Summary
Pathogenic viruses induce metabolic changes in host cells to secure the availability of biomolecules and energy to propagate. Influenza A virus (IAV) and severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) both infect the human airway epithelium and are important human pathogens. The metabolic changes induced by these viruses in a physiologically relevant human model and how this affects innate immune responses to limit viral propagation are not well known. Using an ex vivo model of pseudostratified primary human airway epithelium, we here demonstrate that infection with both IAV and SARS-CoV-2 resulted in distinct metabolic changes including increases in lactate dehydrogenase A (LDHA) expression and LDHA-mediated lactate formation. Interestingly, LDHA regulated both basal and induced mitochondrial anti-viral signaling protein (MAVS)-dependent type I interferon (IFN) responses to promote IAV, but not SARS-CoV-2, replication. Our data demonstrate that LDHA and lactate promote IAV but not SARS-CoV-2 replication by inhibiting MAVS-dependent induction of type I IFN in primary human airway epithelium.
Subject areas: Immune response, Virology, Metabolomics
Graphical abstract
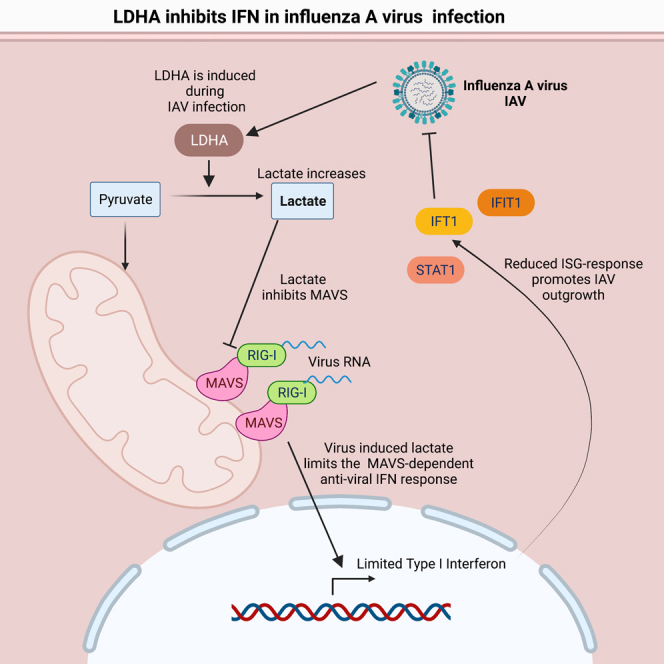
Highlights
-
•
IAV and SARS-CoV-2 infections yield virus-specific changes in glucose metabolism
-
•
IAV and SARS-CoV-2 induce LDHA and lactate production in human airway epithelium
-
•
Lactate is highly pro-viral to IAV but not to SARS-CoV-2
-
•
Lactate decreases IFN production and the following ISG response
Immune response; Virology; Metabolomics
Introduction
Amplified glycolysis accompanied by increases in lactate release is a distinct feature of many cancer cells and is known as the Warburg effect or metabolic reprogramming (DeBeradinis and Thompson, 2012; Parks et al., 2013). Moreover, it is now clear that metabolic reprogramming is an integrated part of innate immune responses to microbial infection. Here, stimulation of immune cells with toll-like receptor (TLR) ligands initiates a Warburg-like shift in cellular metabolism with increases in glycolysis and interference of the tricarboxylic acid (TCA) cycle with subsequent accumulation of distinct metabolites (Liu et al., 2021). As examples hereof, the metabolite itaconate is produced from cis-aconitate through a process that is enhanced by TLR stimuli and catalyzed by the enzyme aconitate decarboxylase (ADOC1) also known as immune-related gene 1 protein (IRG1) in macrophages (Michelucci et al., 2013; Cordes et al., 2016; Lampropoulou et al., 2016). Itaconate and its cell-permeable derivative 4-octyl-itaconate have both been shown to have anti-inflammatory properties. 4-Octyl itaconate has been demonstrated to have anti-inflammatory capacities through its binding to Kelch-like ECH-associated protein 1 (KEAP1)—the suppressor of nuclear factor erythroid 2-related factor 2 (NRF2) (Lampropoulou et al., 2016; Mills et al., 2018). The mechanism behind this suppression seems to be at least partly due to an NRF2-mediated suppression of IL-1β (Bambouskova et al., 2021) and of stimulator of interferon signaling (STING) (Mills et al., 2018; Olagnier et al., 2018).
Recent advances within this area of research have demonstrated that metabolic reprogramming is not reserved for stimulated immune cells. Human pathogenic viruses induce metabolic reprogramming in their target cells to secure the availability of metabolites and energy consequently preserving carbon-carbon bonds to promote anabolism and production of progeny virus (Thyrsted and Holm, 2021a, 2021b). Interestingly, in addition to securing viral propagation, evidence is emerging that metabolic reprogramming can be used actively by the virus to regulate anti-viral immunity. This could be the case for the metabolite lactate, which has recently been added to the list of metabolites that directly affect cellular innate immune responses. More precisely, lactate inhibits the release of type I interferons (IFNs) induced by cytosolic dsRNA (Zhang et al., 2019). Mechanistically, lactate produced by lactate dehydrogenase A (LDHA) directly interacts with the adaptor molecule mitochondrial anti-viral signaling protein mitochondrial anti-viral signaling protein (MAVS), which signals downstream of the cytosolic RNA sensors retinoic-acid-inducible gene I (RIG-I) and melanoma-differentiation-associated protein 5 (MDA-5) (Seth et al., 2005). The binding of lactate to MAVS decreases the association between RIG-I and MAVS and prevents MAVS aggregation, which is important for downstream MAVS signaling (Zhang et al., 2019). Recently, lactate was demonstrated to play a role during infections with the human pathogen Hepatitis B virus (Zhou et al., 2021) in primary hepatic cell lines. If the immunoregulatory properties of lactate are important in pulmonary infections such as influenza A virus (IAV) or severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) in a physiological setting is, however, not yet explored.
Influenza viruses are important human pathogens causing 5-700.000 deaths and several million hospitalizations and have tremendous negative impact on public health worldwide annually (Cassini et al., 2018). Human pathogenic influenza types include influenza A, B, and C and belong to the family Orthomyxoviridae of enveloped negative-strand RNA viruses, with IAV currently considered the most important human pathogen. SARS-CoV-2 is a newly emerged member of the enveloped, positive-strand RNA virus family of Coronaviridae. Discovered in Wuhan in 2019, SARS-CoV-2 has since caused a worldwide pandemic of severe acute respiratory disease, overtaking SARS and Middle East respiratory syndrome (MERS) as the Coronaviridae with the highest number of deaths worldwide. IAV and SARS-CoV-2 both replicate in the epithelial cells of the upper and lower airways. To counter viral infection, the airways are equipped with several defense mechanisms including the ability to release anti-viral type I and type III IFNs. Release of IFNs in response to RNA viruses is mainly through the engagement of the RIG-I/MDA-5 pathway (Opitz et al., 2007; Loo et al., 2008) or in dendritic cells by toll-like receptor 7 (TLR7) which signals through MyD88 (Diebold et al., 2004). Deficiencies in IFN signaling lead to severe and often fatal progression of viral infections, highlighting the importance of IFNs in fighting viral infections (Levin and Hahn, 1985).
Here we utilize a pseudostratified ex vivo model of primary human airway epithelium (HPAE-model) (Gray et al., 1996; Pezzulo et al., 2011) to investigate the metabolic changes that occur during infection with IAV and SARS-CoV-2 in a physiologically relevant setting. Additionally, this model was also used to determine whether such metabolic reprogramming regulate anti-viral immunity.
We demonstrate that IAV and SARS-Cov-2 induce distinct metabolic reprogramming patterns, which include increases in LDHA expression and consequently increases in lactate formation. Furthermore, we show that LDHA inhibits the release of type I IFNs as a consequence of lactate formation to promote viral outgrowth of IAV but not of SARS-CoV-2.
