Abstract
The great majority of clinical isolates of Streptococcus pneumoniae carry prophages that may be identified through their hybridization with a DNA probe specific for the pneumococcal lytA gene (M. Ramirez, E. Severina, and A. Tomasz, J. Bacteriol. 181:3618–3625, 1999). We now show that the lytA hybridization pattern of chromosomal SmaI digests is stable for a given strain during extensive serial culturing in the laboratory; the pattern is specific for the strain’s clonal type, as defined by pulsed-field gel electrophoretis (PFGE) pattern, and variations in PFGE subtypes may be explained by changes in the number and chromosomal localization of this prophage(s). These observations indicate that the lytA hybridization pattern may be used as a molecular epidemiological marker that offers additional resolution of the genetic background of S. pneumoniae strains.
In a recent study, we examined a large number of Streptococcus pneumoniae clinical isolates with a DNA probe specific for lytA (14), the genetic determinant of the pneumococcal autolysin (N-acetyl muramic acid-l-alanine amidase) (6, 7). Since the lytA sequence has no recognition site for SmaI (accession no. Z34303), it was surprising to find that the great majority of isolates showed additional lytA-hybridizing bands in excess of the host autolysin gene, which was identified on an SmaI fragment of approximately 90 kb in most isolates. A number of additional tests indicated that the lytA-hybridizing band(s) in excess of the host autolysin gene represented prophages, which carry sequences highly homologous with the host lytA gene (14). The purpose of the studies described here was to test if the pattern generated by prophage carriage, identified through the number and molecular size of lytA-hybridizing bands in SmaI digests of total DNA, could be useful in molecular typing studies.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. pneumoniae isolates are from the Rockefeller University collection. Liquid cultures of strains were grown in a semisynthetic medium (9) at 37°C without aeration. Viable titers of bacteria were determined, and serial passage of strains was done, by plating on tryptic soy agar (Difco, Detroit, Mich.) supplemented with 3% sterile sheep blood incubated at 37°C.
DNA probes.
A DNA probe for the lytA gene was generated by PCR with DNA from strain R36A as template. The probe encompassed an 890-nucleotide (nt) internal fragment of lytA from which the first 57 and last 10 nt of the lytA gene were missing (14).
The PCR products were purified by using the Wizard DNA Cleanup kit (Promega, Madison, Wis.) before being labeled with the ECL direct labeling kit (Amersham, Little Chalfont, United Kingdom).
Probes for the lytA gene.
The probe for the lytA gene was obtained from plasmid pGL80 (7). Plasmid DNA was prepared by using the Wizard Midiprep kit (Promega), digested with HindIII (New England Biolabs, Beverly, Mass.), and separated by agarose gel electrophoresis. The 1.2-kb fragment from pGL80 contained the entire lytA gene (957 nt), a fragment of upstream sequence (199 nt), and 51 nt downstream of the gene. The 1.2-kb fragment containing the lytA gene was purified from the gel by using the Wizard DNA Cleanup kit. A second DNA probe was generated by PCR to include an 890-nt internal fragment of lytA from which the first 57 and last 10 nt of the lytA gene were missing. The 890-bp product was generated by using primers Lytd-1 and Lytr-1 (14) with DNA from strain R36A as template. The PCR product was purified by using the Wizard DNA Cleanup kit before being labeled with the ECL direct labeling kit.
Pulsed-field gel electrophoresis (PFGE).
Agarose disks for PFGE were prepared as previously described (17). Restriction of total DNA with SmaI (New England Biolabs) was performed as follows: one agarose disk was transferred into a microcentrifuge tube containing 500 μl of the commercially supplied buffer (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 1 mM dithiothreitol, pH 7.9) and incubated for at least 1 h at 25°C. The buffer was then removed, and 50 μl of a solution of SmaI in commercially supplied reaction buffer was added and incubated at 25°C for a period of 14 to 16 h. The reaction was stopped by the addition of EDTA to a final concentration of 0.08 M, and the fragments were separated and analyzed as previously described (17).
Southern blot hybridization.
DNA fragments separated by PFGE were transferred to nylon membranes (Hybond-N+; Amersham) with the Vacuum Gene System (Pharmacia LKB Biotech, Uppsala, Sweden), according to the manufacturer’s instructions. Membranes were hybridized to the lytA-specific DNA probe labeled with the ECL direct labeling system. Hybridization conditions were as recommended by the manufacturer, with a sodium chloride concentration of 0.5 M. The molecular sizes of the hybridization signal(s) (15) and the corresponding SmaI fragments were determined.
RESULTS
Variation of the lytA hybridization pattern with clonal type.
