Abstract
Study objective:
Machine-learning algorithms allow improved prediction of sepsis syndromes in the emergency department (ED), using data from electronic medical records. Transfer learning, a new subfield of machine learning, allows generalizability of an algorithm across clinical sites. We aim to validate the Artificial Intelligence Sepsis Expert for the prediction of delayed septic shock in a cohort of patients treated in the ED and demonstrate the feasibility of transfer learning to improve external validity at a second site.
Methods:
This was an observational cohort study using data from greater than 180,000 patients from 2 academic medical centers between 2014 and 2019, using multiple definitions of sepsis. The Artificial Intelligence Sepsis Expert algorithm was trained with 40 input variables at the development site to predict delayed septic shock (occurring greater than 4 hours after ED triage) at various prediction windows. We then validated the algorithm at a second site, using transfer learning to demonstrate generalizability of the algorithm.
Results:
We identified 9,354 patients with severe sepsis, of whom 723 developed septic shock at least 4 hours after triage. The Artificial Intelligence Sepsis Expert algorithm demonstrated excellent area under the receiver operating characteristic curve (>0.8) at 8 and 12 hours for the prediction of delayed septic shock. Transfer learning significantly improved the test characteristics of the Artificial Intelligence Sepsis Expert algorithm and yielded comparable performance at the validation site.
Conclusion:
The Artificial Intelligence Sepsis Expert algorithm accurately predicted the development of delayed septic shock. The use of transfer learning allowed significantly improved external validity and generalizability at a second site. Future prospective studies are indicated to evaluate the clinical utility of this model.
INTRODUCTION
Background
Sepsis remains a significant public health burden, with more than 1,700,000 cases diagnosed in the United States each year.1 Mortality associated with sepsis remains high, particularly for individuals who develop organ failure and shock, despite considerable investment in improving care for these patients. The majority of cases of severe sepsis or septic shock are identified at or near triage in the emergency department (ED). However, approximately 10% of patients with a sepsis syndrome progress to septic shock after triage in the ED.2–4 Prior research has shown that progression to septic shock is associated with worse outcomes.5–7 Although significant efforts have been made to determine mortality risk in patients with sepsis in the ED, there has been little investigation into identifying which septic patients will progress to shock.8,9
Importance
Patients with delayed onset of septic shock from the ED have up to a 20% higher mortality rate compared with septic patients who do not develop shock or are initially admitted to the ICU.10,11 Earlier identification of patients at risk for progression to septic shock may thus help with appropriate triage and use of resources. We currently lack reliable models to identify patients at high risk of a delayed progression to septic shock. Prior investigations into this have been limited by small numbers, single-center design, or insufficient test characteristics.3,12 Machine-learning techniques allow a more data-driven and comprehensive approach to diagnosis and prognostication compared with traditional statistical methods. Machine-learning methods have been previously used in the detection of sepsis in a variety of clinical scenarios,13–15 with some providing explanations for model outputs, thus ensuring interpretability.15 Transfer learning, a machine-learning method that allows fine-tuning (the process of optimizing parameters in a neural network) of a previously trained model during external validation at new site, has the ability to improve site-specific test characteristics from a general model16 and has not yet been described in the ED, to our knowledge.
Goals of This Investigation
The primary objective of this multicenter cohort study was to describe the use of machine-learning techniques to predict the development of delayed septic shock from a cohort of ED septic patients with end-organ damage (defined as severe sepsis according to Centers for Medicare & Medicaid Services [CMS], or the Sepsis-3 international definition); and demonstrate the feasibility of transfer learning to show improved performance and generalizability at a second clinical site.
MATERIALS AND METHODS
Study Design and Setting
This was a retrospective multicenter cohort study of all adult patients (≥ 18 years) who were admitted from the ED with sepsis between 2014 and 2019 from 2 large urban academic health centers, the University of California–San Diego and Emory University in Atlanta. Institutional review board approval of the study was obtained at both sites with a waiver of informed consent. Emory University has an estimated 192,500 annual ED visits, whereas the University of California–San Diego has an estimated 70,000 annual ED visits. Throughout the article, we refer to the respective hospital systems as the development and the validation cohorts. Within in cohort, we refer to 2 subgroups: a training subgroup and a testing subgroup.
Selection of Participants
All adult patients who were admitted to the hospital from the 2 EDs during the study period were evaluated for suspected sepsis by automated query of the electronic medical record (Epic, Verona, WI; and Cerner, Kansas City, MO). Data were abstracted into a clinical data repository (MicroStrategy, Tyson Corner, VA) and included vital signs, laboratory values, Sequential [Sepsis-related] Organ Failure Assessment (SOFA) scores, comorbidity data (including Charlson Comorbidity Index scores), length of stay, and outcomes. We used 2 definitions of sepsis: the CMS criteria for severe sepsis (“CMS severe sepsis”) and the most recent international consensus criteria for assessment of sepsis (Sepsis-3) from electronic health records.17–19 We chose to focus on CMS severe sepsis as the primary criteria because they can be calculated in real time and are a national quality metric. Sepsis-3 data were used for sensitivity analysis. Additionally, we chose severe sepsis rather than simple sepsis from the CMS guidelines because patients meeting the criteria have an end-organ damage profile similar to that of the sepsis definition in Sepsis-3. We defined CMS severe sepsis as the time at which the patient had a culture taken with subsequent antibiotic administration (excluding prophylactic use), the presence of 2 of 4 systemic inflammatory response syndrome criteria, and evidence of organ dysfunction (eg, lactate level >2 mmol/L) as defined in the most recent CMS Severe Sepsis and Septic Shock Management Bundle guidelines. The onset of CMS severe sepsis (t-sepsis) was the latest time stamp associated with these 3 events, all of which had to occur within a 6-hour window while the patient was in the ED. Sepsis-3 was defined according to the clinical operationalization of the 2016 international consensus definition as the first of 2 points: suspected infection and the presence of end-organ damage.18 The suspicion of sepsis was defined as blood culture tests and antibiotic initiation (for at least 3 days, excluding prophylactic use) within 24 or 72 hours, depending on whether culturing or antibiotic administration occurred first, respectively. End-organ damage was defined by the 2016 international definition of sepsis as a 2-point increase in the subject’s SOFA score. This included the time of a 2-point increase in the SOFA score from up to 24 hours before to up to 12 hours after the suspicion of sepsis (time of a 2-point increase in the SOFA score+24 hours>suspicion of sepsis>time of a 2-point increase in the SOFA score–12 hours). The time of septic shock was defined as the earliest point a patient met criteria for sepsis and had use of a titratable vasoactive medication (eg, norepinephrine).
As with prior research into the identification of patients who develop delayed septic shock after the diagnosis of sepsis, we included patients who developed septic shock between 4 hours after ED triage up to 48 hours after diagnosis of severe sepsis.8 However, for the purpose of prediction of shock, the machine-learning algorithm made predictions only from the time of severe sepsis to time of shock or end of ED visit. We excluded patients who were discharged from the ED, those with an ED length of stay less than 3 hours, and those who developed septic shock within 4 hours of ED triage. Patients who developed severe sepsis after transferring out of the ED were also excluded.
