Abstract
Background: Cancer-derived extracellular vesicles (EVs) are regarded to have significant function in most steps during cancer progression. This meta-analysis aims to investigate the accuracy of EVs as a biomarker in cancer diagnosis.
Methods: The diagnostic efficacy of EVs for different cancers was assessed using pooled sensitivity and specificity, diagnostic odds ratio (DOR), and overall area under the curve (AUC) of the summary receiver operating characteristic (SROC). The positive likelihood ratio (PLR) and negative likelihood ratio (NLR) were verified to estimate the diagnostic efficacy of EV at a clinical level.
Results: In all, 6,183 cancer patients and 2,437 healthy controls from 75 eligible studies reported in 42 publications were included in the study. The overall pooled sensitivity, specificity, PLR, NLR, and DOR were 0.62 (95% CI: 0.60–0.63), 0.76 (95% CI: 0.75–0.78), 3.07 (95% CI: 2.52–3.75), 0.34 (95% CI: 0.28–0.41), and 10.98 (95% CI: 7.53–16.00), respectively. Similarly, the AUC of the SROC was 0.88, indicating a high conservation of EVs as an early diagnostic marker. Furthermore, subgroup analysis suggested that the use of small EVs as a biomarker was more accurate in serum-based samples of nervous system cancer (p < 0.001). As a result, ultracentrifugation and quantification and size determination methods, such as Western blotting and ELISA were the most reliable identification methods for EV detection. We also indicated that increased secretion of EVs made them a capable biomarker for diagnosing cancer in elderly European individuals.
Conclusions: Our study provides evidence that EVs are a promising non-invasive biomarker for cancer diagnosis. Well-designed cohort studies should be conducted to warrant the clinical diagnostic value of EVs.
Keywords: extracellular vesicles, biomarker, cancer diagnostics, exosomal miRNA, meta-analysis
Introduction
Cancer is still the second-leading cause of death worldwide, with an estimated 10 million cancer-related deaths reported in 2020 (Miller et al., 2019; Sung et al., 2021). According to the data reported by Globocan in 2020, lung cancer accounted for 18% of cancer-related deaths, followed by colorectal cancer (9.4%), liver cancer (8.3%), stomach cancer (7.7%), and breast cancer (6.9%) (Sung et al., 2021). Early screening for cancer in both healthy and high-risk populations can help detect cancer early, which greatly improves the opportunity for intervention and reduces mortality. Currently, most solid tumor diagnoses rely on imaging, and are then confirmed by tissue biopsy. However, the information obtained from a biopsy is influenced by the location from which the tumor tissue is taken and may not reflect the heterogeneity of the tumor. Moreover, because of the invasive nature of tissue biopsy, it cannot be performed routinely for monitoring prognosis. In addition, traditional tumor markers are also limited and often unsatisfactory for clinical use (Ju et al., 2014). Therefore, there is an urgent need for a novel non-invasive detection method that can fully clarify tumor characteristics and screen for early detection of cancer or accurately assess treatment efficacy (Ju et al., 2014; Marrugo-Ramírez et al., 2018). Compared with traditional tissue biopsy, liquid biopsy and blood-based biomarkers can detect tumor-related genetic changes and better identify disease recurrence or acquired resistance before clinical symptoms appear (Skotland et al., 2017; Wang et al., 2017; Marrugo-Ramírez et al., 2018).
As a circulating biomarker for cancer, extracellular vesicles (EVs) have gradually attracted wide attention in recent years (Petersen et al., 2020; Chen et al., 2021). EVs are cell-secreted nanoparticles involved in diverse pathological processes of tumors (He et al., 2018). Hypoxic, acidic, and inflammatory tumor microenvironments can enhance EV secretion (Parolini et al., 2009; Ramteke et al., 2015; Atretkhany et al., 2016). Generally, EVs reflect the genetic information of their original cancer cells, and can be isolated from various bodily fluids (Pisitkun et al., 2004; Michael et al., 2010). They have various constituents, including proteins, lipids, carbohydrates, DNA, and different RNA subtypes (Fujita et al., 2016). The abundance and composition of EVs often vary in different cancer types. Importantly, the miRNA in EVs obtained from blood samples and the protein profile of EVs show specificity and heterogeneity of tumors, suggesting that EVs may be promising biomarkers for various cancers (Conde-Vancells et al., 2008; Mitchell et al., 2008). Tumor-derived EVs deliver genetic information and further regulate recipient cells toward a pro-oncogenic phenotype (Salido-Guadarrama et al., 2014; Squadrito et al., 2014; Whiteside, 2016). Notably, growing evidence suggests that EVs are associated with the diagnosis of many malignant human cancers, including lung (Cazzoli et al., 2013), pancreatic (Madhavan et al., 2015), colorectal (Ogata-Kawata et al., 2014), prostate (Bryzgunova et al., 2016), and ovarian cancers (Meng et al., 2016). Although EVs have a strong potential for disease diagnosis and may aid therapeutic decision-making, further research efforts are required for viable clinical applications of EVs. In the field of clinical interpretation, detection of cancer-driving EVs in patients with no previous cancer history may dictate chemoprevention for cancer prevention or suppression, more frequent follow-ups for early cancer detection, as well as risk-reducing prophylactic surgeries such as appendectomy, colectomy, or mastectomy (Umwali et al., 2021; Xiao et al., 2021). Therefore, we conducted a quantitative systematic review along with a comprehensive meta-analysis based on published literature to confirm the diagnostic value of EVs in cancer patients in comparison with healthy subjects to suggest its potential as a non-invasive diagnostic tool for early cancer detection. Besides, we tried to document the evidence for using EVs as a diagnostic marker to predict other clinicopathological feature outcomes of cancer.
Methods
Search Strategy
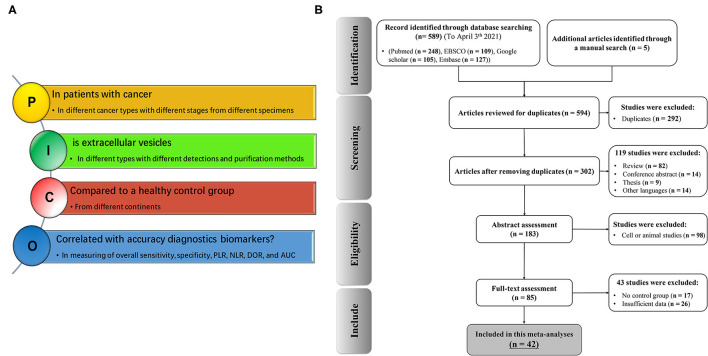
We set up a comprehensive systematic search strategy and defined clinical issues according to the Population, Intervention, Comparison, Outcomes (PICOs) principle. The selection of clinical outcomes after using EVs as an early cancer biomarker is detailed in Figure 1A according to the PICOs principles. We conducted a comprehensive systematic literature search of studies published in English from inception to April 3, 2021 using Medline electronic databases (including PubMed, ISI Web of Science, Google Scholar, Vendor Information Page Database, and Embase) to identify all relevant studies. Our search included the following terms: “extracellular vesicles or EVs or EV or exosome or microvesicle,” “cancer or tumor or neoplasm or carcinoma,” and “prognosis or survival or outcome.” Alternative spellings and synonyms were combined by applying the Boolean “OR,” and main terms were linked using the Boolean “AND” to identify all relevant studies. The reference lists of all relevant articles were further reviewed for additional eligible publications.
Figure 1.
Flow chart of study selection according to the PICOs principles (A) and PRISMA (B) in the meta-analysis (n = number of studies).
Study Inclusion/Exclusion Criteria
The current meta-analysis was conducted in accordance with the PICOs principle. Studies that met the following criteria were considered eligible: (i) cancer was confirmed by immunohistochemical or histochemical analysis; (ii) levels of EVs in tissue, plasma, serum, or other body fluids were measured; and (iii) relationship between EV secretion level and the diagnosis and prognosis of cancer, such as sensitivity and specificity, were reported in case-control study or could be measured from the provided data. The following studies were excluded: (i) non-English articles; (ii) reviews studies, conference abstracts, meetings, comments, letters, or experiments on cell line and animal models (iii); duplicate articles or continued studies of previous publications. (iv) studies with unqualified key data such as true positive (TP), false positive (FP), false negative (FN), true negative (TN), odds ratios (ORs) with 95% confidence intervals (CIs), and inadequate P-value; and (v) studies including cancer patients with additional systematic disorders and co-morbidities such as inflammations and/or autoimmune diseases.
Data Extraction
Information from all eligible studies was separately recorded by two investigators (S. L. and Y. L.). The following key component data were collected from the included studies: first author's name, publication year, country origin, continent, cancer type, cancer stage, cancer category, specimen type, patient age group, therapy type, sampling size, EV purification and identification, detection methods, EV biomarkers, and true and false positives and negatives. Any inconsistency or disagreement in the research process was resolved through a debate and consultations. If a consensus could not be reached, a third investigator (S.I.) resolved these disagreements based on the original data. We also sent e-mails to the corresponding authors of qualified studies and asked for the original data, as well as any missing or additional information, required for our meta-analysis. Table 1 and Supplementary Table 1 show the summarized main demographics, clinicopathological information, and characteristics of EVs from all selected articles.
Table 1.
