Abstract
Metabolic syndrome (MetS) is highly prevalent worldwide. In the United States, estimates show that more than 30% of the adult population has MetS. MetS consists of multiple phenotypes, including obesity, dyslipidemia, and impaired glucose tolerance. Therefore, identifying the molecular mechanisms to explain this complex disease is critical for diagnosing and treating MetS. We previously showed 70 increased genes and 20 decreased genes in whole blood in MetS participants. The present study aimed to identify blood-based DNA methylation biomarkers in non-MetS versus MetS participants. The present study analyzed whole blood DNA samples from 184 adult participants of Latino descent from the Arizona Insulin Resistance (AIR) registry. We used the National Cholesterol Education Program Adult Treatment Panel III (NCEP: ATP III) criteria to identify non-MetS (n = 110) and MetS (n = 74) participants. We performed whole blood methylation analysis on select genes: ATP Synthase, H+ Transporting mitochondrial F1 Complex, Epsilon Subunit (ATP5E), Cytochrome C Oxidase Subunit VIc (COX6C), and Ribosomal Protein L9 (RPL9). The pyrosequencing analysis was a targeted approach focusing on the promoter region of each gene that specifically captured CpG methylation sites. In MetS participants, we showed decreased methylation in two CpG sites in COX6C and three CpG sites in RPL9, all p < 0.05 using the Mann-Whitney U test. There were no ATP5E CpG sites differently methylated in the MetS participants. Furthermore, while adjusting for age, gender, and smoking status, logistic regression analysis reaffirmed the associations between MetS and mean methylation within COX6C and RPL9 (both p < 0.05). In addition, Spearman’s correlation revealed a significant inverse relationship between the previously published gene expression data and methylation data for RPL9 (p < 0.05). In summary, these results highlight potential blood DNA methylation biomarkers for the MetS phenotype. However, future validation studies are warranted to strengthen our findings.
Introduction
Metabolic syndrome (MetS) is a collection of metabolic phenotypes associated with diseases such as obesity, type 2 diabetes mellitus, dyslipidemia, and cardiovascular disease [1–3]. Classification of the MetS is based on the following National Cholesterol Education Program Adult Treatment Panel III (NCEP: ATP III) criteria: waist circumference (>102 cm men / >88 cm women), fasting plasma glucose (≥110 mg/dL), triglycerides (≥150 mg/dL), high-density lipoprotein cholesterol (<40mg/dL men / <50 mg/dL women) and high blood pressure (≥130/≥80 mmHg) [4]. In recent decades, it has become increasingly important to understand the molecular mechanisms of MetS because over a third of all United States adults met the NCEP: ATP III criteria [5].
The pathophysiological mechanisms of MetS are complex and are not entirely known. There have been many proposed physiological mechanisms, including insulin resistance, neurohormonal activation, and chronic low-grade inflammation [6]. Early diagnosis of the MetS is critical so that physicians can intervene and manage the disease to delay the progression of these physiological mechanisms. Identifying non-invasive blood-based biomarkers for MetS may aid in diagnosing and screening those patients at risk of developing MetS. Importantly, these biomarkers may help in the management and treatment of patients with this disease.
A systemic review by Srikanthan et al. highlighted several inflammatory and anti-inflammatory blood biomarkers for MetS [7]. In addition, a review by O’Neill et al. described novel blood-based biomarkers for MetS, including DNA genetic variants, microRNA biomarkers, protein biomarkers, and mRNA biomarkers [8]. Previous work from our laboratory showed differential mRNA expression in MetS in whole blood samples from the Arizona Insulin Resistance (AIR) registry [9]. In that study, we showed ninety altered circulating mRNAs with MetS, which could serve as potential biomarkers for the disease [9]. In more recent years, there has been a focus on identifying blood-based DNA methylation biomarkers for MetS.
Several studies showed an association of circulating DNA methylation biomarkers from either peripheral blood mononuclear cells (PBMCs), peripheral blood leukocytes (PBLs), or buffy coats with MetS [10–13]. These studies identified a number of DNA methylation biomarkers, including suppressor of cytokine signaling 3 (SOCS3) [11], ATP-binding cassette sub-family G member 1 (ABCG1) [10], Paternally Expressed 3 (PEG) [13], and Fatty Acid Binding Protein 3 (FABP3) [12]. Importantly, they linked the DNA methylation biomarker with the gene expression data. However, these aforementioned studies focused on Europeans and African Americans. Therefore, the role of whole blood DNA methylation biomarkers in MetS participants of Latino descent is less known and warrants investigation.
The present study aimed to measure whole blood DNA methylation in non-MetS versus MetS participants of Latino descent in the AIR registry. Specifically, we measured blood DNA methylation in three candidate genes: ATP Synthase, H+ Transporting mitochondrial F1 Complex, Epsilon Subunit (ATP5E), Cytochrome C Oxidase Subunit VIc (COX6C), and Ribosomal Protein L9 (RPL9). We focused on ATP5E, COX6C, and RPL9 because they were three of the top changing mRNAs (>1.5-fold change) in our previously published gene expression study of MetS participants in the AIR registry [9]. Moreover, these differentially expressed genes (ATP5E, COX6C, and RPL9) are part of the oxidative phosphorylation and ribosome pathways. We previously showed normalization of skeletal muscle protein levels for several ribosomal proteins in obese patients before and after bariatric surgery [14]. Moreover, we showed decreased mRNA expression for oxidative phosphorylation genes in skeletal muscle tissue of individuals with MetS characteristics [15, 16]. Therefore, these oxidative phosphorylation and ribosome genes/pathways are of interest to understand metabolic syndrome mechanisms.