Similar to PhysioNet Sepsis Challenge 2019, we included a total of 40 most commonly measured input variables from the electronic health record for model development (Table E1, available online at http://www.annemergmed.com).20 Data sampling began at the first measurement in the ED. Data were sampled hourly and handling of multiple measurements within an hour or missing values was performed similar to that in our previously published work.15 All data used were available to emergency physicians at or ahead of model entry. Values for static demographic information were kept constant for all time bins in an encounter. The median value was used for time-variant predictor features such as vital signs and laboratory tests if a bin contained multiple values. Feature values from prior bins were retained (sample-and-hold interpolation) if data were missing in new bins. Mean values were imputed for features that were missing when there were no prior values. All population-level statistics (eg, mean values for features that were missing) were calculated on development site training data, and were fixed and then applied to the testing data and validation site data. Table E1 (available online at http://www.annemergmed.com) includes the frequencies of missing data of input features. For each input feature, we provide the percentage of missingness during a 24-hour interval, divided into 24 bins, each 1 hour in duration. An input feature (eg, total bilirubin level) checked only once within the first 24 hours would be present 4.1% of the time (1/24), or, alternatively, be missing 95.9% of the time. Prediction of septic shock started when CMS severe sepsis was identified. In the analysis using the Sepsis-3 definition of sepsis, prediction of septic shock began when the patient met criteria for sepsis.
Data were then used to train a modified Weibull-Cox proportional hazards model (the Artificial Intelligence Sepsis Expert) to predict the onset of septic shock hourly, starting from the time of sepsis up to the time of shock. The modification to the Artificial Intelligence Sepsis Expert model was the inclusion of a 2-layer feed-forward neural network (of size 10 each) before calculations of risk score by the Weibull-Cox model. The neural network was introduced to capture multiplicative risk factors (eg, age and temperature, in which in elderly patients hypothermia may be more of a risk factor than in young adults). Both the neural network and the Artificial Intelligence Sepsis Expert parameters were initialized randomly and jointly optimized with an algorithm known as gradient descent.21
During external evaluation, the pretrained Artificial Intelligence Sepsis Expert model derived from data only at the development cohort was fine-tuned on the training subgroup at the validation site, using 20 iterations of the gradient descent algorithm. The Artificial Intelligence Sepsis Expert model was then evaluated on the testing subgroup of the validation cohort. The purpose of the external evaluation step was to show that the model can be tailored to the characteristics of each local population and provide accurate predictions. This approach falls under the framework of transfer learning in machine-learning literature and has been shown to improve prediction performance as opposed to a model trained from scratch on external cohorts, in particular when only limited data are available for training.22 In this framework, knowledge gained by solving a classification problem on the development cohort is stored in the weights of the neural network and is carried forward to the target cohort (Figure 1). As such, learning of an accurate and generalizable classifier on the target cohort can be achieved with fewer training data. This approach has been used to fine-tune a neural network model for detection of diabetic retinopathy in retinal fundus photographs.23
Figure 1.
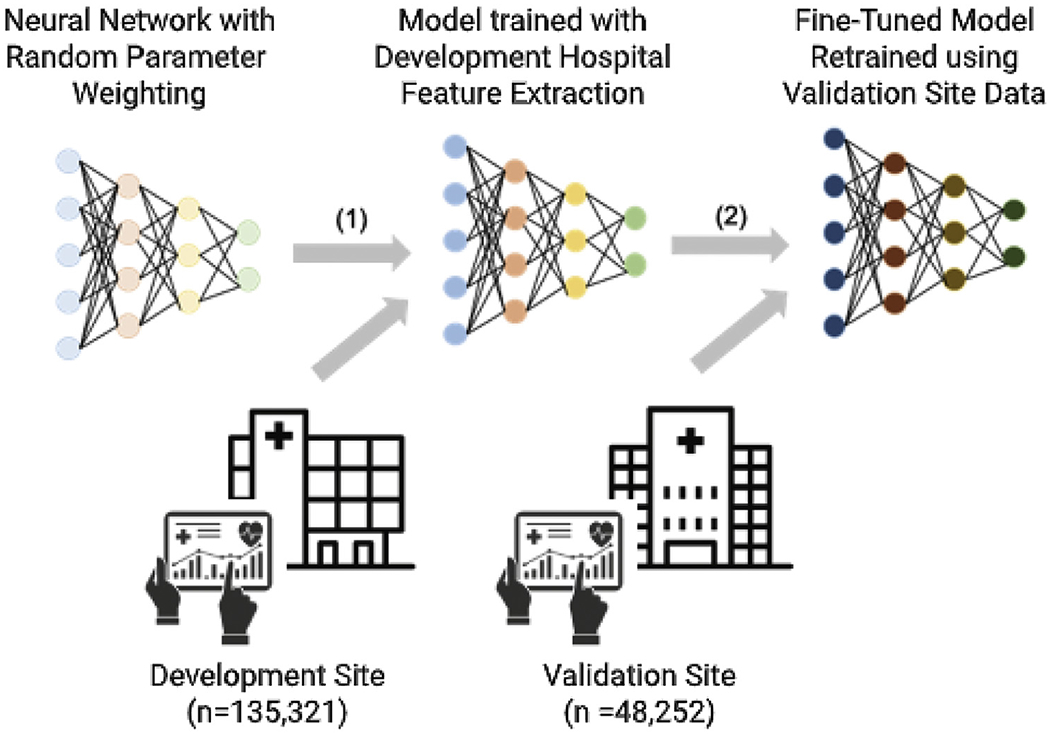
Transfer learning as implemented in this study. In part 1, a prediction model neural network (such as Artificial Intelligence Sepsis Expert) is initialized with random weights and is trained with data from the development site. In part 2, starting with a pretrained model from the development site, the model weights are fine-tuned (or retrained) with a relatively small amount of data from the validation site before application to the validation cohort. This procedure is called transfer learning.
Data Collection and Processing and Primary Data Analysis
Features in the development cohort set first underwent normality transformations and then were standardized by subtracting the mean and dividing by SD. All the remaining data sets were normalized with the mean and SD computed from the development cohort training set. All continuous variables are reported as medians with 25% and 75% interquartile ranges. Binary variables are reported as percentages. We used 10-fold bootstrap cross validation with an 80% to 20% random split within each fold for training and testing subgroups. We report median and interquartile values of the performance statistics, including area under the curve (AUC) and specificity (reported at 85% sensitivity) on both the training and testing cohorts. We elected to use a sensitivity of 85% for our model to reflect the need of a screening tool. For the derivation and validation sites, Artificial Intelligence Sepsis Expert classification for 8-, 12-, 16-, 24-, and 36-hour prediction horizons are reported with AUC receiver operating characteristic (ROC) statistics for both the training and the testing folds, as well as specificity (1–false alarm rate).
