Abstract
Homologous recombination and DNA repair are important for genome maintenance. Genetic variations in essential homologous recombination genes, including BRCA1 and BRCA2 results in homologous recombination deficiency (HRD) and can be a target for therapeutic strategies including poly (ADP-ribose) polymerase inhibitors (PARPi). However, response is limited in patients who are not HRD, highlighting the need for reliable and robust HRD testing. This manuscript will review BRCA1/2 function and homologous recombination proficiency in respect to breast and ovarian cancer. The current standard testing methods for HRD will be discussed as well as trials leading to approval of PARPi’s. Finally, standard of care treatment and synthetic lethality will be reviewed.
Keywords: Ovarian cancer, Breast cancer, PARP inhibitor, Homologous recombination, Homologous recombination deficient, Homologous recombination proficient, BRCA
Background
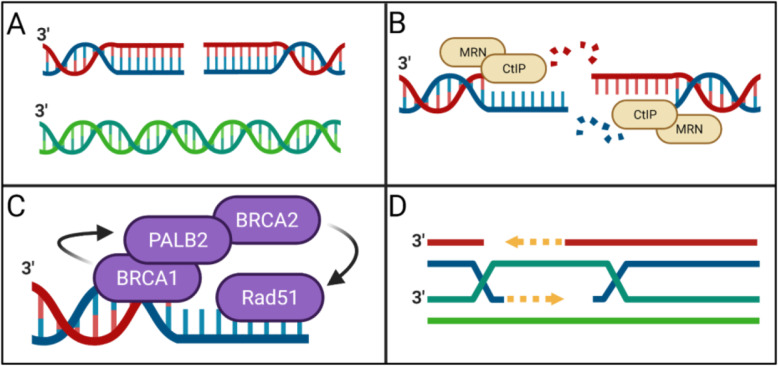
DNA damage is inevitable, multifactorial, and dangerous. Whether initiated by exogenous or endogenous sources, inappropriate alterations to the human genome may result in far-reaching, pathological consequence unless quickly and accurately corrected. Homologous recombination DNA repair (HRR) is a critically important mechanism by which DNA damage can be corrected. Homologous recombination DNA repair is a process by which double-stranded DNA breaks and interstrand crosslinks use sister chromatid as a template for repair [1] (Fig. 1A). In this way, DNA damage is removed in an error-free fashion [2]. Additionally, during DNA replication, HRR pathways support the recovery of stalled replication forks [3]. Successful HRR depends on several properly functioning proteins, with BRCA1 and BRCA2 proteins playing particularly pronounced roles [4]. BRCA1 is a tumor suppressor protein central to several macromolecular complexes which drive HRR and cell cycle progression [5]. After MRN and CtIP mediated DNA resection (Fig. 1B), BRCA1 travels to sites of double-stranded DNA breaks where it participates in DNA damage signaling and coordinates DNA damage repair [5, 6]. While the role of BRCA1 in HRR is well established, emerging evidence suggests BRCA1 also regulates cellular selection of double-strand break repair pathways. By doing so, BRCA1 may influence a cell’s choice between HRR and non-homologous DNA end joining (NHEJ) DNA double-strand break repair mechanisms [6, 7]. During the synthesis phase of normal cell cycle progression, if DNA becomes damaged, the BRCA1 protein complexes recruit BRCA2 protein complexes to initiate strand invasion and/or homology-directed repair [5, 6, 8] (Fig. 1C, D). BRCA-dependent DNA double-strand break repair mechanisms can compensate for dysfunctional DNA single-strand break repair mechanisms. When DNA single-strand breaks accumulate and are converted to double-strand breaks, HRR can repair DNA lesions and maintain cell viability [9].
Fig. 1.
Homologous recombination DNA repair (HRR). A double-stranded DNA break and sister chromatid that will be used as a template for repair. B MRN and CtIP are involved in DNA resection. C HRR repair complex. D Schematic representation of BRCA1/2 mediated HRR
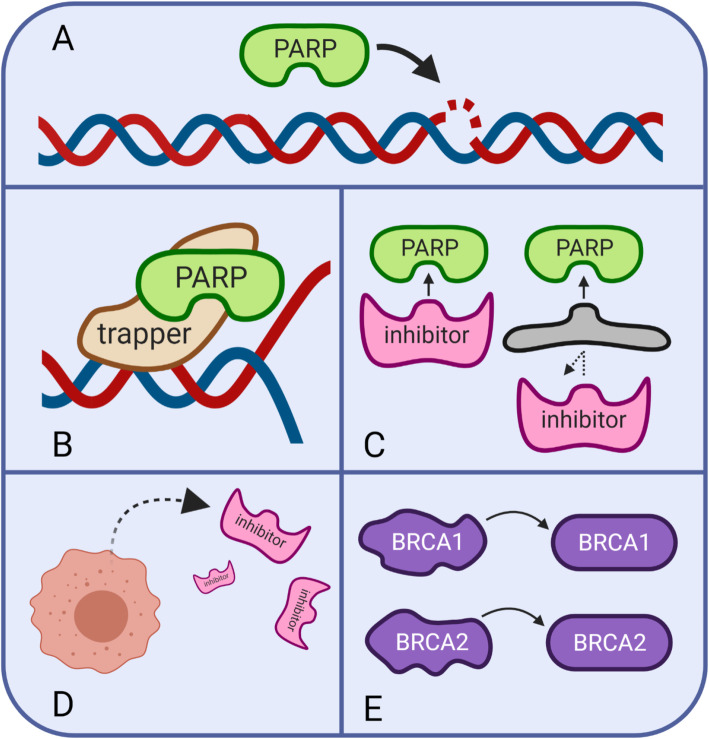
Poly (ADP-ribose) polymerase proteins (PARPs) are nuclear enzymes integral to the base excision repair pathway of single-strand DNA repair [10]. PARPs travel to sites of single-strand DNA breaks (Fig. 2A) where they synthesize polymeric adenosine diphosphate ribose chains for post-translational modification of nuclear proteins [10, 11], in turn promoting downstream single stranded repair processes [12]. Clinically, dysfunctional DNA single-strand break repair may be pharmacologically elicited with PARPis. It is thought that these single-stranded breaks are converted to double-stranded breaks during replication. In cells with BRCA1/2 mutations resulting in the inability to repair double-stranded breaks, treatment with PAPRis results in synthetic lethality. Beyond its role in single-stranded DNA repair, PARP1 is also involved in the alternative end-joining (alt-EJ) double-strand break repair pathway [13]. PARPs also play a role in homologous recombination, although this role may be relegated to homologous recombination mediated recovery of stalled replication forks, rather than double-stranded break repair [14].
Fig. 2.
PARP inhibitors (PARPi) and mechanisms of PARPi resistance. A PARPs travel to DNA single-strand breaks. B PARP inhibitors can trap the enzyme and prevent it from properly functioning. C-E Potential mechanisms of resistance to PARPi therapy. C PARPi may be blocked, D effluxed, or E reversion of BRCA1/2 mutations
Main text
Homologous recombination DNA repair proficiency (HRP) and Cancer
The ability of cancer cells to successfully perform HRR is frequently used as a basis for patient stratification. By grouping patients according HRR status, researchers and clinicians can optimize disease treatments, improve outcome prognostication, and design more informatic clinical trials [15]. Cancer cells that demonstrate homologous recombination DNA repair proficiency (HRP) and cancer cells that demonstrate homologous recombination deficiency (HRD) may be treated using different therapeutic strategies. While HRR status may have important implications for the clinical care of pancreatic cancer patients [16], liver cancer patients [17], lung cancer patients [18], and renal cancer patients [19], its relatively increased incidence in breast and ovarian cancer provides the most robust data regarding treatment effects in this pathway. Indeed, in a sample of 3504 patients with metastatic cancer, genomic footprints indicative of HRD were found in only 6% of cancer cases, while approximately 30% of ovarian cancers and 13% of breast cancers were HRD [20]. In epithelial ovarian cancer (EOC), approximately 50% of cases involve HRD due to alterations of HRR pathway genes [21]. While HRD ovarian cancer cells usually harbor mutations in BRCA1, BRCA2 or other genes with similar features (traits which are collectively referred to as “BRCAness”) [22–24], HRP ovarian cancer cells are often driven by genetic alterations involving other pathways contributing to cell cycle dysregulation, such as cyclin E1 (CCNE1) genes [21]. Indeed, CCNE1 amplification events occur alongside BRCA inactivation at remarkably low frequency [25], with Gorski et al. describing CCNE1 amplification and homologous recombination pathway mutations as “nearly mutually exclusive” [22]. CCNE1 is an important factor in G1/S cell cycle transition, as it activates CDK2 thereby allowing the cell to enter S phase [26]. The amplification of cyclin E1 increases the speed by which cancer cells pass from the G1 to S phase. This can lead to genomic instability and drive the dysregulation of genes responsible for proliferation and cellular survival [22, 27]. Cyclin E1 amplification occurs in 19.1% of all ovarian cancers [22, 25] and 3.4% of breast cancers [28].