For the present study, we measured promoter DNA methylation of ATP5E, COX6C, and RPL9. We focused on DNA methylation since this epigenetic modification can affect how genes are expressed [17–19]. We hypothesized that methylation would be lower in the promoters of ATP5E, COX6C, and RPL9 in the MetS participants. Therefore, we hypothesized that there would be an inverse relationship between the previously published mRNA expression data from the AIR registry [9] with the methylation data for the genes of interest.
Materials and methods
Participants
All participants were of Latino descent from the Arizona Insulin Resistance (AIR) registry. Shaibi et al. previously described the goals of the biorepository [20]. Briefly, 667 participants took part in the AIR registry to establish a biobank for future investigation into diabetes and obesity-related health conditions in the Latino community. We selected a subset of the 667 participants for this present study (n = 184). We studied the same 184 participants that we had published global gene expression data for [9], in part, to look for an association with the methylation data. The 184 participants were selected randomly from the 667 participants. The only criteria used were age (≥18 years old). We realize that we may have inadvertently introduced a selection bias by not studying all the participants in the registry but a random subset aged 18 years or more.
We defined MetS as per the National Cholesterol Education Program Adult Treatment Panel III (NCEP: ATP III) [4]. Participants consented to the collection of anthropometric, demographic, family history, and medical history information. Measurements for weight (kg), body mass index (BMI) (kg/m2), waist circumference (cm) and seated blood pressure (mmHg) were collected. Following a 12-hour fast, participants agreed to blood samples to assess various metabolic phenotypes, including glucose, triglycerides, and high-density lipoprotein.
The Institutional Review Board (IRB) at Arizona State University approved the initial AIR registry study under protocol #0804002873. We obtained written consent from all participants. For the minors recruited into the AIR registry [although not studied as part of this project], we obtained written consent from their parents or guardians. Written consent was obtained to bank serum, DNA, and RNA and to use de-identified data and biospecimens for future studies, like the one described herein. The University of Arizona approved the present study under protocol #1703255156. The present study was considered exempt by the ethics committee at the University of Arizona since it utilized de-identified information of previously consented banked samples. Therefore, we made no recontact with these participants.
Isolation of genomic DNA
Whole blood samples from the AIR registry participants were stored in PAXgene Blood Collection DNA Tubes (Qiagen Inc, Germantown, MD, USA) at -80°C until processed. We performed DNA extractions using the PAXgene Blood DNA Processing kit (Qiagen Inc, Germantown, MD, USA), as per the manufacturer’s instructions. The quality and quantity of the DNA were measured using a NanoDrop 2000 Spectrophotometer (ThermoFisher, Waltham, Massachusetts). The average DNA absorbance ratio (A260/A280) was 1.85 ± 0.05.
Bisulfite conversion
As per the manufacturer’s instructions, the isolated genomic DNA was treated with bisulfite conversion using the EZ DNA Methylation-Lightning Kit (ZYMO, Irvine, CA, USA). The quantity of the bisulfite converted DNA was determined using the NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Primer selection
Pyrosequencing primer sets were pre-designed and readily available for our candidate genes (ATP5E, COX6C, and RPL9) of interest (Qiagen Inc, Germantown, MD, USA). S1 Table provides the Qiagen catalog number, the target sequence to analyze, and the chromosomal location. The pre-designed primer sets were within the promoter of the gene of interest and were near the gene’s transcriptional start site. In addition, we selected these primer sets as the CpG coverage was between 4 to 6 per gene, which allowed us to perform CpG individual site methylation analysis and averaged analysis across the promoter of each gene. Lastly, we selected these pyrosequencing primer sets as computational PROMO analysis [21] revealed enrichment for potential transcription factors (S2 Table) that may bind in the target sequence of the candidate genes.
PCR amplification and pyrosequencing
We added the bisulfite converted DNA (10 ng/μL) to the Qiagen PyroMark CpG assay PCR primer set along with the ZymoTaq Premix (ZYMO, Irvine, CA, USA). Samples were PCR amplified in a Veriti® 96-Well Fast Thermal Cycler. We verified the quality of the PCR product with DNA Gel Electrophoresis using a Thermo Fisher Low Mass Ladder (ThermoFisher, Waltham, MA, USA). The amplified DNA was pyrosequenced on the Q48 Autoprep using the associated sequencing primer set from the PyroMark CpG assay kit (Qiagen Inc, Germantown, MD, USA), as per the manufacturer’s instructions. The PyroMark Q48 Advanced Software (Qiagen Inc, Germantown, MD, USA) calculates the percent of methylation. Specifically, it calculates methylation percentage from the ratio of the heights of a cytosine peak (methylated signal) and the sum of cytosine and thymine peaks (methylated and unmethylated signal) for each cytosine at the CpG site [22]. Methylated and unmethylated bisulfite converted DNA served as controls (Qiagen Inc, Germantown, MD, USA). In addition, we included non-template controls. Samples were performed in duplicate to confirm the reproducibility of methylation data.
