al. CD4+CD25+Foxp3+ Tregs resolve experimental lung injury in mice and are present in humans with acute lung injury. J. Clin. Invest. 2009, 119, 2898–2913. 10.1172/JCI36498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggi J. L.; et al. Granzyme B PET Imaging of Immune Checkpoint Inhibitor Combinations in Colon Cancer Phenotypes. Mol. Imaging Biol. 2020, 22, 1392–1402. 10.1007/s11307-020-01519-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aide N.; et al. FDG PET/CT for assessing tumour response to immunotherapy: Report on the EANM symposium on immune modulation and recent review of the literature. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 238–250. 10.1007/s00259-018-4171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Romano P.; et al. Evidence of pseudoprogression in patients treated with PD1/PDL1 antibodies across tumor types. Cancer Med. 2020, 9, 2643–2652. 10.1002/cam4.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J. W.; et al. CD8(+) T-Cell Density Imaging with (64)Cu-Labeled Cys-Diabody Informs Immunotherapy Protocols. Clin. Cancer Res. 2018, 24, 4976–4987. 10.1158/1078-0432.CCR-18-0261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freise A. C.; et al. ImmunoPET Imaging of Murine CD4(+) T Cells Using Anti-CD4 Cys-Diabody: Effects of Protein Dose on T Cell Function and Imaging. Mol. Imaging Biol. 2017, 19, 599–609. 10.1007/s11307-016-1032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T.; Campbell D. O.; Satyamurthy N.; Czernin J.; Radu C. G. Stratification of nucleoside analog chemotherapy using 1-(2’-deoxy-2’-18F-fluoro-beta-D-arabinofuranosyl)cytosine and 1-(2’-deoxy-2’-18F-fluoro-beta-L-arabinofuranosyl)-5-methylcytosine PET. J. Nucl. Med. 2012, 53, 275–280. 10.2967/jnumed.111.090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald J. A.; et al. A PET Imaging Strategy to Visualize Activated T Cells in Acute Graft-versus-Host Disease Elicited by Allogenic Hematopoietic Cell Transplant. Cancer Res. 2017, 77, 2893–2902. 10.1158/0008-5472.CAN-16-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Donk P. P.; et al. Molecular imaging biomarkers for immune checkpoint inhibitor therapy. Theranostics 2020, 10, 1708–1718. 10.7150/thno.38339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivry S. L.; et al. Global Protease Activity Profiling Provides Differential Diagnosis of Pancreatic Cysts. Clin. Cancer Res. 2017, 23, 4865–4874. 10.1158/1078-0432.CCR-16-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo E. C.; et al. Global Protease Activity Profiling Identifies HER2-Driven Proteolysis in Breast Cancer. ACS Chem. Biol. 2021, 16, 712. 10.1021/acschembio.0c01000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.