Homologous recombination DNA repair deficiency (HRD) and Cancer
Most HRD cancer cells have direct deficiency in a gene or group of genes responsible for homologous recombination DNA repair, although upregulation of miRNAs can also cause HRD [21]. Generally, the etiology of HRD can be attributed to pathogenic germline variants, somatic mutations, or epigenetic changes in HRR pathway genes. Pathogenic or likely pathogenic germline BRCA1 or BRCA2 variants are present in 18% of ovarian cancer cases [29]. In breast cancer, pathogenic or likely pathogenic germline BRCA1 or BRCA2 variants are present in 6.1% of all cases [30–36], and 10–20% of triple-negative breast cancer cases [37]. Pathogenic or likely pathogenic germline BRCA variants are more often associated with the development of cancer at a younger age and present with more aggressive disease phenotypes with worse prognoses when compared to cancers caused by somatic BRCA mutations [38, 39]. In Somatic BRCA1 and BRCA2 mutations are identified in 3 % of high-grade serous ovarian carcinoma cases [40]. In ovarian, fallopian tube, and peritoneal carcinomas, the most common somatic mutations in HR genes are BRCA1 (54% of somatic mutations) and BRCA2 (17% of somatic mutations) [41]. Somatic mutations of BRCA1 and BRCA2 in breast cancer are positively correlated with cancer survival [42–44]. Identification of somatic mutations in cancer is generally associated with a better prognosis then cancers involving pathogenic or likely pathogenic germline variants. Additionally, epigenetic causes of HRD involve the silencing of BRCA genes by up or down regulated miRNA activity or BRCA promoter hypermethylation [45]. Analogous miR-1255b, miR-148b, and miR-193b miRNA molecules targeting BRCAness genes have been described in ovarian cancer [46]. In breast cancer, miR-182 overexpression induces HRD by targeting BRCA1 [47]. Furthermore, BRCA expression may be inhibited by epigenetic hypermethylation. In 11% of high-grade ovarian carcinomas, BRCA1 expression is silenced by hypermethylation [40]. Additionally, hypermethylation of BRCA promoter regions are associated with more aggressive disease. Hypermethylation of BRCA1 in ovarian cancers correlate with significantly shorter median survival (n = 11, 35.6 months) compared to germline BRCA1 (n = 22, 78.6 months) and wild-type BRCA1 (n = 30, 63.3 months) [40]. HRD etiology may play a critical role in clinical selection of therapeutics as well as overall patient prognosis.
Role of BRCA1/2 in homologous recombination DNA repair (HRR)
BRCA1 and BRCA2 mutations put individuals at higher risk for developing certain malignancies, particularly ovarian and breast cancer. The chance of developing ovarian cancer if an individual has a BRCA1 mutation is 39–46% [48–52]. In women with a BRCA1 mutation, the probability of developing breast cancer over her lifetime is 57–65% [50, 53–55]. For BRCA2 mutations, the chance of developing breast cancer is 45–49%, and for ovarian cancer it is 11–18% [39, 50, 53]. Further, patients demonstrate high prevalence of BRCA mutations in triple negative breast cancer (TNBC), which is negative for estrogen receptors (ER-), progesterone receptors (PR-), and excess human epidermal growth factor receptor 2 (HER2) proteins. In fact, 80% of women with a pathogenic mutation in BRCA1 who develop breast cancer have triple negative disease [56, 57].
Within the genes BRCA1 or BRCA2, cancer cluster regions are genetic regions containing a disproportion amount of gene mutations. A risk hazard ratio (RHR) quantifying the chances of developing cancer is used below to evaluate breast versus ovarian cancer for different cancer cluster regions. In ovarian cancer, there is a single ovarian cancer region: c.1380 to c.4062 (RHR = 0.62; 95% CI, 0.56–0.70; P = 9 × 10− 17) in BRCA1 [58]. In breast cancer, there are three breast cancer cluster regions in BRCA1: c.179 to c.505 (RHR = 1.46; 95% CI, 1.22–1.74; P = 2 × 10− 6), c.4328 to c.4945 (RHR = 1.34; 95% CI, 1.01–1.78; P = .04), and c.5261 to c.5563 (RHR = 1.38; 95% CI, 1.22–1.55; P = 6 × 10− 9). There are also three identified breast cancer cluster regions in BRCA2: c.1 to c.596 (BCCR1; RHR = 1.71; 95% CI, 1.06–2.78; P = .03), c.772 to c.1806 (BCCR13; RHR = 1.63; 95% CI, 1.10–2.40; P = .01), and c.7394 to c.8904 (BCCR2; RHR = 2.31; 95% CI, 1.69–3.16; P = .00002). The cancer cluster region for BRCA2 in ovarian cancer is located from c.3249 to c.5681 (RHR = 0.51; 95% CI, 0.44–0.60; P = 6 × 10− 17) and c.6645 to c.7471 (; RHR = 0.57; 95% CI, 0.41–0.80; P = .001) [58]. These breast and ovarian cancer cluster regions are shown below in Table 1. The probability of developing cancer and which type of cancer depends, in part, on the particular cancer cluster region affected within the BRCA genes.
Table 1.
Location of cancer cluster regions in BRCA1 and BRCA2 for breast and ovarian cancer [58]
| BRCA 1 | Exon | Domain | BRCA 2 | Exon | Domain | ||
|---|---|---|---|---|---|---|---|
| Breast Cancer Cluster Region | c.179 to c.505 | 5 | RING | c.1 to c.596 | 1 | – | |
| c.4328 to c.4945 | 13 | Serine Cluster | c.772 to c.1806 | 3 | – | ||
| c.5261 to c.5563 | 19 | BRCT | c.7394 to c.8904 | 14 | DNA binding | ||
| Ovarian Cancer Cluster Region | c.1380 to c.4062 | 11 | Part of region contains Serine Cluster | c.3249 to c.5681 | 11 | BRC | |
| c.6645 to c.7471 | 11 | BRC |
There are also some regions of BRCA that are more commonly mutated in certain populations. The founder mutations, including the BRCA2 region of c.3249 to c.5681 associated with the c.5946del mutation, is common in patients with Ashkenazi Jewish ancestry [39, 58, 59]. Individuals with this mutation are more prone to develop ovarian cancer than breast cancer. Another founder mutation is the c.5266dupC mutation in BRCA1. It is also associated with individuals of Ashkenazi Jewish ancestry as well as European ancestry. This mutation is associated with higher risk of ovarian cancer [39, 60]. The third founder mutation is c.68_69delAG which is located in exon 2 of BRCA1; this is more commonly seen in patients with Ashkenazi Jewish ancestry as well as Indian ancestry; which occurs at a frequency of 16.4% in these populations [39]. The mutation is associated with increased sensitivity of ovarian cancer cells to cisplatin therapy, independent of wild-type BRCA1 alleles [61, 62]. One possible mechanism for this involves the BRCA c.68_69delAG mutation increasing expression of maspin, a mammary serine protease inhibitor, as it is a novel downstream target of the BRCA c.68_69delAG mutation [62]. The increased expression of maspin causes a decreased expression of inhibitor of apoptosis proteins [63]. Increased expression of maspin is associated with increased response to cisplatin therapy; as such, it is also associated with a more favorable prognosis in ovarian cancer [39, 62].
Dysfunction in “BRCAness” genes, such as RAD51 and CDK12, can also cause HRD and is an important pathway in the development of ovarian [21] or breast [64] cancer. CDK12, which promotes transcription in several HR pathway genes, such as BRCA1, is one of the most frequently mutated genes in ovarian cancer [65, 66]. Inactivation of CDK12 leads to suppression of HRR. Between BRCA1 exons 11 and 13, there is a binding site for RB1, PALB2, and RAD51 [39, 67]. Interaction between BRCA1 and BRCA2 is mediated by PALB2. This interaction is critical in the RAD51-mediated HRR of damaged DNA [39]. RAD51’s activity is modulated by BRCA2 and the correlation between RAD51 and BRCA2 is important for the repair of double-strand DNA breaks [68].
Homozygous loss of PTEN and amplification of EMSY may also be involved in HRD, although this is debated as there is not enough information to classify PTEN or EMSY as a HRD or HRP related defect. EMSY colocalizes with BRCA2 at sites of DNA damage and is located at 11q13 [69]. Most sources do describe EMSY amplification or overexpression as a mechanism underlying HRD, although this is controversial and varies from source to source [21]. An EMSY amplification is associated with a poor prognosis in breast and ovarian cancer [70]. Amplification or overexpression of the EMSY transcriptional repressor leads to BRCA2 silencing in approximately 20% of high-grade ovarian carcinoma cases [40]. PTEN acts as a tumor suppressor gene in its regulation of the cell cycle [71]. It is not clear whether PTEN is involved in HRD [21, 41]. PTEN deficiency has been shown to be synthetically lethal with PARP inhibition but this may be due to the downregulation of RAD51, a gene that assists in the repair of double strand DNA breaks [72, 73]. PARP-1 recognizes DNA breaks and is involved in early recruitment of factors facilitating double-strand break repair, and inhibition of PARP can cause cell death as PARP can no longer recruit factors for cell-repair [74].
HRR status determination
BRCA testing
Given the significance of BRCA1/2 gene integrity in HRR functionality, BRCA1/2 are often used as a metric for determining a tumor’s HRR status [75–77]. Because the nature of a given BRCA1/2 variation may predict patient response to certain therapies, it has been shown to be beneficial to differentiate between pathogenic germline variants, somatic mutations, or epigenetic changes. Assessments of pathogenic germline variants require blood or saliva samples, while somatic mutations require direct biopsy or circulating tumor DNA analysis [29]. Epigenetic testing typically relies on formalin-fixed paraffin-embedded (FFPE) tissue samples [78]. However, recent studies are exploring the use of blood samples [79] to identify BRCA1/2 epigenetic changes, with one such study successfully identifying epigenetic changes using hair, buccal mucosa, and blood samples [80].