Statistical analysis
Statistical analyses were performed using STATA 14 (StataCorp, College Station, USA). Participant characteristic data was expressed as mean ± SEM, and categorical data were expressed as counts. The participant characteristic data was organized by MetS status, and statistical comparisons across the groups were performed using an independent sample t-test. The Shapiro-Wilk test was used to test for data normality. The pyrosequencing methylation data were not normally distributed. Therefore, the nonparametric Mann-Whitney U test was utilized. Categorical variables were analyzed using Pearson’s chi-square test. In addition, associations between MetS and DNA methylation were analyzed using logistic regression while controlling for age, gender, and smoking status in the model. The MetS group was the dependent variable with averaged gene methylation, age, gender (females used as the reference), and smoking status (nonsmokers used as the reference) as independent variables in the logistic regression analysis. Finally, Spearman’s correlation was performed to measure the strength and direction of the association between the methylation data and the previously published gene expression data. We used Partial Spearman’s Rank Correlation to adjust for age, gender, and smoking status for the methylation and gene expression correlation. Significance was set at p < 0.05 for all tests.
Results
Participants
Of the 184 participants in the present study, 74 met the criteria for MetS. The remaining 110 participants classified as non-MetS. As expected, MetS participants had significantly higher blood pressure measures, waist circumference, triglycerides and fasting plasma glucose. Moreover, the MetS group showed lower high-density lipoprotein levels (Table 1). However, there was no significant gender or smoking status difference between the non-MetS versus MetS (Table 1).
Table 1. Phenotypes from non-MetS versus MetS participants in the Arizona Insulin Resistance (AIR) registry.
| Phenotype | Non-MetS | MetS | P Value |
|---|---|---|---|
| Gender | 36M/74F | 25M/49F | NS |
| Age | 35 ± 1 | 39 ± 1 | <0.01 |
| Systolic Blood Pressure (mmHg) | 114.7 ± 1.1 | 128.4 ± 2.4 | <0.0001 |
| Diastolic Blood Pressure (mmHg) | 75.3 ± 0.8 | 81.5 ± 1.2 | <0.0001 |
| Waist Circumference (cm) | 95.1 ± 1.2 | 109.1 ± 1.7 | <0.0001 |