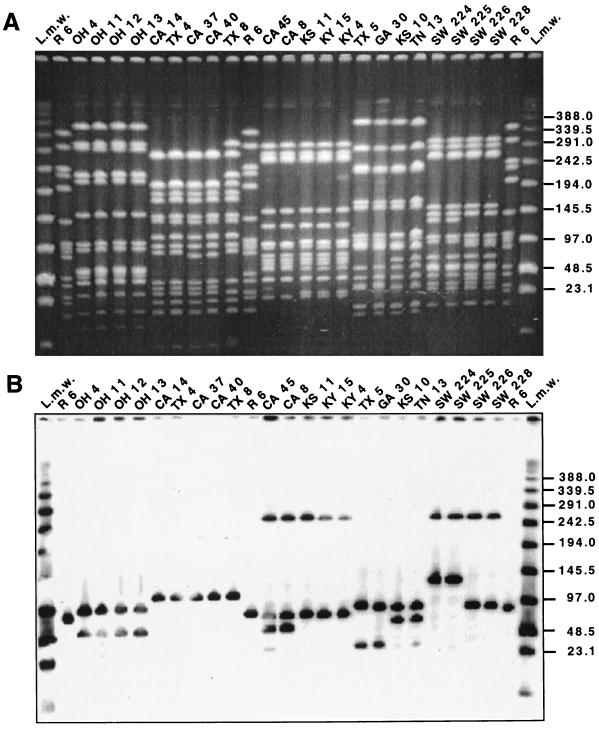
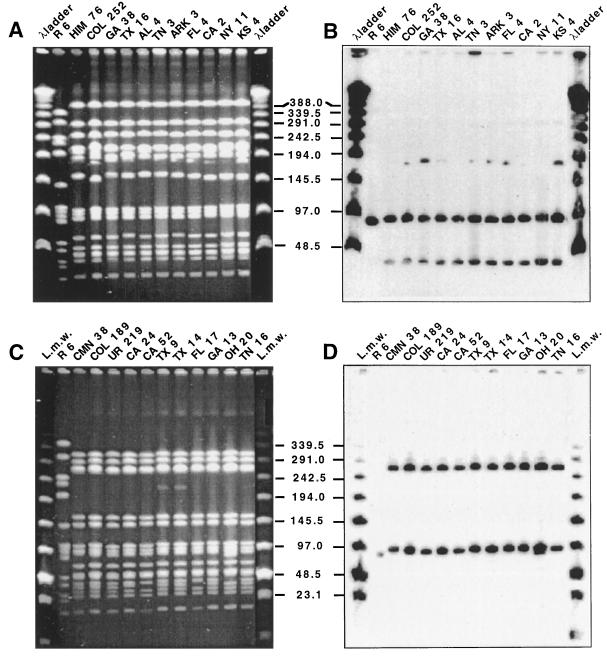
A number of clinical isolates representing a variety of serotypes and antibiotypes and six different PFGE types and 18 different PFGE subtypes were selected to test their lytA hybridization patterns. The strains included five different serogroups (and one nontypeable strain) and a variety of antibiotic resistance patterns and isolation sites from several countries in Latin America and Europe as well as the United States (Table 1). Figure 1 shows the PFGE patterns and corresponding lytA hybridization patterns of 22 S. pneumoniae isolates representing five PFGE types and 11 distinct subtype variants. Figure 2 illustrates the same for an additional 22 strains, which include 11 isolates representing the widely spread Spanish-United States epidemic clone (Fig. 2A and B) and 11 isolates representing the French-Spanish epidemic clone (Fig. 2C and D). In both cases, clonal types expressing different serogroups, antibiotypes, and PFGE subtypes are included in the selection (Table 1). Hybridization patterns, i.e., the number and molecular sizes of SmaI fragments hybridizing with the lytA DNA probe, paralleled variations in PFGE type and subtype. However, not all observable PFGE subtype variations could be explained in this manner.
TABLE 1.
