ABSTRACT
Adipogenesis regulation is crucial for mature adipocyte function. In obesity, a major driver of type 2 diabetes (T2D), this process is disrupted and remains poorly characterized. Here we identified altered DNA methylation profiles in diabetic obese patients, during three adipocytes differentiation stages. We isolated mesenchymal cells from visceral adipose tissue of obese patients with and without T2D to analyse DNA methylation profiles at 0, 3, and 18 days of ex vivo differentiation and documented their impact on gene expression. Methylation and gene expression were analysed with EPIC and Clarion S arrays, respectively. Patients with T2D had epigenetic alterations in all the analysed stages, and these were mainly observed in genes important in adipogenesis, insulin resistance, cell death programming, and immune effector processes. Importantly, at 3 days, we found six-fold more methylated CpG alterations than in the other stages. This is the first study to document epigenetic markers that persist through all three adipogenesis stages and their impact on gene expression, which could be a cellular metabolic memory involved in T2D. Our data provided evidence that, throughout the adipogenesis process, alterations occur in methylation that might impact mature adipocyte function, cause tissue malfunction, and potentially, lead to the development of T2D.
KEYWORDS: Adipogenesis, type 2 diabetes, obesity, DNA methylation
Introduction
White adipose tissue (WAT) is mainly composed of adipocytes; indeed, adipocytes form >90% of WAT mass. Adipocytes store energy in the form of fat, mainly triglycerides, and contribute to the modulation of whole-body metabolism by secreting endocrine and paracrine factors [1,2]. Adipocyte physiology drastically changes during obesity. WAT expansion can arise from adipogenesis, where new adipocytes are formed by precursor differentiation (hyperplasia), or from an increase in adipocyte size (hypertrophy) [3,4]. Although low-grade inflammation is associated with obesity, it has been shown that structural cell changes are related to insulin resistance even in the absence of inflammation [5]. Obesity has important implications for the development of cardiovascular risk diseases, like hypertension, insulin resistance, dyslipidemia, and type 2 diabetes (T2D). Physio pathological studies have shown that obesity and these pathologies are highly interrelated by an exceeded ability to build up adequate fat stores and altered adipogenesis [6–10].
The number of adipocytes in a body is established in childhood. In adulthood, about 10% of the adipocytes are renewed per year, to maintain the WAT compartment [11,12]. Thus, maintaining a balance between metabolism and adipocyte differentiation is of paramount importance for health [13]. Adipocytes are generated from mesenchymal stem cells (MSCs) [14], which undergo differentiation in two phases. The first phase promotes the stem cell transformation into a preadipocyte. During this phase, many epigenetic events occur to guarantee that preadipocytes can achieve the second phase, where preadipocytes acquire mature adipocyte features [15]. This process requires complex integration between the cytoarchitecture and signalling pathways, which involves more than 2000 regulatory transcription factors [16].
Epigenetic factors can modulate accessibility to the chromatin, and thus, they can regulate the genes that participate in the development of mature adipocytes [17]. However, changes induced in DNA methylation patterns can potentially persist long after the original stimuli is eliminated. This persistence results in a mitotically heritable memory, which can contribute to chronic diseases, like T2D [18–20]. Taken together, these findings have suggested that it is relevant to understand adipogenesis and its role in the regulation of metabolic processes.
In the present study, to extend our understanding of the role of the adipocyte in T2D, we sought to explore DNA methylation dynamics in-depth, by tracking its progress through three stages of adipogenesis. We cultured adipocytes from patients that were obese without T2D (OND) or obese with T2D (OD) and compared their methylation dynamics during adipogenesis. Moreover, we aimed to identify markers that persisted or appeared during the different phases of differentiation.
Results
Patients were classified as controls (OND; n = 9) and cases (OD; n = 8). We matched the two groups in terms of body mass index (BMI), age, and biochemical parameters. As expected, we observed differences between the two groups in the HbA1c (p = 0.0002) and serum glucose levels (p = 0.01) (Table 1).
Table 1.
Clinical characteristics of obese patients, with and without diabetes
| Parameter | OD | OND | p-value |
|---|---|---|---|
| n | 8 | 9 | |
| Age (years ± SD) | 45.3 ± 5.94 | 39.11 ± 4.62 | 0.07 |
| BMI (kg/m2 ± SD) | 45.4 ± 7.07 | 44.40 ± 6.02 | 0.68 |
| Glucose (mg/dl ± SD) | 131.7 ± 33.73 | 97.11 ± 8.22 | 0.01 |
| HbA1c (%± SD) | 6.79 ± 0.72 | 5.56 ± 0.54 | 0.0002 |
| HOMA (%B ± SD) | 7.47 ± 4.15 | 5.21 ± 3.90 | 0.15 |
| Insulin (μU/ml ± SD) | 22.28 ± 10.42 | 21.07 ± 14.25 | 0.54 |
| Triglycerides (mg/dl± SD) | 168.7 ± 70.50 | 154.27 ± 65.96 | 0.96 |
| HDL-Cholesterol (mg/dl ± SD) | 48.52 ± 16.64 | 59.6 ± 63.74 | 0.23 |
| Total Cholesterol (mg/dl ±SD) | 180.7 ± 35.63 | 152.25 ± 69.94 | 0.39 |
| Systolic blood pressure (mmHg ± SD) | 133.3 ± 22.24 | 133.11 ± 15.64 | 0.9 |
| Diastolic blood pressure (mmHg ± SD) | 74.6 ± 11.24 | 74.22 ± 10.91 | 0.96 |
| LDL (mg/dl ± SD) | 118.27 ± 25.71 | 107.27 ± 53.64 | 0.78 |
OD: obese patients with diabetes; OND: obese patients without diabetes; HDL: high-density lipoprotein.
Differential DNA methylation
To establish the dynamics of the alterations in methylation observed during adipogenesis, we obtained DNA from MSC cultures (day 0), preadipocyte cultures (PreA, day 3), and mature adipocyte cultures (MA, day 18). We then analysed the DNA methylation profiles in OD and OND samples. After quality control and data normalization, we could detect 755,382 CpG sites. No significant differences in global methylation were observed between the OD and OND samples (Supplemental Figure S1). However, the epigenetic remodelling pattern during adipogenesis was different between OD and OND samples (Supplemental Tables S1-S4 and Supplemental Figure S2). We also compared the beta values for the three different stages between groups to identify differentially methylated CpG sites (DMCs; Figure 1 and Supplemental Tables S5-S7). We identified 115 DMCs that persisted during all three adipogenesis stages in the OD samples. In addition, OD samples showed hypomethylation in the MSC and MA stages, and hypermethylation in the PreA stage (Figure 2 and Supplemental Figure S3). In OD samples, epigenomic alterations were six-fold higher in the PreA stage than in the MSC and MA stages. Interestingly, in the PreA stage these alterations included DMCs mainly in master genes that regulate adipogenesis, such as PPARG, GATA3, JAK2, and STAT5A.
Figure 1.
