Abstract
Mutations in emerging severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineages can interfere with laboratory methods used to generate viral genome sequences for public health surveillance. We identified 20 mutations that are widespread in variant of concern lineages and affect widely used sequencing protocols by the ARTIC network and Freed et al. Three of these mutations disrupted sequencing of P.1 lineage specimens during a recent outbreak in British Columbia, Canada. We provide laboratory validation of protocol modifications that restored sequencing performance. The study findings indicate that genomic sequencing protocols require immediate updating to address emerging mutations. This work also suggests that routine monitoring and protocol updates will be necessary as SARS-CoV-2 continues to evolve. The bioinformatic and laboratory approaches used here provide guidance for this kind of assay maintenance.
KEYWORDS: SARS-CoV-2, Amplicon sequencing, COVID-19, Variant of concern, Viral genomics, Genomic surveillance
Genomic sequencing of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has played a crucial role in managing the ongoing coronavirus disease 2019 (COVID-19) pandemic. This is especially true for variant of concern (VOC) lineages that have emerged globally since December 2020 (Chand et al., 2020; Cherian et al., 2021; Faria et al., 2021; Tegally et al., 2021a, 2021b; Rambault et al., 2020). Genomic sequencing has been instrumental in detecting and characterizing these lineages, tracking their global spread, and identifying local cases to control transmission.
Due to low quantities of viral genomic material in typical clinical specimens, the SARS-CoV-2 genome must be enriched for high-throughput sequencing. This is commonly done by multiplex PCR with numerous primer pairs tiled across the viral genome (Freed et al., 2020; Quick et al., 2017; Tyson et al., 2020). The performance of these primer schemes can be disrupted by mutations arising in emerging viral lineages. These mutations inhibit the amplification of certain amplicons and reduce the amount of sequencing data generated across affected areas of the genome. At best, this necessitates over-sequencing of specimens to compensate for areas with reduced sequencing depth, thereby increasing costs and reducing throughput. At worst, this causes inaccurate nucleotide base calls or gaps in the genome sequence. This undermines public health surveillance by interfering with lineage identification and obscuring genomic linkages between outbreak specimens.
A previously published bioinformatic pipeline called PCR_strainer was used to assess whether current VOC lineages contain mutations impacting two widely used sequencing primer schemes (Kuchinski et al., 2020). A total of 27 244 recent, complete, high-quality genome sequences from lineages B.1.1.7, B.1.351, P.1, and B.1.617+ submitted to GISAID from laboratories around the world were analyzed. All primer site variants present in at least 15% of their lineage's genome sequences are reported in Table 1 . Fifteen primer site variants affecting the popular, commercially available ARTIC version 3 protocol (Tyson et al., 2020) were identified. Five mutated primer sites affecting the protocol by Freed et al., favored by some laboratories for its longer amplicon size (Freed et al., 2020), were also identified. The B.1.1.7 lineage had the fewest primer site variants (n = 1), followed by the B.1.351 lineage (n = 4), then the B.1.617+ lineage (n = 7), and the P.1 lineage (n = 8). Many of these primer site variants were predominant globally within their lineage, with 14 of them being present in at least 85% of their lineage's sequences.
Table 1.
Nucleotide mismatches between SARS-CoV-2 variant of concern lineages and primers from the ARTIC version 3 and Freed et al. protocols. Global sequences from variant of concern lineages were downloaded from GISAID, comprising 8353 B.1.1.7 sequences, 3025 B.1.351 sequences, 9202 P.1 sequences, and 6664 B.1.617+ sequences. All sequences were categorized as complete and high-coverage by GISAID (>29 kb, <1% Ns, <0.05% novel amino acid substitutions, and verified insertion/deletions only). All sequences were submitted before May 22, 2021. Lineage B.1.351, B.1.617+, and P.1+ sequences were collected after April 1, 2021, and lineage B.1.1.7 sequences were collected after May 7, 2021. Sequence details and submitting laboratories, who are gratefully acknowledged for their contributions, are provided in Supplementary Material File S1. Primer site variants were identified using the PCR_strainer pipeline with default parameters. Primer site variants were only reported if they were present in at least 15% of their lineage's sequences. A longer table is provided as Supplementary Material Table S2, which reports all variants present in at least 1% of genomes within a lineage. Primer site variant sequences are provided in ‘oligo sense’, i.e. the reverse complement of the primer site and the sequence that the perfectly identical primer would have for the targeted location. Lower case bases in the primer site variant sequences indicate mismatches with the assay's original primer sequence.