Results
Infection of primary human epithelial cells with IAV or SARS-CoV-2 affects glucose metabolism
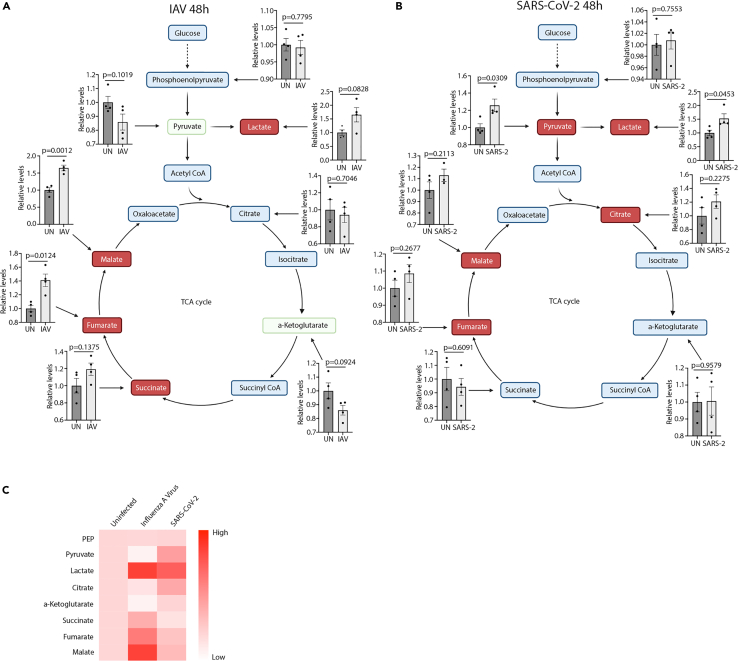
To investigate the metabolic responses to infections with IAV or SARS-CoV-2, we utilized human primary airway epithelium (HPAE-model) using the air-liquid interphase (ALI) setup (de Jong et al., 1994) (Figure S1A). This model allows for establishment of a fully differentiated human airway epithelium via the isolation of epithelial and basal cells from the nasal cavity of healthy donors. Initially, we infected HPAE cultures with either IAV (PR8) or SARS-CoV-2. Spent media samples were used to perform a large-scale metabolomic screen using LC-MS at 48 hours (Table S1) and 96 hours (Table S1) postinfection. Here, we found big changes in key carbohydrate metabolites in infected cells compared to uninfected controls. At 48 hours of infection, distinct changes in several intermediates in both glycolysis and the citric acid cycle (Figures 1A–1C) were observed in response to infection. IAV infection (Figure 1A) led to increased formation of lactate, succinate, fumarate, and malate, while the amounts of pyruvate and alpha-ketoglutarate were decreased. We detected no distinct changes to phosphoenolpyruvate and citrate. These results indicated an increased metabolic flux from pyruvate to lactate and from alpha-ketoglutarate to succinate, fumarate, and malate. Additionally, increased amounts of glutamate, glutamine, and glutathione during IAV infection were observed, suggesting increased glutamine anaplerosis (Figures S1B–S1D). The changes to glycolysis and citric acid cycle metabolites appeared to follow distinct kinetics as there were no observable differences in pyruvate, succinate, and fumarate levels, while lactate concentration was decreased after 96 hours of infection (Figures S1E and S1F). The changes observed during IAV infection were in contrast to those in SARS-CoV-2 infected cells where we observed increased amounts of pyruvate, lactate, citrate, and fumarate, while there were no observed changes in the abundance of phosphoenolpyruvate, alpha-ketoglutarate, succinate, and malate (Figure 1B). At 96 h however, amounts of alpha-ketoglutarate and malate were elevated along with a decrease in succinate (Figures S1E and S1F). As glutamine levels were also decreased in SARS-CoV-2-infected cultures, increases in alpha-ketoglutarate could stem from an increased influx through glutaminolysis (Figure S1G). Together, these results indicate changes in carbohydrate metabolism with virus-specific kinetics.
Figure 1.
IAV and SARS-CoV-2 infection cause specific changes in abundance of key carbohydrate metabolites
See also Figure S1 and Tables S1 and S2.
(A and B) HPAE-ALI cultures either uninfected or infected with IAV (A) or SARS-CoV-2 (B) for 48 hours. Measurements conducted on baso-lateral medium by MS-Omics.
(C) Heatmap presentations of data shown in (A) and (B). Data displayed in each figure represent one experiment with n = 4. Error bars represent standard error of the mean (s.e.m.). Unless otherwise stated, statistical analyses by Welch's t test.
IAV and SARS-CoV-2 induce increased LDHA activity and lactate production in human airway epithelium
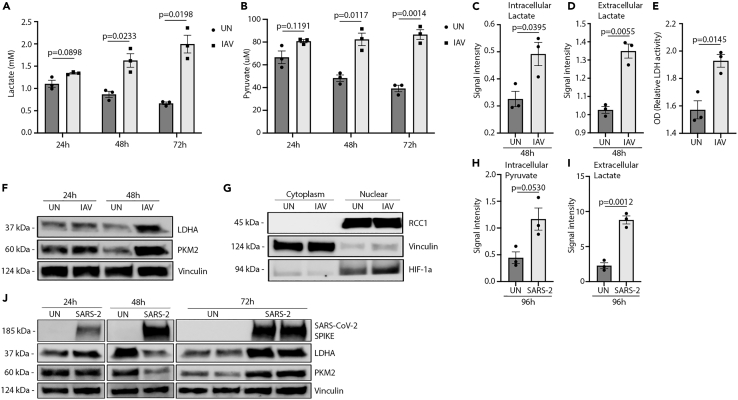
As analysis by mass spectrometry indicated that increased lactate production was shared between IAV and SARS-CoV-2, we aimed to validate these findings and to investigate the underlying mechanisms for this increase. Initially, we infected HPAE cultures with IAV and measured extracellular levels of lactate and pyruvate after 24, 48, and 72 hours by microdialysis. Interestingly, over the course of infection, we could detect a large increase in lactate secretion (Figure 2A) to the cell supernatant accompanied by an increased secretion of pyruvate (Figure 2B). These observations were confirmed by gas chromatography mass spectrometry (GC-MS) which showed increased lactate in the cellular compartment along with increased extracellular lactate at 48 hours (Figures 2C and 2D). In parallel, using an assay to determine enzymatic activity, we could observe increased cellular enzymatic LDH activity in the HPAE cultures infected with IAV (Figure 2E), explaining the increased formation of lactate. Following the observations of increased amounts of lactate and pyruvate production in infected HPAE cultures, we wanted to investigate the expression of LDHA and pyruvate kinase isozyme M2 (PKM2) as these factors are known regulators of lactate (Uyeda, 2013; Adeva-Andany et al., 2014). Here, we observed an increase in both LDHA and PKM2 expression after 24 hours of infection compared with un-infected cells (Figure 2F). Of note, this increase was even more pronounced at 48 hours, again supporting the increased production of lactate and pyruvate. These changes did not seem to be inducible by the stimulation of RNA sensors as treatment of cells with poly I:C did not lead to observed changes in LDHA expression (Figure S2A). Subcellular fractionations further revealed an increase in nuclear hypoxia-inducible factor 1-alpha (HIF-1α), an important transcription factor known to be involved in lipopolysaccharide (LPS)-induced metabolic reprogramming, in IAV-infected A549 cells compared with infected controls (Figure 2G). The changes in pyruvate and lactate were not distinct to IAV as infection with SARS-CoV-2 resulted in highly increased amounts of intracellular pyruvate (Figure 2H) along with increased amounts of extracellular lactate (Figure 2I). Investigating total protein expression, we observed no changes in LDHA and PKM2 expression at 24 hours postinfection, while we observed a decrease at 48 hours countered by a large increase in the expression of both proteins at 72 hours postinfection (Figure 2J). Together, these results demonstrate that the infection of HPAE cultures with IAV and SARS-CoV-2 induce increases in LDHA and PKM2 expression accompanied by increased production of pyruvate and lactate.
Figure 2.