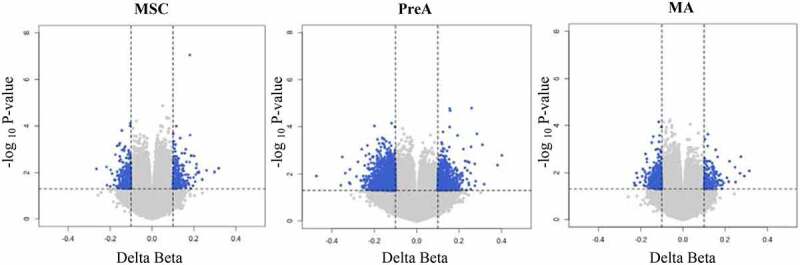
Differential DNA methylation profiles in MSC, PreA and MA in OD patients. Blue points indicate DMCs with |Delta-Beta|> 0.1 and p-value<0.05
Figure 2.
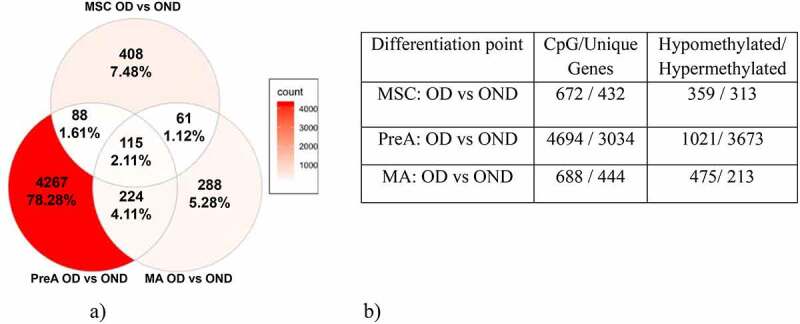
(a) Venn diagram shows a comparison of DMCs from OD and OND samples in the different stages. The background colour represents the number of DMCs; the percentage was calculated based on the total number of DMCs in all three stages. (b) Table shows a comparison of DMC numbers observed in OND and OD samples in all three stages. DMC: differential methylation CpG sites; OD: samples from obese patients with diabetes; OND: samples from obese patients without diabetes
In a principal component analysis (PCA), the beta values of the 115 persistent DMCs could separate patients with diabetes from patients without diabetes (Figure 3). Furthermore, we found a significant correlation between principal component 1 (PC1) and the Hb1Ac values (Supplemental Figure S4). To determine the potential roles of genes with persistent DMCs, we performed an enrichment pathway analysis with a database for annotation, visualization, and integrated discovery (DAVID). We found enriched pathways related to the regulation of caspase activity, endopeptidase activity, and programmed cell death, in addition to pathways involved in immune effector processes (Supplemental Table S8).
Figure 3.
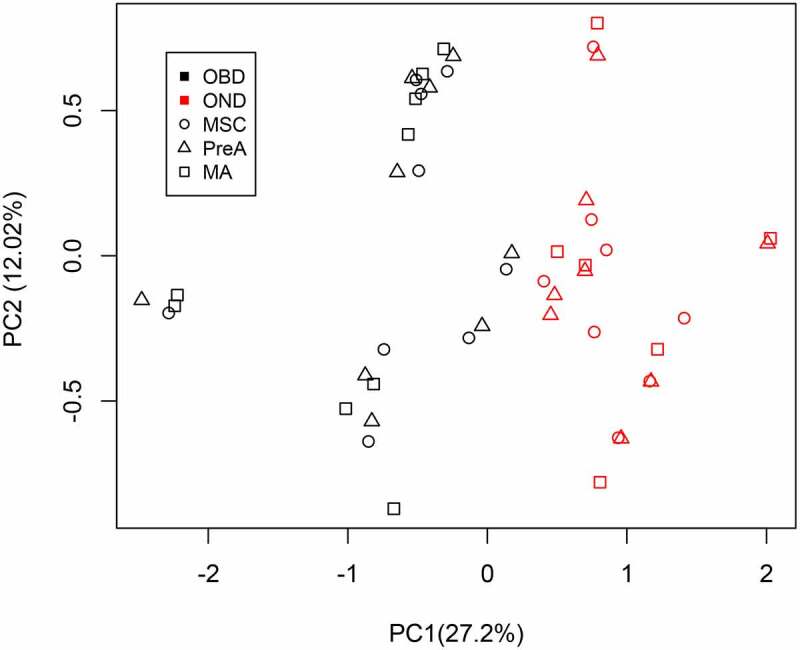
PCA analysis results, based on 115 persistent DMCs, among OD (red symbols) and OND (black symbols) samples. DMC: differential methylation CpG site; OD: samples from obese patients with diabetes; OND: samples from obese patients without diabetes
We found alterations in each differentiation stage. The top 20 DMCs identified in each stage are shown in (Table 2). To investigate enrichments in the functional pathways, we also used DAVID to examine all observed DMCs in each stage (Supplemental Tables S9-S11). Although DMCs were observed on particular genes in specific stages, notably, some pathways were shared in all three stages (Table 3). These pathways included biological adhesion, negative regulation of macromolecule biosynthetic processes, regulation of apoptosis, and regulation of programmed cell death. We also found that some alterations persisted from MSC to PreA; these alterations represented pathways involved in cell–cell adhesion, haemopoietic or lymphoid organ development, homophilic cell adhesion, immune system development, phosphorylation, protein amino acid phosphorylation, and regulation of small GTPase mediated signal transduction. The largest number of altered shared pathways were found in both PreA and MAin particular,: cell–cell signalling, embryonic morphogenesis, negative regulation of biosynthetic processes, positive regulation of gene expression, tissue morphogenesis and cellular carbohydrate catabolic processes. The last one showing the highest fold enriched-level regulation. Our findings provide evidence that differences in methylation affected both specific and common pathways in the different stages of adipogenesis in patients with T2D.
Table 2.
