Abstract
Background
Seed flooding stress is one of the threatening environmental stressors that adversely limits soybean at the germination stage across the globe. The knowledge on the genetic basis underlying seed-flooding tolerance is limited. Therefore, we performed a genome-wide association study (GWAS) using 34,718 single nucleotide polymorphism (SNPs) in a panel of 243 worldwide soybean collections to identify genetic loci linked to soybean seed flooding tolerance at the germination stage.
Results
In the present study, GWAS was performed with two contrasting models, Mixed Linear Model (MLM) and Multi-Locus Random-SNP-Effect Mixed Linear Model (mrMLM) to identify significant SNPs associated with electrical conductivity (EC), germination rate (GR), shoot length (ShL), and root length (RL) traits at germination stage in soybean. With MLM, a total of 20, 40, 4, and 9 SNPs associated with EC, GR, ShL and RL, respectively, whereas in the same order mrMLM detected 27, 17, 13, and 18 SNPs. Among these SNPs, two major SNPs, Gm_08_11971416, and Gm_08_46239716 were found to be consistently connected with seed-flooding tolerance related traits, namely EC and GR across two environments. We also detected two SNPs, Gm_05_1000479 and Gm_01_53535790 linked to ShL and RL, respectively. Based on Gene Ontology enrichment analysis, gene functional annotations, and protein-protein interaction network analysis, we predicted eight candidate genes and three hub genes within the regions of the four SNPs with Cis-elements in promoter regions which may be involved in seed-flooding tolerance in soybeans and these warrant further screening and functional validation.
Conclusions
Our findings demonstrate that GWAS based on high-density SNP markers is an efficient approach to dissect the genetic basis of complex traits and identify candidate genes in soybean. The trait associated SNPs could be used for genetic improvement in soybean breeding programs. The candidate genes could help researchers better understand the molecular mechanisms underlying seed-flooding stress tolerance in soybean.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-021-03268-z.
Keywords: Soybean, Tolerance to submergence stress, QTL mapping, Candidate genes
Background
Soybean [Glycine max (L.) Merr.] is one of the most economically important legume crops, providing protein, oil, carbohydrates, minerals, vitamins, folic acid, fibre, isoflavones and other nutrients for humans and animals [1, 2]. The increased importance of soybeans has resulted in a massive expansion of soybean production in the world. In recent decades, soybean production in the world has increased dramatically from 268.58 million metric tons in 2012/2013 to 360.07 million metric tons in 2018/2019 (Statista 2020; https://www.statista.com). However, sustainable soybean production is threatened by various abiotic stresses, including flooding. The soybean yield is reduced by flooding in the various stages of growth [3–6]. Effect of flooding on soybean includes foliar chlorosis, necrosis, stunted growth, defoliation, reduction in nitrogen fixation and plant death [7–9]. Plants undergo different mechanisms, including morphological, physiological, and biochemical adaptations under flooding stress at germination and seedling stages [10, 11]. Therefore, comprehending the variation in flood tolerance among the genotypes and their underlying genetic architecture is important to develop an effective breeding strategy.
Over the past decades, extensive progress has been made in identifying essential parameters for assessing soybean flooding tolerance/susceptibility at germination or seedling stages [12–14]. Flooding is a complex quantitative trait influenced by multiple quantitative trait loci (QTLs)/genes with both major and minor effects and are largely influenced by the environment. Several QTLs for soybean flooding tolerance have been reported using bi-parental mapping populations [14–18]. So far, 27 QTLs have been detected and reported on SoyBase, which are scattered across the 20 chromosomes in the soybean genome, while several are recently detected [19, 20]. For instance, Dhungana et al. (2020) recently identified 20 QTLs with phenotypic variation explained (R2) and log of odd (LOD) in the range of 5.8–33.3% and 3.59–19.73%, respectively. Out of these, chromosomes 10, 12, and 13 harbored relatively more stable QTLs. However, most of the earlier reported QTLs were detected by linkage mapping strategy with several limitations, making them challenging to use for breeding program. Genome-wide association study (GWAS) provides more extensive ancestral recombination events due to high density of SNP markers. Therefore, to overcome the limitations associated with bi-parental mapping, GWAS has proven to be more effective and efficient in unraveling the genetic architecture of complex traits in soybean [21–25].
In soybean, there are only two independent studies on flooding tolerance with GWAS [26, 27]. In the study of Yu et al. (2019), 25 and 21 quantitative trait nucleotides (QTNs) were detected by the mixed linear model (MLM) and multi-locus random-SNP-effect mixed linear model (mrMLM) of GWAS, respectively. Out of these, QTN13 on chromosome Chr13 detected by the two distinct models for germination rate (GR), electrical conductivity (EC), and normal seedling rate (NSR). From QTN13, one candidate gene, Glyma.13 g248000 (GmSFT) was predicted, which was found to have a nonsynonymous mutation in the seed flooding tolerant genotypes, resulting in an amino acid alternation in the protein. Wu et al. (2020) examined flooding tolerance when more than 50% of plants were in the reproductive growth stage with a blooming flower. Fourteen SNPs distributed on 4 chromosomes viz., Chr03 (8), Chr04 (1), Chr07 (2), Chr13 (1) and Chr19 (2) were detected simultaneously by 4 single-locus models.
From the above, it is clear that less progress has been made in identifying genomic regions associated with soybean flooding tolerance, which is the first step toward identifying candidate genes. In order to speed up the genetic improvement of flooding tolerance in soybean, it is necessary to uncover the molecular mechanisms and develop gene-based functional markers suitable for marker-assisted selection (MAS). The present study used 243 diverse soybean plant introductions (PIs) obtained from the SoySNP50K BeadChip project available on SoyBase (http:www.soybase.org) [28] and utilized two distinct models of GWAS to identify SNPs linked to soybean seed flooding tolerance indices at the germination stage. In addition, putative candidate genes within the stable regions were predicted. The findings may be helpful for MAS to develop high-yielding and seed flood-tolerant soybean cultivars.