Variation in lytA hybridization patterns of penicillin-resistant S. pneumoniae isolatesa
| Strain | Serotype | PFGE type | PEN MIC (μg/ml) | Susceptibility or resistance
|
SmaI hybridization pattern (kb)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | TET | CMP | SXT | ermB | mef | tetM | cat | lytAb | ||||
| OH 4 | 6 | 9 | 2 | R | S | S | R | — | 225 | — | — | 100/60 |
| OH 11 | 6 | 9 | 2 | R | S | S | R | — | 225 | — | — | 100/60 |
| OH 12 | 6 | 9 | 2 | R | S | S | R | — | 225 | — | — | 100/60 |
| OH 13 | 6 | 9 | 2 | R | S | S | R | — | 225 | — | — | 100/60 |
| CA 14 | 19 | 3 | 2 | R | R | S | I | — | 115 | 115 | — | 115 |
| TX 4 | 19 | 3 | 2 | R | R | S | R | — | 115 | 115 | — | 115 |
| CA 37 | 19 | 3 (2) | 3 | R | R | S | R | — | 115 | 115 | — | 115 |
| CA 40 | 19 | 3 (2) | 2 | R | R | S | R | — | 115 | 115 | — | 115 |
| TX 8 | 19 | 3 (3) | 2 | R | R | S | I | — | 115 | 115 | — | 115 |
| CA 45 | 23 | 5 (3) | 1.5 | S | S | S | I | — | — | — | — | 260/85/70 |
| CA 8 | 23 | 5 (2) | 2 | S | S | S | R | — | — | — | — | 260/85/70 |
| KS 11 | 23 | 5 | 4 | S | S | S | R | — | — | — | — | 260/85 |
| KY 15 | 23 | 5 | 6 | S | S | S | I | — | — | — | — | 260/85 |
| KY 4 | 23 | 5 | 2 | S | S | S | I | — | — | — | — | 260/85 |
| TX 5 | 14 | 16 | 6 | R | R | S | R | 390 | — | 390 | — | 95/40 |
| GA 30 | NT | 16 | 8 | R | R | S | R | 390 | — | 390 | — | 95/40 |
| KS 10 | 14 | 16 (2) | 6 | R | R | S | R | 390 | — | 390 | — | 95/70 |
| TN 13 | 14 | 16 (2) | 3 | R | R | S | R | 390 | — | 390 | — | 95/70 |
| SW 224 | 9 | B (6) | 1 | S | S | S | R | ND | ND | ND | ND | 260/130 |
| SW 225 | 9 | B (6) | 1 | S | S | S | R | ND | ND | ND | ND | 260/130 |
| SW 226 | 9 | B | 1 | S | S | S | R | ND | ND | ND | ND | 260/95 |
| SW 228 | 9 | B | 1 | S | S | S | R | ND | ND | ND | ND | 260/95 |
| HIM 76 | 23 | A | 2 | R | R | R | R | ND | ND | 390 | ND | 90/30 |
| COL 252 | 23 | A (3) | 2 | S | R | R | I | ND | ND | 390 | ND | (175)/90/30 |
| GA 38 | 23 | A (2) | 2 | R | R | R | R | 390 | — | 390 | 390 | (180)/90/30 |
| TX 16 | 23 | A (2) | 2 | R | R | R | R | 390 | — | 390 | 390 | (180)/90/30 |
| AL 4 | 23 | A | 1.5 | R | R | R | R | 390 | — | 390 | 390 | 90/30 |
| TN 3 | 23 | A (2) | 1.5 | R | R | R | I | 390 | — | 390 | 390 | (180)/90/30 |
| Ark 3 | 23 | A (2) | 3 | R | R | R | R | 390 | — | 390 | 390 | (180)/90/30 |
| FL 4 | 23 | A (2) | 4 | R | R | R | I | — | 390 | 390 | 390 | (180)/90/30 |
| CA 2 | 23 | A (3) | 2 | R | R | R | R | — | 390 | 390 | 390 | (175)/90/30 |
| NY 11 | 23 | A | 2 | R | R | R | R | — | 390 | 390 | 390 | 90/30 |
| KS 4 | 23 | A (3) | 3 | R | R | R | R | — | 390 | 390 | 390 | (175)/90/30 |
| CMN 38 | 9A | B (4) | 2 | S | S | S | R | ND | ND | ND | ND | 260/95 |
| Col 189 | 9V | B | 2 | S | S | S | R | ND | ND | ND | ND | 260/95 |
| UR 219 | 14 | B | 2 | S | S | S | R | ND | ND | ND | ND | 260/95 |
| CA 24 | 9 | B (3) | 2 | S | S | S | R | — | — | — | — | 260/95 |
| CA 52 | 9 | B (2) | 1.5 | S | S | S | R | — | — | — | — | 260/95 |
| TX 9 | 9 | B | 1.5 | S | S | S | R | — | — | — | — | 260/95 |
| TX 14 | 9 | B | 3 | S | S | S | R | — | — | — | — | 260/95 |
| FL 17 | 9 | B (2) | 3 | S | S | S | R | — | — | — | — | 260/95 |
| GA 13 | 9 | B | 1.5 | S | S | S | R | — | — | — | — | 260/95 |
| OH 20 | 9 | B (5) | 1.5 | S | S | S | R | — | — | — | — | 260/95 |
| TN 16 | 9 | B | 1.5 | S | S | S | R | — | — | — | — | 260/95 |
Strain numbers and PFGE assignments are as in references 4 and 18. —, no hybridization signal. Abbreviations: PEN, penicillin; ERY, erythromycin; TET, tetracycline; CMP, chloramphenicol; SXT, trimethoprim-sulfamethoxazole; NT, nontypeable; ND, not done; S, susceptible; R, resistant; I, intermediate.
Numbers in parentheses refer to unstable bands.
