Sir,
The unprecedented severity of the COVID-19 pandemic caused by a novel human coronavirus SARS-CoV-2 has posed one of the most gigantic multidimensional challenges to the public health systems globally1. One major challenge has been the urgent need to find easily executable and economical means of deactivating the virus in the environment such as surface decontamination, commonly used and shared public utility items and non-disposable personnel protective gear. Several techniques ranging from radiation exposure, chemical inactivation, electrostatic and heat treatment have been experimented with and reported for other coronaviruses2. While the detailed structural organization of the SARS-CoV-2 remains incompletely understood, identifying the sensitivity of the virus to environmental factors such as heat and relative humidity has emerged as a key research focus in the context of virus inactivation. This is not only from an economic feasibility viewpoint for developing low-cost mass-scale thermal-based inactivation application but also the fact that climatologic parameters such as temperature and humidity may be important factors in the pandemic dynamics and geographical variations, if any, as suggested by others3.
In the present study, SARS-CoV-2 was exposed in the solution phase to different temperatures and its ultrastructure was studied using negative staining transmission electron microscopy (TEM) as described earlier4 to characterize the nature of morphological changes seen in the virus particle with temperature variation. The study was conducted at the Indian Council of Medical Research (ICMR)-National Institute of Virology, Pune, India. Briefly, the SARS-CoV-2 strain (NIV-2020-770) isolated earlier in this centre and was treated at 2, 4, 12, 36, 45, 50, 65 and 80°C for 30 min in a temperature-controlled dry bath (Eppendorf Company, Hamburg, Germany). Heat-treated virus suspensions were fixed with three per cent glutaraldehyde, negative stained4, and examined under 120 kilovolts (kV) in a TEM (Technai 12 BioTwin™, Thermofisher Scientific, Waltham, MA, USA). Images were digitally recorded and analyzed using a side-mounted 2K × 2K CCD camera4 (MegaView III, Olympus, GmbH, 1748149 Muenster, Germany). A parallel heat-treated, unfixed aliquot of the same was used to infect Vero CCL-81 cells as described earlier5 and monitored for cytopathic effect (CPE). The findings of the present study are shown in the Figure and the Table. SARS-CoV-2 has morphology of a typical coronavirus. It is an enveloped particle with a size variation from 75 to 200 nm. It has a typical fringe of club-shaped peplomers as imaged by a TEM6,7. Each peplomer, also known as the envelope spike glycoprotein, has three segments: a large ectodomain stalk approximately 20 nm in length that is connected into the virion as a single-pass transmembrane anchor and the terminal end of the stalk is 7 nm wide having a trimeric organization of the receptor-binding S proteins8. Recent studies on cryo-electron tomography of intact SARS-CoV-2 with high-resolution structure, conformational flexibility, in situ distribution of peplomers on the virion surface and morphology of infected cell using TEM has shown importance of ultrastructural imaging9,10.
Figure.
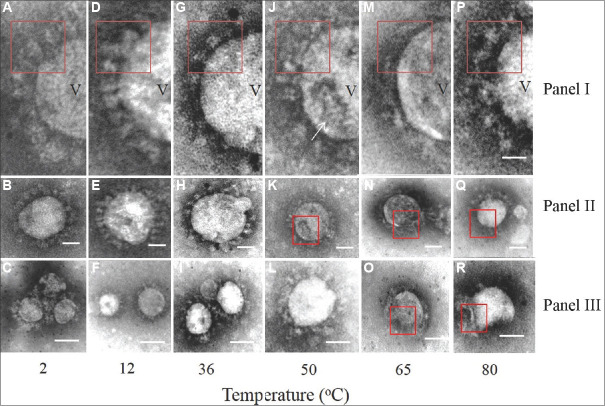
Representative transmission electron microscopy images of the SARS-CoV-2 at different temperatures. Panel I shows the highly magnified images of the envelope glycoprotein projections from selective areas of the virus particles shown in Panel II. Panel C shows representative images from other fields at respective temperature treatment points. (A-C) Images of virus particles treated at 2°C. The envelope spike glycoprotein was prominent as shown in red box outline (A) (D-F) images of virus treated at 12°C. Variation in particle size shown in (F) and distinct envelope projection (D) representative intact virus particles treated at 36°C are shown in (I) with distinct club-shaped terminal knob of the envelope projection shown in red box inset (G). Single virus particles at 36°C and 50°C with intact envelope projections are shown in (H) and (L) respectively. Treatment at 50°C increased the proportion of damaged particles majority of which showed collapsed surfaces (K) and disorganized envelope (J). At 65°C and 80°C, the virus particles were damaged (O,N,R). However, some intact particles were observed (Q). The envelope projections at these temperatures were significantly disorganized in Panel I (M, P). The magnification bar in Panel I, II is 20 nm and in Panel III it is 100 nm. In Panel I, V represents SARS-CoV-2 virion particle; the arrow indicates presence of “moth eaten appearance” on virion particles when subjected to higher temperature.
