Abstract
Inv(16)(p13.1q22) in acute myeloid leukemia (AML) is a common chromosomal abnormality. It leads to the core-binding factor ß-subunit (CBFβ)/smooth muscle myosin heavy chain 11 (MYH11) fusion gene. Different breakpoints were observed in the CBFβ gene at 16q22 and the MYH11 gene at 16p13.1. For this reason, different CBFβ/MYH11 fusion genes are generated, with more than 13 types having been reported to date. Type I CBFβ/MYH11 fusion transcripts are very rare, with only 10 cases being reported to date. This case report describes a primary AML patient with inv(16)(p13.1q22) and a rare type I CBFβ/MYH11 fusion gene. The morphological analysis did not conform to the typical M4eo. Abnormal eosinophils were less than 5%, and there was obvious dysgranulopoiesis. The patient was in hematological and genetic remission for 487 days after the initial chemotherapy cycles. However, the CBFβ/MYH11 fusion had been constantly positive. Moreover, the presence of non-type A fusions may affect its biology and clinical prognosis. Therefore, further studies on understanding its biological and prognostic significance are essential.
Keywords: Acute myeloid leukemia, Type I CBFβ/MYH11, Inv(16)(p13.1q22), Case report
Introduction
Acute myeloid leukemia (AML) is a malignant clonal disease of hematopoietic stem/progenitor cells (HSPCs). The inv(16)(p13.1q22)/t(16;16)(p13.1;q22), CBFβ/MYH11 AML is an independent subtype in the WHO classification of AML. The inv(16)/t(16;16) produces CBFβ/MYH11 fusion gene, formed by the juxtaposition of a 5' sequence from CBFβ at 16q22 and the 3' sequence from MYH11 at 16p13.1. The fusion sites between the CBFβ and MYH11 genes usually differ, which results in the generation of different CBFβ/MYH11 transcripts. To date, at least thirteen different fusion transcripts have been reported (1- 119). The three most reported types include type A (79-87%), type E (5-9%), and type D (3-10%) (3,4). The remaining types have only been reported in case studies. The type I CBFβ/MYH11 fusion transcript has rarely been described, and to our knowledge, only 10 cases have been reported (1-7). This report describes a de novo AML patient with inv(16)(p13.1q22) showing a rare type I CBFβ/MYH11 fusion transcript.
Case Report
A 50-year-old male farmer complained of fatigue and chest tightness with no obvious cause since March 20, 2019. After resting, his condition improved slightly, but then gradually worsened. The patient visited a local hospital for diagnosis on April 2, 2019. Peripheral blood film (PBF) showed white blood cells (WBC) of 5.6×109/L, red blood cells (RBC) of 1.43×1012/L, hemoglobin (Hb) of 53.2 g/L, and platelets (PLT) of 8×109/L. The attending physician recommended a transfer to a tertiary hospital for diagnosis and treatment. The patient was transferred to our hospital on April 4, 2019. Physical examinations showed enlarged bilateral submandibular lymph nodes without hepatosplenomegaly. PBF indicated WBC of 4.20×109/L with 43% neutrophils, 26% lymphocytes, 26% monocytes, and 5% blasts. PBF also revealed RBC of 1.12×1012/L with anisocytosis, Hb of 41 g/L, and PLT of 6×109/L. Bone marrow (BM) aspirates revealed a significant increase in bone marrow cellularity; 18% were myeloblasts with regular or irregular nuclei, fine chromatin, visible nucleoli, and reduced blue plasma. Nuclear-cytoplasmic dyssynchrony, nuclear malformations, binuclear, and Pelger malformed granulocytes accounted for 13% of the granulocytes, and 3% abnormal eosinophils were observed. Nucleated red blood cells were significantly reduced and accounted for 1.5%. Petal-nucleated red blood cells and anisocytosis were also observed; 18% of the aspirates were monocytes. Two granular megakaryocytes were observed on the whole slide, and the number of platelets was significantly reduced. Results from the bone marrow biopsy showed hypercellularity of the bone marrow tissue, with an increase in immature cells. Immunophenotyping of bone marrow cells showed 5.65% blasts, expressing CD34, CD117, CD13, HLA-DR, lack of CD7, CD10, CD38, CD33, CD11C, CD11b, CD64, CD15, CD19, CD123, CD56, CD36, CDCR4, CD14, CD300e, CD4, and CD2 (Supplementary Figure S1).