IAV infection induces a metabolic shift in human primary airway cultures
See also Figures S2 and S3.
(A and B) Metabolite measurements on HPAE-ALI cultures infected with 60 HAU/106 cells IAV infected for 72 hours.
(C and D) HAE-ALI cultures infected with 300 HAU/106 cells IAV. Intracellular and extracellular lactate measured by GC-MS and represented as the area under the curve (AUC).
(E) Cell cultures from (A and B) harvested at 72 h and examined for LDH activity.
(F) Western blot of HPAE-ALI cells infected with 300 HAU/106 cells IAV and harvested at 24 or 48 h.
(G) Western blot of A549 cells infected with 120 HAU/106 IAV for 24 h and fractionated intro cytoplasmic and nuclear fractions. One band on each side was cropped out from the original blot as these were non-essential to this figure.
(H and I) HPAE-ALI cultures infected with MOI 0.5 of SARS-CoV-2 for 96 hours. Intracellular levels of pyruvate and extracellular lactate measured by GC-MS and represented as the area under the curve.
(J) Western blot of HPAE-ALI cells infected with MOI 0.05 of SARS-CoV-2 for 24, 48, and 72 hours. Data displayed in (A, B, E) represent one experiment with n = 3, (C and D) one experiment with n = 3, (F) multiple experiments, (G) repeated twice, and (H–J) one experiment. Error bars represent standard error of the mean (s.e.m.). Unless otherwise stated, statistical analyses by Welch's t test.
Lactate promotes replication of IAV, but not SARS-CoV-2, in human airway epithelium
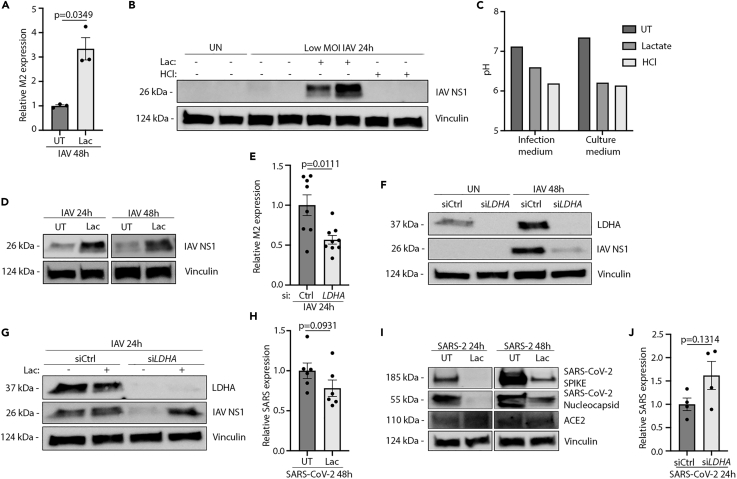
To test if lactate can affect IAV replication, we first infected HPAE cultures either in the presence or in the absence of lactate in the cell culture medium. Here, we observed that IAV-infected HPAE cultures pretreated with lactate contained more viral M2 RNA than untreated controls after 48 hours of infection (Figure 3A). This was backed by increased expression of the viral NS1 protein in lactate-treated cultures as determined by immunoblotting (Figure 3B). As IAV is dependent on acidification of endosomes for the intracellular release of the viral genome (Li et al., 2014), we aimed at investigating if a decrease in pH of the extracellular space was sufficient to promote viral replication. To do this, we used hydrochloric acid (HCl) in concentrations closely mimicking the pH drop seen when using lactate. Notably, treatment with HCl did not lead to increases in IAV replication, indicating that shifts in extracellular pH alone are not sufficient to promote viral outgrowth (Figure 3C). The pro-viral effects of lactate were detectable as early as 24 hours postinfection and still present at 48 hours postinfection (Figure 3D). The effects of lactate pre-treatment were dose dependent as increased concentrations yielded increased viral RNA and protein expression (Figures S2B–S2D). To ensure changes in viral titer were not owing to changes in the structural integrity or lactate-induced cell death of our HPAE cultures, we investigated the cultures for macroscopical changes, changes in barrier integrity, and overall survival via flow cytometry. Pre-treatment with lactate did not precipitate structural changes or changes in barrier integrity at 10 and 21 hours posttreatment (Figures S3A and S3B). Infection with IAV yielded a higher flux than uninfected. A similar pattern was seen in the pre-treated and infected cultures (Figures S3A and S3B). Additionally, we found no change in overall survival during lactate treatment compared with untreated cultures (Figure S3C), nor did we find changes in cell death during infections with neither IAV (Figure S3D) nor SARS-CoV-2 (Figure S3E). As the presence of lactate increased IAV outgrowth in HPAE cultures and because IAV induced LDHA expression and lactate secretion, we hypothesized that the expression of LDHA is important for IAV replication. To test this hypothesis, we first silenced LDHA in non-differentiated primary basal airway epithelial cells using siRNA (Figures 2E–2G). Interestingly, we found that the cells treated with LDHA siRNA (siLDHA) contained lower levels of IAV M2 RNA (Figure 3E) along with a decreased expression of NS1 protein (Figure 3F) than cells treated with control siRNA (siCtrl). Replenishing lactate in siLDHA-treated cells reversed the effects of LDHA silencing (Figure 3G), bringing the level of viral NS1 protein back to the levels of siCtrl cells—further indicating the importance for IAV of LDHA during infection and also demonstrating that most, if not all, of the pro-viral effect of LDHA is mediated through lactate formation. As we observed substantial pro-viral effects of lactate in HPAE-ALI cultures infected with IAV, we also investigated the effects of lactate on SARS-CoV-2 replication. Surprisingly, we found that pretreatment with lactate limited SARS-CoV-2 replication as determined by quantitative polymerase chain reaction (qPCR) (Figure 3H) and immunoblotting for SARS-CoV-2 SPIKE and nucleocapsid protein (Figure 3I). Furthermore, in contrast to IAV infection, silencing of LDHA before SARS-CoV-2 infection led to increased levels of SARS-CoV-2 RNA (Figure 3J). Collectively, these data suggest a highly pro-viral role of LDHA and lactate in IAV but not in SARS-CoV-2 infections.
Figure 3.
Lactate pretreatment is pro-viral to IAV but not to SARS-CoV-2
See also Figure S3.
(A) RT-qPCR for IAV M2 RNA in HPAE basal cell cultures either UT or pre-treated with 10 mM lactate and infected with 300 HAU/106 cells IAV for 48 hours.
(B) WB for IAV NS1 protein in HPAE-ALI cultures either untreated or pretreated with 10 mM Lactate or 10 mM hydrochloric acid and infected with 60 HAU/106 cells IAV.
(C) pH values from pretreatment in (B).
(D) Western blot for HPAE-ALI cultures either untreated or pre-treated with 10 mM lactate and infected with 120 HAU/106 cells IAV.
(E) RT-qPCR for IAV M2 on siRNA-treated HPAE basal cell cultures infected with IAV at 300 HAU/106 cells for 24 h.
(F) WB for IAV NS1 on siRNA-treated HPAE basal cell cultures infected with 300 HAU/106 cells IAV for 48 h.
(G) WB for IAV NS1 on siRNA-treated HPAE basal cell cultures pretreated with 10 mM lactate and infected with 300 HAU/106 cells IAV for 24 h.
(H) RT-qPCR for SARS-CoV-2 RNA (NR) in HPAE-ALI cultures either UT or pre-treated with 10 mM lactate and infected with MOI 0.05 SARS-CoV-2 for 48 hours.
(I) WB for SARS-CoV-2 Spike protein in HPAE-ALI cultures either UT or pre-treated with 10 mM lactate and infected with MOI 0.05 SARS-CoV-2 for 48 hours.
(J) RT-qPCR for SARS-CoV-2 RNA (NR) on siRNA-treated HPAE basal cell cultures infected with MOI 0.05 SARS-CoV-2 for 24 h. Data displayed represents; (A) two independent experiments with n = 3, (B–D) multiple experiments, (E) one experiment with n = 3, (F) two independent experiments, (G) one experiment, (H) two independent experiments with n = 6, (I) one experiment, (J) one experiment with n = 4. Error bars represent standard error of the mean (s.e.m.). Unless otherwise stated, statistical analyses by Welch's t test.