Results
Phenotypic variation and correlation among seed flooding tolerance parameters
To evaluate the phenotypic variation under seed flooding stress in the 243 soybean PIs accessions, the seeds were treated for 72 h under flooding stress and well-watered control. Four traits viz., electrical conductivity (EC), germination rate (GR), shoot length (ShL), and root length (RL) linked to seed flooding tolerance were measured at the germination stag. The descriptive statistics, h2, and ANOVA of each trait for the 243 soybean accessions in 2018 and 2019 are presented in Table 1. The h2 for the four traits ranged from 0.78–0.99 (Table 1). EC was significantly affected by genotype (G), environment (E), and their interaction (GE) in the joint analysis. GR was influenced considerably by only G, whereas ShL and RL were affected by both G and GE (Table 1). There was a significant negative correlation coefficient (r2) with EC and the other three traits. On the other hand, significant positive r2 existed among the GR, ShL and RL (Table 1). The results revealed that these four traits are more under genetic control and can be used to screen and select for seed flooding tolerance.
Table 1.
Statistics for seed flooding tolerance traits of 243 PI soybean accessions
| Traita | Env.b | Mean ± SDc | Min | Max | h2d | F values from ANOVAe | Pearson correlation | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| G | E | G × E | GR | ShL | RL | ||||||
| EC | 2018 | 492.45 ± 294 | 120.00 | 1858.5 | 0.79 | 667.51*** | 21.67*** | 1.97*** | - 0.40* | - 0.50* | - 0.50* |
| 2019 | 485.69 ± 295 | 106.05 | 1867.5 | ||||||||
| GR | 2018 | 0.76 ± 0.15 | 0.19 | 0.95 | 0.98 | 21.48*** | 0.24 ns | 3.24ns | 0.36* | 0.38* | |
| 2019 | 0.76 ± 0.14 | 0.23 | 0.97 | ||||||||
| ShL | 2018 | 0.78 ± 0.15 | 0.27 | 0.99 | 0.98 | 30.94*** | 0.88ns | 2.17*** | 0.73* | ||
| 2019 | 0.79 ± 0.14 | 0.26 | 0.99 | ||||||||
| RL | 2018 | 0.72 ± 0.20 | 0.2 | 0.98 | 0.99 | 35.26*** | 0.78ns | 5.05*** | |||
| 2019 | 0.72 ± 0.19 | 0.24 | 0.98 | ||||||||
aEC Electrical conductivity (μS·cm−1/g−1), GR Germination rate, ShL Shoot length (cm), RL Root length (cm)
bEnvironment
cAverage of 2 biological repeats ± standard deviation
dBroad-sense heritability
eG-Genotype; E-Environment; GE-Genotype by Environment interaction
***indicates significance level at p < 0.001; ns indicates non-significant
*correlation is significant at the 0.05 level (2-tailed)
Distribution of SNPs, population structure and linkage disequilibrium decay
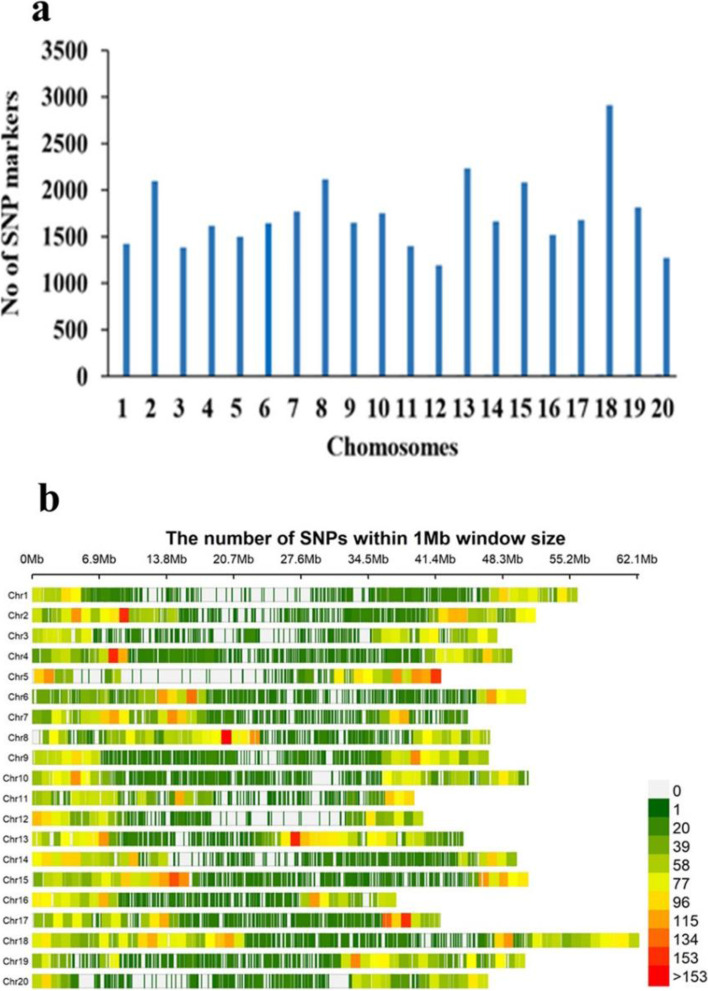
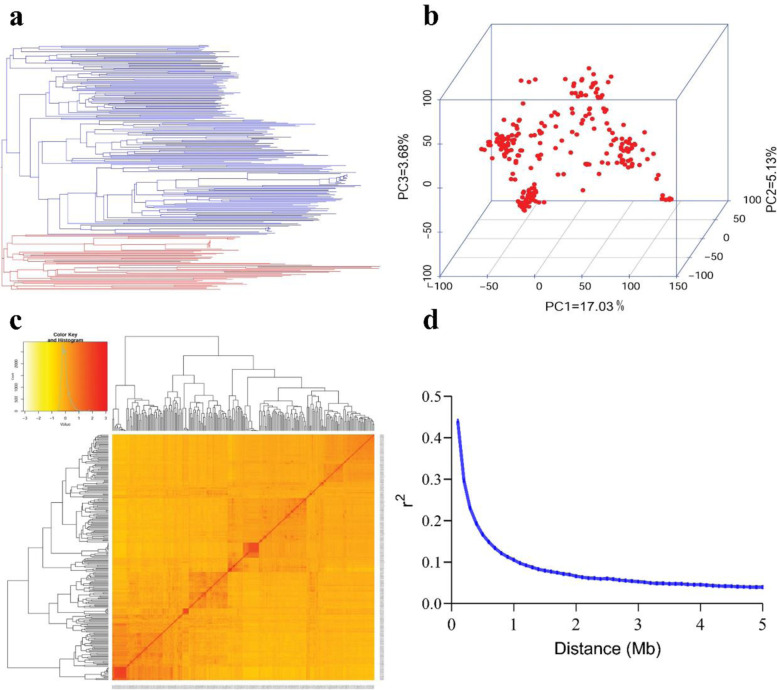
A total of 243 soybean PIs accessions were genotyped with 42,449 SNP markers; however, 34,718 SNPs with MAF ≥ 0.05 were used for further analyses. These SNPs were found on 20 chromosomes with an average of 1736 SNPs on each chromosome (Chr) with a maximum (2912) on the Chr18 and a minimum (1192) on Chr12 (Fig. 1a). Chr13 had the highest SNP marker density (50.49 SNPs/Mb), and Chr01 had the least marker density (11.84 SNPs/Mb) (Fig. 1b). In the present study, the population structure obtained by phylogenetic analysis of 34,718 SNPs divided 243 accessions into 2 clusters (Fig. 2a). The first three principal components (PCs) accounted for 25.84% of the total genetic variation (Fig. 2b). Based on the distribution of the pairwise relative kinship coefficients, the 243 tested PI accessions had a lower level of genetic relatedness, which aligned with the phylogenetic tree (Fig. 2c). Furthermore, r2 declined with increasing distance, and the average LD decay distance by 34,718 SNPs for LD analysis was about 400 kb, with r2 dropping to half of its highest value (Fig. 2d).
Fig. 1.
Distribution analysis of 34,718 SNPs across 243 soybean PIs. a Distribution of SNPs on 20 chromosomes. b The density of SNPs on each chromosome. The horizontal axis shows chromosome length (Mb); the vertical axis gives the chromosome number and the different colors depict SNP density (the number of SNPs per window)
Fig. 2.
Genetic features of the mapping population. a A maximum likelihood neighbor-joining tree of the tested 243 lines. b Population structure of soybean germplasm. c A heatmap of the kinship matrix of the 243 soybean PIs accessions. d LD decay of the genome-wide association study (GWAS) population
GWAS analysis with two distinct models
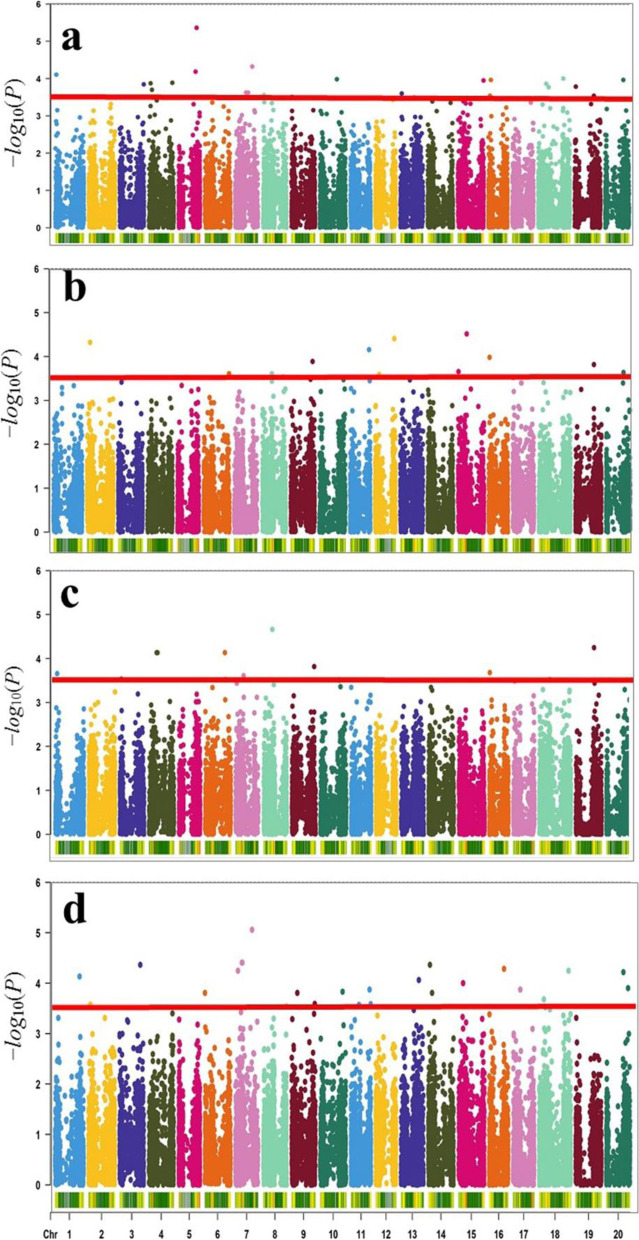
Two models comprising one single-locus model (MLM) and one multi-locus model (mrMLM) were used to perform SNP-trait association to identify significant SNP associated with EC, GR, ShL and RL. In MLM with a threshold of -log10(p) ≥ 3.5, a total of 20 SNPs were significantly associated with EC in 2018, 2019, plus the average across two environments on 4 chromosomes viz. Chr04, Chr08, Chr14, and Chr17 (Additional File 1: Table S1; Fig. 3a-d). We also identified 40, 4, and 9 SNPs associated with GR, ShL, and RL, respectively, via MLM in all environments (2018, 2019 and with the average of the 2 years) (Additional File 1: Table S1; Fig. 3a-d). Out of these, Gm_08_11971416 which associated with EC (log10(P) = 5.16) and GR (log10(P) = 5.06) met the Bonferroni correction criteria (≈ 4.54). SNPs related with GR Gm_08_46236506, Gm_08_46239716, and Gm_08_46242569 met the Bonferroni correction criteria as well (Additional File 1: Table S1). A total of 27, 17, 13, and 18 SNPs were detected by mrMLM for EC, GR, ShL, and RL traits, respectively, in the individual and joint environments (Additional File 2: Table S2). These SNPs were detected on all chromosomes with the exception of Chr06 and Chr17 with R2 (2.27–12.13%) and LOD (3.01–9.24, thus, −log10(P) = 3.71–10.17) (Additional File 2: Table S2).
Fig. 3.
Manhattan plots for GWAS of the 243 soybean accessions for Electrical conductivity (EC) (a); Germination rate (GR) (b); Shoot length (ShL) (c); Root length (RL) (d) using MLM (PCA + K). The threshold of 3.5 was adopted with a red line in the Manhattan plots. The X-axis represents chromosome number and Y-axis represents −log10(P)
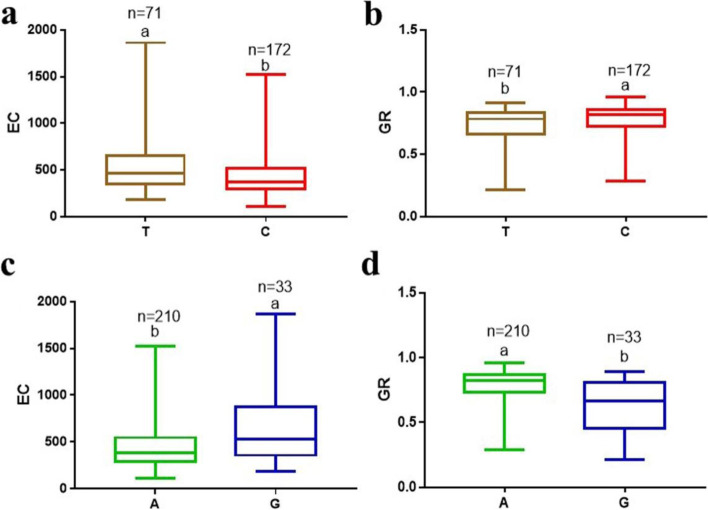
In the comparison of results from MLM and mrMLM, two major SNPs (Gm_08_11971416 and Gm_08_46239716) on Chr08 were detected concurrently for EC and GR (Table 2). In the same direction, Gm_08_11971416 was found to be associated with ShL and RL. The alleles at Gm_08_11971416 (C/T) and Gm_08_46239716 (A/G) caused significant variation in EC and GR (Fig. 4a-d). Moreover, Gm_05_1000479 and Gm_01_53535790 SNPs were identified for ShL and RL, respectively, in both models (Table 2). These four major SNPs on Chr01, Chr05, and Chr08 were considered as major and stable loci and used for mining potential candidate genes underlying seed-flooding tolerance related traits in this study.
