Epitope-imprinted polymers are increasingly recognized as cost-effective artificial receptors for biomacromolecules.
Abstract
Molecular imprinting (MI) has been explored as an increasingly viable tool for molecular recognition in various fields. However, imprinting of biologically relevant molecules like proteins is severely hampered by several problems. Inspired by natural antibodies, the use of epitopes as imprinting templates has been explored to circumvent those limitations, offering lower costs and greater versatility. Here, we review the latest innovations in this technology, as well as different applications where MI polymers (MIPs) have been used to target biomolecules of interest. We discuss the several steps in MI, from the choice of epitope and functional monomers to the different production methods and possible applications. We also critically explore how MIP performance can be assessed by various parameters. Last, we present perspectives on future breakthroughs and advances, offering insights into how MI techniques can be expanded to new fields such as tissue engineering.
INTRODUCTION
Molecular recognition is an essential mechanism in a variety of settings (1, 2). In the biomedical field, antibodies are fundamental tools owing to their high affinity and selectivity toward targeted biomolecules of interest. As such, they have been applied for multiple purposes, from laboratory immunoassays [enzyme-linked immunosorbent assay (ELISA), immunocyto/histochemistry, and Western blot] (3) to targeted therapeutics for neurodegenerative diseases (4) or cancer (5). Yet, their expensive cell culture–based production processes, high lability, and possible immunogenic risk make them less than ideal (6–8). This has led to increased research on alternative binding partners capable of specific recognition, such as peptides or oligonucleotides (9, 10). However, their biological nature similarly raises stability concerns. Furthermore, their discovery and production processes are long and possibly inefficient, sometimes leading to molecules with limited selectivity and affinity (11, 12). Therefore, the development of selective, stable, scalable, and cost-effective alternatives is highly sought (13). To that end, purely synthetic receptors with abiotic affinity for biomolecules based on molecular imprinting (MI) have been proposed for numerous biological applications during the last few decades (14–16).
What is molecular imprinting?
MI refers to the creation of specific recognition sites in polymer networks by cross-linking in the presence of a template molecule, which represents the target to be recognized (17, 18). This process is performed by mixing a solution of one (or several) monomer(s) with the template, thereby forming temporary interactions between the two. Subsequent cross-linking and polymerization, followed by removal of the template, lead to the formation of a polymer structure with embedded complementary cavities for the superstructure of the imprinted molecule. These nanocavities preserve not only the shape and size but also the molecular interactions necessary for the recognition of the target. The resulting molecularly imprinted polymers (MIPs) are thus able to selectively recognize and bind the target via a “lock and key” mechanism similar to those found in biological systems (e.g., antibodies and enzymes).
These biomimetic polymeric networks can be prepared by designing interactions between the building blocks of a biocompatible network and the desired specific ligand and stabilizing these interactions by a three-dimensional (3D) structure. These structures are at the same time flexible enough to allow for diffusion of solvent and ligand into and out of the network. Synthetic networks that can be designed to recognize and bind biologically relevant molecules are of great importance and influence a number of emerging technologies (13). These artificial materials can be used either as unique systems or incorporated into existing technologies that can aid in the removal or delivery of biomolecules, so that the natural profiles of compounds in the body can be restored (19, 20).
Such developments are expected to have a major impact on diseases, such as diabetes and atherosclerosis, which are caused by increased levels of certain compounds in the blood. The monitoring and “on-demand” removal of such detrimental compounds by polymeric systems are highly desirable, and these can be achieved by biomimetic networks (21).
The most important problems to be solved in the design of synthetic recognition-based networks are the following:
1) to obtain reproducible interactions between ligand molecule and network, and an ability to differentiate between the ligand and similar compounds;
2) to create a network compatible with aqueous solutions;
3) to reduce diffusional limitations of ligand into the bulk network; and
4) to establish a platform by which recognition-based polymers can be used as biomimetic systems.
Several inherent advantages have rendered MIPs an object of increasing interest. More than two-thirds of the papers on the subject of MI have been published over the 2010s alone, according to MIPdatabase.com (22) (unfortunately, this database, which constituted a good repository of MIP literature, has not been updated since early 2019). Among the subjects discussed are their affinity and selectivity, allowing them to compete with natural biomolecules like antibodies (14); greater stability over time and in different environments, which allows not only their application in a wider range of conditions unsuitable for biomolecules but also their prolonged storage without refrigeration (23); and comparatively simpler, faster, and cheaper production methods, making them more easily available (24). Another attractive comparative advantage is the possibility of developing receptors for molecules with unknown natural binding partners and even unexplored structures (12, 25). This set of features has led to a growing use of MIPs for purposes such as biosensing (15), separation technology (26), food and environmental decontamination (27), or targeted drug delivery (28).
What are the challenges of imprinting large biomacromolecules?
Creation of wholly synthetic systems capable of recognizing and binding proteins would have significant impact and may lead to the production of sensors, media for affinity chromatography, detoxifying agents, or as biomaterials with molecularly tailored surfaces (29). However, while imprinting small molecules has become standard practice, targeting macromolecules (such as proteins or polysaccharides) has proven more challenging. Proteins have proven exceedingly difficult to imprint, despite the great number of surface-exposed functionalities. The main limitations associated with imprinting biological macromolecules have been thoroughly discussed by Culver and Peppas (16) and Li et al. (30).
Several fundamental differences between these two different types of molecules synergistically contribute to render the initially proposed MI strategies inefficient in the development of synthetic binding partners for large targets. For instance, biological macromolecules exist mostly in highly hydrated environments, such as the cytosol, the extracellular matrix, or the blood and lymph streams, being optimized to function in a strict physiological range of conditions (temperature, pH, ionic strength, etc.). On the contrary, most of the initially proposed MI protocols used organic solvents, in which proteins are generally insoluble or unstable (30, 31). Besides, these protocols required operation in extreme polymerization conditions that led to denaturation of the template. This meant that cavities formed in MIPs did not correspond to the native structure of the template, either in shape or in spatial organization of functional groups.
In the first notable example of protein imprinting, Hjertén and coworkers (32, 33) exploited protocols based on water-soluble monomers for applications in the field of affinity separation of proteins. These methods used (meth)acrylates and (meth)acrylamides and took place in mild aqueous conditions compatible with protein native structures. The resulting desirable MIPs are then behaving as hydrogels, exhibiting the corresponding swelling behavior (16, 34). One of the major problems identified in these systems is the low diffusivity of macromolecules through bulk polymer networks, due to their large size (35).
As discussed further ahead, earlier imprinting methods resulted in the formation of binding sites distributed throughout the bulk of the material. To retain effective binding sites, densely cross-linked materials need to be produced. Thus, mesh sizes are often smaller than protein diameters, which greatly hinders rebinding, since protein access to imprinted sites in the polymer bulk is very difficult. Moreover, the same problem affects the removal of the template after polymerization. This leads to a need for harsh washing conditions to achieve high efficiency, possibly compromising the quality of imprinted sites during this process (30).
This limitation has been, to a certain extent, mitigated with the development of surface imprinting protocols, in which MI occurs only at the surface of thin polymer layers or particles, instead of in bulk (36). This not only allows free template diffusion in solution but also potentially reduces the amounts needed for the imprinting process. Nonetheless, bulk imprinting can still be a useful strategy for some applications (37), although the drawbacks associated with the applicability of MIPs for large biomolecules remain in these cases.
In addition, the strong hydrogen bonding nature of water often disrupts the interactions between template and monomer, a fact well documented in MI literature, which decreases imprinting efficiency (31, 38, 39). This is especially challenging when the target molecule includes a high number of hydroxyl and other strongly hydrophilic groups (e.g., oligosaccharides in glycoproteins), which are strongly solvated by water. Binding these molecules thus requires paying a significant desolvation penalty, rendering the process thermodynamically unfavorable. This is reflected in the comparatively low affinity of natural sugar receptors like lectins, commonly in the range of 103 to 104 M−1 (40, 41).
Another inherent characteristic contributing to the challenging imprinting of proteins is their structural complexity (31, 42). On the one hand, the vast number of different chemical functionalities spread over distinct portions of the protein potentially leads to the generation of heterogeneous binding sites (31). On the other hand, the conformational flexibility reinforces this variability, leading to production of inadequate cavities for later recognition (42). In practice, this results in a higher loss of entropy on rebinding, and thus less efficiency in the process (43). Last, a usually overlooked limitation for the large-scale production of MIPs targeting biologically relevant proteins is their prohibitive costs, making them irrational options to be used as molecular templates in most imprinting settings (12, 16, 44).
As discussed below, several attempts have been made at creating MIPs for the recognition of epitopes, peptides, and proteins. Our laboratories have come up with novel methods for the production of materials capable of binding and recognizing specific proteins (44–46). To minimize the diffusive limitation of a material imprinted in the bulk, we have focused on surface adsorption (47–49). Correspondingly, our methods have been designed for the production of micro- or nanoparticles (NPs), to produce materials with large surface-to-volume ratios. In addition, a main design concern was in the use of solvents amenable for protein stability. To these ends, we have developed a surface imprinting technique whereby the template molecule is adsorbed at the interface between two distinct phases. Agitation during polymerization results in the formation of droplets, with subsequent polymerization yielding microparticles or NPs (48).
Epitopes as alternative imprinting templates
First proposed by Rachkov and Minoura in 2000 (43), the epitope approach attempted to solve these issues, particularly those related to molecular complexity and flexibility, and high costs. This approach is analogous to the way antibodies work, by recognizing a small surface-exposed fragment of a larger molecule—the epitope (31, 50). For example, in the case of proteins, epitopes correspond to oligopeptides (37).
The versatility of this approach was quickly noticed by the MI community and different groups started to explore it, for example, for developing imprinting protocols in aqueous media (51) or to enable the oriented immobilization of templates for synthesis of MIPs with surface-confined binding sites (52, 53). However, this approach remained initially restricted to recognition of small peptides (54). Building on these seminal works, by 2005 and 2006, the first surface epitope–imprinted sensors for the identification of full proteins were reported (55, 56). Around the same time, both organic- and inorganic-based polymers imprinted with C-terminal sequences or surface-exposed amino acids of whole proteins were shown to be effective materials for targeting model proteins such as cytochrome c (57) or lysozyme (58). Not long after, the bioactive potential of epitope-imprinted NPs was demonstrated in in vitro (59, 60) and in vivo (61, 62) assays.
By using a smaller compound as template, the structural and cost issues previously associated with larger molecules can be circumvented while retaining selective recognition and affinity for the larger target (Fig. 1). Their smaller size decreases intramolecular flexibility and the number of exposed functional groups, thus contributing to greater homogeneity of imprinted cavities. Furthermore, peptides do not denature like proteins (since they lack tertiary and quaternary structures) and can be synthesized to be soluble in other solvents besides water. Thus, they present greater versatility in terms of conditions of MIP preparation (31).
Fig. 1. Rationale of epitope imprinting concept.