Top 20 DMCs, grouped by adipogenesis stage
| OD vs OND | CpG ID | OND Beta mean | OD Beta mean | Delta Beta | Chromosome | Gene symbol | Genomic region | P value |
|---|---|---|---|---|---|---|---|---|
| MSC | cg02332293 | 0.710 | 0.889 | 0.180 | 5 | IGR | 7.44E-08 | |
| cg26480858 | 0.305 | 0.201 | 0.103 | 7 | THSD7A | Body | 7.42E-05 | |
| cg05170452 | 0.713 | 0.829 | 0.115 | 5 | IGR | 9.31E-05 | ||
| cg16626954 | 0.513 | 0.406 | 0.107 | 3 | IGR | 0.00015 | ||
| cg24056885 | 0.500 | 0.356 | 0.144 | 3 | ABCC5AS1 | TSS200 | 0.00015 | |
| cg12771637 | 0.555 | 0.657 | 0.101 | 11 | IGR | 0.00021 | ||
| cg25635251 | 0.531 | 0.664 | 0.133 | 11 | IGR | 0.00028 | ||
| cg13482309 | 0.393 | 0.571 | 0.178 | 9 | OR1B1 | 1stExon | 0.00028 | |
| cg18830697 | 0.747 | 0.849 | 0.102 | 6 | RIMS1 | TSS200 | 0.00052 | |
| cg11086466 | 0.762 | 0.608 | 0.154 | 6 | RNF182 | 5’UTR | 0.00054 | |
| cg18454042 | 0.581 | 0.694 | 0.113 | 7 | SDK1 | Body | 0.00063 | |
| cg18116804 | 0.525 | 0.368 | 0.156 | 10 | PLXDC2 | Body | 0.00064 | |
| cg18860301 | 0.634 | 0.466 | 0.168 | 6 | RNF182 | 5’UTR | 0.0007 | |
| cg06121514 | 0.576 | 0.677 | 0.101 | 15 | IPW | Body | 0.00071 | |
| cg14439102 | 0.544 | 0.442 | 0.102 | 9 | GNAQ | Body | 0.00075 | |
| cg06104343 | 0.761 | 0.631 | 0.130 | 17 | IGR | 0.00082 | ||
| cg03915559 | 0.592 | 0.455 | 0.136 | 10 | LGI1 | Body | 0.00082 | |
| cg00435063 | 0.449 | 0.553 | 0.104 | 7 | PTPRN2 | Body | 0.0009 | |
| cg10284704 | 0.773 | 0.645 | 0.128 | 15 | IGR | 0.0009 | ||
| cg03962019 | 0.799 | 0.689 | 0.110 | 1 | IGR | 0.00095 | ||
| PreA | cg17292337 | 0.092 | 0.351 | 0.258 | 12 | IGR | 1.62E-05 | |
| cg20790798 | 0.456 | 0.611 | 0.156 | 5 | IGR | 1.99E-05 | ||
| cg24517467 | 0.444 | 0.602 | 0.158 | 7 | IGR | 2.12E-05 | ||
| cg14950747 | 0.915 | 0.796 | 0.118 | 19 | ZFR2 | Body | 7.18E-05 | |
| cg24449463 | 0.480 | 0.280 | 0.200 | 1 | DCAF6 | Body | 9.36E-05 | |
| cg23084986 | 0.748 | 0.645 | 0.103 | 1 | RAB42 | TSS1500 | 0.00011 | |
| cg07415266 | 0.562 | 0.686 | 0.124 | 2 | IL18RAP | Body | 0.00017 | |
| cg27216788 | 0.266 | 0.459 | 0.193 | 16 | ST3GAL2 | 3’UTR | 0.00025 | |
| cg06821828 | 0.262 | 0.548 | 0.286 | 14 | IGR | 0.00026 | ||
| cg20253542 | 0.872 | 0.718 | 0.155 | 7 | IGR | 0.00026 | ||
| cg06259934 | 0.48 | 0.322 | 0.162 | 10 | ZMIZ1 | Body | 0.0003 | |
| cg02721751 | 0.837 | 0.651 | 0.186 | 6 | SLC44A4 | Body | 0.00032 | |
| cg27614534 | 0.855 | 0.710 | 0.145 | 18 | ALPK2 | TSS1500 | 0.00035 | |
| cg07342647 | 0.847 | 0.669 | 0.178 | 16 | IGR | 0.00041 | ||
| cg04051335 | 0.641 | 0.479 | 0.162 | 5 | C5orf62 | TSS1500 | 0.00045 | |
| cg17071479 | 0.736 | 0.608 | 0.128 | 16 | SRL | Body | 0.00046 | |
| cg26457868 | 0.495 | 0.340 | 0.155 | 10 | SUFU | Body | 0.00053 | |
| cg17045772 | 0.859 | 0.738 | 0.122 | 1 | CTPS | TSS1500 | 0.00053 | |
| cg15397200 | 0.618 | 0.738 | 0.119 | 2 | IGR | 0.00053 | ||
| cg05867245 | 0.563 | 0.872 | 0.310 | 20 | ZBTB46 | Body | 0.00055 | |
| MA | cg22960347 | 0.361 | 0.247 | 0.115 | 7 | IGR | 7.03E05 | |
| cg00216549 | 0.631 | 0.529 | 0.102 | 1 | DDAH1 | Body | 0.00011 | |
| cg05146544 | 0.109 | 0.227 | 0.118 | 2 | IGR | 0.00023 | ||
| cg24224080 | 0.331 | 0.220 | 0.111 | 5 | IGR | 0.00026 | ||
| cg24449463 | 0.451 | 0.310 | 0.140 | 1 | DCAF6 | Body | 0.00029 | |
| cg10354380 | 0.573 | 0.458 | 0.115 | 6 | IL17F | TSS200 | 0.00032 | |
| cg01911494 | 0.290 | 0.177 | 0.113 | 16 | LMF1 | Body | 0.00049 | |
| cg27082921 | 0.803 | 0.655 | 0.147 | 19 | ATP8B3 | Body | 0.00053 | |
| cg11613005 | 0.340 | 0.443 | 0.103 | 5 | GPR151 | 1stExon | 0.00068 | |
| cg19534681 | 0.120 | 0.237 | 0.117 | 2 | IGR | 0.00073 | ||
| cg17487381 | 0.391 | 0.272 | 0.119 | 9 | IGR | 0.00098 | ||
| cg25635251 | 0.497 | 0.652 | 0.156 | 11 | IGR | 0.00099 | ||
| cg23989344 | 0.719 | 0.591 | 0.128 | 10 | ST8SIA6 | Body | 0.00107 | |
| cg00034755 | 0.785 | 0.678 | 0.107 | 22 | IGR | 0.00114 | ||
| cg22891588 | 0.700 | 0.594 | 0.105 | 5 | LRRC70 | 1stExon | 0.00131 | |
| cg04676645 | 0.820 | 0.704 | 0.116 | 8 | CLU | Body | 0.00166 | |
| cg16246747 | 0.722 | 0.607 | 0.115 | 2 | ACOXL | 5’UTR | 0.00176 | |
| cg00945409 | 0.400 | 0.222 | 0.177 | 10 | ZMIZ1AS1 | Body | 0.0019 | |
| cg19868364 | 0.560 | 0.663 | 0.103 | 14 | IGR | 0.00203 | ||
| cg06159404 | 0.188 | 0.352 | 0.164 | 10 | IGR | 0.00217 |
DMC: differential methylation CpG sites,
Transcription starts sites (TSS 200 pb1500 pb), intergenic regions (IGR)
Table 3.