Basic characteristics of the studies included in this meta-analysis.
| First author (Ref.) | Year | Country | Continent | Cancer | Stage* | Cancer category | Specimen | Age (median) | Therapy type | Sample size | NOS*** | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||||||
| Que et al. (2013) | 2013 | China | Asia | PDAC | I–IV | DSC | Serum | NR | NR | 22 | 27 | 6 |
| Cazzoli et al. (2013) | 2013 | Italy | Europe | NSCL | I | RSC | Plasma | 70.5 | NR | 50 | 30 | 6 |
| Wang et al. (2014) | 2014 | China | Asia | LSCC | I–III | RSC | Serum | 59 | SURG | 52 | 49 | 7 |
| Madhavan et al. (2015) | 2014 | Germany | Europe | PDAC | III–IV | DSC | Serum | 64.8 | NR | 131 | 30 | 8 |
| Ogata-Kawata et al. (2014) | 2014 | Spain | Europe | CRC | II–IV | DSC | Serum | NR | SURG | 88 | 11 | 6 |
| Matsumura et al. (2015) | 2015 | Japan | Asia | CRC | I–IV | DSC | Serum | 65 | SURG | 209 | 16 | 7 |
| Melo et al. (2015) | 2015 | Germany | Europe | BC | I–IV | ESC | Serum | NR | NR | 32 | 100 | 7 |
| Butz et al. (2016) | 2015 | Canada | America | RCC | NA | USC | Urine | NR | NR | 109 | 51 | 6 |
| Chiam et al. (2015) | 2015 | Australia | Europe | ESCC | III–IV | DSC | Serum | 59.5 | SURG + CT + RT | 18 | 29 | 8 |
| Zhou et al. (2017) | 2016 | China | Asia | NSCL | I–IV | RSC | Plasma | 60 | NR | 141 | 124 | 5 |
| Bryzgunova et al. (2016) | 2016 | Russian | Asia | PDAC | III | DSC | Urine | 72 | SURG | 14 | 20 | 6 |
| Samsonov et al. (2016) | 2016 | Russian | Asia | PC | I–III | USC | Urine | 65 | SURG + CT | 35 | 35 | 6 |
| Liu et al. (2016) | 2016 | USA | America | CRC | II–III | DSC | Serum | 57 | SURG | 57 | 27 | 8 |
| Zhang et al. (2018) | 2016 | China | Asia | RCC | I–III | USC | Serum | 41 | NR | 82 | 80 | 8 |
| Meng et al. (2016) | 2016 | Germany | Europe | EOC | I–IV | ESC | Serum | 60 | SURG + CT | 163 | 20 | 9 |
| Liu et al. (2017) | 2016 | China | Asia | NSCL | I–IV | RSC | Plasma | 54.5 | SURG + CT + RT | 196 | 21 | 6 |
| Machida et al. (2016) | 2016 | Japan | Asia | HCC | I–IV | DSC | Saliva | 65 | NR | 12 | 13 | 6 |
| Sandfeld-Paulsen et al. (2016) | 2016 | Denmark | Europe | NSCL | I–IV | RSC | Plasma | 68.6 | NR | 107 | 54 | 7 |
| Lea et al. (2017) | 2017 | USA | America | EOC | II–III | ESC | Plasma | NR | NR | 34 | 10 | 8 |
| Qu et al. (2017) | 2017 | China | Asia | HCC | I–IV | DSC | Serum | 54.3 | SURG | 30 | 10 | 7 |
| Lan et al. (2018) | 2017 | China | Asia | GBM | I–IV | NSC | Serum | 46.2 | SURG | 60 | 43 | 8 |
| Wang et al. (2017) | 2017 | China | Asia | ESCC | NA | DSC | Serum | 60.8 | NR | 20 | 20 | 8 |
| Skotland et al. (2017) | 2017 | Norway | Europe | PC | NA | ESC | Urine | NR | NR | 15 | 13 | 6 |
| Rodriguez et al. (2017) | 2017 | Norway | Europe | PDAC | I–III | DSC | Urine | 67.2 | CT** | 28 | 19 | 5 |
| Lai et al. (2017) | 2017 | USA | America | PDAC | NA | DSC | Plasma | NA | NA | 3 | 6 | 7 |
| Jin et al. (2017) | 2017 | China | Asia | NSCL | I | RSC | Plasma | 61.3 | NA | 47 | 13 | 6 |
| Yan et al. (2017) | 2017 | China | Asia | CRC | I–IV | DSC | Serum | 58.6 | CT | 192 | 39 | 8 |
| Shi et al. (2018) | 2017 | China | Asia | HCC | I–IV | DSC | Serum | 65.7 | SURG | 126 | 21 | 7 |
| Shiromizu et al. (2017) | 2017 | Japan | Asia | CRC | I | DSC | Serum | NR | NR | 107 | 54 | 8 |
| Arbelaiz et al. (2017) | 2017 | Spain | Europe | HCC | NA | DSC | Serum | 64.3 | SURG + CT | 43 | 32 | 7 |
| Tsukamoto et al. (2017) | 2017 | Japan | Asia | CRC | I–IV | DSC | Plasma | NA | NR | 326 | 30 | 6 |
| Xu et al. (2017) | 2017 | China | Asia | PC | I–IV | USC | Urine | 69.45 | NA | 60 | 61 | 7 |
| Yan et al. (2018a) | 2018 | China | Asia | CRC | I–IV | DSC | Serum | 58.6 | CT | 168 | 20 | 6 |
| Goto et al. (2018) | 2018 | Japan | Asia | PDAC | I–IV | DSC | Serum | 64.1 | SURG + CT + RT | 32 | 22 | 7 |
| Pan et al. (2018) | 2018 | Germany | Europe | EOC | I–IV | ESC | Plasma | 60.3 | SURG + CT | 106 | 29 | 7 |
| Wang et al. (2018) | 2018 | China | Asia | RCC | I–IV | USC | Serum | 50.7 | SURG | 45 | 30 | 9 |
| Yan et al. (2018b) | 2018 | China | Asia | CRC | I–IV | DSC | Serum | 59.6 | SURG + CT + RT | 142 | 40 | 6 |
| Kanaoka et al. (2018) | 2018 | Japan | Asia | NSCL | I–III | RSC | Plasma | 72.5 | SURG | 285 | 24 | 7 |
| Takahasi et al. (2018) | 2018 | Japan | Asia | PDAC | I–II | DSC | Plasma | NA | NR | 50 | 20 | 7 |
| Xu et al. (2019) | 2019 | China | Asia | NSCL | II–IV | RSC | Plasma | 65.6 | SURG + CT | 43 | 20 | 7 |
| Yu et al. (2019) | 2019 | China | Asia | GBM | I–IV | NSC | Serum | 65.6 | CT | 12 | 32 | 8 |
| Sakaue et al. (2019) | 2019 | Japan | Asia | PDAC | NA | DSC | Ascites | 66.9 | CT | 19 | 49 | 8 |
NSCLC, non-small-cell lung cancer; LSCC, laryngeal squamous cell carcinoma; PDAC, pancreatic ductal adenocarcinoma; PC, prostate cancer; CRC, colorectal cancer; BC, breast cancer; RCC, renal cell carcinoma; EOC, epithelial ovarian cancer; HCC, hepatocellular carcinoma; GBM, glioblastoma; ESCC, esophageal squamous cell carcinoma; RSC, respiratory system cancer; NSC, neuron system cancer; DSC, digestive system cancer; ESC, endocrine system cancer; USC, urinary system cancer; NR, not reported; NA, not available and/or not measurable; SURG, surgery; CT, chemotherapy; RT; Radiotherapy; NOS, the newcastle-ottawa scale.
Malignant tumors classified according the tumor-node-metastasis (TNM) stage.
Neoadjuvant chemotherapy and adjuvant therapy are categorized as chemotherapy.
The specific item information of NOS is available from http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
Quality Assessment
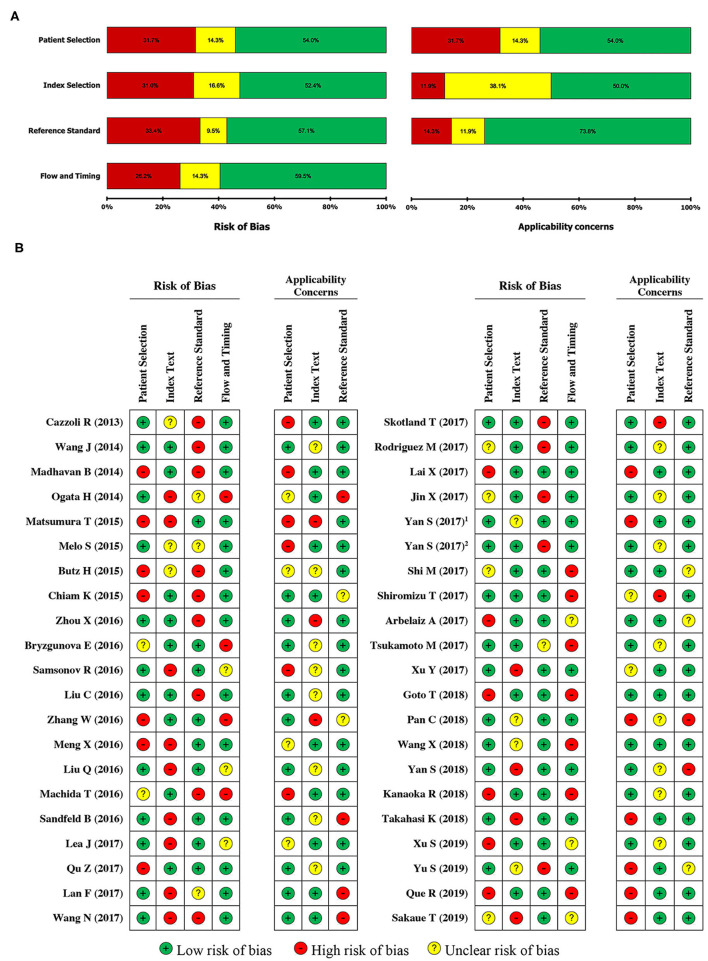
The systematic review and meta-analysis were conducted according to the recommendations of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (46). In case of controversial judgement, the diagnostic accuracy of each study was evaluated independently by three authors using the Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) tool, which consists of four domains: patient selection, index text, reference standard, and flow timing (47). Any discrepancies were resolved through a discussion. Each assessment was subjected to seven questions that were answered with “yes,” “no,” or “unclear.” The answer of “yes” indicated that a study's risk of bias was low, whereas “no” and “unclear” indicated a high risk of bias.
