Abstract
Background
Methicillin-resistant Staphylococcus aureus (MRSA) is a major nosocomial pathogen subdivided into lineages termed sequence types (STs). Since the 1950s, successive waves of STs have appeared and replaced previously dominant lineages. One such event has been occurring in China since 2013, with community-associated (CA-MRSA) strains including ST59 largely replacing the previously dominant healthcare-associated (HA-MRSA) ST239. We previously showed that ST59 isolates tend to have a competitive advantage in growth experiments against ST239. However, the underlying genomic and phenotypic drivers of this replacement event are unclear.
Methods
Here, we investigated the replacement of ST239 using whole-genome sequencing data from 204 ST239 and ST59 isolates collected in Chinese hospitals between 1994 and 2016. We reconstructed the evolutionary history of each ST and considered two non-mutually exclusive hypotheses for ST59 replacing ST239: antimicrobial resistance (AMR) profile and/or ability to colonise and persist in the environment through biofilm formation. We also investigated the differences in cytolytic activity, linked to higher virulence, between STs. We performed an association study using the presence and absence of accessory virulence genes.
Results
ST59 isolates carried fewer AMR genes than ST239 and showed no evidence of evolving towards higher AMR. Biofilm production was marginally higher in ST59 overall, though this effect was not consistent across sub-lineages so is unlikely to be a sole driver of replacement. Consistent with previous observations of higher virulence in CA-MRSA STs, we observed that ST59 isolates exhibit significantly higher cytolytic activity than ST239 isolates, despite carrying on average fewer putative virulence genes. Our association study identified the chemotaxis inhibitory protein (chp) as a strong candidate for involvement in the increased virulence potential of ST59. We experimentally validated the role of chp in increasing the virulence potential of ST59 by creating Δchp knockout mutants, confirming that ST59 can carry chp without a measurable impact on fitness.
Conclusions
Our results suggest that the ongoing replacement of ST239 by ST59 in China is not associated to higher AMR carriage or biofilm production. However, the increase in ST59 prevalence is concerning since it is linked to a higher potential for virulence, aided by the carriage of the chp gene.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13073-021-00992-x.
Keywords: Methicillin-resistant Staphylococcus aureus (MRSA), Molecular evolution, Lineage replacement, Phylogeny, Phylogeography, Virulence
Background
Methicillin-resistant Staphylococcus aureus (MRSA) is ranked amongst the most serious multidrug-resistant threats [1] and are a major cause of nosocomial infections, ranging from superficial wound infections and food poisoning to pneumonia, infective endocarditis, bacteraemia and other systemic infections [2]. Methicillin resistance is conferred by the mecA gene, which is carried on a mobile genetic element (MGE) known as the SCCmec chromosomal cassette [3]. A divergent homologue of mecA first observed in 2011, mecC, can also confer methicillin resistance within a different SSCmec [4]. One prominent feature of MRSA is the strong population structure driven by the presence of multiple, essentially clonal, lineages of highly related strains. This population structure is typically described using sequence types (STs), defined as strains sharing the same sequence at a set of housekeeping genes used in multilocus sequence type (MLST) classification schemes [5].
Distinct MRSA lineages (STs) are found at markedly different prevalence globally. For example, ST239 is common in China [6–9], Singapore [10], Thailand [11] and other Asian countries [12–14], but rare in Europe and the Americas [15–17]. ST225 reaches a prevalence of between 60 and 80% of S. aureus in Central Europe but is virtually absent from the USA [18]. ST8 (USA300) is prevalent in the USA but is rarely observed in the rest of the world [16, 19]. ST22 is the most common lineage in the UK [15, 16, 20, 21] and has been identified at differing prevalence in parts of Asia [22–24].
Although some MRSA lineages remain at high prevalence in certain global regions for decades, the geographic distribution of many lineages is dynamic. Since the use of penicillin in the mid-1940s, multiple waves of new MRSA lineages have emerged and replaced previously established ones [25–28]. More recently, community-associated MRSA (CA-MRSA) have been replacing hospital-associated MRSA (HA-MRSA) as the dominant epidemic strains, with USA300 providing a prominent example [19, 29]. The mechanisms driving these phase transitions are poorly understood and remain best characterized for lineages frequently associated to nosocomial infections rather than commensal carriage in the wider population. Typical hospital-associated lineages include the ST239 and ST5 lineages, which carry the type II and III SCCmec elements, often lack the Panton–Valentine leukocidin (PVL) toxin genes, have relatively low virulence and usually infect people with poor immunity and/or during long-term hospitalization [30]. Conversely, community-associated lineages such as ST8, ST30 and ST59 carry the type IV and V SCCmec elements, often harbour the PVL toxin, and commonly infect young, otherwise healthy adults [19, 31–34].
Globally, ST239 isolates show signatures of convergent adaptation to hospital settings, including increased resistance to antimicrobials, increased ability to persist in healthcare environments and attenuated virulence [35, 36]. ST239 has historically been the dominant lineage in hospital infections in mainland China, where MRSA remains a particularly acute problem despite being in decline. Our multi-centre MRSA molecular surveillance schemes, together with work from Xiao et al. 2013 which considered different administrative regions, previously found that hospital-associated ST239 was the dominant MRSA lineage in China between 2005 and 2011, accounting for 50–80.8% of MRSA [6–8, 37], but has subsequently been declining in frequency (4.3%), with a general replacement of HA-MRSA STs by CA-MRSA STs [38]. CA-MRSA lineages, including ST59, have been increasing in prevalence since 2013 [24, 38–40], with recent work suggesting that between 2015 and 2017, ST59 has become the predominant MRSA clone (33%, 154/471) with ST59 accounting for more than half of CA-MRSA isolates [41]. The replacement of ST239 has been observed in other countries [42–44]. For example, Hsu et al. analysed the replacement of ST239 in Singapore by another HA-MRSA (ST22), likely after a single introduction in 2001, using phylogenetic reconstruction and comparative genomics [10]. However, the replacement of ST239 by CA-MRSA STs in China may have different drivers. In contrast to the situation in Singapore, the multiple STs involved in replacement events in China suggest the potential for more complex competitive interactions.
To understand the drivers of the ongoing replacement of MRSA clonal lineages in China, we have focused on the contrast between ST59 and ST239. We have previously shown that ST59-t437 has a fitness advantage over ST239-t030 in vitro [38], potentially contributing to this replacement. However, the same may not be true for other ST59 sub-lineages, for example, previously designated as the ‘Taiwan’ and ‘Asian-Pacific’ clones. Here, we investigate the comparative genomics of ST59 and ST239 in detail to better understand this replacement event and its potential consequences. We selected 132 ST239 and 72 ST59 S. aureus isolates from our surveillance network across 14 provinces for whole-genome sequencing (WGS). We reconstructed the evolutionary histories of these isolates through phylogenetic and accessory genome analyses. We additionally phenotyped all strains for their ability to produce biofilms and for their cytolytic activity, an indicator of virulence potential in a host. An association study using the presence and absence of virulence genes identified the chemotaxis inhibitory protein (chp) gene, a known contributor to human immune evasion in S. aureus [45], as amongst the strongest candidates for higher cytolytic activity in ST59. We experimentally validated the role of chp as a virulence factor in ST59 by creating Δchp knockout mutants. Our results suggest that the ongoing MRSA lineage replacement in mainland China is linked to the higher virulence of ST59 strains relative to ST239, likely mediated through the carriage of the immune evasive chp gene, which is potentially of concern for future infections.
Methods
Bacterial isolates
We investigated the pathogen spectrum of bloodstream infections (BSI), hospital-acquired pneumonia (HAP) and intra-abdominal infections (IAI), prioritizing for this study ST59 and ST239 strains. We collected a total of 132 ST239 and 72 ST59 Chinese S. aureus isolates from 14 Chinese provinces between 1994 and 2016, sampled to cover a wide temporal and geographic range (Additional file 1: Table S1). The oldest 48 isolates were collected from a hospital in Beijing, because this was the only hospital participating in the surveillance effort that had stored bacterial strains prior to 2000. The remaining 156 isolates were collected from 30 hospitals across 14 provinces in China from 2002 to 2016.
All of the 132 ST239 isolates belonged to MRSA and carried mecA located on a type III SCCmec. Amongst the 72 ST59 isolates, 14 (19.4%) were characterised as MSSA, and 58 (80.6%) were characterised as MRSA. Of the 58 ST59-MRSA isolates, eight (11.1%) carried the type V SCCmec, and the remaining 50 (88.9%) isolates carried the type IV SCCmec (Fig. 1).