On the other hand, although the smaller size of epitopes minimizes problems associated with diffusion constraints for template removal, when used for bulk imprinting, it does not avoid the fact that rebinding of the corresponding target macromolecule is still limited by polymer mesh size. This makes epitope imprinting a more attractive option for strategies of surface imprinting and/or imprinting of nanomaterials, where binding sites are more easily accessible, as pioneered by Titirici (52, 53).
Smaller templates are also cheaper and easier to produce, since they can be chemically synthesized (36). This allows the imprinting of biologically relevant molecules at reasonable prices that would not be possible with the whole macromolecule. This is especially attractive in an era of increasingly numerous commercial suppliers of synthetic peptides, which facilitate the development and widen the availability of this technology.
The growing interest in MI, particularly for protein recognition, has been discussed by some excellent recent reviews covering this topic (14, 63–66). However, most of these have focused on discussing how this technology has been leveraged by some emerging applications. While generic MIP synthesis procedures are also addressed in these reviews, they focus mostly on the imprinting step per se. The critical earlier processes of reagent and material selection, chemical, and thermodynamic considerations are often overlooked, as are the postimprinting evaluation steps [with the exception of the review by Culver and Peppas (16)]. Moreover, the epitope approach is generally just briefly mentioned or included as a subsection of a larger topic, without particular focus on discussing its specificities. However, with the growing body of literature on epitope imprinting in multiple directions, it is important at this point to perform a thorough discussion and systematization of the most relevant developments in this field over the past decade.
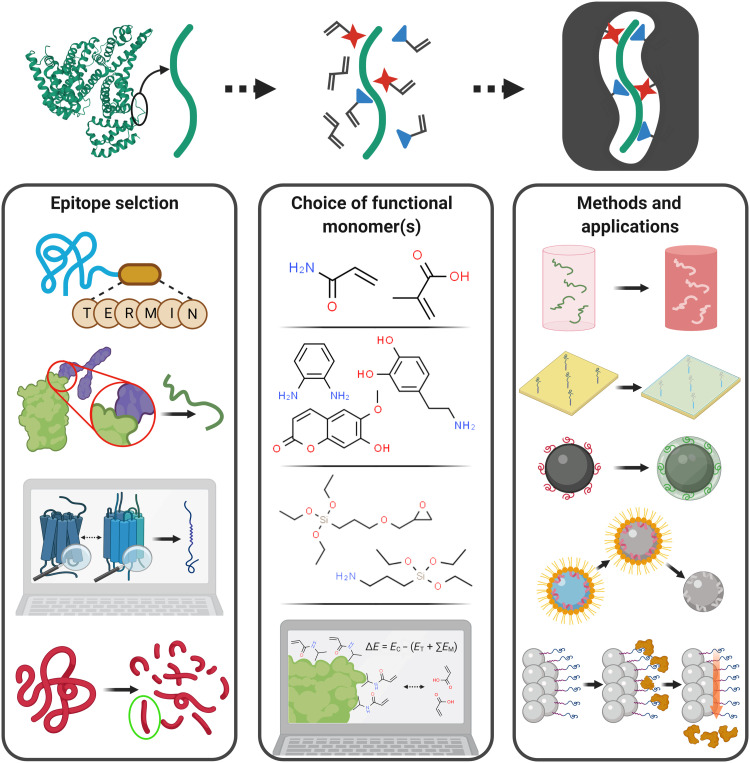
Since its inception, epitope imprinting has been followed for different types of applications, in combination with various synthesis methods (67–73). However, despite its advantages, it also brings new concerns that must be addressed to make it an effectively useful approach. Questions regarding the kind of epitope to use as template, the best functional monomers to couple with it, and which preparation method to follow must all be answered considering the specificities of the molecule (or other entity) to be targeted and the ultimate function it is to fulfil (Fig. 2). Here, we review the latest publications on the subject and discuss the ways in which researchers have addressed these questions.
Fig. 2. Major steps in the process of epitope imprinting.
Each one presents an array of options that must be carefully considered to optimize MIP efficacy considering the target application.
IMPORTANT PARAMETERS FOR ANALYSIS OF MIP PERFORMANCE
Before exploring how the epitope imprinting approach has been applied, it is important to discuss how the recognitive potential of MIPs can be analyzed (66). To be able to evaluate and compare the performance of different MIP formulations, a common set of parameters needs to be established (Fig. 3). The binding of template and target to the MIP may be measured by adsorption experiments. The most common imprinting procedure consists in mixing the prepared MIP in a solution containing the target molecule, incubating for a set amount of time, followed by separation and measuring of the remaining concentration in the supernatant (16). The adsorption capacity Q (usually expressed in milligrams of target per gram of MIP) is then calculated by Eq. 1
| (1) |
where C0 (mg ml−1) is the initial concentration, Ct is the measured concentration after time t, V (ml) is the solution volume, and m (g) is the mass of polymer incubated in it. It is also a common procedure to repeat this test with various incubation times and concentrations, plotting the value of Q according to each variable. The resulting graphs represent, respectively, the adsorption kinetics and the binding isotherms. The value of Q at saturation point (which should be similar in both) is the maximum adsorption capacity (Qmax).
Fig. 3. Parameters commonly used to assess MIP performance.
In addition, the degree to which the imprinting process affects the material binding affinity for the target should be determined. To that end, the same polymer formulation, with (MIP) and without undergoing the imprinting process [nonimprinted polymer (NIP)], should be compared. Thus, the concept of imprinting factor (IF) was created to quantify this effect. IF consists of a ratio between the ability of a given MIP and its NIP counterpart to bind the target molecule. Different authors use different parameters to make this calculation, but the most common one is Q. IF is then calculated as per Eq. 2
| (2) |
with QMIP being a value of Q measured (at any equilibrium concentration of free solute) for the MIP and QNIP being its equivalent value for the corresponding NIP (16). Other works have adapted this concept to the corresponding application. For instance, to evaluate a microextraction monolith, Ji et al. (74) used the ratio of enrichment factors in place of Q, while works on fluorescent sensors take the ratio of Stern-Volmer constants instead (75, 76).
Furthermore, works using solid-phase imprinting methods to produce MI NPs (explored below) usually do not calculate IF values, since suitable NIPs cannot be prepared using this protocol (the affinity separation step cannot be performed in the absence of an immobilized template). As a compromise, NPs imprinted with a different molecule are used as alternative controls and their performances are compared based on kinetic binding constants instead of IF values (73, 77).
The concept of IF has been widely applied to describe the success of an imprinting procedure, since it is practical to measure and simple to understand. However, it has also been questioned, since it is relatively prone to manipulation (78). IF depends exclusively on Q, and this parameter varies differently for MIPs and NIPs, along with the concentration of target molecule. Thus, by choosing the values of Q that yield the highest IF, it is possible to inflate this parameter.
As complementary or alternative methods, binding studies over a significant concentration range have been proposed and are being increasingly adopted. As widely applied for the characterization of antibodies, binding studies are among the most reliable tests to characterize the affinity and specificity of an imprinted material for its target. There are several methods to perform this type of characterization, but most of them typically imply the determination of the equilibrium dissociation constant, KD (24). Some of the techniques and instruments commonly used for these studies are surface plasmon resonance (SPR) and quartz crystal microbalance (QCM). Despite being based on different physical phenomena, both techniques can detect very fine variations of mass at the surface of their sensors. A typical experimental workflow can be summarized as follows. First, either the target molecule or the MIP is immobilized on the sensor surface. Subsequently, the binding partner is injected in solution/suspension at different concentrations. Then, the binding curves recorded by the instrument can be analyzed by different models to calculate either affinity or kinetic parameters. In the first case, the protocol needs to allow binding events to proceed to equilibrium, with the KD being directly calculated. In the latter, association (kon) and dissociation (koff) rate constants are calculated instead. These not only allow the determination of KD by Eq. 3 but also provide information on the velocity and stability of the binding interaction.
| (3) |
There are several models to describe binding/adsorption interactions that can be used to interpret the raw data and determine KD (79). Among the more common equations that can be applied to binding isotherms are the saturation equation for specific binding, and the Langmuir and Freundlich equations. Another method to simplify the calculation of KD is to trace a Scatchard plot from the concentration-response curves. This consists in plotting the response/concentration ratio against the response, to obtain a linear curve, where the slope is −1/KD. An in-depth review of these concepts is presented by Ansell (80).
It is also crucial to assess the selectivity of the material, which is done by comparing its behavior when in contact with the target molecule versus other substances. This can be performed separately (one compound at a time) or in a competitive environment (mixture with multiple macromolecules) (47, 48). From these experiments, it is possible to calculate another parameter, the selectivity coefficient (usually represented by the Greek letter α), relative to each competitor molecule, given by Eq. 4
| (4) |
where QT is the Q value for the target molecule and QC is the equivalent value calculated for a given competitor (16, 80). Competitive experiments can also be performed with varying target-to-competitor ratios, to assess the degree of interference of the latter in the binding process (16, 47). Selection of adequate competitors is also relevant, in this case. Molecules with known similarities to the target should be chosen, so as to test the influence of characteristics such as isoelectric point, size, or structure in the adsorption to the MIP (67). By excluding these factors, it is possible to achieve a better understanding of the effectiveness of MI in generating selectivity in the polymer (47). In the particular case of peptide epitopes, the most common selectivity test consists of challenging MIPs with a “scrambled” version of the template (69). This experimental test ensures that the MIP can specifically distinguish the target amino acid sequence, instead of simply binding any peptide with a similar composition based on charge or length, for example.
Other properties commonly analyzed in MI studies refer to the particular envisioned application. For example, MIPs applied in sample purification can be evaluated for the percentage of recovery of a given substance from a complex mixture. Considering the scope of this review, these usually consist of biological fluids, such as blood (81), plasma (42), serum (75), cerebrospinal fluid (74), or urine (76). This is performed by preparing several dilutions of the mixture, spiking them with a known concentration of the analyte, and incubating them with the MIP. After MIP removal, either the MIP is analyzed to assess the amount of bound analyte or the remaining target in the mix is quantified and subtracted from the initial spiked concentration.
In the case of MI biosensors, parameters such as the linear range and the limit of detection (LOD) are routinely determined to characterize its functioning. The latter can be defined as three times the SD of the blank signal (signal-to-noise ratio of 3) (72). Last, two important properties for both bioseparation and biosensing applications are reusability and reproducibility. The first refers to the ability of the MIP to match the original performance after repeated use, while the latter corresponds to the precision of results between different measurements or batches. Reusability is assessed in terms of either adsorption capacity or recovery percentage retained after a certain number of cycles of use and regeneration (82), while reproducibility is given by the SD between replicates (83).
The parameters discussed here are the ones most commonly assessed in works published in the field. Regrettably, they are often not found together within the same study, precluding suitable comparisons between epitopes and preparation techniques/methods. This may inherently bias the results of a given work and thereby contribute to slowing the progress on the field, as has been previously discussed (78). Hence, the adoption of standard evaluation procedures, based primarily on binding studies and calculation of KD, would represent a significant improvement in epitope imprinting research.