Statistical Analysis
The systematic meta-analysis was conducted by using the R software version 4.0.1 (R Foundation for Statistical Computing, Vienna, Austria) and package “mada” version 0.5.9. Pooled sensitivity, pooled specificity, negative likelihood ratio (NLR), positive likelihood ratio (PLR), and diagnostic odds ratio (DOR) were calculated with corresponding 95% CIs to evaluate the diagnostic value of EVs. Heterogeneity of studies in meta-analysis refers to the variation in study outcomes between studies (Gavaghan et al., 2000). In order to interpret our results, we used the Stats Direct calls statistics model for measuring heterogeneity in meta-analysis “non-combinability” statistics (Higgins et al., 2003). Correspondingly, the heterogeneity of the combined DOR was assessed via Cochran's Q test and Higgins I2 statistic from non-threshold effect (Hedges and Pigott, 2001). Subgroup analysis was conducted to determine the source of the existing heterogeneity between EVs and the available sub-analysis parameters such as ethnicity, cancer type, specimen type, age group, therapy type, EV type, EV purification and identification methods, and EV secretion level. p < 0.05 and/or I2 > 50% indicated statistical heterogeneity. Data are reported as mean ± standard deviation (SD) or median (range), including a description of qualitative variables such as numbers and percentages. The diagnosis accuracy of included studies was presented on the summary receiver operating characteristic (SROC) curve. Subgroup analysis was conducted to determine the source of heterogeneity. Publication bias was evaluated by a funnel plot and Egger's regression test. A value of “Pr > |z|” <0.05 was considered to indicate potential publication bias. All the reported p values were two-sided, and a p < 0.05 was considered to indicate statistical significance.
Results
Selection of Studies
A schematic PRISMA flowchart describing the screening and inclusion and exclusion criteria is shown in Figure 1B. The literature search yielded 594 potentially relevant publications, which were related to the topic of cancer biomarkers and included EVs. Of these, 302 studies eligible for inclusion were confirmed after excluding duplicate studies (292 studies). Subsequently, 119 studies with unrelated content were excluded. The remaining full-text articles of 183 studies were assessed for suitability. However, 98 studies were dismissed because of apparent irrelevance, and 43 studies were removed because of insufficient data. Finally, 42 articles with 75 studies were included in this meta-analysis. In detail, eight articles (Que et al., 2013; Wang et al., 2014; Chiam et al., 2015; Melo et al., 2015; Bryzgunova et al., 2016; Machida et al., 2016; Zhang et al., 2018; Yu et al., 2019) analyzing of two different EVs, four articles (Meng et al., 2016; Samsonov et al., 2016; Liu et al., 2017; Goto et al., 2018) analyzing of three EVs, one research article (Pan et al., 2018) with analyzing of four different EVs, two articles (Arbelaiz et al., 2017; Shiromizu et al., 2017) analyzing five different EVs, and one article with evolution of seven different EVs (Ogata-Kawata et al., 2014) were studied to determine the diagnostic accuracy of EVs in cancer patients (Supplementary Table 1). The detailed process is shown in Figure 1.
Study Characteristics and Quality Assessment
The main characteristics of all included literature are presented in Table 1 in the order of year of publication. In total, 8,620 subjects (6,183 cancer patients and 2,437 healthy controls) from studies reported between 2013 and 2019 were included in this meta-analysis, and histologically classified into five types of cancers: digestive system cancer (n = 22), respiratory system cancer (n = 8), endocrine system cancer (n = 5), urinary system cancer (n = 5), and nervous system cancer (n = 2). We did not find any study between 2019 and 2021 according to the inclusion/exclusion criteria and PICOs principle. The studies were conducted mostly in Asia (27 studies, 64%) and Europe (11 studies, 26%), and four studies were conducted in America (10%), with no study conducted in the African continent. Among these studies, EV secretion levels were measured in serum samples (n = 22), plasma samples (n = 12), urine samples (n = 6), saliva sample (n = 1), and ascites sample (n = 1). The major characteristics of EVs in the included studies are shown in Supplementary Table 1. All 42 studies were methodologically assessed based on the Newcastle-Ottawa scale (NOS) and QUADAS-2 protocols. In general, the average NOS score was about 7 out of 12, which could almost be classified as the high-quality group. Table 1 showed the NOS score of each study. Furthermore, QUADAS-2 results suggested that significant bias did not exist in the current meta-analyses (Figure 2).
Figure 2.
Risk of bias graph. The overall risk of bias was regarded as low in all qualified studies as per the QUADAS-2 assessment. The reviewers' decisions about each risk of bias and applicability concerns graph (A) presented as percentages across of single selected studies (B). “+” low risk of bias; “–” high risk of bias; “?” unclear risk of bias.
Outcome of the Meta-Analysis
The main results of this meta-analysis with regard to the relation between EV secretion and cancer risk are shown in Table 2. A random effect model was used to calculate statistically significant combined OR and 95% CIs (Table 2). Accordingly, we tried to explain the sources of heterogeneity from random sample sources to accurately determine the significance of EVs. The threshold effect of spearman correlation coefficient was the main reason of heterogeneity in the test accuracy studies (Zamora et al., 2006). To clarify the source of heterogeneity, we further performed subgroup analyses.
Table 2.
Meta-analysis results for the association between extracellular vesicles and cancer risk.
| No. of studies | Sample size (cases/controls) | x 2 | I 2 (%) | Pooling model | Pooled | OR (95% CI) | P -value | |
|---|---|---|---|---|---|---|---|---|
| Sensitivity | 75 | 6183/2437 | 1689.52 | 95.60 | R | 0.62 | 0.60-0.63 | <0.001 |
| Specificity | 75 | 6183/2437 | 380.38 | 80.50 | R | 0.76 | 0.75-0.78 | <0.001 |
| PLR | 75 | 6183/2437 | 429.81 | 82.80 | R | 3.07 | 2.52-3.75 | <0.001 |
| NLR | 75 | 6183/2437 | 1572.96 | 95.30 | R | 0.34 | 0.28-0.41 | <0.001 |
| DOR | 75 | 6183/2437 | 514.13 | 85.60 | R | 10.98 | 7.53-16.00 | <0.001 |
OR, odds ratio; CI, confidence interval; PLR, positive likelihood ratio; NLR, negative likelihood ratio; DOR, diagnostic odds ratio; x2, chi-squared; R, randomize model.
Diagnostic Accuracy
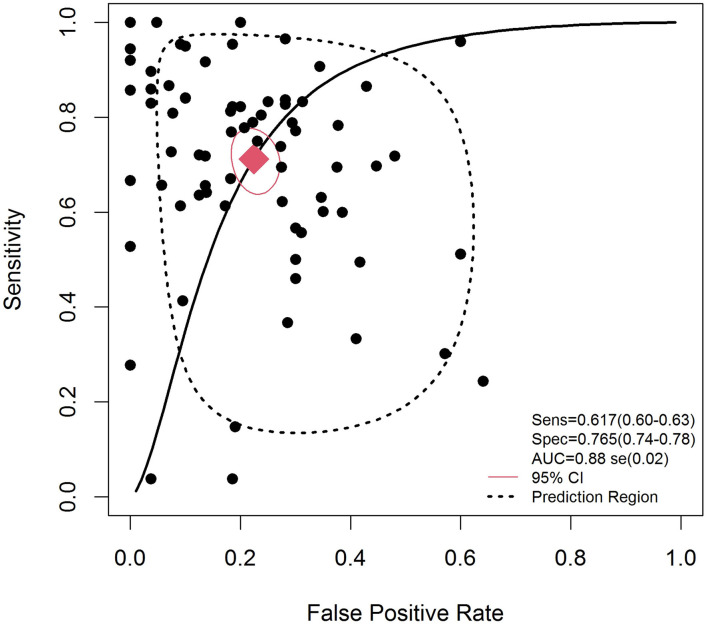
The effect of heterogeneity on the diagnostic threshold was evaluated based on the Spearman correlation coefficient. Figure 3 demonstrates the forest plots of pooled sensitivity, specificity, and DOR with their 95% CIs for individual studies. Based on the results, the overall pooled results for sensitivity, specificity, PLR, NLR, and DOR with their 95% CIs were 0.62 (95% CI: 0.60–0.63), 0.76 (95% CI: 0.75–0.78), 3.07 (95% CI: 2.52–3.75), 0.34 (95% CI: 0.28–0.41), and 10.98 (95% CI: 7.53–16.00), respectively, showing that there was no heterogeneity from the threshold effect of sensitivity and specificity (p = 0.004). The SROC curve for the included studies is indicated in Figure 3 with an overall AUC of 0.88.
Figure 3.
Summary receiver operating characteristic curve for EVs in the diagnosis of cancer. Weights are from random effect analysis.