For years, Sanger sequencing was the gold-standard to detect single-nucleotide alterations, insertions, and deletions in BRCA1/2 genetic sequences, while large genomic alterations were detected using multiplex ligation-dependent probe amplification (MLPA) [81]. However, Sanger sequencing and MLPA are time-consuming and costly. Today, samples are typically analyzed using massive parallel sequencing (MPS) [82]. Multiple studies report that using MPS to analyze BRCA1/2 mutations yields comparable results to Sanger sequencing but on a faster time scale and more cost effective. These factors have contributed to MPS becoming standard practice in recent years [83, 84]. The high-throughput process of MPS allows for the discovery of tiny variants in an individual’s DNA. These variants can be categorized on a spectrum from benign to pathogenic, or they can be categorized as variants of unknown significance (VUS). The identification of VUS results pose a dilemma for both clinicians and patients: additional screening and testing based on a VUS can lead to overtreatment and mismanagement, while patients carrying VUS may experience additional anxiety about potential implications of the variants. In general, known VUS are disclosed to patients, although VUS alone should not change the management of a patient. Attempts are typically made to classify VUS as either benign or pathogenic, although further research and guidelines are needed to determine how best to proceed following the discovery of VUS [85].
Regardless of how mutations are sequenced, the high prevalence of BRCA1/2 mutations necessitate genetic testing in individuals at risk for ovarian or breast cancer. In high-grade serous ovarian cancer (HGSOC), pathogenic germline variants and somatic BRCA1/2 mutations can be found in 17–25% of patients, and 18–30% of all BRCA1/2 variations are somatic in origin [41, 86, 87]. However, germline DNA tests are more sensitive and less invasive than somatic DNA tests, and therefore germline testing is prioritized [21]. However, if germline DNA testing is negative for BRCA variants, the American Society of Clinical Oncology (ASCO) guidelines recommend a tumor sample be harvested and tested for somatic mutation [75, 88, 89]. .At the present time, most laboratories do not test for epigenetic changes such as BRCA promoter hypermethylation, although there is evidence supporting an improved response to certain therapeutic agents for such mutations [90].
Determining a tumor’s HRR status can aid clinicians and patients in the selection of potential therapeutic strategies, but inconsistencies in laboratory testing procedures detract from their overall utility. Although laboratories across the world have been evaluating BRCA1/2 genes for over two decades, the extent to which BRCA1/2 variations are measured, consideration of non-coding DNA regions in genetic sequencing, and which technologies are used to identify large genetic rearrangements all fluctuate drastically between laboratories [82]. Improved institutional guidelines standardizing BRCA1/2 testing may improve the sensitivity of these tests, particularly in identifying mutations that might not have been discovered without uniform guidelines.
Clinically, BRCA testing is vastly underutilized. Only 20% of women eligible for genetic testing based on age at diagnosis and family history per 2017 NCCN guidelines have been tested [79, 91], and it is estimated that over 97% of BRCA carriers in the population remain unidentified [80]. Various reasons for low detection rates include small family size, which makes it difficult to identify patterns in hereditary genetic variants, lack of consideration for paternally inherited genetic variants, incomplete penetrance, population migration, limited public awareness about BRCA, and poor referral guideline implementation by both primary care providers and oncologists. Other limitations include socioeconomic and geographic factors including limited access to genetic counseling in rural areas, lack of insurance coverage and reimbursement for genetic counseling services, and the time requirement to counsel patients [92–94].
While BRCA mutations are more commonly associated with breast and ovarian cancers, men are also affected by the presence of a BRCA mutation. Male breast cancer is overall rare, with a lifetime risk of 0.1%. However, the lifetime risk of breast cancer is significantly higher in men with BRCA1 or BRCA2 mutation, with a incidence of 1.2% in men who carry a BRCA1 mutation and 6.8% in men who carry a BRCA2 mutation [95]. .Men with BRCA mutations are also at increased risk for other cancers, such as prostate, pancreatic, and gastric cancers, as well as melanoma of the skin and eye [96]. .By improving and increasing access for BRCA testing, men and women alike can be informed about their potential risks of developing cancer.
Non-BRCA testing
The need for HRR tests evaluating non-BRCA1/2 genetic abnormalities is growing with our understanding that HRD may be caused by other dysfunctional “BRCAness” proteins, such as RAD51. Indeed, ovarian and breast cancer patients without BRCA1/2 mutations demonstrate positive, yet variable, clinical responses to therapeutic agents that target HRD [25]. This supports non-BRCA1/2 etiologies for HRD and suggests that identification of non-BRCA1/2 etiologies may provide relevant information for therapeutic selection and may thereby impact clinical prognosis. Improved laboratory testing procedures which evaluate other genes implicated in HRR could result in the development of more personalized, comprehensive treatment plans for patients with cancer. RAD51, for instance, is critically involved in HRR processes [97]. RAD51 mutations are implicated in the development and progression of ovarian and breast cancer [98–100]. Mutational analysis of RAD51 in 125 families from 12 countries across Europe and North American found an association between RAD51C/D mutations and increased risk of ovarian cancer (RAD51C: p < 0.001, RAD51D: p < 0.001) [101]. Pre-clinical models also suggest RAD51 mutations may be associated with resistance to anti-cancer therapeutics that target the HRR pathway [102]. Data such as these suggest that pathogenic germline RAD51 variants may be an effective biomarker for HRD. Accordingly, many, but not all, breast and ovarian cancer panels include assessment of RAD51 [103]. Inclusion of genes such as RAD51 in HRR testing procedures may increase detection of HRD in patients without BRCA1/2 mutations [104] and act as a determinant of therapeutic efficacy and prognosis [105].
Other testing strategies
As an alternative to gene-specific testing practices, HRR can also be evaluated by measuring genomic scarring—broadly defined as genomic aberrations of a known origin [106]. More specifically, genomic scarring can be defined here as HRD-related genomic aberrations or large-scale DNA alterations [107]. These scars are associated with unrepaired damage within a patient’s genome resulting from an inability to successfully repair double-strand breaks [107]. This method is used by Myriad in the MyChoice® CDx assay, which is approved as a companion diagnostic for ovarian cancer treatment with olaparib and niraparib. MyChoice® CDx evaluates genomic scarring by measuring loss of heterozygosity (LOH), telomeric allelic imbalances, and large-scale state transitions. A tumor is considered HRD if there is a BRCA1/2 mutation, or a genomic instability score (GIS) of ≥42. In the QUADRA trial, patients with recurrent platinum sensitive HRD ovarian cancer who had undergone at least 3 lines of prior therapy without prior exposure to PARP inhibitor demonstrated a significant response to niraparib (ORR 29%; 95% CI 16–44; p = 0.0003) [108]. Patients with ovarian cancer who received frontline niraparib also demonstrated an improved PFS of 21.9 months versus 10.4 months (HR = 0.43; 95% CI 0.50–0.76; p = < 0.001) for tumors which were HRD. However, veliparib, another PARP inhibitor which was evaluated in combination with chemotherapy in first-line ovarian cancer, used a threshold score of ≥33 after retrospective analysis of this cutoff in triple negative breast cancer, and demonstrated increased sensitivity [109, 110]. When veliparib is combined with first line chemotherapy followed by veliparib maintenance, patients demonstrated a progression free survival benefit of 31.9 versus 20.5 months (HR 0.57; 95% CI 0.43–0.76; p = < 0.001) in the HRD ≥33 cohort [109].
Without focusing on a specific gene, these metrics approximate the burden caused by dysfunctional repair pathways and allow indirect identification of HRD. Therefore, choosing a cutoff may be therapy- or assay-dependent. For instance, Foundation Medicine identifies HRD through mutation in BRCA1/2 or an LOH score of > 16. While Foundation Medicine is not an FDA-approved companion diagnostic for PARPi, it may be used to inform decision making.
Genomic scarring acts as an objective indicator of genomic abnormalities, compared to gene mutations which can be influenced by a variety of factors. Thus, genomic scar biomarkers have strong negative predictive value (NPV) for response to HR-deficiency therapies--meaning individuals without genomic scarring biomarkers will likely not benefit from HR deficiency-targeting drugs. However, they are also poor positive predictive value (PPV) biomarkers, as high levels of genomic instability do not account for mutations that may restore HR proficiency [106]. While genomic scarring has been used to predict HRD in conjunction with other companion diagnostics, improved screening methods and appropriate definition of HRD positivity using genomic scarring represent an active area of research.
Homologous recombination ability and cancer therapeutics
Standard of care for breast and ovarian cancer
Improved mechanistic understanding and higher resolution laboratory identification of BRCA1/2 genetic variation subtypes and non-BRCA1/2 genetic aberrations contribute to the clinical care that breast and ovarian cancer patients receive. In fact, of the 5–10% of breast cancer cases that are related to genetic mutations, 67% of those cases are due to BRCA1/2 mutations [108]. Broadly, the standard of care for breast and ovarian cancer patients depends on many factors. The standard of care for breast cancer depends on classification, receptor status, and whether it has become invasive or metastatic. Based on these factors, recommendations for breast cancer patients may include a surgical excision and lymph node evaluation, radiation and medical therapy [111]. The standard of care for patients with epithelial ovarian cancer includes maximal surgical cytoreduction and systemic platinum-based chemotherapy [112]. The chemotherapeutic agent and whether chemotherapy is undergone prior to surgery are both determined by the stage and histology of the tumor [113] Stage IIIC and IV ovarian cancers are treated with chemotherapy, either after surgery or before, as neoadjuvant therapy [114]. In both diseases, medical therapy is almost always initiated [111, 113]. However, clinical selection of chemotherapeutic agents depends on various factors, one of which being the HRR status of the cancer cells.