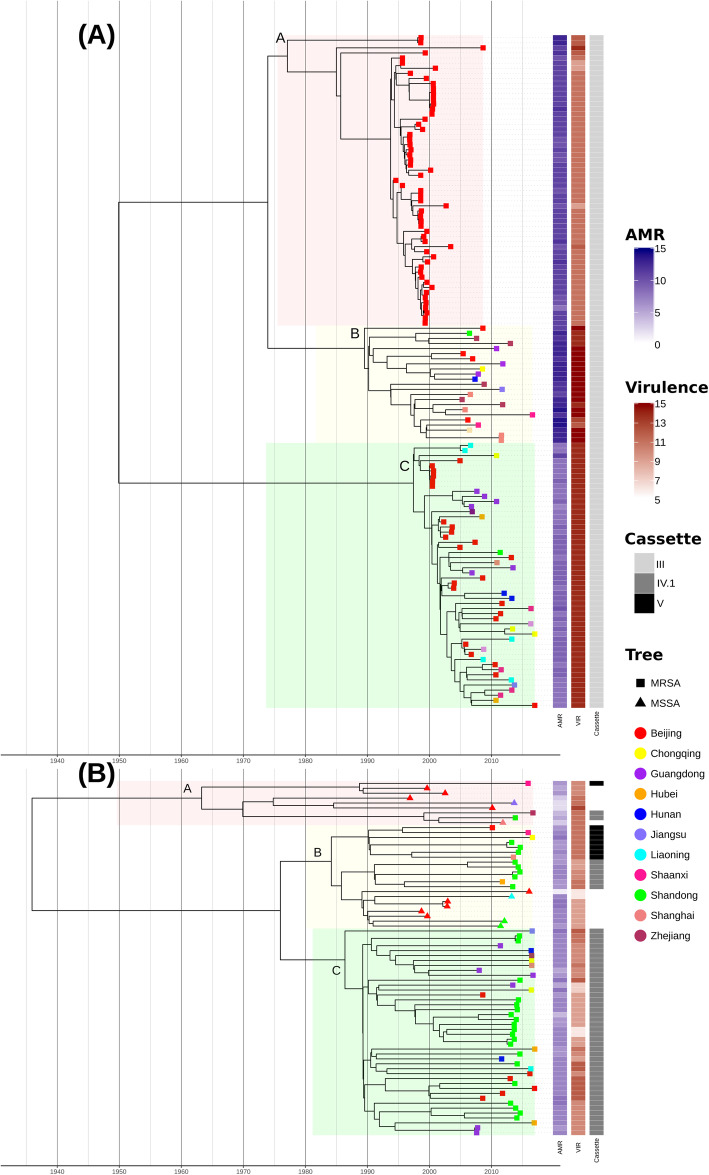
Fig. 1.
Maximum clade credibility (MCC) timed tree of Chinese ST239 (A) and ST59 (B) isolates. BEAST-inferred temporal phylogeny of Chinese S. aureus isolates. Tips are coloured according to the province of sampling. The highlighted boxes denote within ST sub-lineage structure for ST239 (ST239-A, ST239-B, ST239-C) and ST59 (ST59-A, ST59-B, ST59-C). The panel on the right provides the number (colour) of AMR and virulence (VIR) genes carried per isolate, together with the inferred cassette type for MRSA isolates
In order to investigate the global geographic affinity of Chinese ST239 and ST59, we identified relevant genomes made available in published works as well as those released as Pathogenwatch projects associated to each ST type. In total, we included 153 global ST239 genome assemblies (downloaded from Pathogenwatch, https://pathogen.watch and published in previous studies [13, 44]) and 114 ST59 genomes. Of the 114 ST59 genomes, 13 were available via Pathogenwatch projects, and the remaining 101 genomes were downloaded from a previous study by Ward et al. [46].
Whole-genome sequencing and multilocus sequence type assignment
Our 204 new S. aureus isolates were sequenced using 150-bp paired-end protocols on an Illumina NextSeq 500. All genomes were assembled de novo using SPAdes v3.10.0 [47]. The resultant contigs were inspected and then annotated using Prokka v1.12 [48]. MLST were assigned using PubMLST (https://pubmlst.org/saureus/).
Preliminary phylogenetic analyses
The core genome was extracted from the annotated assemblies using Panaroo v1.1.2 [49]. Alignments were screened for recombination using ClonalFrameML [50], and the putative recombinant regions were removed before further phylogenetic analyses. Maximum likelihood phylogenetic trees were constructed in RAxML v8.2.10 using a GTR model and 1000 bootstrap replicates to assess the confidence at each node of the final tree [51] (Additional file 2: Fig. S1). The chromosomes of S. aureus isolates N315 (ST5) (GenBank accession no. NC_002745) and MW2 (ST1) (NC_003923) were used as outgroups to root the respective phylogenies as these represent the most closely related outgroups to Chinese and global ST239 and ST59. We tested for a temporal signal in the data (i.e. measurable evolution over the course of the sampling timespan) with a correlation between root-to-tip distances and the year of sampling in TempEst [52] and assessed significance following 10,000 date randomisations using the roototip() function implemented in BactDating [53] (Additional file 2: Fig. S2).
In parallel, we employed the same phylogenetic approach to construct a recombination pruned phylogenetic tree over the larger global datasets of ST239 and ST59 (Additional file 2: Fig. S3).
Phylogeographic analyses
We used BEAST v2.4.7 [54] to estimate a timed phylogeny from the recombination-free core genome alignments of ST239 (3090 SNPs, 132 isolates) and ST59 (3423 SNPs, 72 isolates), including both tip dates and geographic information for each isolate. For both SNP alignments, the TN93 substitution model was selected based on the evaluation of all possible substitution models in bModelTest [55], and the number of invariant A, T, C and Gs sets to calibrate the clock rate estimation. All combinations of molecular clock models (strict, relaxed lognormal, relaxed exponential) and two population model priors (coalescent skyline, coalescent exponential) were evaluated. For each combination, three independent chains were run for 200,000,000 iterations sampling every 20,000 steps. Convergence of the Markov chain Monte Carlo (MCMC) chain was inspected in Tracer v1.6.0 and through evaluation of the effective sample sizes (ESS) and parameter value traces. Only runs with ESS > 200 were considered. The best-supported model following path sampling [56] was a skyline population model with a strict clock (Additional file 2: Table S2). The maximum clade credibility (MCC) tree under each model was generated in TreeAnnotator and plotted in ggtree [57].
Accessory genome analyses of ST239 and ST59
The accessory genome was extracted using Panaroo v1.1.2 [49] (Additional file 2: Figs. S4-S5), and SCCmec types were determined as previously described [58]. Firstly, the specific SCCmec typing primer sequences were detected using BLAST, and then different combinations of cassette chromosome recombinases (ccr) gene complexes and the mec gene complex were analysed according to the definitions of the International Working Group on the Staphylococcal Cassette Chromosome Elements (IWG-SCC; www.sccmec.org). Prophage sequences within our assemblies were identified and characterized using the PHAge Search Tool web-server PHAST [59]. Contigs were defined as putative plasmids based on the presence of the rep gene or a previously identified plasmid sequence [60], and insertion sequences (IS) were identified and annotated using ISFinder [61]. Other MGEs, including mobile pathogenicity islands (SaPIs), genomic islands, mobile element structures (MES) and transposons, were determined using local ‘blastn’ searches, requiring a minimum query coverage value of 80% and a similarity threshold value of 95% (Additional file 2: Fig. S6, Additional file 2: Table S3). AMR and virulence genes were identified using ResFinder [62], VirulenceFinder [63] and the Virulence Factors DataBase [64] (VFDB), with a minimum query coverage value of 80% and a similarity threshold value of 90% (Fig. 1, Additional file 2: Figs. S7-S8). Resistance gene carriage and temporal patterns in the number of AMR genes carried was assessed using the Wilcoxon signed-rank test and linear regression (Additional file 2: Figs. S9-S12). The presence and phylogenetic association of the surface protein sasX in ST239 were also assessed (Additional file 2: Fig. S13).
Methicillin resistance acquisition analyses
We employed nucleotide blast to identify the location of the SCCmec attachment site as defined in Noto et al. [65] in our ST59 Chinese S. aureus isolates (Additional file 2: Fig. S14). We extracted a maximal alignable region 500 bp upstream of, and including, the attachment site and aligned this across isolates using Clustalo v1.2.2 [66]. Hierarchical clustering was applied to the differences over this alignment using hclust in R [67].
Biofilm formation test
A total of 204 S. aureus strain was cultured overnight at 37 °C, and 0.5 McFarland bacterial suspension was prepared with sterile saline. Twenty microliters of the prepared bacterial suspension and 180 μl Tryptic Soy Broth (TSB) were added to each well of a 96-well microplate. Three parallel wells were prepared for each strain. TSB without bacteria suspension were used as a blank control. The 96-well microplate was placed at 37 °C for 24 h. The supernatant was gently discarded, and the 96-well plate was washed slowly twice with distilled water. The biofilm adhered to the microwells was fixed with 150 μl methanol for 20 min and then stained with 150 μl of 2% gentian violet stain for 15 min at room temperature. The 96-well plate was slowly washed twice with distilled water. Each well was decolorized by adding 150 μl of 95% ethanol for 30 min, and the absorbance at 570 nm of each well was measured using a spectrophotometer (Fig. 2, Additional file 1: Table S1).