Altered common pathways in MSC, PreA and MA adipogenesis stages in diabetic obese patients (ODs) by DMCs enrichment pathway analysis
| Go term | Gene number in OD-MSC (p-value) |
Gene number in OD-PreA (p-value) |
Gene number in OD-MA (p-value) |
|---|---|---|---|
| biological adhesion | 30(0.00002) | 114(0.0003) | 23(0.012) |
| negative regulation of macromolecule biosynthetic process | 17(0.03) | 93(0.0003) | 19(0.014) |
| regulation of apoptosis | 25(0.01) | 123(0.002) | 25(0.015) |
| regulation of cell death | 25(0.01) | 126(0.001) | 25(0.018) |
| regulation of programmed cell death | 25(0.01) | 124(0.002) | 25(0.017) |
| cell-cell adhesion | 14(0.001) | 46(0.016) | |
| haemopoiesis | 10(0.02) | 47(0.0004) | |
| haemopoietic or lymphoid organ development | 12(0.007) | 50(0.0006) | |
| homophilic cell adhesion | 9(0.002) | 27(0.005) | |
| immune system development | 12(0.01) | 50(0.002) | |
| phosphorylation | 24(0.01) | 140(1.73E-06) | |
| protein amino acid phosphorylation | 21(0.01) | 122(8.98E-07) | |
| regulation of small GTPase mediated signal transduction | 10(0.03) | 55(1.03E-05) | |
| cell-cell signalling | 106(2.30E-05) | 19(0.033) | |
| embryonic morphogenesis | 61(6.28E-05) | 13(0.012) | |
| epithelial tube morphogenesis | 14(0.04) | 5(0.036) | |
| epithelium development | 37(0.04) | 11(0.010) | |
| forebrain development | 31(0.003) | 10(0.002) | |
| induction of apoptosis | 50(0.03) | 12(0.038) | |
| morphogenesis of an epithelium | 25(0.0006) | 8(0.002) | |
| negative regulation of biosynthetic process | 101(4.09E-05) | 19(0.022) | |
| negative regulation of nitrogen compound metabolic process | 87(0.0007) | 17(0.034) | |
| negative regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 86(0.0007) | 17(0.031) | |
| negative regulation of RNA metabolic process | 62(0.002) | 13(0.039) | |
| positive regulation of gene expression | 91(0.004) | 19(0.025) | |
| positive regulation of macromolecule metabolic process | 138(0.0001) | 25(0.031) | |
| positive regulation of nitrogen compound metabolic process | 105(0.0006) | 21(0.018) | |
| positive regulation of transcription | 89(0.004) | 18(0.036) | |
| positive regulation of transcription from RNA polymerase II promoter | 59(0.01) | 15(0.010) | |
| regulation of carbohydrate catabolic process | 7(0.007) | 3(0.035) | |
| regulation of carbohydrate metabolic process | 11(0.01) | 4(0.036) | |
| regulation of cellular carbohydrate catabolic process | 7(0.007) | 3(0.035) | |
| regulation of transcription from RNA polymerase II promoter | 112(0.002) | 25(0.004) | |
| tissue morphogenesis | 34(0.007) | 9(0.0019) | |
| tube development | 40(0.006) | 10(0.022) | |
| tube morphogenesis | 24(0.02) | 7(0.032) | |
| anterior/posterior pattern formation | 7(0.04) | 7(0.047) | |
| cell adhesion | 30(2.35E-05) | 23(0.012) | |
| negative regulation of cellular biosynthetic process | 17(0.04) | 19(0.018) | |
| negative regulation of transcription | 15(0.03) | 17(0.012) |
*yellow = low enrichment; green = high enrichment; DMC: differential methylation CpG sites; MSC: mesenchymal stem cell; PreA: preadipocyte; MA: mature adipocyte.
Differentially methylated regions in adipogenesis
We analysed differentially methylated regions (DMRs) with the Bumphunter library provided in the Chip Analysis Methylation Pipeline (ChAMP) package to compare OND and OD samples. We found 17 DMRs distributed as follows: 8 in MSC, 4 in PreA, and 5 in MA. Moreover, among the 17 DMRs, 10 were associated with a single gene, including: HOXA11, CCHCR1, BLK, CAT, and NSF in the MSC stage; HOXA5 and RPH3AL in the PreA stage; and CCHCR1, HOXA5, and RPH3AL in the MA stage (Table 4). Interestingly, the HOX gene family had DMRs in all adipogenesis stages.
Table 4.
DMRs obtained from three adipogenesis stages in OD samples
| Stage | DMR | Chromo some |
DMR start | DMR end | P value | Related gene |
|---|---|---|---|---|---|---|
| MSC | DMR 1 | 7 | 27,183,946 | 27,185,512 | 0.0002 | HOXA11 |
| DMR 2 | 12 | 115,134,148 | 115,135,333 | 0.0005 | ||
| DMR 3 | 6 | 31,148,332 | 31,148,666 | 0.002 | CCHCR1, PSORS1C3 | |
| DMR 4 | 1 | 119,531,625 | 119,532,352 | 0.006 | ||
| DMR 5 | 8 | 11,560,299 | 11,560,851 | 0.008 | BLK | |
| DMR 6 | 11 | 34,460,182 | 34,461,028 | 0.008 | CAT | |
| DMR 7 | 8 | 65,492,280 | 65,492,936 | 0.009 | ||
| DMR 8 | 17 | 46,651,722 | 46,652,501 | 0.03 | NSF | |
| PreA | DMR 1 | 7 | 27,142,204 | 27,143,585 | 0.0009 | HOXA5 |
| DMR 2 | 8 | 65,492,280 | 65,492,936 | 0.008 | ||
| DMR 3 | 4 | 111,532,996 | 111,533,951 | 0.02 | ||
| DMR 4 | 17 | 259,755 | 260,058 | 0.03 | RPH3AL | |
| MA | DMR 1 | 6 | 31,148,332 | 31,148,666 | 0.008 | CCHCR1 |
| DMR 2 | 7 | 27,142,799 | 27,143,788 | 0.01 | HOXA5 | |
| DMR 3 | 10 | 729,204 | 729,956 | 0.02 | ||
| DMR 4 | 8 | 65,492,280 | 65,492,846 | 0.02 | ||
| DMR 5 | 17 | 259,755 | 260,058 | 0.02 | RPH3AL |
DMR: differentially methylated region
Alterations of DNA methylation correlated with gene expression levels
In order to identify the functional consequences of DNA methylation alterations, a correlation between the beta value of 115 DMCs persisting during adipogenesis with gene expression was performed in the MSC and MA stages. A total of 14 DMCs (located on 11 genes) showed a significant correlation with their gene expression: 7 in MSC (CLU/cg04676645, DAXX/cg00117005, FRK/cg26893134, LCLAT1/cg04618327, LPP/cg21354037, MICA/cg25285646 and PRDM16/cg11113363), 4 in MA (DIP2C/cg10064922, MYO10/cg02895120, PLD1/cg05771324 and THSD7A/cg26480858), and 3 located on FRK, which were shared in the two stages (cg26557270, cg18764771 and cg15226275; Supplemental Table S12). Using these 11 genes, Fc gamma R-mediated phagocytosis and Glycerophospholipid metabolism pathway, were identified as significantly enriched (Supplemental Table S13).
Discussion
Tissue homoeostasis is largely maintained by stem cells, which self-renew and give rise to mature cells to build different types of tissues [21]. It has been reported that alterations in MSCs can lead to the development of diseases [22]. When MSCs are activated, there is a huge regulation to express or turn off transcription factors to establish cellular commitment to the final lineage [12]. Several studies have shown that epigenetic alterations in WAT play a fundamental role in the development of diseases, such as T2D and metabolic syndrome. Epigenetic remodelling can occur during differentiation processes and in different metabolic states. It has been shown that the epigenetic alterations can be heritable and reversible [23–27]. However, it remains unclear how epigenetic alterations in DNA that occur during adipogenesis impact metabolic commitments. Identifying altered epigenetic markers during the differentiation process might provide a deeper understanding of the role of epigenetics in metabolic diseases.
In this study, we analysed the MSC, PreA, and MA stages of adipogenesis. A previous expression study showed that, in adipogenesis, the step between MSC and MA stages was crucial for establishing a specialized cell phenotype [28]. Consistent with the findings from previous studies [23,27,29], when we compared global methylation in OD and OND tissue samples, we found small, statistically insignificant differences (Supplemental Figure 1). However, we found specific CpG methylation alterations during the three adipogenesis stages in the OD samples. These alterations were similar to those documented previously in MSCs and WAT [23,30]. To the best of our knowledge, this study was the first to compare differential DNA methylation during three stages of the adipogenesis process, from ex vivo MSC differentiation obtained from obese patients with and without diabetes.