Medical therapyis an important component of breast and ovarian cancer management.. Because HRD cancer cells are more sensitive to certain anti-cancer drugs, such as platinum chemotherapy, the HRR status of a patient’s tumor may influence chemotherapeutic selection. Past research has shown that ovarian cancer patients with BRCA mutations are more susceptible to platinum-based chemotherapeutic agents. These agents, such as carboplatin, damage DNA and induce double-strand breaks, which HRD cancer cells cannot repair, which lead to apoptosis [115]. However, platinum-based agents are not without their drawbacks. These agents are associated with significant neurotoxicity, ematogenecity, and marrow suppression which can impact a patient’s quality of life [116]. Like platinum-based agents, other chemotherapeutic agents also function by inducing double-strand breaks, whether it be directly, like doxorubicin, or by crosslinking DNA through alkylation, like cyclophosphamide. One study involving triple negative breast cancer demonstrated that patients with HRD biomarkers were more susceptible to adjuvant doxorubicin and cyclophosphamide combination therapy. Moreover, the HRD patients undergoing the combination chemotherapy demonstrated better disease-free survival than those who were not HRD [117]. .Given evidence of HRD cancers manifesting increased sensitivity to chemotherapy agents that target malignant cell defective repair mechanisms, the identification of biomarkers for HRD could lead to more effective treatment for this subset of patients.
While PARP inhibition has shown promising results in the treatment of HRD cancers, more research is needed in order to establish optimal treatment regimens for HRP cancers. Gemogenovatucel-T, or Vigil, is a vaccine composed of autologous tumor cells, transfected with a plasmid containing GM-CSF and bi-shRNA to decrease furin activity. Decreased furin expression subsequently down-regulates TGF-β1 and TGF-β2 expression [118]. Safety and efficacy of Vigil has been demonstrated in numerous solid tumors [119–123]. A recent study exploring the efficacy of gemogenovatucel-T in ovarian cancer patients demonstrated significant differences in RFS and OS in patients with BRCA wild-type tumors when compared to those who had BRCA mutations [124]. Subsequent analysis of patients who were HRP versus HRD revealed further RFS (10.6 vs 5.7 months; HR = 0.386 90% CI 0.199–0.750 p = 0.007) and OS (NR vs 26.9 months; HR = 0.342 90% CI 0.141–0.832 p = 0.019) benefit [125]. To compare, BRCA-wt/HRP patients treated with niraparib who had a response to first line chemotherapy in the PRIMA trial had a PFS of 8.1 versus 5.4 months for placebo (HR 0.68 95% CI 0.49–0.94 p = 0.020) [126]. Patients with HRP or unknown HR status treated with olaparib and bevacizumab as first line maintenance had a median PFS of 16.9 months vs 16.0 months in placebo treated (HR 0.92 95% CI 0.72 to 1.17) [77]. Vigil demonstrated improved clinical benefit compared to niraparib or olaparib and bevacizumab with no grade 3/4 adverse events reported in patients receiving Vigil. Both niraparib and olaparib plus bevacizumab result in a large amount of drug related grade 3/4 adverse events (65.3 and 57.0% respectively) and dose discontinuation (14.7 and 41.0% respectively) [77, 126]. Additionally, there has been some concern regarding the rate of treatment related myelodysplastic syndrome and acute myeloid leukemia (tMDS/AML) following PARPi. In a recent meta-analysis, risk of tMDS/AML was increased in patients receiving PARPi compared to placebo (Peto OR 2.63 95% CI 1.13–6.14; p = 0.026) [127]. Prognosis for tMDS/AML is typically poor, with a 5 year survival of less than 10% [110]. Therefore, Vigil is an attractive therapeutic option for frontline ovarian cancer maintenance with improved efficacy and robust safety.
The mechanism for Vigil benefit in HRP patients remains unclear; however, the level of clonal neoantigens present in cells that are capable of homologous recombination may play a role. Colon cancer tumors with mismatch repair deficiency (MMR) have a high proportion of neoantigens. Neoantigens have, therefore, been investigated as a predictive biomarker for response to immunotherapies [128, 129]. However, the amount of neoantigens present in a tumor may not be the sole predictor of response to immunotherapies, and may differ based on immunotherapy mechanisms. Activated and primed T cells may not recognize all neoantigens with the same affinity. McGranahan et al. found that T cells recognize clonal neoantigens compared to subclonal neoantigens preferentially to target the tumor [130]. Vigil has also shown the ability to increase circulating CD3+/CD8+ T cells in advanced cancer patients [131]. Likely these CD3+/CD8 + T cells have been primed to the relevant clonal neoantigens present in higher quantity and concentration in HRP tumors. Therefore, tumors that are HRP may have more clonal neoantigens and may derive clinical benefit from immune based therapies such as vaccinations which increase the primed CD8+ T cell population.
Synthetic lethality
Research shows that identification of HRD tumors leads to more effective chemotherapeutic regimens for this subset of patients. Moreover, the understanding of the molecular mechanisms by which HRD cells are defective also gives way to the use of drugs which exploit the phenomenon of synthetic lethality. In this context, synthetic lethality refers to situations in which a single genetic aberration or chemical perturbagen is individually tolerated by a cell, but becomes lethal when combined with another genetic aberration or chemical perturbagen [132]. First described by Bridges in 1922 [133, 134], synthetic lethality now serves as the basis of pharmacological strategies targeting HRD tumor cells. Synthetic lethality may be pharmacologically realized in a number of ways. One of the most well studied involves the genetic aberrations that drive HRD, and PARP inhibitors that suppress single strand break repair mechanisms. While PARP functions in single-strand break repair, homologous recombination repairs double-strand breaks. Therefore, by inhibiting both mechanisms, the cancer cell is effectively unable to repair DNA damage, which then leads to apoptosis [135]. The therapeutic implications of successful HRD-PARPi synthetic lethality reinforces the need for enhanced definition of HRD biomarkers.
PARP inhibitors are being studied and are currently approved for used in the management of breast and ovarian cancer. There are multiple PARP inhibitors that have been approved by FDA for use in cancer treatment, including olaparib, niraparib, rucaparib, and talazoparib. Three of the four drugs, olaparib, rucaparib, and niraparib, are approved for the treatment of ovarian cancer. PARP inhibitors, such as the ones mentioned above, function by way of PARP-trapping. The PARP inhibitors can act on both PARP1 and PARP2 at the location of DNA damage, effectively trapping the enzymes from functioning (Fig. 2B). Since the PARP enzyme is non-functional, it can no longer recruit any other enzymes to repair the damage, and cell death ensues. The trapping mechanism makes PARP inhibition more effective than a knockout of the PARP enzyme because the PARP1-DNA complex demonstrates more cytotoxicity than the original unrepaired single-strand break [136]. Resistance to PARP inhibitors may develop via a number of mechanisms [56, 137] (Fig. 2C, D, E).
While the therapeutic value of PARP inhibitors is often determined based on BRCA1/2 mutational status, other clinically important tumor suppressor genes also contribute to synthetic lethality. One such example is RAD51, another enzyme involved in double-strand break repair. BRCA2 signals RAD51 to travel to sites of DSBs. At these sites, RAD51 will eventually signal strand invasion and subsequent homologous strand exchange for successful damage repair. A study combining the PARP inhibitor olaparib with BRCA2-RAD51 disruption showed synthetic lethality [138]. Another clinically significant relationship is that of DNA-PK, an enzyme that has an essential role in non-homologous end joining. When the subunit Ku on DNA-PK binds to double-strand breaks, it recruits a host of NHEJ proteins that can function to repair the DNA. The combined inactivation of DNA-PK and BRCA1 also results in synthetic lethality. Research has shown that the DNA-PK inhibitor, AZD7648, used in combination with olaparib, leads to cell death [139]. The synergistic effect between so many of these genes and the BRCA proteins expands the possibility of ovarian and breast cancer combination therapies.
Conclusions
Proficiency or deficiency in HRR is a critical metric of therapeutic selection and prognosis for ovarian and breast cancer patients. Techniques for determining HRR status in patients are currently underutilized, inconsistently implemented, and produce results that are often reduced to binary HRD or HRP designations in clinical practice. The designation of a patient’s tumor for individual HRR status assessment is important because therapeutic efficacy or patient prognosis vary according to the identification of genetic variations (e.g. BRCA1/2; RAD51, CDK12), the nature of these variations (i.e. pathogenic germline variant, somatic mutation, epigenetic change), and the site of these mutations (e.g. c.179 to c.505, c. 4328 to c. 4945). Optimal clinical outcomes require testing which consistently generate these data, and careful consideration of each patient’s tumor’s unique HRR status and etiology. Evidence of therapeutic impact based on HRR status are established in ovarian cancer and likely will have further impact in several other solid tumors.
Acknowledgements
The authors would like to acknowledge Brenda Marr for her competent and knowledgeable assistance in the preparation of the manuscript and Christina Egan for facilitating the collaboration among authors. Figures created with BioRender.com.
Abbreviations
- HRD
Homologous recombination deficiency
- PARP
Poly (ADP-ribose) polymerase
- PARPi
Poly (ADP-ribose) polymerase inhibitor
- HRR
Homologous recombination DNA repair
- HRP
Homologous recombination proficient
- NHEJ
Non-homologous DNA end joining
- Alt-EJ
Alternative end-joining
- EOC
Epithelial ovarian cancer
- RHR
Risk hazard ratio
- FFPE
Formalin-fixed paraffin-embedded
- ASCO
American Society of Clinical Oncology
- NPV
Negative predictive value
- PPV
Positive predictive value
- tMDS
Treatment related myelodysplastic syndrome
- tAML
Treatment related acute myeloid leukemia
- MMR
Mismatch repair deficiency
- DSB
Double stranded break
Authors’ contributions
JFC was involved in the conception, organization, writing and editing of the manuscript. NSN, KRE, CEG, were involved in the research, writing, and editing of the manuscript. DMH and LD contributed to the writing and editing of the manuscript. LS supervised and providing writing and editing support. JN conceived the paper topic, and oversaw the writing and editing of the manuscript. All authors approved the submission of the final manuscript.