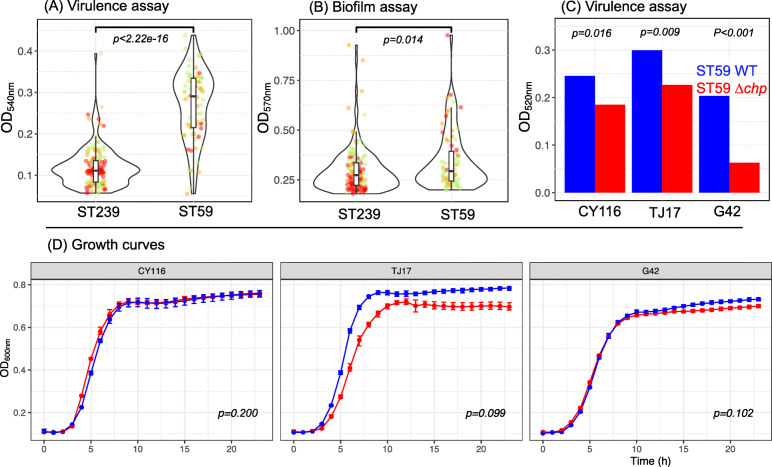
Fig. 2.
Phenotypic assays. A The overall virulence (OD540, y-axis) of 132 ST239 and 72 ST59 strains evaluated by lysis of erythrocytes by culture filtrates. Values for isolates of sub-lineage A, B and C are shown in red, orange and green respectively. B Biofilm formation ability (OD570; y-axis) of all 132 ST239 and 72 ST59 strains with sub-lineages coloured as in Fig. 1. C Cytolytic activity measured by lysis of erythrocytes by culture filtrates for wild-type (WT; blue) and Δchp (chp knockout mutants; red) ST59 strains. Values provide the mean over three replicates per sample. D Growth curves for WT (blue) and Δchp (red) ST59 strains. p-values are provided following the Wilcoxon signed-rank test (A–C) and following the permutation test (D), see the ‘Methods’ section
Lysis of erythrocytes by culture filtrates
A total of 204 S. aureus strains were cultured for 15 h in TSB. The bacterial culture was centrifuged at 5000 rpm for 5 min, and 10 μl of the supernatant was added into a 96-well plate containing 90 μl of PBS buffer per well. Then, 100 μl of human red blood cells (2% v/v in Dulbecco’s phosphate-buffered saline) was added to the supernatant and incubated at 37 °C for 1 h with gentle shaking. The culture was centrifuged at 1500 rpm for 12 min at 4 °C, without disturbing the cells. One hundred microlitres of the supernatant was transferred to a new 96-well plate, and the absorbance at 540 nm of the supernatant was measured using a spectrophotometer. This procedure was repeated twice to assess the concordance (Fig. 2, Additional file 1: Table S1).
Pan-GWAS for ST59 virulence
To identify genes putatively associated with the potential for virulence in ST59, we fitted generalized linear models (GLMs) to the presence and absence matrix of virulence-associated genes (identified by VFDB) against the measurements obtained from the lysis of erythrocyte assay. Detected associations were reported for all genes passing a significance threshold of < 0.05 following Bonferroni correction (Additional file 2: Fig. S15, Additional file 2: Table S4).
chp inactivation
S. aureus chp gene inactivation mutants were obtained for ST59 isolates spanning each sub-lineage: CY116, TJ17 and G41, using the pnCasSA-BEC genome editing system according to methods described previously [68]. Briefly, annealed chp spacer oligos were first inserted into the BsaI sites of the pnCasSA-BEC plasmid by Golden Gate assembly. Then, the chp spacer-introduced pnCasSA-BEC plasmid was transformed into the wild-type S. aureus RN4220 strain using electroporation (2.1 kV mm−1, 100 Ω, 25 μF, 2-mm cuvette). The colonies were selected on a TSB agar plate containing 5 mg/L chloramphenicol at 30 °C. The chp spacer-introduced pnCasSA-BEC plasmid isolated from the RN4220 strain was transformed into target S. aureus CY116, TJ17 and G42 strains by electroporation (2.1 kV mm−1, 100 Ω, 25 μF, 2-mm cuvette). The colonies were also selected on a TSB agar plate containing 5 mg/L chloramphenicol at 30 °C. The successful chp inactivation strain was confirmed by PCR and sequencing. After confirmation, the chp spacer-introduced pnCasSA-BEC plasmid was cured by culturing the cells at 42 °C for 12 h without adding chloramphenicol.
Growth assay
S. aureus wild-type and chp inactivated strains were cultured in TSB to achieve an OD600 of 0.6. The culture was diluted to OD600 = 0.01 and then cultured at 37 °C with agitation at 200 rpm for 24 h. The OD600 was measured every 30 min to obtain a growth curve. Growth curves were compared using a permutation test implemented by the compareGrowthCurves() function within statmod v1.434 (https://CRAN.R-project.org/package=statmod), specifying 50,000 permutations (Fig. 2, Additional file 2: Fig. S16).
In vitro competition test
In vitro competition tests were performed as described previously [38]. Three pairs of ST59 wild strains (CY116, G42, TJ17) and chp knockout mutants (CY116-Δchp, G42-Δchp, TJ17-Δchp), together with two randomly selected ST239 strains (W3200, 09B38), were diluted to 0.5 × 107 colony-forming units (CFU)/ml. Paired, diluted ST59 and ST239 suspensions were mixed 1:1 in equal volumes. Then, 10 μl of the mixture was added to 20 ml of LB broth and cultured at 37 °C with shaking at 200 rpm. The 10-μl subcultures were transferred to fresh LB broth every 24 h. At the same time, 10-μl subcultures were inoculated on drug-free and 1 μg/ml rifampicin MH agar plates. The colonies of ST59 (susceptible to rifampicin) and ST239 (resistant to rifampicin) were counted after incubation overnight. After 96 h, the adaptive difference (S), relative adaptive fitness (F) and fitness cost (C) were calculated according to the following formulas (Additional file 2: Table S5).
Note: st is the number of sensitive colonies, and rt is the number of resistant colonies.
Results
ST239 and ST59 have acquired extensive diversity since their emergence
Our sequenced S. aureus isolates comprise 132 ST239 and 72 ST59 collected between 1994 and 2016 from hospitals spanning 14 Chinese provinces (Additional file 1: Table S1). Phylogenetic analysis of the recombination-free core genome alignments of the ST239 and ST59 isolates identified within-ST diversity over 2313 and 2244 core genes, respectively. ST59 could be resolved into three phylogenetic sub-lineages which we term for reference ST59-A, ST59-B and ST59-C (Fig. 1). Comparing these to more traditional spa typing points to a rough agreement between sub-lineages and dominant spa type (ST59-A: t163, ST59-B: t437, ST59-C: t437; see Additional file 2: Fig. S1). Similarly, ST239 had three major sub-lineages (dominant spa types ST239-A: t037, ST239-B: t037, ST239-C: t030; see Additional file 2: Fig. S1), though with some within sub-lineage diversity in spa type assignment. The phylogenetic sub-lineage structure could not be fully explained by geographic sampling in either ST, with isolates collected from hospitals located in different Chinese provinces interspersed in the phylogeny (Fig. 1). The exception was the oldest sub-lineage of ST239 (ST239-A/ST239-t037), which comprised a set of isolates collected from the same hospital in Beijing between 1994 and 2000. Across the remaining isolates, the lack of a strong phylogenetic structure based on the location of the hospital suggests frequent acquisition and transmission events between sampled provinces.
Using the date of isolate collection to calibrate the respective phylogenies, we obtained a strong correlation between the time of sampling and the root to tip distances which were highly significant following 10,000 date randomisations (ST239: R2 = 0.84; ST59: R2 = 0.50; Additional file 2: Fig. S2). This allowed us to infer genome-wide substitution rates for the non-recombining portion of the ST239 and ST59 alignments of 1.8 × 10−6 (1.7 × 10−6–2.0 × 10−6, 95% HPD) and 1.2 × 10−6 (1.0 × 10−6–1.4 × 10−6) substitutions per nucleotide site per year, respectively (see the ‘Methods’, Additional file 2: Table S2). While these estimates indicate a slightly faster rate in ST239, both fall largely in line with previous evolutionary rates reported for S. aureus which range between 1.2 × 10−6 and 4.0 × 10−6 substitutions per nucleotide site per year [13, 18, 21, 28, 35, 44]. Bayesian inference of a timed phylogeny across isolates [54] suggests that both ST groups have been in joint circulation from the early to mid-twentieth century, prior to the clinical introduction of methicillin. The sampled Chinese ST239 sub-lineages were estimated to last share a common ancestor in approximately 1950 (ST239: 95% HPD 1943–1956) (Fig. 1A), and the three sampled sub-lineages of 72 ST59 isolates were estimated to have diverged from each other in the mid-1930s (ST59: 1936, 95% HPD 1923–1948; Fig. 1B).