THE FIRST STEP: CHOOSING THE EPITOPE
Since the target molecule is replaced by a smaller template, choosing the best epitope candidate from the structure of its macromolecular source is the first crucial step in epitope imprinting. Figure 4 offers a concise and integrated overview of the available strategies for selection, as will be discussed ahead. So far, these strategies have mainly been applied for the recognition of proteins, since they offer the greatest number of specific biological interactions and effects (e.g., cell surface receptors, growth factors, and circulating proteins in blood). Therefore, the majority of selected epitopes in imprinting protocols consist of oligopeptides, although there is a growing interest for saccharide imprinting in recent years. The imprinting of oligosaccharide epitopes has shown potential as an effective strategy for the recognition of specific glycoproteins (40, 84), as well as aberrant glycosylation sites on cells (85, 86). This approach has opened a new path of research in the field, where significant developments can be expected in coming years.
Fig. 4. Epitope selection strategies.
Peptide epitopes (blue box) are by far the most commonly used type of templates in the field, although emerging strategies using saccharides or small molecules (yellow box) have also been shown to be potential alternatives. Complementary strategies have been increasingly applied for rational peptide selection (red box). Whether they can be applied to aid the selection of other types of epitope remains to be demonstrated. a.a., amino acid.
Terminal protein sequences
Most works on peptide imprinting opt for a linear peptide that corresponds to a small part of the protein sequence (primary structure). Following the strategies proposed by the seminal works on the subject (43, 56), the majority of papers on epitope imprinting use the C-terminal portion of proteins as templates. This is based on the fact that this site is a less frequent target for posttranslational modifications (87–89), thus increasing the chances of direct correspondence between the template peptide and the sequence of the protein in real environments. This choice also presents other important advantages that make it attractive in the synthesis of MIPs: (i) A linear peptide is cheaper to produce and avoids the complexity of secondary and tertiary structures, which, as described above, can decrease imprinting efficiency; (ii) selected sites are frequently exposed at protein surfaces, which is essential for their recognition by MIPs (56, 73); (iii) they can be quickly selected since they can be identified just considering the amino acid sequence and structure of the protein (67, 90); and (iv) they can be easily modified with either a single amino acid [e.g., histidine, terminal cysteine (73, 91)] or a custom-designed peptide sequence (69, 89) to facilitate particular bioconjugation reactions.
N termini have not been as commonly used as C termini (92–96). In works where both are used as templates, recognition results were shown to be superior to the use of a single template, since the corresponding MIP can bind multiple sites of the protein, rendering it more versatile (76, 97). On the other hand, comparing the imprinting performance of N- and C-terminal epitopes of the same protein has yielded mixed results (72, 76, 97). Overall, drawing a one-size-fits-all conclusion and optimal criteria for the selection of protein epitopes is probably impossible since many physicochemical factors are at play in the imprinting process. In a notable example of this multifactorial interplay, Urraca et al. (90) attempted to imprint the C-terminal hexapeptide of β-amyloid, but this sequence presents very poor water solubility, even with an acetylated N terminus, thus constituting a barrier to the process.
MIPs prepared against this epitope in dimethyl sulfoxide (DMSO) demonstrated relatively high binding properties, but with an IF ≈ 1, i.e., no different from the corresponding NIP. To circumvent this limitation, the authors transformed the previous peptide into its tetrabutylammonium salt, which significantly improved its solubility, allowing the imprinting to take place in less competitive solvents. This second series of MIPs proved to be considerably more selective, with several formulations having an IF > 1.
Another important criterion to consider when choosing a linear peptide is how long the sequence should be. Whereas overly short peptides are unable to determine a specific protein, excessively long peptide chains acquire flexibility and can generate intramolecular interactions, leading to structural conformations that may impair the imprinting and rebinding processes (88). According to Nishino et al. (56), over half of the works that used protein epitopes as templates chose nonapeptides, since they represent a near-unique code in unstructured domains for the identification of specific proteins. While this represents the likely minimum number of amino acids that can determine protein specificity, higher numbers should, in principle, lead to a more unique recognition. Nonapeptides have been found to produce MIPs with less specificity (IF) (88) and affinity (KD) (98) when compared to slightly longer peptides. Moreover, there appears to be a closer relationship between the molecular weight of the peptide (instead of length) and the resulting affinity (98). A study verified that MIPs produced with a 9-mer peptide had a KD of 10 nM, whereas those imprinted against a longer but lighter 10-mer peptide had a KD of 20 nM. Furthermore, a heavier weight 14-mer was a slightly better selection than the lighter one with the same length (3 versus 5 nM). Although not definitely proven, the authors argue that heavier peptides may tend to maintain a more stable conformation during the polymerization process, thus creating more uniform imprinted cavities. At the other end of the spectrum, another study analyzed peptides of different lengths and concluded that chains lengthier than 16 amino acids may start to undergo intramolecular interactions, negatively influencing their interaction with the polymers (99).
Broadening the choice
Despite the number of reports on the use of C-terminal linear peptides, most of these focus on the same tried-and-tested model proteins, such as cytochrome c (76, 82, 100, 101) or serum albumins (42, 75, 81, 88, 97, 102, 103), which provide only limited practical applications for this technique. However, to develop MIPs with affinity toward proteins with relevant interest as biomarkers or therapeutic targets (that invariably have prohibitive costs), more rational approaches have to be devised. Recent efforts to widen the spectrum of target proteins have begun to use different strategies for selecting the template. Table 1 systematizes these strategies, providing the rationale behind the choice of each epitope, as well as the target protein, corresponding application, and reported performance.
Table 1. Epitopes chosen for molecular imprinting according to the applied selection criteria, their respective target molecule, application, and reported performance.
NR, not reported.
|
Epitope selection
criteria |
Target molecule | Epitope sequence/molecule | Application | Performance* | Ref. |
| C terminus | Human serum albumin (HSA) |
AASQAALGL | Bioseparation | Q = 46.6 mg g−1 | (42) |
| IF = 4.9 | |||||
| Bioseparation | Q = 103.67 mg g−1 | (81) | |||
| IF = 2.57 | |||||
| Sensing | LOD = 26 ng ml−1 | (102) | |||
| IF = 6.9 | |||||
| KLVAASQAALGL | Sensing | LOD = 44.3 nmol ml−1 | (88) | ||
| IF = 2.65 | |||||
| Bovine serum albumin | VVSTQTALA | Sensing | KD = 0.0254 mg ml−1 | (75) | |
| IF = 4.8 | |||||
| Sensing | LOD = 0,02 ng ml−1 | (103) | |||
| β-Amyloid | GGVVIA (isoform 42) | Bioseparation | IF = 2.6 | (90) | |
| KA = 89 mM−1 | |||||
| Q = 4.7 μmol g−1 | |||||
| MVGGVV (isoform 40) | IF = 5.3 | ||||
| KA = 154 mM−1 | |||||
| Q = 4.8 μmol g−1 | |||||
| Immunoglobulin G (IgG) heavy chain |
QKSLSLSPGK | Bioseparation | Q = 30 μg cm−2 | (106) | |
| Bioseparation | KD = 27.4 nM | (168) | |||
| Insulin | NR | Sensing | LOD = 7.24 fM | (197) | |
| Epidermal growth factor receptor (EGFR) |
SLNITSLGLRSLKEISDG | Sensing | LOD = 3 nM | (167) | |
| Targeted drug delivery | KD = 3.6 nM | (77) | |||
| Human VEGF | IKPHQGQHI | Targeted drug delivery | KD = 1.78 nM | (73) | |
| β2 microglobulin (B2M) | RVNHVTLSQPKIVKW | Targeted drug delivery and bioimaging |
KD = 52 nM | (169) | |
| KIVKWDRDM | Bioseparation | KD = 208 nM | (172) | ||
| IF = 6.5 | |||||
| Myoglobin | NYKELGFQG | Bioseparation | IF = 5.6 | (172) | |
| Cytochrome c (Cyt c) | YLKKATNE | Sensing | LOD = 3.6 ng ml−1 | (222) | |
| IF = 3 | |||||
| AYLKKATNE | Bioseparation | Q = 86.47 mg g−1 | (100) | ||
| IF = 3.48 | |||||
| Q = 780 mg g−1 | (82) | ||||
| IF = 11.7 | |||||
| Q = 67.6 mg g−1 | (101) | ||||
| IF = 4.54 | |||||
| Q = 1.32 mg g−1 | (223) | ||||
| IF = 3.94 | |||||
| Sensing | LOD = 89 nM | (76) | |||
| IF = 4.24 | |||||
| Porcine serum albumin (PSA) |
VIEIRGILA | Bioseparation | Q = 45.06 mg g−1 | (97) | |
| IF = 4.5 | |||||
| N terminus | Folate receptor α | QTRIAWARTELLNVAMNAKH | Targeted drug delivery | KD = 5.72 nM | (94) |
| IF = 6 | |||||
| Cyt c | GDVEKGKKI | Sensing | LOD = 89 nM | (76) | |
| IF = 1.88 | |||||
| PSA | RGVFRRDTY | Bioseparation | Q = 45.06 mg g−1 | (97) | |
| IF = 4.5 | |||||
| Outer membrane lipoprotein |
NQATAKARANLAANLKSTLQKDLENEKTRTVDA | Bacterial binding and eradication |
NR | (92) | |
| EGFR | EEKKVCQGT | Targeted bioimaging | LOD = 0.73 μg ml−1 | (95) | |
| IF = 2.76 | |||||
| CD59 | YNCPNPTADCK | Targeted bioimaging and drug delivery |
IF = 3.5 | (96) | |
| IF = 5.46 | (199) | ||||
| Q = 40.4 mg g−1 | |||||
| Hyaluronan-binding protein 1 (HABP1/ p32) |
LHTDGDKAF | Targeted bioimaging and drug delivery |
IF = 5.1 | (198) | |
| Q = 337.6 mg g−1 | |||||
| Solubility and distinctiveness between homologous proteins |
Regenerating protein 1B (REG1B) |
SCSGFKKWKDESCEKK (p2) | Sensing | KD = 25.17 pg ml−1 | (104) |