RESULTS
Characteristics of Study Subjects
A total of 135,321 patients were admitted to the hospital from the first hospital system (the development cohort) and 48,252 patients were admitted from the second hospital system site (the validation cohort) during the study period. Of these, 8,499 patients met criteria for CMS severe sepsis in the development cohort and 6,409 in the validation cohort. We identified 643 patients with septic shock in the development cohort and 715 with septic shock in the validation cohort, of whom 365 (4.3%) and 358 (5.9%) developed septic shock 4 hours after triage and 48 hours after development of severe sepsis, respectively (Figure 2). Patients in the development cohort who developed delayed septic shock had similar age (66 years versus 66 years), higher SOFA scores (3.8 versus 2.6), and higher Charlson Comorbidity Index scores (4.0 versus 3.0) than patients who did not develop delayed septic shock (Table 1). Similar trends were noted within the validation cohort. The median time from sepsis to the development of shock in the development cohort was 9.9 hours (interquartile range 5.5 to 18.8 hours) and 7.6 hours (interquartile range 4.8 to 13.7 hours) in the validation cohort. Inpatient mortality of patients with CMS severe sepsis, but without shock, and those with delayed septic shock in the development cohort were 5.5% and 24.9%, respectively. Likewise, inpatient mortality in the validation cohort with only CMS severe sepsis and those with delayed septic shock was 5.5% and 24.3%, respectively. Patients in the development cohort had higher rates of transfers to inpatient hospice care (6.3% and 10.3%) (for patients with severe sepsis and delayed septic shock) than those in the validation cohort (0.7% and 1.7%, respectively). Table E1 (available online at http://www.annemergmed.com) provides data for similar demographic information using Sepsis-3 definitions for the development and validation cohorts for model development using this definition.
Figure 2.
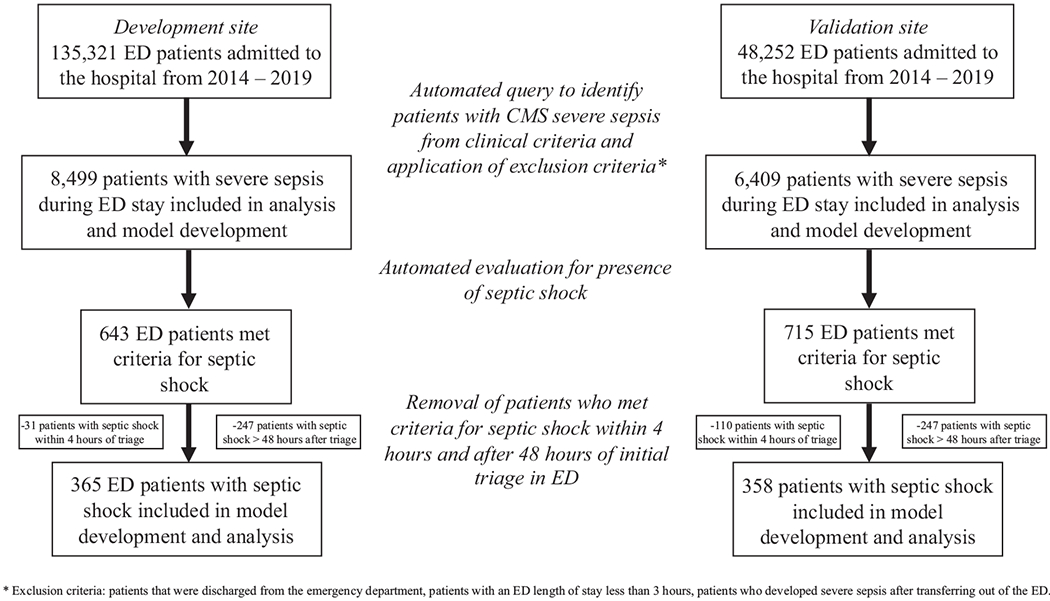
Inclusion of subjects used in the development and validation cohorts using CMS sepsis definitions.
Table 1.
Demographic comparisons of the derivation and validation sites using CMS severe sepsis definition.*
| Development Site After 4 Hours From ED Arrival Until 48 Hours After T0 of Severe Sepsis |
Validation Site After 4 Hours From ED Arrival Until 48 Hours After T0 of Severe Sepsis |
|||
|---|---|---|---|---|
| Demographics | Severe Sepsis Without Shock† | Severe Sepsis With Shock‡ | Severe Sepsis† Without Shock | Severe Sepsis With Shock‡ |
| Patients, n | 4,951 | 365 | 4,403 | 358 |
|
| ||||
| Age (SD), y | 62 (48-73) | 65 (54-74) | 60.5 (47.4-72.1) | 62.2 (53.1-72.9) |
|
| ||||
| Men, % | 53.2 | 49.3 | 57.2 | 57.3 |
|
| ||||
| Race, % | ||||
| White | 42.5 | 44.7 | 53.7 | 56.4 |
| Black | 50.9 | 50.7 | 9.5 | 9.8 |
| Asian | 3.2 | 1.6 | 8.9 | 6.9 |
| Other | 3.4 | 3.0 | 27.9 | 26.9 |
|
| ||||
| ICU LOS (IQR), h | 0 (0-46.5) | 95.1 (51.3-195.5) | 0 (0-16.5) | 87.9 (46.1-169.6) |
|
| ||||
| ED LOS (IQR), h | 6.5 (5.0-8.8) | 5.9 (4.6-7.9) | 9.4 (6.7-16.1) | 8.2 (6.2-11.2) |
|
| ||||
| CCI score, No. (IQR) | 4 (2-6) | 4 (2-7) | 5 (3-8) | 5 (3-8) |
|
| ||||
| SOFA, n (IQR) | 2.9 (1.8-4) | 3.8 (2-5.2) | 1.6 (0.5-2.8) | 2.8 (3.4-2) |
|
| ||||
| Inpatient mortality, % | 5.5 | 24.9 | 5.5 | 24.3 |
|
| ||||
| Transfers to hospice, % | 6.3 | 10.9 | 0.7 | 1.7 |
|
| ||||
| Time from ED admission to shock§ (IQR), h | – | 11.9 (7.0-21.0) | – | 10.0 (6.8-16.9) |
|
| ||||
| Time from sepsis to shock (IQR), h | – | 9.9 (5.5-18.8) | – | 7.6 (4.8-13.7) |
|
| ||||
LOS, Length of stay; IQR, interquartile range; CCI, Charlson Comorbidity Index.