We expanded both datasets to include isolates from worldwide publicly available collections [13, 44, 46] and released as projects on Pathogenwatch (Additional file 2: Fig. S3, see the ‘Methods’ section). Within these global phylogenies, we again observed considerable substructure, highlighting that within-ST diversity is not a unique property of our sampled collection of strains from China but a more general feature of the ST59 and ST239 S. aureus lineages. Across the global core genome phylogenies, we observed that our considered Chinese isolates recapitulated a proportion of the diversity observed globally (Additional file 2: Fig. S3A, Additional file 2: Fig. S3B) consistent with multiple introductions from and into China, likely with distinct phylogeographic origins.
Accessory genome structure
The accessory genome represents an important component of the total S. aureus genome, harbouring elements involved in mediating virulence, immune evasion and resistance, which can be acquired by horizontal gene transfer. The accessory genome is not necessarily conserved within a particular ST and includes genes such as SCCmec, prophages, S. aureus mobile pathogenicity islands (SaPIs), insertion sequence (IS) elements, transposons and plasmids [69]. Having characterized the core genome of our Chinese S. aureus isolates, we compared the accessory genomes of each ST in more depth.
Our ST239 and ST59 isolates had large and variable accessory genomes of 3206 and 3026 genes, respectively (Additional file 2: Fig. S4). For ST239, the shared accessory genome was clustered into three groups, corresponding to the sub-lineages identified by the core genome phylogeny (Additional file 2: Fig. S5A). Variation in the accessory genome (presence/absence) suggests changes took place in the accessory genome of lineages of ST239 during its more recent evolution (Additional file 2: Fig. S5A). Though all ST239 isolates in our dataset carried the SCCmec of type III together with a Tn5801 like a transposon and an IS256 (Fig. 1A, Additional file 2: Fig. S6A, Additional file 2: Table S3). The presence of MGEs in the ST59 Chinese isolates was less variable across sub-lineages than for ST239 and did not recapitulate the core genome structure (Additional file 2: Fig. S5B, Additional file 2: Fig. S6B). The majority (n = 50) of ST59 isolates carried SCCmec of type IV.1, while a further eight carried SCCmec of type V and 14 had no SCCmec (Fig. 1B).
Evolutionary dynamics of antimicrobial resistance in ST239 and ST59
Given that isolates were sampled from hospitals across China, a possible hypothesis for the different trajectories observed between STs could be susceptibility to antimicrobial treatment. For instance, one a priori hypothesis would be that the decline in the prevalence of ST239 in China [6, 7, 31] may derive from a lesser ability to survive antimicrobial treatment. The latter for instance has been shown to have contributed to the success of different clonal complexes, for instance, the replacement of CC30 by CC22 following the emergence of fluoroquinolone resistance [21]. We considered the presence, absence and total carriage of AMR genes in both of our ST239 and ST59 cohorts over the 22-year surveillance period (Fig. 1, Additional file 2: Fig. S7, Additional file 2: Figs. S9-10). We previously showed that a subset of ST239 isolates was more phenotypically resistant than ST59 isolates [38]. This finding was recapitulated from comparative genomics: ST239 isolates instead carried significantly more AMR elements on average than the ST59 isolates (mean: 10.3 vs. 6.3), a trend which held when considering only MRSA isolates (Fig. 1, Additional file 2: Fig. S9).
There was however considerable heterogeneity in the carriage of genes conferring resistance to macrolides (erm(A)) and aminoglycosides (ant(6)-Ia and aph(3’)-III) within ST239 (Additional file 2: Fig. S7A), though based on current sampling, we detected an overall trend of decreasing resistance element carriage over time largely driven by elevated AMR carriage in ST239-A (adj. R2 = 0.05; p = 0.008) (Fig. 1A, Additional file 2: Fig. S10a). Conversely, we found no change in the number of AMR genes carried by ST59 over the sampling period (Additional file 2: Fig. S10a).
All our ST239 isolates were methicillin-resistant. However, the ST59 dataset included a small subset of methicillin-susceptible S. aureus (MSSA) isolates (Fig. 1B, Additional file 2: Fig. S6B, Additional file 2: Fig. S14A). MSSA and MRSA (cassette types IV.1 and V) isolates were interspersed across the core genome phylogeny, consistent with ST59-MRSA evolving through repeat acquisitions of the SCCmec element. Alternatively, some ST59-MRSA strains may have secondarily lost the SCCmec element. To formally test this, we extracted and aligned a maximal region extending 500 bp upstream of the SCCmec attachment site [65]. The diversity in this flanking region formed three clear clusters which matched the cassette type assignments (Additional file 2: Fig. S14B-C). The most parsimonious scenario for this clustering requires at least five independent acquisitions of the SCCmec given our dataset of 58 MRSA ST59. We also infer one likely instance of secondary gene loss (Additional file 2: Fig. S14A), due to the lack of SCCmec in a single isolate clustered within a well-supported (100% bootstrap support) MRSA phylogenetic clade.
Virulence and biofilm production of ST59 and ST239 strains
In silico prediction of virulence potential is more challenging than for AMR profiles [70], with counts of putative virulence factors likely a poor proxy for phenotypic virulence. The number of known virulence factors detected in ST239 was highly structured by core genome sub-lineage (Fig. 1A, Additional file 2: Fig. S8). Given our sampling, ST239 isolates harboured on average significantly more putative virulence genes than ST59 (mean 12.8 vs. 10.0; Wilcoxon signed-rank test p < 0.0001; Additional file 2: Fig. S11), and the total number of virulence genes carried across ST239 significantly increased over sampling time (adj. R2 = 0.62; p < 0.0001), with ST239-A carrying significantly fewer virulence factors than each of ST239-B or ST239-C (Additional file 2: Figs. S11-S12). We observed no such temporal trend in ST59 (Additional file 2: Fig. S12B). Within ST59, only a subset of isolates carried the PVL cytotoxin encoded by lukF-PV and lukS-PV (Additional file 2: Fig. S8B). The S. aureus surface protein SasX has also been shown to enhance nasal colonization and virulence in skin and lung infection models [71]. All isolates in both lineage ST239-A and ST239-B carried sasX, while only one strain of lineage ST239-C (the more recent lineage of ST239) carried sasX (Additional file 2: Fig. S13).
We next assessed the potential for virulence in the host using lysis of erythrocyte assay and the ability to form biofilms via a Tryptic Soy Broth (TSB) assay (see the ‘Methods’ section). Phenotypes were recorded for all the 132 ST239 and 72 ST59 strains that we sequenced (Additional file 1: Table S1). Despite carrying on average fewer virulence-associated genes compared to ST239, ST59 strains had significantly higher cytolytic activity in vitro than their ST239 counterparts (Kruskal-Wallis test, p < 0.0001; Fig. 2A). Conversely, the ability to produce biofilms was only slightly increased in ST59 (Fig. 2B).
Identification and functional validation of chp as a driver of virulence
To identify candidate genes associated to the increased cytolytic activity in the ST59 lineage, we performed an association study using the presence/absence of virulence genes and results from the lysis of erythrocyte assay. We identified, amongst the top five genes most strongly associated to the erythrocyte lysis phenotype, members of the immune evasion complex and, in particular, the chemotaxis inhibitory protein (chp) gene (Additional file 2: Fig. S15, Additional file 2: Table S4). The chp gene encodes the chemotaxis inhibitory protein of staphylococci (CHIPS), a protein which blocks neutrophil chemotaxis and contributes to the evasion of the innate immune response [72]. CHIPS has been identified as a candidate for virulence in ST59 strains [73–75], and the ability to evade immune defence, through the carriage of chp, offers a possible explanation for the increased relative fitness of a lineage. Hence, we performed functional analyses to validate the statistical association between the carriage of the chp gene and the phenotypic measurements in our ST59 dataset.
We selected three ST59 strains, representing the diversity of sub-lineages (annotated ST59-A, ST59-B, ST59-C), to construct chp knockout mutants, and compared the virulence between wild-type strains and chp knockout mutants. Cytolytic activity of all three strains decreased significantly after chp knockout (Fig. 2C), supporting its role in enhancing the virulence potential of ST59 compared to ST239. Knockout of the chp gene led to no significant change in the growth rates of the mutants, though we observe a small, though not significant (p = 0.10), decrease in the growth rate of TJ17-Δchp (Fig. 2D). We next measured the relative fitness of the three knockout Δchp strains and their wild-type progenitors in competition assays against two S. aureus ST239 strains and observed no statistically significant differences in colony counts between wild-type and knock-out strains following 96 h of growth (Additional file 2: Fig. S16, Additional file 2: Tables S5). These results suggest that carriage of chp has no detectable fitness cost to ST59.