It has been proposed that DNA methylation might participate in epigenetic memory and that alterations in methylation were associated with complex diseases, such cancer [31]. However, this notion has been poorly addressed in T2D [32] Here, we provided evidence that altered epigenetic memory participates in T2D. In this study, 115 DMCs persisted throughout the three adipogenesis stages in OD samples (Figure 1a). Thus, we could identify pathways common to all three stages studied that were involved in the functional regulation of adipogenesis. These pathways were involved in programmed cell death, immune effector processes, cell adhesion, macromolecular biosynthesis, and differentiation (Table 3 and Supplemental Table S8). Although these pathways have been widely associated with the development of diabetes, few studies have shown that altered methylation profiles can affect these pathways during adipogenesis in patients with diabetes. The only previous study that documented altered DNA methylation in MSCs obtained from obese patients with T2D suggested that adipose tissue could retain epigenetic memory after a stimulus [30].
Our PCA analysis of the 115 DMCs showed that the OD and OND samples were in-dependently clustered (Figure 2). Interestingly, there was a positive correlation between the %Hb1Ac and PC1 (Supplemental Figure 4). Furthermore, 14 of the persisting 115 DMCs shown a possible role in the regulation of gene expression (Supplemental Table 12). From these, MYO10 and PLD1 were related to Fc gamma R-mediated phagocytosis, as well as PLD1 and LCLAT1 were to Glycerophospholipid metabolism in enrichment pathways analysis (Supplemental Table 13). Both pathways have been related to the development of T2D. The first pathway is associated to the crosstalk between adipocytes and macrophages during clearance of dead cells [33]. The second one, glycerophospholipids, has been broadly reported as a participant in the dysregulation of insulin sensitivity [34]. Otherwise, FRK is shown to have three DMCs in both MSC and MA. These DMCs were located in the gene promoter region, with a significant correlation with gene expression. Our findings support a previous report that documented that FRK expression can be modulated by site-specific promoter methylation in breast cancer cells [35]. FRK is a potent positive regulator of PTEN [36], which has recently been reported as a critical regulator of leptin-sympathetic loops and it can be modulated in adipose tissue by external stimuli such as environmental enrichment in animal models [37,38]. Furthermore, in our analysis FRK shows hypermethylation and under-expression in OD patients. All this suggests a role in adipose metabolism due to persistent DNA methylation alteration, although further studies are needed to corroborate it.
Additionally, we found altered methylation in specific genes that occurred in two of the three differentiation stages. For example, the HOXA5 gene displayed DMRs in PreA and MA. Of note, a DMR was also found in the HOXA11 in MSC. The HOX family influences fat cell development, and it is activated through bone morphogenetic protein signalling and the WNT pathway in early differentiation phases [39].
We also identified DMCs on specific genes in each of the stages analysed. In the MSC stage, an enrichment analysis of the genes that contained DMCs revealed involved terms in the gamma aminobutyric acid (GABA) signalling pathway (Supplemental Table S5). This finding was consistent with findings in a recent study that suggested that adipose-derived stem cells responded to GABA by suppressing macrophage infiltration and enhancing insulin action in an obesity model. Interestingly, in the present study, the top gene with DMCs in the MSC stage was THSD7A, which was previously shown to be involved in the interaction between obesity and T2D. This gene participates in angiogenesis by promoting endothelial cell migration and tube formation [40]. Other interesting genes that were among the top 20 genes with DMCs in the MSC stage were RIMS1 [41], RNF186 [42], and PTPRN2 [43], which were previously associated with impaired insulin secretion. Another gene on this list was LGI3 [44], which was proposed to be an adipogenesis suppressor. This finding suggests that the alterations initiated in the MSC stage might affect the maintenance of adipose tissue.
The next step in adipogenesis, the PreA stage, showed greater epigenetic alterations in the OD group. In this stage, the top genes with significant DMCs participated in pathways relevant to adipocyte functions, such as fat accumulation (DCAF6 [45], SUFU [46], and ZFR2 [47]), insulin resistance (ZMIZ1 [48]), and inflammation (IL18RAP [49]). We also identified master regulators of adipogenesis, including genes in the JAK-STAT cascade, PPARG, GATA3, JAK2, and STAT5A [50,51]. Thus, the PreA stage appeared to be a preparation for the commitment phase, where cells would take on the functions of a mature adipocyte. Our results also suggest that DNA methylation alterations in the PreA stage play a fundamental role in the dysregulation of adipocyte function in patients with diabetes.
In the last stage of differentiation, the MA stage, our enrichment analysis showed that DMCs were identified in pathways involved in cell adhesion and embryonic morpho genesis [52,53]. In the MA stage, we identified new alterations in key pathways for adipocyte metabolism, including pathways implied by the regulation of transcription and the negative regulation of macromolecule biosynthesis. The top 20 genes altered by DMCs were related to the regulation of adipogenesis (DDAH1) [54], the inflammatory response (ST8SIA6 and IL17F) [55,56], obesity phenotypes (GPR151 and ZMIZ1-AS1) [57,58] hypertriglyceridaemia (LMF1) [59], thermogenesis (ACOXL) [60], and insulin resistance (CLU) [61]. These findings suggest that, in obese patients with diabetes, pathways involved in crucial metabolic functions in WAT, such as lipid metabolism or catabolism and endocrine roles were dysregulated by epigenetic alterations.
Among the top 20 genes altered with DMCs in each differentiation stage, we also found new candidate genes related to diabetes. For example, in the MSC stage, DMCs were found on ABCC5-AS1 [62], IPW [63], GNAQ [64], and PLXDC2 [65] In the PreA stage, DMCs were found on ST3GAL2 [58] and ALPK2 [59]. In the MA stage, DMCs were found on LRRC70 [66]. There is limited evidence on the function of these genes in human adipose tissue. However, there is evidence that has suggested the involvement of these genes in T2D pathogenesis. For example, serum levels of insulin, glucose, or free fatty acids were related to OR1B1, ABCC5-AS1, and ST3GAL2; adiposity was related to IPW and ABCC5-AS1; and lipid or adipogenesis signalling pathways were related to GNAQ, ALPK2, and LRRC70. Further studies on these genes are needed to delve into their roles in human adipose tissue and T2D.
Taken together, the results presented here indicate that some alterations in methylation persisted through the adipocyte differentiation process. The main alterations were observed in key genes involved in the regulation of adipocyte differentiation and function (i.e., lipid metabolism and insulin sensitivity), immune effector processes, and apoptosis signalling. Additionally, this study revealed specific methylated sites in each of the three analysed stages that affected particular pathways. Thus, both transient and persistent alterations in methylation could contribute to impairments in mature adipocyte function and lead to the development of T2D. Specifically, in the MSC stage, altered methylation occurred mainly in the GABA signalling pathway. In the PreA stage, where we found the highest number of genes with altered methylation, the alterations were mainly found in the glucose transport and JAK-STAT pathways. In the MA stage, altered methylation occurred mainly in genes involved in the negative regulation of macromolecule metabolic processes and energy generation. Thus, these data have bridged some knowledge gaps regarding the role of DNA methylation remodelling during adipogenesis in diabetes. Future analyses are needed to deepen our understanding of the pathophysiology of these metabolic processes.
