Abstract
Prevalence of hepatitis B virus (HBV), hepatitis C virus (HCV), hepatitis G virus (HGV), and hepatitis E virus (HEV) was investigated among 574 healthy blood donors in Bolivia. HCV RNA and HGV RNA in the serum were identified by a nested reverse transcription-PCR using primers derived from the 5′ untranslated region (5′ UTR). We also tested for hepatitis B surface antigen (HBsAg) and for the antibody to HEV. The results revealed that HGV RNA was present in 84 of 574 (14.6%) tested blood donors, whereas HBsAg was detected in only 2 (0.3%) donors, and no individuals positive for HCV RNA were found. Anti-HEV immunoglobulin G (IgG) was detected in 93 (16.2%) individuals and anti-HEV IgM was found in 10 (1.7%) individuals among the same population. Phylogenetic analysis of 44 HGV isolates in the 5′ UTR showed that 27 (61%) isolates were genotype 3 (Asian type) and the remaining 17 (39%) isolates were genotype 2 (United States and European type). Moreover, we obtained a full-length nucleotide sequence of the HGV genome (designated HGV-BL230) recovered from a Bolivian blood donor. The BL230 was composed of 9,227 nucleotides and had a single open reading frame, encoding 2,842 amino acid residues. Interestingly, the BL230 belonged to genotype 2 of HGV at the level of a full-length sequence, although this was classified as genotype 3 by a phylogenetic analysis based on the 5′ UTR sequence. The BL230 differed from previously reported HGV/hepatitis GB virus type C isolates by 12 to 13% of the nucleotide sequence and 4% of the amino acid sequence. Our data indicate a high prevalence of HGV in native Bolivians, and the major genotype of HGV was type 3.
Viral hepatitis exists throughout the world and is a major global public health problem. Although sensitive and specific tests for the detection of known hepatitis viruses are available, other as-yet-unidentified hepatitis viruses may also be responsible for acute and chronic hepatitis. These viruses may or may not be related to known agents of hepatitis virus types A through E. In 1996, novel RNA viruses were identified from the sera of patients with liver disease by two different American groups: these possible agents have been named hepatitis G virus (HGV) and hepatitis GB virus type C (GBV-C), respectively (15, 34). Molecular characterization of these two agents has shown them to be different isolates of the same virus. They are single-stranded RNA viruses approximately 9.4 kb long with high homology (96%) at the amino acid level (39). The agents also have characteristics of a flavivirus-like genome as in the case of the hepatitis C virus (HCV); however, they represent a new genus in the family Flaviviridae, including flaviviruses and pestiviruses (34). The genetic distance between HGV/GBV-C and HCV is too far to consider HGV/GBV-C a different genotype of HCV. It is also known that HGV/GBV-C has genetic heterogeneity corresponding to the three major genotypes (types 1 to 3) so far identified; we recently identified a novel genotype of HGV (18, 19, 21, 25, 31). Although there have been extensive investigations of HGV/GBV-C since its discovery, the nature of HGV/GBV-C and its real pathogenic role remain controversial. To approach these problems, we are working on the molecular-based epidemiology of hepatitis viruses and their pathogenesis in different geographic regions. Here we report the prevalence of hepatitis viruses (including types B, C, G, and E) in Bolivia, where there has been no epidemiological information on HGV and hepatitis E virus (HEV) infections in the past.
MATERIALS AND METHODS
Study population.
We used serum samples obtained from 574 healthy blood donors in Bolivia. All were native Bolivians ranging in age from 17 to 56 years. The male/female ratio was 2.5:1. Most individuals who received a health checkup did not appear to have any serious health problems. The donors were residents of Santa Cruz, Bolivia, or its suburbs. Informed consent was obtained from each individual for participation in this study. These serum samples were collected between 1992 to 1998 and stored at −20°C or below until analysis.
Extraction of nucleic acids and detection of HCV RNA and HGV RNA.