This study was approved by the Ethics Committee of the Traditional Chinese Medical Hospital of Langfang City, China.
Conventional cytogenetics and fluorescent in situ hybridization
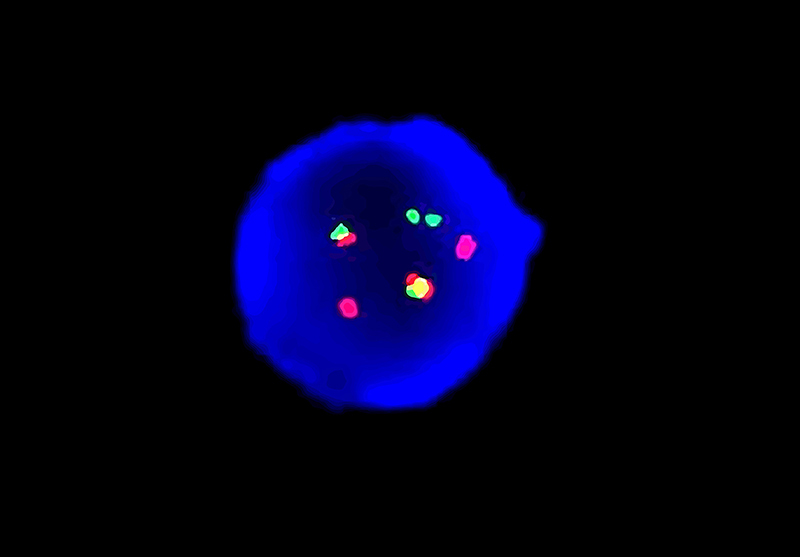
Bone marrow nucleated cells (1-3×106/mL) were cultured in Gibco bone marrow cell culture media (USA) for 48 h and then analyzed by G-banding based on an international system for Human Cytogenomic Nomenclature (ISCN 2016). Karyotype analysis demonstrated 46,XY,inv(16)(p13.1q22)[10] (Figure 1). Fluorescence in situ hybridization (FISH) was performed using the Vysis dual-color separation CBFβ probe (Abbott, USA), green fluorescent labeled 3'CBFβ (16q22) probe, and the red fluorescent-labeled 5'CBFβ probe. The rearrangement of the CBFβ gene fusion was detected on chr16 (Figure 2).
Figure 1. Representative karyotype showing 46,XY,inv(16)(p13.1q22)[10].
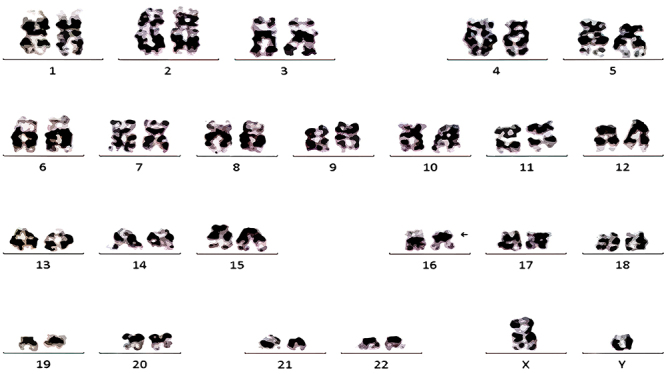
Figure 2. FISH result using the double color separation CBFβ probe (VYSIS), green fluorescent-labeled 3'CBFβ (16q22) probe, and red fluorescent-labeled 5'CBFβ probe. Split CBFβ abnormal signals on chromosome 16 (2R and 2G) are observed in interphase tetraploid cell nuclei.