Materials and methods
Subject recruitment and clinical evaluation
We included 17 women with a BMI ≥35 kg/m2 that were undergoing bariatric surgery at the Integral Clinics for Obesity and Metabolic Diseases in Tláhuac Hospital, Mexico City. All patients were clinically and biochemically evaluated before surgery. Visceral WAT was obtained during surgery, as previously described. Patients were diagnosed without (control) or with diabetes, according to the American Diabetes Association criteria. Patients with hypothyroidism were excluded.
Isolation of adipose-derived stem cells
MSCs were isolated from visceral adipose tissue as was previously described by Zuk P. et ale 2019 [67]. Briefly, a fresh WAT biopsy (median: 2–3 g) was cut into small pieces, washed with phosphate-buffered saline (PBS, Life Technologies, USA) and transferred to a sterile digestion solution, containing HAM- DMEM/F12 medium (Life Technologies, CA, USA), 1 mg/ml collagenase II (Sigma–Aldrich, St. Louis, MO), and 10 mg/ml bovine serum albumin (BSA, Sigma-Aldrich, St. Louis, MO). Samples were incubated at 37°C for 45 min. The digested tissue was centrifuged at 1200 × g for 8 min. The cell suspension (containing stromal vascular cells) was filtered through a cell strainer (Nalgene, Rochester, NY), and cells were transferred to culture dishes for MSC proliferation in culture medium DMEM (Life Technologies, CA, USA) supplemented with 10% foetal bovine serum (Gibco, Fisher Scientific, USA), streptomycin (10,000 μg/ml, Gibco, Fisher Scientific, USA), and penicillin (10,000 units/ml, Gibco, Fisher Scientific, USA). Cells were grown in a humidified incubator with 5% CO2 at 37°C. Medium was changed every day. All cells were expanded until passage four. The MSCs were verified in a flow cytometer (FACSARIA-BD) using a set of MSC surface markers (CD90, CD44, CD105, and CD29) for cells that did not express CD45 (Abcam ab93758, USA) and it was also corroborated by gene expression (Supplemental Figure S5A-B). These cells were stimulated to differentiate into mature adipocytes.
Adipogenic differentiation
To analyse the DNA methylation profiles we prepared 51 culture plates of cells derived from 17 patients. The cells were seeded at a density of 1,000,000 by plate (60 mm diameter). Day 0 was defined as the day the cells reached 80–90% confluency (MSCs). At this point, 17 culture dishes were harvested to examine stage 1. In the remaining 34 plates, differentiation was induced with an adipogenic medium that contained DMEM supplemented with 250 nM dexamethasone (Sigma Aldrich, USA), 0.5 mM 3-isobutyl-1 methylxanthine (Sigma Aldrich, USA), and 10 µg/ml insulin (Sigma Aldrich, USA). The differentiation medium was replaced every 72 h, and adipocytes were grown until day 3 (PreA). At this point, 17 culture dishes were prepared to examine stage 2. The remaining 17 plates were allowed to mature until day 18 (MA). The MAs displayed adipocyte morphology with massive lipid droplet accumulation, identified with oil red stain (Supplemental Figure S6). Similar to the methylation analysis, 22 culture plates were prepared to analyse the gene expression profile at the beginning (day 0, n = 11) and at the end (day 18, n = 11) of the differentiation process, corresponding to 11 patients randomly selected (6 OD and 5 OND). MAs displayed an increased gene expression of adipogenic markers such as: FABP4, PPARG and PPARGC1A (Supplemental Figure S5 C).
DNA extraction
DNA was extracted with the QIAamp DNA Mini Kit (Qiagen, Valencia CA) from cells in all three differentiation stages (MSC, PreA, and MA). DNA was quantified with the NanoDrop® ND-1000 v3.5.2. (NanoDrop Technologies Inc.). During the DNA extraction process, two PreA samples were eliminated because DNA concentration was not enough.
RNA extraction
RNA was obtained from 11 MSC and 11 MA cultures using TRIzol® (Invitrogen, Life Technologies, CA, USA), according to the manufacturer’s instructions. RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, USA) and all samples had an acceptable RIN score >8. Gene expression analysis was performed using the Clariom S Human Expression Assay (Affymetrix, USA).
Genome methylation analysis
We analysed DNA methylation with the Infinium Methylation EPIC Bead Chip kit (Illumina, Inc., San Diego, CA). This assay covers over 850,000 methylation sites per sample at single nucleotide resolution. OD and OND controls were placed randomly among the Bead Chips. We performed the Illumina Infinium methylation assay protocol, according to manufacturer’s instructions, and we used iScan to scan the arrays. Methylation data were visualized and analysed with Genome Studio software version 2011.1 (Illumina) and the Methylation Module. Only samples that passed quality control evaluations were used in the analysis. Methylation beta values were estimated as the ratio of the signal intensity of the methylated probe to the sum of methylated and unmethylated probes. The beta values ranged from 0 to 1.
Statistical analysis
Clinical data are reported as the mean and standard deviations (SD). Differences in biochemical and anthropometric data between OND and OD groups were tested with the unpaired Wilcox test (Table 1). Descriptive statistics for genome-wide methylation were calculated in R, version 3.1 [68]. The ChAMP package was used to perform the control quality (C) evaluation and QC included detection of p-value cut-offs (>0.01), the raw intensities normalization of DNA methylation and Single Variation Deviation analysis. The package was also used to perform the differential methylation analysis (DMCs and DMRs). The Delta-beta value was obtained by subtracting the average Beta value of the OD group from the average Beta value of the OND group for each locus. A DMC was defined when p-value <0.05 and absolute mean Delta-Beta >0.1 [69]. The oligo package was used to normalize the intensity of the gene expression [70]. The Pearson’s correlation between DNA methylation and gene expression was performed comparing the Beta values of CpGs and the gene expression where the CpGs were located.
Pathway analyses
DAVID V.6.8 was used to analyse potentially altered pathways from the list of genes that were differentially methylated between OD and OND samples [71].
Supplementary Material
Acknowledgments
Elaheh Mirzaeicheshmeh is a doctoral student from the programa de doctorado en ciencias biomedicas Universidad Nacional Autonoma de Mexico (UNAM) and has received CONACyT fellowship 280204.
Funding Statement
This work was supported by the Consejo Nacional de Ciencia y Tecnología, Mexico [280204].
Abbreviations
- Type 2 diabetes (T2D)
- White adipose tissue (WAT)
- Mesenchymal stem cells (MSCs)
- Obese without T2D (OND)
- Obese with T2D (OD)
- Preadipocyte (PreA)
- Mature adipocyte (MA)
- Differentially methylated CpG sites (DMCs)
- Differentially methylated regions (DMRs)
- Principal component analysis (PCA)
- Gamma-aminobutyric acid (GABA)
Author contributions
Conceptualization, LO, FBO, FCC, and EM.; Clinical Research and Metabolic Phenotyping, CZ and ECM; Methodology, EM and PB; formal analysis, LO, FBO, HGO and EM; resources, LO and CZ; data curation, EM and FBO; writing – original draught preparation, LO and EM.; writing review and editing, EM, LO, FBO, CZ, FCC, PBP, HGO, AM, MGSM, YSA, and ECM; visualization, EM, LO, FBO, CZ, FCC, PBP, HGO, AM, MGSM, YSA, and ECM; supervision, LO and FBO; project administration, LO, CZ and AM; funding acquisition LO and CZ. All authors have read and agreed to the published version of the manuscript.