FIG. 1.
Association of PFGE pattern and lytA hybridization profile. Lanes marked L.m.w were loaded with low-range PFGE markers (New England Biolabs). Strain properties are listed in Table 1. SmaI fragments generated from the laboratory strain R6 served as additional molecular size standards. Numbers at right indicate molecular sizes in kilobases. (A) SmaI digest of total DNA separated by PFGE. (B) Hybridization of a Southern blot of the gel in panel A with the lytA gene probe.
FIG. 2.
Variation in PFGE pattern and lytA hybridization profile of two major epidemic clones. Lanes marked λ ladder were loaded with lambda ladder PFGE markers (New England Biolabs), whereas lanes marked L.m.w were loaded with low-range PFGE markers. Strain properties are listed in Table 1. Numbers in the center indicate molecular sizes in kilobases. (A) SmaI digest of total DNA separated by PFGE of members of the Spanish-United States clone. (B) Hybridization of a Southern blot of the gel in panel A with the lytA gene probe. (C) SmaI digest of total DNA separated by PFGE of members of the French-Spanish clone. (D) Hybridization of a Southern blot of the gel in panel C with the lytA gene probe.
Stability of lytA hybridization pattern during serial passage in vitro.
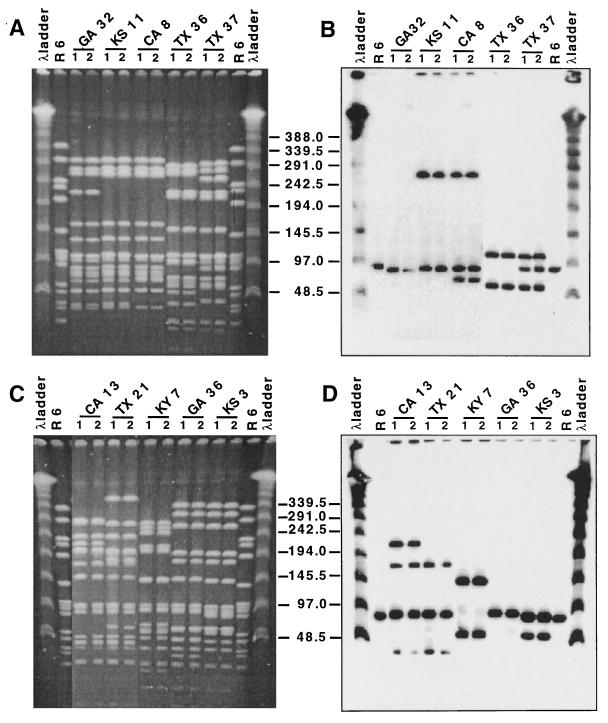
Ten S. pneumoniae isolates from a collection of strains from the United States (4), representing distinct PFGE and lytA patterns, were selected to test the stability of their lytA pattern during serial passage in vitro. Bacteria grown in liquid medium were spread onto blood agar plates and incubated as described in Materials and Methods. The following day, single colonies were picked and restreaked on new plates, and this procedure was repeated 15 to 20 times, yielding an estimated number of over 500 generations. The initial culture and the culture generated from a single colony isolated after 10 consecutive passages on blood agar plates were grown in liquid medium, and their PFGE and lytA hybridization patterns were compared. Figure 3 shows that both PFGE and lytA hybridization patterns have remained stable under these conditions.
FIG. 3.
Stability of lytA hybridization pattern during extensive serial cultivation of strains in vitro. Lanes marked λ ladder were loaded with lambda ladder PFGE markers. All isolates are from the United States and were described recently (4). Lanes 1 represent the isolate before extensive serial passage in vitro. Lanes 2 represent the isolate after in vitro passage, as described in Materials and Methods. Numbers in the center indicate molecular sizes in kilobases. (A and C) SmaI digest of total DNA separated by PFGE. (B and D) Hybridization of Southern blots of the gels in panels A and C, respectively, with the lytA gene probe.
Nature of PFGE subtype variation and variation in lytA hybridization pattern observed in some S. pneumoniae isolates belonging to the multiresistant Spanish-United States clone.