| Triglycerides (mg/dL) | 107.7 ± 4.3 | 204.7 ± 12.2 | <0.0001 |
| Fasting Plasma Glucose (mg/dL) | 92.5 ± 1.8 | 112.8 ± 4.8 | <0.0001 |
| High Density Lipoprotein (mg/dL) | 47.8 ± 1.1 | 37.6 ± 1.0 | <0.0001 |
| Smoking status | 91 No/18 Yes/1 UNK | 63 No/11 Yes | NS |
Data are means ± SEM. UNK = unknown.
Differential DNA methylation between MetS and non-MetS groups
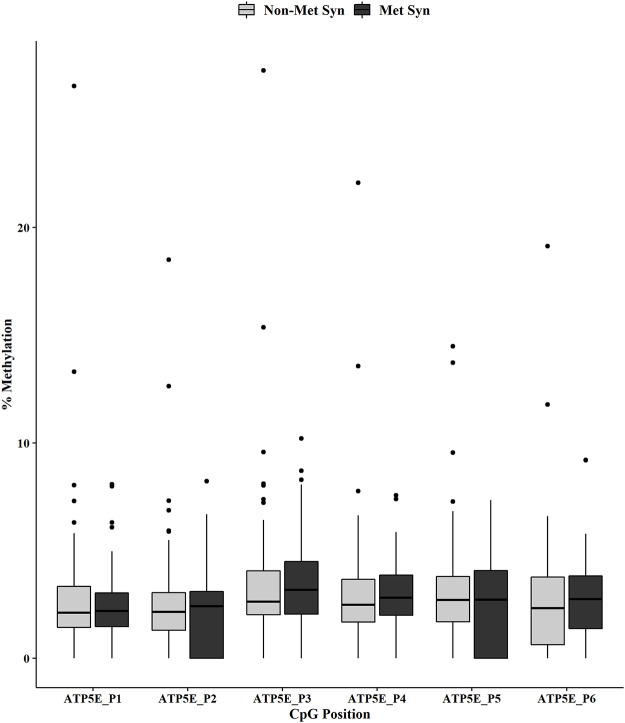
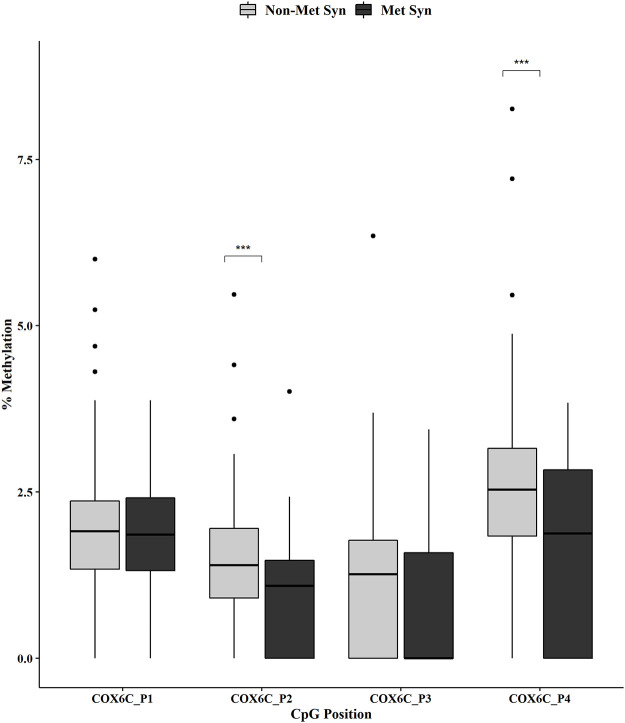
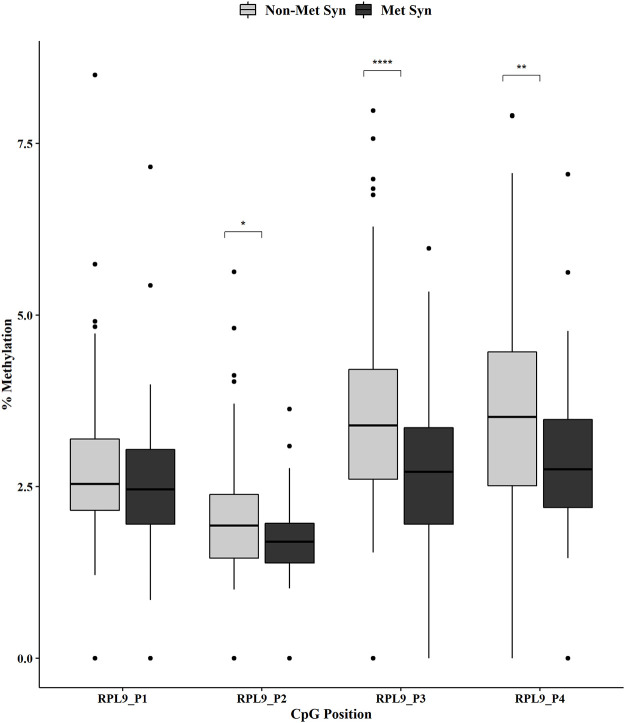
Individual CpG methylation positions within each gene were analyzed by MetS status using the Mann-Whitney U test. We showed no change in methylation for the ATP5E CpG positions across the non-MetS versus MetS groups (Fig 1). However, we did observe a significant decrease in methylation for COX6C at CpG positions 2 and 4 in the MetS group (Fig 2, both p < 0.001). In addition, the RPL9 methylation analysis revealed a significant decreased methylation for positions 2 (p < 0.05), 3 (p < 0.0001) and 4 (p < 0.01) in the MetS group (Fig 3).
Fig 1. ATP5E promoter DNA methylation by MetS status.
DNA methylation at six CpGs within the promoter of ATP5E was measured in non-MetS (n = 110) and MetS (n = 74). Mann-Whitney U test was used for the statistical comparisons between the non-MetS versus MetS.
Fig 2. COX6C promoter DNA methylation by MetS status.
DNA methylation at four CpGs within the promoter of COX6C was measured in non-MetS (n = 110) and MetS (n = 74). Mann-Whitney U test was used for the statistical comparisons between the non-MetS versus MetS. ***p < 0.001.
Fig 3. RPL9 promoter DNA methylation by MetS status.
DNA methylation at four CpGs within the promoter of RPL9 was measured in non-MetS (n = 110) and MetS (n = 74). Mann-Whitney U test was used for the statistical comparisons between the non-MetS versus MetS. *p < 0.05, **p < 0.01, ****p < 0.0001.
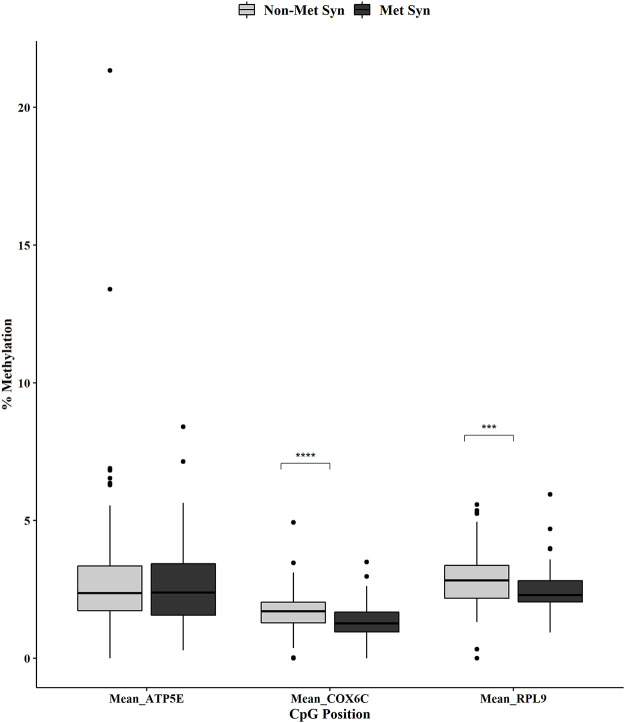
We used the Mann-Whitney U test to determine mean methylation differences when averaging across the CpG positions for ATP5E, COX6C, and RPL9. The averaged methylation within the promoter of ATP5E was not significantly different between the non-MetS and MetS groups (Fig 4). However, the averaged methylation across the promoters for COX6C and RPL9 were both significantly decreased (p < 0.0001 and p < 0.001, respectively, Fig 4).
