Abstract
SPT5 and its binding partner SPT4 regulate transcriptional elongation by RNA polymerase II. SPT4 and SPT5 are involved in both 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB)-mediated transcriptional inhibition and the activation of transcriptional elongation by the human immunodeficiency virus type 1 (HIV-1) Tat protein. Recent data suggest that P-TEFb, which is composed of CDK9 and cyclin T1, is also critical in regulating transcriptional elongation by SPT4 and SPT5. In this study, we analyze the domains of SPT5 that regulate transcriptional elongation in the presence of either DRB or the HIV-1 Tat protein. We demonstrate that SPT5 domains that bind SPT4 and RNA polymerase II, in addition to a region in the C terminus of SPT5 that contains multiple heptad repeats and is designated CTR1, are critical for in vitro transcriptional repression by DRB and activation by the Tat protein. Furthermore, the SPT5 CTR1 domain is a substrate for P-TEFb phosphorylation. These results suggest that C-terminal repeats in SPT5, like those in the RNA polymerase II C-terminal domain, are sites for P-TEFb phosphorylation and function in modulating its transcriptional elongation properties.
Regulation of transcriptional elongation is a critical process in the control of viral and cellular gene expression (reviewed in references 3 and 28). A number of cellular factors that regulate transcriptional elongation have been defined using both biochemical and genetic techniques. These factors include the general transcription factors TFIIF and TFIIS, as well as other factors including the elongin and ELL proteins (20, 41, 48).
In addition, cellular kinases play an important role in the control of transcriptional elongation based on their ability to phosphorylate the RNA polymerase II C-terminal domain (CTD) (27). One of these kinases, CDK-activating kinase (CAK), is composed of the CDK7 kinase in addition to cyclin H and MAT1. CAK is contained in the multiprotein TFIIH complex and is involved in modulating promoter clearance of specific promoters (13, 45, 47). A second kinase complex, P-TEFb, is composed of cyclin T1 and CDK9 and also phosphorylates the RNA polymerase II CTD and stimulates transcriptional elongation (18, 32, 33, 36, 64). The Tat protein, which is a potent stimulator of transcriptional elongation, interacts with P-TEFb to stimulate human immunodeficiency virus type 1 (HIV-1) gene expression (4, 7, 17–19, 25, 26, 30, 31, 55, 56, 62, 64).
SPT4 and SPT5 are highly conserved proteins which are present in a variety of species from yeast to humans and are involved in the regulation of transcriptional elongation (23, 53, 58, 60, 61). Genetic assays in yeast demonstrate that SPT5 conditional mutants can be suppressed by mutations in the genes encoding two largest subunits of RNA polymerase II (23). Furthermore, SPT5 interacts directly with RNA polymerase II via a domain in SPT5 that has homology to the Escherichia coli transcription elongation factor NusG (23, 53, 61). The human homologues of the SPT4 and SPT5 proteins have also been characterized (8, 9, 22, 49). These proteins were also isolated independently by two groups based on their ability to either mediate the inhibition of transcriptional elongation in the presence of the ATP analogue 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (10, 53) or rescue Tat activation in fractionated HeLa extract that does not otherwise support this process (58). Although SPT4 and SPT5 are required for DRB-mediated inhibition of transcriptional elongation, these proteins also can stimulate transcriptional elongation in in vitro transcription assay mixtures containing limiting concentrations of ribonucleoside triphosphates (53). Thus, SPT4 and SPT5 can regulate transcriptional elongation in both a positive and negative manner depending on the experimental conditions.
The mechanism by which SPT4 and SPT5 regulate transcriptional elongation has recently been investigated. SPT5 contains a number of distinct domains including an acidic amino terminus, four KOW repeats that have homology to the E. coli transcriptional regulator NusG (23, 53, 61), and two C-terminal repeat elements designated CTR1 and CTR2 (49). These latter domains contain multiple amino acid repeats that are rich in serine and threonine residues and may serve as potential sites for phosphorylation by cellular kinases. Recent data indicate that SPT4 and SPT5 function at an early step in the transcriptional elongation process that is regulated by P-TEFb (37, 54). For example, immunodepletion of P-TEFb from HeLa nuclear extract greatly reduces the production of full-length transcripts in in vitro transcription assays, while immunodepletion of both P-TEFb and SPT5 restores transcription to control levels. However, the addition of SPT4 and SPT5 to extract that is immunodepleted of both SPT5 and P-TEFb results in transcriptional repression. The subsequent addition of P-TEFb to this extract is able to alleviate the inhibitory effect of the SPT4 and SPT5 proteins (54). Therefore, immunodepletion of P-TEFb from HeLa nuclear extract results in a similar effect to the addition of DRB, demonstrating the negative effect of the SPT4-SPT5 complex on transcriptional elongation. These data suggest that P-TEFb is probably the target of DRB-inhibitory effects on transcriptional elongation mediated by SPT4 and SPT5.
In this study, the functional interaction between SPT4 and SPT5 and P-TEFb in DRB-mediated transcriptional repression and Tat activation was studied. We demonstrated that SPT5 domains that bind SPT4 and RNA polymerase II in addition to the CTR1 domain are critical for mediating DRB inhibition and Tat activation. Furthermore, we found that P-TEFb phosphorylates the CTR1 domain in SPT5. These studies suggest that P-TEFb phosphorylation of both RNA polymerase II and SPT5 may be critical for the regulation of transcriptional elongation.
MATERIALS AND METHODS
Materials.
DRB was purchased from CalBiochem and dissolved in 50% ethanol–10 mM HEPES (pH 7.9) at 10 mM. Prior to the addition of DRB to in vitro transcription reaction mixtures, a 10 mM stock was diluted with 10% ethanol–10 mM HEPES (pH 7.9) to a concentration of 1.5 mM.
Nuclear-extract preparation.
Nuclear extract was prepared as previously described (12). It was then fractionated on a phosphocellulose column as previously described (58, 63). The column was washed with buffer A (20 mM Tris HCl [pH 7.9], 20% glycerol, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM dithiothreitol [DTT]) containing 0.1 M KCl and 0.5 M potassium acetate and eluted with buffer containing 0.1 M KCl and 1.0 M potassium acetate. The eluate was dialyzed against buffer A containing 0.1 M KCl and used in partially reconstituted transcription reactions as a fraction that can support basal transcription but not DRB-sensitive transcription or Tat transactivation.
In vitro transcription assays.
In vitro transcription reactions assaying the sensitivity of transcription to DRB were carried out as described previously (53) with modifications. For the partially reconstituted in vitro transcription reactions, 15 μl (∼15 μg) of 1.0 M potassium acetate phosphocellulose eluate supplemented with recombinant human TATA binding protein (25 to 50 ng) purified from E. coli was incubated with 250 ng of circular pTF3-6C2AT template (a generous gift of H. Handa). This template contained a 380-bp G-less cassette immediately following the promoter. These reactions were performed in the presence of 4 mM MgCl2 and 10 μl of purified SPT4 and SPT5 for 45 min at 30°C. Reaction mixtures with nuclear extract contained 6.5 μl (∼30 μg) of unfractionated nuclear extract. DRB was then added to a final concentration of 50 μM, and this was followed by the addition of 38 μl of TRX buffer (25 mM HEPES [pH 7.9], 10% glycerol, 50 mM KCl, 6 mM MgCl2, 0.5 mM DTT, 0.5 mM PMSF, 60 μM ATP, 5 μM CTP, 600 μM GTP, 600 μM UTP) containing [α-32P]CTP (NEN) (800Ci/mmol; 1 μl per reaction). After a 10-min reaction, 250 U of RNase T1 (GIBCO) was added and the incubation was continued at 37°C for an additional 10 min. Reactions were then stopped by the addition of 350 μl of termination mixture (0.1 M Tris HCl [pH 7.5], 7 M urea, 0.35 M NaCl, 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], 25 μg of yeast tRNA per ml). Samples were extracted with a mixture of phenol and chloroform, ethanol precipitated, dissolved in gel-loading buffer (8 mM EDTA, 0.3 mg of bromphenol blue per ml, and 0.3 mg of xylene cyanol per ml in deionized formamide), heat treated for 3 min at 100°C, and separated on a 6% denaturing gel. Autoradiography was performed at −70°C with two intensifying screens for 12 to 24 h.
For in vitro transcription reactions assaying the effect of Tat on the HIV-1 long terminal repeat (LTR), 15 μl of 1.0 M potassium acetate phosphocellulose eluate was supplemented with recombinant human TBP (25 to 50 ng) and Sp1 (15 ng) purified from E. coli (58) and 200 ng each of circular templates containing either wild-type HIV-1 sequences (−454 to +1092) or similar HIV-1 sequences with a mutation in the Tat activation response element (TAR) RNA loop sequences between +31 and +34 as described previously (58). For the wild-type template, a 380-bp G-less cassette was inserted between nucleotides 1092 and 1587, while the mutant template contained a 110-bp G-less cassette inserted in the same position. Preincubation mix (15 μl) was added to the reaction mixtures containing 450 ng of poly(I-C), 75 ng of poly(dG-dC), 25 mM creatine phosphate, 17.5 mM MgCl2, 1.6 mM ATP, 32.5 mM HEPES (pH 7.9), and 17.5 mM DTT. The reaction mixtures were supplemented with recombinant Tat or glutathione S-transferase (GST) purified from E. coli (25 ng). Preincubation was carried out for 45 min at 30°C and was followed by the addition of 2 μl of nucleotide mix (125 μM CTP, 5 mM GTP, 5 mM UTP) and 1 μl of [α-32P]CTP. The reactions were allowed to proceed for an additional 30 min at 30°C, and the mixtures were then given a 10-min incubation at 37°C with 250 U of RNase T1.
Construction of GST fusion expression vectors.
The full-length SPT5 cDNA (58) was cloned into pGEX-KG vector at the EcoRI site. An StuI-EcoRI fragment containing CTR2 sequences (encoding amino acids [aa] 852 to 1087), a SmaI-StuI fragment containing CTR1 repeats (encoding aa 760 to 852), and an SmaI-EcoRI fragment containing CTR1 and CTR2 repeats (encoding aa 760 to 1087) were each individually subcloned into the pGEX-KG vector. To make a construct devoid of CTR sequences at the C terminus, a nonsense mutation substituting an amber stop codon (TGA) for a Ser codon (TCA) at position 755 was introduced by PCR mutagenesis using a QuickChange site-directed mutagenesis kit (Stratagene). Vectors encoding the GST fusion proteins were transformed into the BL21 pLysE strain of E. coli, and proteins were induced and purified by affinity chromatography on a glutathione-agarose column as described previously (26).