Table 2.
Four SNPs associated with electrical conductivity (EC), germination rate (GR), shoot length (ShL), and root length (RL) on MLM and mrMLM models
| Traits a | SNP IDb | Chrc | Position (bp) | Allele | MLMd | mrMLMf | ||
|---|---|---|---|---|---|---|---|---|
| −log10 (p) | R2 (%)e | LODg | R2 (%)e | |||||
| EC | Gm_08_11971416 | 8 | 11,971,416 | C/T | 5.16–5.25 | 8.80–8.99 | 7.99–9.25 | 7.78–9.20 |
| Gm_08_46239716 | 8 | 46,239,716 | A/G | 3.98–4.08 | 6.48–6.66 | 3.78–5.53 | 4.88–7.26 | |
| GR | Gm_08_11971416 | 8 | 11,971,416 | C/T | 4.82–5.08 | 8.29–8.84 | 4.93–5.84 | 5.19–6.89 |
| Gm_08_46239716 | 8 | 46,239,716 | A/G | 4.88–5.79 | 8.26–10.12 | 7.14–7.24 | 9.54–12.13 | |
| ShL | Gm_05_1000479 | 5 | 1,000,479 | A/C | 3.73–4.31 | 6.08–6.87 | 7.38 | 9.92 |
| RL | Gm_01_53535790 | 1 | 53,535,790 | A/G | 3.78–3.88 | 6.09–6.28 | 3.69 | 6.07 |
aEC Electrical conductivity (μS·cm−1/g−1), GR Germination rate, ShL Shoot length (cm), RL Root length (cm)
bSingle nucleotide polymorphism
cChromosome
dMixed Linear Model
ePhenotypic variation explained (%)
fmulti-locus random-SNP-effect mixed linear model (mrMLM)
glogarithm of odds
Fig. 4.
Effect of alleles of Gm_08_11971416_C/T) and Gm_08_46239716_ A/G) on electrical conductivity (EC) expressed as μS·cm− 1 g− 1 and Germination rate (GR). a C/T on EC. b C/T on GR. c A/G on EC. d A/G on GR. The line in the middle of each box represents mean. The boxes in each plot with different alphabets on top indicate significant difference with Post-hoc means separation by Duncan Multiple Range Test at 5% probability. The n represents the number of accessions with the allele in focus
Allele effect of SNPs associated with seed flooding tolerance
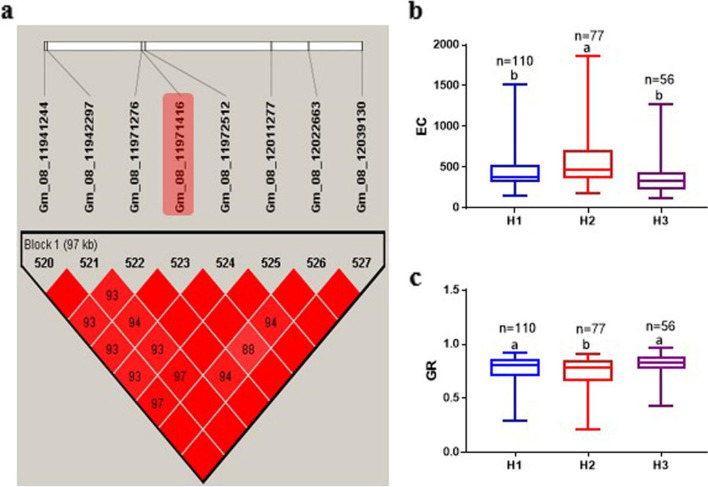
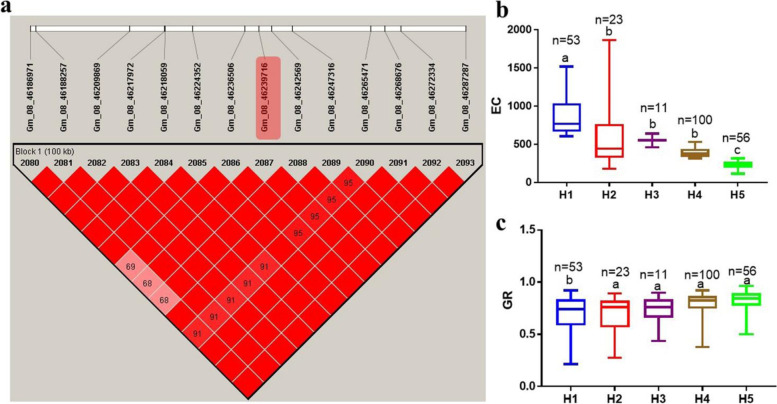
Haplotype block analyses were performed for Gm_08_11971416 and Gm_08_46239716 SNPs associated with EC and GR using Haploview 4.2 software. The distance within each block ranged 97–100 kb with range of 8–14 SNPs. The 243 soybean PIs accessions were grouped into three haplotypes (H1-H3) around Gm_08_11971416 (Fig. 5a). The mean ± standard error values of EC for H1, H2 and H3 varied significantly (P < 0.05), thus, 476.60 ± 25.46, 594.1 ± 39.25, and 365.4 ± 28.26 μS·cm− 1 g− 1, respectively (Fig. 5b). Similarly, groupings of the accessions on the basis of GR differed significantly (0.77 ± 0.01, 0.72 ± 0.02 and 0.81 ± 0.01 for H1, H2 and H3, respectively) (Fig. 5c). On the other hand, haplotype block around Gm_08_46239716 divided the 243 accessions into five groups (H1-H5) (Fig. 6a-b). The EC and GR of these groups differed significantly (P < 0.05). The highest EC was observed in H1 (873.7 ± 35.11 μS·cm− 1 g− 1), while H3 had the least EC (226.3 ± 7.25 μS·cm− 1 g− 1) (Fig. 6b). In terms of GR, the highest and least were observed in H5 (0.83 ± 0.01) and H2 (0.68 ± 0.04), respectively (Fig. 6c).
Fig. 5.
Haplotype block around Gm_08_11971416 and phenotypic variation analysis. a Haplotype block. b Haplotype groupings and their variation in electrical conductivity (EC, expressed as μS·cm− 1 g− 1). c Haplotype groupings and their variation in germination rate (GR). Groupings H1, H2 and H3 have TCTCATCC, TCCTGTCC and CTTCACTA, respectively. The line in the middle of each box represents mean. The boxes in each plot with different alphabets on top indicate significant difference with Post-hoc means separation by Duncan Multiple Range Test at 5% probability. The n represents the number of accessions with the alleles combined
Fig. 6.
Haplotype block around Gm_08_46239716 and phenotypic variation analysis. a Haplotype block. b Haplotype groupings and their variation in electrical conductivity (EC, expressed as μS·cm− 1 g− 1). c Haplotype groupings and their variation in germination rate (GR). Groupings H1, H2, H3, H4 and H5 have AGGTAAATCTATT, CAAGCCGCTCCCC, CAAGCAATCTATT, CAAGCCGCCTATT, and AGGTCAATCTATT, respectively. The line in the middle of each box represents mean. The boxes in each plot with different alphabets on top indicate significant difference with Post-hoc means separation by Duncan Multiple Range Test at 5% probability. The n represents the number of accessions with the alleles combined
Candidate and hub genes prediction underlying seed flooding tolerance
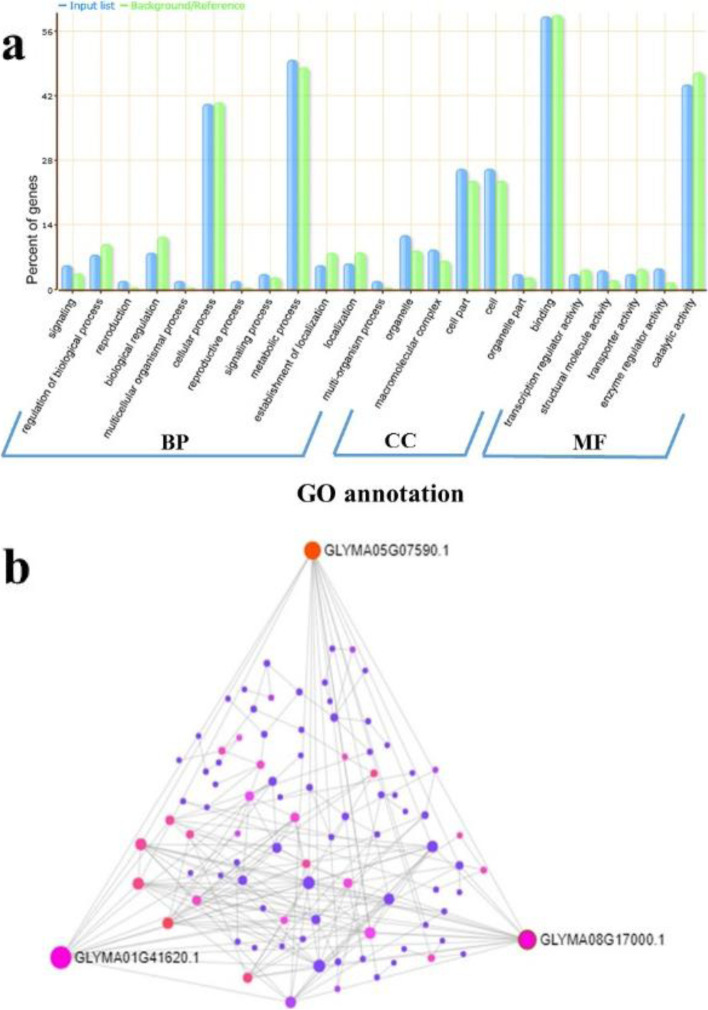
Based on the physical position of four SNPs viz., Gm_08_11971416, Gm_08_46239716, Gm_05_1000479, and Gm_01_53535790 linked with seed flooding tolerance, we performed the candidate gene prediction analysis with the SNP position ±500 kb. A total of 483 model genes were found within the regions of the four SNPs using Glyma2.0 models in SoyBase (Additional File 3: Table S3-S6). To further clarify the potential functions of these genes, various functional groups were categorized based on GO enrichment analysis (http://bioinfo.cau.edu.cn/agriGO). Of these genes, 61 had no functional annotations (representing NA) (Additional File 3: Table S3-S6). A total of 422 genes were assigned to one of the three GO categories: biological processes (BP), molecular function (MF) and cellular components (CC). The highest percentage of genes were connected with the GO terms cellular process (GO:0009987), metabolic process (GO:0008152), cell part (GO:0044464), cell (GO:0005623), binding (GO:0005488) and catalytic activity (GO:0003824) (Fig. 7a). Hence, these GO terms may play roles in regulating mechanisms in soybean seed-flooding stress.