| KSWDTGSPSSANAGYCAS (p4) | |||||
| IF = 2.9 (p2) | |||||
| KESSTDDSNVWIG (p6) | |||||
| IF = 2.6 (p4) | |||||
| IF = 3.0 (p6) | |||||
| NEDRETWVDADLY (p1) | Sensing and bioseparation |
1 < IF < 4 | (99) | ||
| KESGTDDFNVWIG (p3) | |||||
| KSWGIGAPSSVNPGYCVS (p5) | |||||
| SSTGFQKWKDVPCEDK (p7) | |||||
| Previously verified immunogenic epitopes or bioinformatics- predicted regions |
Class 3 outer membrane protein allele of N. meningitidis |
KGLVDDADI | Sensing | LOD = 15.71 ng ml−1 | (111) |
| IF = 3.34 | |||||
| fbpA periplasm protein of N. meningitidis |
KPYAKNSVALQAV | Sensing | LOD = 1.39 ng ml−1 | (112) | |
| IF = 12.27 | |||||
| Proline-tRNA ligase protein of M. leprae |
LDIYTTLARDMAAIP | Sensing | LOD = 0.161 nM | (109) | |
| IF = 8.28 | |||||
| HIV-1 glycoprotein 41 (gp41) |
RILAVERYLKDQQLLGIWGCSGKLICTTAVPWNAS | Sensing |
KD = 3.17 nM (for epitope, not protein) |
(108) | |
| CGSWSNKSC | Targeted drug delivery | KD = 85.4 nM | (117) | ||
| Troponin T (TnT) isoform 6 |
MSDIEEVVEE (1–10) | Sensing | LOD = 14.8 nM | (72) | |
| EEAKEAEDGPM (50–60) | |||||
| EQQRIRNEREKERQN (136–150) | |||||
| GKAKVTGRWK (279–288) | |||||
| Anthrax-protective antigen (PA) 83 |
VKKSDEYTF (71–79) | Sensing | KD = 20 pM | (98) | |
| RYDMLNISSLRQDG (659–672) | KD = 10 pM | ||||
| YNDKLPLYISNPNY (681–694) | KD = 20 pM | ||||
| DKLPLYISNPNY (683–694) | KD = 30 pM | ||||
| NGDTSTNGIK (713–722) | KD = 200 pM | ||||
| Neuron-specific enolase (NSE) |
AMRLGAEVYHTL | Sensing | KD = 0.23 nM (Cys) | (91) | |
| IF = 8.8 (Cys) | |||||
| KD = 0.03 nM (His) | |||||
| IF = 11 (His) | |||||
| Human epidermal growth factor receptor 2 (HER2) |
C*PLHNQEKCSKPC*ARV | Targeted drug delivery | NR | (118) | |
| HABP1/p32 | C1NC2KAPETADC1AFVC2FLS | Targeted drug delivery |
NR | (71) | |
| Transforming growth factor–β3 (TGF-β3) |
C1NC2KAPETALC1TNYC2FRN | Tissue engineering | NR | (48) | |
| NSE | LKAVDHINST | Sensing | KD = 53 pM | (113) | |
| CKGVLKAVDHINSTIAPC | Sensing | KD = 26 pM | (89) | ||
| 9 < IF < 10 | |||||
| Transferrin | CGLVPVLAENYNK | Bioseparation | KD = 2.0 nM | (190) | |
| IF = 1.6 | |||||
| Cardiac troponin I | NIDALSGMEGR | Bioseparation | KD = 23 nM | (190) | |
| IF = 1.9 | |||||
| KD = 1 nM | (224) | ||||
| IF = 6 | |||||
| Common sequence in peptide family |
Cholecystokinins | WMDF | Bioseparation |
Q = 0.73 mg g−1 and IF = 24.5 (CCK5) |
(74) |
|
Q = 1.1 mg g−1 and IF = 28.3 (CCK8) | |||||
| Enkephalins | YGGF | Bioseparation |
Q = 1.2 mg g−1 and IF = 32.2 (Met enkephalin) |
(68) | |
|
Q = 0.94 mg g−1 and IF = 33.2 (Leu enkephalin) | |||||
| Angiotensins I and II | DRV | Bioseparation | LOD = 0.07 ng ml−1 and IF = 4.9 (ang I) |
(93) | |
| LOD = 0.06 ng ml−1 and IF = 5.2 (ang II) | |||||
| Amanitins | WC | Bioseparation | IF = 6.5 | (105) | |
| KD = 112.7 μg liter−1 | |||||
| Q = 0.408 mg g−1 | |||||
| Chemical properties (pI, hydropathy, and solubility) |
Atrial natriuretic peptide |
RMDRIGAQSG | Sensing | KD = 20 pM | (107) |
| Brain natriuretic peptide |
FGRKMDRISS | Sensing | KD = 2 pM | (107) | |
| Single amino acid | IgG | l-lysine | Bioseparation | Q = 75.1 mg g−1 | (142) |
| IF = 1.93 | |||||
| Structurally similar small molecule |
Proteins containing phosphorylated tyrosine residues |
PPA | Bioseparation | Q = 7.35 mg g−1 | (154) |
| IF = 2.5 | |||||
| Q = 23 mg g−1 | (153) | ||||
| IF = 1.8 | |||||
| Q = 66 mg g−1 | (156) | ||||
| IF = 3.6 | |||||
| Q = 32 mg g−1 | (70) | ||||
| IF = 4.59 | |||||
| Sensing | LOD = 0.37 μM | (155) | |||
| IF = 2.68 | |||||
| Monosaccharides | Glycosylation sites | N-acetylneuraminic acid, glucuronic acid | Targeted bioimaging | IF = 3.1 (glucuronic acid) |
(157) |
| IF = 2.4 (N- acetylneuraminic acid) | |||||
| Targeted bioimaging |
KD = 141 μM (glucuronic acid) |
(158) | |||
|
KD = 24 μM (N- acetylneuraminic acid) | |||||
| N-acetylneuraminic acid | Targeted bioimaging | KD = 1.7 × 10−4 M | (85) | ||
| Hyaluronic acid (HA) | d-glucuronic acid | Targeted bioimaging | KD = 800 nM | (159) | |
| IF = 3.2 | (86) | ||||
| Monosaccharide and single amino acid |
Telavancin | Mannose-tryptophan | Bioseparation | IF = 3.0 | (160) |
| Q = 6.26 mg g−1 | |||||
| Teicoplanin | Mannose-tryptophan | Bioseparation | KD = 1.865 mg liter−1 | (161) | |
| IF > 2 | |||||
| Digested glycans from glycoprotein surface |
RNase B | Man5GlcNAc2, Man6GlcNAc2 | Bioseparation | KD = 24.89 μM | (139) |
| IF = 8.4 | |||||
| Man7GlcNAc2, Man8GlcNAc2 | |||||
| Transferrin | NR | Bioseparation | IF = 21.8 | (139) | |
| HER2 | NR | Targeted drug delivery bioimaging |
IF = 8.02 | (84) | |
| Erythropoietin | NR | Targeted drug delivery bioimaging |
NR | (84) | |
| Digested peptides from protein surface |
Hemoglobin | VLSPADK, VHLTPEEK (among others not specified) |
Bioseparation | IF < 2 | (67) |
| Pro-gastrin releasing peptide (ProGRP) |
NLLGLIEAK | Bioseparation | KD = 3.4 × 10−6 M | (140) | |
| Q = 59 × 10−3 mol g−1 | |||||
| KD = 7.1 × 10−6 M | (141) | ||||
| Q = 39 × 10−6 mol g−1 |
*Imprinting factor (IF) is given for all works where it is reported. Dissociation constant (KD) is the preferred performance parameter and is provided for all works that report it. When KD is not available, the most adequate parameter reported in each study is provided, namely, limit of detection (LOD) for sensing and imaging applications, and adsorption capacity (Q) for bioseparation.
For example, to detect regenerating protein 1B (REG1B), a biomarker for pancreatic ductal adenocarcinoma, the protein sequence was scanned alongside other proteins of the same family, such as REG3, REG4, and aggrecan (99, 104). Multiple peptides were selected from various portions of the protein (named peptide nos. 1 to 7), considering properties of interest for the specific work, namely, physicochemical characteristics (such as solubility in an adequate solvent) and a sufficient number of different amino acids between the proteins to avoid cross-reactivity. In their first work, despite the best theoretical peptide (no. 2) being coincident with the C-terminal 16 amino acids of REG1B, the authors show that sensors imprinted with peptide no. 4 show a higher accuracy, likely due to its greater exposure at the surface of the protein (104). In the follow-up work, peptide no. 7–imprinted sensors showed the best performance, in this case, thanks to the good solubility provided by the six charged and three polar amino acids in the 13-mer sequence (99).
While the previous works sought to avoid cross-reactivity with proteins of the same family, others have looked for the opposite response. Another epitope selection method that has been successfully applied consists in identifying small sequences common to a target family of peptidic molecules, such as neurotransmitters, hormones, or toxins [e.g., Asp-Arg-Val for angiotensins I and II (93), Trp-Asp-Met-Phe for cholecystokinins (74), Tyr-Gly-Gly-Phe for enkephalins (68), and Trp-Cys for amanitins (105)]. This allows the resulting MIP to bind multiple molecules that share the same amino acid sequence, proving to be versatile tools with unmatched IF values (24.5 to 33.2). The same principle was applied to prepare membranes for monoclonal antibody purification from cell culture supernatants (106). Immunoglobulin G (IgG) molecules are composed of a variable antigen binding (Fab) region and a constant fragment crystallizable (Fc) region. By imprinting the C-terminal decapeptide of the IgG heavy chain (part of the Fc region), the prepared membranes could rebind full IgG molecules regardless of their target antigen, thereby constituting promising versatile tools for commercial production of antibodies.
In another study, seeking to build a platform for detection of natriuretic peptide hormones in blood samples, nonapeptide epitopes were selected on the basis of the similarity of isoelectric point compared to the original hormone and to have a balanced hydrophobic to hydrophilic amino acid ratio (107). This template choice led to impressive KD values between 2 and 20 pM, an LOD in the same range, and little to no reported cross-reactivity. It is thus possible to observe that a more varied and careful epitope selection, considering physicochemical properties, has been shown to elevate the performance of resulting MIPs. This is of particular importance when adding to the imprinting equation the functional monomers that will compose the polymer, as will be discussed ahead.
Approaching biology: Looking for naturally occurring epitopes
Another interesting strategy for rational epitope selection goes back to the root of the epitope approach: immune recognition by antibodies. If the concept of epitope is itself inspired by this interaction, why not follow it to its logical conclusion and use already established immunogenic regions? That is what several studies have tested. In one of these, the authors first looked for known binding sites on the anthrax protective antigen 83, an essential protein for this bacterium to infect mammalian cells (98). Two regions were picked, one identified as responsible for interacting with cell receptors and another that was recognized by antibodies. QCM sensors were then covered with a layer of MIP produced against each one of the templates. Both MIP sensors were found to bind the respective domain, as well as the whole protein, at particularly high affinities (KD in the dozens of picomolar range).
Similarly, a QCM sensor for HIV was imprinted with a sequence corresponding to a major immunodominant region of HIV-1 glycoprotein 41 (gp41; residues 579 to 613), resulting in a KD similar to that of monoclonal antibodies (3.17 nM) and a detection limit for gp41 comparable to ELISA (2 ng ml−1) (108). The same methodology was also followed to prepare MIP-coated QCM sensors for detection of Mycobacterium leprae, attaining an order of magnitude lower LOD (0.161 nM) (109). The epitope selected by the authors had previously been shown to stimulate interferon-γ secretion by T cells from infected patients, thus being identified as a natural antigen (110).