Discussion
MRSAs remain a major nosocomial problem [76], in particular, in China [77]. Globally, the distribution of MRSA STs is highly structured and characterized by serial emergence and replacement events of new strains [30]. Until recently, HA-MRSA lineages such as ST239 have dominated in Asia [78], likely aided by their ability to evade treatment by methicillin [79]. However, in China, ST239 MRSAs have been in marked decline over the past two decades, while CA-MSSA and CA-MRSA lineages including ST59 are becoming more prevalent [31, 38]. In vitro characterisations have indicated a lower competitive ability of ST239 relative to CA-MRSA lineages ST30, ST8 and ST59 [35, 38], though somewhat counter-intuitively, with ST239 less susceptible to antibiotic treatment than replacing lineages such as ST59 [38]. As a result, the specific genotypic and phenotypic drivers underlying the displacement of ST239 by CA-MRSA lineages in China, and what the consequences of these may be for pathogenicity, remain poorly understood.
In this work, we generated paired genomic and phenotypic data for ST239 and CA-MRSA ST59 isolated from patients in Chinese hospitals spanning 22 years, covering the time course of lineage replacement. We estimated the age of emergence of both STs to the early to mid-twentieth century, suggesting both evolved prior to the first clinical use of methicillin before going on to acquire extensive pan-genome diversity (Fig. 1). Consistent with prior phenotypic observations, we found that the declining ST239 carried significantly more resistance elements than ST59, reinforcing that susceptibility to treatment is not an important driver of changing patterns of CA-MRSA prevalence in this case. This mirrors the situation in Singapore where a less drug-resistant lineage, ST22, is increasing in prevalence [44]. Consistently, we also detected no trend of increasing genotypic AMR carriage in ST59 over the roughly 20-year sampling period.
We therefore tested two hypotheses to explain the dynamics of ST replacement in China: increased transmission through longer persistence on surfaces due to biofilm production or higher virulence, for instance, through the ability to evade early innate immune defences. Despite the identification of large and highly structured accessory genomes, we found no evidence for biofilm production driving lineage replacement (Fig. 2). Conversely, we observed that ST59 strains had markedly enhanced cytolytic activity in vitro compared to ST239, consistent with the higher virulence of CA-MRSA strains more broadly (Fig. 2). We identified the chp gene as a strong contributor to the enhanced virulence potential of ST59 in our dataset. In support of the mechanistic role of chp in ST59, we found that the presence of chp, when compared to chp knockouts, increased the expression of other S. aureus virulence factors, including other members of the innate immune evasion cluster (sea, sak, scn).
We suggest that enhanced virulence is likely to play an important role in the success of ST59 in China. However, we note that virulence is a complex and context-dependent trait. For example, a simple sum of the number of putative virulence elements is unlikely to be a good proxy for the total infective potential of a CA-MRSA or HA-MRSA S. aureus. Consistently, we observed no relationship between the decrease in the prevalence of ST239 in Chinese hospitals and the number of predicted virulence elements over the sampling period. In fact, as with the number of resistance elements, ST239 carried on average a larger number of putative virulence factors than ST59, with the number of virulence determinants tending to increase over time, while remaining constant in ST59 over the same period. Our results therefore do not suggest a mere count of elements previously associated with clinically relevant phenotypes can explain differing patterns of ST incidence in China.
We do however highlight the importance of taking the variable accessory genome into account for the identification of virulence-associated factors and AMR determinants. For instance, although we saw no overall trend towards increasing resistance gene carriage (count), we inferred that ST59 independently acquired methicillin resistance at least five times in the 1980s (Additional file 2: Fig. S14). While the acquisition of methicillin resistance has been detected prior to the therapeutic use of methicillin [31] in the 1960s, our estimates of the times of SCCmec acquisition by MSSA ST59 is more consistent with a scenario where horizontal transfer was driven by the selective pressure of clinical drug use. This is also consistent with reports of high mobility of SCCmec between MSSA and MRSA strains in a recent hospital outbreak [80].
Virulence as a measurable trait ultimately requires evaluation using in vivo models though cytolytic activity, which can be readily measured using lysis of erythrocyte assays, offers a good proxy given hemolysins are well-established virulence factors [81]. Higher cytolytic activity as a virulence mechanism in our ST59 isolates is consistent with pronounced measurements of virulence in ST59 lineages [82] and the role of the innate immune evasion cluster in mouse infection models [83]. In our dataset, we identified, through a knock-out experiment, that the mechanism enhancing the virulence of ST59 in China can be well explained by the carriage of a single gene, encoding CHIPS (chp). chp is located on the β-hemolysin (hlb)-converting bacteriophages, which drives instant immune evasion by S. aureus [72]. While fitness costs have been associated to the acquisition of resistance genes [27, 35, 84–86], including large SCCmec elements, we detect no such effect with chp when comparing the growth rates of wild-type to knockout ST59 strains.
Enhanced virulence activity has recently been suggested as a contributor to the increasing prevalence of another CA-MRSA ST5 in China, including in MSSA [83]. Our findings lend further support to this hypothesis, with the carriage of chp as a potentially major contributor to the ongoing prevalence of ST59 strains in China, likely at the expense of ST239. This is concerning as CHIPS ability to evade the early immune response through both inhibitions of chemotaxis and phagocytosis [72], suggests a potential for more severe and longer-term infections. However, it is probable that other factors also contribute. For instance, it is important to remember that surveillance data informing the replacement of MRSA lineages is primarily based on severe infections in hospitals caused by different MRSA lineages. MRSAs are also found at fairly high prevalence in the nares of healthy carriers [87], possibly at different frequencies than in nosocomial infections. As such, the high relative virulence of the ST59 lineage may more directly reflect its incidence in nosocomial MRSA infection, rather than its prevalence in the wider population. In addition, there may be some stochasticity in which STs rise in prevalence when previously dominant STs decline. One possibility is that documented loss of fitness of ST239 [35] has also played an important role in the dynamics of which CA-MRSA are now being observed.
Our work highlights the full potential of WGS in microbial genomics when analysed in conjunction with associated phenotypes. Far too often, genomic sequence data is deposited on publicly available databases without the associated metadata, even when available. This greatly limits the potential of genomic sequence data reuse to address further questions. We hope that this work will contribute to encouraging others to make their phenotypic data fully accessible. In principle, similar, larger studies could be conducted on publicly available data to address pressing questions on the genetic basis of other key phenotypic traits in major human pathogens.
Conclusions
By analysing the core and accessory genomes of a carefully phenotyped set of isolates collected in China over the last two decades, we were able to identify the likely driver of the replacement of HA-MRSA lineages by their CA-MRSA counterparts in China. We found no evidence for a role of AMR or the ability to colonise and persist in the environment. Conversely, we found that the expanding ST59 lineage was far more virulent than its previously widespread nosocomial ST239 counterpart. We could further show that this difference in virulence is likely largely driven by the carriage of the chp gene by ST59, an important contributor to immune evasion. We surmise that the ongoing lineage replacement observed in Chinese hospitals is primarily driven by differences in virulence of different MRSA lineages, which can in part be explained by the carriage of the chp gene by the ST59 lineage.
Supplementary Information
Additional file 1: Table S1. 132 ST239 and 72 ST59 Chinese isolates included in the phylogenetic analysis. MLST, multilocus sequence type.
Additional file 2: Fig. S1. Core genome phylogeny for ST239 and ST59. Fig. S2. Regression of root-to-tip distance against sampling time for core genome alignments. Fig. S3. Global core genome phylogenies for ST239 and ST59. Fig. S4. Shared core and accessory genes amongst Chinese ST239 and ST59 isolates. Fig. S5. Conservation of accessory genome homology groups (HGs) across the Chinese ST239 and ST59 isolates. Fig. S6. Presence/absence of mobile genetic elements (MGEs) amongst Chinese ST239 and ST59 isolates. Fig. S7. Presence/absence of antimicrobial resistance genes amongst Chinese ST239 and ST59 isolates. Fig. S8. Presence/absence of virulence genes amongst Chinese ST239 and ST59 isolates. Fig. S9. Number of resistance (AMR) genes carried between ST sub-lineages. Fig. S10. Changing trends of antimicrobial resistance (AMR) mutations/elements within ST239 and ST59 lineages over time. Fig. S11. Number of virulence genes carried between ST sub-lineages and within ST239 and ST59 sub-lineages. Fig. S12. Trends of virulence mutations/elements within Chinese ST239 and ST59 lineages over time. Fig. S13. Core genome phylogeny for ST239 annotated for sasX. Fig. S14. Methicillin resistance acquisition analyses. Fig. S15. Presence absence of virulence genes against cell lysis phenotype. Fig. S16. Colony counts at 0 hours and 96 hours for three ST59 isolates (wild-type and knock-out) compared to two possible ST239 control strains. Table S2. Inferred tMRCA, clock rates and marginal likelihoods inferred by Bayesian dating analyses. Table S3. The distribution of mobile genetic elements (MGEs) in different sub-lineages. Table S4. Bonferroni corrected p-values following fitting of a generalized linear model (GLM) for VFDB virulence gene presence / absence against cell lysis phenotype. Table S5. Relative adaptive difference (S), fitness (F) and fitness cost (C) estimated from the in vitro competition experiment between ST59-WT and ST59 Δchp.