Patients from the overall cohort were included in the study cohorts (derivation and validation cohorts) if they met all of the following criteria: (1) met sepsis criteria according to CMS guidelines, defined as the time at which the patient had a culture taken with antibiotic administration of at least 3 days, and met systemic inflammatory response syndrome criteria, and had a lactate level greater than 2 mmol/L or other signs of organ dysfunction, as defined in the CMS guidelines18); (2) were admitted to the ED for at least 3 hours and no more than 7 days; (3) developed shock after the first 4 hours of ED admission, if applicable; and (4) developed shock after 2 hours of developing sepsis, if applicable.
Sepsis is defined as the time at which the patient had a culture taken with antibiotic administration of at least 3 days, and met systemic inflammatory response syndrome criteria, and had a lactate level greater than 2 mmol/L or other signs of organ dysfunction, as defined in the CMS guidelines. The time of sepsis onset was the earliest time stamp associated with these events.
Septic shock was defined as patients with sepsis who required vasopressor administration.
Time until septic shock, defined as the time of vasopressor administration in an individual with sepsis.
The AUC ROC at various prediction windows after ED sepsis triage to detect delayed septic shock at the development site and validation site is provided in Figure 3. Regardless of definition of sepsis, the AUC ROC was 0.80 or higher up to 12 hours in advance in the development cohort. The AUC ROC of the ability of the algorithm to predict delayed septic shock at the validation site was greater than 0.8 at 8 and 12 hours with both the Sepsis-3 and CMS definitions. The specificity of the algorithm at various prediction windows is also provided in Figure 3 for both the development and validation cohorts at 8, 12, 16, 24, and 36 hours. We found that as the prediction window was extended past 16 hours, the AUC ROC in both the development and validation cohorts decreased, regardless of definition of sepsis used. Additionally, the AUC ROC of the development cohort was greater than that of the validation cohort at almost all prediction windows.
Figure 3.
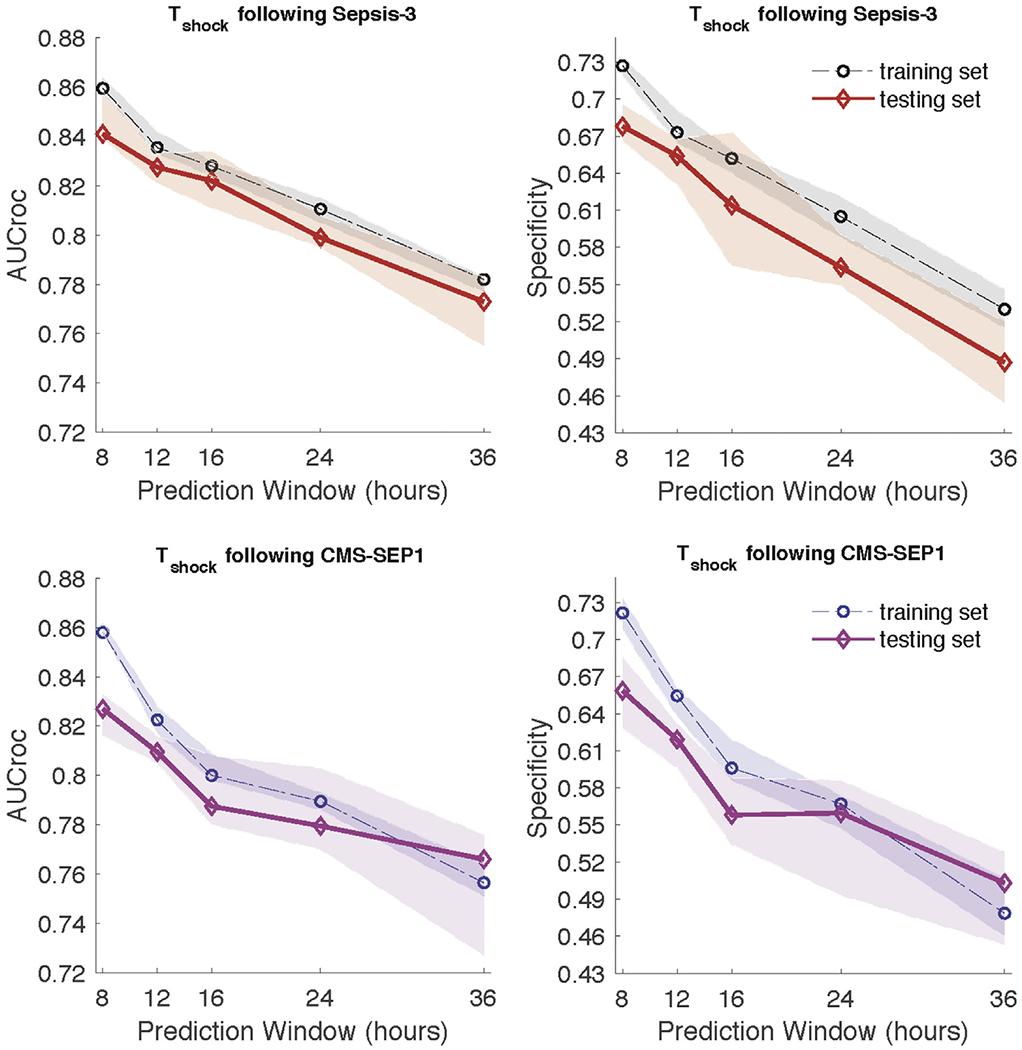
Comparison of AUC and specificity (calculated at 85% sensitivity) of models predicting septic shock at different prediction windows at the validation site. Ten-fold cross validation was performed and median and interquartile ranges are presented with lines and shaded areas, respectively.
The most weighted input features of the algorithm, in order of importance for the detection of septic shock, were systolic blood pressure, blood urea nitrogen level, respiratory rate, temperature, and change in blood pressure in the cohort of patients when CMS sepsis definitions were used. We used a collection of vital sign measurements at a given time (with carry-forward or sample-and-hold values for missing values) to make a prediction, and then moved forward by 1 hour and make another prediction. Table 2 provides the remainder of the top 20 most important features for the prediction of septic shock in the development cohort and the corresponding change in the AUC ROC if removed from the model. In the cohort of patients identified with Sepsis-3, the 5 most important variables were, in order, systolic blood pressure, respiratory rate, pulse rate, blood urea nitrogen, and diastolic blood pressure. Table E2 (available online at http://www.annemergmed.com) provides the top 20 most important features for the prediction of septic shock and corresponding change in AUC ROC if removed from the model.
Table 2.