Subgroup Analysis
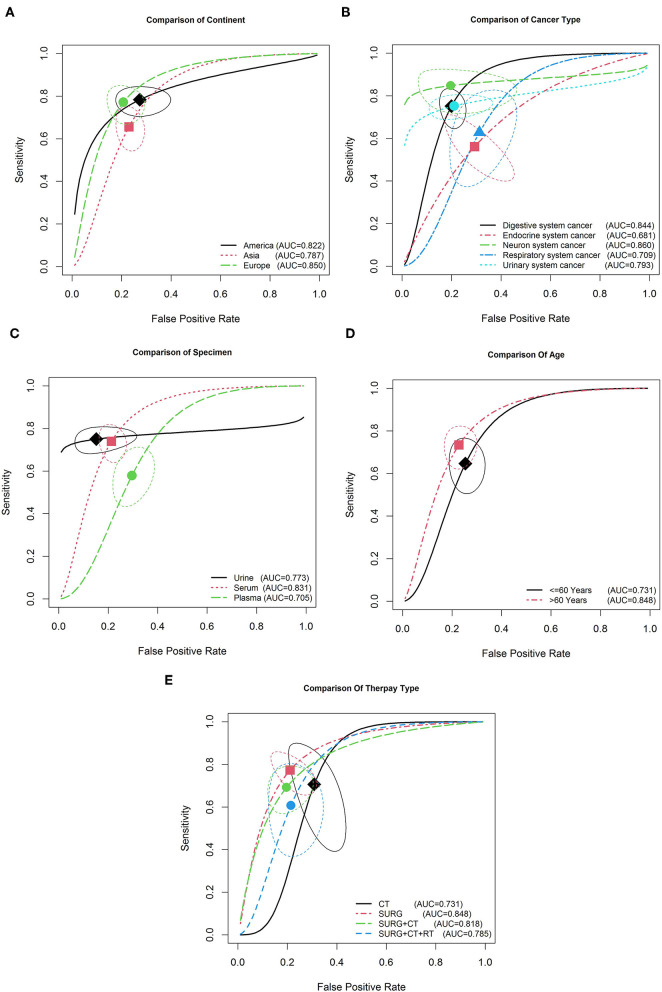
Subgroup analysis was conducted to determine the source of heterogeneity between the EVs and available sub-analysis parameters. Table 3 reveals that none of the above mentioned covariates contributed to heterogeneity (all p > 0.05). Therefore, based on these covariates, the pooled sensitivity, specificity, and AUC for important sub-analysis parameters were measured. The results showed higher accuracy in American than in Asian and European populations, with a sensitivity of 0.79 (95% CI: 0.72–0.84), specificity of 0.74 (95% CI: 0.64–0.83), and AUC of 0.822 (Figure 4A). As shown in Figure 4B and Table 3, EVs are a potentially accurate diagnostic biomarker in nervous system cancers (P = 0.24, 95% CI: 0.15–0.35) compared with the other four types, with a sensitivity of 0.85 and specificity of 0.80. With regard to specimen type, EVs showed a higher diagnostic accuracy for cancer detection in urine samples (with an 0.79 sensitivity and 0.82 specificity) than in plasma (with an 0.50 sensitivity and 0.72 specificity) and serum (with an 0.66 sensitivity and 0.77 specificity) samples, with an AUC of 0.78 (Table 3 and Figure 4C). Furthermore, subgroups analysis was performed based on age (≤60 vs. >60). The sensitivity and specificity of the population aged over 60 years (33 studies with 2,013 cases) were 0.65 (95% CI: 0.63–0.68) and 0.77 (95% CI: 0.74–0.8) respectively, whereas those of the population aged <60 years (20 studies with 2,216 cases) were 0.58 (95% CI: 0.56–0.6) and 0.73 (95% CI: 0.7–0.76), respectively. As a result, a higher diagnostic accuracy for cancer detection was observed in individuals aged over 60 years than in those aged <60 years, with AUCs of 0.848 vs. 0.731 (Figure 4D). Furthermore, subgroup analysis of different therapy types showed a higher accuracy of EVs in patients undergoing surgery (Figure 4E). Interestingly, our results show that EVs isolated from patients undergoing surgery show a higher accuracy than those isolated from patients undergoing chemotherapy or radiotherapy (Figure 4E). As seen in Table 3, the pooled sensitivity and specificity of EVs isolated from patients undergoing only surgery (18 studies with 1,560 cases) were 0.68 (95% CI: 0.66–0.71) and 0.79 (95% CI: 0.75–0.83), respectively. However, EVs isolated from patients undergoing surgery and chemotherapy (16 studies with 1,248 cases) showed a close diagnostic accuracy to that of EVs isolated from patients undergoing only surgery (with a sensitivity of 0.71 and specificity of 0.77).
Table 3.
Subgroup analysis of the included studies.
| Subgroup analyses | No. | Case/control | Sensitivity (95% CI) | Specificity (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) | |
|---|---|---|---|---|---|---|---|---|
| Ethnicity | America | 4 | 203/94 | 0.79 (0.72–0.84) | 0.74 (0.64–0.83) | 2.92 (2.11–4.20) | 0.29 (0.21–0.38) | 10.32 (5.92–18.01) |
| Asia | 41 | 3685/1493 | 0.55 (0.53–0.56) | 0.77 (0.74–0.79) | 2.70 (2.09–3.50) | 0.40 (0.30–0.52) | 7.64 (4.70–12.41) | |
| Europe | 30 | 2295/850 | 0.70 (0.68–0.72) | 0.76 (0.73–0.79) | 3.92 (2.72–5.66) | 0.28 (0.21–0.39) | 20.24 (10.10–40.57) | |
| Cancer type | DSC | 44 | 3366/1252 | 0.65 (0.64–0.67) | 0.79 (0.77–0.81) | 3.62 (2.68–4.90) | 0.27 (0.19–0.38) | 15.69 (8.99–27.41) |
| ESC | 10 | 994/299 | 0.55 (0.52–0.58) | 0.70 (0.64–0.76) | 2.66 (1.24–5.73) | 0.54 (0.39–0.74) | 5.57 (1.70–18.25) | |
| NSC | 3 | 84/107 | 0.85 (0.76–0.92) | 0.80 (0.71–0.87) | 4.40 (1.67–11.63) | 0.17 (0.01–0.28) | 26.31 (6.92–99.99) | |
| RSC | 10 | 1258/372 | 0.49 (0.46–0.51) | 0.69 (0.64–0.74) | 1.79 (1.26–2.54) | 0.52 (0.36–0.75) | 4.04 (1.88–8.70) | |
| USC | 8 | 483/407 | 0.76 (0.72–0.80) | 0.76 (0.71–0.80) | 3.40 (2.22–5.20) | 0.32 (0.25–0.39) | 12.36 (6.49–23.57) | |
| Specimen | Urine | 9 | 345/289 | 0.79 (0.74–0.83) | 0.82 (0.77–0.86) | 5.50 (2.78–10.90) | 0.29 (0.20–0.42) | 26.19 (9.26–74.10) |
| Serum | 47 | 3804/1617 | 0.66 (0.64–0.67) | 0.77 (0.74–0.79) | 3.43 (2.60–4.54) | 0.29 (0.22–0.38) | 14.22 (8.30–24.37) | |
| Plasma | 16 | 1991/456 | 0.50 (0.47–0.52) | 0.72 (0.67–0.76) | 1.87 (1.42–2.45) | 0.58 (0.47–0.72) | 3.64 (2.21–5.93) | |
| Other | 3 | 43/75 | 0.67 (0.52–0.81) | 0.73 (0.62–0.83) | 2.83 (1.09–7.35) | 0.44 (0.28–0.68) | 7.24 (1.84–28.49) | |
| Age | < = 60 | 20 | 2216/772 | 0.58 (0.56–0.6) | 0.73 (0.7–0.76) | 2.52 (1.91–3.32) | 0.42 (0.32–0.53) | 7.05 (4.15–11.97) |
| >60 | 33 | 2013/934 | 0.65 (0.63–0.68) | 0.77 (0.74–0.8) | 3.16 (2.32–4.31) | 0.3 (0.21–0.42) | 13.48 (7.27–25) | |
| Therapy | SURG | 18 | 1560/386 | 0.68 (0.66–0.71) | 0.79 (0.75–0.83) | 3.42 (2.39–4.91) | 0.26 (0.17–0.41) | 15.52 (7.43–32.42) |
| SURG + CT* | 16 | 1248/461 | 0.71 (0.68–0.73) | 0.77 (0.73–0.8) | 3.49 (2.29–5.3) | 0.33 (0.24–0.44) | 14.39 (6.86–30.17) | |
| SURG + CT + RT | 9 | 862/227 | 0.44 (0.41–0.47) | 0.81 (0.75–0.86) | 2.77 (1.68–4.58) | 0.48 (0.34–0.69) | 6.3 (2.62–15.13) | |
| Type of EVs** | lEVs | 41 | 5064/1681 | 0.57(0.56–0.59) | 0.76(0.73–0.78) | 2.67(2.10–3.34) | 0.39(0.31–0.51) | 8.42(5.34–13.28) |
| sEVs | 34 | 1119/756 | 0.67(0.65–0.69) | 0.81(0.75–0.85) | 3.71(2.62–5.25) | 0.29(0.22–0.40) | 14.61(7.71–27.68) | |
| EVs purification | Isolation kit*** | 26 | 2074/875 | 0.56(0.53–0.58) | 0.73(0.7–0.76) | 2.52(1.9–3.34) | 0.4(0.31–0.53) | 7.41(4.31–12.74) |
| UC# | 49 | 4109/1562 | 0.65 (0.63–0.66) | 0.78(0.76–0.8) | 3.51(2.66–4.64) | 0.31(0.24–0.41) | 13.92(8.3–23.34) | |
| EVs identification | NA | 11 | 703/312 | 0.56 (0.52–0.6) | 0.78(0.73–0.83) | 2.59(1.44–4.68) | 0.36(0.14–0.89) | 8.13(2.64–25.06) |
| Microscopic## | 31 | 2579/1075 | 0.54 (0.52–0.56) | 0.72(0.69–0.75) | 2.61(1.98–3.43) | 0.41(0.32–0.53) | 8.2(4.61–14.6) | |
| Exosomal surface biomarkers### | 20 | 1817/378 | 0.68(0.66–0.7) | 0.8(0.77–0.83) | 3.6(2.09–6.18) | 0.26(0.13–0.51) | 14.86(8.36–26.41) | |
| Quantification and size determination& | 13 | 1084/672 | 0.74 (0.71–0.76) | 0.82(0.77–0.85) | 4.35(2.85–6.62) | 0.31(0.24–0.41) | 16.37(5.91–45.3) | |
| EVs secretion level | Increase | 62 | 5123/2095 | 0.63 (0.61–0.64) | 0.77(0.76–0.79) | 3.22(2.61–3.98) | 0.32(0.26–0.4) | 12.31(8.19–18.5) |
| Decrease | 11 | 1032/302 | 0.56 (0.53–0.59) | 0.69(0.63–0.74) | 1.99(1.2–3.3) | 0.58(0.41–0.81) | 4.21(1.73–10.23) | |
EVs, extracellular vesicles; DSC, digestive system cancer; ESC, endocrine system cancer; NSC, neuron system cancer; RSC, respiratory system cancer; USC, urinary system cancer; SURG, surgery; CT, chemotherapy; RT; Radiotherapy; lEVs, large EVs; sEVs, small EVS; UC, ultracentrifuge.
Neoadjuvant chemotherapy and adjuvant therapy are categorized as chemotherapy.
EVs sedimenting at 100,000 × g into is categorized as a small EVs (sEVs) and EVs sedimenting at 2,000 × g is categorized as a large EVs (lEVs, large fragments of cells, large apoptotic bodies) (Mateescu et al., 2017; Slomka et al., 2018).
Isolation kit refer to the standard total extracellular vesicles isolation kit media and urine for biomarker analysis.
Using the differential ultracentrifugation range (>100,000 ×g (100,000–200,000 ×g) for 2 h) for isolation of different exosome isolation methods yield different amount of exosomes (Patel et al., 2019).
Microscopic methods for EVS identification is contended the transmission electron microscope, atomic force microscopy and selected reaction monitoring methods.