Acknowledgements
We additionally wish to thank Dr. Bernadette Young for useful the discussions on virulence mechanisms in S. aureus.
Authors’ contributions
HC, YY, HG, JY, FC, SS, XW, SL, YZ and HW collected the data and performed the phenotypic testing and competition assays. Bioinformatic analyses were performed by HC, LvD, LPS, MA and RAF. FB and HW designed the study. LvD and FB wrote the paper with contributions from all authors. The authors read and approved the final manuscript.
Funding
This work was supported by the China NSFC grant (81991533 and 81625014) and the Newton Fund UK-China NSFC initiative (grants MR/P007597/1 and 81661138006). FB acknowledges the support from the BBSRC GCRF scheme. All authors acknowledge the UCL Biosciences Big Data equipment grant from BBSRC (BB/R01356X/1). LvD is supported by a UCL Excellence Fellowship. RAF is part of the Medical Research Council Centre for Medical Mycology MR/N006364/2 and is supported by a Wellcome Trust Seed Award (215239/Z/19/Z). LPS is supported by a Sir Henry Wellcome Postdoctoral Fellowship (220422/Z/20/Z). HC acknowledges financial support from a 111 Talent Discipline Planning of Peking University People’s Hospital (PKUPH) award for a 1-year visit at University College London.
Availability of data and materials
Raw Illumina short-read data from the 204 newly generated S. aureus isolates are available on NCBI with BioProject ID: PRJNA491594 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA491594) [88]. Full accompanying metadata and phenotypic information are available in Additional file 1: Table S1.
Declarations
Ethics approval and consent to participate
Bacterial strains were collected in clinical routine diagnostics, and ethical approval and consent to participate from patients were not required in accordance with Decree No. 11 of the National Health Commission of the People’s Republic of China (Operational Guideline for the Ethic Review of Biomedical Research Involving Human Subject, approved on September 30, 2016, implemented from December 1, 2016).
The analysis of de-identified patient demographic data was approved by the Peking University People’s Hospital Institutional Review Board (No. 2020PHB193-01). The research conformed to the principles of the Helsinki Declaration and its later amendments.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hongbin Chen, Yuyao Yin and Lucy van Dorp contributed equally to this work.
Contributor Information
Lucy van Dorp, Email: lucy.dorp.12@ucl.ac.uk.
Hui Wang, Email: wanghui@pkuph.edu.cn.
Francois Balloux, Email: f.balloux@ucl.ac.uk.
References
- 1.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, Aboderin AO, al-Abri SS, Awang Jalil N, Benzonana N, Bhattacharya S, Brink AJ, Burkert FR, Cars O, Cornaglia G, Dyar OJ, Friedrich AW, Gales AC, Gandra S, Giske CG, Goff DA, Goossens H, Gottlieb T, Guzman Blanco M, Hryniewicz W, Kattula D, Jinks T, Kanj SS, Kerr L, Kieny MP, Kim YS, Kozlov RS, Labarca J, Laxminarayan R, Leder K, Leibovici L, Levy-Hara G, Littman J, Malhotra-Kumar S, Manchanda V, Moja L, Ndoye B, Pan A, Paterson DL, Paul M, Qiu H, Ramon-Pardo P, Rodríguez-Baño J, Sanguinetti M, Sengupta S, Sharland M, Si-Mehand M, Silver LL, Song W, Steinbakk M, Thomsen J, Thwaites GE, van der Meer JWM, van Kinh N, Vega S, Villegas MV, Wechsler-Fördös A, Wertheim HFL, Wesangula E, Woodford N, Yilmaz FO, Zorzet A. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–32. 10.1056/NEJM199808203390806. [DOI] [PubMed]
- 3.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44(6):1549–55. 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed]
- 4.Paterson GK, Harrison EM, Holmes MA. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014:42–7. 10.1016/j.tim.2013.11.003. [DOI] [PMC free article] [PubMed]
- 5.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A. 2002;99(11):7687–92. 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed]
- 6.Liu Y, Wang H, Du N, Shen E, Chen H, Niu J, et al. Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob Agents Chemother. 2009;53(2):512–8. 10.1128/AAC.00804-08. [DOI] [PMC free article] [PubMed]
- 7.He W, Chen H, Zhao C, Zhang F, Li H, Wang Q, et al. Population structure and characterisation of Staphylococcus aureus from bacteraemia at multiple hospitals in China: association between antimicrobial resistance, toxin genes and genotypes. Int J Antimicrob Agents. 2013;42(3):211–9. 10.1016/j.ijantimicag.2013.04.031. [DOI] [PubMed]
- 8.Xiao M, Wang H, Zhao Y, Mao LL, Brown M, Yu YS, et al. National surveillance of methicillin-resistant Staphylococcus aureus in China highlights a still-evolving epidemiology with 15 novel emerging multilocus sequence types. J Clin Microbiol. 2013;51(11):3638–44. 10.1128/JCM.01375-13. [DOI] [PMC free article] [PubMed]
- 9.Xu BL, Zhang G, Ye HF, Feil EJ, Chen GR, Zhou XM, et al. Predominance of the Hungarian clone (ST 239-III) among hospital-acquired meticillin-resistant Staphylococcus aureus isolates recovered throughout mainland China. J Hosp Infect England. 2009;71(3):245–55. 10.1016/j.jhin.2008.10.029. [DOI] [PubMed]
- 10.Hsu L-YY, Koh T-HH, Singh K, Kang M-LL, Kurup A, Tan B-HH. Dissemination of multisusceptible methicillin-resistant Staphylococcus aureus in Singapore. J Clin Microbiol. 2005;43(6):2923–5. 10.1128/JCM.43.6.2923-2925.2005. [DOI] [PMC free article] [PubMed]
- 11.Jariyasethpong T, Tribuddharat C, Dejsirilert S, Kerdsin A, Tishyadhigama P, Rahule S, et al. MRSA carriage in a tertiary governmental hospital in Thailand: emphasis on prevalence and molecular epidemiology. Eur J Clin Microbiol Infect Dis. 2010;29(8):977–85. 10.1007/s10096-010-0954-7. [DOI] [PubMed]
- 12.Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, et al. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother. 2006;50(3):1001–12. 10.1128/AAC.50.3.1001-1012.2006. [DOI] [PMC free article] [PubMed]
- 13.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuang Y-YY, Huang Y-CC. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis. 2013;13(8):698–708. 10.1016/S1473-3099(13)70136-1. [DOI] [PubMed]
- 15.Johnson AP. Methicillin-resistant Staphylococcus aureus: the European landscape. J Antimicrob Chemother. 2011;66(Suppl 4):IV43–8. 10.1093/jac/dkr076. [DOI] [PubMed]
- 16.Diekema DJ, Richter SS, Heilmann KP, Dohrn CL, Riahi F, Tendolkar S, et al. Continued emergence of USA300 methicillin-resistant Staphylococcus aureus in the United States: results from a nationwide surveillance study. Infect Control Hosp Epidemiol. 2014;35(3):285–92. 10.1086/675283. [DOI] [PubMed]
- 17.Aanensen DM, Feil EJ, Holden MTG, Dordel J, Yeats CA, Fedosejev A, et al. Whole-genome sequencing for routine pathogen surveillance in public health: a population snapshot of invasive Staphylococcus aureus in Europe. MBio. 2016;7(3). 10.1128/mBio.00444-16. [DOI] [PMC free article] [PubMed]
- 18.Nübel U, Dordel J, Kurt K, Strommenger B, Westh H, Shukla SK, et al. A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog. 2010;6:e1000855. . [DOI] [PMC free article] [PubMed]
- 19.Tenover FC, Goering RV. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J Antimicrob Chemother. 2009;64(3):441–6. 10.1093/jac/dkp241. [DOI] [PubMed]
- 20.Johnson AP, Aucken HM, Cavendish S, Ganner M, Wale MC, Warner M, et al. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS) J Antimicrob Chemother. 2001;48(1):143–144. doi: 10.1093/jac/48.1.143. [DOI] [PubMed] [Google Scholar]
- 21.Holden MTG, Hsu L-YY, Kurt K, Weinert LA, Mather AE, Harris SR, et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013;23(4):653–64. 10.1101/gr.147710.112. [DOI] [PMC free article] [PubMed]
- 22.Huang Y-CC, Chen C-JJ. Community-associated meticillin-resistant Staphylococcus aureus in children in Taiwan, 2000s. Int J Antimicrob Agents. 2011;38(1):2–8. 10.1016/j.ijantimicag.2011.01.011. [DOI] [PubMed]
- 23.Pang R, Wu S, Zhang F, Huang J, Wu H, Zhang J, et al. The genomic context for the evolution and transmission of community-associated Staphylococcus aureus ST59 through the food chain. Front Microbiol. 2020;11:422. 10.3389/fmicb.2020.00422. [DOI] [PMC free article] [PubMed]
- 24.Sun C, Wang Q, Li W-T, Wen D-N, Chen C-H, Yang X, et al. Molecular characteristics and antimicrobial susceptibility of Staphylococcus aureus among children with respiratory tract infections in southwest China. World J Pediatr. 2020;16(3):284–92. 10.1007/s12519-019-00317-4. [DOI] [PubMed]