| Protocol | Lineage | Primer name | Primer site variant sequence (5′ to 3′) | Prevalence within lineage (%) |
|---|---|---|---|---|
| ARTIC v3 | B.1.1.7 | nCoV-2019_93_LEFT | TGAGGCTGGTTCTtAATCACCCA | 47.11 |
| ARTIC v3 | B.1.351 | nCoV-2019_76_LEFT | AGGGCAAACTGGAAAtATTGCT | 97.09 |
| ARTIC v3 | B.1.351 | nCoV-2019_76_LEFT_alt3 | GGGCAAACTGGAAAtATTGCTGA | 97.09 |
| ARTIC v3 | B.1.351 | nCoV-2019_86_LEFT | TtAGGTGATGGCACAACAAGTC | 96.86 |
| ARTIC v3 | B.1.351 | nCoV-2019_74_LEFT | ACATCACTAGGTTTCAAACTTTACaTag | 92.99 |
| ARTIC v3 | B.1.617 | nCoV-2019_93_RIGHT | AGGcCTTCCTTGCCATGTTGAG | 87.03 |
| ARTIC v3 | B.1.617 | nCoV-2019_81_LEFT | GCACTTGGAAAACTTCAAaATGTGG | 85.98 |
| ARTIC v3 | B.1.617 | nCoV-2019_72_RIGHT | gaataaActcCACTTTCCATCCAAC | 82.28 |
| ARTIC v3 | B.1.617 | nCoV-2019_73_LEFT | CAATTTTGTAATGATCCATTTTTGGaTGT | 63.61 |
| ARTIC v3 | B.1.617 | nCoV-2019_64_LEFT | TCGATAGATATCCTGtTAATTCCATTGT | 49.29 |
| ARTIC v3 | P.1 | nCoV-2019_12_RIGHT | TTCACTCTTCATTTCCAAAAAGCTTaA | 99.54 |
| ARTIC v3 | P.1 | nCoV-2019_92_RIGHT | AGGTTtCTGGCAATTAATTGTAAAAGG | 99.41 |
| ARTIC v3 | P.1 | nCoV-2019_73_LEFT | CAATTTTGTAATtATCCATTTTTGGGTGT | 97.14 |
| ARTIC v3 | P.1 | nCoV-2019_76_LEFT_alt3 | GGGCAAACTGGAAcGATTGCTGA | 94.27 |
| ARTIC v3 | P.1 | nCoV-2019_76_LEFT | AGGGCAAACTGGAAcGATTGCT | 94.19 |
| Freed | B.1.617 | SARSCoV_1200_5_LEFT | ACCTACTAAAAAGtCTGGTGGC | 49.32 |
| Freed | B.1.617 | SARSCoV_1200_27_RIGHT | TGTTCGTTTAGGCGTGACAAaT | 49.2 |
| Freed | P.1 | SARSCoV_1200_24_LEFT | GCTGAAtATGTCAACAACTCATATGA | 99.47 |
| Freed | P.1 | SARSCoV_1200_21_RIGHT | GCAGaGGGTAATTGAGTTCTGt | 99.25 |
| Freed | P.1 | SARSCoV_1200_25_LEFT | TGCTGCTAtTAAAATGTCAGAGTGT | 98.87 |
The corollary is that some region-specific primer site variants exist within VOC sub-lineages, and public health laboratories should assess their sequencing protocols against locally circulating VOC sequences. This can direct limited laboratory resources towards correcting the most locally relevant primer site variants, which is especially important when global databases are biased by differences in genomics capacity and surveillance priorities between submitting jurisdictions.
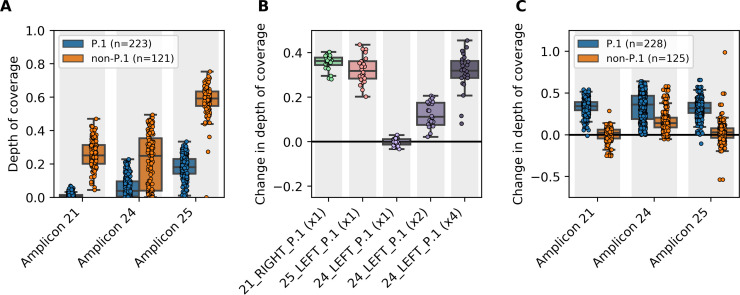
This kind of assessment was done by the British Columbia Centre for Disease Control Public Health Laboratory. While investigating P.1 lineage outbreaks in March and April of this year, significantly reduced depths of coverage were observed across three amplicons covering parts of the orf1ab and spike genes (Figure 1 A). The affected amplicons were amplicon 21, amplicon 24, and amplicon 25 from the Freed et al. primer scheme, which all had mutated primer sites identified by PCR_strainer in global P.1 lineage sequences (Table 1). Compared to non-P.1 lineage specimens, median depths of coverage across these amplicons were reduced up to 32-fold. Genome sequences from 907 local P.1 lineage specimens were analyzed with PCR_strainer, confirming that the same mutations identified above in primers 21_LEFT, 24_RIGHT, and 25_RIGHT were present in 99.4%, 100%, and 95.8% of local sequences, respectively.