RNA were extracted from 100 μl of serum by using the SepaGene RV-R kit (Sanko Junyaku Co., Ltd., Tokyo, Japan), precipitated with isopropanol, and washed in ethanol. The resulting pellet was resuspended in 50 μl of RNase-free water. The sequences of PCR primers were as follows. For HCV (5′ untranslated region [UTR]), the sequences were 5′-GCGACACTCCACCATAGAT-3′ (primer 19, sense primer, nucleotides [nt] 2 to 20) and 5′-GCTCATGGTGCACGGTCTA-3′ (primer 20, antisense primer, nt 312 to 330) for the outer primer pairs (329 bases) and 5′-CTGTGAGGAACTACTGTCT-3′ (primer 21, sense primer, nt 28 to 46) and 5′-ACTCGCAAGCACCCTATCA-3′ (primer 22, antisense primer, nt 277 to 295) for the inner primer pairs (268 bases). For HGV (5′ UTR), the sequences were 5′-GGTCGTAAATCCCGGTCACC-3′ (HG1, sense; nt 139 to 158) and 5′-CCCACTGGTCCTTGTCAACT-3′ (HG1R, antisense; nt 381 to 400) for the outer primer pairs (262 bases) and 5′-TAGCCACTATAGGTGGGTCT-3′ (HG2, sense; nt 163 to 182) and 5′-ATTGAAGGGCGACGTGGACC-3′ (HG2R, antisense; nt 331 to 350) for the inner primer pairs (188 bases). The nucleotide positions were deduced from HC-J1 (24a) for HCV and from HGV-PNF2161 (15) for HGV.
HCV RNA and HGV RNA were amplified by nested reverse transcription (RT)-PCR. Actually, the first PCR was combined with the RT step (one-step method) in the same tube as previously described (3). We used AmpliTaq Gold (Perkin-Elmer, Norwalk, Conn.) as a thermostable DNA polymerase for PCR to obtain an automatic hot-start reaction. Amplification conditions included preincubation at 95°C for 10 min followed by 40 cycles of the first-round PCR (94°C for 20 s, 55°C for 20 s, and 72°C for 30 s, with a final 7-min, 72°C extension). For the second-round PCR, the annealing temperature was set to 60°C instead of 55°C. PCR products were analyzed by electrophoresis on 2% agarose gels, stained with ethidium bromide, and observed under UV light.
Nucleotide sequencing and genotyping of HGV.
PCR products, estimated to consist of 188 bases and obtained by a combination of primers with HG2/HG2R in the 5′ UTR of HGV, were sequenced, and a phylogenetic analysis was performed in order to classify the genotypes. We also cloned to obtain full-length nucleotide sequencing of a Bolivian HGV isolate. For this purpose, overlapping PCR products were generated by using primers deduced from the prototype HGV-PNF2161. We used a serum sample obtained from a native Bolivian blood donor and designated it HGV-BL230. The BL230 was divided into 12 fragments (covering the full-length genome, but without the short 5′-terminal sequence only) as previously described (10). For the terminal sequence of the 3′ UTR, extracted RNA from serum was tailed with a polyadenosine [poly(A)] with poly(A) polymerase (TaKaRa Biochemicals, Tokyo, Japan) and then isolated with a rapid amplification of the cDNA ends (RACE) kit (5′/3′ RACE kit; Boehringer Mannheim), because HGV/GBV-C does not have a poly(A) tail. This method is based on a single-sided PCR. HGV cDNA was amplified by nested RT-PCR as described above. PCR products were separated by 1 to 2% agarose gel electrophoresis and purified using the QIAquick gel extraction kit (Qiagen Inc., Chatsworth, Calif.). The recovered PCR products from agarose gels were subcloned by using a pBluescript II SK(−) vector (Stratagene, La Jolla, Calif.) through the EcoRV site. We determined the sequence of three independent clones. Alternatively, purified PCR products for short sequencing in the 5′ UTR were subjected to direct sequencing from both directions using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer). Sequences of amplified cDNA were determined by using a sequencer (ABI model 373A; Applied Biosystems, Foster City, Calif.).
Reference sequences from database.
A total of 18 full or near-full genome sequences covering the open reading frame (ORF) of HGV/GBV-C isolates obtained from GenBank databases were used to compare the sequences of the isolates in the present study. The isolates, accession numbers, and references of the reported sequences were as follows: HGV-PNF2161 and HGV-R10291 from the United States, U44402 and U45966, respectively (15); GBV-C from West Africa, U36380 (13); GBV-C(EA) from East Africa, U63715 (8); HGV-GA128 from Ghana, AB013500 (30); HGV-C964 from China, U75356 (38); HGV Iw from Japan, D87255 (33); GT110 and GT230 from Japan, D90600 and D90601, respectively (24); GSI85 and GSI93 from Japan, D87262 and D87263, respectively (22); HGV IM71 from Japan, AB008342 (10); BG1HC, CG01BD, CG07BD, CG12LC, G05BD, G13HC from Japan, AB003288, AB003289, AB003290, AB003291, AB003292, and AB003293, respectively (36).
