Sir,
Coronavirus disease 2019 (COVID-19) is an acute infection of the respiratory tract that emerged in December 2019 in Wuhan, China. The pandemic of SARS-CoV-2 as of March 31, 2021 has affected 220 countries and territories around the world including India1. The number of reported SARS-CoV-2 cases in India is 12,039,644 with 1,62,114 deaths2. Pune city remains one of the hotspot cities of SARS-CoV-2 from western India.
There are reports from China on SARS-CoV-2 infection in a family setting with person-to-person transmission3,4. There are no studies on family cluster reported from India which is the leading mode of human-to-human transmission3,4,5. In the present study, we report the clinical and laboratory findings, and a transmission pattern of four patients in a family cluster from Pune city. All four members presented with different clinical features after being infected with COVID-19. The index case contracted this infection from the hospital and subsequently infected other members in the family.
The present study was a part of a hospital-based study initiated at the Indian Council of Medical Research-National Institute of Virology (ICMR-NIV), Pune, to investigate the SARS-CoV-2 shedding in excreta of COVID-19 patients during treatment and after recovery. The study was approved by the ethics committees of the institute and the hospital. The family that had four members in total was admitted for COVID-19 treatment. Written informed consent was obtained before the study.
The index case was a 37 yr old male (index), a healthcare worker, who had worked in the COVID-19 ward. He was living with his wife (34 yr old) and two children (8 and 6 yr old). Being a healthcare worker, index had been exposed to multiple laboratory-confirmed COVID-19 patients. The index patient presented with fever, nasal discharge and generalized weakness on the onset of symptoms. He was admitted in the hospital and diagnosed with COVID-19 by real-time reverse transcriptase–polymerase chain reaction (rRT-PCR) using throat/nasal swabs as per the protocol published by the WHO6.
On the fourth day of onset of symptoms, his wife (case 1) developed sore throat and rhinorrhoea. Case 1 along with two children (cases 2-3) was admitted on the sixth day of onset of symptoms in the index case, as they were confirmed positive for COVID-19 by rRT-PCR. Both the children were asymptomatic throughout the disease. An asymptomatic case was defined as a laboratory-confirmed COVID-19 infection case who was afebrile and well. None of the patients had diarrhoea as a part of their symptoms. All family members were discharged from the hospital by day 14 of the admission of the index case.
Stool and urine samples were collected from all these patients on admission to check if the virus could be detected in specimens from non-respiratory sites and 14 days after collection of the first specimens (based on the incubation period of the virus). Additional stool samples were collected from the index case on 21, 32, 40, 48 and 55 days after the onset of symptoms to determine the duration of excretion of this novel virus in faeces. The viral nucleic acids were extracted from 30 per cent (w/v) suspensions in phosphate-buffered saline (pH 7.2-7.4) using spin columns (Qiagen, Hilden, Germany) as per manufacturer's instructions and determined by rRT-PCR targeting the genes, E (envelope), RdRp (RNA dependent RNA polymerase), ORF-1b-nsp14 (Open Reading Frame) and RNaseP (human RNase P) gene as internal control7. Viral copy numbers were quantified using E quantitative PCR standards developed at ICMR-NIV in ten-fold serial dilutions to generate a standard curve8.
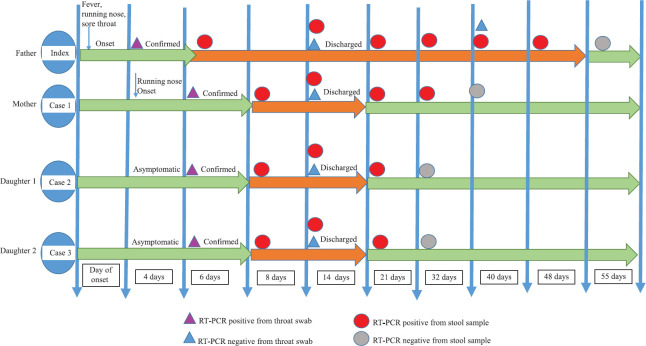
The family cluster of four (index case and cases 1-3) was infected with SARS-CoV-2. The timeline of contact and the duration of transmission of disease within the family as measured by rRT-PCR are indicated in Fig. 1. All the cases showed positivity in stool samples on days 8, 14 and 21 where higher viral loads corresponding to lower Ct values for E and ORF-1b-nsp14 genes were observed and no significant difference in Ct values of symptomatic and asymptomatic patients was observed (Fig. 2). However, on day 32, cases 2 and 3 became negative and case 1 was negative on the fortieth day. Furthermore, the index patient continued to excrete virus in stool until 48 days despite throat/nasal swab was found negative (Fig. 3). The viral shedding profile (Fig. 3) showed that the shedding increased slightly on day 6, peaked on the day 14 after the onset of illness and then dropped gradually to lower levels on day 55. Urine specimens of all the patients were negative. In general, the Ct values of ORF-1b-nsp14 gene for all specimens were lower compared to that of RdRp gene confirming earlier studies9 that ORF-1b-nsp14-based assay performed well as a confirmatory assay as compared to RdRp-based assays.