Figure 1.
(A) Predominant mutations in the P.1 lineage reduced sequencing depth across three amplicons. A total 373 clinical specimens, collected during March and April 2021 in British Columbia, Canada, were sequenced using the Freed et al. primers. Normalized mean depths of coverage are shown for three amplicons affected by mutations in primer sites (the mean depth of coverage across each library's most deeply sequenced amplicon was used for normalization). (B) Amplicon drop-out was corrected by spiking in supplemental primers. Twenty-four clinical specimens were sequenced with non-spiked and spiked primer pools, then changes in normalized mean depth of coverage were calculated for the three affected amplicons. Supplemental primers were spiked in at the same molarity as the original primers. Primer 24_LEFT_P.1 was also assayed at two-times and four-times molarity after one-times molarity did not significantly improve performance. (C) Spiked primer pools had no detrimental effects on non-P1 lineages. A total 373 clinical specimens were sequenced with non-spiked and spiked primer pools containing primers 21_RIGHT_P.1 and 25_LEFT_P.1 at one-times molarity and primer 24_LEFT_P.1 at four-times molarity. Changes in normalized mean depth of coverage were calculated for the three affected amplicons.
Supplemental primers were designed by copying the three mutant primer site sequences identified by PCR_strainer: 5′-GCAGAGGGTAATTGAGTTCTGT-3′, 5′-GCTGAATATGTCAACAACTCATATGA-3′, and 5′-TGCTGCTATTAAAATGTCAGAGTGT-3′, which we called 21_RIGHT_P.1, 24_LEFT_P.1, and 25_LEFT_P.1 respectively. Primers 21_RIGHT_P.1 and 25_LEFT_P.1 were spiked into existing Freed et al. primer pools at the same molarity as the rest of the primers. Primer 24_LEFT_P.1 was spiked in at four-times molarity after a titration experiment on 24 clinical specimens showed that spiking in at one-times molarity did not significantly improve performance (Figure 1B). We then sequenced 373 clinical specimens with both non-spiked and spiked primer pools. Using these paired data, changes in depth of coverage across the affected amplicons were calculated for each specimen. It was observed that the spiked primers pools significantly improved depth of coverage for all three impacted amplicons without detrimental effects on non-P.1 lineages (Figure 1C).
From these analyses, it was concluded that the established amplicon sequencing protocols, like ARTIC version 3 and Freed et al., need immediate updates to address numerous prevalent mutations in VOC lineages. We have shown that spike-in primers can restore performance for impacted amplicons, but we caution that these spike-in primers are likely a temporary measure. As new SARS-CoV-2 lineages continue to emerge, multiplex PCR amplification-based sequencing protocols will need to evolve alongside their viral target.
This principle was demonstrated in the months following the work presented above. B.1.617+ lineages, collectively renamed ‘delta’ by the World Health Organization, went from representing only a handful of cases in British Columbia to being predominant. We assessed 388 specimens from local delta lineage specimens with PCR_strainer (SupplementaryMaterial Table S1), which confirmed the presence of the two Freed et al. primer site mutations previously identified in B.1.617+ sequences from GISAID (Table 1). It was also noted that an additional, less prevalent primer site mutation had emerged in delta lineages in the intervening months. The mutation in the left primer of amplicon 5 had no meaningful impact on depth of coverage, which was not surprising given its location in the middle of the oligo (Supplementary Material Figure S1). The mutations in the left and right primers of amplicon 27 both caused reduced depth of coverage for these amplicons, but not to the same magnitude as the primer site mutations impacting P.1 lineages discussed above (Supplementary Material Figure S1).
Emerging lineages must be monitored for disruptive mutations so that primer schemes can be updated routinely. As demonstrated here, spike-in primers can be effective stopgaps to maintain performance between major redesigns. The PCR_strainer pipeline can facilitate both tasks by screening tens-of-thousands of genome sequences and identifying relevant mutations.
Acknowledgements
We thank the dedicated staff at the British Columbia Centre for Disease Control Public Health Laboratory for processing and sequencing SARS-CoV-2 clinical specimens, especially the Molecular Microbiology and Genomics program for optimizing genomics methods, and the Bacteriology and Mycology program for routine sequencing of clinical specimens. We also thank the analytical staff for routine bioinformatic analysis.