In another study, SPR sensors were simultaneously imprinted with four peptide residues of cardiac troponin T (TnT), along with sensors imprinted with each single epitope (72). These corresponded to the N and C termini of TnT and to two epitopes recognized by commercial antibodies. In this case, only the C terminus–imprinted sensor significantly detected TnT, and its performance was close to the sensor imprinted with the four peptides. Thus, the authors concluded that only that fragment was responsible for the specificity of the MIP. Considering the two epitopes recognized by antibodies, the authors suggest that molecular constraints of the inner portions of the protein could represent an obstacle to properly match the shape produced by linear peptides in the MIP. Instead, for the N-terminal epitope, unfavorable electrostatic interactions between the highly acid peptide and negative dopamine monomers were suggested as responsible for the failure of its MIP. From these studies, it is then possible to conclude that multiple factors, besides the type of epitope per se, affect the imprinting efficiency. These will be discussed in more detail further ahead.
Using previously verified antigenic determinants may also be unfeasible when the target protein has not yet been extensively studied. In these cases, bioinformatics tools can help visualize critical domains and predict candidate epitopes. This approach has been used to develop sensors to target the bacteria Neisseria meningitidis, by targeting exposed sequences of certain surface proteins (111, 112). Nonapeptides from exposed immunogenic loop VII of class 3 outer membrane protein (111) and a 13-mer epitope of iron requisition protein (112) were separately tested as MI templates. Both MIP-coated sensors were successful in detecting the target bacteria in blood samples of patients with brain fever.
In a similar strategy, the x-ray crystallographic structure of neuron-specific enolase (NSE) was analyzed to select a surface-exposed 12-mer sequence as template for this cancer biomarker (91). Recently, molecular dynamic simulations have been performed to predict the most structurally stable epitopes after screening the structure of NSE (89, 113). Sensors imprinted with the best candidate epitope achieved an affinity of 53 pM, significantly higher than similar sensors imprinted with the C-terminal nonapeptide of cytochrome c, for example. Furthermore, when sensors were imprinted with the peptide predicted to be the least stable, their binding to NSE produced signals three to five times lower than sensors imprinted with the most stable peptide (113), demonstrating that this computational analysis can be a very useful tool for refining the epitope search.
Beyond a linear amino acid chain
Another rather recent innovation on epitope design is that of conformational epitopes. As described above, the vast majority of available literature reports the use of linear peptides. Notwithstanding, the prevailing mode of recognition mechanisms in nature is instead based on interactions with secondary and tertiary structures of proteins (particularly for antibody-antigen interactions, the inspiration for epitope MIPs) (71, 114, 115).
To recapitulate these mechanisms, Cenci et al. (116) used a cyclic peptide (CC9ox) mimicking the apical portion of the β-hairpin hormone hepcidin-25 as imprinting template. The selected epitope maintained a stable circular structure thanks to a cysteine bridge between its C- and N-terminal cysteine residues. In this breakthrough work, the authors show that MIP NPs (MINPs) could selectively recognize the full hormone molecule at high affinity (KD = 9 ± 1 nM), while having no measurable interaction with the linear scrambled version of the template peptide. These MINPs could act as “artificial chaperones” by favoring the folding of the linear version of the template peptide into its structured form, opening exciting prospects for future treatment of diseases involving protein misfolding. In a similar approach, MINPs were developed to serve as plastic antibodies against HIV infection (117). The authors used molecular modeling to design a cyclic peptide that contained part of a natural epitope of the target gp41 (SWSNKS), which is usually used to produce antibodies. Although this sequence is smaller than the recommended eight amino acids, KD for the MINP/gp41 interaction was 85.4 nM, while cross-reactivity varied between 0.1% (transferrin) and 1.6% (human serum albumin). The cyclic design of the peptide, mimicking the natural conformation of this epitope, might have played an important part in these results, according to the authors.
A different group devised a refined conformational epitope design workflow to target human epidermal growth factor receptor 2 (HER2), an overexpressed receptor in a significant number of cancer cells (118). Starting with the use of the Immune Epitope Database server (119, 120) to perform epitope mapping, PyMOL (121) and ElliPro software (122, 123) were then applied to design four candidate conformational epitopes. Protein-protein BLAST (BLASTP) (124) was then used to score the best candidate, ensuring the highest correspondence to the target protein HER2 while having the lowest possible cross-reactivity with other unrelated proteins. The produced MINPs (loaded with doxorubicin) were able to significantly increase the drug’s concentration in the tumor, while reducing its off-target distribution in healthy tissues. They also more than doubled cumulative survival of mice in the experimental group, compared to controls.
Another innovative approach was taken by Zhang et al. (71), where the apamin peptide toxin was used as a scaffold to recreate the exposed N-terminal α helix of hyaluronan-binding protein 1 (HABP1/p32, another overexpressed surface receptor in cancer cells), which serves as a specific recognition motif. Following the previously proposed strategy for conformational epitope design (125), the original apamin residues were replaced with topologically equivalent residues from p32 (Fig. 5A). However, the ninth alanine (that exhibits the highest α helix propensity) and two cysteines (for disulfide bond stabilization) were maintained to preserve the peptidic structure. MINPs were able to efficiently target different tumor tissues at different anatomical sites, leading to substantial reduction in their growth.
Fig. 5. Construction of conformational epitopes using apamin as molecular scaffold.
Adapted with permission (48, 71). Copyright 2015 (A) and 2020 (B), Wiley.
The previous strategies have recently been combined by our groups to design a conformational epitope for transforming growth factor–β3 (TGF-β3) (48). The protein structure was scanned for surface-exposed domains that did not interfere in receptor binding using The US Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) (126, 127) and PyMOL software, followed by BLASTP to score their uniqueness. Since the N-terminal α helix portion showed the best results, apamin was used as a molecular scaffold to prepare the epitope, as previously demonstrated (71, 125). The structure of the novel hybrid peptide was predicted and compared to the native epitope using PEP-FOLD 3 De novo peptide structure prediction server (128, 129). It showed a spatial distribution of side chains similar to native protein epitope structures (Fig. 5B). Acrylamide-based MINPs were then produced by inverse microemulsion polymerization (130) and were shown to selectively bind the full TGF-β3 from a complex biological fluid (platelet lysate) despite being present at a significantly reduced concentration compared to proteins with similar structure (TGF-β1) and isoelectric point [vascular endothelial growth factor (VEGF)].
A key concept for the success of the previous works was the use of bioinformatic tools for a process named fingerprint analysis. Originally proposed by Bossi et al. (131), this streamlined process for the rational selection of an idiotypic (i.e., unique) peptide epitope consists of the following basic steps: (i) select a target protein sequence and obtain it from the database [National Center for Biotechnology Information (132), UniProt (133), RCSB PDB, etc.]; (ii) cut it in silico into peptides by choosing a suitable cutting agent (e.g., trypsin) using ExPASy PeptideCutter (134, 135); following previously mentioned studies (56, 88, 98, 99), peptides that are too small or too long should be discarded; (iii) peptides of adequate length (8 to 15 amino acids) are aligned to the whole database using BLASTP; the best epitope is the one that best matches the parental protein (highest S score) while simultaneously having the lowest E value (representing the number of distinct alignments that occur in the database by chance). While this workflow has been proposed for linear peptides, our work (48) shows that it can also be adapted to conformational peptides, by including 3D visualization, surface functionality assessment, and secondary structure prediction steps, using platforms such as Mol* (/´molstar/) (136), PyMOL, or PEP-FOLD 3.
Direct epitope extraction from target macromolecule digestion
A recently introduced alternative strategy circumvents the step of choosing the epitope a priori (67). In this work, the target protein hemoglobin was immobilized on the surface of aldehyde-modified silica NPs (SNPs), which served as a solid-phase support. Then, these were subjected to denaturating conditions and digested with trypsin, with only the covalently bound fragments remaining at the surface of the particles. In this manner, a variety of surface-exposed peptides from hemoglobin could be imprinted, which, theoretically, should increase the effectiveness of the process, compared to the imprinting of a whole protein or a single epitope. Nonetheless, this approach requires the use of the full protein with its associated disadvantages, particularly regarding cost of implementation when working with expensive recombinant proteins as molecular targets. Moreover, despite a certain degree of selectivity being observed, the IF and selectivity coefficients were lower than 2, some of the lowest among the present selection of works. This indicates that this strategy still needs further refinement before becoming a viable option.
A similar approach that has achieved comparatively improved results is termed boronate affinity–controllable oriented surface imprinting (137). This method likewise relies on the digestion of the protein to isolate epitopes for imprinting. However, instead of peptides, glycans are used. Although it limits the applicability to glycated proteins, this is often the case for extracellular or membrane-associated proteins with extracellular portions (138), which should be the ones most accessible for MIP applications. Moreover, these glycan sequences are exposed at the surface of proteins, as necessary for interaction with the polymer. By using various oligosaccharide sequences from the same protein [e.g., ribonuclease B (RNase B), transferrin (139), HER2, and erythropoietin (84)], a near-unique MIP can be prepared, achieving impressive selectivity (coefficients > 3.5). The use of saccharides as templates also offers an easy route for their immobilization onto a solid-phase support for polymerization, by exploring their affinity for boronic acids. These covalent bonds are established at relatively high pH values and reversibly dissociate at acidic pH, therefore proving to be an efficient method for removal of the template after imprinting. The major drawback in these systems appears to be the relatively low affinity for the target molecule (KD in the micromolar range), although this is to be expected for saccharide binding in aqueous media, as discussed before. Furthermore, the NP composition might also have played a role. Because tetraethyl orthosilicate (TEOS) was used as the only functional monomer, perhaps it did not provide the most adequate interactions with the template molecules for high MIP affinity.
Protein digestion can also be used as a preliminary procedure to identify proteotypic peptides for later epitope imprinting (140, 141). This approach has been used to improve the speed and effectiveness of proteomic analysis, allowing precise identification of the signature nonapeptide of pro-gastrin–releasing peptide (ProGRP) in tryptic digests of serum samples.
It has been shown that smaller molecules can also be used as templates to target some specific proteins. For example, on the basis of the frequency of l-lysine residues in Fc fragments of IgG molecules, l-lysine–imprinted cryogels were prepared for IgG adsorption (142). Since a single amino acid cannot be nearly as idiotypic as a peptide, the values of IF and selectivity coefficients were, understandably, relatively low (<2). Nonetheless, it performed relatively well in real sample analysis, with up to 90% of IgG being removed from diluted serum. Similarly, the Sellergren group (143–149) has explored the use of fluorenylmethyloxycarbonyl-protected phosphorylated single amino acids as template epitopes for the selective separation of phosphorylated peptides, with a focus on phosphotyrosine (pY). This strategy has been successfully validated against the commonly used approaches based on TiO2 and anti-pY antibodies (145) and has been used in combination with TiO2 to selectively enrich a protein extracts from cancer cells in phosphopeptides before mass spectrometry analysis (146), suggesting that this method can be a valuable alternative for applications in phosphoproteomics research. This strategy has also been recently expanded to selectively bind phosphoserine (150, 151) and phosphohistidine (152) residues, demonstrating its versatility.