Fig 4. ATP5E, COX6C and RPL9 overall promoter DNA methylation by MetS status.
Mean methylation within the promoters of ATP5E, COX6C and RPL9 in non-MetS (n = 110) and MetS (n = 74). Mann-Whitney U test was used for the statistical comparisons between the non-MetS versus MetS. ***p < 0.001, ****p < 0.0001.
Association of promoter methylation with MetS status
Logistic regression analysis assessed whether averaged promoter methylation for ATP5E, COX6C, and RPL9 was associated with MetS when controlling for potential confounding factors such as age, gender, and smoking status. As expected, we observed no association for the mean ATP5E methylation with MetS. However, mean methylation for the promoter of COX6C was significantly associated with MetS when controlling for age, gender, and smoking status [the adjusted odds ratio: 0.36, 95% confidence interval: 0.21–0.60], p < 0.001. Similarly, the mean methylation for the promoter of RPL9 was significantly associated with MetS when controlling for age, gender, and smoking status [the adjusted odds ratio: 0.56, 95% confidence interval: 0.38–0.82], p < 0.01.
Correlation analysis of gene expression data with methylation data
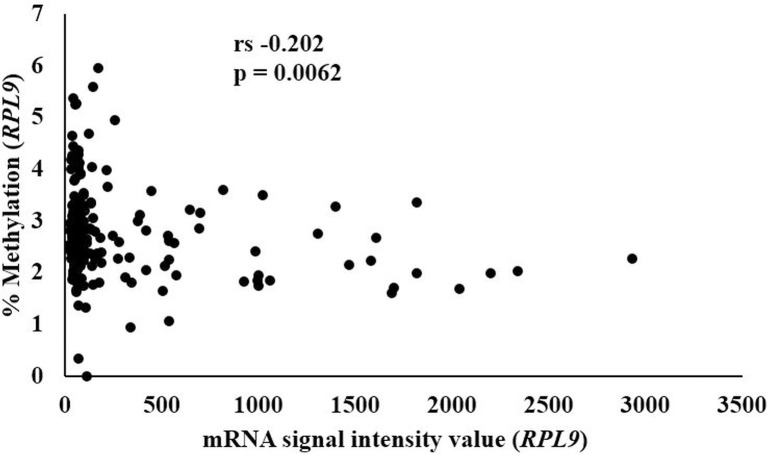
We performed a Spearman’s correlation between the previously published gene expression data [9] with the averaged methylation for ATP5E, COX6C, and RPL9. ATP5E gene expression was not correlated to the mean methylation as demonstrated with a Rho (rs) value of 0.117, p = NS. Likewise, the gene expression data for COX6C did not correlate to the mean methylation (rs -0.088, p = NS). However, the averaged methylation for RPL9 showed a significant inverse correlation with the previously published gene expression data (rs -0.202, p = 0.0062, Fig 5). When we adjusted for age, sex, and smoking status using the Partial Spearman’s Rank Correlation, the significance was p = 0.0030.
Fig 5. Correlation analysis between RPL9 mRNA and RPL9 methylation.
Spearman’s correlation was used to calculate the strength and direction of the relationship between the RPL9 mRNA and RPL9 methylation data. Rho (rs) value of rs -0.202, p = 0.0062.
Discussion
This study set out to identify potential DNA methylation biomarkers in whole blood samples from participants of Latino descent from the AIR registry. In particular, we focused on select genes (ATP5E, COX6C, and RPL9) in non-MetS versus MetS participants. We focused on these genes since we previously showed altered gene expression of ATP5E, COX6C, and RPL9 in MetS participants from the AIR registry [9]. In that study, we demonstrated an increased gene expression for ATP5E, COX6C, and RPL9 in the MetS group [9]. In the present study, we showed a decreased methylation of COX6C and RPL9 in participants with MetS while controlling for potential confounding variables such as age, gender, and smoking status. We found no association of ATP5E DNA methylation with MetS in the AIR registry samples. Our findings provide evidence for reduced methylation of cytosines in the promoter region of our select candidate genes, specifically COX6C and RPL9. Taken collectively, COX6C and RPL9 may be useful as potential blood-based DNA methylation biomarkers for MetS.
ATP Synthase, H+ Transporting mitochondrial F1 Complex, Epsilon Subunit (ATP5E) was the first gene-targeted in this blood-based DNA methylation biomarker study. ATP5E is a subunit of the mitochondrial ATP synthase and is essential for oxidative phosphorylation. We previously showed a collection of whole blood oxidative phosphorylation genes that were increased in the MetS participants [9]. Furthermore, in type 2 diabetic mouse liver models, the oxidative phosphorylation gene ATP5E was upregulated in the diabetic livers compared to control livers [23]. Based on these aforementioned studies, we pursued this gene as a blood-based DNA methylation biomarker. We showed no difference in methylation between the non-MetS group versus the MetS group in the ATP5E gene. In this study, we focused on 6 CpG sites within the promoter of ATP5E. Based on these findings, we suggest that we did not target enough CpG sites within the ATP5E. Moreover, the target sequence we measured was likely not important for gene regulation.