Phylogenetic analysis.
Nucleotide sequences were multiply aligned using CLUSTAL W, version 1.4. The distance matrix of the nucleotide substitutions in each clone was estimated by the eight-parameter method (29), and phylogenetic trees were constructed by the neighbor-joining method (32) from the matrix. These procedures were computed using PHYLO_WIN, version 1.2 (9) on a DEC alpha 2000 server, and the trees were drawn by TreeView, version 1.5 (26). The reliability and topology of each tree branch were tested by bootstrap analysis (6) of the data of 100 bootstrap resamplings of the columns in the 5′ UTR and full genome sequence alignment.
Assay for HBsAg and antibody to HEV.
Hepatitis B surface antigen (HBsAg) was assayed by the particle agglutination method (Serodia-HBs; Fujirebio Inc., Tokyo, Japan). Furthermore, immunoglobulin G (IgG) and IgM antibodies to HEV were measured by enzyme-linked immunosorbent assay (ELISA). The ELISA to detect anti-HEV using virus-like particles expressed by a recombinant baculovirus was performed as reported previously (14).
Statistical analyses.
Statistical analyses were performed by the chi-square test or Fisher’s exact test. A difference with a P value of <0.05 was considered significant.
Nucleotide sequence accession numbers.
The nucleotide sequences of Bolivian HGV isolates reported in this paper have been submitted to the DDBJ, EMBL, and GenBank databases under accession no. AB013501 for HGV-BL230, AB013190 for BL148, AB013191 for BL303, AB013194 for BL265, AB013195 for BL249, AB013196 for BL214, AB013216 for BL274, and AB013226 for BL285.
RESULTS
Prevalence of hepatitis virus infections.
HGV RNA was detected in 84 of 574 (14.6%) healthy native Bolivian blood donors. None of the HGV-infected individuals had an abnormality in their serum alanine aminotransferase levels. In contrast, no HCV RNA-positive individuals and only two (0.3%) HBsAg-positive individuals were seen among the population examined. Anti-HEV IgG was detected in 93 (16.2%) individuals and anti-HEV IgM was detected in 10 (1.7%) individuals. The infection rate of HGV reached a peak at an age range of 20 to 39 years; 15.1% of the individuals of this group were infected (Table 1). Moreover, 8.8% of the age group under 19 years and 5.3% of the age group over 40 years were HGV RNA positive. On the other hand, the prevalence of anti-HEV IgG reached 20% in the age group under 19 years. We also determined the relationship between the occurrence of Chagas’ disease, which is hyperendemic in Bolivia, and HGV infection. However, the result showed the absence of a correlation between two (Table 2).
TABLE 1.
Age-specific prevalence of HGV and anti-HEV in healthy Bolivian individuals
| Virus or antibody | No. (%) of individuals with virus or antibody who were aged (yr):
|
||
|---|---|---|---|
| ≤19 (n = 34) | 20–39 (n = 279) | ≥40 (n = 57) | |
| HGV RNA | 3 (8.8) | 42 (15.1) | 3 (5.3) |
| Anti-HEV (IgG) | 7 (20.6) | 50 (17.9) | 10 (17.5) |
TABLE 2.
Relationship between HGV infection and Chagas’ disease
| Chagas’ disease | No. of subjects | No. (%) of HGV RNA positive subjects |
|---|---|---|
| Yes | 59 | 6 (10.2)a |
| No | 173 | 18 (10.4)a |
Not statistically significant.
Nucleotide sequence and genotyping of HGV.
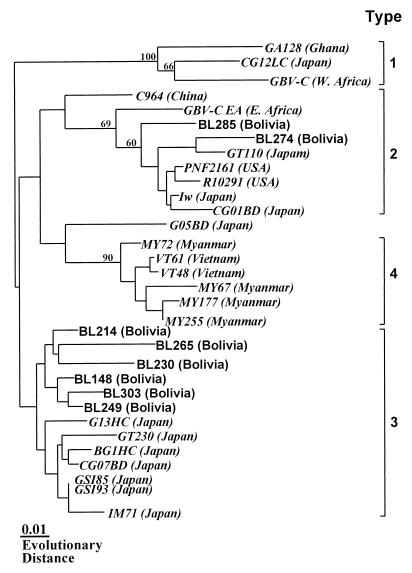
We determined the nucleotide sequences at the 5′ UTR of HGV from 44 Bolivian blood donors. These sequences were then compared by phylogenetic analysis to previously reported representative sequences. The results revealed that 27 (61%) isolates were genotype 3 and 17 (39%) isolates were genotype 2 (Fig. 1). No type 1 or 4 isolates were seen in the present study.