Molecular analysis
RT-PCR was used to detect the following gene fusions based on a previous study(20). All of these genes were negative: BCR-ABL, SIL-TAL1, E2A-HLF, TEL-AML1, MLL-AF4, E2A-PBX1, AML1-ETO, MLL-AF9, PML-RARα, PLZF-RARα, STAT5b-RARα, MLL-AF6, MLL-AF10, MLL-ELL, MLL-ENL, NPM-MLF1, TEL-PDGFRB, FIP1L1-PDGFRA, AML1-MDS1/EVI1, DEK-CAN, TEL-ABL, ETV6-PDGFRA, NUP98-HoxA13, NUP98-HoxC11, NUP98-HoxD13, NUP98-HoxA9, NUP98- HoxA11, NUP98- PMX1, TEL-JAK2, MLL-AF17, MLL- AF1q, MLL- AF1p, MLL-AFX, MLL-SEPT6, (NPM, FIP1L1, PRKAR1A, NUMA1)-RARα, NPM-ALK, SET-CAN, TLS-ERG, and AML1-MTG16. WT1 was 11.7%. A rare fusion transcript of CBFβ/MYH11 was detected, and electrophoresis of the PCR amplified product showed a positive band between 200-300bp (Figure 3A). Additional analysis of the PCR amplified product by capillary sequencing was performed. The sequence was as follows: TTTCAGAATTTTGAAGGCTCCCATGATTCTGAATGGAGTCTGTGTTATCTGGAAAGGCTGGATTGATCTCCAAAGACTGGATGGTATGGGCTGTCTGGAGTTTGATGAGGAGCGAGCCCAGCTTCACGAGTATGAGACGGAACTGGAAGACGAGCGAAAGCAACGTGCCCTGGCAGCTGCAGCAAAGAAGAAGCTGGAAGGGGACCTCTTCTAAA.
Figure 3. Gel electrophoresis and sequence analysis of the CBFβ/MYH11 transcript. A, Gel electrophoresis 111/121 and 112/122 primer pairs used for amplification of the CBFβ/MYH11 transcript. E2A is the internal reference primer. M1 is the DNA molecular weight marker 1 (100-600 bp, Tiangen, China). 111/121 amplified product showed a specific band between 200-300bp. B, Sequence analysis of the CBFβ/MYH11 transcript showed a breakpoint between exon 4 of the CBFβ gene and exon 34 of the MYH11 gene.
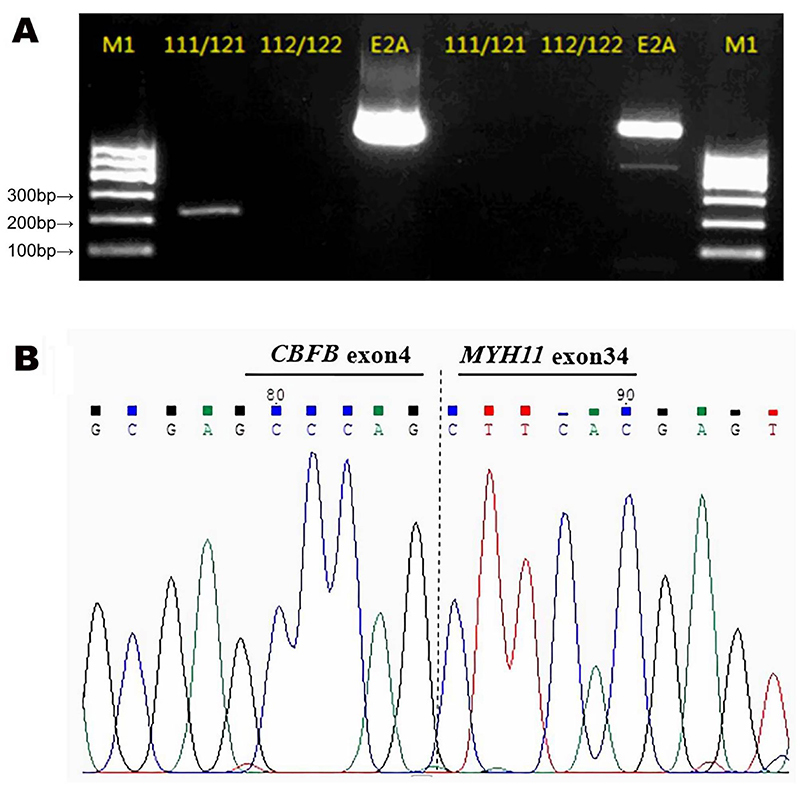
G denoted the end of the CBFβ exon4 and C denoted the beginning of the MYH11 exon34 according to NM_002474.3 and CCDS_10565.1. This confirmed that CBFβ exon4 and MYH11 exon34 were fused to form the fusion transcript. Comparing the CCDS sequence of the NCBI database, the CBFβ/MYH11 fusion transcript belonged to a rare type I fusion transcript (Figure 3B) (6,10,12). The patient was then diagnosed with inv(16)(p13.1q22); CBFβ/MYH11 (type I) AML. Next-generation sequencing (NGS) analysis was used to detect additional prognostic mutational genes, including FLT3, NPM1, KIT, CEBPA, DNMT3A, IDH1, IDH2, TET2, EZH2, AML1, ASXL1, PHF6, TP53, SF3B1, SRSF2, U2AF1, ZRSR2, NRAS, CBL, SETBP1, ETV6, and JAK2. No mutations in these genes were observed.