Another oxidative phosphorylation gene that we targeted was cytochrome C oxidase subunit 6c (COX6C). COX6C is a subunit of the Complex IV protein structure in the mitochondrial respiratory chain [24], and it regulates the formation or activity of Complex IV in the electron transport chain [25]. Many studies have implicated a role for COX6C in metabolic diseases. For example, a microarray study of the aorta and adipose tissue showed a decreased expression for COX6C in insulin-resistant and atherosclerotic obese mouse models [26]. Moreover, an analysis of type 2 diabetic Goto-Kakizaki mice revealed a differential gene expression of COX6C in liver samples [27]. The present study’s findings supported our overall hypothesis, wherein we observed a decreased methylation in COX6C in whole blood in the AIR registry MetS participants. However, we found no significant correlation when we performed Spearman’s correlation between the methylation data with the previously published gene expression data [9]. The lack of association with the mRNA data suggests that the methylation sites we measured were not important for gene regulation.
The human ribosomal protein L9 (RPL9) gene codes for a subunit of the 60S subunit or the large ribosomal subunit [28]. Ribosomal proteins, like RPL9, serve to stabilize specific rRNAs by binding directly and assist in ensuring correct structural alterations to rRNA during ribosome assembly [29]. We chose to study the RPL9 gene for multiple reasons. Our laboratory showed a role for ribosomal proteins in obese patients before and after bariatric surgery [14]. In addition, there have been many studies implicating ribosomal genes/proteins in metabolic diseases. For example, a transcriptomic analysis of samples from non-steatotic and steatotic livers of severely obese adolescents showed a significant downregulation for RPL9 in the steatotic population [30]. In addition, a study in liver mitochondrial fractions showed an increase of >1.5-fold in RPL9 in those mice fed a western diet versus a standard diet [31]. In the present study, we showed significant changes in DNA methylation in the MetS participants. In particular, we showed that the methylation was decreased significantly in 3 out of 4 of the CpG sites captured.
We hypothesized that there would be a significant inverse relationship between the previously published gene expression data [9] with the methylation data for the genes of interest. Spearman’s correlation showed that averaged methylation across the RPL9 promoter was significantly associated with the previously published gene expression data. The RPL9 findings support our hypothesis that the reduction of methylation in the promoter region would correlate with the previously observed increased RPL9 gene expression data [9]. Although we did not directly measure it, we can hypothesize there is an increase in the cellular content of ribosomes in the blood of MetS participants. The increase in cellular content of ribosomes may increase protein synthesis to account for the increased energy demands observed in MetS participants [32]. A study by Hoppe et al. showed a direct correlation between energy demands and ribosomal RNA synthesis [33]. Their findings suggest that ribosomal RNA synthesis increases when mitogenic conditions are optimal, for example, an overabundance of available nutrients. Taken together, RPL9 is a promising candidate gene, and future studies are warranted for understanding the exact role of ribosomal proteins in whole blood and skeletal muscle in metabolic diseases.
Some limitations apply to this study that warrants a discussion in the context of our findings. Firstly, we did not experimentally measure whether the RPL9 methylation sites directly alter gene expression. Future studies are warranted to address this wherein we would perform an in vitro methylation luciferase assay to measure the effect of methylation in the promoters of the select genes (ATP5E, COX6C, and RPL9) on gene expression levels [34]. Secondly, another limitation is the target sequences presented in this study captured only 4–6 CpG sites for each gene studied. In comparison, another study analyzed over 20 CpG sites for one target gene [12]. Measuring more CpGs would provide more insight into the epigenetic landscape of the promoter of these target genes on the MetS phenotype. A future study would be to design additional CpG assays covering the promoter region for each gene. Thirdly, our laboratory studies metabolic disease in whole blood and skeletal muscle, and we have previously shown contradictory findings between these two tissues [9]. This study was unable to measure skeletal muscle methylation of these candidate genes. A future study would be to perform methylation analysis of ATP5E, COX6C, and RPL9 in skeletal muscle of MetS and non-MetS participants. Fourthly, we did not validate our findings in an independent cohort nor other populations. Validating these DNA methylation biomarkers in larger studies would be critical for determining if they would be suitable to function as biomarkers, and ultimately be utilized in clinical practice for screening and diagnosing MetS. Lastly, another limitation of this study is the random selection of the 184 participants used for the previously published gene expression study [9] and the present study. The age criteria may have introduced a bias into the analysis.
In conclusion, we showed decreased whole blood DNA methylation for COX6C and RPL9 in participants with MetS. Moreover, we provide evidence of a significant inverse relationship between the gene expression and methylation data for RPL9. Our findings contribute to the field of whole blood-based DNA methylation biomarkers [10–13], specifically highlighting MetS biomarkers in Latino participants. Our novel potential epigenetic biomarkers should be measured in other populations and importantly validated in larger cohorts.