FIG. 1.
Phylogram generated by neighbor-joining analysis of genetic distances in the 5′ UTR sequence of HGV/GBV-C isolates. Isolates determined in this study are presented in roman typeface. The sources of database-derived isolates with the accession numbers are given in the text. The percentages of bootstrap replicates supporting these branches are shown.
Full-length sequence of Bolivian HGV isolate.
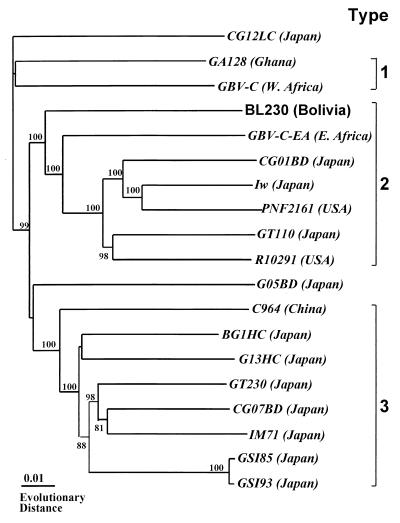
We cloned the full-length sequence of the HGV genome (designated HGV-BL230) recovered from the healthy Bolivian blood donor. The BL230 was composed of 9,227 bases and contained a long ORF spanning 8,526 nt and coding for 2,842 amino acids (aa) flanked by a putative 5′ UTR (nt 1 to 388, not including the short terminal sequence) and a 3′ UTR (nt 8915 to 9227) without a poly(U) stretch or poly(A) tail. Based on the predicted cleavage sites indicated by Leary et al. (13) and Muerhoff et al. (20), we obtained the following sizes of each region of the polyprotein in the isolate: core, 48 nt, 16 aa; E1, 564 nt, 188 aa; E2, 1,161 nt, 387 aa; NS2, 843 nt, 281 aa; NS3, 2,031 nt, 677 aa; NS4, 945 nt, 315 aa; NS5a, 1,245 nt, 415 aa; and NS5b, 1,689 nt, 563 aa. The genomic organization was similar to those of the 18 previously reported isolates. As well as the prototypes of the HGV/GBV-C isolates, the putative core gene in BL230 was not clearly defined because of the existence of only 16 aa residues. Compared to other previously reported HGV/GBV-C isolates with full genome sequences, BL230 showed an overall identity of 87 to 88% at the nucleotide level and 96% at the amino acid level, thereby indicating that they were the same virus (Table 3). HGV/GBV-C isolates with fully sequenced genomes, including database-derived sequences, were grouped into three major genotypes by phylogenetic analysis. Based on this analysis, the HGV-BL230 isolate belonged to genotype 2 of HGV (Fig. 2). Interestingly, the phylogenetic analysis showed that BL230 belonged to type 3 in the 5′ UTR sequence of HGV although this isolate appeared to be type 2 at the level of the entire nucleotide sequence.
TABLE 3.
Percentages of nucleotide and amino acid identity in entire nucleotide sequence between HGV-BL230 and three reported HGV/GBV-C isolates
| Isolate (genotype) | % Identity of:
|
|
|---|---|---|
| Nucleotides | Amino acids | |
| GBV-C (type 1) | 87 | 96 |
| HGV-PNF2161 (type 2) | 88 | 96 |
| HGV-IM71 (type 3) | 88 | 96 |
FIG. 2.
Phylogram generated by neighbor-joining analysis of genetic distances in an entire sequence of HGV/GBV-C isolates. Isolates determined in this study are presented in roman typeface. The sources of database-derived isolates with the accession numbers are given in the text. The full-length sequence of HGV type 4 isolate is not yet available. The percentages of bootstrap replicates supporting these branches are shown.