- 25.Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, et al. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc Natl Acad Sci U S A. 2008;105(4):1327–32. 10.1073/pnas.0710217105. [DOI] [PMC free article] [PubMed]
- 26.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7(9):629–41. 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed]
- 27.Knight GM, Budd EL, Whitney L, Thornley A, Al-Ghusein H, Planche T, et al. Shift in dominant hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) clones over time. J Antimicrob Chemother. 2012;67(10):2514–22. 10.1093/jac/dks245. [DOI] [PubMed]
- 28.McAdam PR, Templeton KE, Edwards GF, Holden MTG, Feil EJ, Aanensen DM, et al. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2012;109(23):9107–12. 10.1073/pnas.1202869109. [DOI] [PMC free article] [PubMed]
- 29.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298(15):1763–71. 10.1001/jama.298.15.1763. [DOI] [PubMed]
- 30.Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 2019;17(4):203–18. 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed]
- 31.Diep BA, Sensabaugh GF, Somboona NS, Carleton HA, Perdreau-Remington F, Somboonna N, et al. Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J Clin Microbiol. 2004;42(5):2080–4. 10.1128/JCM.42.5.2080-2084.2004. [DOI] [PMC free article] [PubMed]
- 32.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–87. 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed]
- 33.Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. In: Gottesman S, Harwood CS, editors. Annu Rev Microbiol, vol. 64. Palo Alto: Annual Reviews; 2010. p. 143–62. 10.1146/annurev.micro.112408.134309. [DOI] [PubMed]
- 34.Chen C-JJ, Unger C, Hoffmann W, Lindsay JA, Huang Y-CC, Götz F, et al. Characterization and comparison of 2 distinct epidemic community-associated methicillin-resistant Staphylococcus aureus clones of ST59 lineage. PLoS One. 2013;8(9):e63210. 10.1371/journal.pone.0063210. eCollec. [DOI] [PMC free article] [PubMed]
- 35.Gill JL, Hedge J, Wilson DJ, MacLean RC. Evolutionary processes driving the rise and fall of Staphylococcus aureus ST239, a dominant hybrid pathogen. bioRxiv. 2021:2021.01.10.426095. [DOI] [PMC free article] [PubMed]
- 36.Baines SL, Holt KE, Schultz MB, Seemann T, Howden BO, Jensen SO, et al. Convergent adaptation in the dominant global hospital clone ST239 of methicillin-resistant Staphylococcus aureus. mBio. 2015;6(2):e00080. 10.1128/mBio.00080-15. [DOI] [PMC free article] [PubMed]
- 37.Chen H, Yin Y, Li X, Li S, Gao H, Wang X, et al. Whole-genome analysis of livestock-associated methicillin-resistant Staphylococcus aureus sequence type 398 strains isolated from patients with bacteremia in China. J Infect Dis. 2020;221:S220–8. 10.1093/infdis/jiz575. [DOI] [PubMed]
- 38.Li S, Sun S, Yang C, Chen H, Yin Y, Li H, et al. The changing pattern of population structure of Staphylococcus aureus from bacteremia in China from 2013 to 2016: ST239-030-MRSA replaced by ST59-t437. Front Microbiol. 2018;9:332. 10.3389/fmicb.2018.00332. [DOI] [PMC free article] [PubMed]
- 39.Song Q, Wu J, Ruan P. Predominance of community-associated sequence type 59 methicillin-resistant Staphylococcus aureus in a paediatric intensive care unit. J Med Microbiol. 2018;67(3):408–14. 10.1099/jmm.0.000693. [DOI] [PubMed]
- 40.Wu S, Huang J, Zhang F, Wu Q, Zhang J, Pang R, et al. Prevalence and characterization of food-related methicillin-resistant Staphylococcus aureus (MRSA) in China. Front Microbiol. 2019;10:304. 10.3389/fmicb.2019.00304. [DOI] [PMC free article] [PubMed]
- 41.Chen Y, Sun L, Ba X, Jiang S, Zhuang H, Zhu F, et al. Epidemiology, evolution and cryptic susceptibility of methicillin-resistant Staphylococcus aureus in China: a whole-genome-based survey. Clin Microbiol Infect. 2021;S1198-743X(21)00264-0. Advance online publication. 10.1016/j.cmi.2021.05.024. [DOI] [PubMed]
- 42.Amorim ML, Faria NA, Oliveira DC, Vasconcelos C, Cabeda JC, Mendes AC, et al. Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J Clin Microbiol. 2007;45(9):2881–8. 10.1128/JCM.00603-07. [DOI] [PMC free article] [PubMed]
- 43.Conceição T, Aires-de-Sousa M, Füzi M, Tóth A, Pászti J, Ungvári E, et al. Replacement of methicillin-resistant Staphylococcus aureus clones in Hungary over time: a 10-year surveillance study. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2007;13:971–9. [DOI] [PubMed]
- 44.Hsu L-Y, Harris SR, Chlebowicz MA, Lindsay JA, Koh T-H, Krishnan P, et al. Evolutionary dynamics of methicillin-resistant Staphylococcus aureus within a healthcare system. Genome Biol. 2015;16(1):81. 10.1186/s13059-015-0643-z. [DOI] [PMC free article] [PubMed]
- 45.Thammavongsa V, Kim HK, Missiakas D, Schneewind O. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol. 2015;13(9):529–543. doi: 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward MJ, Goncheva M, Richardson E, McAdam PR, Raftis E, Kearns A, et al. Identification of source and sink populations for the emergence and global spread of the East-Asia clone of community-associated MRSA. Genome Biol. 2016;17(1):160. doi: 10.1186/s13059-016-1022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed]
- 48.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014:btu153 Oxford Univ Press. [DOI] [PubMed]
- 49.Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180. 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed]
- 50.Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11(2):e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed]
- 52.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016;2:vew007. 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed]
- 53.Didelot X, Croucher NJ, Bentley SD, Harris SR, Wilson DJ. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res. 2018;46:e134. 10.1093/nar/gky783. [DOI] [PMC free article] [PubMed]
- 54.Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu C-HH, Xie D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. Prlic A, editor. PLoS Comput Biol. 2014;10:e1003537. 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed]
- 55.Bouckaert RR, Drummond AJ. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol Biol. 2017;17:42. 10.1186/s12862-017-0890-6. [DOI] [PMC free article] [PubMed]
- 56.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 2012;29:2157–67. 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed]
- 57.Yu G, Smith DK, Zhu H, Guan Y, Lam TTT-Y. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. McInerny G, editor. Methods Ecol Evol. 2017;8:28–36. http://doi.wiley.com/10.1111/2041-210X.12628.
- 58.Strauß L, Stegger M, Akpaka PE, Alabi A, Breurec S, Coombs G, et al. Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. Proc Natl Acad Sci U S A. 2017;114(49):E10596–604. 10.1073/pnas.1702472114. [DOI] [PMC free article] [PubMed]
- 59.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39(suppl):W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jamrozy D, Coll F, Mather AE, Harris SR, Harrison EM, MacGowan A, et al. Evolution of mobile genetic element composition in an epidemic methicillin-resistant Staphylococcus aureus: temporal changes correlated with frequent loss and gain events. BMC Genomics. 2017;18(1):684. 10.1186/s12864-017-4065-z. [DOI] [PMC free article] [PubMed]
- 61.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–6. 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed]
- 62.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4. https://doi.org/10.1093/jac/dks261 . [DOI] [PMC free article] [PubMed]
- 63.Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, et al. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 2014;52(5):1501–10. 10.1128/JCM.03617-13. Epub 2014 Feb. [DOI] [PMC free article] [PubMed]
- 64.Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47(D1):D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noto MJ, Kreiswirth BN, Monk AB, Archer GL. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J Bacteriol. 2008;190(4):1276–83. 10.1128/JB.01128-07. [DOI] [PMC free article] [PubMed]
- 66.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed]
- 67.Müllner D. fastcluster: fast hierarchical, agglomerative clustering routines for R and Python. J Stat Softw. 2013;53:1–18.