Top 20 most important features in septic shock prediction.
| No. | Feature |
|---|---|
| Development cohort and corresponding change in AUC ROC if removed | |
| 1 | SBP (0.062) |
| 2 | BUN (0.019) |
| 3 | Resp rate (0.012) |
| 4 | Temp (0.008) |
| 5 | ΔSBP (0.007) |
| 6 | Hct (0.007) |
| 7 | WBC (0.007) |
| 8 | Lactate (0.007) |
| 9 | Creatinine (0.007) |
| 10 | HR (0.006) |
| 11 | Calcium (0.004) |
| 12 | DBP (0.003) |
| 13 | Potassium (0.002) |
| 14 | O2 sat (0.001) |
| 15 | ΔO2 sat (0.001) |
| 16 | ΔTemp (0.001) |
| 17 | MAP (0.001) |
| 18 | Δetco2 (0.001) |
| 19 | ΔBase excess (0.001) |
| 20 | ΔHCO3 (0.001) |
|
| |
| Validation cohort | |
| 1 | SBP (0.049) |
| 2 | HR (0.015) |
| 3 | Temp (0.015) |
| 4 | BUN (0.013) |
| 5 | Hct (0.011) |
| 6 | Lactate (0.009) |
| 7 | Resp rate (0.005) |
| 8 | Bilirubin total (0.005) |
| 9 | Creatinine (0.003) |
| 10 | DBP (0.003) |
| 11 | Alkaline phos (0.003) |
| 12 | Magnesium (0.002) |
| 13 | Age (0.002) |
| 14 | Time in ED (0.002) |
| 15 | ΔSBP (0.001) |
| 16 | pH (0.001) |
| 17 | ΔBUN (0.001) |
| 18 | ΔWBC (0.001) |
| 19 | ΔFibrinogen (0.001) |
| 20 | Men (0.001) |
SBP, Systolic blood pressure; Resp,, respiratory; Temp, temperature; HR, pulse rate; DBP, diastolic blood pressure; O2 sat, oxygen saturation; ETCO2, end tidal carbon dioxide; HCO3, bicarbonate.
The numbers in parentheses show the amount of change in AUC if a feature is treated as missing in the model.
Table E3 (available online at http://www.annemergmed.com) provides the mortality rate and percentage of patients who underwent transition to comfort measures according to the algorithm’s prediction for the development of delayed septic shock with a prediction window of 12 hours or less in the development cohort. As the probability of delayed septic shock increased, so did the chance of mortality or transition to hospice, particularly if the probability was greater than 0.8 when combined hospice and mortality rates were 22.6%.
There were only 18 false-negative predictions by the algorithm (ie, patients with septic shock who were misclassified as not having this), as shown in Table E3 (available online at http://www.annemergmed.com). Among these 18 patients, all had probabilities of developing delayed septic shock less than 0.4 and none died or were transitioned to hospice. In other words, when the algorithm said the patient was not at risk for septic shock, there was a very high likelihood that the person was actually not at risk of septic shock and, even if misclassified, had a very low chance of mortality. Patients with a false-positive prediction (ie, when the algorithm predicted septic shock, but the patient did not develop it) had a high combined mortality and hospice rate, particularly with a probability between 0.8 and 1.0 (21.3%). So even if patients may not develop shock within the 48-hour window, they are more likely to die and merit physician evaluation.
The AUC ROC in the development cohort at 12 hours in the training and testing subgroups was 0.822 and 0.833, respectively, for patients with CMS severe sepsis (Figure 4). At the 12-hour prediction window in the validation cohort, before transfer learning, the AUC was 0.778 (sensitivity 0.85, specificity 0.549) in the cohort of patients with CMS severe sepsis. The positive predictive value of the algorithm is also provided in Figure 4. Negative predictive value of the algorithm for the prediction of delayed septic shock was greater than 0.99. At the same prediction window and with the same patient population after applying transfer techniques, we found an AUC ROC of 0.85 (sensitivity 0.85, specificity 0.678) in the testing subgroup at the validation site (DeLong test, P<.001). The 5 most important features in the algorithm after transfer learning was performed were systolic blood pressure, pulse rate, temperature, blood urea nitrogen level, and hematocrit level. The remainder of the top 20 most important input features with corresponding AUC change are provided in Table 2. We then evaluated the AUC ROC in the development cohort at a 12-hour prediction window in the training at testing subgroups with Sepsis-3 definitions. Using a 12-hour prediction window, we found that the AUC ROC in the training subgroup of the validation cohort before transfer learning was 0.792. After the application of transfer learning in the training subgroup, the AUC ROC was 0.838 (P <.001 from the DeLong test). In the cohort of patients identified with Sepsis-3, the 5 most important variables were, in order, systolic blood pressure, respiratory rate, pulse rate, blood urea nitrogen level, and diastolic blood pressure. Table E2 (available online at http://www.annemergmed.com) provides the top 20 most important features for the prediction of septic shock and corresponding change in AUC ROC if removed from the model.
Figure 4.
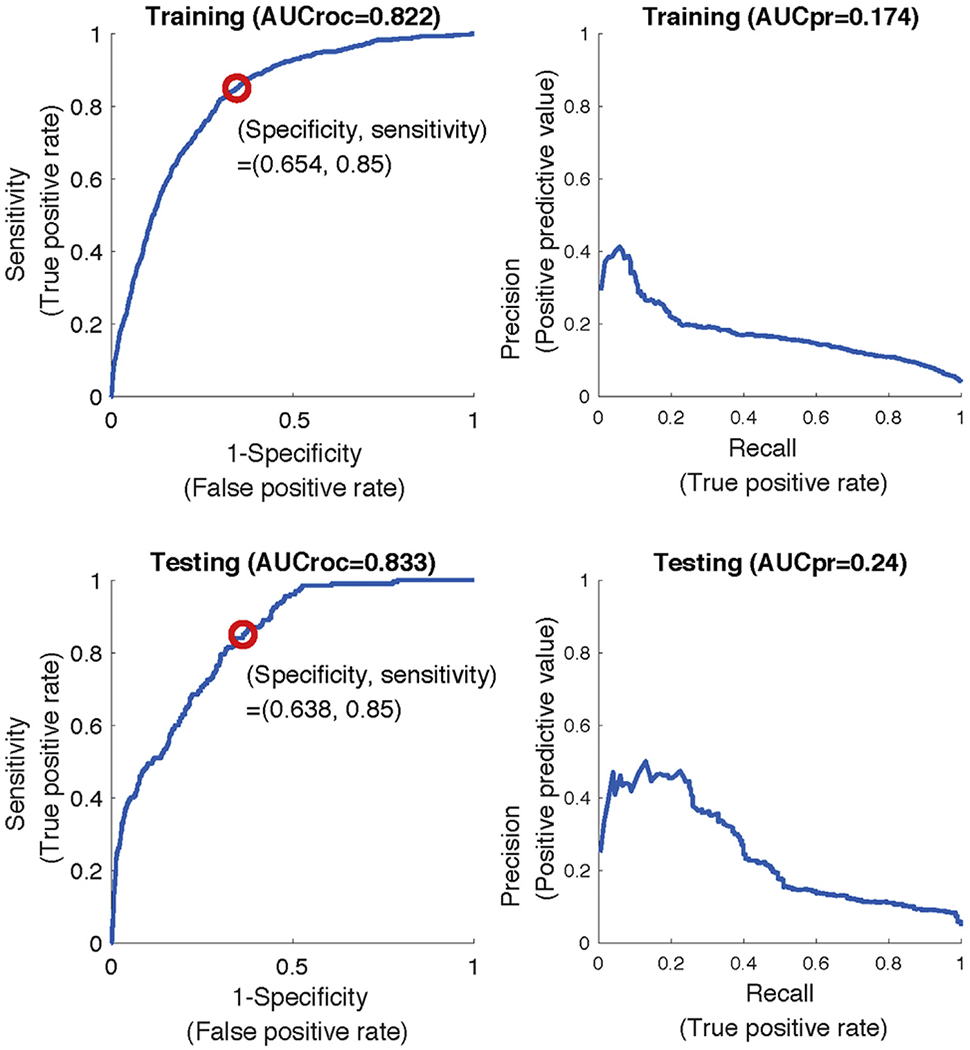
AUC ROC and precision-recall curves for predicting t-sepsis 12 hours in advance at the development site using CMS severe sepsis definition. AUCroc, Area under the receiver operating characteristic; AUCpr, AUC precision-recall curve.