DISCUSSION
HGV/GBV-C is a recently discovered human RNA virus (15, 34). HGV may be transmitted by transfusion of blood or blood products and has a worldwide distribution. The presence of HGV/GBV-C has been detected in the blood of asymptomatic individuals and in patients with liver diseases (2, 15, 16, 23). On the other hand, it has also been reported that HGV/GBV-C infection does not induce significant liver damage; hence, the nature of HGV/GBV-C and its real pathogenic role are controversial (4, 5). Although there have been many reports on HGV/GBV-C, most epidemiological studies of HGV/GBV-C have been conducted in developed countries. Accordingly, information on HGV infection in developing countries, and particularly in isolated communities, is scarce. In Bolivia, no information about the epidemiology of HGV infection has been available. In this study, we performed molecular-based seroepidemiology of HGV infection in Bolivia and found a high prevalence of this virus in this country. Surprisingly, no HCV RNA-positive individuals and very few HBV-infected individuals (0.3%) were found in the same study population. These findings suggest that HCV and HBV are not endemic in Bolivia, although the existence of anti-HBc and anti-HBs remains to be examined. Regarding the prevalence of HBV in native Bolivians, a similar finding was reported by Sugimura et al. (35) in 1990 and by Miyoshi et al. (17) in 1993. It is known that in individuals infected with hepatitis G, coinfections with HCV, HBV, and HGV alone are rare (2, 16, 23, 25). However, all of the HGV-infected individuals in the present study were seronegative for HCV, although the existence of anti-HCV remains to be examined. These data indicate that HGV has been independently widespread among native Bolivians. In general, the route of viral infection in tropical areas is not clear. The routes and factors involved also were not identified in the present study. The highest prevalence of HGV was seen in the age group of 20 to 39 years, suggesting a role of sexual transmission of HGV in this population. A similar finding of age-specific prevalence of HGV examined in Brazil has been reported by Lampe et al. (11). The high prevalence of HGV infection among human immunodeficiency virus-infected patients has previously been reported, and it has been suggested that HGV may be transmitted via sexual contact (1, 30). However, sexual transmission could not explain the overall spread of HGV observed in this study in view of the large number of infected individuals. There was no evidence of massive use of intramuscular and/or intravenous injection or blood transfusion. Based on this background, we predict that the parenteral route may not be the principal mode of HGV transmission in Bolivia. Interesting, but difficult to explain, is a high prevalence of HGV alone among the isolated population of Santa Cruz, Bolivia. Low socioeconomic status and poor hygienic conditions occurring in developing countries may contribute to HGV infection.
Bolivia is an area in which Chagas’ disease is hyperendemic (12, 28). This disease is caused by the protozoan parasite Trypanosoma cruzi, which is transmitted from sylvatic and domestic mammals to humans by hematophagous reduviids. We determined the relationship between the occurrence of Chagas’ disease and HGV infection. The result showed the absence of a correlation between the two.
Although HGV/GBV-C is not characterized by genome variability as great as that of HCV, several studies have suggested the existence of three different genotypes (18, 19, 25, 31). Recently, the existence of a novel genotype of HGV in Southeast Asian countries was reported, designated genotype 4 (21). Therefore, HGV can be classified now into four different genotypes corresponding to geographic distribution: strains from genotype 1 would be predominant in West Africa, including Ghana, while genotype 2 has been found in the United States and Europe, genotype 3 in Asia, and genotype 4 in Southeast Asia. Based on this classification, we found that the major genotype of HGV in infected Bolivians is type 3, followed by type 2. This fact may be correlated to the movements of Mongoloid migrants who migrated from Asia to South America, including Bolivia, a long time ago. In fact, Mongoloid people account for more than half the general population in Bolivia.
Since the discovery of the HGV/GBV-C genome in human serum in 1996, 18 HGV/GBV-C isolates with entire and/or nearly full nucleotide sequences have been reported. In this study, we cloned the entire sequence of the HGV genome (HGV-BL230) recovered from a Bolivian blood donor and compared the results with previously reported full-length isolates. The results revealed that HGV-BL230 had a high level of similarity to previous database-derived HGV/GBV-C isolates. BL230 had an incomplete putative core protein consisting of only 16 aa residues and no poly(A) tail, compared to the other reported isolates of HGV/GBV-C. To obtain the terminal sequence of the BL230, we designed poly(A)-tailed HGV RNA using poly(A) polymerase for application to RACE. By this method, poly(A) should be added only at the 3′ end of RNA. The genome obtained showed there was no poly(A) tail in the 3′ UTR and this strongly suggested that the BL230 does not have a further extended sequence such as the HCV 3′ X tail (37). Phylogenetic analysis using the full genome sequence of HGV/GBV-C isolates revealed that they were classified into three major genotypes, and the BL230 isolated in the present study belonged to type 2. Interestingly, phylogenetic analysis showed that the 5′ UTR sequence of BL230 belonged to type 3 although this isolate appeared to be type 2 at the level of the entire nucleotide sequence. This suggests that BL230 appeared to be a recombinant between type 2 and 3 strains of HGV. The biological function of the putative structural and nonstructural regions of the HGV/GBV-C genome is still unknown. Future studies of this viral pathogenesis will need to take these considerations into account.