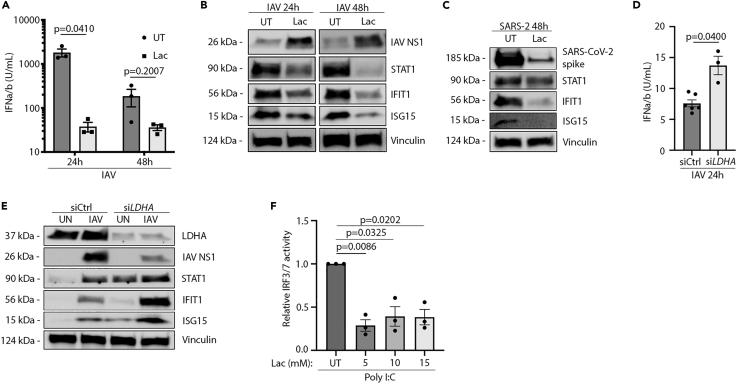
Lactate is a regulator of the IFN response to IAV infection
A recent report by Zhang et al. documents that lactate inhibits IFN formation in response to cytosolic RNA. For this reason, we wanted to investigate if the increase in LDHA expression and lactate formation could regulate the IFN response to influenza virus infection in the human airway epithelium. To test this, we infected HPAE cultures that were either untreated or pre-treated with lactate. Interestingly, type I IFN levels were clearly decreased in cells pre-treated with lactate compared with untreated controls (Figure 4A). Furthermore, the decrease in IFN production was of sufficient magnitude to affect the expression of known IFN-inducible genes (ISGs) including STAT1, IFIT1, and ISG15 as measured by Western blot (Figure 4B). Interestingly, dampening of the IFN response with lactate pre-treatment was not sufficient to promote SARS-CoV-2 replication (Figure 4C). Following these observations, we wanted to investigate whether or not endogenous LDHA activity is sufficient to inhibit the IFN response to IAV infection and if this could explain the changes in viral replication. By siRNA-mediated knockdown, we could observe that cells lacking LDHA expression had a higher IFN response to IAV infection than wild-type cells expressing LDHA (Figure 4D). Interestingly, when we compared the protein expression in uninfected cultures with siRNA-mediated knockdown of LDHA with siRNA-control-treated cells, the basal expression of a series of ISGs were increased when LDHA expression was inhibited (Figure 4E). This indicates an important role of LDHA in controlling basal IFN levels and thus control of ISG expression at homeostasis.
Figure 4.
Lactate decreases IFN production and ISG expression in human airway epithelium
See also Figure S4.
(A) HEK-Blue IFN bioassay on supernatant from 60 HAU/106 cells IAV infected HPAE-ALI cells either pre-treated with 10 mM lactate for 1 hour or untreated.
(B) Western blot on HPAE-ALI cells either untreated or pretreated for 1 hour with 10 mM lactate followed by infection with 120 HAU/106 cells IAV.
(C) Western blot on HPAE-ALI cells either untreated or pretreated for 1 hours with 10 mM lactate followed by infection with MOI 0.05 SARS-CoV-2.
(D) HEK-Blue IFN bioassay on siRNA-treated HPAE basal cell cultures infected with 300 HAU/106 cells IAV for 24 h or 48 h.
(E) Western blot on siRNA-treated HPAE basal cell cultures either pretreated for 1 hour with 10 mM lactate or left untreated followed by infection with 300 HAU/106 cells IAV for 24 h.
(F) A549-Dual reporter cell line either pretreated with 10 mM lactate for 1 hour or left untreated followed by stimulation with transfected Poly I:C. Data displayed represents; (A) one experiment with n = 3, (B) multiple experiments, (C) one experiment, (D) one experiment, (E) multiple experiments, (F) two independent experiments with n = 3. Error bars represent standard error of the mean (s.e.m.). Unless otherwise stated, statistical analyses by Welch's t test.
When infected, we again observed a lower expression of viral NS1 protein in siLDHA-treated cells along with highly increased protein levels of the ISGs STAT1, IFIT1, and ISG15 (Figure 4E) when compared with siCtrl cells. To further investigate the effects of lactate on IFN signaling, we utilized A549 dual reporter cells. Here, we saw that pre-treatment with lactate followed by transfected Poly I:C treatment yielded significantly lower IRF3/IRF7 activity, thus hinting at an inhibitory effect upstream of IRF3/7 (Figure 4F). Additionally, in THP-1 WT cells, the decrease in IFN signaling was unique to lactate (Figures S4A–S4C) and could not be observed to the same extent with HCl pre-treatment (Figures S4D–S4F). Furthermore, the effects of lactate were unique for RNA sensing as the ISG patterns of dsDNA treatment were unchanged with lactate treatment (Figures S4G and S4H). The unchanged pattern of ISG expression with Poly I:C treatment could be due to some signaling through TLR3 or an indication of large amounts of RNA being able to overcome the regulatory effects of LDHA and lactate. These results hint to an important interplay between LDHA and IFN signaling, both in homeostasis and during infection. Together, these data indicate a role of lactate as a regulator of the IFN response in human airway epithelium both during steady state and during infections with IAV.
Discussion
Increased blood lactate levels have long been associated with increased severity and mortality during viral infections. In fact, blood LDH levels are directly associated with poor outcome in patients with viral infections (Trzeciak et al., 2007). In this study, we demonstrate that lactate produced by LDHA functions as a regulator of the IFN response during infection with IAV. This matches the observations made by Zhang et al. who demonstrated lactate as a natural suppressor of RLR signaling by targeting MAVS (Zhang et al., 2019). In accordance, we find that pre-treatment with lactate causes significant decreases in IRF3 signaling along with decreased type I IFN production. Additionally, knockdown of LDHA proved highly anti-viral toward IAV and resulted in increased production of type I IFN. These effects were evident not only during infection but also during homeostasis. Our results thus demonstrate a role of LDHA in regulating both induced and basal IFN responses. These observations point toward LDHA and lactate as central regulators of IFN responses. Furthermore, we find that increased concentrations of extracellular lactate are highly pro-viral for IAV but not for SARS-CoV-2. In fact, increased lactate appears to instead limit replication of SARS-CoV-2. One explanation could be that SARS-CoV-2 infection induces relatively low amounts of IFN (Dalskov et al., 2020). This would make the inhibitory effects of lactate on IFN less important for SARS-CoV-2 than for IAV, which induce high levels of IFN.
Altogether, this study highlights the metabolic changes occurring during infection of the human airway epithelium with IAV or SARS-CoV-2 virus and how these changes can affect the replication of these viruses. Using the highly relevant HPAE-ALI model, we identify specific changes to glucose metabolism and intermediate metabolite amount in the extracellular spent medium. Despite not highlighting if these changes to extracellular metabolite amounts are induced by cell death or active transport by live cells, the pattern of glucose metabolism remains specific during each viral infection. One common feature of changes to glucose metabolism was increased production of lactate during both IAV and SARS-CoV-2 infection. As previously mentioned, lactate has inhibitory effects on the IFN response and it can therefore be speculated that an increased production and release of lactate will have effects on both the producing cells and the by-stander cells in close contact with the high extracellular amounts of lactate. Therefore, lactate will have effects during both cell-death-mediated release and active release by live cells. Currently, multiple viruses are known to induce changes to glucose metabolism (Munger et al., 2006; Ritter et al., 2010; Vastag et al., 2011). Despite these discoveries, not much is known about the mechanisms and effects of these changes. Additionally, many of these studies were performed in cancer cell lines and not in primary cells or organoids.
In addition, we show that increased expression of LDHA and PKM2 along with increased production of lactate and pyruvate are common features of infection with IAV and SARS-CoV-2. The question remains, however, if these changes in metabolic profile are a host- or pathogen-induced mechanism. One example of direct virus-induced metabolic changes is the adenovirus gene E4ORF1. E4ORF1 has the ability to drive increased glucose metabolism through enhancement of the host transcription factor MYC (Thai et al., 2014), currently highlighting one of the only known mechanisms of direct viral manipulation of host metabolism. Interestingly, as IAV and SARS-CoV-2 showed different patterns of metabolic changes, a hypothesis could be raised if the changes are virus specific.