Fig. 7.
Candidate gene analysis. a Diagram showing Gene Ontology (GO) enrichment analysis for Biological processes (BP), Cellular components (CC), and Molecular function (MF) using agriGO toolkit (b) Graphical depiction of the selective hub module of sub-network-1. Hub genes are highlighted in red and pink. Edges of the hub are shown in gray
The annotated 422 genes were related to plant growth regulation, flavonoids biosynthesis, phytohormone signaling pathways, stress signal perception and transduction, seed germination, flowering and senescence, embryo development, secondary wall synthesis, and plant hormonal responses, such as gibberellin responses, and ethylene biosynthesis (Additional File 3: Table S3-S6). By considering the results of GO enrichment analysis as well as gene functional annotations from the SoyBase, and Phytozome databases, we predicted eight stress responsive genes, Glyma.01G198000 (Transcription factor bHLH18), Glyma.01G206300 (G-Type Lectin S-Receptor-Like Serine/Threonine-Protein Kinase), Glyma.05G006700 (Protein kinase superfamily protein) Glyma.05G008000 (CCCH-type zinc finger family protein), Glyma.08G152800 (Leucine-rich repeat family protein), Glyma.08G152900 (Tetratricopeptide repeat like superfamily protein), Glyma.08G344500 (GATA Transcription factor 26), and Glyma.08G348500 (UDP-glycosyltransferase) from the 422 genes (Table 3). The results suggest that flood-response mechanisms in soybean are complex and may be governed by several signal pathways.
Table 3.
A subset of candidate and hub genes adjacent four SNPs associated with soybean seed flooding tolerance at the germination stage
| Candidate Gene ID a | Position (bp)b | Homolog in Arabidopsis | Functional annotationc |
|---|---|---|---|
| Glyma.01G198000 | 53,200,980–53,202,684 | AT4G37850 | Transcription factor bHLH18 |
| Glyma.01G206300 | 53,862,540–53,862,540 | AT5G60900 | G-Type Lectin S-Receptor-Like Serine/Threonine-Protein Kinase |
| Glyma.05G006700 | 608,574–621,690 | AT5G14720 | Protein kinase superfamily protein |
| Glyma.05G008000 | 749,837–753,271 | AT2G02160 | CCCH-type zinc finger family protein |
| Glyma.08G152800 | 11,760,404–11,762,319 | AT3G12610 | Leucine-rich repeat (LRR) family protein |
| Glyma.08G152900 | 11,780,677–11,784,610 | AT5G40410 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| Glyma.08G344500 | 45,957,501–45,962,417 | AT4G17570 | GATA Transcription factor 26 |
| Glyma.08G348500 | 46,292,645–46,301,440 | AT2G15490 | UDP-glycosyltransferase |
| Hub genes | |||
| Glyma.01G207700 | 53,959,688–53,961,931 | AT4G16720 | Ribosomal protein L23/L15e family protein |
| Glyma.05G016800 | 1,491,261–1,493,536 | AT5G08180 | Ribosomal protein L23/L15e family protein |
| Glyma.08G159800 | 12,386,775–12,390,398 | AT3G04770 | 40s ribosomal protein SA B |
aGenes position in Glyma2.0 version obtained from SoyBase while those with bolded font are hub genes (highly connected genes in the network)
bGenes start and end positions in Glyma2.0 version
cPossible functional annotation obtained from SoyBase (https://soybase.org/) and Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html)
To construct possible protein-protein interaction (PPI) networks associated with seed flooding tolerance, we used STRING online software. The possible network obtained from STRING was subsequently visualized in the Network Analyst platform. From PPI network analysis, we identified three hub genes viz., Glyma.01G207700 (Ribosomal protein L23/L15e family protein), Glyma.05G016800 (Ribosomal protein L23/L15e family protein) and Glyma.08G159800 (40s ribosomal protein SA B) that may be associated with soybean seed-flooding tolerance (Fig. 7b).
We retrieved RNA-Seq data of these candidate genes from SoyBase (www.soybase.org). Based on RNA-seq analysis, all the predicted candidate genes showed significantly higher gene expression in the root tissues, root nodule, young leaf, flower, and pod shell as well as seed developmental stages except Glyma.08G344500 (Additional File 4: Figure S1). In addition, the predicted candidate and hub genes were found to possess phytohormone Cis-elements such as auxin-responsive element (TGA-element and AuxRR-core), jasmonic acid (CGTCA-motif), abscisic acid (ABRE), salicylic acid (TCA-element) and others (Additional File 5: Table S7). Hence, these candidate and hub genes might be associated with soybean seed-flooding tolerance. However, they need further functional validation to check their actual roles in seed flooding tolerance.
Discussion
Genetic variation and correlation of traits evaluated for seed flooding tolerance in soybean
Seed flooding stress is a severe abiotic stress that reduces soybean seed yields by affecting seed germination, seedling growth, and development [27, 29]. Since seed-flooding tolerance is a complex quantitative trait, it is imperative to understand the genetic basis and genes involved in seed-flooding tolerance and this has been a major focus in soybean for a breeding program targeted at developing flood-tolerant cultivars. To date, the genetic mechanisms controlling seed flooding tolerance in soybean are not well understood at the germination stage. Seed germination is governed by different phytohormones and environmental conditions [30]. The strong correlation between seed-flooding tolerance and its related traits, viz., EC, GR, NSR, ShL, and RL in soybean has been previously reported [12–14, 26, 31]. Therefore, we examined EC, GR, ShL, and RL as the most important determinants to better estimate soybean seed-flooding tolerance. These traits are primarily regulated by genetic factors, as shown in ANOVA (Table 1) which are in consonance with earlier reports [27, 31]. According to a previous study, seed flooding stress caused seed material leakage and subsequent seed damage due to rapid water imbibition, which was confirmed by EC measurements [14]. As a result, we examined the EC of 243 soybean PIs accessions to better assess soybean seed-flooding tolerance. Furthermore, significant positive correlations among GR, ShL, and RL were observed in the soybean PIs accessions, and EC had a negative correlation with GR, ShL, and RL (Table 1). Overall, the comprehensive analysis of EC, GR, ShL, and RL is an important consideration in determining seed-flooding tolerance during germination in soybean.
Population structure and linkage disequilibrium in the panel
As evident from the phylogenetic tree and kinship plot, the population structure divided the 243 lines into 2 sub-groups (Fig. 2a, c). The sub-group 1 (blue) comprised mainly accessions from United States, Brazil, and Zimbabwe etc. whereas sub-group 2 (red) consisted accessions from mainly from Asia (China, Nepal, Bhutan, India, and Vietnam etc.). This is not surprising as soybean is noted to have originated and domesticated in East Asia [32]; thus, possibly the accessions used may have their lineage from Asia, specifically China. These findings pinpoint the grouping of accessions using molecular markers that showed a sub-structure based on the geographical origin of accessions. The extent of LD is a crucial determinant of association analysis efficiency [33]. In this study, the LD decay of the soybean genome was estimated to be about 400 kb which was considerably higher than that of other plants like rice (75 kb in indica rice) [34] and Arabidopsis thaliana (10 kb) [35]. This is related to the cleistogamous features of soybeans, which might have a significant impact on genomic homogeneity and lower genomic variation, and this character may become more sensitive to domestication practices due to low genetic diversity and high LD [36, 37].
According to previous studies, there are 27 QTLs associated with flooding tolerance in the soybean genome, distributed across 15 chromosomes, suggesting that flooding tolerance is a complex quantitative trait regulated by several genes with both substantial and modest impacts in soybean.
Significant SNPs and previously reported regions
Limited QTL mapping studies have been undertaken to detect the genomic regions linked with flooding tolerance using different bi-parental mapping populations. According to previous studies, there are 27 QTLs associated with flooding tolerance in the soybean genome, distributed across 15 chromosomes, suggesting that flooding tolerance is a complex quantitative trait and regulated by multiple genes with both major and minor effects in soybean [14–18]. However, most of these QTL mapping studies were conducted using low-density genetic maps based on linkage mapping. GWAS leverages on LD in the natural population and helps to overcome the limitations in linkage mapping. Hence, the present study used diverse soybean germplasm from 22 different countries to perform two distinct models of GWAS. There are numerous single-locus models, however MLM is more popular possibly because of its accounts of population structure and familial relatedness among the study population to avoid possible spurious marker-trait associations [38, 39]. However, the single-locus models are limited in detecting marginal effects QTNs influenced by the polygenic background and stringent Bonferroni correction [40]. For example, out of the numerous SNPs studied, only Gm_08_11,971,416 connected to EC and GR, Gm_08_46236506, Gm_08_46239716, and Gm_08_46242569 linked to GR met the Bonferroni correction criteria (≈ 4.54). Therefore, we adjusted the threshold in MLM to > 3.5 [41, 42]. Briefly, it has been established that a Bonferroni threshold (example P = 2.88E-05 or -log10(1/34,718) = 4.54, in the current study) is overly strict when the linkage disequilibrium among genetic markers is large, which is generally the case with soybean [43, 44]. Therefore, we adjusted to P ≤ 0.0003 or -log10(P) > 3.5 in the current study was used, which is less stringent than the Bonferroni-corrected threshold, but more stringent than the threshold used in the study by Wu et al. [27] (−log10(P) > 2.5) and Song et al. [45] (−log10(P) > 3.0) with accessions and SNPs from SoySNP50K BeadChip. The adopted threshold in study was similar to Kaler et al. (2017) who utilized 373 accessions and 31,260 SNPs and Chamarthi et al. (2021) with 200 accessions and 34,680 SNPs from SoySNP50K BeadChip to map genomic regions associated with soybean canopy wilting.