Institutional review board statement:
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Ethics Committee of Instituto Nacional de Medicina Genómica (C1_29/2011).
Informed consent statement:
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement:
The methylation dataset can be found in https://www.ebi.ac.uk/ with accession number: E-MTAB-10178.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Cohen P, Spiegelman BM.. Cell biology of fat storage. MBoC. 2016;27(16):2523–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Arner P, Bernard S, Appelsved L, et al. Adipose lipid turnover and long-term changes in body weight. Nat Med. 2019;25(9):1385–1389. [DOI] [PubMed] [Google Scholar]
- [3].Ghaben AL, Scherer PE. Adipogenesis and metabolic health. Nat Rev Mol Cell Biol. 2019;20(4):242–258. [DOI] [PubMed] [Google Scholar]
- [4].Arner P, Arner E, Hammarstedt A, et al. Genetic predisposition for type 2 diabetes, but not for overweight/ obesity, is associated with a restricted adipogenesis. PLoS ONE. 2011;6(4):e18284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kim JI, Huh JY, Sohn JH, et al. Lipid-overloaded enlarged adipocytes provoke insulin resistance independent of inflammation. Mol Cell Biol. 2015;35(10):1686–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jiang S-Z, Lu W, and Zong X-F, et al. Obesity and hypertension Pharmacological Research. 2016. ;122 :1–7. [Google Scholar]
- [7].Zhu J, Su X, Li G, et al. Systematic review/meta-analysis the incidence of acute myocardial infarction in relation to overweight and obesity: a meta-analysis. aoms. 2014;5:855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Guilherme A, Henriques F, Bedard AH, et al. Molecular pathways linking adipose innervation to insulin action in obesity and diabetes mellitus. Nat Rev Endocrinol. 2019;15(4):207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Langin D. In and out: adipose tissue lipid turnover in obesity and dyslipidemia. Cell Metab. 2011;14(5):569–570. [DOI] [PubMed] [Google Scholar]
- [10].Castro AM, L. E. M-DLC, Pantoja Meléndez CA. Low-grade inflammation and its relation to obesity and chronic degenerative diseases. Revista Médica del Hospital General de México. 2017;80(2):101–105. [Google Scholar]
- [11].Spalding KL, Arner E, Westermark PO, et al. Dynamics of fat cell turnover in humans. Nature. 2008;453(7196):783–787. [DOI] [PubMed] [Google Scholar]
- [12].Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12(11):722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Reusch JEB, Colton LA, Klemm DJ. CREB activation induces adipogenesis in 3T3-L1 cells. Mol Cell Biol. 2000;20(3):1008–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Majka SM, Barak Y, Klemm DJ. Concise review: adipocyte origins: weighing the possibilities. STEM CELLS. 2011;29(7):1034–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Christodoulides C, Lagathu C, Sethi JK, et al. Adipogenesis and WNT signalling. Trends Endocrinol Metab. 2009;20(1):16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7(12):885–896. [DOI] [PubMed] [Google Scholar]
- [17].Nic-Can GI, Rodas-Junco BA, Carrillo-Cocom LM, et al. Epigenetic regulation of adipogenic differentiation by histone lysine demethylation. IJMS. 2019;20(16):3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Reddy MA, Zhang E, Natarajan R. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia. 2015;58(3):443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kennedy EM, Powell DR, Li Z, et al. Galactic cosmic radiation induces persistent epigenome alterations relevant to human lung cancer. Sci Rep. 2018;8(1):6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rønningen T, Shah A, Reiner AH, et al. Epigenetic priming of inflammatory response genes by high glucose in adipose progenitor cells. Biochem Biophys Res Commun. 2015;467(4):979–986. [DOI] [PubMed] [Google Scholar]
- [21].Via AG, Frizziero A, Oliva F. Biological properties of mesenchymal stem cells from different sources. 9. [PMC free article] [PubMed] [Google Scholar]
- [22].Kornicka K, Houston J, Marycz K. Dysfunction of mesenchymal stem cells isolated from metabolic syndrome and type 2 diabetic patients as result of oxidative stress and autophagy may limit their potential therapeutic use. Stem Cell Rev Rep. 2018;14(3):337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Barajas-Olmos F, Centeno-Cruz F, Zerrweck C, et al. Altered DNA methylation in liver and adipose tissues derived from individuals with obesity and type 2 diabetes. BMC Med Genet. 2018;19(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Togliatto G, Dentelli P, Gili M, et al. Obesity reduces the pro-angiogenic potential of adipose tissue stem cell-derived extracellular vesicles (EVs) by impairing mir-126 content: impact on clinical applications. Int J Obes. 2016;40(1):102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nilsson E, Jansson PA, Perfilyev A, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63(9):2962–2976. [DOI] [PubMed] [Google Scholar]
- [26].Broholm C, Olsson AH, Perfilyev A, et al. Human adipogenesis is associated with genome-wide DNA methylation and gene-expression changes. Epigenomics. 2016;8(12):1601–1617. [DOI] [PubMed] [Google Scholar]
- [27].Castellano-Castillo D, Moreno-Indias I, Sanchez-Alcoholado L, et al. Altered adipose tissue DNA methylation status in metabolic syndrome: relationships between global DNA methylation and specific methylation at adipogenic, lipid metabolism and inflammatory candidate genes and metabolic variables. JCM. 2019;8(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].van de Peppel J, Strini T, Tilburg J, et al. Identification of three early phases of cell-fate determination during osteogenic and adipogenic differentiation by transcription factor dynamics. Stem Cell Reports. 2017;8(4):947–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kirchner H, Sinha I, Gao H, et al. Altered DNA methylation of glycolytic and lipogenic genes in liver from obese and type 2 diabetic patients. Mol Metab. 2016;5(3):171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Andersen E, Ingerslev LR, Fabre O, et al. Preadipocytes from obese humans with type 2 diabetes are epigenetically reprogrammed at genes controlling adipose tissue function. Int J Obes. 2019;43(2):306–318. [DOI] [PubMed] [Google Scholar]
- [31].Kim M, Costello J, Methylation: DNA. An epigenetic mark of cellular memory. Exp Mol Med. 2017;49(4):e322–e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sundararajan J. Epigenetic mechanisms of metabolic memory in diabetes. Circulation Research. 2012;110: 1039–1041 . [DOI] [PubMed] [Google Scholar]
- [33].Kuroda M, Sakaue H. Adipocyte death and chronic inflammation in obesity. J Med Invest. 2017;64(3.4):193–196. PMID: 28954980. [DOI] [PubMed] [Google Scholar]
- [34].Chang W, Hatch GM, Wang Y, et al. The relationship between phospholipids and insulin resistance: from clinical to experimental studies. J Cell Mol Med. 2019Feb;23(2):702–710. Epub 2018 Nov 6. PMID: 30402908; PMCID: PMC6349352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bagu ET, Miah S, Dai C, et al. Repression of Fyn-related kinase in breast cancer cells is associated with promoter site-specific CpG methylation. Oncotarget. 2017Feb14;8(7):11442–11459. PMID: 28077797; PMCID: PMC5355277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yim EK, Peng G, Dai H, et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell. 2009Apr7;15(4):304–314. PMID: 19345329; PMCID: PMC2673492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huang W, Queen NJ, McMurphy TB, et al. Adipose PTEN regulates adult adipose tissue homeostasis and redistribution via a PTEN-leptin-sympathetic loop. Mol Metab. 2019Dec; 30: 48–60. Epub 2019 Sep 28. PMID: 31767180; PMCID: PMC6812328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Huang W, Queen NJ, McMurphy T, et al. (2020). Adipose PTEN acts as a downstream mediator of a brain-fat axis in environmental enrichment. [DOI] [PMC free article] [PubMed]