Purification of recombinant proteins from baculovirus.
Baculovirus expression vectors containing either CDK9, CDK9, and cyclin T1, CDK9 and cyclin T2a, or CDK9 and cyclin T2b were a generous gift from David Price (36). Recombinant His6-tagged proteins were purified by Ni2+-nitrilotriacetic acid (NTA)-agarose and S-Sepharose chromatography as described previously (36). Baculoviruses expressing CDK7, cyclin H, and MAT1, were a generous gift of David Morgan. Cyclin H had an amino-terminal His6 tag. Trimeric CAK was purified by chromatography on Ni2+-NTA agarose, Q-Sepharose, and Superdex 200 columns as described previously (11).
Baculoviruses expressing the SPT5 and SPT4 proteins were derived by cloning full-length cDNA sequences of the respective proteins into pVL1392 vector. These constructs each had C-terminal influenza virus hemagglutinin epitope and His6 tags. Sf9 cells were coinfected with the two baculoviruses, and recombinant proteins were purified by chromatography on Ni2+-NTA agarose and S-Sepharose columns by the same procedures used to isolate the baculovirus-derived recombinant CDK9 and cyclin T1 proteins.
In vitro kinase assays.
RNA polymerase II used as a substrate in kinase reactions was purified from HeLa cells as described previously (26, 34, 39). GST-SPT5 fusion proteins, which were purified as described elsewhere (26), were also analyzed in kinase assays. Kinase assays (in a 15-μl volume) were performed for 2 h at room temperature in reaction buffer containing 20 mM HEPES (pH 7.9), 15% glycerol, 0.15 mM EDTA, 7 mM MnCl2, 2 mM DTT, 0.7 mM PMSF, 7 mM NaF, 0.7 mM Na3VO4, 2 μM ATP, and 0.04 μl of [γ-32P]ATP (ICN) (7,000 Ci/mmol).
Immunoprecipitation of SPT5 from nuclear extract.
Rabbit polyclonal antibody was generated against a GST-SPT5 fusion protein (aa 1 to 73) or GST-CTR2 (aa 852 to 1087) (58). Antibodies were purified following two fractionation steps on a protein A-Sepharose column and covalently coupled to a protein A-Sepharose matrix using dimethylpimelimidate (21). Nuclear extract (0.5 ml) was incubated with 75 μl of the beads for 2 h at 4°C with gentle shaking. The beads were washed three times with 10 bed volumes of buffer containing 20 mM Tris HCl (pH 7.9), 1 mM EDTA, 100 mM NaCl, 1% NP-40, 1 mM DTT, and 0.5 mM PMSF. They were then resuspended in 150 μl of SDS-polyacrylamide gel electrophoresis (PAGE) gel-loading buffer. One-third of the sample (50 μl) was separated on an SDS-polyacrylamide gel. Western blot analysis was performed using rabbit polyclonal antibody against CDK9 (Santa Cruz) or mouse monoclonal antibody against the CTD of the largest subunit of RNA polymerase II (8WG16; BAbCo). To deplete nuclear extract of endogenous SPT5, 1 mg of anti-CTR2 rabbit polyclonal affinity-purified antibody was chemically coupled to 200 μl of protein A-containing beads. Nuclear extract (1 ml) was incubated with the beads overnight and then with fresh beads for an additional 4 h.
Construction of SPT5 and SPT4 expression vectors.
A wild-type SPT5 cDNA was cloned into the EcoRI and SalI endonuclease restriction sites of pCMV2-pFlag (Sigma). C-terminal deletion mutants of SPT5 were prepared using a QuickChange site-directed mutagenesis kit with pCMV2-pFlag-SPT5 as a template. SPT5 constructs containing aa 1 to 837, 1 to 754, 1 to 518, and 1 to 229 were constructed using oligonucleotide primers 5′-GAA GAA TAT GAG TAG GCT TTC GAT GAT-3′, 5′-CAC GGT GGG CTG ACG GCG CCC GGG CGG CA-3′, 5′-CCG GGA CCT GTA GCT CTG CTC AGA GAC AG-3′, and 5′-GTG GAG GCC TAG AAG CAG ACC CAC GTG A-3′, respectively, and complementary oligonucleotides. In each case, a stop codon (underlined) was inserted in the sequence. The N-terminal deletion mutants of SPT5 extending from aa 176 to 1087, 519 to 1087, 755 to 1087, and 838 to 1087 were constructed using the 5′ PCR primers 5′-CGG AAT TCC GAT CCC AAT CTG TGG ACT GTC AA-3′, 5′-CGG AAT TCC CAG CTC TGC TCA GAG ACA GCA TCA-3′, 5′-CGG AAT TCC TCA CGG CGC CCG GGC GGC ATG AC-3′, and 5′-CGG AAT TCC TAT GCT TTC GAT GAT GAG CCC ACC-3′, respectively, and the 3′ PCR primer 5′-GCG TCG ACT CAG GCT TCC AGG AGC TTC CCC AG-3′.
Internal fragment mutants of SPT5 containing aa 313 to 542, 313 to 755, and 421 to 755 were constructed as follows: The SPT5 aa 313 to 542 construct was made using oligonucleotide primers 5′-CGG AAT TCC TAC GAT CGC ATC AAG GCC CG-3′ and 5′-GCG TCG ACT CAC TGC ACC AGC TCG CCC CAT-3′; the SPT5 aa 313 to 755 construct was made using oligonucleotide primers 5′-CGG AAT TCC TAC GAT CGC ATC AAG GCC CG-3′ and 5′-GCG TCG ACT CAG CCC ACC GTG GTG AGC CGC TG-3′; and the SPT5 aa 421 to 755 construct was generated by PCR with oligonucleotide primers 5′-GCG TCG ACT CAG CCC ACC GTG GTG AGC CGC TG-3′ and 5′-CGG AAT TCC CAA CCT GGG GAC AAC GTG GA-3′.
The SPT5 internal deletion mutants Δ(176–314), Δ(314–516), and Δ(758–837) were generated by recombinant PCR (24). To prepare the SPT5 deletion mutant Δ(176–314), PCR with oligonucleotide primers 5′-CTG CTC CCA GGA GTC AAG CGC ATC AAG GCC CGC ATG A-3′ and 5′-GCG TCG ACT CAG GCT TCC AGG AGC TTC CCC AG-3′ was performed to generate aa 315 to 1087 of SPT5. Next, aa 1 to 175 of SPT5 were generated by PCR with oligonucleotide primers 5′-CCG AAT TCC ATG TCG GAC AGC GAG GAC AGC AAC T-3′ and 5′-T CAT GCG GGC CTT GAT GCG CTT GAC TCC TGG GAG CAG-3′. These two PCR products were then joined to generate SPT5 Δ(176–314) in a PCR without a template. To generate SPT5 Δ(314–516), a similar strategy was used. Oligonucleotide primers 5′-ATC CCA CGC ATC GAC TAC GAC CTG CAG CTC TGC TCA-3′ and 5′-GCG TCG ACT CAG GCT TCC AGG AGC TTC CCC AG-3′ were used to produce aa 517 to 1087 of SPT5, and aa 1 to 314 of SPT5 were generated using the oligonucleotide primers 5′-CCG AAT TCC ATG TCG GAC AGC GAG GAC AGC AAC T-3′ and 5′-TGA GCA GAG CTG CAG GTC GTA GTC GAT GCG TGG GAT-3′. The two PCR products were used in a no-template PCR to produce the final product SPT5 Δ(314–516).
To prepare SPT5 Δ(755–837), oligonucleotide primers 5′-G CGG CTC ACC ACG GTG GGC TAT GCT TTC GAT GAT GAG CCC-3′ and 5′-GCG TCG ACT CAG GCT TCC AGG AGC TTC CCC AG-3′ were used to produce aa 838 to 1087 of SPT5. The product of this PCR was used as a primer for a second PCR in conjunction with an oligonucleotide primer 5′-CCG AAT TCC ATG TCG GAC AGC GAG GAC AGC AAC T-3′ to generate the SPT5 Δ(758–837) construct. All SPT5 mutants were subject to DNA sequence analysis and cloned into pCMV2-pFlag. The SPT4 cDNA was cloned into pCMV2-pFlag or pCMV5-Myc1.
Affinity purification of Flag-tagged SPT5 proteins from COS cells.
Expression vectors containing Flag-tagged SPT5 cDNA either alone or in combination with expression vectors containing Myc-tagged SPT4 cDNA were transfected into COS cells using Fugene 6 (Roche). At 48 h posttransfection, the cells were harvested and homogenized in Tris-buffered saline (50 mM Tris HCl [pH 7.4], 150 mM NaCl). After centrifugation at 3,000 rpm for 10 min in a CS-6R Beckman centrifuge, supernatant was applied to an anti-Flag M2 affinity gel column (Sigma) as specified by the manufacturer. Flag-tagged fusion proteins were eluted with a Tris-buffered saline solution containing Flag peptide (Sigma) at 100 μg/ml. Proteins were dialysed against buffer D and used in the in vitro transcription assays.
Immunoprecipitation and Western blotting.
SPT5 and SPT4 expression vectors were transfected into COS cells with Fugene 6 (Roche). At 48 h posttransfection, the cells were harvested in phosphate-buffered saline and subjected to centrifugation for 2 min at 2,000 rpm at 4°C in a CS-6R Beckman centrifuge. The cell pellets were resuspended in 600 μl of buffer B (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, and 0.5 mM PMSF) and allowed to swell on ice for 15 min. Cells were then lysed by being rapidly pushed through a 25-gauge 5/8 BD hypodermic needle eight times. The cell homogenates were then centrifuged for 20 s at 12,000 rpm in a 5415 C-Eppendorf microcentrifuge. The lysates were incubated with anti-Flag M2 monoclonal antibody (Sigma) or anti-HA monoclonal antibody (Roche) for 1 h at 4°C. Then 20 μl of protein G-agarose beads was added and mixed for 1 h at 4°C. After the mixture was washed three times with ELB buffer (50 mM HEPES [pH 7.9], 250 mM NaCl, 5 mM EDTA, 1% NP-40, 0.5 mM DTT, and 0.5 mM PMSF), the immunoprecipitates were analyzed on an SDS-polyacrylamide gel. Western blotting was performed using an enhanced chemiluminescence agent (Amersham).