Fig. 1.
Time course of real-time reverse transcriptase–polymerase chain reaction (RT-PCR) test results for viral RNA in throat and stool specimens of COVID-19 patients.
Fig. 2.
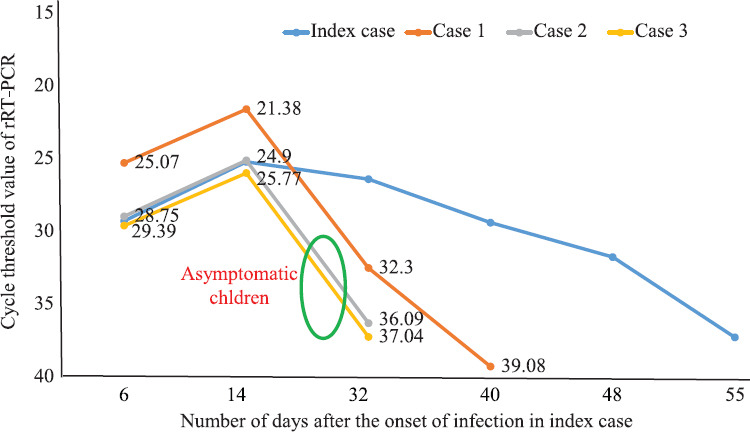
Cycle threshold values for all cases within the family, positive for COVID-19 from stool specimen.
Fig. 3.
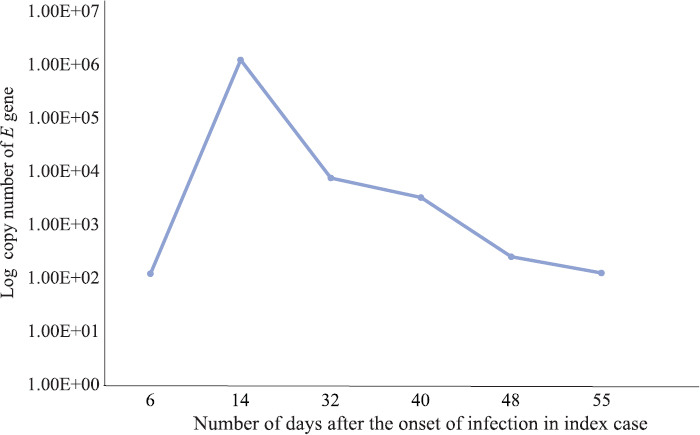
Copy number of E gene of SARS-CoV-2 from stool specimens collected at different time intervals from the index case using real-time (RT-PCR).
The clinical features were diverse across the family; both the children were asymptomatic, the wife with mild symptoms and the index case had more severe respiratory symptoms. It was observed in this study that the transmission of SARS-CoV-2 infection occurred during the incubation period. A study done by Zou et al10 found that the viral loads of symptomatic and asymptomatic patients were similar and asymptomatic patients could infect others. Correlation of extended viral shedding in faeces in severe cases needs to be determined. Our observations are in concordance with reports that children develop mild or even asymptomatic illness compared to adults and elderly persons, and they may fare better when they have contracted the virus4,5,11.
Viral RNA was not detectable in urine specimens of these patients. However, improved methods of testing of urine samples are warranted. A rather surprising finding from the study was detection of viral RNA from the faecal specimens of the patients, though none of them had diarrhoea as part of their symptoms.