Together, this study highlights an anti-inflammatory highly pro-viral effect of metabolic reprogramming during IAV infection in the human airway epithelium. This sheds new light upon the question of lactate’s role during severe IAV infections and helps explain the correlation between high amounts of lactate and increased severity and mortality. Additionally, this study adds new and important knowledge of how virus is utilizing metabolic reprogramming as a means of immune evasion. Targeting these metabolic changes could prove to be a new and novel way of treating critically ill patients with influenza.
Limitations of the study
With this study we aimed to highlight the metabolic changes happening in a primary human airway epithelium during infection with IAV and SARS-CoV-2. One limitation to this study is the lack of other airway-associated cell types in our cultures, such as alveolar macrophages, which might have an impact on the overall metabolic changes during infections. Introducing these additional cells types into this already-complex ex vivo model would further increase the relevance of this model. The complexity of working with the HPAE-ALI model serves as another limitation as certain experiments needed to be performed in conventional cell lines. Being able to perform, for example, si-mediated knockdown in the HPAE-ALI cultures would have been more relevant to the story. Finally, we were not able to identify the potential specific viral proteins generating these metabolic changes and thus cannot specifically determine if these changes are induced by the virus or the host.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-Spike | GeneTex | Cat# GTX632604; RRID:AB_2864418 |
| anti-IFIT1 | Cell Signaling | Cat# 14769; RRID:AB_2783869 |
| anti-ISG15 | Cell Signaling | Cat# 2758; RRID:AB_2126200 |
| anti-Viperin | Cell Signaling | Cat# 13996; RRID:AB_2734772 |
| anti-STAT1 | Cell Signaling | CAt# 9172; RRID:AB_2198300 |
| anti-Vinculin | Cell Signaling | Cat# 18799; RRID:AB_2714181 |
| anti-HIF-1a | Cell Signaling | Cat# 3716; RRID:AB_2116962 |
| anti-LDHA | Cell Signaling | Cat# 2012; RRID:AB_2137173 |
| anti-PKM2 | Cell Signaling | Cat# 3198; RRID:AB_2252325 |
| anti-p-PKM2 | Cell Signaling | Cat# 3827; RRID:AB_1950369 |
| anti-NOS2 | R&D Systems | Cat# MAB9502; RRID:AB_2152874 |
| anti-NS1 | Santa Cruz | Cat# sc-130568; RRID:AB_2011757 |
| anti-RCC1 | Cell Signaling | Cat# 3589; RRID:AB_2269600 |
| Bacterial and virus strains | ||
| Influenza A/PR/8/34 (H1N1) | Charles River | 10100374 |
| SARS-CoV-2 strain #291.3 FR-4286 | Prof. George Koch | x |
| Chemicals, peptides, and recombinant proteins | ||
| L(+)-Lactic acid solution | Fluka BioChemika | 69773 |
| Lipofectamine RNAiMax | Invitrogen | 12778-150 |
| Critical commercial assays | ||
| BCA protein assay kit | Thermo Scientific | 23225 |
| High Pure RNA Isolation Kit | LifeScience | 11828665001 |
| Thermo ScientificTMPierceTMLDH Cytotoxicity Assay Kit | Fischer Scientific | P188953 |
| Nuclear Extraction Kit | Active Motif | 40010 |
| QUANTI-Blue | InvivoGen | Rep-qlc1 |
| Experimental models: cell lines | ||
| A549 cells | ||
| A549-Dual reporter cell line | InvivoGen | A549d-nfis |
| HEK-BlueTMIFN-α/β cells | InvivoGen | Hkb-ifnab |
| Oligonucleotides | ||
| SiLDHA | Santa Cruz biotechnology | Sc-43893 |
| SiCtrl | Santa Cruz biotechnology | Sc-37007 |
| Software and algorithms | ||
| Prism | Graph Pad | 9 |
| Other | ||
| Airway Epithelial Cells Basal Medium | PromoCell | C-21260 |
| Airway Epithelial Cell Growth Medium Supplement | PromoCell | C-21260 |
| Penicillin/Streptomycin | Gibco | 10378 |
| Bovine type I collagen solution | Sigma-Aldrich | 804592 |
| 6,5 mm Transwell membranes | Corning | 3470 |
| DMEM-low glycose | Sigma-Aldrich | D5921 |
| Hydrocortisone | StemCell | 07925 |
| Heparin | StemCell | 07980 |
| Pneumacult ALI medium kit | StemCell | 5001 |
| ALI medium supplement | StemCell | 5001 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Christian K. Holm (Holm@biomed.au.dk).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Air-liquid interface model
Primary nasal cells were isolated using a nasal brush (Dent-O-Care, #620B) inserted into the nasal turbinations and twisted. Cells was isolated from the brush by gently expelling monolayer medium (Airway Epithelial Cells Basal Medium, PromoCell, #C-21260 + 1 pack of Airway Epithelial Cell Growth Medium Supplement, PromoCell, #39160 + 100 U/ml Penicillin/Streptomycin, Gibco #10378) and PBS to wash of cells. Cells were cultured in monolayer culture in tissue culture flask (Sarstedt, TC: Standard #83.3911) coated with 0,1 mg/ml Bovine type I collagen solution (Sigma-Aldrich, #804592, diluted in sterile ddH2O). Monolayer cultures were split using 1x Trypsin mixed with 0.3 mM EDTA (10x Trypsin (2,5 %), Gibco, #15090, diluted to working concentration in PBS + UltraPure 0.5 M EDTA, Invitrogen, #15575) at approx. 80% confluency. At passage two, cells were seeded at 2-3x10ˆ4 cells on 6,5 mm Transwell membranes (Corning, #3470) coated with 30 ug/ml Bovine type I collagen solution (Sigma-Aldrich, #804592, diluted in sterile ddH2O). Cells were seeded and submerged in 2x P/S (200 U/ml Pen/Strep) DMEM-low glycose (Sigma-Aldrich, D5921) mixed one to one with 2x Monolayer medium (Airway Epithelium Cell Basal Medium, (PromoCell, #C-21260) supplemented with 2 packs of Airway Epithelial Cell Growth Medium Supplement (PromoCell, #C-39160) without triiodothyronine + 1 ml of 1.5 mg/ml BSA). When cultures reach full confluency ALI (=Air-liquid interface) is introduced and medium is changed to ALI medium (Pneumacult ALI medium kit (StemCell, #5001) with ALI medium supplement (StemCell, #5001) and 100 U/ml Pen/strep) supplemented with 24 ug of hydrocortisone (StemCell, #07925) and 0.2 mg heparin (StemCell, #07980). Membranes were allowed at least 21 days of differentiation verified by extensive cilia beating and mucus covering, both hallmarks of the human airway epithelium. Through all culture development cells were kept at 37°C, 5% CO2. Both male and female donors where used in this study. Upon initiation of infection, membranes were washed for 5 minutes using DMEM-ø (low glycose, no additives) and the baso-lateral medium changed for fresh ALI medium. At time of infection 100 ul DMEM-ø with or without virus was added to the apical compartment and left in a 37°C incubator for one hour. Following the one-hour infection, all liquid was removed from the apical compartment and membranes were placed in an incubator at 37°C, 5% CO2 until harvest.
At time of harvest, baso-lateral medium was removed and 500 ul Trypsin/EDTA was added bao-laterally and 100 ul was added apically. This way of harvesting was evaluated as the most efficient when comparing cells survival and harvest time (Figure S3F). After cell detachment (5-15 minutes) cells were harvested using 5% FBS/DMEM (low glycose). Cell pellets were isolated and lysed according to read-out of choice.