Construction of SPT5 GST-CTR1 mutants.
Sequences encoding two of the CTR1 repeats (aa 772 to 787) were incorporated into the 3′ oligonucleotide primer that was used in the PCR to amplify a fragment of polylinker of pGEX-KG vector. For the wild-type construct, the sequence of the 3′ primer was TTC CCA AGC TTA [GTA CAT GGG TGT TCG GGA GCC AGA GCC ATA CAT GGG CGT CTG GGA GCC] ACC CAT GGA GTC TAG. The portion of the primer corresponding to the CTR1 repeat is shown in brackets. The 5′ portion of the primer contains a HindIII site and the stop codon (UAA) immediately following the coding sequence. The 5′ oligonucleotide primer CCG CGT GGA TCC CCG GGA anneals to pGEX-KG and contains a BamHI site. The PCR product was then digested with HindIII and BamHI and cloned into pGEX-KG vector. Mutant CTR1 repeats were also generated which contained alanine in place of either threonine (at aa 775 and 784) or serine (at aa 773 and 782). All the constructs were confirmed by DNA sequencing. The construct encoding GST fused to two of the RNA polymerase II CTD repeats was generated using a similar approach and was described previously (26).
Phosphorylation of SPT5.
Recombinant SPT4-SPT5 complex purified from baculovirus was incubated with recombinant P-TEFb also expressed in baculovirus in a total volume of 20 μl. Kinase reactions were performed in buffer (15 mM HEPES [pH 7.9], 0.1 mM EDTA, 1.5 mM DTT, 0.5 mM DTT, 10% glycerol) supplemented with 5 mM MnCl2 and 2.5 mM MgCl2 in either the presence or absence of 5 mM ATP at room temperature for 30 min. Samples were then dialyzed against buffer D at 4°C to remove excess ATP. In vitro transcription reactions were performed as described above, except that DRB was added at the start of the incubation.
RESULTS
Domains in the SPT5 protein.
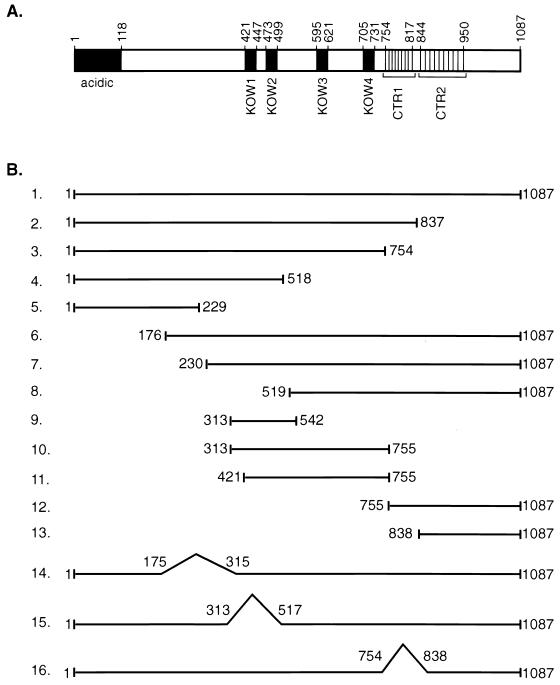
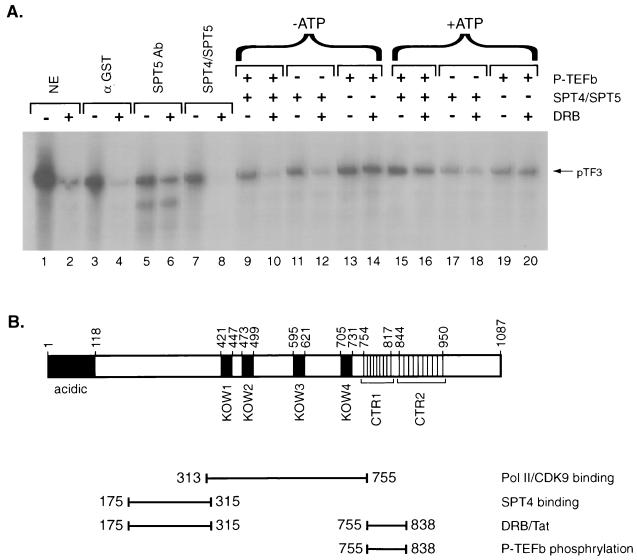
Several distinct domains in the human SPT5 protein have been delineated based on their homology to the yeast SPT5 protein and the E. coli transcriptional regulator NusG (Fig. 1A). The amino terminus of SPT5 extending between aa 1 and 118 is highly charged, being abundant in the acidic amino acid residues aspartic acid and glutamic acid (53, 58). Four regions in the middle of the SPT5 protein extending between residues 421 and 447, 473 and 499, 595 and 621, and 705 and 731 have homology to KOW motifs present in the procaryotic transcription termination and antitermination factor NusG (23, 53). There are two clusters of different repeats in the C-terminal 280 residues of SPT5 which are rich in serine, threonine, proline, and tyrosine but are unrelated to C-terminal repeat motifs in the yeast SPT5 protein (49). The first motif, which has been designated CTR1, contains 9 repeats of the consensus sequence GS(R/Q)TPXY, while a less highly conserved motif, designated CTR2, contains 10 repeats of the consensus sequence P(T/S)PSP(Q/A)(S/G)Y, which has weak homology to the repeats found in the CTD of RNA polymerase II.
FIG. 1.
Schematic of the SPT5 protein. (A) A schematic of the SPT5 protein with the positions of the acidic domain, the four KOW domains that have homology to the E. coli NusG protein, and two domains, CTR1 and CTR2, each of which has multiple amino acid repeats, is shown. (B) A variety of contructs with mutations in different domains of the SPT5 protein are shown. These mutants were analyzed for their function using both in vivo interaction and in vitro transcription assays.
Mutagenesis of SPT5 was performed in an attempt to better define the functions of the different domains in this protein. Mutant SPT5 proteins were generated to test for their ability to interact with SPT4, RNA polymerase II, and P-TEFb (Fig. 1B). In addition, we assayed the function of these purified proteins in in vitro transcription assays for their ability to mediate DRB-mediated inhibition of transcription and Tat activation of the HIV-1 LTR. First, a series of carboxy-terminal deletion mutants were constructed which contained truncated SPT5 proteins that extended from aa 1 to 837, 754, 518, or 229. These constructs allowed us to evaluate the function of CTR1 and CTR2 and the KOW domains. Next, a series of amino-terminal deletion mutants were constructed which removed either 176, 230, or 519 aa from the amino terminus of SPT5. A series of internal fragments from the SPT5 protein which extended from aa 313 to 542, 313 to 755, 421 to 755, 755 to 1087, and 838 to 1087 were also assayed. Finally, a series of internal deletions were constructed which resulted in SPT5 proteins that lacked potential binding sites for SPT4 (aa 175 to 315) and RNA polymerase II (aa 313 to 517) (61) in addition to the CTR1 motif (aa 754 to 838).
Domains in SPT5 that interact with SPT4.
Previously, in vitro interaction studies with a GST-SPT4 fusion protein and in vitro-translated SPT5 proteins labeled with [35S]methionine were used to map SPT5 interactions with SPT4 (61). To map the in vivo interactions between these proteins, we used expression vectors containing a Myc-tagged SPT4 protein and Flag-tagged SPT5 proteins. Following transfection of COS cells with these epitope-tagged SPT4 and SPT5 constructs, extracts were prepared and the association of SPT4 and SPT5 was assayed using immunoprecipitation followed by Western blot analysis.
The extracts prepared from transfected COS cells were first analyzed in Western blot analysis with a monoclonal antibody directed against the Myc epitope. This analysis indicated that there were similar levels of expression of the epitope-tagged SPT4 construct (Fig. 2A, upper panel). Next, immunoprecipitation of SPT5 protein with a monoclonal antibody directed against Flag was performed followed by Western blot analysis with Myc antibody to detect SPT4 association (lower panel). SPT4 was immunoprecipitated in the presence of the wild-type SPT5 protein (Fig. 2A, lane 1) but not in its absence (lane 2). SPT5 constructs aa 1 to 754 and aa 1 to 518, which deleted CTDs including both CTR1 and CTR2, associated with SPT4 (lanes 3 and 4). However, an SPT5 construct that contained only the amino-terminal 229 aa (lane 5) and a construct that deleted the amino-terminal 519 aa (lane 7) were unable to interact with SPT4.
FIG. 2.
SPT5 association with SPT4, RNA polymerase II, and CDK9. (A) Expression vectors containing either a Myc-tagged SPT4 construct or the indicated Flag-tagged SPT5 constructs were cotransfected into COS cells. Extracts were prepared from each of these transfections, and the SPT4 levels were analyzed by Western blot analysis with antibody directed against the Myc epitope (upper panel). Extracts prepared from the SPT4-SPT5-cotransfected cells were immunoprecipitated with M2 monoclonal antibody to isolate the Flag-tagged SPT5 proteins, and then Western blot analysis was performed with monoclonal antibody directed against the Myc epitope to detect the epitope-tagged SPT4 protein (lower panel). In lane 2, an expression vector containing the SPT4 cDNA was transfected into COS cells alone without cotransfection of the SPT5 cDNA. (B) Western blot analysis of COS cell extracts prepared following transfection of expression vectors containing Flag-tagged SPT5 cDNA constructs (lanes 1 to 17) or a Flag-tagged SPT4 cDNA (lane 18) was performed using the M2 monoclonal antibody. In lane 1, an expression vector containing SPT4 was cotransfected with the wild-type SPT5 construct. MW, molecular weight in thousands. (C and D) Flag-tagged SPT5 constructs were each transfected into COS cells, and extracts were prepared. Immunoprecipitation of these extracts was performed with antibodies directed against either the RNA polymerase II CTD (C) or CDK9 (D). Western blot analysis with the M2 monoclonal antibody was then performed to detect the Flag-tagged SPT5.