Figure 5 shows the corresponding differences in the AUC ROC for prediction of delayed septic shock at 12 hours with the Artificial Intelligence Sepsis Expert algorithm trained from scratch at the validation site compared with the algorithm developed with transfer learning with the initial model trained at the development site, using an increased number of patient encounters in model development.
Figure 5.
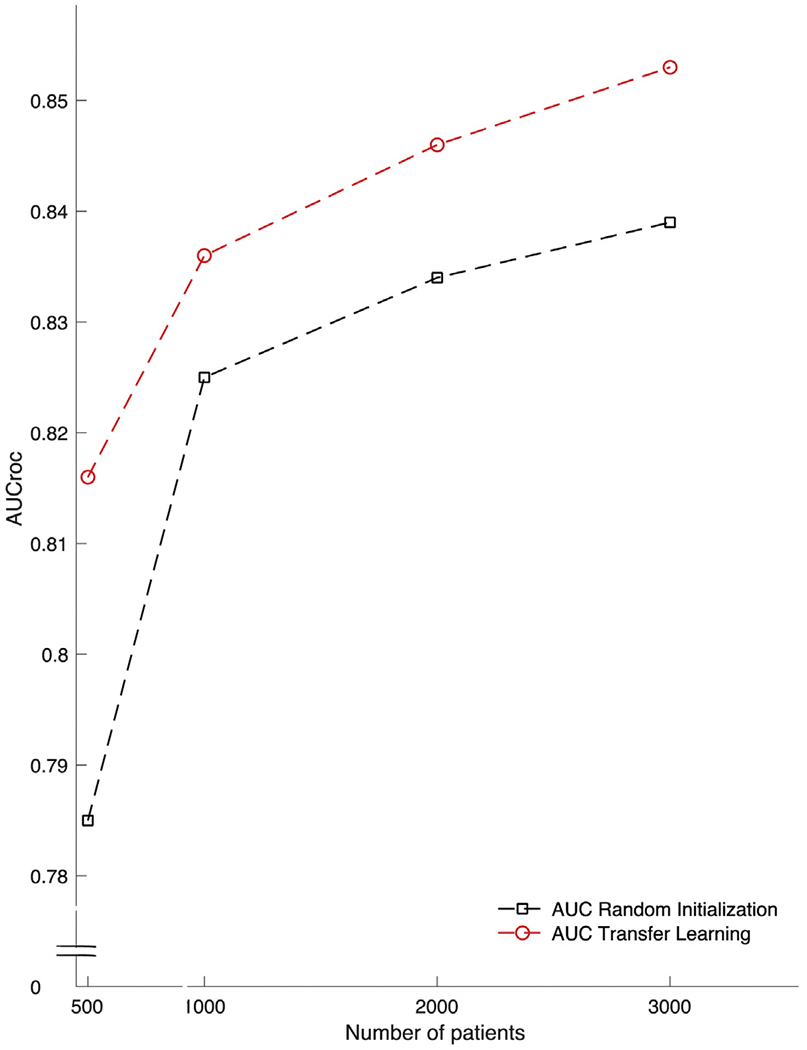
AUC ROC of the ability of the Artificial Intelligence Sepsis Expert algorithm to detect septic shock 12 hours ahead of time in the validation cohort with and without transfer learning (red and black dashed lines, respectively) based on increasing amounts of patient encounters in model development.
LIMITATIONS
We report results from a single validation site, and although we are confident in the use of transfer learning to improve external validity, results should be tested on a more geographically diverse patient population. The definitions for sepsis, severe sepsis, and septic shock used in this article are based on previously described criteria from CMS and international guidelines and have been optimized for analysis of large clinical databases, but accurate diagnosis of sepsis often requires clinical chart view and adjudication by multiple reviewers.24 Additionally, we did not include patients who had simple sepsis (eg, 2/4 systemic inflammatory response syndrome+suspected infection) without any end-organ damage and thus may have not included some patients who developed delayed septic shock without preceding severe sepsis. Patient populations and the diagnostic approach to sepsis may change in the coming years and such algorithms may require revisions, although the proposed transfer-learning framework is well suited for model fine-tuning, and this forms the basis of future studies. Furthermore, we limited the number of input features to 40 most commonly measured vital signs and laboratory tests. Although this was done to increase portability of the algorithm and allow seamless electronic medical record agnostic deployment by Health Level Seven standard protocols, we acknowledge there are additional variables that could be used to improve the prediction of delayed septic shock. Smaller clinical sites may lack experience or the infrastructure at this point for implementation of such algorithms. However, major electronic health record distributors are interested in such technologic advances and may make implementation easier for interested sites. As we have shown in Figure 5, clinical sites with fewer patients benefit more from model development with transfer learning. Finally, application of this study to broader clinical use will require further validation with prospective, randomized clinical trials assessing patient-centered outcomes.25
DISCUSSION
We report a retrospective multicenter study that demonstrates the feasibility of a machine-learning algorithm to predict the development of delayed septic shock from 135,321 patients and multiple definitions of sepsis. We showed that the machine-learning algorithm can provide excellent predictive characteristics at a separate site through the use of transfer learning. The clinical implications of this are multiple. First, we highlighted the need to identify patients at high risk for delayed decompensation because these patients have a significant increase in mortality compared with those who do not progress to septic shock. We then showed favorable test characteristics of machine-learning algorithms for the identification of patients at high risk of delayed septic shock. Finally, we provided evidence that transfer learning can be used in the ED to boost test characteristics of machine-learning algorithms at external sites and improve generalizability and portability. These results can help inform the development and implementation of future machine-learning algorithms to predict the development of conditions or outcomes that are inherently complex and challenging for physicians in the ED.