- 68.Gu T, Zhao S, Pi Y, Chen W, Chen C, Liu Q, et al. Highly efficient base editing in Staphylococcus aureus using an engineered CRISPR RNA-guided cytidine deaminase. Chem Sci. 2018;9:3248–53. [DOI] [PMC free article] [PubMed]
- 69.Lindsay JA, Holden MTG. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 2004;12(8):378–85. 10.1016/j.tim.2004.06.004. [DOI] [PubMed]
- 70.Balloux F, Brønstad Brynildsrud O, van Dorp L, Shaw LP, Chen H, Harris KA, et al. From theory to practice: translating whole-genome sequencing (WGS) into the clinic. Trends Microbiol. 2018;26:1035–48. [DOI] [PMC free article] [PubMed]
- 71.Liu Q, Du X, Hong X, Li T, Zheng B, He L, et al. Targeting surface protein SasX by active and passive vaccination to reduce Staphylococcus aureus colonization and infection. Infect Immun. 2015;83:2168–74. [DOI] [PMC free article] [PubMed]
- 72.Rooijakkers SHM, Ruyken M, van Roon J, van Kessel KPM, van Strijp JAG, van Wamel WJB. Early expression of SCIN and CHIPS drives instant immune evasion by Staphylococcus aureus. Cell Microbiol. 2006;8(8):1282–93. 10.1111/j.1462-5822.2006.00709.x. [DOI] [PubMed]
- 73.Delauné A, Dubrac S, Blanchet C, Poupel O, Mäder U, Hiron A, Leduc A, Fitting C, Nicolas P, Cavaillon JM, Adib-Conquy M, Msadek T. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect Immun. 2012;80(10):3438–3453. doi: 10.1128/IAI.00195-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blomfeldt A, Aamot HV, Eskesen AN, Monecke S, White RA, Leegaard TM, et al. DNA microarray analysis of Staphylococcus aureus causing bloodstream infection: bacterial genes associated with mortality? Eur J Clin Microbiol Infect Dis. 2016;35:1285–95. [DOI] [PubMed]
- 75.Grousd JA, Rich HE, Alcorn JF. Host-pathogen interactions in Gram-positive bacterial pneumonia. Clin Microbiol Rev. 2019;32(3). 10.1128/CMR.00107-18. [DOI] [PMC free article] [PubMed]
- 76.Lee AS, de Lencastre H, Garau J, Kluytmans J, Malhotra-Kumar S, Peschel A, et al. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Prim. 2018;4:18033. [DOI] [PubMed]
- 77.Zhang Y, Yao Z, Zhan S, Yang Z, Wei D, Zhang J, et al. Disease burden of intensive care unit-acquired pneumonia in China: a systematic review and meta-analysis. Int J Infect Dis. 2014;29:84–90. [DOI] [PubMed]
- 78.Feil EJ, Nickerson EK, Chantratita N, Wuthiekanun V, Srisomang P, Cousins R, et al. Rapid detection of the pandemic methicillin-resistant Staphylococcus aureus clone ST 239, a dominant strain in Asian hospitals. J Clin Microbiol. 2008;46(4):1520–2. 10.1128/JCM.02238-07. [DOI] [PMC free article] [PubMed]
- 79.Gray RR, Tatem AJ, Johnson JA, Alekseyenko AV, Pybus OG, Suchard MA, et al. Testing spatiotemporal hypothesis of bacterial evolution using methicillin-resistant Staphylococcus aureus ST239 genome-wide data within a Bayesian framework. Mol Biol Evol. 2011;28(5):1593–603. 10.1093/molbev/msq319. [DOI] [PMC free article] [PubMed]
- 80.Weterings V, Bosch T, Witteveen S, Landman F, Schouls L, Kluytmans J. Next-generation sequence analysis reveals transfer of methicillin resistance to a methicillin-susceptible Staphylococcus aureus strain hat subsequently caused a methicillin-resistant Staphylococcus aureus outbreak: a descriptive study. J Clin Microbiol. 2017;55(9):2808–16. 10.1128/JCM.00459-17. [DOI] [PMC free article] [PubMed]
- 81.DeLeo FR, Diep BA, Otto M. Host defense and pathogenesis in Staphylococcus aureus infections. Infect Dis Clin N Am. 2009;23(1):17–34. 10.1016/j.idc.2008.10.003. [DOI] [PMC free article] [PubMed]
- 82.Li M, Dai Y, Zhu Y, Fu C-L, Tan VY, Wang Y, et al. Virulence determinants associated with the Asian community-associated methicillin-resistant Staphylococcus aureus lineage ST59. Sci Rep. 2016;6(1):27899. 10.1038/srep27899. [DOI] [PMC free article] [PubMed]
- 83.Jian Y, Zhao L, Zhao N, Lv H-Y, Liu Y, He L, et al. Increasing prevalence of hypervirulent ST5 methicillin susceptible Staphylococcus aureus subtype poses a serious clinical threat. Emerg Microbes Infect. 2021;10(1):109–22. 10.1080/22221751.2020.1868950. [DOI] [PMC free article] [PubMed]
- 84.Brockhurst MA, Harrison E, Hall JPJ, Richards T, McNally A, MacLean C. The ecology and evolution of pangenomes. Curr Biol. 2019;29(20):R1094–R1103. doi: 10.1016/j.cub.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 85.Vogwill T, MacLean RC. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol Appl. 2015;8(3):284–295. doi: 10.1111/eva.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee SM, Ender M, Adhikari R, Smith JMB, Berger-Bächi B, Cook GM. Fitness cost of staphylococcal cassette chromosome mec in methicillin-resistant Staphylococcus aureus by way of continuous culture. Antimicrob Agents Chemother. 2007;51(4):1497–9. 10.1128/AAC.01239-06. [DOI] [PMC free article] [PubMed]
- 87.Wu M, Tong X, Liu S, Wang D, Wang L, Fan H. Prevalence of methicillin-resistant Staphylococcus aureus in healthy Chinese population: a system review and meta-analysis. PLoS One. 2019;14(10):e0223599. 10.1371/journal.pone.0223599. [DOI] [PMC free article] [PubMed]
- 88.Chen H, Yen Y, van Dorp L, Shaw LP, Gao H, Acman M, et al. Drivers of methicillin resistant Staphylococcus aureus (MRSA) lineage replacement in China. BioProject PRJNA491594, NCBI Sequence Read Archive. 2021. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA491594. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. 132 ST239 and 72 ST59 Chinese isolates included in the phylogenetic analysis. MLST, multilocus sequence type.
Additional file 2: Fig. S1. Core genome phylogeny for ST239 and ST59. Fig. S2. Regression of root-to-tip distance against sampling time for core genome alignments. Fig. S3. Global core genome phylogenies for ST239 and ST59. Fig. S4. Shared core and accessory genes amongst Chinese ST239 and ST59 isolates. Fig. S5. Conservation of accessory genome homology groups (HGs) across the Chinese ST239 and ST59 isolates. Fig. S6. Presence/absence of mobile genetic elements (MGEs) amongst Chinese ST239 and ST59 isolates. Fig. S7. Presence/absence of antimicrobial resistance genes amongst Chinese ST239 and ST59 isolates. Fig. S8. Presence/absence of virulence genes amongst Chinese ST239 and ST59 isolates. Fig. S9. Number of resistance (AMR) genes carried between ST sub-lineages. Fig. S10. Changing trends of antimicrobial resistance (AMR) mutations/elements within ST239 and ST59 lineages over time. Fig. S11. Number of virulence genes carried between ST sub-lineages and within ST239 and ST59 sub-lineages. Fig. S12. Trends of virulence mutations/elements within Chinese ST239 and ST59 lineages over time. Fig. S13. Core genome phylogeny for ST239 annotated for sasX. Fig. S14. Methicillin resistance acquisition analyses. Fig. S15. Presence absence of virulence genes against cell lysis phenotype. Fig. S16. Colony counts at 0 hours and 96 hours for three ST59 isolates (wild-type and knock-out) compared to two possible ST239 control strains. Table S2. Inferred tMRCA, clock rates and marginal likelihoods inferred by Bayesian dating analyses. Table S3. The distribution of mobile genetic elements (MGEs) in different sub-lineages. Table S4. Bonferroni corrected p-values following fitting of a generalized linear model (GLM) for VFDB virulence gene presence / absence against cell lysis phenotype. Table S5. Relative adaptive difference (S), fitness (F) and fitness cost (C) estimated from the in vitro competition experiment between ST59-WT and ST59 Δchp.
Data Availability Statement
Raw Illumina short-read data from the 204 newly generated S. aureus isolates are available on NCBI with BioProject ID: PRJNA491594 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA491594) [88]. Full accompanying metadata and phenotypic information are available in Additional file 1: Table S1.