To overcome the limitation in MLM, we implemented mrMLM (Multi-locus model) with the LOD = 3 which has been recommended in multi-locus GWAS to balance high power and low false positive rate [46]. Multi-locus GWAS has been developed as a multidimensional genome scan method in which the effects of all markers are estimated at the same time [47]. mrMLM is emerged as a powerful tool for genetic dissection of complex traits [40].
Previous studies reported that GWAS is an effective and efficient strategy for detecting the genetic loci and candidate genes associated with complex quantitative traits [23, 48, 49]. MLM effectively controls genomic inflation and is widely used in genome-wide association analysis to identify QTL associated with soybean traits [50–52]. In the present study, a total of 73 and 75 significantly-associated SNPs of EC, GR, ShL, and RL traits were detected by MLM (Additional File 1: Table S1) and mrMLM, respectively (Additional File 2: Table S2). Among these SNPs, two major and stable SNPs, Gm_08_11971416 and Gm_08_46239716, were detected across the environments for EC and GR traits at the significance threshold -log10 (P) ≥ 3.5 (Table 2). In addition, Gm_05_1000479 and Gm_01_53535790 SNPs were identified for ShL and RL, respectively, in both models.
The stability of QTL is a requisite for their use in a practical breeding program such as MAS. Therefore, the major and stable SNPs (Gm_01_53,535,790, Gm_05_1,000,479, Gm_08_11,971,416, and Gm_08_46,239,716) (Table 2) could further be validated and exploited further for their use in the breeding program. Also, some associated SNPs in this study overlapped with a number of earlier reported regions; Gm_01_3260769 linked to ShL detected by mrMLM co-located with the physical region of Flood tolerance 6–1 (1031087–7,729,201 bp) [53]. Similarly, SNP at 10789902 bp overlapped with Flood tolerance 7–2 [14] on Chr08. Also, two SNPs; Gm_13_20995641 and Gm_14_4550573 co-located within the genomic regions of Flood tolerance 6–3 [53] and Flood tolerance 4–5 [16]. All the SNPs detected on Chr17 except for Gm_17_8735403 co-located within a recent QTL (qSFT_17–52) reported by Dhungana et al. [19]. Last but not the least, Flood tolerance 6–2 [53] harbored 2 SNPs on Chr19 in this study (Gm_19_40771881 & Gm_19_40783256). These results give credence to the findings of this study; however, the new SNPs/loci detected need further verification for their use in the breeding program. Also, the haplotype alleles detected on the major loci on Chr08 (Figs. 5 and 6) could be exploited for haplotype-based breeding [54, 55].
Candidate genes analysis
It is of great interest to identify the candidate genes underlying the genomic region for practical plant breeding. To date, the SoyBase has reported on 27 QTLs related to soybean flooding tolerance. However, none was used to mine putative candidate genes with the exception of recent studies [26, 27]. For example, GmSFT (Glyma.13 g248000) showed significantly higher expression levels in tolerant genotypes than sensitive genotype under seed flooding stress [27]. In this study, candidate genes were predicted in the genomic region surrounding four major and stable SNPs, viz., Gm_08_11971416, Gm_08_46239716, Gm_05_1000479, and Gm_01_53535790 (Table 2). Most of the genes are related with the terms cellular process, metabolic process, cell part, cell, binding, and catalytic activity as per GO enrichment analysis, and these GO terms are reported to be essential in seed flooding tolerance mechanisms [56].
Based on the GO enrichment analysis, and gene functional annotations we predicted eight genes as the possible candidate genes (Table 3). The gene (Glyma.01G198000), located on Chr01, encodes a basic helix-loop-helix (bHLH) transcription factor that is involved in plant adaptive responses to various abiotic stresses, including drought, salinity, heavy metal stress, oxidative stress, iron deficiency, low temperature stresses, and osmotic stress [57–59]. On Chr01, there is also a gene (Glyma.01G206300) that codes for G-Type Lectin S-Receptor-Like Serine/Threonine-Protein Kinase and plays a crucial role in plant response to salt stress [60]. Gene (Glyma.05G006700) encodes a protein kinase that is involved in plant responses to drought, salt, and cold stress [61]. Gene (Glyma.05G008000) located on Chr05, encodes a CCCH-type zinc finger protein. CCCH genes contribute to seed germination by regulating abscisic acid (ABA), light and gibberellic acid in Arabidopsis [62]. The previous study in rice reported that OsCCCH-Zn-1 is induced under hypoxia, submergence, and drought stresses [63]. On Chr08, Glyma.08G152800 encodes a leucine-rich repeat protein. Overexpression of the leucine-rich repeat receptor-like kinase gene (LRK2) increased drought tolerance in rice [64]. Gene (Glyma.08G152900) is also located on Chr08 which encodes for a tetratricopeptide repeat like protein (TPR) involved in the plant hormonal regulation, such as ethylene biosynthesis, gibberellic acid and cytokinin responses [65, 66]. TPR proteins have also been found to be involved in the regulation of ABA signaling and abiotic stress responses [67]. The recent study demonstrated that AtTPR10 functions as a molecular chaperone to protect plants from diverse abiotic stresses, such as low temperature, drought, and salinity [68].
Gene (Glyma.08G344500) encodes a GATA Transcription factor 26 that is involved in the regulation of growth processes and various environmental stresses, including salinity and drought [69]. A recent study by Zhao et al. (2021) revealed that overexpression of SlGATA17 (Solyc05g056120.2.1) enhances drought tolerance with increased activation of the phenylpropanoid biosynthetic pathway in transgenic tomato compared to the wild type [70]. In addition, Glyma.08G348500 encodes a UDP-glycosyltransferase that is involved in the regulation of grain size and multiple abiotic stress tolerance (salinity, drought and heat stress) in rice [71]. Furthermore, our study identified three hub genes (Glyma.01G207700, Glyma.05G016800, and Glyma.08G159800) as ribosomal protein genes in the functional co-expression network generated by network analyst that might regulate abiotic stress tolerance (Table 3). Previous studies reported that ribosomal protein genes enhance tolerance to drought, and heavy metal stress [72–74]. From the available RNA-seq data, the predicted candidate and hub genes expressed significantly higher gene expression in the various tissues and developmental stages, giving indication of their possibility of regulating soybean response to seed-flooding tolernace [75]. Hub genes (highly connected genes) are reported to modulate expression of large number of genes in a functional network of genes [23, 76]. In addition, with exception of Glyma.08 g152900, all the predicted candidate and hub genes contain Cis-elements which are essential in regulating plant response under flooding/waterlogging conditions (Additional File 5: Table S7). For example, Glyma.01 g198000, Glyma.01 g206300 and Glyma.05 g006700 contain auxin responsive element. It has been demonstrated that auxin accumulation in the stem triggers additional ethylene synthesis which stimulates a flux of auxin towards flooded parts of the plants [77, 78]. In addition, auxin accumulation in the base of the plant induces growth of pre-formed root initials thereby responding by new root system capable of replacing the original one when it has been damaged by submergence [77]. Moreover, Salicylic acid (SA) participates in the waterlogging-tolerance of plants [79]. SA content in waterlogging-tolerant soybean lines increased significantly after waterlogging for 5 or 10 days compared to non-waterlogging conditions while SA content in sensitive lines exhibited no significant change, implying that SA mediates waterlogging-tolerance of soybean through regulating the formation of aerenchyma or adventitious roots. One gene, Glyma.08 g348500 possesses SA responsive element (TCA-element). These warrant functional validation of the eight candidate and three hub genes to unravel their actual regulatory mechanism in seed flooding tolerance.
Conclusions
In this study, two distinct models of GWAS were used to decipher the genetic architecture underlying seed flooding stress tolerance in soybean. The major SNPs, Gm_08_11971416, Gm_08_46239716, Gm_05_1000479, and Gm_01_53535790 were identified to be associated with seed flooding tolerance related traits, viz., EC, GR, ShL and RL. Based on GO enrichment analysis, gene functional annotations, and PPI network analysis, we predicted eight candidate genes and three hub genes with functions directly or indirectly connected to stress defense mechanisms. However, further genetic and molecular analyses are required to validate the functional importance of the putative candidate genes in adaptation to seed flooding stress in soybean. Taken together, these findings provide valuable insight on the genetic basis of soybean seed flooding tolerance, and they could assist MAS in determining the molecular mechanism of seed flooding tolerance in soybean.
Materials and methods
Plant materials
A panel of 243 PIs originated from 22 different countries globally was selected from the United States Department of Agriculture, Soybean Germplasm Collection. The seeds of the selected PIs were obtained from the National Center for Soybean Improvement, Nanjing Agricultural University, Nanjing, China. These accessions were planted in 2018 and 2019 at Jiangpu Experimental Station (latitude 32.12°N; longitude 118.37°E) of Nanjing Agricultural University in Nanjing, Jiangsu Province of China. All the accessions were grown in a randomized complete block design with three replications every year. Additional File 6: Table S8 gives details information about the 243 soybean PIs accessions used in this study.
Phenotypic evaluation for seed-flooding tolerance
Soybean PIs accessions were phenotypically evaluated for seed flooding stress following a previously described method [12]. Briefly, good quality and healthy seeds were surface sterilized by soaking in 70% ethanol for 10 s to remove the contaminants. These seeds were then rinsed in distilled water three times. Twenty seeds of each accession were dipped into 350 ml plastic cups containing 50 ml distilled water covered with sterilized petri dishes for 72 h of seed flooding stress and incubated in a germination cabinet at 25 °C. The experiment was conducted with two replications arranged in a completely randomized design. All accessions were phenotypically evaluated for four traits viz., EC, GR, ShL, and RL at the germination stage (Additional File 7: Table S9). Immediately after the treatment, a conductivity meter (model: DDS-307A) was utilized to record the EC of steep-water. Germination experiment was carried out using a paper roll method in which seeds were grown for 5 days under normal conditions. The germination of seeds with a radicle length of more than 1 cm was considered. For the control, seeds without seed flooding treatment were grown under the same conditions. Relative values of each trait were obtained by dividing the treatment of each accession by its control.