HEV, previously referred to as enterically transmitted non-A, non-B hepatitis, is a major cause of epidemic hepatitis and of acute, sporadic hepatitis in developing countries (27). Many outbreaks of HEV-induced hepatitis have been reported in India, Southeast and central Asia, Africa, and Mexico (7). Although antibodies to HEV have been found in sera from individuals in developing countries, data from South America are scarce. Our data showed that HEV is an important etiological agent in Bolivia. This is not unexpected considering the socioeconomic and climatic environment of this part of Bolivia, which facilitates the dissemination and spread of viral diseases via oral-fecal transmission.
In conclusion, we found a high prevalence of HGV and anti-HEV in a healthy population of native Bolivians. HGV genotype 3, which is seen frequently in Asian countries, was predominant in the study population. Further investigation will be needed to elucidate the origin and transmission routes of HGV in these isolated communities.
ACKNOWLEDGMENTS
We thank Takeshi Kurata, Teiichiro Shiino, Naokazu Takeda, Tamiko Kaneko, Hideo Naito, and Yoshihiro Edamoto; the National Institute of Infectious Diseases, Tokyo; and Mari Yamaguchi and Mitsugu Usui, Sanko Junyaku Co., Ltd., Tokyo, for their kindly cooperation during this study.
This study was supported in part by grants-in-aid for science research from the Ministry of Education, Science and Culture of Japan and the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Abe K, Naito H, Kurata T, Sato H, Takebe Y, Zhang D Y. Prevalence of HCV and HGV in patients infected with HIV: isolation of a new HGV genotype. Hepatology. 1998;28:368A. [Google Scholar]
- 2.Abe K, Moriyama M, Hayashi S, Nakai K, Miyauchi I, Edamoto Y, Saito T, Fukushima S, Shimizu T, Matsumura H, Arakawa Y. Prevalence of hepatitis G virus infection among patients with liver diseases in Japan. Int Hepatol Commun. 1997;6:239–248. [Google Scholar]
- 3.Abe K, Edamoto Y, Park Y N, Nomura A M Y, Taltavull T C, Tani M, Thung S N. In situ detection of hepatitis B, C, and G virus nucleic acids in human hepatocellular carcinoma tissues from different geographic regions. Hepatology. 1998;28:568–572. doi: 10.1002/hep.510280239. [DOI] [PubMed] [Google Scholar]
- 4.Alter H J, Nakatsuji Y, Melpolder J, Wages J, Wesley R, Shih W-K, Kim J P. The incidence of transfusion-associated hepatitis G virus infection and its relation to liver disease. N Engl J Med. 1997;336:747–754. doi: 10.1056/NEJM199703133361102. [DOI] [PubMed] [Google Scholar]
- 5.Alter M J, Gallagher M, Morris T T, Moyer L A, Meeks E L, Krawczynski K, Kim J P, Margolis H S. Acute non-A-E hepatitis in the United States and the role of hepatitis G virus infection. N Engl J Med. 1997;336:741–746. doi: 10.1056/NEJM199703133361101. [DOI] [PubMed] [Google Scholar]
- 6.Billis D M, Bull J J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- 7.Bradley D W. Hepatitis E: epidemiology, aetiology, and molecular biology. Rev Med Virol. 1992;2:19–28. [Google Scholar]
- 8.Erker J C, Simons J N, Muerhoff A S, Leary T P, Chalmers M L, Desai S M, Mushahwar I K. Molecular cloning and characterization of a GB virus C isolate from a patient with non-A-E hepatitis. J Gen Virol. 1996;77:2713–2720. doi: 10.1099/0022-1317-77-11-2713. [DOI] [PubMed] [Google Scholar]
- 9.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 10.Kaneko T, Hayashi S, Arakawa Y, Abe K. Molecular cloning of full-length sequence of hepatitis G virus genome isolated from a Japanese patient with liver disease. Hepatol Res. 1998;12:207–216. [Google Scholar]