Authors’ information
No information provided.
Funding
The authors have no funding to disclose.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have no competing interests to disclose.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477(1–2):131–153. doi: 10.1016/S0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 3.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18(1):99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pellegrino B, Musolino A, Llop-Guevara A, Serra V, De Silva P, Hlavata Z, et al. Homologous recombination repair deficiency and the immune response in breast cancer: a literature review. Transl Oncol. 2020;13(2):410–422. doi: 10.1016/j.tranon.2019.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang Q, Greenberg RA. Deciphering the BRCA1 tumor suppressor network. J Biol Chem. 2015;290(29):17724–17732. doi: 10.1074/jbc.R115.667931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Lu LY. BRCA1 and homologous recombination: implications from mouse embryonic development. Cell Biosci. 2020;10(1):49. doi: 10.1186/s13578-020-00412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daley JM, Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol. 2014;34(8):1380–1388. doi: 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Lu LY, Yu X. The role of BRCA1 in DNA damage response. Protein Cell. 2010;1(2):117–123. doi: 10.1007/s13238-010-0010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cetin B, Wabl CA, Gumusay O. The DNA damaging revolution. Crit Rev Oncol Hematol. 2020;156:103117. doi: 10.1016/j.critrevonc.2020.103117. [DOI] [PubMed] [Google Scholar]
- 11.Herceg Z, Wang ZQ. Functions of poly (ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat Res. 2001;477(1–2):97–110. doi: 10.1016/S0027-5107(01)00111-7. [DOI] [PubMed] [Google Scholar]
- 12.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9(8):619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 13.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34(21):6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol. 2012;84(2):137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Hoppe MM, Sundar R, Tan DSP, Jeyasekharan AD. Biomarkers for homologous recombination deficiency in cancer. J Natl Cancer Inst. 2018;110(7):704–713. doi: 10.1093/jnci/djy085. [DOI] [PubMed] [Google Scholar]
- 16.O'Kane GM, Jang GH, Denroche R, Zhang A, Picardo SL, Grant RC, et al. Outcomes and immunogenicity of pancreatic cancer stratified by the HRDetect score. J Clin Oncol. 2020;38(15_suppl):4630. [Google Scholar]
- 17.Chen J, Huang T, Liu M, Liao W, Fu M, Yang B, et al. The prognostic value of HRD mutations in liver cancer. J Clin Oncol. 2020;38(15_suppl):e13546-e. [Google Scholar]
- 18.Jiang Y, Dang S, Yang L, Han Y, Zhang Y, Mu T, et al. Association between homologous recombination deficiency and tumor mutational burden in lung cancer. J Clin Oncol. 2020;38(15_suppl):e21043-e. [Google Scholar]
- 19.Pilie PG, Zhou L, Peterson CB, Peng Y, Peng G, Jonasch E. Homologous repair deficiency in VHL-mutated clear cell renal cell carcinoma. J Clin Oncol. 2018;36(6_suppl):585. [Google Scholar]
- 20.Nguyen L, J WMM. Van Hoeck A, Cuppen E. Pan-cancer landscape of homologous recombination deficiency. Nat Commun. 2020;11(1):5584. doi: 10.1038/s41467-020-19406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D'Andrea AD. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 2015;5(11):1137–1154. doi: 10.1158/2159-8290.CD-15-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorski JW, Ueland FR, Kolesar JM. CCNE1 amplification as a predictive biomarker of chemotherapy resistance in epithelial ovarian cancer. Diagnostics (Basel) 2020;10(5):279. doi: 10.3390/diagnostics10050279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muggia F, Safra T. ‘BRCAness’ and its implications for platinum action in gynecologic cancer. Anticancer Res. 2014;34(2):551–556. [PMC free article] [PubMed] [Google Scholar]
- 24.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4(10):814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 25.Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lew DJ, Dulic V, Reed SI. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66(6):1197–1206. doi: 10.1016/0092-8674(91)90042-W. [DOI] [PubMed] [Google Scholar]
- 27.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24(17):2776–2786. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 28.Lee C, Fernandez KJ, Alexandrou S, Sergio CM, Deng N, Rogers S, et al. Cyclin E2 promotes whole genome doubling in breast cancer. Cancers (Basel) 2020;12(8):2268. doi: 10.3390/cancers12082268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alldredge J, Randall L. Germline and somatic tumor testing in gynecologic cancer care. Obstet Gynecol Clin N Am. 2019;46(1):37–53. doi: 10.1016/j.ogc.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beitsch PD, Whitworth PW, Hughes K, Patel R, Rosen B, Compagnoni G, et al. Underdiagnosis of hereditary breast cancer: are genetic testing guidelines a tool or an obstacle? J Clin Oncol. 2019;37(6):453–460. doi: 10.1200/JCO.18.01631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250(4988):1684–1689. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 32.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4(4):511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 33.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40(1):17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 34.John EM, Miron A, Gong G, Phipps AI, Felberg A, Li FP, et al. Prevalence of pathogenic BRCA1 mutation carriers in 5 US racial/ethnic groups. JAMA. 2007;298(24):2869–2876. doi: 10.1001/jama.298.24.2869. [DOI] [PubMed] [Google Scholar]
- 35.Malone KE, Daling JR, Doody DR, Hsu L, Bernstein L, Coates RJ, et al. Prevalence and predictors of BRCA1 and BRCA2 mutations in a population-based study of breast cancer in white and black American women ages 35 to 64 years. Cancer Res. 2006;66(16):8297–8308. doi: 10.1158/0008-5472.CAN-06-0503. [DOI] [PubMed] [Google Scholar]
- 36.Bodily WR, Shirts BH, Walsh T, Gulsuner S, King MC, Parker A, Roosan M, Piccolo SR. Effects of germline and somatic events in candidate BRCA-like genes on breast-tumor signatures. PLoS One. 2020;15(9):e0239197. doi: 10.1371/journal.pone.0239197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pogoda K, Niwińska A, Sarnowska E, Nowakowska D, Jagiełło-Gruszfeld A, Siedlecki J, et al. Effects of BRCA germline mutations on triple-negative breast cancer prognosis. J Oncol. 2020;2020:8545643. doi: 10.1155/2020/8545643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musolino A, Bella MA, Bortesi B, Michiara M, Naldi N, Zanelli P, et al. BRCA mutations, molecular markers, and clinical variables in early-onset breast cancer: a population-based study. Breast. 2007;16(3):280–292. doi: 10.1016/j.breast.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Mylavarapu S, Das A, Roy M. Role of BRCA mutations in the modulation of response to platinum therapy. Front Oncol. 2018;8:16. doi: 10.3389/fonc.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moschetta M, George A, Kaye SB, Banerjee S. BRCA somatic mutations and epigenetic BRCA modifications in serous ovarian cancer. Ann Oncol. 2016;27(8):1449–1455. doi: 10.1093/annonc/mdw142. [DOI] [PubMed] [Google Scholar]
- 41.Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20(3):764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Yang W, Li D, Yang JY, Guan R, Yang MQ. Toward the precision breast cancer survival prediction utilizing combined whole genome-wide expression and somatic mutation analysis. BMC Med Genet. 2018;11(Suppl 5):104. doi: 10.1186/s12920-018-0419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong Q, Peng HL, Zhao X, Zhang L, Hwang WT. Effects of BRCA1- and BRCA2-related mutations on ovarian and breast cancer survival: a meta-analysis. Clin Cancer Res. 2015;21(1):211–220. doi: 10.1158/1078-0432.CCR-14-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang T, Xu Y, Sheng S, Yuan H, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Xie Y. HER2 somatic mutations are associated with poor survival in HER2-negative breast cancers. Cancer Sci. 2017;108(4):671–677. doi: 10.1111/cas.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashraf AZ, Afroze SH, Osuji GA, Kayani SY, Colon N, Pantho AF, et al. Epigenetic modifications in ovarian cancer: a review. J Cancer Treat Diagn. 2020;4(2):17–35.