Supporting information
(PDF)
(PDF)
Acknowledgments
We thank the volunteers of the AIR registry and are grateful for their participation and cooperation. In addition, we thank the research coordinators and staff who facilitated recruitment and study in the AIR registry. We thank Dr. Lawrence J. Mandarino for access to the Arizona Insulin Resistance registry.
Data Availability
All relevant data are within the manuscript and its Supporting information files.
Funding Statement
This study was supported, in part by the National Institutes of Health Grants R01DK094013. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Dawn K. Coletta (DKC) received salary support from the National Institutes of Health Grants R01DK094013.
References
- 1.Samson SL, Garber AJ. Metabolic syndrome. Endocrinol Metab Clin North Am. 2014;43(1):1–23. Epub 2014/03/04. doi: 10.1016/j.ecl.2013.09.009 . [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, Cui X, Li F, Wang S, Liu X, Hui L, et al. Association between diabetes mellitus with metabolic syndrome and diabetic microangiopathy. Exp Ther Med. 2014;8(6):1867–73. Epub 2014/11/06. doi: 10.3892/etm.2014.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L, Miura K, Fujiyoshi A, Kadota A, Miyagawa N, Nakamura Y, et al. Impact of metabolic syndrome on the risk of cardiovascular disease mortality in the United States and in Japan. Am J Cardiol. 2014;113(1):84–9. Epub 2013/10/31. doi: 10.1016/j.amjcard.2013.08.042 . [DOI] [PubMed] [Google Scholar]
- 4.National Cholesterol Education Program Expert Panel on Detection E, Treatment of High Blood Cholesterol in A. Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106(25):3143–421. Epub 2002/12/18. . [PubMed] [Google Scholar]
- 5.Moore JX, Chaudhary N, Akinyemiju T. Metabolic Syndrome Prevalence by Race/Ethnicity and Sex in the United States, National Health and Nutrition Examination Survey, 1988–2012. Prev Chronic Dis. 2017;14:E24. Epub 2017/03/17. doi: 10.5888/pcd14.160287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rochlani Y, Pothineni NV, Kovelamudi S, Mehta JL. Metabolic syndrome: pathophysiology, management, and modulation by natural compounds. Ther Adv Cardiovasc Dis. 2017;11(8):215–25. Epub 2017/06/24. doi: 10.1177/1753944717711379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srikanthan K, Feyh A, Visweshwar H, Shapiro JI, Sodhi K. Systematic Review of Metabolic Syndrome Biomarkers: A Panel for Early Detection, Management, and Risk Stratification in the West Virginian Population. Int J Med Sci. 2016;13(1):25–38. Epub 2016/01/28. doi: 10.7150/ijms.13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Neill S, Bohl M, Gregersen S, Hermansen K, O’Driscoll L. Blood-Based Biomarkers for Metabolic Syndrome. Trends Endocrinol Metab. 2016;27(6):363–74. Epub 2016/05/07. doi: 10.1016/j.tem.2016.03.012 . [DOI] [PubMed] [Google Scholar]
- 9.Tangen SE, Tsinajinnie D, Nunez M, Shaibi GQ, Mandarino LJ, Coletta DK. Whole blood gene expression profiles in insulin resistant Latinos with the metabolic syndrome. PLoS One. 2013;8(12):e84002. Epub 2013/12/21. doi: 10.1371/journal.pone.0084002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akinyemiju T, Do AN, Patki A, Aslibekyan S, Zhi D, Hidalgo B, et al. Epigenome-wide association study of metabolic syndrome in African-American adults. Clin Epigenetics. 2018;10:49. Epub 2018/04/13. doi: 10.1186/s13148-018-0483-2 neutral with regard to jurisdictional claims in published maps and institutional affiliations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali O, Cerjak D, Kent JW Jr., James R, Blangero J, Carless MA, et al. Methylation of SOCS3 is inversely associated with metabolic syndrome in an epigenome-wide association study of obesity. Epigenetics. 2016;11(9):699–707. Epub 2016/08/27. doi: 10.1080/15592294.2016.1216284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Kent JW 2nd, Lee A, Cerjak D, Ali O, Diasio R, et al. Fatty acid binding protein 3 (fabp3) is associated with insulin, lipids and cardiovascular phenotypes of the metabolic syndrome through epigenetic modifications in a Northern European family population. BMC Med Genomics. 2013;6:9. Epub 2013/03/21. doi: 10.1186/1755-8794-6-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Otterdijk SD, Binder AM, Szarc Vel Szic K, Schwald J, Michels KB. DNA methylation of candidate genes in peripheral blood from patients with type 2 diabetes or the metabolic syndrome. PLoS One. 2017;12(7):e0180955. Epub 2017/07/21. doi: 10.1371/journal.pone.0180955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell LE, Langlais PR, Day SE, Coletta RL, Benjamin TR, De Filippis EA, et al. Identification of Novel Changes in Human Skeletal Muscle Proteome After Roux-en-Y Gastric Bypass Surgery. Diabetes. 2016;65(9):2724–31. Epub 2016/05/22. doi: 10.2337/db16-0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coletta DK, Mandarino LJ. Mitochondrial dysfunction and insulin resistance from the outside in: extracellular matrix, the cytoskeleton, and mitochondria. Am J Physiol Endocrinol Metab. 2011;301(5):E749–55. Epub 2011/08/25. doi: 10.1152/ajpendo.00363.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100(14):8466–71. Epub 2003/07/02. doi: 10.1073/pnas.1032913100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noble D. Conrad Waddington and the origin of epigenetics. J Exp Biol. 2015;218(Pt 6):816–8. Epub 2015/03/20. doi: 10.1242/jeb.120071 . [DOI] [PubMed] [Google Scholar]