All primary cells where collected and cultures as approved by The Science Ethical Committee (Den Videnskabsetiske Komitéer) for the Region of Midtjylland with case number 1-10-72-182-19. Due to our system of anonymity for donors the project lead was not familiar with the sex of the donors used in this project. A range of different donors was used to ensure diversity in sampling.
Additional cell cultures
For some specific experiments alternative cell cultures were used. When using non-differentiated primary basal cells, the cells were harvested and cultured as described above. Differently though, when the cells reach approx. 80% confluency they were plated in 24 well plates coated with 1 mg/ml Bovine type I collagen solution at approx. 37,5∗10ˆ3 cells pr. well in monolayer medium. Cells were allowed to rest for 24 hours before being used as regular cells lines. To fraction infected cells into cytoplasmic and nuclear fraction we utilized A549 cells. A549 is a well-established cell line originating form an Adenocarcinoma human alveolar basal epithelial cell. To investigate IRF3 activation we utilized the A549-Dual reporter cell line (InvivoGen, #a549d-nfis). This cell line expresses two inducible reporter constructs under the control of NF-kB (Secreted embryonic alkaline phosphatase, SEAP) and IRF3/7 (Lucia Luciferase). These reporter proteins become measurable by using QUANTI-Blue (InvivoGen, #rep-gbs) or Quanti-Luc (InvivoGen, #rep-qlc1), thus allowing for a quantification of signaling through these transcription factors.
For investigations of MAVS dependency on IFN signaling during IAV and IFN treatment we used THP-1 cells. THP-1 cells are a human monocytic cell line derived from an acute monocytic leukemia patient. Here we used both THP-1 wildtype and a THP-1 cell line with a knockout of MAVS.
Cell lines were cultured in cell-optimized medium Dulbecco's modified Eagle medium (Sigma, #D6429) or RPMI-1640 (Sigma, #R8758) supplemented with 10% heat inactivated fetal bovine serum (MERCK, F7524)), 1000 Units/ml Penicillin, 10 mg/ml Streptomycin and 29,2 mg/ml Glutamine (Life Technologies) and kept at 37°C, 5% CO2.
Viruses
To perform infection studies, we used the Influenza A/PR/8/34 (H1N1) strain (Charles River #10100374) or the SARS-CoV-2 strain #291.3 FR-4286 (a kind donation by Professor Georg Koch, Freiburg).
Method details
Western Blot
For detection of proteins in cell lysate samples we used Western Blotting.
To isolate protein from our culture's cells were lysed using a lysis buffer consisting of Pierce RIPA lysis buffer (Thermo, #89901) supplemented with 5 IU/mL Benzonase (Sigma, #E1014), 10 mM Sodium Fluoride and 1x complete protease inhibitor cocktail (Roche, #5892988001). To determine protein concentration a BCA protein assay kit (Thermo, #23225) was used and samples equalized using lysis buffer. Samples were denatured for 2 min at 98°C diluted in XT Sample Buffer (Bio-Rad, #1610791) and XT Reducing Agent (Bio-Rad, #161-0762). Samples were separated by SDS-Page with 10–30 ug of reduced samples. This was done using a 4–20% Criterion TGX precast gradient gel (Bio-Rad, #5671094). The SDS-Page was run at 70V for approx. 20 min followed by approx. 40–60 min at 110V. To transfer protein to a PVDF membrane (Bio-Rad, #1704157) a trans-Blot Turbo Transfer was used. Blocking of membranes was done in Phosphate buffered saline (Biowest, #MS00FS100F) supplemented with 0.05% Tween 20 (Sigma-Aldrich, P1379) (PBST) with 5% skim-milk powder added (Sigma Aldrich, #70166). Blocking was done for at least one hour at room temperature. Immediately following blocking, membranes was cut according to the size of the protein of interest given by the manufacturer. Cut membranes was incubated with one of the following specific primary antibodies diluted in PBST: anti-Spike (GeneTex, #GTX632604, 1:100), anti-IFIT1 (Cell Signaling, #14769, 1:1000), anti-ISG15 (Cell Signaling, #2758, 1:1000), anti-Viperin (Cell Signaling, #13996, 1:1000), anti-STAT1 (Cell Signaling, #9172, 1:1000), anti-Vinculin (Cell Signaling, #18799, 1:1000), anti-HIF-1a (Cell Signaling, #3716, 1:1000), anti-LDHA (Cell Signaling, #2012, 1:1000), anti-PKM2 (Cell Signaling, #3198, 1:1000), anti-p-PKM2 (Cell signaling, #3827, 1:1000), anti-NOS2 (R&D Systems, #MAB9502, 1ug/ml), anti-NS1 (Santa Cruz, #sc-130568, 1:100) and anti-RCC1 (Cell Signaling, #3589). Anti-vinculin and anti-RCC1 was used as loading control. Membranes was incubated with primary antibodies over night at 4°C. Following incubation with primary antibodies, membranes was washed three times using PBST. Hereafter secondary antibodies, peroxidase-conjugated F(ab)2 donkey anti-mouse IgG (Jackson, #715-036-150, 1:10,000) or peroxidase-conjugated F(ab)2 donkey anti-rabbit IgG (Jackson, #711-036-152, 1:10,000), was added. Secondary antibodies were diluted in PBST +1% skim milk powder and left on membranes to incubate for approx. 1.5 hours at room temperature. Before imaging, membranes were washed thrice with PBST. Imaging was done using an Image Quant LAS4000 mini imager (GE Healthcare). For exposing either SuperSignal West Pico PLUS chemiluminescent substrate (Thermo, #3577) or the SuperSignal West Femto maximum sensitivity substrate (Thermo, #34095) was used.
Reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR)
To quantify transcription of specific target genes we implemented the RT-qPCR technique.
For RNA extraction a High Pure RNA Isolation Kit (LifeScience, #11828665001) was used. Cells were lysed and processed as per manufacturers instructions. RNA concentrations and quality of isolation was determined using a NanoDrop spectrometer (Thermo Fisher). Specific gene expression was determined using quantitative real-time PCR. Using TaqMan RNA-to-Ct 1-step kit (Applied Biosystems) we measured mRNA levels of hβ-Actin (Thermo Fisher, Assay ID; Hs01060665_g1) along with IAV M protein and SARS-CoV2 with specially designed primers:
IAV – seq7(M): Forward 5′-ATTTGCCTATGAGCCGATGCT, Reverse 5′-AGGATGGGGGCTGTGACC
SARS-CoV-2 – N: Forward 5′-AAATTTTGGGGACCAGGGAAC, Reverse 5′- TGGCACCTGTGTAGGTCAAC.
Lactate dehydrogenase (LDH) activity assay
To measure LDH activity, a Thermo ScientificTM PierceTM LDH Cytotoxicity Assay Kit (Fischer Scientific, #PI88953) was used. ALI cells were harvested as mentioned earlier and lysed using lysis buffer from LDH Cytotoxicity Assay Kit. Absorbance at 490nm and 680nm was measured using a Synergy HTX Multi-Mode Reader (BioTek Instruments). Data was generated using the Gen5 software.
Type I interferon bioassay
To measure excreted active IFN produced by cells of interest we utilized the interferon reporter cell-line HEK-Blue TM IFN-α/β cells from InvivoGen (#hkb-ifnab). These cells contain an inducible reporter gene, secreted embryonic alkaline phosphatase (SEAP), which is expressed and secreted upon interferon mediated signaling through IFNAR. SEAP is quantifiable in the supernatant by measuring optical density on a plate reader after adding QUANTI-Blue (InvivoGen, #rep-qbs). Comparing own samples with samples of known IFN amount allows for precise calculations of IFN amount based on SEAP levels. HEK-Blue cells were seeded in 96-well plates in 150 ul test medium with 30.000 cells pr. well. After incubation overnight, 50 ul of test sample was added along with a twofold dilution of a known IFN stock (500 U/mL to approx. 2 U/mL). Cells were incubated for 24 hours after which 150 ul medium was harvested. As this medium contains the potentially produced SEAP, QUANTI-Blue was added and SEAP levels were measured on a plate reader at 655 nm.