We have shown a significantly higher inpatient mortality in patients with delayed septic shock (24.6%) than in those with CMS severe sepsis but without shock (5.5%). Prior investigations demonstrated similar findings across a variety of conditions.10,26,27 Although this mortality difference is well described, the ability to identify these patients in the ED has been limited by single-center design and small sample sizes of previous studies. Capp et al3 evaluated a cohort of patients (1,336) who progressed to septic shock (111 patients) at least 4 hours after ED arrival and found that female sex, nonpersistent hypotension, bandemia, lactate level greater than 4.0 mmol/L, and history of coronary artery disease were associated with delayed presentation of septic shock. Holder et al28 reported that in a cohort of 582 patients, serum albumin level less than 3.5 g/dL and triage diastolic blood pressure less than 52 mm Hg were also associated with decompensation in 108 patients. Although these findings are important, the clinical utility is challenging to implement because a majority of patients will meet at least one of the above criteria and will likely not progress to septic shock. Use of early warning systems (eg, Modified Early Warning Score, National Early Warning Score, and VitalPAC early warning score) for the prediction of adverse events from patients with sepsis has been described in the literature.29–32 These scoring systems have performed better than the quick SOFA and systemic inflammatory response syndrome for the prediction of adverse events, but their use has been limited by moderate test performance characteristics in septic patients.12
The field of machine learning refers to a subset of artificial intelligence that automates analytic model building to identify patterns in data to predict outcomes. In particular, machine-learning algorithms are powerful tools for the detection of complicated and nonlinear outcomes when traditional statistical methods (eg, linear regression, recursive partitioning) are overrun by a large number of variables. Recently, machine-learning algorithms have shown superior test characteristics in the prediction of sepsis in the ED and may decrease mortality.13,14 The Artificial Intelligence Sepsis Expert algorithm used in this study has previously been shown to have good performance in the detection of sepsis in the ICU at various prediction windows (4, 6, 8, and 12 hours), using 65 readily available variables that yield easily interpretable scores and can be calculated in real time.15 Using the same algorithm, but with decreased number of variables for better portability across hospital systems, we found that regardless of the definition of sepsis used, an AUC ROC greater than 0.8 for the prediction of delayed septic shock was found for both the development and validation site up to 12 hours in advance. We found that the majority of patients who experienced a delay in the development of septic shock did so at a median of 7.6 hours (validation site) and 9.9 hours (development site), the majority of which occurred after transfer out of the ED.
Given the excellent test characteristics of the Artificial Intelligence Sepsis Expert algorithm at these points, the utility of our findings is clinically relevant because implementation may decrease unanticipated ICU transfers, prompt a change in management (eg, fluids, broadening of antibiotics), and raise situational awareness of physicians for these high-risk patients. We also found that the false-positive predictions by the Artificial Intelligence Sepsis Expert algorithm (patients predicted to develop septic shock but who did not) had mortality and transition to hospice rates similar to true-positive predictions. This was maintained across sites that had a significant difference in the proportion of septic patients transferred to hospice care. Although inappropriate identification for delayed septic shock worsens test characteristics of the Artificial Intelligence Sepsis Expert algorithm, these patients likely benefit from additional evaluation and potential interventions to prevent decompensation and potentially death.
Transfer learning is a technique used in machine learning to translate patterns and extracted features learned in one setting and generalize those patterns in another setting. This allows machine-learning models developed from a large robust data set to be fine-tuned onto a more sparse data set.33 We found that the use of transfer-learning techniques significantly improved the AUC ROC for the prediction of delayed septic shock at a hospital system geographically distinct and consisting of different patient populations with an algorithm trained at an external site (Figure 1). Although others have demonstrated similar results in medical applications of machine learning, to our knowledge, this is the first instance of a machine-learning algorithm using transfer learning for an application in the ED.34–36 Previously, decision rules used in emergency medicine have shown impressive test characteristics at the institutions or regions that developed them, but attempts at external validation have yielded lower sensitivity and specificity, limiting general use.37,38 Transfer-learning techniques are positioned to further adoption and generalizability of prediction rules and machine-learning algorithms in external environments, allowing portability of an algorithm from site to site while maintaining superior test characteristics that can be available in real time to physicians.33,39,40
Transfer learning is best applied when a source data set’s features and outcomes of interest are generalizable, as are the variables used by the Artificial Intelligence Sepsis Expert algorithm. Whereas some emphasis in machine learning has been focused on knowledge distillation from large implemented machine-learning algorithms to simpler models that can be applied broadly (for example, a rule-based decision tree), transfer-learning techniques allow complicated models such as the one evaluated in this study to be applied to clinical scenarios in more resource-limited environments with smaller target data sizes. One strong advantage of this particular implementation and algorithm is privacy and data security. Whereas previously reported methods commingle data from multiple institutions to yield stronger test characteristics, this approach may infringe on a covered entity’s autonomy over protected health care data and terms of patient data privacy.41–43 We report a machine-learning process that uses transfer learning in such a way that it affords hospital systems the ability to maintain data privacy and does not require a data transfer to a central location for fine-tuning, but rather can be completed at an individual site.
Patients who develop septic shock 4 hours after triage in the ED have in hospital mortality approximately 5 times that of those who do not progress to septic shock. The Artificial Intelligence Sepsis Expert algorithm was trained to provide excellent AUC ROC in identifying patients at risk of delayed development of septic shock, particularly at prediction windows of 8 to 12 hours. The most important features in the detection of delayed septic shock were systolic blood pressure, blood urea nitrogen level, respiratory rate, temperature, and change in blood pressure. Transfer learning was effective at augmenting prediction performance at a second, distinct clinical site. Future prospective studies are required to validate the use of such techniques in clinical practice.
Supplementary Material
Editor’s Capsule Summary.
What is already known on this topic
Septic shock causes high mortality and is best treated early, before entrenchment.
What question this study addressed
Can a machine-learning approach help predict who will develop septic shock?
What this study adds to our knowledge
Using data from 180,000 patients, 723 who eventually had septic shock, the authors derived and validated an algorithm using available clinical data that predicted progression with modest accuracy.
How this is relevant to clinical practice
If tested and found usable in practice, this could aid sepsis outcomes by allowing earlier action to prevent or mitigate shock.
Funding and support:
By Annals policy, all authors are required to disclose any and all commercial, financial, and other relationships in any way related to the subject of this article as per ICMJE conflict of interest guidelines (see www.icmje.org). Dr. Wardi is supported by the National Foundation of Emergency Medicine and funding from the Gordon and Betty Moore Foundation (GBMF9052). He has received speaker’s fees from Thermo-Fisher and consulting fees from General Electric. Dr. Holder is supported by the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1TR002378 and KL2TR002381). Dr. Nemati is funded by the National Institutes of Health (K01ES025445), National Science Foundation (1822378), Biomedical Advanced Research and Development Authority (HHSO100201900015C), and the Gordon and Betty Moore Foundation (GBMF9052).