Selection of non–peptide-based epitopes
Last, some authors have also explored epitope imprinting using other types of template molecules beyond peptides and amino acids. In several works, phenylphosphonic acid (PPA) was used as an alternative dummy template for detection and isolation of peptides containing pY residues, since PPA and pY have remarkable similarity. This method has been optimized over recent years, achieving steadily improving results in terms of affinity, adsorption capacity, and selectivity (70, 153–156).
Monosaccharides such as glucuronic acid and N-acetylneuraminic acid have also demonstrated their effectiveness as epitope templates. NPs imprinted with these epitopes were successfully used to target and fluorescently label specific glycosylation sites in proteoglycans or glycans such as hyaluronic acid (HA), based on the relative abundance of these sugar monomers in these macromolecules (157–159). Furthermore, they showed no cross-reactivity with other monosaccharides commonly found at the terminal end of sugar chains from proteoglycans or glycoconjugates existing on cell surfaces. Last, a combination of a single monosaccharide (mannose) and a single amino acid (tryptophan) was shown to be enough to imprint microspheres capable of identifying the glycopeptide antibiotic telavancin, with an IF of 3.0 (160). However, a more recent study used the exact same template to produce similar microspheres for the antibiotic teicoplanin (161). The authors analyze the selectivity of this MIP when challenged with telavancin, demonstrating insignificant recovery from solution (≈10%), contrasting with the previous results. Although the chemical composition of the MIP was slightly different, it is hard to explain this apparent contradiction, a fact that is also not commented by the authors.
As described over the course of this section (and summarized in Fig. 4), the selection of the right epitope for MI is not a linear process. One must take into account a range of factors, from the specificities of the target molecule (proteins having been the most exploited), to the accessibility of the corresponding fragment (surface-exposed regions), the size of the dummy template (if too long, it might hinder the MIP synthesis; if too short, it might not confer specificity), or its physicochemical (solubility; pI) and structural properties (linear versus conformational). However, as mentioned above, the choice of epitope itself is only one of the parameters of this technique, and its choice must simultaneously consider other MIP design factors, as will be discussed in the next section.
FUNCTIONAL MONOMERS FOR EPITOPE IMPRINTING
MI involves the polymerization of a functional monomer solution in interaction with the template. The final affinity of the produced MIP is influenced by the strength of these interactions, which are crucial for the formation of recognition sites and, consequently, their selectivity (162). As such, the monomers chosen, alongside their solvent, are also important parameters in the imprinting equation (43). The epitope approach presents a significant advantage in this matter. Since macromolecules of interest are replaced by their smaller epitope templates, which do not have complex structures, the limitations of water solubility and stability are generally minimized, and a greater variety of synthesis protocols can be applied and compared for imprinting efficiency.
Acrylic acid, acrylamide, and derivatives
By far, the most common option is the use of acrylic acid, acrylamide, or their derivatives (30, 43, 44, 75, 84, 91, 159, 163). All of these monomers offer the ability to interact via hydrogen bonds, versatile noncovalent interactions with a binding energy between 1 and 40 kJ mol−1 (164). H-bonding is especially prevalent between organic compounds, which correspond to the chemical nature of the epitope templates discussed in the previous section, making it a desirable feature for the backbone monomer of imprinted polymers. Moreover, acrylic acid and acrylamide derivatives are used in a variety of fields, and their free-radical polymerization process is well studied and established, which helped to repurpose these compounds for MI (165).
Following the strategy proposed by Hoshino et al. (166), in recent years, N-isopropylacrylamide (NIPAAm), alone or mixed with other acrylates, has also become a popular base monomer for MIP synthesis (73, 77, 96, 159, 167–169). In general, these works follow protocols of polymerization in mild conditions, ideal for biological macromolecules. Phosphate-buffered saline (PBS) is adopted as the solvent, thus avoiding harmful organic solvents, with reactions usually being carried out at room temperature, without any need for sophisticated and expensive equipment. Furthermore, poly-NIPAAm also has another well-known useful attribute: thermoresponsiveness. It undergoes a phase transition at 32°C (its lower critical solution temperature) from a swollen hydrated state to a shrunken dehydrated state (76). This intrinsic property enables its exploitation for the binding and release of target molecules at select temperature intervals, which is useful, for example, in drug delivery (117, 169) or sample purification (81, 156, 168).
Another widely selected monomer derived from acrylic acid is zinc acrylate, due to its ability to establish metal chelation interactions and form heterocycles with the template (81, 82, 97, 102, 154, 170, 171). Several works have shown that this introduction of an extra type of possible interactions beside the common H-bonds can increase the strength of the template-functional monomer complex. This results in a higher number of binding sites and, consequently, improved affinity and binding capacity of the MIP (81, 97).
Other organic monomers
Among other commonly used monomers is dopamine (72, 108, 118). Mainly known as a neurotransmitter, in alkaline conditions, the catechol group is oxidized to quinone form, which self-polymerizes into an adhesive polymer with high stability, hydrophilicity, and biocompatibility (108). Dopamine is especially advantageous when interacting with tendentially basic peptides, since the quinone moiety preferentially interacts with basic amino acid residues (72). O-phenylenediamine (103) and scopoletin (89, 91) are two interesting monomers that have also been tested in MI protocols because they can be processed by electropolymerization. This process offers precise control over the thickness of the MIP layer, depending on the applied voltage and number of cycles. This feature makes them particularly attractive for the development of MIP-coated gold sensors for QCM or SPR, for example, although other types of sensors such as gold wires have also been successfully imprinted (89, 91).
In a combination of techniques, 3-sulfopropyl methacrylate/benzyl methacrylate MINPs were first produced, followed by electropolymerization of 4-aminothiophenol to bind the particles together, coating a QCM sensor (109). The new MIP achieved remarkable results, with an IF of 8.28, a selectivity coefficient of 4.34 against the same peptide with just two amino acids switching positions in the sequence, and a subnanomolar LOD. Other less commonly used polymers such as poly(ethylene-co-vinyl)alcohols (EVALs) have recently been applied for epitope imprinting, owing to their tunable ethylene to vinyl ratios that can be optimized for each given template (99, 104). Furthermore, EVAL films can be prepared by simple evaporation of the solvent (namely, DMSO), which also makes them attractive for sensor coating, for example. In addition, it remains to be studied how MIP production in different solvents affects their interaction with target macromolecules in aqueous environments. This is particularly relevant because, for example, peptide templates could acquire aberrant conformations that mask key amino acids from the native protein.
Given the limited set of possible molecular interactions offered by the most commonly used monomers, Zhang et al. synthesized a brand-new functional monomer simultaneously combining the properties of ionic liquids and β-cyclodextrins, which might be considered promising MIP monomer candidates due to their designability and facile functionalization (101). This new monomer could establish various types of interactions with the template in aqueous medium, including H-bonding, π-π stacking, electrostatic, hydrophobic, dipole-dipole, and steric effect. This was reflected on the higher binding energy predicted between the template and the new monomer than between template and other commonly used monomers that preferentially establish H-bonding interactions. Empirical results then confirmed the prediction, with the new MIPs achieving an IF of 4.54 and selectivity coefficients of up to 9.66.
Inorganic MIPs
An alternative to the previously discussed organic polymers is the preparation of silica structures from silane precursors. SNPs have attracted attention for a number of interesting properties, such as suggested biological inertness, the possibility of varied surface modifications, precisely controllable size depending on the cross-linking time, the creation of mesoporous structures, or their transparency to visible light (70, 75, 84, 100, 139). This has allowed their utilization for different applications.
For example, mesoporous silica NPs (MSNs) are structures with a greatly increased surface area, leading to higher adsorption capacities, placing them at the forefront of bioseparation and purification strategies (70, 100). In MSN production protocols, it is important to choose a surfactant that allows the mesopores to be big enough for suitable molecular diffusion. This choice needs to be done in a case-by-case basis, according to the size of the target macromolecule (100). Alternatively, imprinted SNPs can also work as coating for a magnetic core, to facilitate bioseparation processes by applying a magnetic field (139, 172). The transparency of SNPs also allows their coupling with smaller fluorescent NPs [such as carbon or other quantum dots (QDs) (75, 155)]. This combination can allow, for example, tracking of NP localization in living tissues, bioimaging, or efficient detection of target molecule binding through fluorescence quenching. Moreover, silica’s purported inertness is essential for the safe use of some of these fluorescent NPs in vivo by forming a biocompatible shell that impedes leakage of toxic heavy metals (155).
Another advantage of SNPs is the ease of surface modification. While polymeric MINPs, in principle, need to incorporate specific monomers during polymerization to acquire particular surface groups, silica-based MINPs can be grafted specific functionalities post factum by silanization (173). One downside of this strategy is that binding sites may become sterically hindered or even compromised by the subsequent modification. Moreover, if the various functional silanes are not included in the initial imprinting mixture, the interaction between template and silica MINPs could be relatively weak [as previously mentioned (84, 139)].
Choice of cross-linker
Depending on the aimed MIP application, the choice of cross-linker might be as important as the choice of monomers, but the reality is that this design parameter has generally not been a major concern of most studies. The vast majority has used the same classical reagents, such as the homobifunctional N,N′-methylenebisacrylamide (48, 71, 73, 77, 167, 174) and ethylene glycol dimethacrylate (36, 42, 43, 47, 84, 98, 156, 168, 169), each having two vinyl groups that can cross-link acrylate-based monomers, or TEOS, a homotetrafunctional silica-based monomer (70, 75, 84, 139, 172). Despite their well-known properties and effectiveness, these molecules do not provide any particular functionality or advantage to synthesized MIPs.
However, this untapped potential hidden in cross-linkers has been increasingly noticed and realized. For example, polyethylene glycol diacrylate has been used for its hydrophilic properties, to reduce nonspecific protein adsorption (153, 154), while divinyl benzene was used in a different work to introduce π-π stacking interactions with another functional monomer (161). A recent study has taken cross-linker selection a step ahead, developing MINPs for targeted drug delivery using a monomer and cross-linker that selectively degrade in tumoral tissues (96). In this elaborated strategy, the active targeting potential provided by epitope imprinting of a cancer cell overexpressed surface biomarker (CD59) ensured their selective tumor accumulation and cellular uptake. The following MINP biodegradation in tumor tissues through the combined actions of glutathione over the cross-linker N,N′-diacrylylcystamine and the lower pH over the monomer dimethylaminoethyl methacrylate enabled the localized release of a chemotherapeutic drug, doxorubicin, at the tumor site, protecting healthy cells from its cytotoxic effects. This led to an effective reduction in the growth of tumor volume and weight, which were less than 40% than those observed with pure doxorubicin treatment.