Our study had a few limitations. SARS-CoV-2 was not isolated by the culture method from the samples that tested positive for the SARS-CoV-2 E gene by rRT-PCR. Real-time PCR detects viral RNA genome which can represent a replicating or a non-viable non-replicating virus. Angiotensin-converting enzyme 2, the receptor for spike protein of SARS-CoV-2, is expressed in large numbers on the brush border of intestinal enterocytes. Experimental models using human small intestinal organoids have demonstrated that SARS-CoV-2 can infect, replicate and produce significant titres of infectious viral particles in enterocytes12. Gastrointestinal symptoms in COVID-19 patients can be explained by this.
In summary, our findings showed consistent person-to-person transmission of this novel coronavirus in hospital and family settings and also prolonged viral excretion by COVID-19 patients after negative conversion of pharyngeal swabs. The presence of SARS-CoV-2 RNA was demonstrated in the faeces of COVID-19 patients and suggested the possibility of SARS-CoV-2 transmission via the faecal-oral route. Faeces of the patients can be a potential source of transmission of the virus that may have important public health implications. Further studies are needed to ascertain if this would be a possible route of transmission.
Acknowledgment:
The authors thank Dr Priya Abraham, Director ICMR-NIV, Pune, for the support during the study. The authors are grateful to Dr M L Chaudhary (Influenza Group, ICMR-NIV, Pune) for coordination with hospital laboratory authority for clinical sample from known SARS-CoV-positive patient. The assistance provided by Servshri P.S. Jadhav and Santosh (Deenanath Mangeshkar Hospital, Pune) during sample collection from the hospital and technical help by Shri R. Doiphode is duly acknowledged. The authors are thankful to NIC Team for supplying reagents for testing. Smt Veena Vipat is acknowledged for providing technical support for viral load detection.
Footnotes
Financial support & sponsorship: The authors acknowledge the ICMR-NIV, Pune and ICMR, New Delhi, for financial support.
Conflicts of Interest: None.
References
- 1.Worldometer. COVID-19 coronavirus pandemic. [accessed on March 30, 2021]. Available from: https://www.worldometers.info/coronavirus/
- 2.World Health Organization. Coronavirus disease (COVID-2019) situation reports WHO. 2020. [accessed on March 30, 2021]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports .
- 3.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet. 2020;395:514–23. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song R, Han B, Song M, Wang L, Conlon CP, Dong T, et al. Clinical and epidemiological features of COVID-19 family clusters in Beijing, China. J Infect. 2020;81:e26–30. doi: 10.1016/j.jinf.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun D, Zhu F, Wang C, Wu J, Liu J, Chen X, et al. Children infected with SARS-CoV-2 from family clusters. Front Pediatr. 2020;8:386. doi: 10.3389/fped.2020.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Coronavirus disease (COVID-19) technical guidance: Laboratory testing for 2019-nCoV in humans. [accessed on February 9, 2020]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance/
- 7.Gupta N, Potdar V, Praharaj I, Giri S, Sapkal G, Yadav P, et al. Laboratory preparedness for SARS-CoV-2 testing in India: Harnessing a network of Virus Research & Diagnostic Laboratories. Indian J Med Res. 2020;151:216–25. doi: 10.4103/ijmr.IJMR_594_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary ML, Vipat V, Jadhav S, Basu A, Cherian S, Abraham P, et al. Development of in vitro transcribed RNA as positive control for laboratory diagnosis of SARS-CoV-2 in India. Indian J Med Res. 2020;151:251–4. doi: 10.4103/ijmr.IJMR_671_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alagarasu K, Choudhary ML, Lole KS, Abraham P, Potdar V. Evaluation of RdRp & ORF-1b-nsp14-based real-time RT-PCR assays for confirmation of SARS-CoV-2 infection: An observational study. Indian J Med Res. 2020;151:483–5. doi: 10.4103/ijmr.IJMR_1256_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–9. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian G, Yang N, Ma AH, Wang L, Li G, Chen X, et al. A COVID-19 transmission within a family cluster by presymptomatic infectors in China. Clin Infect Dis. 2020;71:861–2. doi: 10.1093/cid/ciaa316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–4. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]