- 11.Lampe E, Saback F L, Viazov S, Roggendorf M, Niel C. Age-specific prevalence and genetic diversity of GBV-C/hepatitis G virus in Brazil. J Med Virol. 1998;56:39–43. [PubMed] [Google Scholar]
- 12.Landivar W H C, Nakasa T, Tachibana H, Paz K C, Tateno S. Seropositivity to Trypanosoma cruzi in blood donors in Santa Cruz, Bolivia. J Infect Dis. 1992;166:1464–1465. doi: 10.1093/infdis/166.6.1464. [DOI] [PubMed] [Google Scholar]
- 13.Leary T P, Muerhoff A S, Simons J N, Pilot-Matias T J, Erker J C, Chalmers M L, Schlauder G G, Dawson G J, Desai S M, Mushahwar I K. Sequence and genomic organization of GBV-C: a novel member of the flaviviridae associated with human non-A-E hepatitis. J Med Virol. 1996;48:60–67. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 14.Li T-C, Yamakawa Y, Suzuki K, Tatsumi M, Razak M A A, Uchida T, Takeda N, Miyamura T. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J Virol. 1997;71:7207–7213. doi: 10.1128/jvi.71.10.7207-7213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linnen J, Wages J, Zhang-Keck Z-Y, Fry K, Krawczynski K, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shin J W K, Young L, Piatak M, Hoover C, Fernandez J, Chen S, Zou J-C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 16.Miyakawa Y, Mayumi M. Hepatitis G virus—a true hepatitis virus or an accidental tourist? N Engl J Med. 1997;336:795–796. doi: 10.1056/NEJM199703133361109. [DOI] [PubMed] [Google Scholar]
- 17.Miyoshi C, Tanabe Y, Takahashi K, Kawai S, Matsuda T, Hujisawa K, Kaminura Y, Hashimoto M, Katayama T. Screening test for blood transfusion in Santa Cruz General Hospital, Bolivia. Jpn J Transfusion Med. 1993;39:952–958. [Google Scholar]
- 18.Muerhoff A S, Smith D B, Leary T P, Erker J C, Desai S M, Mushahwar I K. Identification of GB virus C variants by phylogenetic analysis of 5′-untranslated and coding region sequences. J Virol. 1997;71:6501–6508. doi: 10.1128/jvi.71.9.6501-6508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muerhoff A S, Simons J N, Leary T P, Erker J C, Chalmers M L, Pilot-Matias T J, Dawson G J, Desai S M, Mushahwar I K. Sequence heterogeneity within the 5′-terminal region of the hepatitis GB virus C genome and evidence for genotypes. J Hepatol. 1996;25:379–384. doi: 10.1016/s0168-8278(96)80125-5. [DOI] [PubMed] [Google Scholar]
- 20.Muerhoff A S, Leary T P, Simons J N, Pilot-Matias T J, Dawson G J, Erker J C, Chalmers M L, Schlauder G G, Desai S M, Mushahwar I K. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J Virol. 1995;69:5621–5630. doi: 10.1128/jvi.69.9.5621-5630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naito H, Win K M, Abe K. Identification of a novel genotype of hepatitis G virus in Southeast Asia. J Clin Microbiol. 1999;37:1217–1220. doi: 10.1128/jcm.37.4.1217-1220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakao H, Okamoto H, Fukuda M, Tsuda F, Mitsui T, Masuko K, Iizuka H, Miyakawa Y, Mayumi M. Mutation rate of GB virus C/hepatitis G virus over the entire genome and in subgenomic regions. Virology. 1997;233:43–50. doi: 10.1006/viro.1997.8615. [DOI] [PubMed] [Google Scholar]
- 23.Nakatsuji Y, Shih J W-K, Tanaka E, Kiyosawa K, Wages J, Jr, Kim J P, Alter H J. Prevalence and disease association of hepatitis G virus infection in Japan. J Viral Hepat. 1996;3:307–316. doi: 10.1111/j.1365-2893.1996.tb00103.x. [DOI] [PubMed] [Google Scholar]
- 24.Okamoto H, Nakao H, Inoue T, Fukuda M, Kishimoto J, Iizuka H, Tsuda F, Miyakawa Y, Mishiro M. The entire nucleotide sequence of two GB virus C/hepatitis G virus isolates of distinct genotypes from Japan. J Gen Virol. 1997;78:737–745. doi: 10.1099/0022-1317-78-4-737. [DOI] [PubMed] [Google Scholar]