- 46.Choi YE, Pan Y, Park E, Konstantinopoulos P, De S, D'Andrea A, et al. MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. eLife. 2014;3:e02445. doi: 10.7554/eLife.02445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K, Weinstock DM, Gorospe M, Harris AL, Helleday T, Chowdhury D. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41(2):210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.King M-C, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302(5645):643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 49.Satagopan JM, Boyd J, Kauff ND, Robson M, Scheuer L, Narod S, et al. Ovarian cancer risk in Ashkenazi Jewish carriers of BRCA1 and BRCA2 mutations. Clin Cancer Res. 2002;8(12):3776–3781. [PubMed] [Google Scholar]
- 50.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Group ABCS Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br J Cancer. 2000;83(10):1301. doi: 10.1054/bjoc.2000.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G, Yang D, Sun Y, Shmulevich I, Xue F, Sood AK, et al. Differing clinical impact of BRCA1 and BRCA2 mutations in serous ovarian cancer. Pharmacogenomics. 2012;13(13):1523–1535. doi: 10.2217/pgs.12.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25(11):1329–1333. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caminsky NG, Mucaki EJ, Perri AM, Lu R, Knoll JH, Rogan PK. Prioritizing variants in complete hereditary breast and ovarian cancer genes in patients lacking known BRCA mutations. Hum Mutat. 2016;37(7):640–652. doi: 10.1002/humu.22972. [DOI] [PubMed] [Google Scholar]
- 55.Alaofi RK, Nassif MO, Al-Hajeili MR. Prophylactic mastectomy for the prevention of breast cancer: review of the literature. Avicenna J Med. 2018;8(3):67–77. doi: 10.4103/ajm.AJM_21_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keung MYT, Wu Y, Vadgama JV. PARP inhibitors as a therapeutic agent for homologous recombination deficiency in breast cancers. J Clin Med. 2019;8(4):435. doi: 10.3390/jcm8040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lips EH, Mulder L, Oonk A, van der Kolk LE, Hogervorst FB, Imholz AL, et al. Triple-negative breast cancer: BRCAness and concordance of clinical features with BRCA1-mutation carriers. Br J Cancer. 2013;108(10):2172–2177. doi: 10.1038/bjc.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rebbeck TR, Mitra N, Wan F, Sinilnikova OM, Healey S, McGuffog L, Mazoyer S, Chenevix-Trench G, Easton DF, Antoniou AC, Nathanson KL, CIMBA Consortium. Laitman Y, Kushnir A, Paluch-Shimon S, Berger R, Zidan J, Friedman E, Ehrencrona H, Stenmark-Askmalm M, Einbeigi Z, Loman N, Harbst K, Rantala J, Melin B, Huo D, Olopade OI, Seldon J, Ganz PA, Nussbaum RL, Chan SB, Odunsi K, Gayther SA, Domchek SM, Arun BK, Lu KH, Mitchell G, Karlan BY, Walsh C, Lester J, Godwin AK, Pathak H, Ross E, Daly MB, Whittemore AS, John EM, Miron A, Terry MB, Chung WK, Goldgar DE, Buys SS, Janavicius R, Tihomirova L, Tung N, Dorfling CM, van Rensburg E, Steele L, Neuhausen SL, Ding YC, Ejlertsen B, Gerdes AM, Hansen Tv, Ramón y Cajal T, Osorio A, Benitez J, Godino J, Tejada MI, Duran M, Weitzel JN, Bobolis KA, Sand SR, Fontaine A, Savarese A, Pasini B, Peissel B, Bonanni B, Zaffaroni D, Vignolo-Lutati F, Scuvera G, Giannini G, Bernard L, Genuardi M, Radice P, Dolcetti R, Manoukian S, Pensotti V, Gismondi V, Yannoukakos D, Fostira F, Garber J, Torres D, Rashid MU, Hamann U, Peock S, Frost D, Platte R, Evans DG, Eeles R, Davidson R, Eccles D, Cole T, Cook J, Brewer C, Hodgson S, Morrison PJ, Walker L, Porteous ME, Kennedy MJ, Izatt L, Adlard J, Donaldson A, Ellis S, Sharma P, Schmutzler RK, Wappenschmidt B, Becker A, Rhiem K, Hahnen E, Engel C, Meindl A, Engert S, Ditsch N, Arnold N, Plendl HJ, Mundhenke C, Niederacher D, Fleisch M, Sutter C, Bartram CR, Dikow N, Wang-Gohrke S, Gadzicki D, Steinemann D, Kast K, Beer M, Varon-Mateeva R, Gehrig A, Weber BH, Stoppa-Lyonnet D, Sinilnikova OM, Mazoyer S, Houdayer C, Belotti M, Gauthier-Villars M, Damiola F, Boutry-Kryza N, Lasset C, Sobol H, Peyrat JP, Muller D, Fricker JP, Collonge-Rame MA, Mortemousque I, Nogues C, Rouleau E, Isaacs C, de Paepe A, Poppe B, Claes K, de Leeneer K, Piedmonte M, Rodriguez G, Wakely K, Boggess J, Blank SV, Basil J, Azodi M, Phillips KA, Caldes T, de la Hoya M, Romero A, Nevanlinna H, Aittomäki K, van der Hout A, Hogervorst FB, Verhoef S, Collée JM, Seynaeve C, Oosterwijk JC, Gille JJ, Wijnen JT, Gómez Garcia EB, Kets CM, Ausems MG, Aalfs CM, Devilee P, Mensenkamp AR, Kwong A, Olah E, Papp J, Diez O, Lazaro C, Darder E, Blanco I, Salinas M, Jakubowska A, Lubinski J, Gronwald J, Jaworska-Bieniek K, Durda K, Sukiennicki G, Huzarski T, Byrski T, Cybulski C, Toloczko-Grabarek A, Złowocka-Perłowska E, Menkiszak J, Arason A, Barkardottir RB, Simard J, Laframboise R, Montagna M, Agata S, Alducci E, Peixoto A, Teixeira MR, Spurdle AB, Lee MH, Park SK, Kim SW, Friebel TM, Couch FJ, Lindor NM, Pankratz VS, Guidugli L, Wang X, Tischkowitz M, Foretova L, Vijai J, Offit K, Robson M, Rau-Murthy R, Kauff N, Fink-Retter A, Singer CF, Rappaport C, Gschwantler-Kaulich D, Pfeiler G, Tea MK, Berger A, Greene MH, Mai PL, Imyanitov EN, Toland AE, Senter L, Bojesen A, Pedersen IS, Skytte AB, Sunde L, Thomassen M, Moeller ST, Kruse TA, Jensen UB, Caligo MA, Aretini P, Teo SH, Selkirk CG, Hulick PJ, Andrulis I. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA. 2015;313(13):1347–1361. doi: 10.1001/jama.2014.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336(20):1401–1408. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 60.Karami F, Mehdipour P. A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. Biomed Res Int. 2013;2013:928562. doi: 10.1155/2013/928562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Donnell JD, Johnson NC, Turbeville TD, Alfonso MY, Kruk PA. BRCA1 185delAG truncation protein, BRAt, amplifies caspase-mediated apoptosis in ovarian cells. In Vitro Cell Dev Biol Anim. 2008;44(8–9):357–367. doi: 10.1007/s11626-008-9122-0. [DOI] [PubMed] [Google Scholar]
- 62.O'Donnell JD, Linger RJ, Kruk PA. BRCA1 185delAG mutant protein, BRAt, up-regulates maspin in ovarian epithelial cells. Gynecol Oncol. 2010;116(2):262–268. doi: 10.1016/j.ygyno.2009.10.052. [DOI] [PubMed] [Google Scholar]
- 63.Latha K, Zhang W, Cella N, Shi HY, Zhang M. Maspin mediates increased tumor cell apoptosis upon induction of the mitochondrial permeability transition. Mol Cell Biol. 2005;25(5):1737–1748. doi: 10.1128/MCB.25.5.1737-1748.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chartron E, Theillet C, Guiu S, Jacot W. Targeting homologous repair deficiency in breast and ovarian cancers: biological pathways, preclinical and clinical data. Crit Rev Oncol Hematol. 2019;133:58–73. doi: 10.1016/j.critrevonc.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 65.Bajrami I, Frankum JR, Konde A, Miller RE, Rehman FL, Brough R, et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014;74(1):287–297. doi: 10.1158/0008-5472.CAN-13-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joshi PM, Sutor SL, Huntoon CJ, Karnitz LM. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly (ADP-ribose) polymerase inhibitors. J Biol Chem. 2014;289(13):9247–9253. doi: 10.1074/jbc.M114.551143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clark SL, Rodriguez AM, Snyder RR, Hankins GD, Boehning D. Structure-function of the tumor suppressor BRCA1. Comput Struct Biotechnol J. 2012;1(1):e201204005. doi: 10.5936/csbj.201204005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lord CJ, Ashworth A. RAD51, BRCA2 and DNA repair: a partial resolution. Nat Struct Mol Biol. 2007;14(6):461–462. doi: 10.1038/nsmb0607-461. [DOI] [PubMed] [Google Scholar]
- 69.Brown LA, Irving J, Parker R, Kim H, Press JZ, Longacre TA, Chia S, Magliocco A, Makretsov N, Gilks B, Pollack J, Huntsman D. Amplification of EMSY, a novel oncogene on 11q13, in high grade ovarian surface epithelial carcinomas. Gynecol Oncol. 2006;100(2):264–270. doi: 10.1016/j.ygyno.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 70.Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115(5):523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 71.Chu EC, Tarnawski AS. PTEN regulatory functions in tumor suppression and cell biology. Med Sci Monit. 2004;10(10):RA235–RA241. [PubMed] [Google Scholar]