CD81, CD63, CD64, CD65, and CD66 are a cell surface glycoprotein exosomal cell surface markers that is using mostly for isolation isolating exosomes from tissue culture media and urine for biomarker analysis (Konoshenko et al., 2018).
Quantificative methods for EVS identification is the quantificative and size determination based on the western blot and ELISA.
Figure 4.
Summary receiver operating characteristic curve for EVs and subgroup analysis based on continents (A), cancer type (B), specimen type (C), patient age (D), and therapy type (E).
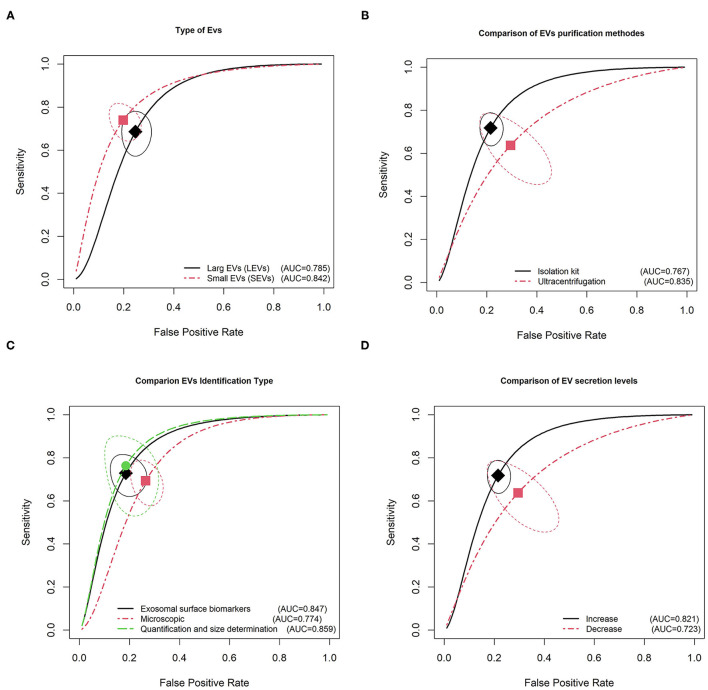
Figure 5 shows the results of the SROC analysis according to the characterization parameters of EVs, such as type of EVs, purification methods, identification methods of EVs, and EV secretion level. We categorized EVs sedimenting at 100,000 × g into small EVs (sEVs), those sedimenting at speeds lower than 20,000 × g into medium EVs (mEVs, microvesicles, ectosomes), and those sedimenting at 2,000 × g into large EVs (lEVs, large fragments of cells, large apoptotic bodies) (Mateescu et al., 2017; Slomka et al., 2018). As shown in Table 3, 5,064/1,681 cases/controls in 41 studies were compared with 1,119/756 cases/controls in 34 studies with sEVs. The sEVs showed higher pooled sensitivities and specificities than lEVs (0.67 vs. 0.57 and 0.81 vs. 0.76, respectively; Figure 5A). Meanwhile, with regard to purification methods, the ultracentrifugation method (range 100,000–200,000 × g for 2 h) showed higher sensitivity and specificity than kit-based isolation methods (0.65 vs. 0.56 and 0.78 vs. 0.73, respectively). The kit-based isolation methods depend on the optimization step for rapidly isolating EVs from the tissue culture media (Figure 5B). In this study, we categorized EV identification methods in three main groups, microscopic methods, identification based on exosomal surface biomarkers, and quantification and size determination methods. Microscopic methods for EV identification use the transmission electron microscope, atomic force microscopy, and selected reaction monitoring methods. Our results show that CD81, CD63, CD64, CD65, and CD66 are cell surface glycoproteins; exosomal cell surface markers were the surface biomarkers mostly used for the identification of exosomes from tissue culture media and urine (Konoshenko et al., 2018). Also, Western blotting and ELISA were the most reported quantification methods for EV identification (Table 3). Our meta-analysis results show that the quantification and size determination methods such as Western blotting and ELISA were the most reliable identification methods with a sensitivity of 0.74 (CIs: 0.71–0.76), specificity of 0.82 (CIs: 0.77–0.85), and AUC of 0.86 (Figure 5C). However, identification based on the EV surface biomarkers has close diagnostic accuracy to EVs identification (with a sensitivity of 0.68 and specificity of 0.80). In univariate logistic regression analysis, increased secretion of EVs showed higher diagnostic accuracy for cancer detection compared with decreased secretion of EVs, with the sensitivities of 0.63 vs. 0.56, specificities of 0.77 vs. 0.69, and AUCs of 0.821 vs. 0.723 (Figure 5D). Therefore, increased secretion of EVs could be a promising biomarker for cancer diagnosis.
Figure 5.
Summary receiver operating characteristic curve for EVs and subgroup analysis based on type of EVs (A), purification methods (B), identification type (C), and secretion level (D).
Publication Bias and Sensitivity Analysis
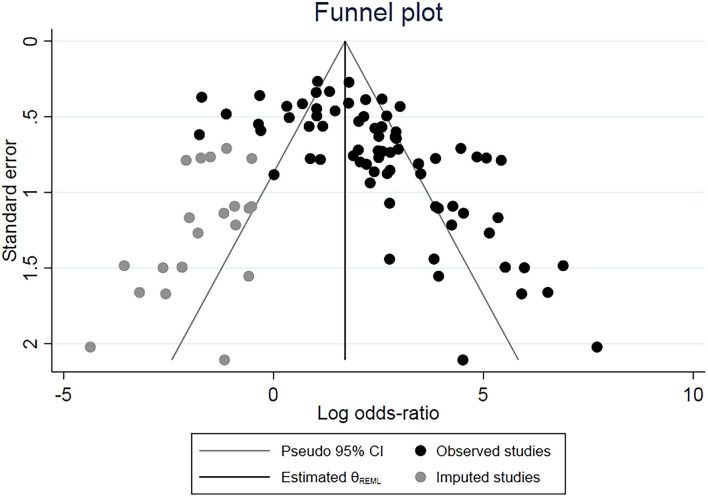
Funnel plots and Egger's test were performed to estimate publication bias. These were conducted by precluding a single study at a time and significant differences between events and hypothesis were observed (Figure 6) (Tobias, 1999). The trim-and-fill method was employed and generally resulted in a confidence interval of 0.09 (95% CI: 0.04–0.14) using the unadjusted random-effect model. Additionally, this test identified 20 articles of all included studies with possible publication bias, but no significant effect on our main findings was found (P = 0.1038) and the high level of heterogeneity (I2 = 69%) remained (Figure 6). The resulting shape of the funnel plot and Egger's test provided no statistical evidence for publication bias (t = 1.16 and p = 0.164). Hence, no noticeable evidence for significant publication bias in our meta-analysis was observed, which indicates that our meta-analysis results are stable and credible.
Figure 6.
Contour-enhanced funnel plots for the detection of publication bias. Each point represents one of the 74 studies for the specified association. The size of each circle is proportional to the percentage weight that each study contributes to the HRs. These plots indicate that some studies were in significant areas where p ≤ 0.01 (solid lines). Solid triangles refer to included studies and X's refer to filled studies. The vertical axis represents the standard error of logarithmic HR limits.
Discussion
It is well-established that cancer cell-derived EVs released into a tumor microenvironment and circulation can promote tumor progression and metastasis by inducing matrix remodeling, angiogenesis, inflammation, and metastatic niche formation (Dong et al., 2002; Hoshino et al., 2015; Guo et al., 2019). Mechanically, cancer cell-derived exosomes promote tumor growth by triggering changes in naïve mesenchymal stem cells (MSCs) to form pro-inflammatory MSC in many solid tumors such as melanoma (Peinado et al., 2012), ovarian cancer (Feng et al., 2019), prostate cancer (Soekmadji et al., 2017), and breast cancers (King et al., 2012). Similarly, exosomal miRNA and lncRNA premetastatic niches develop in many cancers and modulate cell proliferation and motility/invasion (Soekmadji et al., 2017). Similarly, tumor-derived EVs, mostly exosomal miRNA and lncRNA, provide a genetic landscape, which promotes endothelial cell angiogenesis, and creates conditions for tumor growth, making this molecule a promising candidate as a biomarker to reflect various physiological and pathological states of cancer cells (Skog et al., 2008; Keller et al., 2011). This evidence indicates that EV-associated miRNAs are not only promising diagnostic and prognostic biomarkers but also important therapeutic targets as their secretion can reflect the information isolated from millions of oncogenes during tumor aggressiveness, resistance to chemotherapy, and tumor immune-escape. Several studies have identified candidate tumor-derived EV biomarkers that can be considered as cost-effective and non-invasive prospective biomarkers for cancer patients because they can be detected in bio fluid samples of cancer patients (D'Souza-Schorey and Clancy, 2012; Feng et al., 2019). These results propose that tumor-derived EV biomarkers can be used in a clinical setting if they are specific to a particular cancer type, which was partially demonstrated in our study.
With these assumptions and foreground, in this comprehensive study, we collected all available articles and performed a meta-analysis to confirm the diagnostic value of different types of EVs in different cancers. We aimed to understand the relationship of EVs as a diagnostic marker to predict other clinicopathological features and outcomes of different cancers, such as cancer type, specimen type, sample size, and cancer grade. As expected, the pooled HR from our results showed that the EV secretion levels have potential value for the diagnosis of cancer. To the best of our best knowledge, no meta-analysis has investigated the diagnostic value of EVs in cancers by displaying consistent, statistically significant changes in EV secretion level. In our meta-analysis of 42 articles with 75 studies, which included different types of cancers, the pooled sensitivity and specificity were 0.617 and 0.765, respectively; these are statistical measures that help determine the diagnostic value of EVs in cancer. In this comprehensive study, the assessment of diagnostic accuracy of EVs at a clinical level was verified by PLR and NLR tests. The values of PLR and NLR were 3.07 and 0.34, respectively, which demonstrated that the probability of a TP diagnosis is 3.07 times higher than the probability of a FP diagnosis and that there is a 34% error rate in the individuals testing negative. Specifically, the upper left corner SROC curve is the perfect test to evaluate the diagnostic value (Walter, 2002); our overall AUC of SROC curve was 0.88, which is considered to be in a good range of SROC curve statistically (the good range of AUC: 0.75–0.92) (Moher et al., 2009). This indicates that EVs are highly accurate as a biomarker for detecting cancer. Our results clearly illustrated that exosomal miRNA had a better accuracy with regard to cancer detection than exosomal protein, with AUCs of 0.811 vs. 0.810.