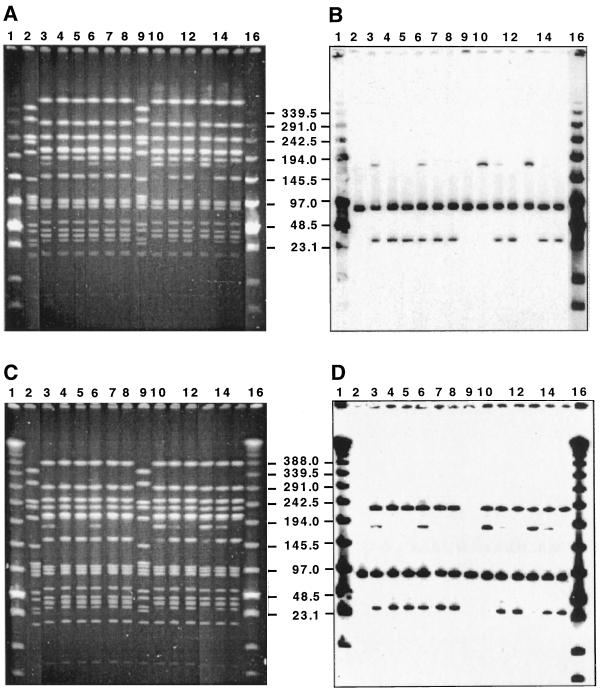
A series of simple tests indicated that the occasional variation in PFGE subtype (paralleled by a variation in lytA type) observed with some isolates belonging to the Spanish-United States clone was related to the concentration of SmaI enzyme used for preparation of the PFGE gels. Two bacterial strains, both belonging to the Spanish-United States clone, were chosen for further testing: strain KY 6 expressed serogroup 19 and was resistant to penicillin, erythromycin, tetracycline, and chloramphenicol (Fig. 4A and B), and strain FL 26 expressed serogroup 23 and was resistant to penicillin, tetracycline, and chloramphenicol (Fig. 4C and D). Each strain was treated with the two lots of SmaI endonuclease as described in Materials and Methods, and the digestion was repeated twice on two different disk preparations for each strain. Each lot was used at three different concentrations (20, 40, and 60 U of SmaI per DNA disk), and the results are presented in Fig. 4. After treatment with the higher concentrations of the restriction enzyme, a 180-kb DNA fragment was no longer detectable either by ethidium bromide (EtBr) staining or by lytA hybridization. The results indicated that the higher concentration of SmaI had split the fragment into a 150- and a 30-kb DNA fragment, the latter showing a strong hybridization signal with the lytA probe. No evidence for similar variation in PFGE profile and/or lytA hybridization pattern was detectable in a number of other S. pneumoniae strains examined.
FIG. 4.
Effect of the concentration of SmaI endonuclease on the PFGE profile and lytA patterns of some strains belonging to the Spanish-United States clone. Lanes 2 and 9 have SmaI fragments from strain R6 used as molecular size standards. Samples in lanes 3 to 8 were treated with SmaI lot 1. Samples in lanes 10 to 15 were treated with SmaI lot 2. Samples in lanes 3, 6, 10, and 13 were treated with 20 U of SmaI per disk. Samples in lanes 4, 7, 11, and 14 were treated with 40 U of SmaI per disk. Samples in lanes 5, 8, 12, and 15 were treated with 60 U of SmaI per disk. Numbers in the center indicate molecular sizes in kilobases. Lanes 1 and 16 in panels A and B were loaded with low-range PFGE markers. Lanes 1 and 16 in panels C and D were loaded with lambda ladder PFGE markers. (A) SmaI digest of total DNA separated by PFGE of strain KY 6 (4). (B) Hybridization of a Southern blot of the gel in panel A with the lytA gene probe. (C) SmaI digest of total DNA separated by PFGE of strain FL 26 (4). (D) Hybridization of a Southern blot of the gel in panel C with the lytA gene probe.
DISCUSSION
Variation of the lytA hybridization pattern with clonal type.
Comparison of the lytA hybridization profiles seen in Fig. 1 and 2 indicates that each one of the strains produced a lytA hybridization pattern which correlated with the corresponding PFGE type. The exceptions were strains representing PFGE subtypes of the Spanish-United States epidemic clone, which occasionally showed variations in a single SmaI fragment detectable both by EtBr stain and as a band hybridizing to the lytA probe. The observations discussed below, however, will offer a possible explanation for these exceptions based on the activity of the SmaI endonuclease against particular sites.
Effect of SmaI concentration on the PFGE pattern of some isolates belonging to the Spanish-United States clone.
The inspection of the patterns in Fig. 4 clearly shows that increasing the concentration of the restriction enzyme from the 20 U per DNA disk normally used in the SmaI restriction of DNA for PFGE gels to three times this amount (60 U per DNA disk) resulted in the elimination of the PFGE band at 180 kb and the intensification of the two fragments detectable in the EtBr stain at 30 and 150 kb. After treatment with the higher concentration of SmaI endonuclease, the lytA hybridization pattern of both strains—KY 6 (Fig. 4B) and FL 26 (Fig. 4D)—was reduced: from three to two hybridizing bands in strain KY 6 (one at 90 kb, representing the host autolytic enzyme, and the second one at 30 kb, which corresponded to a fragment visible in the EtBr stain with the same size) and from four to three hybridizing bands in strain FL 26. These observations suggest that the 180-kb SmaI DNA fragment contains a cryptic SmaI site that can be cut into a 30- and a 150-kb fragment with a sufficiently high concentration of the endonuclease. Only the 30-kb fragment has lytA-hybridizing sequences. Some differences in the effectiveness of the two lots of the SmaI enzyme were also noted: while one lot was completely ineffective in restriction of the 180-kb band at the 20-U-per-DNA-disk concentration, the same concentration of the enzyme from the second lot achieved partial digestion.