Additional SPT5 constructs were also used to define the binding site for SPT4. An SPT5 construct which deleted the amino-terminal 176 aa was able to interact with SPT4 (Fig. 2A, lane 8), while an SPT5 construct with an internal deletion of aa 175 to 315 was unable to interact with SPT4 (lane 9). In contrast, an SPT5 construct with a deletion which removed aa 314 to 516 interacted with SPT4 (lane 10). These in vivo interaction studies are in agreement with previous in vitro interaction studies which indicate that a region of SPT5 extending from aa 176 to 270 is required for interaction with SPT4 (61).
SPT5 interactions with RNA polymerase II and CDK9.
Next we addressed the domains in SPT5 that are responsible for in vivo interactions with RNA polymerase II and potentially CDK9 (Fig. 2B to D). Flag-tagged SPT5 constructs were transfected into COS cells, and their association with both endogenous RNA polymerase II and CDK9 was assayed. Western blot analysis of the COS cell extracts revealed roughly equivalent expression of each of the epitope-tagged SPT5 constructs (Fig. 2B, lanes 1 to 17), except for the SPT5 construct extending from aa 1 to 229 (lane 10), which had consistently low levels of expression. A Flag-tagged SPT4 construct that was transfected alone (lane 18) or cotransfected with SPT5 (lane 1) was also assayed. COS extracts were then immunoprecipitated with antibodies directed against either the CTD of RNA polymerase II (Fig. 2C) or CDK9 (Fig. 2D) followed by Western blot analysis with the M2 monoclonal antibody which detects the Flag-tagged SPT5 proteins. For RNA polymerase II, the monoclonal antibody 8WG16 (anti-CTD), which recognizes both the phosphorylated and unphosphorylated forms of this enzyme, was used for immunoprecipitation. CDK9 was immunoprecipitated with a rabbit polyclonal antibody directed against this kinase.
RNA polymerase II interacted with wild-type SPT5 when it was transfected alone or in the presence of SPT4 (Fig. 2C, lanes 1 and 2). No epitope-tagged SPT5 protein was detected when it was not expressed by transfection (lane 3). A number of SPT5 constructs including those that deleted the amino-terminal 176 aa, the SPT4 binding site (aa 175 to 315), CTR1 (aa 754 to 838), or CTR1 and CTR2 (carboxy terminal to residue 754) were all able to efficiently bind to RNA polymerase II (lanes 4 to 9). However, an SPT5 construct that left intact only the amino-terminal 229 aa, SPT5 truncations that extended between aa 519 and 1087, 755 and 1087, and 838 and 1087, or an SPT5 internal deletion of aa 313 to 517 did not bind to RNA polymerase II (lanes 9 to 14). Finally, SPT5 truncation mutants with deletions that extended between aa 313 and 542, 313 and 755, and 421 and 755 were all able to bind to RNA polymerase II (lanes 15 to 17). These results suggest that the region of SPT5 spanning aa 313 to 755 was capable of binding to RNA polymerase II as previously shown in in vitro binding studies (61).
Similar in vivo association studies with the epitope-tagged SPT5 constructs and endogenous CDK9 were also performed. An identical pattern of CDK9 binding to SPT5 was detected to that seen with RNA polymerase II (Fig. 2D, lanes 1 to 18). These results suggest that the domains in SPT5 that interact with RNA polymerase II are also required for binding to CDK9. This observation would be consistent with the fact that CDK9 and RNA polymerase II directly interact (35) and that SPT5 interactions with RNA polymerase II are required for its association with CDK9.
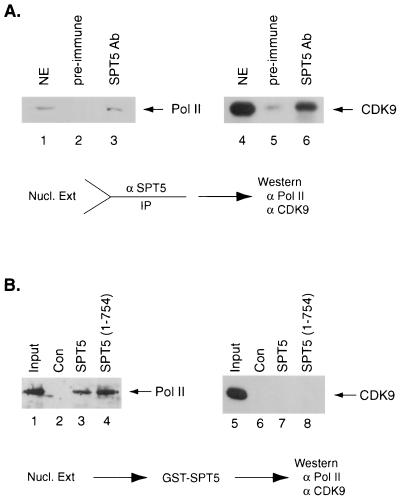
In vitro interaction studies were next performed using HeLa nuclear extract to further explore the association between SPT5, RNA polymerase II, and CDK9. First, we asked whether antibody directed against SPT5 could immunoprecipitate a complex containing SPT5, RNA polymerase II, and CDK9. HeLa nuclear extract was first immunoprecipitated with either preimmune rabbit serum or rabbit polyclonal antibody directed against SPT5. Western blot analysis was performed on these immunoprecipitates with antibodies directed against either CDK9 or RNA polymerase II (Fig. 3A). SPT5 antibody, but not preimmune rabbit serum, was able to immunoprecipitate both RNA polymerase II and CDK9 (Fig. 3A, lanes 3 and 6). These results suggest that a complex composed of SPT5, RNA polymerase II, and CDK9 was present in HeLa nuclear extract. However, only a small portion (10%) of RNA polymerase II or CDK9 present in nuclear extract was found to be associated with SPT5. No CDK9 or RNA polymerase II was detected in the preparation of recombinant SPT4-SPT5 complexes purified from COS cells (see below). Therefore, based on the overall amounts of these factors in nuclear extract, it appears that the SPT5-CDK9-polymerase II complex represents only a minor component.
FIG. 3.
Association of SPT5 with RNA polymerase II and CDK9 in vivo and in vitro. (A) HeLa nuclear extract was immunoprecipitated with either preimmune sera (lanes 2 and 5) or rabbit polyclonal antibody directed against SPT5 (lanes 3 and 6). A total of 20% of the HeLa nuclear extract that was subjected to immunoprecipitation is shown (lanes 1 and 4). Following immunoprecipitation, Western blot analysis was performed with antibody directed against the RNA polymerase II (Pol II) CTD (lanes 1 to 3) or CDK9 (lanes 4 to 6). (B) Purified RNA polymerase II (lanes 1 to 4) or baculovirus-purified CDK9/cyclin T1 (lanes 5 to 8) was either untreated (30% of the input is shown in lanes 1 and 5) or incubated with glutathione-agarose beads containing GST (lanes 2 and 6), GST-SPT5 (lanes 3 and 7), or a GST-SPT5 fusion protein extending from aa 1 to 754 (lanes 4 and 8). Following extensive washing, Western blot analysis was performed with antibodies directed against the RNA polymerase II CTD (lanes 1 to 4) or CDK9 (lanes 5 to 8).
Finally, we analyzed whether purified RNA polymerase II or baculovirus-produced and purified CDK9 and cyclin T1 could directly interact with either GST, GST-SPT5, or a GST fusion protein containing a region of SPT5 extending from aa 1 to 754. Glutathione beads containing these GST fusion proteins were incubated with purified RNA polymerase II or recombinant CDK9-cyclin T1. Following extensive washing, Western blot analysis was performed with antibodies directed against either RNA polymerase II or CDK9. RNA polymerase II interacted with both of the GST-SPT5 fusion proteins (Fig. 3B, lanes 3 and 4) but not with GST (lane 2), while CDK9-cyclin T1 did not interact with either of the SPT5 fusion proteins (lanes 7 and 8). These results indicate that RNA polymerase II, but not CDK-cyclin T1, directly interacts with SPT5 and that the association of SPT5 with CDK9 is mediated by CDF9 interaction with RNA polymerase II.
The CTR1 domain of SPT5 is critical for DRB-mediated repression.
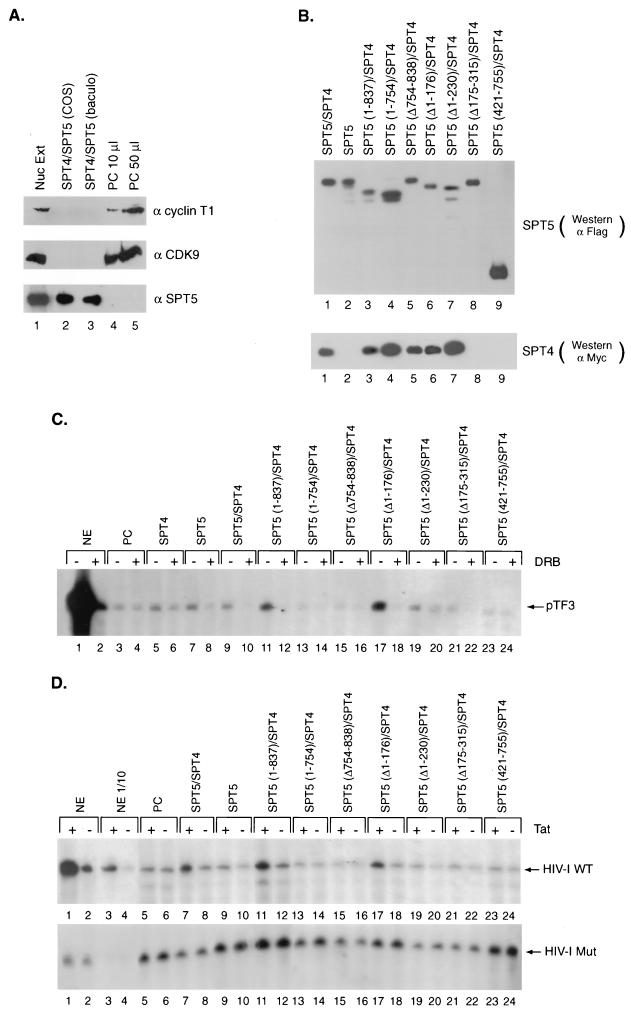
Previously, we and others have demonstrated that fractionation of HeLa nuclear extract on a phosphocellulose column followed by washing with 0.5 M potassium acetate and elution with 1.0 M potassium acetate resulted in extract that is unable to support Tat activation (58, 63). Addition of a highly purified cellular fraction that contained SPT5 restored Tat activation (58). First, we analyzed this 1.0 M potassium acetate fraction for the presence of cyclin T1, CDK9, and SPT5. The 1.0 M potassium acetate phosphocellulose fraction contained the P-TEFb components, cyclin T1 and CDK9, but not SPT5 (Fig. 4A, lanes 4 and 5). Baculovirus-produced and purified SPT4 and SPT5 (lane 3) and affinity-purified SPT4 and SPT5 proteins isolated from COS cells transfected with expression vectors containing these epitope-tagged cDNAs (lane 2) were also assayed by Western blot analysis.
FIG. 4.