Phenotypic data analysis
Analysis of variance (ANOVA) of the phenotypic data was performed using the PROC GLM procedure in SAS 9.4 (SAS Institute, Inc., Cary, NC, United States). Broad-sense heritability (h2) of each trait was estimated for the combined environments as for combined environments, where represents the genotypic variance, is the variance of the genotype-by-environment interaction, is the error variance, n is the number of environments, and r represents the number of replications within each environment [80]. Pearson correlations were also calculated to measure the degree of relationship between each pair of traits, and the individual hypothesis tests of the correlations were performed at α = 0.01 using OriginPro 9.0 software (Origin Lab, Corporation, Northampton, USA).
SNP genotyping and quality control
SNP data of the selected PIs is available at Illumina Infinium SoySNP50K BeadChip database on SoyBase (https://soybase.org/snps/index.php). The details on genotyping and SNP calling procedures for the 42,449 SNPs as described by Song et al. (2013). After removing SNPs with minor allele frequency (MAF) < 0.05, a total of 34,718 SNPs were used for further analysis in this study.
Population structure and linkage disequilibrium analyses
A neighbor-joining tree was constructed together with Principal component analysis (PCA) was calculated using TASSEL 5.0 software [38]. The 34,718 SNPs were also used to calculate kinship matrixes by the identity-by-state (IBS) method implemented in TASSEL 5.0 to infer population stratification and relatedness among individuals. A heatmap of the kinship matrix of the 243 accessions was constructed with the kinship 2 package in R (R Core Team, 2019).
Pairwise linkage disequilibrium (LD) between 34,718 SNPs with a missing rate < 10% and MAF ≥ 0.05 was estimated using squared allele frequency correlations (r2) using the RTM-GWAS V1.1 software [81]. The panel’s LD decay rate was calculated as the chromosomal distance when r2 fell to half its highest value. The average LD decay figure was drawn by GraphPad Prism version 5.01 (GraphPad Software, San Diego California USA, www.graphpad.com) using r2 for SNPs with pairwise distances less than 5 Mb in each chromosome.
Genome-wide association study and haplotype block analysis
An association analysis was performed for four traits viz., EC, GR, ShL, and RL through two distinct models, Mixed Linear Model (MLM) and Multi-Locus Random-SNP-Effect Mixed Linear Model (mrMLM) for each year plus average of phenotypic data across the 2 years. The MLM was run with the TASSEL 5.0 software, and the K matrix serves as a random effect in this model. However, mrMLM was carried out using the R package mrMLM.GUI version 2.1 [82]. The significance threshold used in the present study was set at -log10 (1/m) where m = the number of markers, thus -log10(1/34,718) = 4.54 as the Bonferroni correction line, however this threshold was adjusted to -log10 (P) ≥ 3.5 and logarithm of odd (LOD) = 3 were used to declare SNP-trait association in MLM [41, 42] and mrMLM [46] respectively.
Haplotype block analysis of the stable SNPs was investigated with Haploview software version 4.2 with the four-gamete rule method [83, 84]. Duncan Range Multiple test (pairwise comparison) was used to evaluate variation in seed flooding tolerance among accession groupings in each haplotype block at the significant level of P ≤ 0.05.
Candidate and hub genes identification
The stable SNPs (detected for at least 2 traits by either of the models or both) were used to mine putative candidate genes at 500 kb up- and down-stream of the SNP position by using Glyma2.0 models on SoyBase (http:www.soybase.org). The model genes were retrieved from each of the regions for candidate gene analysis. Gene ontology (GO) enrichment analysis was conducted for all the model genes within the four SNPs (±500 kb) using agriGO (http://bioinfo.cau.edu.cn/agriGO) [85], in which the parameters of singular enrichment analysis (SEA) tool following the default settings and G. max gene model as a reference background. Candidate genes associated with seed flooding tolerance were predicted based on GO enrichment analysis and gene functional annotations from the Phytozome (https://phytozome.jgi.doe.gov) and SoyBase (http://www.soybase.org) databases. These model genes were also used to construct probable protein-protein interaction (PPI) network with a publicly available online database Search Tool for Retrieval of Interacting Genes/Proteins (STRING) [86]. Credible PPI interactions were further visualized with the network analyst 3.0 to identify hub genes [87]. The expression data of predicted candidate genes were obtained from transcriptome profile data publicly available on SoyBase (https://soybase.org/soyseq/) developed by Severin et al. [75] and heatmapped by TBTool [88]. In addition, the promoter regions of predicted genes that may be involved in seed-flooding tolerance were further analyzed following the procedure outlined by with Karikari et al. [23]
Supplementary Information
Additional file 1: Table S1. SNPs markers associated with electrical conductivity (EC), germination rate (GR), Shoot Length (ShL), and Root Length (RL) via MLM.
Additional file 2: Table S2. SNPs markers associated with electrical conductivity (EC), germination rate (GR), Shoot Length (ShL), and Root Length (RL) via mrMLM.
Additional file 3: Table S3. One hundred fifteen (115) model genes were found within the region of Gm_08_11,971,416 SNP using Glyma2.0 models in SoyBase. Table S4. One hundred thirty (130) model genes were found within the region of Gm_08_46,239,716 SNP using Glyma2.0 models in SoyBase. Table S5. One hundred thirteen (113) model genes were found within the region of Gm_05_1,000,479 SNP using Glyma2.0 models in SoyBase. Table S6. One hundred twenty-five (125) model genes were found within the region of Gm_01_53,535,790 SNP using Glyma2.0 models in SoyBase.
Additional file 4: Figure S1. Heat map exhibiting the expression profiles of candidate genes among the different soybean tissues and development stages.
Additional file 5: Table S7. Cis-acting regulating elements related to seed flooding tolerance obtained from PlantCare database.
Additional file 6: Table S8. A core set of 243 soybean plant introductions (PIs) selected from the USDA Soybean Germplasm Collection.
Additional file 7: Table S9. Phenotypic data (average) of 243 PI accessions used in this study.
Acknowledgements
The authors are thankful to all the team- and lab mates and friends who supported us during the research project and writing of the manuscript.
Abbreviations
- GWAS
Genome-wide association study
- MLM
Mixed Linear Model
- mrMLM
Multi-Locus Random-SNP-Effect MLM
- EC
Electrical conductivity
- GR
Germination rate
- ShL
Shoot length
- RL
Root length
- GO
Gene ontology
- PPI
Protein-protein interaction
- QTL
Quantitative trait locus
- QTNs
Quantitative trait nucleotides
Authors’ contributions
T.J.Z. and R.A.S. conceived and designed the experiments. R.A.S., M.R.B. and Z.C. performed the experiments. R.A.S., B.K., F.C., G.M.A., A.H., W. L. and N.B. analyzed the data. R.A.S drafted the manuscript. T.J.Z. & B.K. revised the paper. The authors read and approved the final manuscript.
Funding
This work was supported by grants from the National Key Research and Development Program, National Natural Science Foundation of China (Grant Nos. 31871646, 31571691), the Science and Technology Innovation Project for Hebei Province Modern Seed Industry (21326313D-1), the MOE Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT_17R55), the Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP) Program. The funders had no role in experimental design, data collection and analysis as well as writing of the manuscript.
Availability of data and materials
With the exception of SNP datasets, all data generated or analyzed during this study are included in this article and its supplementary information files. The SNP dataset used in the current study were selected from United States Department of Agriculture, Soybean Germplasm Collection sequenced by Illumina Infinium SoySNP50K BeadChip Technology available on SoyBase (https://soybase.org/snps/index.php). Phenotypic data generated for the 2 years (2018 and 2019) and combined years (CE) have been included in the supplementary Tables (Additional File 5: Supplementary Table 8).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hou A, Chen P, Alloatti J, Li D, Mozzoni L, Zhang B, Shi A. Genetic variability of seed sugar content in worldwide soybean germplasm collections. Crop Sci. 2009;49(3):903–912. doi: 10.2135/cropsci2008.05.0256. [DOI] [Google Scholar]
- 2.Wilson RF. Seed composition. In: Boerma HR, Specht JE, editors. Soybeans: improvement, production, and uses. Madison: ASA-CSSA-SSSA; 2004. pp. 621–677. [Google Scholar]
- 3.Oosterhuis D, Scott H, Hampton R, Wullschleger SD. Physiological responses of two soybean [Glycine max (L.) Merr] cultivars to short-term flooding. Environ Exp Bot. 1990;30(1):85–92. doi: 10.1016/0098-8472(90)90012-S. [DOI] [Google Scholar]
- 4.Rhine MD, Stevens G, Shannon G, Wrather A, Sleper D. Yield and nutritional responses to waterlogging of soybean cultivars. Irrig Sci. 2010;28(2):135–142. doi: 10.1007/s00271-009-0168-x. [DOI] [Google Scholar]
- 5.Wu C, Chen P, Hummer W, Zeng A, Klepadlo M. Effect of flood stress on soybean seed germination in the field. Am J Plant Sci. 2017;8(01):53. doi: 10.4236/ajps.2017.81005. [DOI] [Google Scholar]
- 6.Zhao T, Aleem M, Sharmin RA. Adaptation to Water stress in soybean: morphology to genetics. In: Andjelkovic V, editor. Plant, Abiotic Stress and Responses to Climate Change. Croatia: IntechOpen; 2018. pp. 33–68. [Google Scholar]
- 7.Bacanamwo M, Purcell LC. Soybean dry matter and N accumulation responses to flooding stress, N sources and hypoxia. J Exp Bot. 1999;50(334):689–696. doi: 10.1093/jxb/50.334.689. [DOI] [Google Scholar]