Recently, many studies have demonstrated that EV-packaged mRNAs are the most reported and well-evaluated extracellular RNAs (exRNAs) enriched in EVs that can serve as biomarkers for cancer (Cazzoli et al., 2013; Que et al., 2013; Ogata-Kawata et al., 2014; Wang et al., 2014; Chiam et al., 2015; Madhavan et al., 2015; Matsumura et al., 2015; Melo et al., 2015; Butz et al., 2016). Numerous pro-angiogenic miRNAs present in EVs released from breast cancer (miR-100, miR-222, miR-30a and miR-17), lung cancer (miR-146a, miR-100-5p), and ovarian cancer (miR-21) samples may contribute to new blood vessel formation and have been proposed as a reliable tumor biomarker (Que et al., 2013; Ogata-Kawata et al., 2014; Wang et al., 2017; Pan et al., 2018; Zhang et al., 2018). EV abundance and composition are also altered in individuals with cancer (Hu et al., 2021). As a main type of EVs, sEVs can provide a protective, enriched source of miRNA and increase the stability of endogenous miRNA (Hu et al., 2021). Goto et al. demonstrated that exosomal miRNA191, miRNA21, and miRNA451a are superior to serum circulating miRNAs in establishing a diagnosis of pancreatic neoplasms (Goto et al., 2018). Similarly, there is a possibility that the distinct protein or lipid profiling of sEVs was performed for identifying molecular features with novel biomarker potential diagnostic values, specifically in urine or blood samples of cancer patients (Momen-Heravi, 2017). However, little is known about clinical applicability of using protein or lipid profiling composition of EVs for detecting different stages of cancer.
Other results with different subgroups were relatively consistent with our main findings, indicating that our findings are reliable. Our systematic search clearly indicated that serum-based isolated EVs (AUC: 0.831) are more accurate diagnostic biomarkers than plasma or urine-based EVs. Recently, several studies have reported that the biological fluid source of exRNA is associated with the histological grade of cancer. Surely, most clinical studies have evaluated EVs as cancer biomarkers in whole blood, urine, and cerebrospinal fluids rather than in tissue. For example, serum miR-34a levels were associated with the last stage of breast cancer and serum-based tumor markers were the most effective screening tool for the detection of metastatic breast cancer (Taplin et al., 2008; Imani et al., 2017). Moreover, the secretion level of serum EVs of miR-18a, miR-221, and miR-224 has been suggested to be a potential diagnostic biomarker in hepatocellular carcinoma (Sohn et al., 2015; Lai et al., 2017; Rodriguez et al., 2017). However, no significant association between serum-based EVs and clinicopathological features of tumors, such as hormone receptors and lymph node metastasis, has been observed (Erbes et al., 2015; Mishra et al., 2015).
Owing to the existence of significant heterogeneity, a meta-analysis and subgroup analysis were performed to identify other related factors affecting the heterogeneity. We observed that different isolation methods of EVs even within the same approach for EV isolation interfered with verification and yielded different amounts of EVs (Patel et al., 2019). As expected, our results confirmed that ultracentrifugation techniques of EVs had a higher accuracy for diagnosing cancers. Ultracentrifuges are a classic technique considered as a gold standard for EV collation (Stam et al., 2021). These methods allow for the purification of subpopulations of EVs such as exosomes according to size differentiation (Sunkara et al., 2019; Stam et al., 2021). Also, differential ultracentrifugation is the most available method for EV isolation that has been integrated with mass spectrometry techniques with unique peptides in a multicenter study (Yan et al., 2009). However, the excellence of proper isolation of EVs using ultracentrifugation techniques depends on the materials available in the laboratory, type of EVs, samples, type of cancer, and the required amount of EVs (Momen-Heravi, 2017; Stam et al., 2021). For instance, quantification and size determination showed great accuracy when used to identify exosomal secretion. Considering the advantages and disadvantages of all existing methods for EV isolation and identification, future analytical study is required to find promising techniques and methods that are all characterized by savings in terms of cost, availability, efficiency, labor, and time (Greening et al., 2015; Chen et al., 2019).
It is noteworthy that on comparing different cancer types, a significant diagnostic role of EVs was found in nervous system cancers. Our pooled results provide compelling evidence of a significant positive association between EVs and race. Our results also suggested promising accuracy for EV-mediated diagnosis in Europe compared with that in Asia and America, especially the patients aged more than 60 years. Considering the limitation of the small sample size of the American group, further large-scale studies among American populations should be designed to provide a comprehensive outcome.
We used the Duval and Tweedie's Trim and Fill model to analyses the possible publication bias. The key idea behind of this model is iterative procedure to remove the most extreme small studies from the positive side of the funnel plot, re-computing the effect size at each iteration until the funnel plot is symmetric about the (new) effect size. In theory, this will yield an unbiased estimate of the effect size. Too, major advantage of this approach is finding the best estimate of the unbiased effect size in intuitive visual display (Duval and Tweedie, 2000). Therefore, according incorporate Trim and Fill algorithm, we did not find any noticeable evidence for significant publication bias in our meta-analysis.
This study had some limitations. First, we only included articles published in English language, whereas published papers in other languages were ignored. Fundamentally, the meta-analysis results were based on unadjusted estimates because some studies did not provide detailed information to calculate the adjusted estimates. Definitely, chemotherapy and other therapeutic interventions can also alter the EV profile (Ab Razak et al., 2019; Wang et al., 2021), which has been out of control in reported studies. In this regard, further research should validate the diagnostic value of EV indices in cancer patients who receive different treatments, such as chemotherapy, immunotherapy, or radiotherapy, and differentiate between those treated with chemo radiation and those treated with monotherapy. Similarly, there are several reports showing that the clinical relevance of EVs is different between hematological malignancies and solid tumors (Iaccino et al., 2017; Maisano et al., 2020; Trino et al., 2021). In addition, the populations in the studies were not comprehensively represented, and there was a lack of research in the African populations. Researchers must seek to include a greater diversity of patient populations in future studies to analyze their EV profile. Furthermore, many confounding factors were not controlled for or reported in bias statistical results. Undoubtedly, well-designed large-scale studies with matched case-controls and functional studies are warranted in the future to validate these findings. Similarly, at a clinical level, suitable diagnostic examinations and appropriate randomized comprehensive experiments with different observational studies should be defined, to establish reliable diagnostic EV panel and guidelines for using EVs for early cancer detection.
Conclusion
In conclusion, the data of the present meta-analysis show that high levels of EVs are reliable diagnostics biomarkers for the early detection of cancer. It has been determined that exosomal miRNA EVs are accurate diagnostic biomarkers in serum-based samples of patients with nervous system cancers.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
S-yL, YL, and HH contributed to the data curation, conceptualization and methodology, review, and editing. SI and HH performed data validation, formal analysis, and editing. Q-lW gave the funding acquisition and validation and the final approval of the version to be submitted. SI is responsible for supervision, project administration, visualization, and paper writing—original draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Southwest Medical University (SWMU), Sichuan Province, China grants (No. 18080) that were awarded to SI.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thankfully acknowledge all authors for their continuous collaborations with us in sharing information. We also gratefully acknowledge all authors and coworkers for their contributions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.705791/full#supplementary-material
References
- Ab Razak N. S., Ab Mutalib N. S., Mohtar M. A., Abu N. (2019). Impact of chemotherapy on extracellular vesicles: understanding the chemo-EVs. Front. Oncol. 9:1113. 10.3389/fonc.2019.01113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbelaiz A., Azkargorta M., Krawczyk M., Santos-Laso A., Lapitz A., Perugorria M. J., et al. (2017). Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology 66, 1125–1143. 10.1002/hep.29291 [DOI] [PubMed] [Google Scholar]
- Atretkhany K., Drutskaya M., Nedospasov S., Grivennikov S., Kuprash D. (2016). Chemokines, cytokines and exosomes help tumors to shape inflammatory microenvironment. Pharmacol. Ther. 168, 98–112. 10.1016/j.pharmthera.2016.09.011 [DOI] [PubMed] [Google Scholar]
- Bryzgunova O., Zaripov M. M., Skvortsova T. E., Lekchnov E. A., Grigor'eva A. E., Zaporozhchenko I. A., et al. (2016). Comparative study of extracellular vesicles from the urine of healthy individuals and prostate cancer patients. PLoS ONE 11:e0157566. 10.1371/journal.pone.0157566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz H., Nofech-Mozes R., Ding Q., Khella H. W. Z., Szabo P. M., Jewett M., et al. (2016). Exosomal micrornas are diagnostic biomarkers and can mediate cell-cell communication in renal cell carcinoma. Eur. Urol. Focus 2, 210–218. 10.1016/j.euf.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Cazzoli R., Buttitta F., Di Nicola M., Malatesta S., Marchetti A., Rom W., et al. (2013). MicroRNAs derived from circulating exosomes as non-invasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 8, 1156–1162. 10.1097/JTO.0b013e318299ac32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wu T., Zhu Z., Huang H., Zhang L., Goel A., et al. (2021). An integrated workflow for biomarker development using microRNAs in extracellular vesicles for cancer precision medicine. Semin. Cancer Biol. 74, 134–155. 10.1016/j.semcancer.2021.03.011 [DOI] [PubMed] [Google Scholar]