Reconstitution of DRB inhibition and Tat activation by recombinant SPT5 proteins. (A) Western blot analysis was performed with antibodies directed against cyclin T1, CDK9, and SPT5 using unfractionated HeLa nuclear extract (Nuc Ext) (lane 1), affinity-purified SPT4-SPT5 complex produced following transfection of epitope-tagged SPT4 and SPT5 cDNAs into COS cells (lane 2), baculovirus-expressed and purified SPT4-SPT5 (lane 3), or 10-μl (lane 4) or 50-μl (lane 5) aliquots of a 1.0 M potassium acetate fraction of HeLa nuclear extract obtained following phosphocellulose (PC) chromatography. (B) Western blot analysis of affinity-purified SPT4 and SPT5 proteins. Expression vectors containing the different Flag-tagged SPT5 cDNA constructs and a Myc-tagged SPT4 cDNA construct were cotransfected into COS cells. The Flag epitope-tagged SPT5 and associated SPT4 protein in these extracts were purified by binding to protein G beads containing M2 monoclonal antibody followed by elution with Flag peptide. Western blot analysis was performed with anti-Flag monoclonal antibody to detect SPT5 proteins (top panel) or anti-Myc monoclonal antibody to detect the associated SPT4 (lower panel). (C) In vitro transcription analysis was performed with the pTF3-6C2AT template using unfractionated HeLa nuclear extract (NE) (lanes 1 and 2), or the 1.0 M potassium acetate fraction of HeLa nuclear extract eluted from the phosphocellulose (PC) column (lanes 3 to 24). The 1.0 M potassium acetate fraction was assayed either alone (lanes 3 and 4), with affinity-purified SPT4 added alone (lanes 5 and 6), with affinity-purified SPT5 added alone (lanes 7 and 8), or with both SPT4 and the different affinity-purified SPT5 proteins shown in panel B added as indicated (lanes 9 to 24). DRB was added to the even-numbered lanes in the in vitro transcription assays. (D) In vitro transcription analysis was performed with the HIV-1 LTR wild-type (top panel) or the HIV-1 LTR loop mutant (lower panel) templates using unfractionated HeLa nuclear extract (lanes 1 to 4), the 1.0 M potassium acetate fraction of HeLa nuclear extract eluted from the phosphocellulose (PC) column alone (lanes 5 and 6), or the 1.0 M potassium acetate fraction in the presence of affinity-purified SPT4 and SPT5 proteins (lanes 7 and 8), SPT5 alone (lane 9 and 10), or SPT4 and affinity-purified SPT5 mutants (lanes 11 to 24) as indicated. Tat (25 ng) was added to the odd-numbered lanes and GST (25 ng) was added to the even-numbered lanes in the in vitro transcription assays. Following the in vitro transcription analysis with these templates containing G-less cassettes, the labeled RNAs were digested with RNase T1 and gel electrophoresis and autoradiography were performed.
It was important to address which domains in SPT5 were involved in regulating DRB-mediated repression. For these studies, we used the affinity-purified SPT4 and SPT5 proteins isolated from COS cells. COS cells were transfected with expression vectors containing Flag-tagged SPT5 and Myc-tagged SPT4 cDNAs. The SPT4-SPT5 complex was purified following binding of the COS cell extract to protein G-agarose beads coupled to the M2 monoclonal antibody. SPT5 and the associated SPT4 protein was then eluted from the M2 antibody with Flag peptide. Western blot analysis of these affinity-purified proteins was performed with either the M2 monoclonal antibody directed against the Flag epitope for SPT5 or the Myc epitope for SPT4 (Fig. 4B). The 1.0 M potassium acetate fraction of HeLa nuclear extract was used with the affinity-purified SPT5 and SPT4 proteins in in vitro transcription assays to characterize their role in mediating DRB inhibition and Tat activation.
DRB markedly inhibited in vitro transcription from the pTF3-6C2AT plasmid using unfractionated HeLa nuclear extract (Fig. 4C, lanes 1 and 2). In contrast, DRB was not able to inhibit in vitro transcription from this template using the 1.0 M potassium acetate phosphocellulose fraction which lacked SPT5 (lanes 3 and 4). The addition of affinity-purified SPT4 alone did not result in significant DRB inhibition, while the addition of SPT5 alone resulted in modest DRB inhibition (lanes 5 to 8). This latter result may be due to the association of low levels of endogenous SPT4 with the affinity-purified SPT5. However, the addition of either baculovirus-expressed and purified SPT4 and SPT5 (data not shown) or affinity-purified SPT4 and SPT5 isolated from transfected COS cells (lanes 9 and 10) to the 1.0 M potassium acetate fraction resulted in both a slight stimulation of basal transcription and marked DRB-mediated inhibition. These results confirm the previously described roles of SPT4 and SPT5 in mediating DRB inhibition (53).
SPT5 mutants were then assayed in conjunction with SPT4 for their ability to mediate DRB inhibition. The addition of an SPT5 mutant (aa 1 to 837) that lacked the CTR2 domain increased basal transcription and resulted in marked DRB inhibition (Fig. 4C, lanes 11 and 12). In contrast, an SPT5 mutant (aa 1 to 754) that lacked both the CTR1 and CTR2 domains did not stimulate basal transcription or result in significant DRB-mediated repression (lanes 13 and 14). An SPT5 mutant that lacked the CTR1 domain (aa 755 to 837) alone did not alter either basal transcription or result in DRB-mediated repression (lanes 15 and 16). In contrast, an SPT5 deletion mutant (with aa 1 to 176 deleted) which lacked acidic amino acids in the amino terminus of SPT5, stimulated both basal transcription and DRB-mediated repression (lanes 17 and 18). An SPT5 deletion mutant which lacked the amino-terminal 230 aa of this protein resulted in modest stimulation of basal transcription and DRB-mediated repression (lanes 19 and 20). A deletion mutant which lacked aa 175 to 315 of SPT5, comprising the SPT4 binding site, did not stimulate basal transcription and resulted in only slight DRB-mediated repression (lanes 21 and 22). Finally, a truncated SPT5 protein which extended from aa 421 to 755 and included a portion of the RNA polymerase II binding domain, did not alter basal transcription or function in DRB-mediated repression (lanes 23 and 24). These results indicate that the CTR1 domain of SPT5 is critical for DRB-mediated repression by SPT5.
The CTR1 domain of SPT5 is critical for mediating Tat activation.
The 1.0 M potassium acetate phosphocellulose fraction was next assayed for its ability to support Tat activation. First, we demonstrated, using unfractionated HeLa nuclear extract, that Tat strongly activated in vitro transcription from the wild-type HIV-1 LTR template (Fig. 4D, lanes 1 to 4, upper panel) but not from an HIV-1 template with mutations in the TAR RNA loop sequences (lanes 1 to 4, lower panel) (58). In contrast, there was no Tat activation when the 1.0 M potassium acetate phosphocellulose fraction of HeLa nuclear extract was used (lanes 5 and 6). The addition of the affinity-purified SPT4 and SPT5 resulted in the restoration of low levels of Tat activation for the wild type (lanes 7 and 8, upper panel) but not the HIV-1 loop mutant template (lanes 7 and 8, lower panel). Similar results were seen with the 1.0 M potassium acetate phosphocellulose fraction following the addition of baculovirus-produced SPT4 and SPT5 (data not shown). The addition of SPT5 alone did not markedly stimulate Tat activation in the absence of transfected SPT4 (lanes 9 and 10).
A variety of affinity-purified SPT5 and SPT4 proteins were also assayed for their ability to restore Tat activation when added to the 1.0 M potassium fraction of HeLa nuclear extract. An SPT5 mutant (aa 1 to 837) that lacked the CTR2 domain but not the CTR1 domain stimulated Tat activation of the wild-type HIV-1 LTR (Fig. 4D, lanes 11 and 12, upper panel). However, an SPT5 mutant (aa 1 to 754) that lacked both the CTR1 and CTR2 domains or that lacked the CTR1 domain alone (aa 755 to 837) did not stimulate Tat activation (lanes 13 to 16, upper panel). These SPT5 mutants also did not stimulate Tat activation of the HIV-1 loop mutant (lanes 9 to 16, lower panel). These results support a role for the SPT5 CTR1 domain in regulating Tat activation.
An SPT5 mutant that lacked the amino-terminal 176 aa of SPT5 still mediated Tat activation (Fig. 4D, lanes 17 and 18), while only minimal Tat activation was seen with an SPT5 deletion mutant that lacked the amino-terminal 230 aa (lanes 19 and 20). An SPT5 protein (Δ175–315), which lacked the SPT4 binding site, resulted in only minimal Tat activation (lanes 21 and 22). Finally, a truncated SPT5 protein extends from aa 421 to 755 did not alter Tat activation (lanes 23 and 24). We conclude that the same domains of SPT5 that mediated DRB repression were also required for Tat activation.
P-TEFb phosphorylation of SPT5.
Previous data suggest that P-TEFb is critical for SPT5 function (54). For example, immunodepletion of CDK9 from HeLa nuclear extract prevented SPT5 from mediating DRB repression. Thus, one possible mechanism to explain DRB-mediated repression is that this compound inhibits P-TEFb phosphorylation of RNA polymerase II, which alters its interaction with SPT5 and results in transcriptional inhibition (31). Alternatively, it is possible that DRB could inhibit P-TEFb phosphorylation of SPT5, which may lead to enhanced SPT5 inhibitory effects on transcription. Thus, the CTR1 domain of SPT5 may be a substrate for P-TEFb phosphorylation which is important in the regulation of both DRB-mediated repression and Tat activation.
First, we assayed whether CDK9 could directly phosphorylate SPT5 (Fig. 5). Baculovirus-produced CDK9, either alone or when coexpressed with cyclin T1, cyclin T2a, or cyclin T2b (36), was assayed in in vitro kinase assays for its ability to phosphorylate GST-SPT5. CDK9 alone had very low kinase activity (Fig. 5A, lane 6). The addition of CDK9 with either cyclin T1, cyclin T2a, or cyclin T2b in the presence of the GST-SPT5 substrate resulted in marked phosphorylation of SPT5 (lanes 7 to 9). The phosphorylation of SPT5 by CDK9 was associated with decreased mobility of SPT5 in SDS-PAGE (lanes 6 to 9). Western blot analysis of the baculovirus-produced proteins demonstrated the expression of CDK9 and cyclin T (Fig. 5B). These results indicate that P-TEFb can directly phosphorylate SPT5.
FIG. 5.