The mechanism of less effective restriction at the SmaI cutting site in the 180-kb DNA fragment is not known. The mode of the hypothetical prophage attachment in this fragment may pose some kind of steric hindrance to the restriction enzyme. An alternative, intriguing possibility is that the 180-kb fragment may correspond to unintegrated phage genome, i.e., intracellular replicative intermediates or cell-adsorbed phage particles, since the number of phage particles closely parallels the number of bacteria in some lysogenic strains (2). The presence of free phage DNA in total DNA preparations was reported previously with other systems (2, 11). The difficulty in digesting the 180-kb fragment could then be due to the expression of a methylase by this particular phage during vegetative growth. Several of these enzymes have been described for phages infecting Bacillus species and Escherichia coli (8, 10, 13, 16), some of which show specificity to the core of the SmaI recognition sequence (8). The action of the SmaI endonuclease on methylated sites is impaired to different degrees depending on the type of modification and the position and the number of modified bases (1, 3, 12). A higher concentration of the enzyme could increase the efficiency of cleavage at methylated sites, generating the patterns observed in Fig. 4.
PFGE subtype variation corresponding to variations in lytA hybridization pattern.
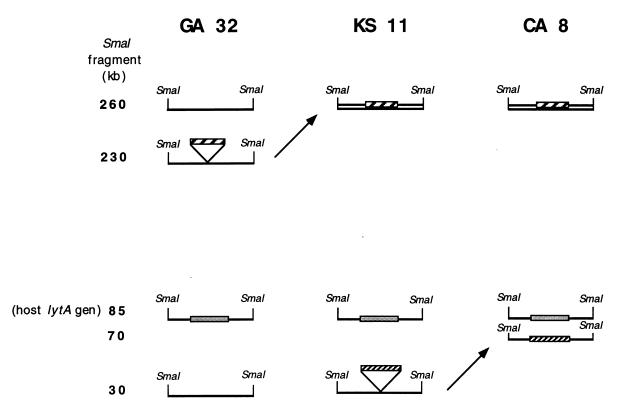
Several S. pneumoniae isolates representing simple subtype variants (i.e., differing in one or two fragments visible in the EtBr stain) were examined with the lytA DNA probe. In the case of the three strains shown in Fig. 3 (GA 32, KS 11, and CA 8), the differences in band patterns could be explained by the acquisition of prophage elements (detected through lytA hybridization) in a manner suggested in Fig. 5. The three strains have a common PFGE type 5 but represent simple subtype variants of it (4). Strain GA 32 has a single lytA-hybridizing band corresponding to the host lytA gene located in an approximately 90-kb fragment. Strain KS 11 has two lytA-hybridizing bands: one corresponding to the host autolysin enzyme and the second associated with a 260-kb DNA fragment. Strain CA 8 has three lytA-hybridizing fragments: one corresponding to the host autolysin gene (about 90 kb), a second one associated with the 260-kb DNA fragment identical to the fragment seen in KS 11, and a third lytA-hybridizing band with a molecular size of 70 kb. The sketch in Fig. 5 provides a hypothetical explanation for the possible origin of both subtype variations in these strains and also how this variation may be envisioned as a result of the acquisition of one (KS 11) or two (CA 8) prophages carrying no SmaI recognition sites. The lytA-hybridizing DNA fragment of 260 kb in KS 11 is assumed to originate in the insertion of an approximately 30-kb prophage into the 230-kb DNA fragment of GA 32. The 70-kb lytA-hybridizing fragment of CA 8, on the other hand, is assumed to have originated by the insertion of a similar size (40-kb) prophage element into the 30-kb DNA fragment of KS 11. Consistent with this scheme, a 30-kb fragment is detectable by EtBr stain in both GA 32 and KS 11 but no longer detectable in CA 8. Similarly, no 70-kb fragment is detectable by EtBr stain in GA 32 and KS 11, but such a band is visible in CA 8, and it carries lytA-hybridizing material. The 230-kb fragment detectable by EtBr stain in GA 32 is not visible either in KS 11 or in CA 8. The relationship between these strains could be envisioned as follows: strain KS 11 may have originated from GA 32 by phage acquisition; subsequently KS 11 acquired another phage, generating strain CA 8.
FIG. 5.