Table.
Effect of temperature on the cytopathic effect (CPE) of SARS-CoV-2 in vitro
| Temperature (°C) | Pre-exposure cytopathic effect | Post-exposure cytopathic effect |
|---|---|---|
| 2 | +++ | +++ |
| 4 | +++ | +++ |
| 12 | +++ | +++ |
| 36 | +++ | +++ |
| 45 | +++ | +++ |
| 50 | +++ | ++ |
| 65 | +++ | No CPE |
| 80 | +++ | No CPE |
Exposure of SARS-CoV-2 to different temperatures and their corresponding cytopathic effect was evaluated in vitro on Vero CCL-81 cells as described in the text. CPE, cytopathogenic effect; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; +++ represents maximum CPE seen; ++ represents reduced CPE; + indicate the presence of CPE; No CPE, absence of CPE (Only qualitative summarization of experimental observations was done)
A total of 50 single-virus particles were examined in triplicate grids for each temperature data point and the observations were averaged. At temperatures of 2°C (Figure A-C), 12°C (Figure D-F) and 36°C (Figure G-I), the virus particles were seen to be well preserved with distinct envelope projections. The mean value of SARS-CoV-2 particle sizes was calculated from randomly identified single-virus particle images (n=5) treated at a particular temperature and plotted against temperature (°C) with standard deviation as error bars using Prism 5.0, GraphPad Software (San Diego, CA, USA). It showed a minimal size variation of approximately ±4 nm. A field showing minor size variation of the virus in the same field is shown in Figure F. At 50°C (Figure J-L) and at higher temperature points of 65°C (Figure M-O) and 80°C (Figure P-R), morphologically damaged particles and size variation were prominent. Damaged particles showing ‘moth-eaten appearance’ due to loss of glycoprotein projections (Figure J and M) and particles with collapsed surfaces were detected. Similar changes were also seen at 50°C. Significantly decreased particle size was observed at 65°C (Figure M-O) with a high fraction of damaged particles. Majority of the particles were damaged at the 80°C treatment point (Figure P-R). The size variation was most marked at 65°C and above (Supplementary Figure). At 80°C treatment, mostly damaged particles with background debris were seen, envelope projections were lost and the particles were broken with collapsed surfaces (Figure P-R). However, at 80°C treatment for 30 min, a few intact virus particles could still be imaged in the fields scanned (approximately 10-12 particles in random areas of the grid scanned). On higher magnification of these particles, the envelope projection appeared disorganized (red inset box in Figure P).
The observation on viability as evidenced by in vitro CPE of SARS-CoV-2 treated at different temperatures was qualitative. Being a novel virus, an accurate titration method of the virus is still under development while earlier established microtitration methods have been adapted for SARS-CoV-2 in viability assays. In our study, we used an assay as described earlier5. A decrease in the CPE was observed at 50°C with complete loss of CPE at 65 and 80°C (Table). In a recent study, the effect of temperature and relative humidity on survival of SARS-CoV-2 on different surfaces was observed and a temperature-sensitive decrease in the virus viability was reported11. However, identifying the precise ‘kill switch’ temperature of the virus is limited by the high biosafety requirement of experimentation and precision of viability assays.
The clear effect due to the heat treatment was a loss of surface projections. The transmembrane spike glycoprotein (S) of SARS-CoV-2 is a homotrimer comprising two subunits: S1 that binds to the host cellular receptor and a S2 subunit that helps in virus fusion to the host cell. The entire projection as measured from TEM imaging is approximately 15-17 nm12. A review of the literature showed limited TEM studies on the whole SARS-CoV-2 and a few cryo-electron microscopy and X-ray crystallography reports on the molecular structure of the cellular receptor binding by the spike protein4,8,9,10. The overall heat-sensitive molecular changes in the virus remain unknown.
Studies addressing the thermal mechanisms of coronavirus inactivation are only a few13. A review of literature on the temperature-sensitive profile of SARS-CoVs revealed several interesting findings. It is a practical difficulty to do environmental sensitivity experiments on highly transmissible and pathogenic human CoVs due to the stringent laboratory biosafety facility requirement. Thus, several surrogate human and animal CoVs, such as 229E and OC43 have been studied as reviewed by Casanova et al14. Porcine transmissible gastroenteritis virus as a representative coronavirus was suggested to have an inactivation temperature of 45°C15. A review by Kampf et al16 showed that treatment at 60°C for 30 min and 65°C for 15 min could significantly reduce coronavirus infectivity by 4 log10.