P-TEFb phosphorylation of SPT5. (A) Baculovirus-produced and purified proteins including CDK9 (lanes 1 and 6), CDK9 and cyclin T1 (lanes 2 and 7), CDK9 and cyclin T2a (lanes 3 and 8), and CDK9 and cyclin T2b (lanes 4 and 8) were assayed in in vitro kinase assays in the absence of the GST-SPT5 substrate (lanes 1 to 4), the GST-SPT5 substrate was assayed alone (lane 5), or the GST-SPT5 substrate was added along with the different CDK9 preparations (lanes 6 to 9). (B) The baculovirus-produced and purified preparations of CDK9 (lane 1), CDK9 and cyclin T1 (lane 2), CDK9 and cyclin T2a (lane 3), and CDK9 and cyclin T2b (lane 4) were assayed in Western blot analysis using antibodies directed against CDK9 (top panel), cyclin T1 (middle panel), or cyclin T2 (lower panel).
Both CTR1 and CTR2 have multiple amino acid repeats which are rich in serine and threonine residues (see below in Fig. 7A). Next, we compared the ability of baculovirus-produced P-TEFb, composed of CDK9-cyclin T1 (31, 36, 55, 64), and CAK, composed of CDK7, cyclin H, and MAT1 (14, 15), to phosphorylate either different GST-SPT5 fusion proteins or RNA polymerase II. Both CAK and P-TEFb phosphorylation of the RNA polymerase II CTD are associated with increases in promoter clearance and transcriptional elongation (reviewed in reference 27). These kinases were also demonstrated to phosphorylate the same amino acids within the CTD (26).
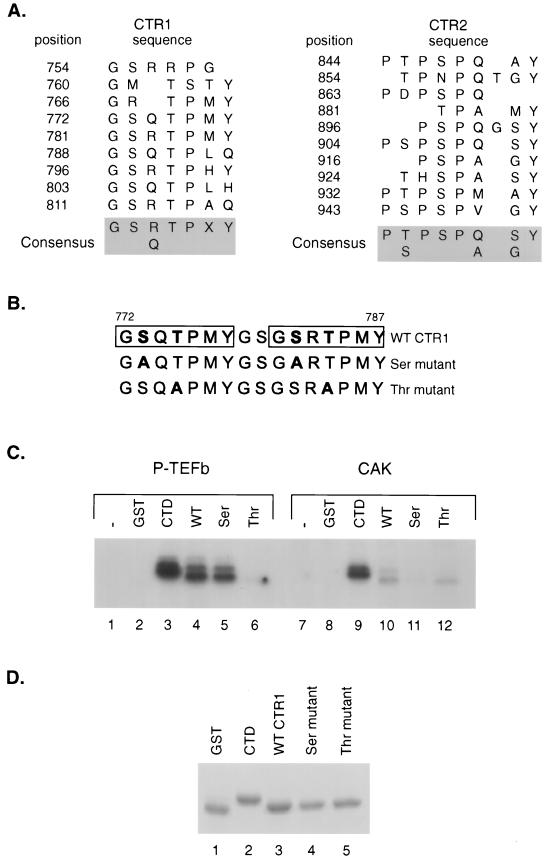
FIG. 7.
Comparison of P-TEFb and CAK phosphorylation of SPT5 CTR1 repeats. (A) Sequences of CTR1 and CTR2 repeats. The positions of the C-terminal repeat motifs in the CTR1 and CTR2 domains of SPT5 are indicated, as is the consensus sequence for each domain, as previously described (49). (B) The sequences of the two CTR1 repeats that were fused to GST and used as substrates in in vitro kinase reactions are shown. The boxes indicate the positions of the repeats, and the mutated amino acids are shown in boldface type. (C) In vitro kinase reactions were performed using baculovirus-produced and purified CDK9-cyclin T1 (lanes 1 to 6) or the baculovirus-produced and purified CAK components including CDK7, cyclin H, and MAT1 (lanes 7 to 12). The kinase reactions were performed without the addition of substrate (lanes 1 and 7) or following the addition of GST (lanes 2 and 8), a GST-CTD fusion protein containing two repeats of the RNA polymerase II CTD (lanes 3 and 9), or GST fusion proteins containing two CTR1 repeats with either the wild-type (WT) sequences (lanes 4 and 10), serine residues substituted for alanine (lanes 5 and 11), or threonine residues substituted for alanine (lanes 6 and 12). (D) A Coomassie blue-stained SDS-polyacrylamide gel of the GST fusion proteins that were used as substrates for kinase assays in panel B is shown.
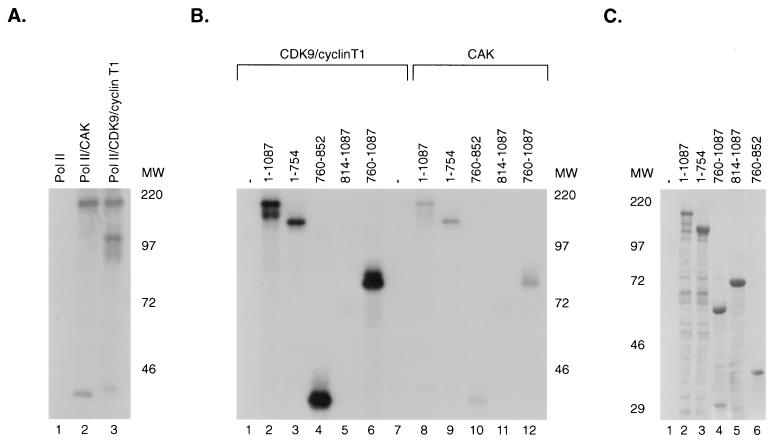
P-TEFb and CAK phosphorylated RNA polymerase II to similar levels (Fig. 6A). CDK9-cyclin T1 was able to phosphorylate wild-type SPT5 (Fig. 6B, lane 2). Both the amino-terminal 754 aa (lane 3) and the carboxy-terminal 327 aa (lane 6) of SPT5 were phosphorylated. The ability of CDK9/cyclin T1 to phosphorylate the CTR1 and CTR2 domains was next assayed. CDK9-cyclin T1 was able to phosphorylate CTR1 (lane 4) but not CTR2 (lane 5). These results indicate that CTR1 domain appears to be a site for phosphorylation by P-TEFb, although there are additional phosphorylation sites in the N-terminal portion of SPT5. CAK phosphorylated similar regions of SPT5, but at a remarkably reduced level compared to P-TEFb (lanes 8 to 12). Therefore, SPT5 serves as a preferred substrate for P-TEFb rather than another CTD kinase that is also involved in the regulation of transcriptional elongation.
FIG. 6.
Comparison of P-TEFb and CAK phosphorylation of SPT5 constructs. (A) Purified RNA polymerase II (Pol II) was assayed in in vitro kinase assays using RNA polymerase II alone (lane 1) or in the presence of baculovirus-produced and purified CAK including CDK7, cyclin H, and MAT1 (lane 2) or CDK9 and cyclin T1 (lane 3). MW, molecular weight in thousands. (B) In vitro kinase assays were performed using baculovirus-produced preparations of CDK9/cyclin T1 (lanes 1 to 6) or CAK (lanes 7 to 12) with either no added substrate (lanes 1 and 7), GST-SPT5 (lanes 2 and 8), GST-SPT5 (aa 1 to 754) (lanes 3 and 9), GST-SPT5 (aa 760 to 852) (lanes 4 and 10), GST-SPT5 (aa 814 to 1087) (lanes 5 and 11), or GST-SPT5 (aa 760 to 1087) (lanes 6 and 12). Phosphorylation was assayed following SDS-PAGE and autoradiography. (C) A Coomassie blue-stained gel of the various GST-SPT5 fusion proteins used in the in vitro kinase assays in part B is shown.
CTR1 amino acid residues phosphorylated by P-TEFb.
Next we determined which amino acids within the CTR1 repeat (Fig. 7A) are phosphorylated by CDK9-cyclin T1. The consensus repeat in CTR1, GS(R/Q)TPXY, contains serine and threonine residues that can potentially be phosphorylated by P-TEFb. We constructed a GST fusion protein containing two CTR1 repeats spanning aa 772 to 787 and then either left this region intact or created mutations of serine or threonine residues in both repeats to alanine (Fig. 7B). The wild-type and mutant GST fusion proteins were then used as substrates in in vitro kinase reactions with baculovirus-produced CDK9-cyclin T1 (Fig. 7C). The wild-type and serine mutant GST-CTR1 fusion proteins were readily phosphorylated by CDK9-cyclin T1 (Fig. 7C, lanes 4 and 5), while the GST-CTR1 threonine mutant was not significantly phosphorylated (lane 6). This result indicated that threonine residues in the CTR1 repeat are probably the preferred amino acids that are phosphorylated by P-TEFb.
When CAK was used as a kinase with these CTR1 substrates, the level of phosphorylation of the wild-type repeat was severalfold lower than the level of phosphorylation seen with P-TEFb (Fig. 7C, lane 10). Both kinases phosphorylated the CTD repeat of RNA polymerase II to similar levels (lanes 3 and 9). Remarkably, replacement of either serine or threonine residues by alanine led to a similar reduction in the level of phosphorylation of the CTR1 repeat by CAK (lanes 10 to 12). This result indicated that CAK does not display a clear preference for threonine residues in the CTR1 repeats as does P-TEFb.
Role of P-TEFb phosphorylation on SPT5 function.
Finally, we tested whether P-TEFb phosphorylation of SPT5 alters its functional properties. The recombinant SPT4-SPT5 complex purified following baculovirus production was incubated with P-TEFb in either the presence or absence of ATP. SPT4-SPT5 was then added to in vitro transcription reaction mixtures with the pTF3-6C2AT template and HeLa nuclear extract depleted of endogenous SPT5. First, we demonstrated that DRB inhibited in vitro transcription of this template by using both unfractionated HeLa nuclear extract and HeLa nuclear extract immunodepleted with GST antibody (Fig. 8A, lanes 1 to 4). In contrast, HeLa nuclear extract immunodepleted of SPT5 exhibited only minimal DRB sensitivity (lanes 5 and 6), which was restored by the addition of recombinant SPT4-SPT5 complex (lanes 7 and 8). Preincubation of the SPT4-SPT5 complex with P-TEFb, in the absence of ATP, did not change its ability to confer DRB sensitivity to in vitro transcription reactions with HeLa extract depleted of SPT5 (lanes 9 to 12). However, when ATP was present during the preincubation of SPT4-SPT5 complex with P-TEFb, little DRB sensitivity was observed (lanes 15 and 16). When SPT4-SPT5 was preincubated with ATP in the absence of P-TEFb, it was still able to confer DRB sensitivity, although to a somewhat lesser extent than was the control lacking ATP (lanes 17 and 18). This result may be explained by the fact that the preparation of recombinant SPT4-SPT5 contains trace amounts of kinases that are capable of phosphorylating SPT5. Thus, phosphorylation of SPT5 by P-TEFb before beginning the in vitro transcription reaction alters SPT5 function and abrogates its ability to mediate DRB inhibition of transcription.