Model to explain PFGE subtype variation in terms of prophage acquisition or loss. Lines represent relevant fragments detected by EtBr staining of a total DNA digestion with SmaI endonuclease separated by PFGE. Grey bars represent the host lytA gene. The two kinds of hatched bars represent hypothetical phage genomes (the two prophages detectable in strain CA 8 are not necessarily the same). Numbers on the left indicate molecular sizes in kilobases. PFGE profiles and lytA hybridization patterns of the three strains GA 32, KS 11, and CA 8 are shown in Fig. 3.
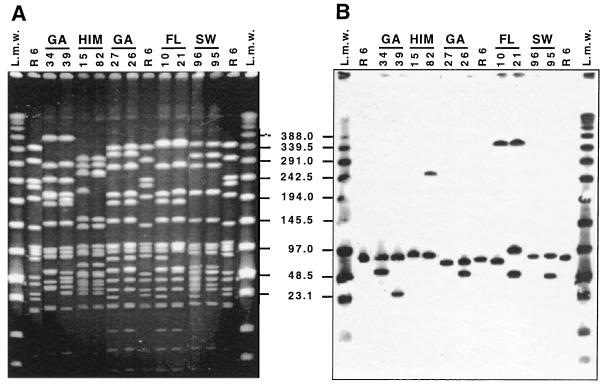
Additional examples of parallel changes in PFGE subtype and lytA hybridization pattern may be seen in Fig. 6. GA 34 and GA 39 represent subtype variants of the same serotype 23F Spanish-United States clone by SmaI PFGE pattern (4). However, only GA 39, not GA 34, has the lytA hybridization pattern typical of this clone. One may assume the following sequence of events: a prophage of about 30 kb is inserted into the 30-kb SmaI fragment detectable by EtBr stain in GA 39 and already containing lytA-hybridizing sequences, generating a new fragment of about 60 kb in strain GA 34, which now reacts with the lytA probe and which is not detectable in strain GA 39 by hybridization. GA 34 has no detectable 30-kb fragment by the EtBr stain. In this case, an alternative process in the opposite direction may also be imagined, namely, the introduction of an SmaI cutting site into the 60-kb fragment of GA 34.
FIG. 6.
Parallel changes in PFGE profile and lytA hybridization pattern in selected strains of S. pneumoniae. Lanes 1 and 15 were loaded with low-range PFGE markers. Lanes 2, 9, and 14 contain SmaI fragments of the laboratory strain R6 used as molecular size markers. Strains GA 34, GA 39, GA 27, GA 26, FL 10, and FL 21 are from a United States strain collection (4). Strains HIM 15 and HIM 82 are from Mexico (5). Strains SW 96 and SW 95 are from Sweden. (A) SmaI digest of total DNA separated by PFGE. (B) Hybridization of a Southern blot of the gel in panel A with the lytA gene probe. Numbers in the center indicate molecular sizes in kilobases.
Strains HIM 15 and HIM 82 are subtype variants of the French-United States clone (5). HIM 15 has one and HIM 82 has two lytA-hybridizing SmaI fragments: both carry a 90-kb band representing the host lytA gene, and in HIM 82, the 260-kb SmaI fragment shows a much more intense fluorescence with EtBr staining than that in HIM 15. This band is also positive for the lytA gene probe in HIM82 but not in HIM 15. One may imagine that the fragment detectable by EtBr staining at 230 kb, present only in HIM 15, acquired a prophage, thus generating the intensely fluorescent 260-kb fragment in strain HIM 82 corresponding to two fragments of similar size, which now also gives a positive hybridization signal with lytA.
GA 26 and GA 27 are subtype variants of PFGE pattern 6 (4); they both contain the 90-kb lytA-hybridizing fragment representing the host autolysin gene. However, GA 26 has an additional lytA-positive band at 60 kb, which may have originated from the fragment of about 20 kb visible by EtBr stain in GA 27: a prophage of about 40 kb may have inserted itself into the 20-kb band, thus generating the 60-kb lytA-positive band of GA 27. Similar interpretations may apply for the pairs of strains FL 10 and FL 21 and SW 96 and SW 95.
While most of the isolates described in this communication were antibiotic-resistant pneumococci, variation in the lytA band pattern was also observed among fully drug-susceptible strains (data not shown). The observations described in this communication indicate that the pattern of lytA-hybridizing bands, presumably representing prophage genomes, may be used as an epidemiological tool providing extra resolution to the genetic background of S. pneumoniae isolates in molecular typing studies as well as providing a rationale for the interpretation of differences observed between the PFGE patterns of some clinical isolates.
ACKNOWLEDGMENTS
These investigations received partial support from a grant from the National Institutes of Health, RO1 AI37275. M.R. received partial support from the Gulbenkian Foundation, the Fundação Luso Americana para o Desenvolvimento, and BPD/20185/99.