Although, thus far, only a few studies leveraged on cross-linker properties to define MIP functionality, cross-linking designs are expected to become a major synthesis parameter for their specific performance. This could be especially attractive, for example, in tissue engineering and regenerative medicine, where biodegradability is a crucial parameter for biomaterial-based scaffold construction (175). Other applications where responses must be limited to a specific time window or confined to a specific site could also benefit from similar degradable polymers. Therefore, the incorporation of novel cross-linking chemistries and molecules in MIP design will further expand the usefulness of MI technology to a greater variety of fields in the near future.
Combining previous scattered design knowledge to evolve MIP performance
Overall, MIP design benefits from the inclusion of different types of noncovalent interactions to decrease the overall binding energy between template and monomers, thereby increasing the affinity and specificity of the final polymer. The greater the number of building blocks used, the more unique the final construct will be. Table 2 summarizes the different monomers used in reviewed works and the molecular interactions they offer. The choice of functional monomers must be made considering the template, so as to adapt and maximize the intermolecular interactions happening between the two. For example, in the case of peptide epitopes, analyzing their amino acid composition in terms of hydropathy and charge (as well as their positioning and spatial distribution for peptides with a secondary structure) can help to select rational monomer candidates for imprinting. Moreover, the optimization of monomer to monomer, cross-linker, and template ratios should be an important initial step in every MI protocol. Accordingly, this aspect has been receiving increasing attention. A common empirical approach is to synthesize a library of MIPs with different polymerization mixture compositions and compare their imprinting effectiveness to select the best combination for that particular application (36, 40, 44, 47, 82, 84, 90, 141, 153, 163). This methodology should also lead to the discovery and optimization of new formulations for MIP preparation, which is needed in this still developing field (99).
Table 2. Functional monomers used for epitope MI and corresponding intermolecular interactions.
| Functional monomer | Molecular interaction(s) | Reference(s) |
| Acrylic acid | H-bonding; electrostatic (−) | (42, 73, 77, 167–169) |
| Methacrylic acid | H-bonding | (67, 74, 76, 90, 93, 105, 106, 111, 140, 190, 224) |
| Methyl methacrylate | H-bonding; hydrophobic | (161, 223) |
| 2-(Trifluoromethyl) acrylic acid | Electrostatic (−) | (90, 94, 96, 117, 140, 198) |
| 2-(Hydroxyethyl) methacrylate | Hydrophilic; H-bonding | (90, 140, 142, 224) |
| 2-(Aminoethyl) methacrylate | H-bonding; electrostatic (+) | |
| Benzyl methacrylate | Hydrophobic; π-π stacking; aromatic rings may aid charge transfer |
(109, 112) |
| 3-Sulfopropyl methacrylate potassium | Electrostatic (−); sulfur may aid grafting on gold surface |
(109, 112) |
| Dimethylaminoethyl methacrylate | Electrostatic (+) | (90, 96, 140) |
| Acrylamide | H-bonding | (48, 67, 71, 92, 94, 95, 160, 171, 190, 199, 224) |
| Methacrylamide | Hydrophilic; H-bonding | (86, 90, 106, 157) |
| 2-(Hydroxyethyl) acrylamide | Hydrophilic; H-bonding | (69) |
| Phenylacrylamide | H-bonding; hydrophobic; π-π stacking | (117) |
| N-tert-butylacrylamide | Hydrophobic | (73, 77, 90, 93, 96, 117, 167–169, 190, 198, 199, 224) |
| N-[3-(dimethylamino)propyl]methacrylamide | Electrostatic (+) | (42) |
| N-(3-aminopropyl)methacrylamide | Electrostatic (+) | (73, 77, 167, 169) |
| N-(2-aminoethyl)methacrylamide | Electrostatic (+) | (90, 140, 141, 225) |
| N-isopropylacrylamide | Hydrophilic; H-bonding | (42, 73, 76, 77, 81, 88, 96, 117, 156, 159, 160, 167–169, 198, 199) |
| 4-Acrylamidophenyl(amino)-methaniminium chloride or acetate |
Electrostatic (+) | (69, 157, 159) |
| N-acrylamido-benzamidine | Electrostatic (+) | (86) |
| N-methacryloyl-l-aspartic acid | Electrostatic (−) | (142) |
| Zinc acrylate | Metal chelation; heterocyclic compound formation |
(81, 82, 97, 102, 154, 170, 171) |
| Vinylphosphonic acid-Ti4+ | Metal coordination | (156) |
| Ti4+-ethylene glycol methacrylate phosphate | Metal coordination; hydrophilic | (153) |
| Vinylphenylboronic acid | Hydrophobic; π-π stacking; boronate affinity for cis-diols |
(85, 160, 161, 170) |
| 4-Formylphenylboronic acid | π-π stacking; boronate affinity for cis-diols | (84) |
| 3-Aminobenzeneboronic acid | Electrostatic (+); π-π stacking; boronate affinity for cis-diols |
(83) |
| 3-Ureidopropyltriethoxysilane | H-bonding; electrostatic (+) | (70, 155, 172) |
| 3-Aminopropyltriethoxysilane | Electrostatic (+) | (75, 100, 172, 222) |
| 3-Isobutyltriethoxysilane | Hydrophobic | (172) |
| 1-Propyltrimethoxysilane-3-methylimidazolium chloride |
Electrostatic (+) | (100) |
| 4-Vinylpyridine | Electrostatic (+) | (90, 105, 140) |
| 2-(-3-(4-Nitrobenzo[c][1,2,5] oxadiazo-7-yl)ureido) ethylmethacrylate |
H-bonding (fluorescence) | (85) |
|
N-3,5-bis (trifluoromethyl)-phenyl-N′-4- vinylphenylurea |
H-bonding | (90, 140) |
As an alternative, a more efficient high-throughput screening procedure has recently been developed for this type of empirical selection process, allowing a simultaneous evaluation of 32 different monomer compositions in triplicate (162). This method combines the advantages of using small sample volumes in a 96-well filtration microplate with the reusability and affinity separation offered by solid-phase synthesis. In this ingenious approach, the template is covalently attached to a solid-phase support and placed in the microplate wells, with each well containing a unique combination of monomers and cross-linker. After polymerization, weak-affinity MINPs are washed away, while high-affinity MINPs remain bound to the template in the wells. The larger the number of MINPs that remain in each well, the more adequate the composition of the corresponding polymerization mixture. In addition, by incorporating a fluorescent reporter monomer in the mix, the results can easily and quickly be acquired using a microplate fluorescence reader. The results of this assay were consistent with SPR affinity analysis, producing MINPs with KD in the nanomolar range, the usual benchmark for natural antibodies. These promising results suggest that case-by-case refinement of polymer composition can soon become an efficient routine step in MI works, contributing to increasing performances at decreased development costs.
Incorporation of computational modeling approaches
Notwithstanding its importance, the process of optimizing the polymer formulation is generally cumbersome. To make this process more efficient, reducing expenditures in materials and reagents, several groups have taken advantage of software tools to model the molecular interactions between monomers, cross-linkers, and templates (176–182). These can be based on various computational approaches, including quantum chemical calculation (QCC), molecular mechanics (MM)/molecular dynamics (MD), and thermodynamic analysis (183, 184). Each approach is based on different assumptions and favors the assessment of different parameters.
QCC, traditionally the most commonly used method, is also the most precise in describing the interactions between smaller templates and functional sites of monomers. It works on the basis of quantum mechanics, making calculations based on electron orbitals or bands at the single-atom level. Its main aim is to solve Eq. 5, minimizing the association energy ΔE
| (5) |
where E refers to Gibbs free energy, EC being that of the monomer-template complex, ET that of the template, and EM that of the isolated monomer(s) (93, 183, 185). However, the high level of detail of these models requires a correspondingly high computational power. This drawback restricts the use of QCC to small molecules and few at a time. Moreover, it does not consider the effects of solvent, time, and temperature (184, 186).
MM, on the other hand, sacrifices precision in predicting molecular structure and binding energy to simplify calculations, making them more efficient and practical, especially for modeling multiple components. It is based instead on force fields, following Eq. 6
| (6) |
with E1,2 corresponding to bond stretching, E1,3 to bond angle bending, E1,4 to torsion of dihedral bond angle, EVdW to van der Waals interactions, and Eel to electrostatic interactions, with the possibility of adding additional terms corresponding to other interactions like H-bonds. The efficiency of these calculations allows a faster analysis of a greater number of elements. Furthermore, it can be combined with (i) the Newtonian laws of motion to include the time factor, (ii) Maxwell-Boltzmann distribution for analysis at different temperatures, and (iii) restriction to a “box” of specified dimensions to control the pressure. These combinations of MM with additional parameters are termed MD. In addition, MM/MD simulations can also be applied for interaction between the template and the final polymer, which can provide different outcomes than those observed for template/monomer complexes (184, 186). These properties have recently prompted an increase in the use of MM/MD compared to QCC, particularly by using the LEAPFROG algorithm in the framework of the SYBYL software suite (Tripos Inc., St. Louis, MO, USA) (177, 178, 180–182, 187, 188). In addition, Busato et al. (179) have developed MIRATE, a freely accessible platform to support and guide the user through the whole process of designing MIPs. Softening the requirement of specific knowledge on molecular modeling techniques while presenting a user-friendly interface, this gateway can make computational modeling tools more accessible to a wider audience, contributing to accelerate MIP development.
Despite their advantages, neither of the previous approaches considers mesoscopic effects, which are also important to determine MIP properties. To analyze these effects, coarse-grained and thermodynamic simulations have proven useful (184, 189). The first simplifies monomers into equal spheres, allowing the simulation of much larger complexes with much less computational power. Consequently, larger-scale properties can be studied in these models. Although not as often, other groups have studied the thermodynamic properties of polymerizing systems, drawing important conclusions on the influence of particle mobility, template size, initiator concentration, or phase separation effects (184). Similar to the epitope choice, using computational modeling tools to assist the prediction of the best functional monomer candidates is a wise strategy that can save resources and time in protocol optimization later on.
In summary, the importance of selecting the most adequate polymeric formulation is recognized more than ever before. As discussed, this process should take into account the target application as well as the specificities of the template molecule. By analyzing available libraries, a careful and rational selection can be undertaken. This can then be refined by in silico models, particularly for optimizing the ratio of each component in the polymerization mixture. Combined with high-throughput testing and the development of novel, more functionally rich monomers, we can foresee a continued growth in the production efficiency of epitope MIP platforms.
IMPRINTING METHODS AND APPLICATIONS
After selecting the reagents—epitope template, functional monomers, and cross-linker—the next step in MIP production is to select a suitable preparation method according to its intended final application. MIPs are available in a variety of formats, such as monoliths, membranes, or NPs, showing tremendous versatility of employment. This section explores the main imprinting methodologies currently applied, making an effort to identify their strengths and weaknesses, the types of MIP that can be produced, and their corresponding applications, to exemplify the fields for which they are best suited.