In conclusion, the findings of the present study showed the possible effect of temperature above 50°C on the survival of SARS-CoV-2. The limitation of the study was the need to have a broader temperature spectrum to identify the accurate thermal ‘kill switch’ inactivation temperature of SARS-CoV-2 with virus morphology changes. Such studies may have significant potential in assisting development of computational predictive models for SARS-CoV-2 infection dynamics and population spread in varied geographical areas of the world17. From a clinical point of view, the possible role of degraded viral proteins and induction of host heat shock protein response needs to be studied for both potential pathophysiology and treatment development.
Acknowledgment:
Authors acknowledge the support from the NIC COVID team and thank Shri Atul Walimbe of the Bioinformatics Group of NIV for his help with the statistical data analysis.
Footnotes
Financial support & sponsorship: Authors acknowledge the Indian Council of Medical Research, New Delhi, for financial support.
Conflicts of Interest: None.
References
- 1.Gorbalenya AE, Baker SC, Baric RS, Groot RJ, Drosten C, Gulyaeva AA, et al. The species Severe Acute Respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–44. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duan SM, Zhao XS, Wen RF, Huang JJ, Pi GH, Zhang SX, et al. Stability of SARS coronavirus in human specimens and environment and its sensitivity to heating and UV irradiation. Biomed Environ Sci. 2003;16:246–55. [PubMed] [Google Scholar]
- 3.Demongeot J, Flet-Berliac Y, Seligmann H. Temperature decreases spread parameters of the new Covid-19 case dynamics. Biology (Basel) 2020;9:94. doi: 10.3390/biology9050094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prasad S, Potdar V, Cherian S, Abraham P, Basu A. ICMR COVID Team. Transmission electron microscopy imaging of SARS-CoV-2. Indian J Med Res. 2020;151:241–3. doi: 10.4103/ijmr.IJMR_577_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarkale P, Patil S, Yadav PD, Nyayanit DA, Sapkal G, Baradkar S, et al. First isolation of SARS-CoV-2 from clinical samples in India. Indian J Med Res. 2020;151:244–50. doi: 10.4103/ijmr.IJMR_1029_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai MM. Coronavirus: Organization, replication and expression of genome. Ann Rev Microbiol. 1990;44:303–33. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- 7.Davies HA, Macnaughton MR. Comparison of the morphology of three coronaviruses. Arch Virol. 1979;59:25–33. doi: 10.1007/BF01317891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shang J, Ye G, She K, Wan Y, Lou C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–4. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ke Z, Oton J, Qu K, Cortese M, Zila V, McKeane L, et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao J, Zhou H, Huang W, Zhou J, Qiu M, Deng Z, et al. Cell morphological analysis of SARS-CoV-2 infection by transmission electron microscopy. J Thorac Dis. 2020;12:4368–73. doi: 10.21037/jtd-20-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biryukov J, Boydston JA, Dunning RA, Yeager JJ, Wood S, Reese AL, et al. Increasing temperature and relative humidity accelerates inactivation of SARS-CoV-2 on surfaces. mSphere. 2020;5:e00441–20. doi: 10.1128/mSphere.00441-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walls CA, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D, et al. Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;180:281–92. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabenau HF, Cinatl J, Morgenstern B, Bauer G, Preiser W, Doerr HW. Stability and inactivation of SARS coronavirus. Med Microbiol Immunol. 2005;194:1–6. doi: 10.1007/s00430-004-0219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casanova LM, Jeon S, Rutala WA, Weber DJ, Sobsey MD. Effect of air temperature and relative humidity on coronaviral survival on surfaces. Appl Environ Microbiol. 2010;76:2712–7. doi: 10.1128/AEM.02291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laude H. Thermal inactivation studies of a coronavirus, transmissible gastroenteritis virus. J Gen Virol. 1981;56:235–40. doi: 10.1099/0022-1317-56-2-235. [DOI] [PubMed] [Google Scholar]
- 16.Kampf J, Voss A, Schiethauer S. Inactivation of coronavirus by heat. J Hosp Inf. 2020;105:348–9. doi: 10.1016/j.jhin.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polgreen PM, Polgreen EL. Infectious diseases, Weather, and Climate. Clin Infect Dis. 2018;66:815–7. doi: 10.1093/cid/cix1105. [DOI] [PubMed] [Google Scholar]