- 72.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1(6–7):315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128(1):157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 74.Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017;18(10):610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379(26):2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 76.Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, Korach J, Huzarski T, Poveda A, Pignata S, Friedlander M, Colombo N, Harter P, Fujiwara K, Ray-Coquard I, Banerjee S, Liu J, Lowe ES, Bloomfield R, Pautier P, Korach J, Huzarski T, Byrski T, Pautier P, Friedlander M, Harter P, Colombo N, Pignata S, Scambia G, Nicoletto M, Nussey F, Clamp A, Penson R, Oza A, Poveda Velasco A, Rodrigues M, Lotz JP, Selle F, Ray-Coquard I, Provencher D, Prat Aparicio A, Vidal Boixader L, Scott C, Tamura K, Yunokawa M, Lisyanskaya A, Medioni J, Pécuchet N, Dubot C, de la Motte Rouge T, Kaminsky MC, Weber B, Lortholary A, Parkinson C, Ledermann J, Williams S, Banerjee S, Cosin J, Hoffman J, Penson R, Plante M, Covens A, Sonke G, Joly F, Floquet A, Banerjee S, Hirte H, Amit A, Park-Simon TW, Matsumoto K, Tjulandin S, Kim JH, Gladieff L, Sabbatini R, O'Malley D, Timmins P, Kredentser D, Laínez Milagro N, Barretina Ginesta MP, Tibau Martorell A, Gómez de Liaño Lista A, Ojeda González B, Mileshkin L, Mandai M, Boere I, Ottevanger P, Nam JH, Filho E, Hamizi S, Cognetti F, Warshal D, Dickson-Michelson E, Kamelle S, McKenzie N, Rodriguez G, Armstrong D, Chalas E, Celano P, Behbakht K, Davidson S, Welch S, Helpman L, Fishman A, Bruchim I, Sikorska M, Słowińska A, Rogowski W, Bidziński M, Śpiewankiewicz B, Casado Herraez A, Mendiola Fernández C, Gropp-Meier M, Saito T, Takehara K, Enomoto T, Watari H, Choi CH, Kim BG, Kim JW, Hegg R, Vergote I. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18(9):1274–1284. doi: 10.1016/S1470-2045(17)30469-2. [DOI] [PubMed] [Google Scholar]
- 77.Ray-Coquard I, Pautier P, Pignata S, Perol D, Gonzalez-Martin A, Berger R, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381(25):2416–2428. doi: 10.1056/NEJMoa1911361. [DOI] [PubMed] [Google Scholar]
- 78.Aref-Eshghi E, McGee JD, Pedro VP, Kerkhof J, Stuart A, Ainsworth PJ, et al. Genetic and epigenetic profiling of BRCA1/2 in ovarian tumors reveals additive diagnostic yield and evidence of a genomic BRCA1/2 DNA methylation signature. J Hum Genet. 2020;65(10):865–873. doi: 10.1038/s10038-020-0780-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Anjum S, Fourkala EO, Zikan M, Wong A, Gentry-Maharaj A, Jones A, et al. A BRCA1-mutation associated DNA methylation signature in blood cells predicts sporadic breast cancer incidence and survival. Genome Med. 2014;6(6):47. doi: 10.1186/gm567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Evans DGR, van Veen EM, Byers HJ, Wallace AJ, Ellingford JM, Beaman G, Santoyo-Lopez J, Aitman TJ, Eccles DM, Lalloo FI, Smith MJ, Newman WG. A dominantly inherited 5′ UTR variant causing methylation-associated silencing of BRCA1 as a cause of breast and ovarian cancer. Am J Hum Genet. 2018;103(2):213–220. doi: 10.1016/j.ajhg.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schenkel LC, Kerkhof J, Stuart A, Reilly J, Eng B, Woodside C, et al. Clinical next-generation sequencing pipeline outperforms a combined approach using sanger sequencing and multiplex ligation-dependent probe amplification in targeted gene panel analysis. J Mol Diagn. 2016;18(5):657–667. doi: 10.1016/j.jmoldx.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 82.Toland AE, Forman A, Couch FJ, Culver JO, Eccles DM, Foulkes WD, et al. Clinical testing of BRCA1 and BRCA2: a worldwide snapshot of technological practices. NPJ Genom Med. 2018;3(1):7. doi: 10.1038/s41525-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park J, Jang W, Chae H, Kim Y, Chi HY, Kim M. Comparison of targeted next-generation and sanger sequencing for the BRCA1 and BRCA2 mutation screening. Ann Lab Med. 2016;36(2):197–201. doi: 10.3343/alm.2016.36.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Park HS, Park SJ, Kim JY, Kim S, Ryu J, Sohn J, et al. Next-generation sequencing of BRCA1/2 in breast cancer patients: potential effects on clinical decision-making using rapid, high-accuracy genetic results. Ann Surg Treat Res. 2017;92(5):331–339. doi: 10.4174/astr.2017.92.5.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoffman-Andrews L. The known unknown: the challenges of genetic variants of uncertain significance in clinical practice. J Law Biosci. 2017;4(3):648–657. doi: 10.1093/jlb/lsx038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang S, Royer R, Li S, McLaughlin JR, Rosen B, Risch HA, et al. Frequencies of BRCA1 and BRCA2 mutations among 1,342 unselected patients with invasive ovarian cancer. Gynecol Oncol. 2011;121(2):353–357. doi: 10.1016/j.ygyno.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 87.Norquist B, Wurz KA, Pennil CC, Garcia R, Gross J, Sakai W, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29(22):3008–3015. doi: 10.1200/JCO.2010.34.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15(8):852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 89.Ledermann JA, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott C, Meier W, Shapira-Frommer R, Safra T, Matei D, Fielding A, Spencer S, Rowe P, Lowe E, Hodgson D, Sovak MA, Matulonis U. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. Lancet Oncol. 2016;17(11):1579–1589. doi: 10.1016/S1470-2045(16)30376-X. [DOI] [PubMed] [Google Scholar]
- 90.Swisher EM, Harrell MI, Lin K, Coleman RL, Konecny GE, Tinker AV, et al. BRCA1 and RAD51C promoter hypermethylation confer sensitivity to the PARP inhibitor rucaparib in patients with relapsed, platinum-sensitive ovarian carcinoma in ARIEL2 part 1. Gynecol Oncol. 2017;145:5. [Google Scholar]
- 91.Childers CP, Childers KK, Maggard-Gibbons M, Macinko J. National estimates of genetic testing in women with a history of breast or ovarian cancer. J Clin Oncol. 2017;35(34):3800–3806. doi: 10.1200/JCO.2017.73.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Manchanda R, Loggenberg K, Sanderson S, Burnell M, Wardle J, Gessler S, Side L, Balogun N, Desai R, Kumar A, Dorkins H, Wallis Y, Chapman C, Taylor R, Jacobs C, Tomlinson I, McGuire A, Beller U, Menon U, Jacobs I. Population testing for cancer predisposing BRCA1/BRCA2 mutations in the Ashkenazi-Jewish community: a randomized controlled trial. J Natl Cancer Inst. 2015;107(1):379. doi: 10.1093/jnci/dju379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gabai-Kapara E, Lahad A, Kaufman B, Friedman E, Segev S, Renbaum P, Beeri R, Gal M, Grinshpun-Cohen J, Djemal K, Mandell JB, Lee MK, Beller U, Catane R, King MC, Levy-Lahad E. Population-based screening for breast and ovarian cancer risk due to BRCA1 and BRCA2. Proc Natl Acad Sci U S A. 2014;111(39):14205–14210. doi: 10.1073/pnas.1415979111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hawkins AK, Hayden MR. A grand challenge: providing benefits of clinical genetics to those in need. Genet Med. 2011;13(3):197–200. doi: 10.1097/GIM.0b013e31820c056e. [DOI] [PubMed] [Google Scholar]
- 95.Tai YC, Domchek S, Parmigiani G, Chen S. Breast cancer risk among male BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2007;99(23):1811–1814. doi: 10.1093/jnci/djm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mohamad HB, Apffelstaedt JP. Counseling for male BRCA mutation carriers: a review. Breast. 2008;17(5):441–450. doi: 10.1016/j.breast.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 97.Baumann P, West SC. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23(7):247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 98.Zhao M, Chen P, Dong Y, Zhu X, Zhang X. Relationship between Rad51 G135C and G172T variants and the susceptibility to cancer: a meta-analysis involving 54 case-control studies. PLoS One. 2014;9(1):e87259. doi: 10.1371/journal.pone.0087259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang BB, Wang DG, Xuan C, Sun GL, Deng KF. Genetic 135G/C polymorphism of RAD51 gene and risk of cancer: a meta-analysis of 28,956 cases and 28,372 controls. Familial Cancer. 2014;13(4):515–526. doi: 10.1007/s10689-014-9729-0. [DOI] [PubMed] [Google Scholar]
- 100.Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7(4):a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang X, Song H, Leslie G, Engel C, Hahnen E, Auber B, Horváth J, Kast K, Niederacher D, Turnbull C, Houlston R, Hanson H, Loveday C, Dolinsky JS, LaDuca H, Ramus SJ, Menon U, Rosenthal AN, Jacobs I, Gayther SA, Dicks E, Nevanlinna H, Aittomäki K, Pelttari LM, Ehrencrona H, Borg Å, Kvist A, Rivera B, Hansen TVO, Djursby M, Lee A, Dennis J, Bowtell DD, Traficante N, Diez O, Balmaña J, Gruber SB, Chenevix-Trench G, Investigators CF, Jensen A, Kjær SK, Høgdall E, Castéra L, Garber J, Janavicius R, Osorio A, Golmard L, Vega A, Couch FJ, Robson M, Gronwald J, Domchek SM, Culver JO, de la Hoya M, Easton DF, Foulkes WD, Tischkowitz M, Meindl A, Schmutzler RK, Pharoah PDP, Antoniou AC. Ovarian and breast cancer risks associated with pathogenic variants in RAD51C and RAD51D. J Natl Cancer Inst. 2020;112(12):1242–1250. doi: 10.1093/jnci/djaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cruz C, Castroviejo-Bermejo M, Gutierrez-Enriquez S, Llop-Guevara A, Ibrahim YH, Gris-Oliver A, et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol. 2018;29(5):1203–1210. doi: 10.1093/annonc/mdy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Easton DF, Pharoah PD, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372(23):2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Blanco A, Gutierrez-Enriquez S, Santamarina M, Montalban G, Bonache S, Balmana J, et al. RAD51C germline mutations found in Spanish site-specific breast cancer and breast-ovarian cancer families. Breast Cancer Res Treat. 2014;147(1):133–143. doi: 10.1007/s10549-014-3078-4. [DOI] [PubMed] [Google Scholar]