- Chen B. Y., Sung C. W., Chen C., Cheng C. M., Lin D. P., Huang C. T., et al. (2019). Advances in exosomes technology. Clin. Chim. Acta 493, 14–19. 10.1016/j.cca.2019.02.021 [DOI] [PubMed] [Google Scholar]
- Chiam K., Wang T., Watson D. I., Mayne G. C., Irvine T. S., Bright T., et al. (2015). Circulating serum exosomal miRNAs as potential biomarkers for esophageal adenocarcinoma. J. Gastrointest. Surg. 19, 1208–1215. 10.1007/s11605-015-2829-9 [DOI] [PubMed] [Google Scholar]
- Conde-Vancells J., Rodriguez-Suarez E., Embade N., Gil D., Matthiesen R., Valle M., et al. (2008). Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 7, 5157–5166. 10.1021/pr8004887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Strome S. E., Salomao D. R., Tamura H., Hirano F., Flies D. B., et al. (2002). Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 8, 793–800. 10.1038/nm730 [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey C., Clancy J. (2012). Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers. Genes Dev. 26, 1287–1299. 10.1101/gad.192351.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval S., Tweedie R. (2000). A non-parametric “Trim and Fill” method of accounting for publication bias in meta-analysis. J. Am. Stat. Assoc. 95, 89–98. 10.1080/01621459.2000.10473905 [DOI] [Google Scholar]
- Erbes T., Hirschfeld M., Rücker G., Jaeger M., Boas J., Iborra S., et al. (2015). Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer 15:193. 10.1186/s12885-015-1190-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Dean D. C., Hornicek F. J., Shi H., Duan Z. (2019). Exosomes promote pre-metastatic niche formation in ovarian cancer. Mol. Cancer 18:124. 10.1186/s12943-019-1049-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Yoshioka Y., Ochiya T. (2016). Extracellular vesicle transfer of cancer pathogenic components. Cancer Sci. 107, 385–390. 10.1111/cas.12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavaghan D. J., Moore A. R., McQuay H. J. (2000). An evaluation of homogeneity tests in meta-analyses in pain using simulations of individual patient data. Pain 85, 415–424. 10.1016/S0304-3959(99)00302-4 [DOI] [PubMed] [Google Scholar]
- Goto T., Fujiya M., Konishi H., Sasajima J., Fujibayashi S., Hayashi A., et al. (2018). An elevated expression of serum exosomal microRNA-191,−21,−451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer 18:116. 10.1186/s12885-018-4006-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening D. W., Xu R., Ji H., Tauro B. J., Simpson R. J. (2015). A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 1295, 179–209. 10.1007/978-1-4939-2550-6_15 [DOI] [PubMed] [Google Scholar]
- Guo Y., Ji X., Liu J., Fan D., Zhou Q., Chen C., et al. (2019). Effects of exosomes on pre-metastatic niche formation in tumors. Mol. Cancer 18:39. 10.1186/s12943-019-0995-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Zheng S., Luo Y., Wang B. (2018). Exosome theranostics: biology and translational medicine. Theranostics 8, 237–255. 10.7150/thno.21945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges L. V., Pigott T. D. (2001). The power of statistical tests in meta-analysis. Psychol. Methods 6, 203–217. 10.1037/1082-989X.6.3.203 [DOI] [PubMed] [Google Scholar]
- Higgins J. P., Thompson S. G., Deeks J. J., Altman D. G. (2003). Measuring inconsistency in meta-analyses. BMJ 327, 557–560. 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A., Costa-Silva B., Shen T. L., Rodrigues G., Hashimoto A., Tesic Mark M., et al. (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335. 10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Wolfram J., Srivastava S. (2021). Extracellular vesicles in cancer detection: hopes and hypes. Trends Cancer 7, 122–133. 10.1016/j.trecan.2020.09.003 [DOI] [PubMed] [Google Scholar]
- Iaccino E., Mimmi S., Dattilo V., Marino F., Candeloro P., Di Loria A., et al. (2017). Monitoring multiple myeloma by idiotype-specific peptide binders of tumor-derived exosomes. Mol. Cancer 16:159. 10.1186/s12943-017-0730-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imani S., Zhang X., Hosseinifard H., Fu S., Fu J. (2017). The diagnostic role of microRNA-34a in breast cancer: a systematic review and meta-analysis. Oncotarget 8, 23177–23187. 10.18632/oncotarget.15520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Chen Y., Chen H., Fei S., Chen D., Cai X., et al. (2017). Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin. Cancer Res. 23, 5311–5319. 10.1158/1078-0432.CCR-17-0577 [DOI] [PubMed] [Google Scholar]
- Ju R.-J., Li X.-T., Shi J.-F., Li X.-Y., Sun M.-G., Zeng F., et al. (2014). Liposomes, modified with PTD(HIV-1) peptide, containing epirubicin and celecoxib, to target vasculogenic mimicry channels in invasive breast cancer. Biomaterials 35, 7610–7621. 10.1016/j.biomaterials.2014.05.040 [DOI] [PubMed] [Google Scholar]
- Kanaoka R., Iinuma H., Dejima H., Sakai T., Uehara H., Matsutani N., et al. (2018). Usefulness of plasma exosomal microRNA-451a as a non-invasive biomarker for early prediction of recurrence and prognosis of non-small cell lung cancer. Oncology 94, 311–323. 10.1159/000487006 [DOI] [PubMed] [Google Scholar]
- Keller S., Ridinger J., Rupp A. K., Janssen J. W., Altevogt P. (2011). Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 9:86. 10.1186/1479-5876-9-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King H. W., Michael M. Z., Gleadle J. M. (2012). Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 12:421. 10.1186/1471-2407-12-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konoshenko M. Y., Lekchnov E. A., Vlassov A. V., Laktionov P. P. (2018). Isolation of extracellular vesicles: general methodologies and latest trends. Biomed Res. Int. 2018:8545347. 10.1155/2018/8545347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X., Wang M., McElyea S. D., Sherman S., House M., Korc M. (2017). A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 393, 86–93. 10.1016/j.canlet.2017.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Qing Q., Pan Q., Hu M., Yu H., Yue X. (2018). Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell Oncol. (Dordr). 41, 25–33. 10.1007/s13402-017-0355-3 [DOI] [PubMed] [Google Scholar]
- Lea J., Sharma R., Yang F., Zhu H., Ward E. S., Schroit A. J. (2017). Detection of phosphatidylserine-positive exosomes as a diagnostic marker for ovarian malignancies: a proof of concept study. Oncotarget 8, 14395–14407. 10.18632/oncotarget.14795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Eng C., Shen J., Lu Y., Takata Y., Mehdizadeh A., et al. (2016). Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget 7, 76250–76260. 10.18632/oncotarget.12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yu Z., Yuan S., Xie W., Li C., Hu Z., et al. (2017). Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 8, 13048–13058. 10.18632/oncotarget.14369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida T., Tomofuji T., Maruyama T., Yoneda T., Ekuni D., Azuma T., et al. (2016). miR1246 and miR4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol. Rep. 36, 2375–2381. 10.3892/or.2016.5021 [DOI] [PubMed] [Google Scholar]
- Madhavan B., Yue S., Galli U., Rana S., Gross W., Müller M., et al. (2015). Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int. J. Cancer 136, 2616–2627. 10.1002/ijc.29324 [DOI] [PubMed] [Google Scholar]
- Maisano D., Mimmi S., Russo R., Fioravanti A., Fiume G., Vecchio E., et al. (2020). Uncovering the exosomes diversity: a window of opportunity for tumor progression monitoring. Pharmaceuticals (Basel) 13:180. 10.3390/ph13080180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrugo-Ramírez J., Mir M., Samitier J. (2018). Blood-based cancer biomarkers in liquid biopsy: a promising non-invasive alternative to tissue biopsy. Int. J. Mol. Sci. 19:2877. 10.3390/ijms19102877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B., Kowal E. J., van Balkom B. W., Bartel S., Bhattacharyya S. N., Buzas E. I., et al. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—an ISEV position paper. J. Extracell. Vesicles 6:1286095. 10.1080/20013078.2017.1286095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T., Sugimachi K., Iinuma H., Takahashi Y., Kurashige J., Sawada G., et al. (2015). Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 113, 275–281. 10.1038/bjc.2015.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo S. A., Luecke L. B., Kahlert C., Fernandez A. F., Gammon S. T., Kaye J., et al. (2015). Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523, 177–182. 10.1038/nature14581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Muller V., Milde-Langosch K., Trillsch F., Pantel K., Schwarzenbach H. (2016). Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 7, 16923–16935. 10.18632/oncotarget.7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael A., Bajracharya S., Yuen P., Zhou H., Star R., Illei G., et al. (2010). Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 16, 34–38. 10.1111/j.1601-0825.2009.01604.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K., Nogueira L., Mariotto A., Rowland J., Yabroff K., Alfano C., et al. (2019). Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 69, 363–385. 10.3322/caac.21565 [DOI] [PubMed] [Google Scholar]
- Mishra S., Srivastava A. K., Suman S., Kumar V., Shukla Y. (2015). Circulating miRNAs revealed as surrogate molecular signatures for the early detection of breast cancer. Cancer Lett. 369, 67–75. 10.1016/j.canlet.2015.07.045 [DOI] [PubMed] [Google Scholar]
- Mitchell P., Parkin R., Kroh E., Fritz B., Wyman S., Pogosova-Agadjanyan E., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518. 10.1073/pnas.0804549105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 6:e1000097. 10.1371/journal.pmed.1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momen-Heravi F. (2017). Isolation of extracellular vesicles by ultracentrifugation. Methods Mol. Biol. 1660, 25–32. 10.1007/978-1-4939-7253-1_3 [DOI] [PubMed] [Google Scholar]