- 18.Russo VEA, Martienssen RA, Riggs AD. Epigenetic mechanisms of gene regulation. Plainview, N.Y.: Cold Spring Harbor Laboratory Press; 1996. xii, 692 p. p. [Google Scholar]
- 19.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38. Epub 2012/07/12. doi: 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaibi GQ, Coletta DK, Vital V, Mandarino LJ. The design and conduct of a community-based registry and biorepository: a focus on cardiometabolic health in Latinos. Clin Transl Sci. 2013;6(6):429–34. Epub 2013/10/15. doi: 10.1111/cts.12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18(2):333–4. Epub 2002/02/16. doi: 10.1093/bioinformatics/18.2.333 . [DOI] [PubMed] [Google Scholar]
- 22.Sestakova S, Salek C, Remesova H. DNA Methylation Validation Methods: a Coherent Review with Practical Comparison. Biol Proced Online. 2019;21:19. Epub 2019/10/05. doi: 10.1186/s12575-019-0107-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F, Xu X, Zhang Y, Zhou B, He Z, Zhai Q. Gene expression profile analysis of type 2 diabetic mouse liver. PLoS One. 2013;8(3):e57766. Epub 2013/03/08. doi: 10.1371/journal.pone.0057766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schweppe DK, Chavez JD, Lee CF, Caudal A, Kruse SE, Stuppard R, et al. Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc Natl Acad Sci U S A. 2017;114(7):1732–7. Epub 2017/01/29. doi: 10.1073/pnas.1617220114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pierron D, Wildman DE, Huttemann M, Markondapatnaikuni GC, Aras S, Grossman LI. Cytochrome c oxidase: evolution of control via nuclear subunit addition. Biochim Biophys Acta. 2012;1817(4):590–7. Epub 2011/08/02. doi: 10.1016/j.bbabio.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno-Viedma V, Amor M, Sarabi A, Bilban M, Staffler G, Zeyda M, et al. Common dysregulated pathways in obese adipose tissue and atherosclerosis. Cardiovasc Diabetol. 2016;15(1):120. Epub 2016/08/27. doi: 10.1186/s12933-016-0441-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng WJ, Nie S, Dai J, Wu JR, Zeng R. Proteome, phosphoproteome, and hydroxyproteome of liver mitochondria in diabetic rats at early pathogenic stages. Mol Cell Proteomics. 2010;9(1):100–16. Epub 2009/08/25. doi: 10.1074/mcp.M900020-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazuruk K, Schoen TJ, Chader GJ, Iwata T, Rodriguez IR. Structural organization and chromosomal localization of the human ribosomal protein L9 gene. Biochim Biophys Acta. 1996;1305(3):151–62. Epub 1996/03/01. doi: 10.1016/0167-4781(95)00201-4 . [DOI] [PubMed] [Google Scholar]
- 29.Baik IH, Jo GH, Seo D, Ko MJ, Cho CH, Lee MG, et al. Knockdown of RPL9 expression inhibits colorectal carcinoma growth via the inactivation of Id-1/NF-kappaB signaling axis. Int J Oncol. 2016;49(5):1953–62. Epub 2016/10/26. doi: 10.3892/ijo.2016.3688 . [DOI] [PubMed] [Google Scholar]
- 30.Sheldon RD, Kanosky KM, Wells KD, Miles L, Perfield JW 2nd, Xanthakos S, et al. Transcriptomic differences in intra-abdominal adipose tissue in extremely obese adolescents with different stages of NAFLD. Physiol Genomics. 2016;48(12):897–911. Epub 2016/10/21. doi: 10.1152/physiolgenomics.00020.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Einer C, Hohenester S, Wimmer R, Wottke L, Artmann R, Schulz S, et al. Data on chow, liver tissue and mitochondrial fatty acid compositions as well as mitochondrial proteome changes after feeding mice a western diet for 6–24 weeks. Data Brief. 2017;15:163–9. Epub 2017/10/17. doi: 10.1016/j.dib.2017.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh S, Dent R, Harper ME, Gorman SA, Stuart JS, McPherson R. Gene expression profiling in whole blood identifies distinct biological pathways associated with obesity. BMC Med Genomics. 2010;3:56. Epub 2010/12/03. doi: 10.1186/1755-8794-3-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoppe S, Bierhoff H, Cado I, Weber A, Tiebe M, Grummt I, et al. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc Natl Acad Sci U S A. 2009;106(42):17781–6. Epub 2009/10/10. doi: 10.1073/pnas.0909873106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han W, Shi M, Spivack SD. Site-specific methylated reporter constructs for functional analysis of DNA methylation. Epigenetics. 2013;8(11):1176–87. Epub 2013/09/06. doi: 10.4161/epi.26195 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files.