Short-Interfering RNA (siRNA)-mediated knock down
For knockdown experiments, approx. 50∗103 primary nasal cells were plated pr. well in a 24 well plate and left for 24 hours. Cells were transfected using approx. 50 nM of human siLDHA (Santa Cruz Biotechnology, #sc-43893) or control siRNA (Santa Cruz Biotechnology, #sc-37007) diluted in DMEM (without serum and antibiotics). As a transfection agent Lipofectamine RNAi Max (Invitrogen, #13778-150) was used in accordance to manufacturer's instructions. Cells were incubated with siRNA and Lipofectamine for 48 hours before being used further.
Poly I:C treatment
To stimulate signaling through RLR receptors we used transfected Poly I:C treatment. Cells were plated in 12 well plates and left for 24 hours before transfection. Cells were transfected using between 30ng/ml and 10 ug/ml Poly I:C (InvivoGen #tlrl-pic), as per manufacturers recommendations, mixed with Lipofectamine 2000 (Invitrogen #11668-027) at a concentration of 0.5 ug/ml.
Cell fractionation
To fraction cells into a cytoplasmic and a nuclear fraction we utilized a Nuclear Extraction Kit from Active Motif (#40010). After treatment cells were washed with ice-cold PBS containing a phosphatase inhibitor and gently collected using a cell scraper. Cells were pelleted by centrifugation at 200xG followed by resuspension in a hypotonic buffer and left to incubate for 15 min on ice. Following incubation, a detergent was added and cells were vigorously vortexed for 10 seconds and pelleted at 14.000xG. Supernatant was stored at −20°C as this now contained the cytoplasmic fraction. The pellet was resuspended in complete lysis buffer and a small volume of detergent was added. Before the 30-minute incubation on ice pellet was vigorously vortexed for 10 seconds. After 30 minutes the lysate was centrifuged for 10 minutes at 14.000xG in a centrifuge chilled for 4°C. Supernatant was transferred to new tubes and stored at −20°C as these contained the nuclear fraction.
Metabolite measurements
Following infection or treatment, medium was isolated for further testing. To measure metabolites in the medium, 15 ul sample was loaded in plastic tubes (CMA Microdialysis, #7431007) and placed in an ISCUSflex Microdialysis Analyzer. As references Reagent Set C (M Dialysis AB, #8002164) containing lactate, glucose and pyruvate along with a calibrator fluid was used.
Metabolomics by MS-Omics
For full scale metabolomics HPAE-ALI cells were harvested using pure MeOH. Medium was removed from cultures and MeOH was added and incubated for 5 min. Cells were scraped of and vortexed for 10 sec. three times. Lysed cells were centrifuged for 15 min at 16.000 G at 4°C. Supernatants where isolated and transported to MS-Omics on dry ice.
Sample analysis was carried out by MS-Omics as follows. The analysis was carried out using a Thermo Scientific Vanquish LC coupled to Thermo Q Exactive HF MS. An electrospray ionization interface was used as ionization source. Analysis was performed in negative and positive ionization mode. The UPLC was performed using a slightly modified version of the protocol described by Catalin et al. (UPLC/MS Monitoring of Water-Soluble Vitamin Bs in Cell Culture Media in Minutes, Water Application note 2011, 720004042en). Peak areas were extracted using Compound Discoverer 2.0 (Thermo Scientific). Identification of compounds were performed at four levels; Level 1: identification by retention times (compared against in-house authentic standards), accurate mass (with an accepted deviation of 3ppm), and MS/MS spectra, Level 2a: identification by retention times (compared against in-house authentic standards), accurate mass (with an accepted deviation of 3ppm). Level 2b: identification by accurate mass (with an accepted deviation of 3ppm), and MS/MS spectra, Level 3: identification by accurate mass alone (with an accepted deviation of 3ppm).
Measurements of epithelial barrier integrity of HPAE-ALI cultures
The FITC-Dextran permeability assay evaluates barrier integrity and proper tight junction formation by studying the paracellular flux of fluorescent labelled hydrophilic tracers. The inserts were gently washed for 5 min using pre-warmed DMEM – low glucose (Sigma-Aldrich, D5921). Next, 90 μl/well (apically) and 500 μl/well (basolateral) of pre-warmed DMEM (Sigma, #D6429) was added and the inserts were allowed to equilibrate for 15 min at 37°. Then, 60 μl of 5 mg/mL FITC-4 kDa Dextran (Sigma-Aldrich, #46944) was added to the apical chamber for the final concentration of 2 mg/mL. Setups were incubated at 37° and 5% CO2. 10 and 21 hrs after addition of FITC-Dextran solution to the apical chamber, 3 × 100 μl of each sample (technical triplicates) from the basolateral chamber was collected to a 96-well plate. The technical triplicates were averaged and FITC-Dextran concentration diffused to the basolateral chamber was determined by extrapolation using a FITC-Dextran standard curve. The FITC fluorescence was read using the Synergy HTX multimode plate reader at excitation/emission spectrums of 485/20 nm and 532/20 nm, respectively.
GC-MS on lactate and pyruvate
HPAE-ALI cultures were washed with 0.9% HCl followed by quenching of metabolites using 1:1 ice-cold LC.MS-grade Methanol (Merck, 1.06035.1000) and Milli-Q water containing 2 ug/ml D6 glutaric acid (CDN isotopes, D-5227) as an internal standard. Cells were scraped of Transwell and added along with Methanol and water to ice-cold HPLC-grade Chloroform (Merck, 650498). Cells lysed by shaking on HulaMixer – Sample Mixer (Invitrogen) at 2000 rpm for 20 min at 4°C. Lysates were centrifuged at 17.000G for 10 min where supernatants where isolated and freeze dried for transport.
Flow cytometry
Following harvest, the HPAE-ALI cells were stained and analyzed by flow cytometry. The cells were stained with Zombie NIR Fixable Viability Kit (BioLegend, cat.no. 423105) for 15 min on ice and washed with PBS. Then, cells were fixed using 1 or 2% vol/vol formaldehyde solution (Sigma-Aldrich, cat.no. F1635, in PBS) for 15 min at RT and washed once with staining buffer (PBS +0.5% BSA). Samples were finally resuspended in staining buffer and stored cold until analysis. The analysis was done on NovoCyte Quanteon analyzer. Gating strategy as follows; cells were first gated on forward (FCS-H) versus side scatter (SSC-H) to exclude debris. Then, doublet exclusion was done, first based on forward scatter area (FSC-A) versus forward scatter height (FSC-H) and secondly based on side scatter area (SSC-A) versus side scatter height (SSC-H). Finally, the percentage of cell alive were estimated. Data were analyzed in FlowJo (version 10.7.1, Tree Star Inc., Ashland, OR, USA).
Quantification and statistical analysis
Statistical analysis
A description of statistical work and parameters can be found in the figure texts. All bars were expressed as the mean ± SEM. Graphs and statistics were computed using Graph Pad Prism 9. An unpaired, Welch's t test was used to determine significance of the difference between control and each experimental condition. p values are indicated above each condition.
Acknowledgments
This research work was supported by the Graduate School of Health Aarhus University, Fhv. Dir. Leo Nielsen & Hustru Karen Margrethe Nielsens Legat for Lægevidenskabelig Grundforskning, Ester M. & Konrad Kristian Sigurdssons Dyreværnsfond, Beckett-fonden, Kong Christian IX & Dronning Louises Jubilæumslegat, Christian Larsen & Dommer Ellen Larsens Legat, Direktør Emil C. Hertz & hustru Inger Hertz’ fond og A.P. Møller Fonden. S.C. and A.W. were supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001206), the UK Medical Council (FC001206), and the Wellcome Trust (FC001206). Flow cytometry was performed at the FACS Core Facility, Aarhus University, Denmark.
Author contributions
J.T. and C.K.H conceived the project. J.T. drafted the manuscript and finalized it with C.K.H. J.T. and C.K.H. planned experiments, analyzed data, and created the figures. J.T., J.S., A.L.H., J.B.C., A.H., A.L.T., A.W., S.C., and K.H. planned and performed experiments.