- Ogata-Kawata H., Izumiya M., Kurioka D., Honma Y., Yamada Y., Furuta K., et al. (2014). Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 9:e92921. 10.1371/journal.pone.0092921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Stevic I., Muller V., Ni Q., Oliveira-Ferrer L., Pantel K., et al. (2018). Exosomal microRNAs as tumor markers in epithelial ovarian cancer. Mol. Oncol. 12, 1935–1948. 10.1002/1878-0261.12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolini I., Federici C., Raggi C., Lugini L., Palleschi S., De Milito A., et al. (2009). Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 284, 34211–34222. 10.1074/jbc.M109.041152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G. K., Khan M. A., Zubair H., Srivastava S. K., Khushman M., Singh S., et al. (2019). Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications. Sci. Rep. 9:5335. 10.1038/s41598-019-41800-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Alečković M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., et al. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18, 883–891. 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E. V., Chudakova D. A., Skorova E. Y., Anikin V., Reshetov I. V., Mynbaev O. A. (2020). The extracellular matrix-derived biomarkers for diagnosis, prognosis, and personalized therapy of malignant tumors. Front. Oncol. 10:575569. 10.3389/fonc.2020.575569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun T., Shen R., Knepper M. (2004). Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A. 101, 13368–13373. 10.1073/pnas.0403453101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z., Wu J., Wu J., Ji A., Qiang G., Jiang Y., et al. (2017). Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 8, 80666–80678. 10.18632/oncotarget.20881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que R., Ding G., Chen J., Cao L. (2013). Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 11:219. 10.1186/1477-7819-11-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramteke A., Ting H., Agarwal C., Mateen S., Somasagara R., Hussain A., et al. (2015). Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol. Carcinog. 54, 554–565. 10.1002/mc.22124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M., Bajo-Santos C., Hessvik N. P., Lorenz S., Fromm B., Berge V., et al. (2017). Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol. Cancer 16:156. 10.1186/s12943-017-0726-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue T., Koga H., Iwamoto H., Nakamura T., Ikezono Y., Abe M., et al. (2019). Glycosylation of ascites-derived exosomal CD133: a potential prognostic biomarker in patients with advanced pancreatic cancer. Med. Mol. Morphol. 52, 198–208. 10.1007/s00795-019-00218-5 [DOI] [PubMed] [Google Scholar]
- Salido-Guadarrama I., Romero-Cordoba S., Peralta-Zaragoza O., Hidalgo-Miranda A., Rodríguez-Dorantes M. (2014). MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. Onco. Targets. Ther. 7, 1327–1338. 10.2147/OTT.S61562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonov R., Shtam T., Burdakov V., Glotov A., Tsyrlina E., Berstein L., et al. (2016). Lectin-induced agglutination method of urinary exosomes isolation followed by mi-RNA analysis: application for prostate cancer diagnostic. Prostate 76, 68–79. 10.1002/pros.23101 [DOI] [PubMed] [Google Scholar]
- Sandfeld-Paulsen B., Aggerholm-Pedersen N., Baek R., Jakobsen K. R., Meldgaard P., Folkersen B. H., et al. (2016). Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 10, 1595–1602. 10.1016/j.molonc.2016.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Jiang Y., Yang L., Yan S., Wang Y. G., Lu X. J. (2018). Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell. Biochem. 119, 4711–4716. 10.1002/jcb.26650 [DOI] [PubMed] [Google Scholar]
- Shiromizu T., Kume H., Ishida M., Adachi J., Kano M., Matsubara H., et al. (2017). Quantitation of putative colorectal cancer biomarker candidates in serum extracellular vesicles by targeted proteomics. Sci. Rep. 7:12782. 10.1038/s41598-017-13092-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J., Würdinger T., van Rijn S., Meijer D., Gainche L., Sena-Esteves M., et al. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476. 10.1038/ncb1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotland T., Ekroos K., Kauhanen D., Simolin H., Seierstad T., Berge V., et al. (2017). Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 70, 122–132. 10.1016/j.ejca.2016.10.011 [DOI] [PubMed] [Google Scholar]
- Slomka A., Urban S. K., Lukacs-Kornek V., Zekanowska E., Kornek M. (2018). Large extracellular vesicles: have we found the holy grail of inflammation? Front. Immunol. 9:2723. 10.3389/fimmu.2018.02723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soekmadji C., Corcoran N. M., Oleinikova I., Jovanovic L., Australian Prostate Cancer Collaboration B., Ramm G. A., et al. (2017). Extracellular vesicles for personalized therapy decision support in advanced metastatic cancers and its potential impact for prostate cancer. Prostate 77, 1416–1423. 10.1002/pros.23403 [DOI] [PubMed] [Google Scholar]
- Sohn W., Kim J., Kang S. H., Yang S. R., Cho J. Y., Cho H. C., et al. (2015). Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp. Mol. Med. 47:e184. 10.1038/emm.2015.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squadrito M., Baer C., Burdet F., Maderna C., Gilfillan G., Lyle R., et al. (2014). Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 8, 1432–1446. 10.1016/j.celrep.2014.07.035 [DOI] [PubMed] [Google Scholar]
- Stam J., Bartel S., Bischoff R., Wolters J. C. (2021). Isolation of extracellular vesicles with combined enrichment methods. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1169:122604. 10.1016/j.jchromb.2021.122604 [DOI] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209-249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Sunkara V., Kim C. J., Park J., Woo H. K., Kim D., Ha H. K., et al. (2019). Fully automated, label-free isolation of extracellular vesicles from whole blood for cancer diagnosis and monitoring. Theranostics 9, 1851–1863. 10.7150/thno.32438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K., Iinuma H., Wada K., Minezaki S., Kawamura S., Kainuma M., et al. (2018). Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J. Hepatobiliary. Pancreat. Sci. 25, 155–161. 10.1002/jhbp.524 [DOI] [PubMed] [Google Scholar]
- Taplin S., Abraham L., Barlow W. E., Fenton J. J., Berns E. A., Carney P. A., et al. (2008). Mammography facility characteristics associated with interpretive accuracy of screening mammography. J. Natl. Cancer Inst. 100, 876–887. 10.1093/jnci/djn172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias A. (1999). Assessing the influence of a single study in the meta-anyalysis estimate. Stata Tech. Bull. 8, 7526–7529. [Google Scholar]
- Trino S., Lamorte D., Caivano A., De Luca L., Sgambato A., Laurenzana I. (2021). Clinical relevance of extracellular vesicles in hematological neoplasms: from liquid biopsy to cell biopsy. Leukemia 35, 661–678. 10.1038/s41375-020-01104-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto M., Iinuma H., Yagi T., Matsuda K., Hashiguchi Y. (2017). Circulating exosomal microRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology 92, 360–370. 10.1159/000463387 [DOI] [PubMed] [Google Scholar]
- Umwali Y., Yue C. B., Gabriel A. N. A., Zhang Y., Zhang X. (2021). Roles of exosomes in diagnosis and treatment of colorectal cancer. World J. Clin. Cases 9, 4467–4479. 10.12998/wjcc.v9.i18.4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S. D. (2002). Properties of the summary receiver operating characteristic (SROC) curve for diagnostic test data. Stat. Med. 21, 1237–1256. 10.1002/sim.1099 [DOI] [PubMed] [Google Scholar]
- Wang J., Zhou Y., Lu J., Sun Y., Xiao H., Liu M., et al. (2014). Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med. Oncol. 31:148. 10.1007/s12032-014-0148-8 [DOI] [PubMed] [Google Scholar]
- Wang M., Lv C. Y., Li S. A., Wang J. K., Luo W. Z., Zhao P. C., et al. (2021). Near infrared light fluorescence imaging-guided biomimetic nanoparticles of extracellular vesicles deliver indocyanine green and paclitaxel for hyperthermia combined with chemotherapy against glioma. J. Nanobiotechnol. 19:210. 10.1186/s12951-021-00907-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Wang L., Yang Y., Gong L., Xiao B., Liu X. (2017). A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem. Biophys. Res. Commun. 493, 1322–1328. 10.1016/j.bbrc.2017.10.003 [DOI] [PubMed] [Google Scholar]
- Wang X., Wang T., Chen C., Wu Z., Bai P., Li S., et al. (2018). Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. J. Cell. Biochem. 120, 1492-1502. 10.1002/jcb.27347 [DOI] [PubMed] [Google Scholar]
- Whiteside T. (2016). Tumor-derived exosomes and their role in cancer progression. Adv. Clin. Chem. 74, 103–141. 10.1016/bs.acc.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Wu K., Yan A., Wang J. G., Zhang Z., Li D. (2021). Intelligent probabilistic system for digital tracing cellular origin of individual clinical extracellular vesicles. Anal. Chem. 93, 10343–10350. 10.1021/acs.analchem.1c01971 [DOI] [PubMed] [Google Scholar]
- Xu S., Li J., Chen L., Guo L., Ye M., Wu Y., et al. (2019). Plasma miR-32 levels in non-small cell lung cancer patients receiving platinum-based chemotherapy can predict the effectiveness and prognosis of chemotherapy. Medicine (Baltimore) 98:e17335. 10.1097/MD.0000000000017335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Qin S., An T., Tang Y., Huang Y., Zheng L. (2017). MiR-145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate 77, 1167–1175. 10.1002/pros.23376 [DOI] [PubMed] [Google Scholar]
- Yan S., Dang G., Zhang X., Jin C., Qin L., Wang Y., et al. (2017). Downregulation of circulating exosomal miR-638 predicts poor prognosis in colon cancer patients. Oncotarget 8, 72220–72226. 10.18632/oncotarget.19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Jiang Y., Liang C., Cheng M., Jin C., Duan Q., et al. (2018a). Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J. Cell. Biochem. 119, 4113–4119. 10.1002/jcb.26609 [DOI] [PubMed] [Google Scholar]
- Yan S., Liu G., Jin C., Wang Z., Duan Q., Xu J., et al. (2018b). MicroRNA-6869-5p acts as a tumor suppressor via targeting TLR4/NF-kappaB signaling pathway in colorectal cancer. J. Cell. Physiol. 233, 6660–6668. 10.1002/jcp.26316 [DOI] [PubMed] [Google Scholar]
- Yan W., Apweiler R., Balgley B. M., Boontheung P., Bundy J. L., Cargile B. J., et al. (2009). Systematic comparison of the human saliva and plasma proteomes. Proteomics Clin. Appl. 3, 116–134. 10.1002/prca.200800140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Wang X. S., Cao K. C., Bao X. J., Yu J. (2019). Identification of CDK6 and RHOU in serum exosome as biomarkers for the invasiveness of non-functioning pituitary adenoma. Chin. Med. Sci. J. 34, 168–176. 10.24920/003585 [DOI] [PubMed] [Google Scholar]
- Zamora J., Abraira V., Muriel A., Khan K., Coomarasamy A. (2006). Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med. Res. Methodol. 6:31. 10.1186/1471-2288-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Ni M., Su Y., Wang H., Zhu S., Zhao A., et al. (2018). MicroRNAs in serum exosomes as potential biomarkers in clear-cell renal cell carcinoma. Eur. Urol. Focus 4, 412–419. 10.1016/j.euf.2016.09.007 [DOI] [PubMed] [Google Scholar]
- Zhou X., Wen W., Shan X., Zhu W., Xu J., Guo R., et al. (2017). A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget 8, 6513–6525. 10.18632/oncotarget.14311 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.