Declarations
Funding source
This work was funded in part by the Canadian COVID genomics network (CanCOGeN), and a CIHR grant (OV4-170361).
Ethical approval
This work was conducted under a surveillance mandate, authorized by the Provincial Health Officer of British Columbia under the Health Act, without requirement for research ethics board review.
Conflict of interest
The authors have no conflicts of interest to declare.
Footnotes
Kevin S. Kuchinski and Jason Nguyen contributed equally to this work
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2021.10.050.
Appendix. Supplementary materials
References
- Chand M, Hopkins S, Dabrera G, Achison C, Barclay W, Ferguson N, Volz E, Loman N, Rambaut A, Barrett J. Investigation of Novel SARS-CoV-2 variant 2020: technical briefing #1. Dec. 21, 2020. Accessed from https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201
- Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, Rakshit P, Singh S, Abraham P, Panda S, NIC team. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv [Preprint]. 2021 May 3. doi: https://doi.org/10.1101/2021.04.22.440932 [DOI] [PMC free article] [PubMed]
- Faria NR, Morales Claro I, Candido D, Moyses Franco LA, Andrade PS, Coletti TM, Silva CAM, Sales FC, Manuli ER, Aguiar RS, Gaburo N, Camilo CdC, Fraiji NA, Esashika Crispim MA, do Perpétuo S. S. Carvalho M, Rambaut A, Loman N, Pybus OG, Sabino EC, on behalf of CADDE Genomic Network. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Jan 12, 2021. Accessed from www.virological.org
- Freed NE, Vlková M, Faisal MB, Silander OK. Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford Nanopore Rapid Barcoding. Biol Methods Protoc. 2020;5(1) doi: 10.1093/biomethods/bpaa014. Jul 18bpaa014PMID: 33029559PMCID: PMC7454405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchinski KS, Jassem AN, Prystajecky NA. Assessing oligonucleotide designs from early lab developed PCR diagnostic tests for SARS-CoV-2 using the PCR_strainer pipeline. J Clin Virol. 2020;131 doi: 10.1016/j.jcv.2020.104581. OctEpub 2020 Aug 21. PMID: 32889496PMCID: PMC7441044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, Oliveira G, Robles-Sikisaka R, Rogers TF, Beutler NA, Burton DR, Lewis-Ximenez LL, de Jesus JG, Giovanetti M, Hill SC, Black A, Bedford T, Carroll MW, Nunes M, Alcantara LC, Jr, Sabino EC, Baylis SA, Faria NR, Loose M, Simpson JT, Pybus OG, Andersen KG, Loman NJ. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12(6):1261–1276. doi: 10.1038/nprot.2017.066. JunEpub 2017 May 24. PMID: 28538739PMCID: PMC5902022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambault A, Loman N, Pybus O, Barclay W, Barrett J, Carabelli A, Connor T, Peacock T, Robertson DL, Volz E, on behalf of COVID-19 Genomic Consortium UK (CoG-UK). Preliminary genomic characterisation of an emergent SARS-CoV-2 lineages in the UK defined by a novel set of spike mutations. Dec 9, 2020. Accessed from www.virological.org
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao NY, Korsman S, Davies MA, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Kosakovsky Pond SL, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592(7854):438–443. doi: 10.1038/s41586-021-03402-9. AprEpub 2021 Mar 9. PMID: 33690265. [DOI] [PubMed] [Google Scholar]
- Tegally H, Wilkinson E, Lessells RJ, Giandhari J, Pillay S, Msomi N, Mlisana K, Bhiman JN, von Gottberg A, Walaza S, Fonseca V, Allam M, Ismail A, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Williamson C, Petruccione F, Sigal A, Gazy I, Hardie D, Hsiao NY, Martin D, York D, Goedhals D, San EJ, Giovanetti M, Lourenço J, Alcantara LCJ, de Oliveira T. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat Med. 2021;27(3):440–446. doi: 10.1038/s41591-021-01255-3. MarEpub 2021 Feb 2. PMID: 33531709. [DOI] [PubMed] [Google Scholar]
- Tyson JR, James P, Stoddart D, Sparks N, Wickenhagen A, Hall G, Choi JH, Lapointe H, Kamelian K, Smith AD, Prystajecky N, Goodfellow I, Wilson SJ, Harrigan R, Snutch TP, Loman NJ, Quick J. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv [Preprint]. 2020 Sep 4:2020.09.04.283077. doi: 10.1101/2020.09.04.283077. PMID: 32908977; PMCID: PMC7480024.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.