- 24a.Okamoto H, Okada S, Sugiyama Y, Yotsumoto S, Tanaka T, Yoshizawa H, Tsuda F, Miyakawa Y, Mayumi M. The 5′-terminal sequence of the hepatitis C virus genome. Jpn J Exp Med. 1990;60:167–177. [PubMed] [Google Scholar]
- 25.Orito E, Mizokami M, Nakano T, Wu R R, Cao K, Ohba K, Ueda R, Mukaide M, Hikiji K, Matsumoto Y, Iino S. GB virus C/hepatitis G virus infection among Japanese patients with chronic liver diseases and blood donors. Virus Res. 1996;46:89–93. doi: 10.1016/s0168-1702(96)01379-2. [DOI] [PubMed] [Google Scholar]
- 26.Page R D M. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 27.Pawlotsky J M, Belec L, Cresenguet G, Deforges L, Bouvier M, Duval J, Dhumeaux D. High prevalence of hepatitis B, C, and E in young sexually active adults from the Central African Republic. J Med Virol. 1995;46:269–273. doi: 10.1002/jmv.1890460318. [DOI] [PubMed] [Google Scholar]
- 28.Pless M, Juranek D, Kozarsky P, Steurer F, Tapia G, Bermudez H. The epidemiology of Chagas’ disease in a hyperendemic area of Cochabamba, Bolivia: a clinical study including electrocardiography, seroreactivity to Trypanosoma cruzi, xenodiagnosis, and domiciliary triatomine distribution. Am J Trop Med Hyg. 1992;47:539–546. doi: 10.4269/ajtmh.1992.47.539. [DOI] [PubMed] [Google Scholar]
- 29.Rzhetsky A N. Tests of applicability of several substitution models for DNA sequence data. Mol Biol Evol. 1995;12:131–151. doi: 10.1093/oxfordjournals.molbev.a040182. [DOI] [PubMed] [Google Scholar]
- 30.Saito T, Ishikawa K, Osei-Kwasi M, Kaneko T, Brandful J A M, Nuvor V, Aidoo S, Ampofo W, Apeagyei F A, Ansah J E, Adu-Sarkodie Y, Nkrumah F K, Abe K. Prevalence of hepatitis G virus and characterization of viral genome in Ghana. Hepatol Res. 1999;13:221–231. [Google Scholar]
- 31.Saito T, Shiino T, Arakawa Y, Hayashi S, Abe K. Geographical characterization of hepatitis G virus genome: evidence for HGV genotypes based on phylogenetic analysis. Hepatol Res. 1998;10:121–130. [Google Scholar]
- 32.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–423. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Shao L, Shinzawa H, Ishikawa K, Zhang X, Ishibashi M, Misawa H, Yamada N, Togashi H, Takahashi T. Sequence of hepatitis G virus genome isolated from a Japanese patient with non-A-E hepatitis: amplification and cloning by long reverse transcription-PCR. Biochem Biophys Res Commun. 1996;228:785–791. doi: 10.1006/bbrc.1996.1732. [DOI] [PubMed] [Google Scholar]
- 34.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushahwar I K. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 35.Sugimura H, Tsugane S, Watanabe S, Nanri S, Ishii H. Hepatitis B virus markers in Japanese immigrants and their descendants in Bolivia and native Bolivians. Gastroenterol Jpn. 1990;25:335–338. doi: 10.1007/BF02779447. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K, Hijikata M, Hino K, Mishiro S. Entire polyprotein-ORF sequence of Japanese GBV-C/HGV isolates: implications for new genotypes. Hepatol Res. 1997;8:139–148. [Google Scholar]
- 37.Tanaka T, Kato N, Cho M-J, Shimotohno K. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem Biophys Res Commun. 1995;215:744–749. doi: 10.1006/bbrc.1995.2526. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Y S, Chen W, Zhao Q M, Zhao H L, Zhang J S, Xu J, Wang H T. cDNA cloning and sequencing of HGV genome from Chinese. Bull Acad Military Med Sci. 1996;20:249–253. [Google Scholar]
- 39.Zuckerman A J. Alphabet of hepatitis viruses. Lancet. 1996;347:558–559. doi: 10.1016/s0140-6736(96)91267-2. [DOI] [PubMed] [Google Scholar]