FIG. 8.
Role of P-TEFb phosphorylation on SPT4-SPT5 function in mediating DRB repression. (A) In vitro transcription assays were performed with the pTF3-6C2AT template in the presence (even-numbered lanes) or absence (odd-numbered lanes) of DRB. Untreated HeLa nuclear extract (lanes 1 and 2), HeLa nuclear extract immunodepleted with GST antibody (lanes 3 and 4), or HeLa nuclear extract immunodepleted with SPT5 antibody (lanes 6 to 20) were used in the in vitro transcription assays. In vitro transcription assay mixtures with the SPT5-immunodepleted HeLa nuclear extracts were supplemented with the SPT4-SPT5 complex purified following baculovirus expression in the absence of other treatment (lanes 7 and 8), the SPT4-SPT5 complex preincubated with P-TEFb (lanes 9 and 10 and lanes 15 and 16), the SPT4-SPT5 complex preincubated without P-TEFb (lanes 11 and 12 and lanes 17 and 18), or P-TEFb alone preincubated in the absence of SPT4-SPT5 (lanes 13 and 14 and lanes 19 and 20). The preincubation procedure was performed in the absence of ATP (lanes 9 to 14) or in the presence of 5 mM ATP (lanes 15 to 20), which was subsequently removed by dialysis. (B) A schematic of the functional domains in the SPT5 protein that were analyzed in this study is shown.
DISCUSSION
The SPT4 and SPT5 proteins are involved in the control of transcriptional elongation (23, 53, 58, 61). Depending on the conditions, such as the concentrations of nucleotides used in the in vitro transcription assay, SPT4 and SPT5 function in both inhibiting and enhancing transcriptional elongation (53). Although SPT5 directly interacts with RNA polymerase II (53, 61), the mechanism by which it alters the processivity of polymerase remains to be determined. One study suggests that SPT5 associates better with the unphosphorylated (IIa) form of RNA polymerase II than with the phosphorylated (IIo) form (54). Therefore, it is possible that P-TEFb may stimulate transcriptional elongation both by increasing the phosphorylation of SPT5 bound to RNA polymerase II and by increasing the phosphorylation of the polymerase CTD. This could result in the release of unphosphorylated SPT5 from the IIo form of the polymerase, while the phosphorylated form of SPT5 stimulates its polymerase processivity. Further studies are needed to test this model. In addition to P-TEFb (54), multiprotein complexes including NELF (59) and Tat-SF1 (29, 35) modulate the SPT4-SPT5, function suggesting that a number of positive and negative factors may regulate its activity.
In the present study, we analyzed domains in SPT5 that are involved in in vivo interactions with SPT4, RNA polymerase II, and CDK9. Furthermore, we characterized domains in SPT5 that are responsible for DRB-mediated repression and Tat activation (Fig. 8B). This analysis defines domains in SPT5 that bind SPT4 and RNA polymerase II that are in agreement with a previous study (61). In contrast to the results of that study, our analysis demonstrates that a CTD of SPT5 with nine conserved heptad motifs, designated CTR1, is critical for DRB-mediated repression. Furthermore, CTR1 is involved in SPT5 modulation of Tat activation and is also a target for P-TEFb phosphorylation. One possible explanation for these results is that the recombinant SPT5 and SPT4 used in the previous study was produced in bacteria and subjected to a denaturation and renaturation protocol that may have altered its activity compared to the SPT4 and SPT5 proteins used in our study.
Our results demonstrate that both DRB inhibition and Tat activation can be restored by the addition of purified SPT4 and SPT5 to fractionated HeLa nuclear extract that is not competent for either DRB inhibition or Tat activation. Our previous work (58) suggested that SPT5 but not SPT4 was critical for Tat activation. However, this result was due to the low affinity of the antibody used to immunodeplete SPT4. The effects of SPT4 and SPT5 are due to both increases in transcriptional repression in the presence of DRB (1.5- to 2-fold) and increases in basal transcription in the absence of DRB (2.5- to 3.5-fold). The effects of SPT4 and SPT5 on Tat activation of the HIV LTR are primarily due to decreases in basal transcription (1.5- to 2-fold), although increases in Tat-induced transcription (1.5-fold) are noted. Similar effects on DRB inhibition and Tat activation are observed with the SPT5 deletion mutants and SPT4 when they are added to HeLa nuclear extract immunodepleted of SPT5 rather than when they are added to the 1.0 M potassium acetate fraction of HeLa nuclear extract (D. Ivanov and R. Gaynor, unpublished observation). However, there are differences in the ability of SPT4 and SPT5 to activate HIV-1 transcription in the SPT5-immunodepleted HeLa extract compared to the 1.0 M potassium acetate fraction. When SPT4 and SPT5 are added to the 1.0 M potassium acetate phosphocellulose fraction, these proteins activate HIV-1 transcription in the presence of Tat and repress basal transcription. However when SPT4 and SPT5 are added to the SPT5-immunodepleted extract, they result primarily in repression of basal transcription rather than stimulation of transcription in the presence of Tat.
The yeast SPT5 homologue, like human SPT5, contains a repetitive structure in its C-terminal portion (50). It is composed of 15 copies of a 6-aa repeat with a consensus consequence S(T/A)WGG(A/G) that is very different from the repeats found in mammalian SPT5 protein. Since no homologue of P-TEFb has been identified in yeast, it is possible that the serine residue in this repeat is phosphorylated by another kinase which recognizes a different consensus sequence. In support of this possibility, partial deletion of these repeats in the yeast SPT5 protein impairs its function while deletion of all 15 of these repeats virtually eliminates SPT5 function (50). Therefore, it is likely that the C-terminal repeats found in both yeast and mammalian SPT5 proteins function to promote protein-protein interactions and regulate SPT5 activity. It is likely that other domains in SPT5, including acidic residues in its amino terminus, function in other regulatory properties such as modulating chromatin structure, which would not be detected in the in vitro transcription assays used in this study.
Our model would suggest that it is probably the unphosphorylated form of SPT5 bound to RNA polymerase II that is initially involved in modulating its transcriptional elongation properties. P-TEFb phosphorylation of SPT5 may alter its interactions with RNA polymerase II or facilitate the interaction with other factors that associate with polymerase. In the absence of the CTR1 domain, SPT5 can bind to RNA polymerase II but is not able to modulate the transcriptional elongation properties of polymerase. SPT5 that is phosphorylated prior to the onset of transcription is unable to mediate DRB inhibition. This result is consistent with the idea that the timing of SPT5 phosphorylation during transcription is critical in regulating its ability to modulate polymerase processivity. Further work is needed to address how phosphorylation of SPT5 leads to changes in its functional properties.
Interestingly, SPT5 protein from mitotic HeLa cells appears to migrate more slowly in SDS-polyacrylamide gels than does SPT5 isolated from interphase cells, and this effect is probably the result of enhanced SPT5 phosphorylation (49). We find that SPT5 phosphorylation by P-TEFb also results in a slower-migrating form of SPT5, probably reflecting the phosphorylation of multiple C-terminal repeats. In this regard, it is remarkable that during mitosis RNA polymerase II aborts transcription and that nascent transcripts dissociate from the template (46). This suggests the existence of mechanisms controlling the elongation phase of transcription during mitosis, which might involve inactivation of specific transcription elongation factors such as SPT5 by enhancing their phosphorylation prior to preventing the transcription of specific genes. In addition, a number of transcription factors such as Sp1 and Oct-1 are phosphorylated during mitosis and demonstrate decreased DNA binding properties (43).
Among the known elongation factors, TFIIF increases the rate of RNA chain synthesis relatively uniformly (2, 16, 38) while TFIIS releases polymerase stalled at intrinsic pause sites (40, 42). Elongin was suggested to facilitate the proper positioning of the 3′-hydroxyl terminus of the nascent transcript with respect to the catalytic site of RNA polymerase II to increase the rate of RNA chain elongation (52). We speculate that the SPT4-SPT5 complex works through a different mechanism by actually decreasing the rate of transcriptional elongation while at the same time preventing the collapse of the RNA polymerase II into a “dead-end” configuration that might happen during the “leap” coinciding with the polymerase leaving an intrinsic pause site (6). This would explain why SPT4-SPT5 can function as both positive and negative transcription factors depending on the experimental conditions. Since the rate of transcriptional elongation is usually assessed by the amount of transcript that reaches a certain point, it is difficult to distinguish between an elongation factor that causes polymerase to increase RNA synthesis and a factor that allows polymerase to move more slowly but extend past intrinsic pause sites such as nucleosomes (1, 5, 50, 51, 57). This model would agree with genetic data indicating that mutations in the two largest subunits of the RNA polymerase II complex, which reduce the efficiency of elongation, can suppress conditional mutations in SPT5.
The existing model of SPT5 action (54) suggests that the sole function of P-TEFb is to relieve the repression caused by SPT4-SPT5. P-TEFb-mediated CTD phosphorylation may prevent SPT4-SPT5 from interacting with RNA polymerase II. Our results indicate that P-TEFb also phosphorylates SPT5 directly and that this phosphorylation is critical for SPT5 function. Thus, P-TEFb, like another CTD kinase, TFIIH, is able to stimulate the activity of other transcriptional regulators (44). The ability of SPT4-SPT5 to stimulate transcription in the absence of DRB suggests that phosphorylation by P-TEFb not only overcomes its negative effect on transcription but also converts it into a positive factor. We propose that phosphorylation of the CTR1 domain in SPT5 may result in recruitment of additional positive regulators of transcriptional elongation. This hypothesis is also supported by the observation that DRB does not function in transcription assay mixtures containing purified RNA polymerase II, recombinant SPT4-SPT5, and P-TEFb (54). In addition, our preliminary results suggest the existence of additional proteins that specifically interact with the phosphorylated CTR1 and function to stimulate transcriptional elongation in the absence of DRB. Further studies are necessary to better understand the mechanism by which SPT5 modulates transcriptional elongation.