- 8.Kim Y, Seo CW, Khan AL, Mun BG, Shahzad R, Ko JW, Yun BW, Park SK, Lee IJ. Exo-ethylene application mitigates waterlogging stress in soybean (Glycine max L.) BMC Plant Biol. 2018;18(1):254. doi: 10.1186/s12870-018-1457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.VanToai TT, Hoa TTC, Hue NTN, Nguyen HT, Shannon JG, Rahman MA. Flooding tolerance of soybean [Glycine max (L.) Merr.] germplasm from Southeast Asia under field and screen-house environments. Open Agric J. 2010;4:38–46. doi: 10.2174/1874331501004010038. [DOI] [Google Scholar]
- 10.Bacanamwo M, Purcell LC. Soybean root morphological and anatomical traits associated with acclimation to flooding. Crop Sci. 1999;39(1):143–149. doi: 10.2135/cropsci1999.0011183X003900010023x. [DOI] [Google Scholar]
- 11.Hasanuzzaman M, Al Mahmud J, Nahar K, Anee TI, Inafuku M, Oku H, Fujita M. Reactive oxygen species and antioxidant systems in plants: role and regulation under abiotic stress. Singapore: Springer; 2017. Responses, adaptation, and ROS metabolism in plants exposed to waterlogging stress; pp. 257–281. [Google Scholar]
- 12.Ali MJ, Yu Z, Xing G, Zhao T, Gai J. Establishment of evaluation procedure for soybean seed-flooding tolerance and its application to screening for tolerant germplasm sources. Legum Res. 2018;41(1):34–40. [Google Scholar]
- 13.Hou FF, Thseng FS. Studies on the screening technique for pre-germination flooding tolerance in soybean. Jpn J Crop Sci. 1992;61(3):447–453. doi: 10.1626/jcs.61.447. [DOI] [Google Scholar]
- 14.Sayama T, Nakazaki T, Ishikawa G, Yagasaki K, Yamada N, Hirota N, Hirata K, Yoshikawa T, Saito H, Teraishi M, Okumoto Y. QTL analysis of seed-flooding tolerance in soybean (Glycine max [L.] Merr.) Plant Sci. 2009;176(4):514–521. doi: 10.1016/j.plantsci.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Cornelious B, Chen P, Chen Y, De Leon N, Shannon J, Wang D. Identification of QTLs underlying water-logging tolerance in soybean. Mol Breed. 2005;16(2):103–112. doi: 10.1007/s11032-005-5911-2. [DOI] [Google Scholar]
- 16.Githiri S, Watanabe S, Harada K, Takahashi R. QTL analysis of flooding tolerance in soybean at an early vegetative growth stage. Plant Breed. 2006;125(6):613–618. doi: 10.1111/j.1439-0523.2006.01291.x. [DOI] [Google Scholar]
- 17.Nguyen V, Vuong T, VanToai T, Lee J, Wu X, Mian M, Dorrance A, Shannon J, Nguyen HT. Mapping of quantitative trait loci associated with resistance to Phytophthora sojae and flooding tolerance in soybean. Crop Sci. 2012;52(6):2481–2493. doi: 10.2135/cropsci2011.09.0466. [DOI] [Google Scholar]
- 18.VanToai TT, St Martin SK, Chase K, Boru G, Schnipke V, Schmitthenner AF, Lark KG. Identification of a QTL associated with tolerance of soybean to soil waterlogging. Crop Sci. 2001;41(4):1247–1252. doi: 10.2135/cropsci2001.4141247x. [DOI] [Google Scholar]
- 19.Dhungana SK, Kim HS, Kang BK, Seo JH, Kim HT, Shin SO, Park CH, Kwak DY. Quantitative trait loci mapping for flooding tolerance at an early growth stage of soybean recombinant inbred line population. Plant Breed. 2020;139(3):626–638. doi: 10.1111/pbr.12790. [DOI] [Google Scholar]
- 20.Ye H, Song L, Chen H, Valliyodan B, Cheng P, Ali L, Vuong T, Wu C, Orlowski J, Buckley B, Chen P. A major natural genetic variation associated with root system architecture and plasticity improves waterlogging tolerance and yield in soybean. Plant Cell Environ. 2018;41(9):2169–2182. doi: 10.1111/pce.13190. [DOI] [PubMed] [Google Scholar]
- 21.Do TD, Vuong TD, Dunn D, Clubb M, Valliyodan B, Patil G, Chen P, Xu D, Nguyen HT, Shannon JG. Identification of new loci for salt tolerance in soybean by high-resolution genome-wide association mapping. BMC Genomics. 2019;20(1):318. doi: 10.1186/s12864-019-5662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ersoz ES, Yu J, Buckler ES. Genomics-assisted crop improvement. Dordrecht: Springer; 2007. Applications of linkage disequilibrium and association mapping in crop plants; pp. 97–119. [Google Scholar]
- 23.Karikari B, Wang Z, Zhou Y, Yan W, Feng J, Zhao T. Identification of quantitative trait nucleotides and candidate genes for soybean seed weight by multiple models of genome-wide association study. BMC Plant Biol. 2020;20(1):404. doi: 10.1186/s12870-020-02604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Li H, Gou Z, Zhang Y, Wang X, Ren H, Wen Z, Kang BK, Li Y, Yu L, Gao H. Genome-wide association study of soybean seed germination under drought stress. Mol Gen Genomics. 2020;295(3):661–673. doi: 10.1007/s00438-020-01646-0. [DOI] [PubMed] [Google Scholar]
- 25.Steketee CJ, Schapaugh WT, Carter TE, Jr, Li Z. Genome-wide association analyses reveal genomic regions controlling canopy wilting in soybean. G3- genes Genom. Genet. 2020;10(4):1413–1425. doi: 10.1534/g3.119.401016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Z, Chang F, Lv W, Sharmin RA, Wang Z, Kong J, Bhat JA, Zhao T. Identification of QTN and Candidate Gene for Seed-flooding Tolerance in Soybean [Glycine max (L.) Merr.] using Genome-Wide Association Study (GWAS) Genes. 2019;10(12):957. doi: 10.3390/genes10120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C, Mozzoni LA, Moseley D, Hummer W, Ye H, Chen P, Shannon G, Nguyen H. Genome-wide association mapping of flooding tolerance in soybean. Mol Breed. 2020;40(1):4. doi: 10.1007/s11032-019-1086-0. [DOI] [Google Scholar]
- 28.Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One. 2013;8(1):e54985. doi: 10.1371/journal.pone.0054985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wuebker EF, Mullen RE, Koehler K. Flooding and temperature effects on soybean germination. Crop Sci. 2001;41(6):1857–1861. doi: 10.2135/cropsci2001.1857. [DOI] [Google Scholar]
- 30.Miransari M, Smith DL. Plant hormones and seed germination. Environ Exp Bot. 2014;99:110–121. doi: 10.1016/j.envexpbot.2013.11.005. [DOI] [Google Scholar]
- 31.Ali MJ, Xing G, He J, Zhao T, Gai J. Detecting the QTL-allele system controlling seed-flooding tolerance in a nested association mapping population of soybean. Crop J. 2020;8(5):781–792. doi: 10.1016/j.cj.2020.06.008. [DOI] [Google Scholar]
- 32.Lee GA, Crawford GW, Liu L, Sasaki Y, Chen X. Archaeological soybean (Glycine max) in East Asia: does size matter? PLoS One. 2011;6(11):e26720. doi: 10.1371/journal.pone.0026720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priolli R, Carvalho C, Bajay M, Pinheiro J, Vello NA. Genome analysis to identify SNPs associated with oil content and fatty acid components in soybean. Euphytica. 2019;215(3):1–14. doi: 10.1007/s10681-019-2378-5. [DOI] [Google Scholar]
- 34.Mather KA, Caicedo AL, Polato NR, Olsen KM, McCouch S, Purugganan MD. The extent of linkage disequilibrium in rice (Oryza sativa L.) Genetics. 2007;177(4):2223–2232. doi: 10.1534/genetics.107.079616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39(9):1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 36.Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, Li J. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42(12):1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 37.Valliyodan B, Ye H, Song L, Murphy M, Shannon JG, Nguyen HT. Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J Exp Bot. 2016;68(8):1835–1849. doi: 10.1093/jxb/erw433. [DOI] [PubMed] [Google Scholar]
- 38.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES. Mixed linear model approach adapted for genome wide association studies. Nat Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang SB, Feng JY, Ren WL, Huang B, Zhou L, Wen YJ, Zhang J, Dunwell JM, Xu S, Zhang YM. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci Rep. 2016;6(1):1–10. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaler AS, Ray JD, Schapaugh WT, King CA, Purcell LC. Genome-wide association mapping of canopy wilting in diverse soybean genotypes. Theor Appl Genet. 2017;130(10):2203–2217. doi: 10.1007/s00122-017-2951-z. [DOI] [PubMed] [Google Scholar]
- 42.Chamarthi SK, Kaler AS, Abdel-Haleem H, Fritschi FB, Gillman JD, Ray JD, et al. Identification and confirmation of loci associated with canopy wilting in soybean using genome-wide association mapping. Front Plant Sci. 2021;12:698116. [DOI] [PMC free article] [PubMed]
- 43.Hyten DL, Choi IY, Song Q, Shoemaker RC, Nelson RL, Costa JM, Specht JE, Cregan PB. Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics. 2007;175(4):1937–1944. doi: 10.1534/genetics.106.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaler AS, Purcell LC. Estimation of a significance threshold for genome-wide association studies. BMC Genomics. 2019;20(1):1–8. doi: 10.1186/s12864-019-5992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB. Fingerprinting soybean germplasm and its utility in genomic research. G3 (Bethesda) 2015;5(10):1999–2006. doi: 10.1534/g3.115.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang YM, Jia Z, Dunwell JM. The applications of new multi-locus GWAS methodologies in the genetic dissection of complex traits. Front Plant Sci. 2019;10:100. doi: 10.3389/fpls.2019.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui Y, Zhang F, Zhou Y. The application of multi-locus GWAS for the detection of salt-tolerance loci in rice. Front Plant Sci. 2018;9:1464. doi: 10.3389/fpls.2018.01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riedelsheimer C, Lisec J, Czedik-Eysenberg A, Sulpice R, Flis A, Grieder C, Altmann T, Stitt M, Willmitzer L, Melchinger AE. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci U S A. 2012;109(23):8872–8877. doi: 10.1073/pnas.1120813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao K, Tung C-W, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J, McClung AM. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun. 2011;2(1):1–10. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen Z, Boyse JF, Song Q, Cregan PB, Wang D. Genomic consequences of selection and genome-wide association mapping in soybean. BMC Genomics. 2015;16(1):671. doi: 10.1186/s12864-015-1872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen Z, Tan R, Yuan J, Bales C, Du W, Zhang S, Chilvers MI, Schmidt C, Song Q, Cregan PB, Wang D. Genome-wide association mapping of quantitative resistance to sudden death syndrome in soybean. BMC Genomics. 2014;15(1):809. doi: 10.1186/1471-2164-15-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Song Q, Cregan PB, Jiang GL. Genome-wide association study, genomic prediction and marker-assisted selection for seed weight in soybean (Glycine max) Theor Appl Genet. 2016;129(1):117–130. doi: 10.1007/s00122-015-2614-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rizal G, Karki S. Research article alcohol dehydrogenase (ADH) activity in soybean (Glycine max [L.] Merr.) under flooding stress. Electron J Plant Breed. 2011;2(1):50–57. [Google Scholar]