REFERENCES
- 1.Rhee C, Dantes R, Epstein L, et al. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009-2014. JAMA. 2017;318:1241–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villar J, Clement JP, Stotts J, et al. Many emergency department patients with severe sepsis and septic shock do not meet diagnostic criteria within 3 hours of arrival. Ann Emerg Med. 2014;64:48–54. [DOI] [PubMed] [Google Scholar]
- 3.Capp R, Horton CL, Takhar SS, et al. Predictors of patients who present to the emergency department with sepsis and progress to septic shock between 4 and 48 hours of emergency department arrival. Crit Care Med. 2015;43:983–988. [DOI] [PubMed] [Google Scholar]
- 4.Wardi G, Wali AR, Villar J, et al. Unexpected intensive care transfer of admitted patients with severe sepsis. J Intensive Care. 2017;5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakr Y, Vincent JL, Schuerholz T, et al. Early- versus late-onset shock in European intensive care units. Shock. 2007;28:636–643. [PubMed] [Google Scholar]
- 6.Arnold RC, Sherwin R, Shapiro NI, et al. Multicenter observational study of the development of progressive organ dysfunction and therapeutic interventions in normotensive sepsis patients in the emergency department. Acad Emerg Med. 2013;20:433–440. [DOI] [PubMed] [Google Scholar]
- 7.Glickman SW, Cairns CB, Otero RM, et al. Disease progression in hemodynamically stable patients presenting to the emergency department with sepsis. Acad Emerg Med. 2010;17:383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor RA, Pare JR, Venkatesh AK, et al. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data-driven, machine learning approach. Acad Emerg Med. 2016;23:269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro NI, Wolfe RE, Moore RB, et al. Mortality in Emergency Department Sepsis (MEDS) score: a prospectively derived and validated clinical prediction rule. Crit Care Med. 2003;31:670–675. [DOI] [PubMed] [Google Scholar]
- 10.Chalfin DB, Trzeciak S, Likourezos A, et al. Impact of delayed transfer of critically ill patients from the emergency department to the intensive care unit. Crit Care Med. 2007;35:1477–1483. [DOI] [PubMed] [Google Scholar]
- 11.Huang CT, Tsai YJ, Tsai PR, et al. Severe sepsis and septic shock: timing of septic shock onset matters. Shock. 2016;45:518–524. [DOI] [PubMed] [Google Scholar]
- 12.Churpek MM, Snyder A, Han X, et al. Quick Sepsis-related Organ Failure Assessment, Systemic Inflammatory Response Syndrome, and Early Warning Scores for detecting clinical deterioration in infected patients outside the intensive care unit. Am J Respir Crit Care Med. 2017;195:906–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delahanty RJ, Alvarez J, Flynn LM, et al. Development and evaluation of a machine learning model for the early identification of patients at risk for sepsis. Ann Emerg Med. 2019;73:334–344. [DOI] [PubMed] [Google Scholar]
- 14.Shimabukuro DW, Barton CW, Feldman MD, et al. Effect of a machine learning–based severe sepsis prediction algorithm on patient survival and hospital length of stay: a randomised clinical trial. BMJ Open Respir Res. 2017;4:e000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemati S, Holder A, Razmi F, et al. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med. 2018;46:547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kermany DS, Goldbaum M, Cai W, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–1131.e9. [DOI] [PubMed] [Google Scholar]
- 17.Rhodes A, Evans LE, Alhazzani W, et al. Surviving Sepsis Campaign: international guidelines for management of sepsis and septic shock: 2016. Crit Care Med. 2017;45:486–552. [DOI] [PubMed] [Google Scholar]
- 18.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315:801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Medicare and Medicaid Services. Severe Sepsis and Septic Shock: Management Bundle (Composite Measure). Available at: https://cmit.cms.gov/CMIT_public/ViewMeasure?MeasureId=1017. Accessed April 30, 2020.
- 20.Reyna MA, Josef CS, Jeter R, et al. Early prediction of sepsis from clinical data: the PhysioNet/Computing in Cardiology Challenge 2019. Crit Care Med. 2020;48:210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313:504–507. [DOI] [PubMed] [Google Scholar]
- 22.Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. [DOI] [PubMed] [Google Scholar]
- 24.Rhee C, Brown SR, Jones TM, et al. Variability in determining sepsis time zero and bundle compliance rates for the Centers for Medicare and Medicaid Services SEP-1 measure. Infect Control Hosp Epidemiol. 2018;39:994–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz Rivera S, Liu X, Chan AW, et al. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Nat Med. 2020;26:1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Churpek MM, Wendlandt B, Zadravecz FJ, et al. Association between intensive care unit transfer delay and hospital mortality: a multicenter investigation. J Hosp Med. 2016;11:757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rincon F, Morino T, Behrens D, et al. Association between out-of-hospital emergency department transfer and poor hospital outcome in critically ill stroke patients. J Crit Care. 2011;26:620–625. [DOI] [PubMed] [Google Scholar]
- 28.Holder AL, Gupta N, Lulaj E, et al. Predictors of early progression to severe sepsis or shock among emergency department patients with nonsevere sepsis. Int J Emerg Med. 2016;9:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleuren LM, Klausch TLT, Zwager CL, et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020;46:383–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prytherch DR, Smith GB, Schmidt PE, et al. ViEWS—towards a national early warning score for detecting adult inpatient deterioration. Resuscitation. 2010;81:932–937. [DOI] [PubMed] [Google Scholar]
- 31.Gardner-Thorpe J, Love N, Wrightson J, et al. The value of Modified Early Warning Score (MEWS) in surgical in-patients: a prospective observational study. Ann R Coll Surg Engl. 2006;88:571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith GB, Prytherch DR, Meredith P, et al. The ability of the National Early Warning Score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation. 2013;84:465–470. [DOI] [PubMed] [Google Scholar]
- 33.Wiens J, Guttag J, Horvitz E. A study in transfer learning: leveraging data from multiple hospitals to enhance hospital-specific predictions. J Am Med Inform Assoc. 2014;21:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med. 2018;1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desautels T, Calvert J, Hoffman J, et al. Using transfer learning for improved mortality prediction in a data-scarce hospital setting. Biomed Inform Insights. 2017;9; 1178222617712994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Topiwala R, Patel K, Twigg J, et al. Retrospective observational study of the clinical performance characteristics of a machine learning approach to early sepsis identification. Crit Care Explor. 2019;1:e0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beam AL, Manrai AK, Ghassemi M. Challenges to the reproducibility of machine learning models in health care. JAMA. 2020;323:305–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein BA, Navar AM, Pencina MJ, et al. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc. 2017;24:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao C, Choi E, Sun J. Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review. J Am Med Inform Assoc. 2018;25:1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desautels T, Calvert J, Hoffman J, et al. Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Inform. 2016;4:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao Q, Jay M, Hoffman JL, et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open. 2018;8: e017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kayaalp M Patient privacy in the era of big data. Balkan Med J. 2018;35:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price WN 2nd, Cohen IG. Privacy in the age of medical big data. Nat Med. 2019;25:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