Treatment and response
The patient underwent chemotherapy with decitabine (DAC), daunorubicin (DNR), and cytarabine (Ara-C) (DAC: 50 mg qod×3; DNR: 40 mg d1-2, 60 mg d3; Ara-C: 200 mg d1-4, 150 mg d5, 200 mg d6-7). In addition, supportive treatment including heart, liver, and stomach protection, antiemetics, RBC, and platelet transfusions, was administered. On the 24th day after chemotherapy, BM cellularity was reduced, hence, supportive treatment was continued. On the 41st day, the cellular composition of the BM aspirates consisted of 10% myeloblasts, 11% monoblasts and pre-monocytes, and 2% abnormal eosinophils. The patient was administered a second cycle of chemotherapy with DAC, homoharringtonine (HHT), Ara-C, and etoposide (VP-16) (DAC: 50 mg qod×3; HHT: 2mg d1-5, 3mg d6-7; Ara-C: 150mg d1-7; VP16 0.1g, d1-7). In addition, supportive treatment was also administered. Seventy-five days after the initial chemotherapy, BM morphology returned to normal, and chromosomes were 46,XY(20). FISH showed CBFβ gene separation and rearrangement. Chemotherapy was continued, and 272 days after the initial chemotherapy, CBFβ gene separation and rearrangement was negative as determined by FISH, and the CBFβ/MYH11 fusion transcript showed positive. After 487 days of the initial chemotherapy, bone marrow morphology showed complete response (CR), with 46,XY(20) chromosomes. FISH was negative for CBFβ gene separation and rearrangement. However, the CBFβ/MYH11 fusion transcript was still observed. At present, the patient is still undergoing regular chemotherapy and follow-up.
Discussion
CBFβ/MYH11 fusions mainly manifest from inv(16)(p13.1q22), and at much lower levels with t(16;16)(p13.1;q22)(5,8). CBFβ/MYH11 fusions account for 5.04% of primary AML. The incidence of CBFβ/MYH11 fusions in infants and children decreases with age, while the fusions increase steadily with age in adults. Fusions are rare in patients 50 years of age and over (20).
CBFβ/MYH11 fusion transcripts are heterogeneous and depend on the different intron breakpoints between the exons of CBFβ and MYH11 genes. To date, over 13 types of fusions have been reported, the majority of which being type A transcripts, with fewer D and E types, while the other types are rare. Type I fusion transcripts are very rare, with only 10 cases reported in the literature to date. Of the 10 cases, only 6 cases have been reported in detail (1,2,5-7).
We treated a primary AML patient with a rare type I fusion transcript. Morphological analysis did not conform to the typical M4eo. Abnormal eosinophils were less than 5%, and there was obvious dysgranulopoiesis. Chromosome karyotype analysis and FISH assays confirmed the presence of inv(16)(p13.1q22). Gene sequencing showed a type I CBFβ/MYH11 fusion transcript. Type I, also known as type S/I, was first reported by Dissing et al. (1), followed by other cases (2- 7). Inv(16)(p13.1q22)/t(16;16)(p13.1;q22) AML with non-type A CBFβ/MYH11 fusion transcripts are more common in t-AML patients. Its occurrence has been associated with exposure to topoisomerase II inhibitors/topoisomerase I inhibitors (1,3,6,8). The other fusions that are rarer show a more atypical cytomorphology, mostly with pathologic eosinophils <5% and lower WBC counts (3,4,9). Atypical changes in chromosome numbers have been reported in the literature with chromosomes 8, 21, and 22. Several reports have suggested that the numerical gains in chromosomes 8, 21, and 22 occur mostly in patients with type A rather than rare fusion types (3). However, it has been reported that non-type A patients frequently have extra +8 and +21 chromosomes, with none having extra +22 chromosomes (4). No additional cytogenetic abnormalities and leukopenia were detected at the initial diagnosis in our patient. However, there were obvious abnormalities in granulocyte morphology. Among the reports published in patients with type I, there were 3 cases with t-AML and 5 cases with de novo AML (1,2,4-7).