- 54.Brinton J, Ramirez-Gonzalez RH, Simmonds J, Wingen L, Orford S, Griffiths S, Haberer G, Spannagl M, Walkowiak S, Pozniak C, Uauy C. A haplotype-led approach to increase the precision of wheat breeding. Commun Biol. 2020;3(1):712. doi: 10.1038/s42003-020-01413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinha P, Singh VK, Saxena RK, Khan AW, Abbai R, Chitikineni A, Desai A, Molla J, Upadhyaya HD, Kumar A, Varshney RK. Superior haplotypes for haplotype-based breeding for drought tolerance in pigeonpea (Cajanus cajan L.) Plant Biotechnol J. 2020;18(12):2482–2490. doi: 10.1111/pbi.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharmin RA, Bhuiyan MR, Lv W, Yu Z, Chang F, Kong J, Bhat JA, Zhao T. RNA-Seq based transcriptomic analysis revealed genes associated with seed-flooding tolerance in wild soybean (Glycine soja Sieb. & Zucc.) Environ Exp Bot. 2020;171:103906. doi: 10.1016/j.envexpbot.2019.103906. [DOI] [Google Scholar]
- 57.Babitha K, Vemanna RS, Nataraja KN, Udayakumar MJ. Overexpression of EcbHLH57 transcription factor from Eleusine coracana L. in tobacco confers tolerance to salt, oxidative and drought stress. PLoS One. 2015;10(9):e0137098. doi: 10.1371/journal.pone.0137098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qian Y, Zhang T, Yu Y, Gou L, Yang J, Xu J, Pi E. Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses. Front Plant Sci. 2021;12:1143. doi: 10.3389/fpls.2021.677611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Q, Ren YR, Wang QJ, Yao YX, You CX, Hao YJ. Overexpression of Mdb HLH 104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnol J. 2016;14(7):1633–1645. doi: 10.1111/pbi.12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun XL, Yu QY, Tang LL, Ji W, Bai X, Cai H, Liu XF, Ding XD, Zhu YM. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J Plant Physiol. 2013;170(5):505–515. doi: 10.1016/j.jplph.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 61.Chen X, Ding Y, Yang Y, Song C, Wang B, Yang S, Guo Y, Gong Z. Protein kinases in plant responses to drought, salt, and cold stress. J Integr Plant Biol. 2021;63(1):53–78. doi: 10.1111/jipb.13061. [DOI] [PubMed] [Google Scholar]
- 62.Bogamuwa S, JANG JC. The a rabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid-and gibberellic acid-mediated regulation of seed germination. Plant Cell Environ. 2013;36(8):1507–1519. doi: 10.1111/pce.12084. [DOI] [PubMed] [Google Scholar]
- 63.Pandey DM, Kim SR. Identification and expression analysis of hypoxia stress inducible CCCH-type zinc finger protein genes in rice. J Plant Biol. 2012;55(6):489–497. doi: 10.1007/s12374-012-0384-4. [DOI] [Google Scholar]
- 64.Kang J, Li J, Gao S, Tian C, Zha X. Overexpression of the leucine-rich receptor-like kinase gene LRK 2 increases drought tolerance and tiller number in rice. Plant Biotechnol J. 2017;15(9):1175–1185. doi: 10.1111/pbi.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D. Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell. 2005;17(1):92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 2005;5(1):1–13. doi: 10.1186/1471-2229-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosado A, Schapire AL, Bressan RA, Harfouche AL, Hasegawa PM, Valpuesta V, Botella MA. The Arabidopsis tetratricopeptide repeat-containing protein TTL1 is required for osmotic stress responses and abscisic acid sensitivity. Plant Physiol. 2006;142(3):1113–1126. doi: 10.1104/pp.106.085191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paeng SK, Kang CH, Chi YH, Chae HB, Lee ES, Park JH, Wi SD, Bae SB, Phan KAT, Lee SY. AtTPR10 containing multiple ANK and TPR domains exhibits chaperone activity and heat-shock dependent structural switching. Appl Sci. 2020;10(4):1265. doi: 10.3390/app10041265. [DOI] [Google Scholar]
- 69.Gupta P, Nutan KK, Singla-Pareek SL, Pareek A. Abiotic stresses cause differential regulation of alternative splice forms of GATA transcription factor in rice. Front Plant Sci. 2017;8:1944. doi: 10.3389/fpls.2017.01944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, Wu T, Li Z, Huang K, Kim NE, Ma Z, Kwon S-W, Jiang W, Du X. OsGATA16, a GATA transcription factor, confers cold tolerance by repressing OsWRKY45–1 at the seedling stage in rice. Rice. 2021;14(1):1–15. doi: 10.1186/s12284-021-00485-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dong NQ, Sun Y, Guo T, Shi CL, Zhang YM, Kan Y, Xiang YH, Zhang H, Yang YB, Li YC, Zhao HY. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat Commun. 2020;11(1):1–16. doi: 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu XD, Xie L, Wei Y, Zhou X, Jia B, Liu J, Zhang S. Abiotic stress resistance, a novel moonlighting function of ribosomal protein RPL44 in the halophilic fungus Aspergillus glaucus. Appl Eviron Microbiol. 2014;80(14):4294–4300. doi: 10.1128/AEM.00292-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mazahar M, Achala B, Anusree S, Kirti PB. Ribosomal proteins and their extra ribosomal functions in abiotic stress tolerance of plants. Proteomics. 2019;12(7):1024–1038. [Google Scholar]
- 74.Xu T, Lee K, Gu L, Kim J-I, Kang H. Functional characterization of a plastid-specific ribosomal protein PSRP2 in Arabidopsis thaliana under abiotic stress conditions. Plant Physiol Biochem. 2013;73:405–411. doi: 10.1016/j.plaphy.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 75.Severin AJ, Woody JL, Bolon YT, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE. RNA-Seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 2010;10(1):160. doi: 10.1186/1471-2229-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Das S, Meher PK, Rai A, Bhar LM, Mandal BN. Statistical Approaches for Gene Selection, Hub Gene Identification and Module Interaction in Gene CoExpression Network Analysis: An Application to Aluminum Stress in Soybean (Glycine max L.) PLoS One. 2017;12(1):e0169605. doi: 10.1371/journal.pone.0169605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vidoz ML, Loreti E, Mensuali A, Alpi A, Perata P. Hormonal interplay during adventitious root formation in flooded tomato plants. Plant J. 2010;63(4):551–562. doi: 10.1111/j.1365-313X.2010.04262.x. [DOI] [PubMed] [Google Scholar]
- 78.Lin C, Sauter M. Polar auxin transport determines adventitious root emergence and growth in rice. Front Plant Sci. 2019;10:444. doi: 10.3389/fpls.2019.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jia W, Ma M, Chen J, Wu S. Plant morphological, physiological and anatomical adaption to flooding stress and the underlying molecular mechanisms. Int J Mol Sci. 2021;22(3):1088. doi: 10.3390/ijms22031088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nyquist WE, Baker RJ. Estimation of heritability and prediction of selection response in plant populations. Crit Rev Plant Sci. 1991;10(3):235–322. doi: 10.1080/07352689109382313. [DOI] [Google Scholar]
- 81.He J, Meng S, Zhao T, Xing G, Yang S, Li Y, Guan R, Lu J, Wang Y, Xia Q, Yang B. An innovative procedure of genome-wide association analysis fits studies on germplasm population and plant breeding. Theor Appl Genet. 2017;130(11):2327–2343. doi: 10.1007/s00122-017-2962-9. [DOI] [PubMed] [Google Scholar]
- 82.Wang J, Chu S, Zhang H, Zhu Y, Cheng H, Yu D. Development and application of a novel genome-wide SNP array reveals domestication history in soybean. Sci Rep. 2016;6(1):1–10. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 84.Wang N, Akey JM, Zhang K, Chakraborty R, Jin L. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet. 2002;71(5):1227–1234. doi: 10.1086/344398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014;43(D1):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou G, Soufan O, Ewald J, Hancock RE, Basu N, Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–W241. doi: 10.1093/nar/gkz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen C, Chen H, Zhang Y, et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant. 2020;13(8):1194–202. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. SNPs markers associated with electrical conductivity (EC), germination rate (GR), Shoot Length (ShL), and Root Length (RL) via MLM.
Additional file 2: Table S2. SNPs markers associated with electrical conductivity (EC), germination rate (GR), Shoot Length (ShL), and Root Length (RL) via mrMLM.
Additional file 3: Table S3. One hundred fifteen (115) model genes were found within the region of Gm_08_11,971,416 SNP using Glyma2.0 models in SoyBase. Table S4. One hundred thirty (130) model genes were found within the region of Gm_08_46,239,716 SNP using Glyma2.0 models in SoyBase. Table S5. One hundred thirteen (113) model genes were found within the region of Gm_05_1,000,479 SNP using Glyma2.0 models in SoyBase. Table S6. One hundred twenty-five (125) model genes were found within the region of Gm_01_53,535,790 SNP using Glyma2.0 models in SoyBase.
Additional file 4: Figure S1. Heat map exhibiting the expression profiles of candidate genes among the different soybean tissues and development stages.
Additional file 5: Table S7. Cis-acting regulating elements related to seed flooding tolerance obtained from PlantCare database.
Additional file 6: Table S8. A core set of 243 soybean plant introductions (PIs) selected from the USDA Soybean Germplasm Collection.
Additional file 7: Table S9. Phenotypic data (average) of 243 PI accessions used in this study.
Data Availability Statement
With the exception of SNP datasets, all data generated or analyzed during this study are included in this article and its supplementary information files. The SNP dataset used in the current study were selected from United States Department of Agriculture, Soybean Germplasm Collection sequenced by Illumina Infinium SoySNP50K BeadChip Technology available on SoyBase (https://soybase.org/snps/index.php). Phenotypic data generated for the 2 years (2018 and 2019) and combined years (CE) have been included in the supplementary Tables (Additional File 5: Supplementary Table 8).