- 105.Xu L, Wu T, Lu S, Hao X, Qin J, Wang J, et al. Mitochondrial superoxide contributes to oxidative stress exacerbated by DNA damage response in RAD51-depleted ovarian cancer cells. Redox Biol. 2020;36:101604. doi: 10.1016/j.redox.2020.101604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Watkins JA, Irshad S, Grigoriadis A, Tutt AN. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16(3):211. doi: 10.1186/bcr3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.do Canto LM, Larsen SJ, Catin Kupper BE, Begnami M, Scapulatempo-Neto C, Petersen AH, et al. Increased levels of genomic instability and mutations in homologous recombination genes in locally advanced rectal carcinomas. Front Oncol. 2019;9(395):395. doi: 10.3389/fonc.2019.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moore K, Secord A, Geller M, Miller D, Cloven N, Fleming G, et al. Abstract NT-101: QUADRA: A PHASE 2, OPEN-LABEL, SINGLE-ARM STUDY TO EVALUATE SINGLE-AGENT NIRAPARIB TREATMENT IN PATIENTS WITH RELAPSED OVARIAN CANCER (ROC) WHO HAVE RECEIVED ≥3 PRIOR CHEMOTHERAPY REGIMENS. Clin Cancer Res. 2019;25(22 Supplement):NT-101-NT. [Google Scholar]
- 109.Coleman RL, Fleming GF, Brady MF, Swisher EM, Steffensen KD, Friedlander M, et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med. 2019;381(25):2403–2415. doi: 10.1056/NEJMoa1909707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rund D, Krichevsky S, Bar-Cohen S, Goldschmidt N, Kedmi M, Malik E, Gural A, Shafran-Tikva S, Ben-Neriah S, Ben-Yehuda D. Therapy-related leukemia: clinical characteristics and analysis of new molecular risk factors in 96 adult patients. Leukemia. 2005;19(11):1919–1928. doi: 10.1038/sj.leu.2403947. [DOI] [PubMed] [Google Scholar]
- 111.Sharma GN, Dave R, Sanadya J, Sharma P, Sharma KK. Various types and management of breast cancer: an overview. J Adv Pharm Technol Res. 2010;1(2):109–126. [PMC free article] [PubMed] [Google Scholar]
- 112.Herzog TJ. Recurrent ovarian cancer: how important is it to treat to disease progression? Clin Cancer Res. 2004;10(22):7439–7449. doi: 10.1158/1078-0432.CCR-04-0683. [DOI] [PubMed] [Google Scholar]
- 113.Goff BA. Advanced ovarian cancer: what should be the standard of care? J Gynecol Oncol. 2013;24(1):83–91. doi: 10.3802/jgo.2013.24.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vergote I, Tropé CG, Amant F, Kristensen GB, Ehlen T, Johnson N, et al. Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. N Engl J Med. 2010;363(10):943–953. doi: 10.1056/NEJMoa0908806. [DOI] [PubMed] [Google Scholar]
- 115.Tan DS, Kaye SB. Chemotherapy for patients with BRCA1 and BRCA2-mutated ovarian cancer: same or different? Am Soc Clin Oncol Educ Book / ASCO Am Soc Clin Oncol Meet. 2015;35:114–121. doi: 10.14694/EdBook_AM.2015.35.114. [DOI] [PubMed] [Google Scholar]
- 116.Pfisterer J, Plante M, Vergote I, du Bois A, Hirte H, Lacave AJ, et al. Gemcitabine plus carboplatin compared with carboplatin in patients with platinum-sensitive recurrent ovarian cancer: an intergroup trial of the AGO-OVAR, the NCIC CTG, and the EORTC GCG. J Clin Oncol. 2006;24(29):4699–4707. doi: 10.1200/JCO.2006.06.0913. [DOI] [PubMed] [Google Scholar]
- 117.Sharma P, Barlow WE, Godwin AK, Pathak H, Isakova K, Williams D, et al. Impact of homologous recombination deficiency biomarkers on outcomes in patients with triple-negative breast cancer treated with adjuvant doxorubicin and cyclophosphamide (SWOG S9313) Ann Oncol. 2018;29(3):654–660. doi: 10.1093/annonc/mdx821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maples P, Kumar P, Yu Y, Wang Z, Jay C, Pappen B, et al. FANG vaccine: autologous tumor cell vaccine genetically modified to express GM-CSF and block production of Furin. Bioprocess J. 2010;8:4–14. [Google Scholar]
- 119.Ghisoli M, Barve M, Schneider R, Mennel R, Lenarsky C, Wallraven G, Pappen BO, LaNoue J, Kumar P, Nemunaitis D, Roth A, Nemunaitis J, Whiting S, Senzer N, Fletcher FA, Nemunaitis J. Pilot trial of FANG immunotherapy in Ewing’s sarcoma. Mol Ther. 2015;23(6):1103–1109. doi: 10.1038/mt.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oh J, Barve M, Matthews CM, Koon EC, Heffernan TP, Fine B, et al. Phase II study of vigil® DNA engineered immunotherapy as maintenance in advanced stage ovarian cancer. Gynecol Oncol. 2016;143(3):504–510. doi: 10.1016/j.ygyno.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 121.Oh J, Barve M, Senzer N, Aaron P, Manning L, Wallraven G, et al. Long-term follow-up of phase 2A trial results involving advanced ovarian cancer patients treated with vigil® in frontline maintenance. Gynecol Oncol Rep. 2020;34:1–3. doi: 10.1016/j.gore.2020.100648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Senzer N, Barve M, Kuhn J, Melnyk A, Beitsch P, Lazar M, et al. Phase I trial of “bi-shRNAi (furin)/GMCSF DNA/autologous tumor cell” vaccine (FANG) in advanced cancer. Mol Ther. 2012;20(3):679–686. doi: 10.1038/mt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Senzer N, Barve M, Nemunaitis J, Kuhn J, Melnyk A, Beitsch P, et al. Long term follow up: phase I trial of “bi-shRNA furin/GMCSF DNA/autologous tumor cell” immunotherapy (FANG™) in advanced Cancer. J Vaccines Vaccin. 2013;4(8):209. [Google Scholar]
- 124.Rocconi RP, Grosen EA, Ghamande SA, Chan JK, Barve MA, Oh J, et al. Gemogenovatucel-T (Vigil) immunotherapy as maintenance in frontline stage III/IV ovarian cancer (VITAL): a randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Oncol. 2020;21(12):1661–1672. doi: 10.1016/S1470-2045(20)30533-7. [DOI] [PubMed] [Google Scholar]
- 125.Rocconi RP, Monk BJ, Walter A, Herzog TJ, Galanis E, Manning L, Bognar E, Wallraven G, Stanbery L, Aaron P, Senzer N, Coleman RL, Nemunaitis J. Gemogenovatucel-T (Vigil) immunotherapy demonstrates clinical benefit in homologous recombination proficient (HRP) ovarian cancer. Gynecol Oncol. 2021;161(3):676–680. doi: 10.1016/j.ygyno.2021.03.009. [DOI] [PubMed] [Google Scholar]
- 126.González-Martín A, Pothuri B, Vergote I, DePont CR, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2019;381(25):2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 127.Morice P-M, Leary A, Dolladille C, Chrétien B, Poulain L, González-Martín A, Moore K, O'Reilly EM, Ray-Coquard I, Alexandre J. Myelodysplastic syndrome and acute myeloid leukaemia in patients treated with PARP inhibitors: a safety meta-analysis of randomised controlled trials and a retrospective study of the WHO pharmacovigilance database. Lancet Haematol. 2021;8(2):e122–ee34. doi: 10.1016/S2352-3026(20)30360-4. [DOI] [PubMed] [Google Scholar]
- 128.Germano G, Lamba S, Rospo G, Barault L, Magrì A, Maione F, Russo M, Crisafulli G, Bartolini A, Lerda G, Siravegna G, Mussolin B, Frapolli R, Montone M, Morano F, de Braud F, Amirouchene-Angelozzi N, Marsoni S, D’Incalci M, Orlandi A, Giraudo E, Sartore-Bianchi A, Siena S, Pietrantonio F, di Nicolantonio F, Bardelli A. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature. 2017;552(7683):116–120. doi: 10.1038/nature24673. [DOI] [PubMed] [Google Scholar]
- 129.Choucair K, Morand S, Stanbery L, Edelman G, Dworkin L, Nemunaitis J. TMB: a promising immune-response biomarker, and potential spearhead in advancing targeted therapy trials. Cancer Gene Ther. 2020;27(12):841–853. doi: 10.1038/s41417-020-0174-y. [DOI] [PubMed] [Google Scholar]
- 130.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Herron J, Smith N, Stanbery L, Aaron P, Manning L, Bognar E, et al. Vigil: personalized immunotherapy generating systemic cytotoxic T cell response. Cancer Sci Res. 2020;3(1):1–4. [Google Scholar]
- 132.Yang H, Cui W, Wang L. Epigenetic synthetic lethality approaches in cancer therapy. Clin Epigenetics. 2019;11(1):136. doi: 10.1186/s13148-019-0734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ferrari E, Lucca C, Foiani M. A lethal combination for cancer cells: synthetic lethality screenings for drug discovery. Eur J Cancer. 2010;46(16):2889–2895. doi: 10.1016/j.ejca.2010.07.031. [DOI] [PubMed] [Google Scholar]
- 134.Bridges CB. The mutants of Drosophila melanogaster. 1944. [Google Scholar]
- 135.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. 2011;5(4):387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72(21):5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li H, Liu Z-Y, Wu N, Chen Y-C, Cheng Q, Wang J. PARP inhibitor resistance: the underlying mechanisms and clinical implications. Mol Cancer. 2020;19(1):107. doi: 10.1186/s12943-020-01227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Falchi F, Giacomini E, Masini T, Boutard N, Di Ianni L, Manerba M, et al. Synthetic lethality triggered by combining olaparib with BRCA2-Rad51 disruptors. ACS Chem Biol. 2017;12(10):2491–2497. doi: 10.1021/acschembio.7b00707. [DOI] [PubMed] [Google Scholar]
- 139.Topatana W, Juengpanich S, Li S, Cao J, Hu J, Lee J, et al. Advances in synthetic lethality for cancer therapy: cellular mechanism and clinical translation. J Hematol Oncol. 2020;13(1):118. doi: 10.1186/s13045-020-00956-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.