REFERENCES
- 1.Brooks J E, Roberts R J. Modification profiles of bacterial genomes. Nucleic Acids Res. 1982;10:913–934. doi: 10.1093/nar/10.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brussow H, Bruttin A. Characterization of a temperate Streptococcus thermophilus bacteriophage and its genetic relationship with lytic phages. Virology. 1995;212:632–640. doi: 10.1006/viro.1995.1521. [DOI] [PubMed] [Google Scholar]
- 3.Butkus V, Petrauskiene L, Maneliene Z, Klimasauskas S, Laucys V, Janulaitis A. Cleavage of methylated CCCGGG sequences containing either N4-methylcytosine or 5-methylcytosine with MspI, HpaII, SmaI, XmaI and Cfr9I restriction endonucleases. Nucleic Acids Res. 1987;15:7091–7102. doi: 10.1093/nar/15.17.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corso A, Severina E P, Petruk V F, Mauriz Y R, Tomasz A. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb Drug Resist. 1998;4:325–337. doi: 10.1089/mdr.1998.4.325. [DOI] [PubMed] [Google Scholar]
- 5.Echaniz-Aviles G, Velazquez-Meza M E, Carnalla-Barajas M N, Soto-Nogueron A, Di Fabio J L, Solorzano-Santos F, Jimenez-Tapia Y, Tomasz A. Predominance of the multiresistant 23F international clone of Streptococcus pneumoniae among isolates from Mexico. Microb Drug Resist. 1998;4:241–246. doi: 10.1089/mdr.1998.4.241. [DOI] [PubMed] [Google Scholar]
- 6.Garcia E, Garcia J L, Ronda C, Garcia P, Lopez R. Cloning and expression of the pneumococcal autolysin gene in Escherichia coli. Mol Gen Genet. 1985;201:225–230. doi: 10.1007/BF00425663. [DOI] [PubMed] [Google Scholar]
- 7.Garcia P, Garcia J L, Garcia E, Lopez R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43:265–272. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- 8.Jentsch S, Gunthert U, Trautner T A. DNA methyltransferases affecting the sequence 5′CCGG. Nucleic Acids Res. 1981;9:2753–2759. doi: 10.1093/nar/9.12.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 10.Lange C, Noyer-Weidner M, Trautner T A, Weiner M, Zahler S A. M.H2I, a multispecific 5C-DNA methyltransferase encoded by Bacillus amyloliquefaciens phage H2. Gene. 1991;100:213–218. doi: 10.1016/0378-1119(91)90369-m. [DOI] [PubMed] [Google Scholar]
- 11.Lina B, Bes M, Vandenesch F, Greenland T, Etienne J, Fleurette J. Role of bacteriophages in genomic variability of related coagulase-negative staphylococci. FEMS Microbiol Lett. 1993;109:273–277. doi: 10.1016/0378-1097(93)90032-w. [DOI] [PubMed] [Google Scholar]
- 12.Nelson P S, Papas T S, Schweinfest C W. Restriction endonuclease cleavage of 5-methyl-deoxycytosine hemimethylated DNA at high enzyme-to-substrate ratios. Nucleic Acids Res. 1993;21:681–686. doi: 10.1093/nar/21.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noyer-Weidner M, Walter J, Terschuren P A, Chai S, Trautner T A. M.phi 3TII: a new monospecific DNA (cytosine-C5) methyltransferase with pronounced amino acid sequence similarity to a family of adenine-N6-DNA-methyltransferases. Nucleic Acids Res. 1994;22:5517–5523. doi: 10.1093/nar/22.24.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramirez M, Severina E, Tomasz A. A high incidence of prophage carriage among natural isolates of Streptococcus pneumoniae. J Bacteriol. 1999;181:3618–3625. doi: 10.1128/jb.181.12.3618-3625.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schaffer H E, Sederoff R R. Improved estimation of DNA fragment lengths from agarose gels. Anal Biochem. 1981;115:113–122. doi: 10.1016/0003-2697(81)90533-9. [DOI] [PubMed] [Google Scholar]
- 16.Schlagman S L, Miner Z, Feher Z, Hattman S. The DNA [adenine-N6]methyltransferase (Dam) of bacteriophage T4. Gene. 1988;73:517–530. doi: 10.1016/0378-1119(88)90516-1. [DOI] [PubMed] [Google Scholar]
- 17.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 18.Tomasz A, Corso A, Severina E P, Echániz-Aviles G, de C. Brandileone M C, Camou T, Castañeda E, Figueroa O, Rossi A, di Fabio J L Members of the PAHO/Rockefeller University Workshop. Molecular epidemiologic characterization of penicillin-resistant Streptococcus pneumoniae invasive pediatric isolates recovered in six Latin-American Countries: an overview. Microb Drug Resist. 1998;4:195–207. doi: 10.1089/mdr.1998.4.195. [DOI] [PubMed] [Google Scholar]