The patient in this study had de novo AML. This suggests that type I is more common in patients with de novo AML. The type of CBFβ/MYH11 fusion transcript is not an independent prognostic factor. No significant differences in overall survival (OS) or event-free survival (EFS) were observed with the type of fusion (3). Previous studies observed no significant differences in CR rate, the cumulative incidence of relapse (CIR), and OS between non-type A patients and type A patients. However, non-type A patients had longer EFS compared to type A patients. This may be related to the presence of KIT mutations rather than the type of fusion transcript. KIT mutations were observed in 24% of type A patients and none in non-type A patients (4). In addition, previous case reports showed that non-type A patients had a high CR rate and a better prognosis (2,5,9,12,13,16). In our patient, next-generation sequencing showed no genetic mutations related to prognosis. After a limited number of chemotherapy cycles, the patient was in hematological and genetic remission 487 days after the initial chemotherapy. However, the CBFβ/MYH11 fusion had been always positive. The patient is currently undergoing regular chemotherapy and follow-up.
The limitation of this study is that the interphase FISH specimens could not be saved due to a long experimental time, and metaphase FISH could not be performed.
It is currently believed that the type of fusion has no effect on the prognosis of patients with inv(16)(p13.1q22)/t(16;16)(p13.1;q22). However, the presence of non-type A fusions, related distinctive clinical and genetic characteristics, and unique gene expression profiles may affect its biology and clinical outcome. Due to the limited number of CBFβ/MYH11 fusion types, understanding its biological and prognostic significance is challenging.
Supplementary Material.
Click here to view [pdf].
References
- 1.Dissing M, Le Beau MM, Pedersen-Bjergaard J. Inversion of chromosome 16 and uncommon rearrangements of the CBFB and MYH11 genes in therapy-related acute myeloid leukemia: rare events related to DNA-topoisomerase II inhibitors? J Clin Oncol. 1998;16:1890–1896. doi: 10.1200/JCO.1998.16.5.1890. [DOI] [PubMed] [Google Scholar]
- 2.Grardel N, Roumier C, Soenen V, Lai JL, Plantier I, Gheveart C, et al. Acute myeloblastic leukemia (AML) with inv (16)(p13;q22) and the rare I type CBFbeta-MYH11 transcript: report of two new cases. Leukemia. 2002;16:150–151. doi: 10.1038/sj.leu.2402332. [DOI] [PubMed] [Google Scholar]
- 3.Schnittger S, Bacher U, Haferlach C, Kern W, Haferlach T. Rare CBFB-MYH11 fusion transcripts in AML with inv(16)/t(16;16) are associated with therapy-related AML M4eo, atypical cytomorphology, atypical immunophenotype, atypical additional chromosomal rearrangements, and low white blood cell count: a study on 162 patients. Leukemia. 2007;21:725–731. doi: 10.1038/sj.leu.2404531. [DOI] [PubMed] [Google Scholar]
- 4.Schwind S, Edwards CG, Nicolet D, Mrózek K, Maharry K, Wu YZ, et al. inv(16)/t(16;16) acute myeloid leukemia with non-type A CBFB-MYH11 fusions associate with distinct clinical and genetic features and lack KIT mutations. Blood. 2013;121:385–391. doi: 10.1182/blood-2012-07-442772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poddighe PJ, Veening MA, Mansur MB, Loonen AH, Westers TM, Merle PA, et al. A novel cryptic CBFB-MYH11 gene fusion present at birth leading to acute myeloid leukemia and allowing molecular monitoring for minimal residual disease. Hum Pathol Case Rep. 2018;11:34–38. doi: 10.1016/j.ehpc.2017.09.001. [DOI] [Google Scholar]