- [39].Bhatlekar S, Fields JZ, Boman BM. Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. 2018;2018:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nizamuddin S, Govindaraj P, Saxena S, et al. A novel gene THSD7A is associated with obesity. Int J Obes. 2015;39(11):1662–1665. [DOI] [PubMed] [Google Scholar]
- [41].Andersson SA, Olsson AH, Esguerra JLS, et al. Reduced insulin secretion correlates with decreased expression of exocytotic genes in pancreatic islets from patients with type 2 diabetes. Mol Cell Endocrinol. 2012;364(1–2):36–45. [DOI] [PubMed] [Google Scholar]
- [42].Tong X, Zhang Q, Wang L, et al. RNF186 impairs insulin sensitivity by inducing er stress in mouse primary hepatocytes. Cell Signal. 2018;52:155–162. [DOI] [PubMed] [Google Scholar]
- [43].Lee S. The association of genetically controlled CpG methylation (Cg158269415) of protein tyrosine phosphatase, receptor type N2 (PTPRN2) with childhood obesity. Sci Rep. 2019;9(1):4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kim HA, Park W-J, Jeong H-S, et al. Leucine-rich glioma inactivated 3 regulates adipogenesis through ADAM23. Biochimica Et Biophysica Acta (BBA) - Mol Cell Biol Lipids. 2012;1821(6):914–922. [DOI] [PubMed] [Google Scholar]
- [45].Groh BS, Yan F, Smith MD, et al. The antiobesity factor WDTC 1 suppresses adipogenesis via the CRL 4 WDTC 1 E3 Ligase. EMBO Rep. 2016;17(5):638–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shi Y, Long F. Hedgehog signaling via Gli2 prevents obesity induced by high-fat diet in adult mice. eLife. 2017;6:e31649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Turcot V, Lu Y, and Highland HM, et al. Implicate pathways that control energy intake and expenditure underpinning obesity. Nat Genet. 2018;50(1); 25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Van De Bunt M, Manning Fox JE, Dai X, et al. Transcript expression data from human islets links regulatory signals from genome-wide association studies for type 2 diabetes and glycemic traits to their downstream effectors. PLoS Genet. 2015;11(12):e1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Martínez-Barquero V, Marco GD, Martínez-Hervas S, et al. Are IL18RAP gene polymorphisms associated with body mass regulation? A cross-sectional study. BMJ Open. 2017;7(11):e017875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Richard AJ, Stephens JM. The role of JAK–STAT Signaling in Adipose Tissue Function. Biochim Biophys Acta (BBA) - Mol Basis Dis. 2014;1842(3):431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Al-Mansoori L, Al-Jaber H, and Madani AY, et al. Suppression of GATA-3 increases adipogenesis, reduces inflammation and t improves insulin sensitivity in 3T3L-1 preadipocytes. Cellular Signalling 2020. ;75 109735 . [DOI] [PubMed] [Google Scholar]
- [52].Eguchi J, Wada J, Hida K, et al. Identification of adipocyte adhesion molecule (ACAM), a novel CTX gene family, implicated in adipocyte maturation and development of obesity. Biochem J. 2005;387(2):343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Minakuchi H, Wakino S, Hosoya K, et al. The role of adipose tissue asymmetric dimethylarginine/dimethylarginine dimethylaminohydrolase pathway in adipose tissue phenotype and metabolic abnormalities in subtotally nephrectomized rats. Nephrology Dialysis Transplantation. 2016;31(3):413–423. [DOI] [PubMed] [Google Scholar]
- [54].Crotts SB, Friedman DJ, and Wang Z. Regulation of the immune response by ST8Sia6. The Journal of Immunology. 2020;204(1). 1 Supplement. 228.18. [Google Scholar]
- [55].Ahmed M, Gaffen SL. IL-17 in obesity and adipogenesis. Cytokine Growth Factor Rev. 2010;21(6):449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tanigawa Y, Li J, Justesen JM, et al. Components of genetic associations across 2,138 phenotypes in the UK biobank highlight adipocyte biology. Nature Communications. 2019;10(1). 10.1038/s41467-019-11953-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Chen -W-W, Yang Q, Li X-Y, et al. Identification of a novel and heterozygous lmf1 nonsense mutation in an acute pancreatitis patient with severe hypertriglyceridemia, severe obesity and heavy smoking. Lipids Health Dis. 2019;18(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhu Z, Guo Y, and Shi H, et al. Shared Genetic and Experimental Link between Obesity-Related Traits and Asthma Subtypes in UK Biobank. J Allergy Clin Immunol. 2020;145(2) 537–549 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bradley D, Blaszczak A, and Yin Z, et al. Clusterin impairs hepatic insulin sensitivity and adipocyte clusterin associates with cardiometabolic risk. Diabetes Care. 2019;42(3):466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cyranka M, Veprik A, McKay EJ, et al. Abcc5 knockout mice have lower fat mass and increased levels of circulating GLP‐1. Obesity. 2019;27(8):1292–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zahova S, Isles AR. The role of the prader-Willi syndrome critical interval for epigenetic regulation, transcription and phenotype. Epigenomes. 2018;2(4):18. [Google Scholar]
- [62].Klenke S, Tan S, Hahn S, et al. A functional GNAQ promoter haplotype is associated with altered gq expression and with insulin resistance and obesity in women with polycystic ovary syndrome. Pharmacogenetics and Genomics. 2010;20(8):476–484. [DOI] [PubMed] [Google Scholar]
- [63].Hosseini SM, Boright AP, and Sun L, et al. The association of previously reported polymorphisms for microvascular complications in a meta-analysis of diabetic retinopathy. Human Genetics. 2015. ;134: 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lopez PH, Hahn S, and Aoki K, et al. Mice lacking sialyltransferase st3gal-ii develop late-onset obesity and insulin resistance Glycobiology. 2016;27(nos. 2):129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ambrosi TH, Scialdone A, Graja A, et al. Adipocyte accumulation in the bone marrow during obesity and aging impairs stem cell-based hematopoietic and bone regeneration. Cell Stem Cell. 2017;20(6):771–784.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wei W, Yan Y, and Lei L, et al. Synleurin, a Novel leucine-rich repeat protein that increases the intensity of pleiotropic cytokine responses Biochem Biophys Res Commun. 2003;305:981–988. [DOI] [PubMed] [Google Scholar]
- [67].Zuk M, Zhu H, Mizuno J, et al. «Multilineage cells from human adipose tissue: implications for cell-based therapies,». Tissue Eng. 2001;7(2):211–226. [DOI] [PubMed] [Google Scholar]
- [68].R Core Team . R: a language and environment for statistical computing. Vienna Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
- [69].Tian Y, Morris TJ, Webster AP, et al. ChAMP: updated methylation analysis pipeline for illumina beadChips. Bioinformatics. 2017;33(24):3982–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using David bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The methylation dataset can be found in https://www.ebi.ac.uk/ with accession number: E-MTAB-10178.