ACKNOWLEDGMENTS
Dmitri Ivanov and Youn Tae Kwak contributed equally to this work.
We thank David Price, Hiroshi Handa, and David Morgan for sharing reagents and procedures, Sharon Johnson for preparation of the manuscript, and Alexjandra Herrera for preparation of the figures.
This work was supported by grants from the NIH, the Welch Foundation, and the Veterans Administration.
REFERENCES
- 1.Basrai M A, Kingsbury J, Koshland D, Spencer F, Hieter P. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2838–2847. doi: 10.1128/mcb.16.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bengal E, Flores O, Krauskopf A, Reinberg D, Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991;11:1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley D L. Regulation of transcriptional elongation by RNA polymerase II. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–65. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 6.Chamberlin M J. New models for the mechanism of transcription elongation and its regulation. Harvey Lect. 1995;88:1–21. [PubMed] [Google Scholar]
- 7.Chen D, Fong Y, Zhou Q. Specific interaction of Tat with the human but not rodent P-TEFb complex mediates the species-specific Tat activation of HIV-1 transcription. Proc Natl Acad Sci USA. 1999;96:2728–2733. doi: 10.1073/pnas.96.6.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang P-W, Fogel E, Jackson C L, Lieuallen K, Lennon G, Qu X, Wang S-Q, Kurnit D M. Isolation, sequencing, and mapping of the human homologue of the yeast transcription factor, SPT5. Genomics. 1996;38:421–424. doi: 10.1006/geno.1996.0646. [DOI] [PubMed] [Google Scholar]
- 9.Chiang P-W, Wang S-Q, Smithivas P, Song W-J, Crombez E, Akhtar A, Im R, Greenfield J, Ramamoorthy S, Van Keuren M, Blackburn C C, Tsai C-H, Kurnit D M. Isolation and characterization of the human and mouse homologues (SUPT4H and Supt4h) of the yeast SPT4 gene. Genomics. 1996;34:368–375. doi: 10.1006/geno.1996.0299. [DOI] [PubMed] [Google Scholar]
- 10.Chodosh L A, Fire A, Samuels M, Sharp P A. 5,6-Dichloro-1-beta-d-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J Biol Chem. 1989;264:2250–2257. [PubMed] [Google Scholar]
- 11.Cujec T P, Okamoto H, Fujinaga K, Meyer J, Chamberlin H, Morgan D O, Peterlin B M. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:2645–2657. doi: 10.1101/gad.11.20.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feaver W J, Svejstrup J Q, Henry N L, Kornberg R D. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 14.Fisher R P, Jin P, Chamberlin H M, Morgan D O. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83:47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- 15.Fisher R P, Morgan D O. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78:713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- 16.Flores O, Maldonado E, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Factors IIE and IIF independently interact with RNA polymerase II. J Biol Chem. 1989;264:8913–8921. [PubMed] [Google Scholar]
- 17.Fujinaga K, Cujec T P, Peng J, Garriga J, Price D H, Grana X, Peterlin B M. The ability of positive transcription elongation factor b to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J Virol. 1998;72:7154–7159. doi: 10.1128/jvi.72.9.7154-7159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gold M O, Yang X, Herrmann C H, Rice A P. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J Virol. 1998;72:4448–4453. doi: 10.1128/jvi.72.5.4448-4453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hampsey M, Reinberg D. RNA polymerase II as a control panel for multiple coactivator complexes. Curr Opin Genet Dev. 1999;9:132–139. doi: 10.1016/S0959-437X(99)80020-3. [DOI] [PubMed] [Google Scholar]
- 21.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 22.Hartzog G A, Basrai M A, Ricupero-Hovasse S L, Hieter P, Winston F. Identification and analysis of a functional human homolog of the SPT4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2848–2856. doi: 10.1128/mcb.16.6.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartzog G A, Wada T, Handa H, Winston F. Evidence that SPT4, SPT5 and SPT6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horton R M, Ho S N, Pullen J K, Hunt H D, Cai Z, Pease L R. Gene splicing by overlap extension. Methods Enzymol. 1993;217:270–279. doi: 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- 25.Isel C, Karn J. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J Mol Biol. 1999;290:929–941. doi: 10.1006/jmbi.1999.2933. [DOI] [PubMed] [Google Scholar]
- 26.Ivanov D, Kwak Y T, Nee E, Guo J, Garcia-Martinez L F, Gaynor R B. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for Tat-activation. J Mol Biol. 1999;288:41–56. doi: 10.1006/jmbi.1999.2663. [DOI] [PubMed] [Google Scholar]
- 27.Jones K A. Taking a new TAK on Tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 28.Jones K A, Peterlin B M. Control of RNA initiation and elongation at the HIV-1 promoter. Annu Rev Biochem. 1994;63:717–743. doi: 10.1146/annurev.bi.63.070194.003441. [DOI] [PubMed] [Google Scholar]
- 29.Kim J B, Yamaguchi Y, Wada T, Handa H, Sharp P A. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol Cell Biol. 1999;19:5960–5968. doi: 10.1128/mcb.19.9.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwak Y T, Ivanov D, Guo J, Nee E, Gaynor R B. Role of the human and murine cyclin T proteins in regulating HIV-1 Tat-activation. J Mol Biol. 1999;288:57–69. doi: 10.1006/jmbi.1999.2664. [DOI] [PubMed] [Google Scholar]
- 31.Mancebo H S Y, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall N F, Peng J, Xie P, Price D H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 33.Marshall N F, Price D H. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J Biol Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 34.Mavankal G, Ou S H I, Oliver H, Sigman D, Gaynor R B. HIV-1 and HIV-2 Tat proteins specifically interact with RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:2089–2094. doi: 10.1073/pnas.93.5.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parada C A, Roeder R G. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 1999;18:3688–3701. doi: 10.1093/emboj/18.13.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ping Y H, Rana T M. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J Biol Chem. 1999;274:7399–7404. doi: 10.1074/jbc.274.11.7399. [DOI] [PubMed] [Google Scholar]
- 38.Price D H, Sluder A E, Greenleaf A L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reinberg D, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of initiation factors IIB and IIE. J Biol Chem. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 40.Reines D, Chamberlin M J, Kane C M. Transcription elongation factor SII (TFIIS) enables RNA polymerase II to elongate through a block to transcription in a human gene in vitro. J Biol Chem. 1989;264:10799–10809. [PubMed] [Google Scholar]
- 41.Reines D, Conaway J W, Conaway R C. The RNA polymerase II general elongation factors. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- 42.Reines D, Mote J J. Elongation factor SII-dependent transcription by RNA polymerase II through a sequence-specific DNA-binding protein. Proc Natl Acad Sci USA. 1993;90:1917–1921. doi: 10.1073/pnas.90.5.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts S B, Segil N, Heintz N. Differential phosphorylation of the transcription factor Oct1 during the cell cycle. Science. 1991;253:1022–1026. doi: 10.1126/science.1887216. [DOI] [PubMed] [Google Scholar]
- 44.Rochette-Egly C, Adam S, Rossignol M, Egly J M, Chambon P. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90:97–107. doi: 10.1016/s0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 45.Roy R, Adamczewski J P, Seroz T, Vermeulen W, Tassan J P, Schaeffer L, Nigg E A, Hoeijmakers J H J, Egly J-M. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell. 1994;79:1093–1101. doi: 10.1016/0092-8674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 46.Shermoen A W, O'Farrell P H. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–10. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiekhattar R, Mermelstein F, Fisher R P, Drapkin R, Dynlacht B, Wessling H C, Morgan D O, Reinberg D. Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature. 1995;374:283–287. doi: 10.1038/374283a0. [DOI] [PubMed] [Google Scholar]
- 48.Shilatifard A. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 1998;12:1437–1446. doi: 10.1096/fasebj.12.14.1437. [DOI] [PubMed] [Google Scholar]
- 49.Stachora A A, Schafer R, Pohlmeier M, Maier G, Ponsting H. Human Supt5h protein, a putative modulator of chromatin structure, is reversibly phosphorylated in mitosis. FEBS Lett. 1997;409:74–78. doi: 10.1016/s0014-5793(97)00486-9. [DOI] [PubMed] [Google Scholar]
- 50.Swanson M S, Malone E A, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanson M S, Winston F. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takagi Y, Conaway J W, Conaway R C. A novel activity associated with RNA polymerase II elongation factor SIII. SIII directs promoter-independent transcription initiation by RNA polymerase II in the absence of initiation factors. J Biol Chem. 1995;270:24300–24305. doi: 10.1074/jbc.270.41.24300. [DOI] [PubMed] [Google Scholar]
- 53.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human SPT4 and SPT5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei P, Garber M E, Fang S-M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 56.Wimmer J, Fujinaga K, Taube R, Cujec T P, Zhu Y, Peng J, Price D H, Peterlin B M. Interactions between Tat and TAR and human immunodeficiency virus replication are facilitated by human cyclin T1 but not cyclins T2a or T2b. Virology. 1999;255:182–189. doi: 10.1006/viro.1998.9589. [DOI] [PubMed] [Google Scholar]
- 57.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 58.Wu-Baer F, Lane W S, Gaynor R B. Role of the human homologues of the yeast transcription factors SPT5 and SPT6 in HIV-1 Tat-activation. J Mol Biol. 1998;277:179–197. doi: 10.1006/jmbi.1997.1601. [DOI] [PubMed] [Google Scholar]
- 59.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi Y, Wada T, Handa H. Interplay between positive and negative elongation factors: drawing a new view of DRB. Genes Cells. 1998;3:9–15. doi: 10.1046/j.1365-2443.1998.00162.x. [DOI] [PubMed] [Google Scholar]
- 61.Yamaguchi Y, Wada T, Watanabe D, Takagi T, Hasegawa J, Handa H. Structure and function of the human transcription elongation factor DSIF. J Biol Chem. 1999;274:8085–8092. doi: 10.1074/jbc.274.12.8085. [DOI] [PubMed] [Google Scholar]
- 62.Zhou Q, Chen D, Pierstorff E, Luo K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998;17:3681–3691. doi: 10.1093/emboj/17.13.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Q, Sharp P A. Novel mechanism and factor for regulation by HIV-1 Tat. EMBO J. 1995;14:321–328. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu Y, Pery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 Tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]