- 6.Yamamoto K, Nishikawa S, Minagawa K, Yakushijin K, Okamura A, Matsui T. Therapy-related myelodysplastic syndrome with inv(16)(p13q22) and I type CBFβ/MYH11 after autologous transplantation: Undetectable fusion transcript in pretransplant progenitor cells. Leuk Res. 2006;30:354–361. doi: 10.1016/j.leukres.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Van der Reijden BA, de Wit L, van der Poel S, Luiten EB, Lafage-Pochitaloff M, Dastugue N, et al. Identification of a novel CBFB-MYH11 transcript: implications for RT-PCR diagnosis. Hematol J. 2001;2:206–209. doi: 10.1038/sj.thj.6200103. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama H, Yamamoto M, Sakashita C, Umezawa Y, Kurosu T, Murakami N, et al. Therapy-related leukemia with Inv(16)(p13.1q22) and type D CBFB/MYH11 developing after exposure to irinotecan-containing chemoradiotherapy. Intern Med. 2015;54:651–655. doi: 10.2169/internalmedicine.54.3535. [DOI] [PubMed] [Google Scholar]
- 9.Albano F, Anelli L, Zagaria A, Coccaro N, Tota G, Impera L, et al. Acute myeloid leukemia with t(16;16) (p13;q22) showing a new CBFB-MYH11 fusion transcript associated with an atypical leukemic blasts morphology. Hum Pathol. 2014;45:643–647. doi: 10.1016/j.humpath.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 10.van Dongen JJ, Macintyre EA, Gabert JA, Delabesse E, Rossi V, Saglio G, et al. Standardized RT-PCR analysis of fusion gene transcripts from chromosome aberrations in acute leukemia for detection of minimal residual disease. Report of the BIOMED-1 Concerted Action: investigation of minimal residual disease in acute leukemia. Leukemia. 1999;13:1901–1928. doi: 10.1038/sj.leu.2401592. [DOI] [PubMed] [Google Scholar]
- 11.Kurata K, Yamamoto K, Okazaki Y, Noguchi Y, Matsui K, Matsumoto H, et al. Detection of a novel CBFB-MYH11 fusion transcript in acute myeloid leukemia M1 with inv(16)(p13q22) Cancer Genet. 2020;241:72–76. doi: 10.1016/j.cancergen.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 12.Rowe D, Strain L, Lowe C, Jones G. A case of acute myeloid leukemia with inv(16)(p13q22) reveals a novel MYH11 breakpoint and a new CBF beta-MYH11 transcript variant. Haematologica. 2007;92:1433–1434. doi: 10.3324/haematol.11536. [DOI] [PubMed] [Google Scholar]
- 13.Park TS, Lee ST, Song J, Lee KA, Lee JH, Kim J, et al. Detection of a novel CBFB/MYH11 variant fusion transcript (K-type) showing partial insertion of exon 6 of CBFB gene using two commercially available multiplex RT-PCR kits. Cancer Genet Cytogenet. 2009;189:87–92. doi: 10.1016/j.cancergencyto.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Shurtleff SA, Meyers S, Hiebert SW, Raimondi SC, Head DR, Willman CL, et al. Heterogeneity in CBF beta/MYH11 fusion messages encoded by the inv(16)(p13q22) and the t(16;16)(p13;q22) in acute myelogenous leukemia. Blood. 1995;85:3695–3703. doi: 10.1182/blood.V85.12.3695.bloodjournal85123695. [DOI] [PubMed] [Google Scholar]
- 15.Opatz S, Bamopoulos SA, Metzeler KH, Herold T, Ksienzyk B, Bräundl K, et al. The clinical mutatome of core binding factor leukemia. Leukemia. 2020;34:1553–1562. doi: 10.1038/s41375-019-0697-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobayashi T, Ichikawa M, Kamikubo Y, Kurokawa M. Acute myeloid leukemia with cryptic CBFB-MYH11 type D. Int J Clin Exp Pathol. 2013;6:110–112. [PMC free article] [PubMed] [Google Scholar]
- 17.Stulberg J, Kamel-Reid S, Chun K, Tokunaga J, Wells RA. Molecular analysis of a new variant of the CBF beta-MYH11 gene fusion. Leuk Lymphoma. 2002;43:2021–2026. doi: 10.1080/1042819021000015989-1. [DOI] [PubMed] [Google Scholar]
- 18.Martinelli G, Ottaviani E, Buonamici S, Isidori A, Malagola M, Piccaluga P, et al. Two more inv(16) acute myeloid leukemia cases with infrequent CBFbeta-MYH11 fusion transcript: clinical and molecular findings. Haematologica. 2002;87:554–555. [PubMed] [Google Scholar]
- 19.Liu PP, Hajra A, Wijmenga C, Collins FS. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia. Blood. 1995;85:2289–2302. doi: 10.1182/blood.V85.9.2289.bloodjournal8592289. [DOI] [PubMed] [Google Scholar]
- 20.Chen X, Wang F, Zhang Y, Wang M, Tian W, Teng W, et al. Panoramic view of common fusion genes in a large cohort of Chinese de novo acute myeloid leukemia patients. Leuk Lymphoma. 2019;60:1071–1078. doi: 10.1080/10428194.2018.1516876. [DOI] [PubMed] [Google Scholar]