Declaration of interests
The authors declare no competing interests.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103300.
Contributor Information
Jacob Thyrsted, Email: thyrsted@biomed.au.dk.
Christian Kanstrup Holm, Email: holm@biomed.au.dk.
Supplemental information
Data and code availability
-
•
Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/cwvnbt2m8p.1
-
•
The paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
References
- Adeva-Andany M., López-Ojén M., Funcasta-Calderón R., Ameneiros-Rodríguez E., Donapetry-García C., Vila-Altesor M., Rodríguez-Seijas J. Comprehensive review on lactate metabolism in human health. Mitochondrion. 2014;17:76–100. doi: 10.1016/j.mito.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Bambouskova M., Potuckova L., Paulenda T., Kerndl M., Mogilenko D.A., Lizotte K., Swain A., Hayes S., Sheldon R., Kim H. Itaconate confers tolerance to late NLRP3 inflammasome activation. Cell Rep. 2021;34:108756. doi: 10.1016/j.celrep.2021.108756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassini A., Colzani E., Pini A., Mangen M.J.J., Plass D., McDonald S.A., Maringhini G., Alies L., Haagsma J., Havelaar A. Impact of infectious diseases on population health using incidence-based disability-adjusted life years (DALYs): results from the burden of communicable diseases in Europe study, European Union and European economic countries, 2009 to 2013. Eurosurveillance. 2018;23:1–20. doi: 10.2807/1560-7917.ES.2018.23.16.17-00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes T., Wallace M., Michelucci A., Divakaruni A.S., Sapcariu S.C., Sousa C., Koseki H., Cabrales P., Murphy A., Hiller K. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J. Biol. Chem. 2016;291:14274–14284. doi: 10.1074/jbc.M115.685792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong P.M., van Sterkenburg M.A., Hesseling S.C., Kempenaar J.A., Mulder A.A., Mommaas A.M. Ciliogenesis in human bronchial epithelial cells cultured at the air-liquid interface. Am. J. Respir. Cell. Mol. Biol. 1994;10:271–277. doi: 10.1165/ajrcmb.10.3.8117445. [DOI] [PubMed] [Google Scholar]
- DeBeradinis R., Thompson C. Cellular metabolism and disease. Cell. 2012;148:1132–1144. doi: 10.1016/j.cell.2012.02.032.Cellular. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold S.S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science (New York, N.Y.) 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Dalskov L., Møhlenberg M., Thyrsted J., Blay-Cadanet J., Poulsen E.T., Folkersen B.H., Skaarup S., Olagnier D., Reinert L., Enghild J. SARS-CoV-2 evades immune detection in alveolar macrophages. EMBO Rep. 2020;21:e51252. doi: 10.15252/embr.202051252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T.E., Guzman K., Davis C.W., Abdullah L.H., Nettesheim P. Mucociliary differentiation of serially passaged normal human tracheobronchial epithelial cells. Am. J. Respir. Cell. Mol. Biol. 1996;14:104–112. doi: 10.1165/ajrcmb.14.1.8534481. [DOI] [PubMed] [Google Scholar]
- Lampropoulou V., Sergushichev A., Bambouskova M., Nair S., Vincent E.E., Loginicheva E., Cervantes-Barragan L., Ma X., Huang S., Griss E. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 2016;24:158–166. doi: 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin S., Hahn T. Interferon deficiency syndrome. Clin. Exp. Immunol. 1985;60:267–273. [PMC free article] [PubMed] [Google Scholar]
- Li S., Sieben C., Ludwig K., Höfer C.T., Chiantia S., Herrmann A., Eghiaian F., Schaap I. pH-Controlled two-step uncoating of influenza virus. Biophys. J. 2014;106:1447–1456. doi: 10.1016/j.bpj.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu R., Gu H., Zhang E., Qu J., Cao W., Huang X., Yan H., He J., Cai Z. Metabolic reprogramming in macrophage responses. Biomarker Res. 2021;9:1. doi: 10.1186/s40364-020-00251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.-M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M., García-Sastre A., Katze M. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/jvi.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelucci A., Cordes T., Ghelfi J., Pailot A., Reiling N., Goldmann O., Binz T., Wegner A., Tallam A., Rausell A. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. U S A. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.L., Ryan D.G., Prag H.A., Dikovskaya D., Menon D., Zaslona Z., Jedrychowski M., Costa A., Higgins M., Hams E. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J., Bajad S.U., Coller H.A., Shenk T., Rabinowitz J.D. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2:e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olagnier D., Brandtoft A.M., Gunderstofte C., Villadsen N.L., Krapp C., Thielke A.L., Laustsen A., Peri S., Hansen A., Bonefeld L. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat. Commun. 2018;9:3506. doi: 10.1038/s41467-018-05861-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz B., Rejaibi A., Dauber B., Eckhard J., Vinzing M., Schmeck B., Hippenstiel S., Suttorp N., Wollf T. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007;9:930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- Parks S.K., Mazure N.M., Counillon L., Pouysségur J. Hypoxia promotes tumor cell survival in acidic conditions by preserving ATP levels. J. Cell. Physiol. 2013;228:1854–1862. doi: 10.1002/jcp.24346. [DOI] [PubMed] [Google Scholar]
- Pezzulo A.A., Starner T.D., Scheetz T.E., Traver G.L., Tilley A.E., Harvey B.G., Crystal R., McCray P., Zabner J. The air-liquid interface and use of primary cell cultures are important to recapitulate the transcriptional profile of in vivo airway epithelia. Am. J. Physiol. Lung Cell Mol. Physiol. 2011;300:25–31. doi: 10.1152/ajplung.00256.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter J.B., Wahl A.S., Freund S., Genzel Y., Reichl U. Metabolic effects of influenza virus infection in cultured animal cells: intra- and extracellular metabolite profiling. BMC Syst. Biol. 2010;4 doi: 10.1186/1752-0509-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Thai M., Graham N.A., Braas D., Nehil M., Komisopoulou E., Kurdistani S.K., McCormick F., Graeber T., Christofk H. Adenovirus E4ORF1-induced MYC activation promotes host cell anabolic glucose metabolism and virus replication. Cell Metab. 2014;19:694–701. doi: 10.1016/j.cmet.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyrsted J., Holm C.K. Virus-induced metabolic reprogramming and innate sensing hereof by the infected host. Curr. Opin. Biotechnol. 2021;68:44–50. doi: 10.1016/j.copbio.2020.10.004. [DOI] [PubMed] [Google Scholar]
- Thyrsted J., Holm C.K. Influenza A induces lactate formation to inhibit type I IFN in primary human airway epithelium. J. Thyrsted et.al. 2021. Mendeley Data. 2021;V1 doi: 10.17632/cwvnbt2m8p.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzeciak S., Dellinger R.P., Chansky M.E., Arnold R.C., Schorr C., Milcarek B., Hollenberg S., Parrillo J. Serum lactate as a predictor of mortality in patients with infection. Intensive Care Med. 2007;33:970–977. doi: 10.1007/s00134-007-0563-9. [DOI] [PubMed] [Google Scholar]
- Uyeda K. In: Encyclopedia of Biological Chemistry (Second Edition) Second Edition. Lennarz W.J., Lane M.D., editors. Academic Press; 2013. Pyruvate Kinase; pp. 719–721. [DOI] [Google Scholar]
- Vastag L., Koyuncu E., Grady S.L., Shenk T.E., Rabinowitz J.D. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7:e1002124. doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Wang G., Xu Z.G., Tu H., Hu F., Dai J., Chang Y., Chen Y., Lu Y., Zeng H. Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell. 2019;178:176–189.e15. doi: 10.1016/j.cell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., He R., Fang P., Li M., Yu H., Wang Q., Zhang Y., Chen A., Peng N., Lin Y. Hepatitis B virus rigs the cellular metabolome to avoid innate immune recognition. Nat. Commun. 2021;12:1–13. doi: 10.1038/s41467-020-20316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Additional Supplemental Items are available from Mendeley Data at https://doi.org/10.17632/cwvnbt2m8p.1
-
•
The paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request