Bulk imprinting
The first works on the field started by following the most straightforward bulk imprinting method (68, 74, 102, 142, 154, 190). This consists in dissolving the template molecule with the functional monomer(s) and cross-linker(s), thoroughly mixing to homogenize, allowing the monomers to interact with the template, followed by triggering the polymerization process (adding an initiator or irradiating with ultraviolet light, for example). This leads to the formation of a macroscale polymeric structure, which can then be ground into particles, if that is the desired final shape (Fig. 6A). This is the simplest method for MIP synthesis and has the advantages of involving the smallest number of different reagents and equipment, while allowing production of a variety of structures such as cryogels (Fig. 6B) (142), monoliths (68, 74), or films (Fig. 6C) (102), besides ground microparticles/NPs (154, 190).
Fig. 6. Bulk molecular imprinting method and representative applications.
(A) Bulk imprinting procedure; bulk MIPs can further be processed into microparticles/nanoparticles by grinding and sieving (dashed arrow). (B) Scanning electron microscopy micrographs of imprinted (MIP C) and nonimprinted (NIP C) poly(2-hydroxyethyl methacrylate-co-N-methacryloyl-l-aspartic acid) cryogels for IgG purification. Reproduced with permission (74). Copyright 2015, Elsevier. (C) Atomic force microscopy (AFM) images of MIP-coated (above) and bare (below) gold chips for human albumin detection. Adapted with permission (111). Copyright 2016, Elsevier.
By using this technique, poly(2-hydroxyethyl methacrylate-co-N-methacryloyl-l-aspartic acid) cryogels were prepared for IgG purification in chromatography (142). The choice for cryogels rested on their microporous structure (shown in Fig. 6B), which was intended to compensate for the diffusion limitations of macromolecules in bulk MIPs. These presented a surface area of 34.2 m2 g−1, with an average pore diameter of 37.2 Å, reaching a Qmax of 75.1 mg of IgG/g of cryogel. In other studies, methacrylate-based micro–solid-phase extraction monolithic columns were developed for sample enrichment in selected physiologically produced peptides, namely, cholecystokinin (74) and encephalin (68). Both columns, prepared with the same protocol but different tetrapeptides, could be used over 100 times in a reproducible manner, with the ability to extract over 90% of target molecule from simulated and real cerebrospinal fluid samples. On the other hand, their adsorption capacity is manifestly inferior when compared, for example, with the surface imprinting approach of Tan et al. (93) (described further ahead) due to shortcomings associated with the diffusional limitation of macromolecules in bulk MIPs.
Since template molecules are randomly distributed in the prepolymerization mixture, a large number of them end up trapped in the polymer matrix. This works well for small molecules, including peptides, but major difficulties arise when applied to biomacromolecules, as previously stated. Their low rates of diffusion through polymer networks due to their large size (or even lack of accessibility depending on the mesh size) means that purification and rebinding are inherently more difficult. This effect is observed even if the resulting monolith is ground and sieved into particulate (87, 191). Moreover, this grinding process is also inefficient, since it damages and destroys binding sites, reducing the rebinding properties of the resulting MINPs. Besides, ground particles are usually heterogeneous in shape and size, which is less than optimal for precision applications (192).
Surface imprinting
To overcome these limitations, the technique known as surface imprinting was developed (193–195) and quickly adapted for the imprinting of epitopes as well. In the first studies exploring this strategy, the authors ingeniously used crude products from solid-phase peptide synthesis to produce what was then named “hierarchically” imprinted materials (52, 53). This technique involves the restriction of the imprinting process, and therefore MIP-target interactions, to a single interface. This decreases the amount of template required, while providing higher binding site accessibility, improved binding kinetics, response speed, and regeneration (47, 87, 192). To that end, template molecules are attached to a suitable supporting surface, which can either be sacrificial or permanent, and polymerization occurs only in a thin layer. On the downside, this technique results in fewer recognition sites, which can lower the binding capacity or the sensitivity in sensor applications, for example, (87, 191). Nevertheless, some strategies have been proposed to overcome these limitations, such as using NPs with large relative surface areas to increase binding capacity (93, 196), enzymatic amplification to improve sensor signals (83, 103), or dual-template imprinting for improvement in both parameters (97). Variations of surface imprinting have spawned since its introduction, with different support structures and immobilization strategies.
Thin-film coatings
A widely used variation of surface imprinting is the production of an imprinted film on gold sensors, usually for SPR or QCM-based applications (72, 89, 91, 98, 108, 111, 112). These analytical techniques allow a direct evaluation of the binding capability of the produced MIP, while also representing promising precursors of point-of-care screening devices. Two imprinting methodologies can be distinguished for this purpose. One consists in covering the sensor with a premix of the epitope template and functional monomers, followed by polymerization. This has been performed through processes based on free radical polymerization (98), self-polymerization of dopamine (72, 108), and electropolymerization (103). The second variant consists of a two-step process: First, the template is organized by self-assembly on the gold surface via, for example, thiol chemistry (usually by using cysteine end-modified peptides); next, the monomer mix is added and polymerization started (Fig. 7A). The first step is thought to promote a single orientation of all template molecules, potentially leading to more uniform and, therefore, more sensitive and selective imprinted cavities. Methacrylate-based radical polymerization (111, 112), solvent evaporation using EVALs (99, 104), and electropolymerization of o-phenylenediamine (197) or scopoletin (89, 91) have been successfully applied to produce MIP films following this procedure. Unfortunately, various studies fail to report on important parameters of MIP evaluation, making detailed comparisons between methodologies difficult. Some representative applications of this technique include detection of HIV in urine samples (108), TnT in blood samples for real-time testing of heart injury (72), or NSE for early diagnosis of small cell lung cancer (91). Cysteine terminal modification and histidine insertion into peptide epitopes have been recently compared as template immobilization strategies (91). The results show that a histidine inserted in the middle of the sequence for attachment to gold wire sensors led to an order of magnitude lower LOD and KD, higher IF, and more than double the selectivity as compared to other peptides (one from another surface-exposed portion of the same protein, and another with a similar α-helical structure and terminal cysteines) and bovine albumin. These suggest that histidine might be a superior alternative to the more common cysteine end-modification of peptides for gold surface tethering in imprinting protocols.
Fig. 7. Surface imprinting on thin flat films.
(A) Surface MI procedure, allowing the creation of surface-accessible binding sites for the target molecule. (B) Generation of an epitope-imprinted biointerface for dynamic cell adhesion and harvesting. Adapted with permission (69). Copyright 2017, the authors.
Another significant advance introduced in MIP-based sensing platforms has been the incorporation of enzymatic amplification of the electrochemical signal by horseradish peroxidase (HRP) catalysis (103). In this variant, an HRP-coupled template is preincubated with the sensor before its application in sample analysis. Then, the target protein competes with the HRP peptide for binding to the MIP; the greater the target concentration, the more HRP peptide will be displaced. Last, it is immersed in a hydroquinone–hydrogen peroxide solution that reacts with HRP, triggering an electric current proportional to the remaining HRP peptide bound to the sensor. This signal is therefore inversely proportional to the target protein concentration, allowing its indirect calculation. This approach managed to increase the sensitivity of imprinted sensors, attaining an LOD of 0.02 nM.
Other strategies of surface coating with MIPs include cellulose membranes for monoclonal antibody purification from complex mixtures [as previously mentioned (106)] or a bioactive interface for cell sheet harvesting (69). The latter (Fig. 7B) consisted of a quartz surface coated with a polymer imprinted with the peptide tag DDDGGDDD (a structurally symmetrical and carboxyl-rich peptide designed as epitope for this work). The surface molecular-recognition sites could be used to bind a bioactive peptide featuring the epitope tag at one terminus and cell-adhesive peptide RGD at the other end. Because of the reversibility of the peptide binding to the MIP cavities, the biointerface exhibited dynamic RGD presentation and, subsequently, controllable cell-adhesive behavior.
Imprinting on NP surfaces
A different way to leverage surface imprinting is by building thin imprinted coating layers on NPs of different kinds. This allows one to take advantage of the inherent physicochemical properties of a specific particle in combination with the molecular recognition potential of MI (Fig. 8A). For example, carbon nanotubes and iron oxide NPs can be used for their magnetic properties to bind and extract a specific constituent from complex samples (81, 82, 93, 97, 101). This property also facilitates the preparation procedure through the application of an external magnetic field, leading to a quick and efficient separation of the NPs. This has been used to isolate angiotensins I and II, important molecules for the maintenance of blood pressure and general homeostasis, which have very low concentrations in human blood (≈20 pg ml−1) (93, 196). These superparamagnetic particles achieved high adsorption capacities (above 70 mg g−1) and high selectivity (IF ≈ 5) and could remove more than 90% of angiotensin from human plasma samples. Iron oxide NPs have also been used to build an immunosensor for ultrasensitive detection of HIV-1 antibody, as an effort to reduce the time between HIV infection and diagnosis (83). In this study, the authors prepared IgG-imprinted NPs, which were able to bind anti–HIV-1 antibody. By using enzymatic amplification [as described above (103)], the resulting sensors achieved a detection limit 200 times lower than existing ELISA assays.
Fig. 8. Surface imprinting on NP thin coatings.
(A) MI procedure with immobilized templates on the surface of an NP. (B to E) Transmission electron microscopy images of (B) magnetic carbon nanotubes (MCNTs), (C) silica-coated MCNTs, (D) MCNTs coated with methacryloxypropyl trimethoxysilane, and (E) MIP-coated MCNTs. Adapted with permission (221). Copyright 2018, Wiley.
SNPs are another type of particles that are especially interesting for surface imprinting, because their surface can easily be modified with silane derivatives, making them a versatile platform for surface functionalization. Moreover, as previously mentioned, SNPs are relatively bioinert and transparent. Thus, they have attracted attention for use in sensing applications, by coupling a silica matrix with smaller QDs, which emit fluorescent light in a specific controllable wavelength (75, 155). These QD/SiO2/MIPNPs are then able to act as sensors that specifically recognize the target molecule thanks to the imprinted cavities in the polymeric shell. At the same time, thanks to the transparency of the SiO2 matrix, the QDs are still able to absorb light and emit a fluorescent signal proportional to this interaction. These systems have also been applied specifically with carbon dots, yielding MINPs with an LOD in the nanomolar range and the ability to work in complex samples, such as diluted urine (76).
A recent innovation that has been coupled with both magnetic and silica NPs is dual-template imprinting. By using two different template molecules simultaneously in the imprinting process, recent studies were able to either (i) increase the sensitivity and selectivity of MIP-coated carbon nanotubes (Fig. 8, B to E) to target albumin, using two epitopes form the same protein (97), or (ii) recognize two separate targets (albumin and transferrin) with the same nanotube formulation, using a single template from each (81).
This strategy has proven useful for targeted drug delivery (198, 199). Using the dual-template approach, poly-dopamine–coated SNPs were imprinted with a conformational epitope of HER2, an overexpressed surface receptor in some cancer cells, and doxorubicin, a widely used chemotherapeutic drug (118). These NPs led to significantly increased doxorubicin concentration in tumor tissues, while slightly reducing its distribution in liver, kidney, and heart. They also led to over 70% cumulative survival, as compared to approximately 30% for free doxorubicin or doxorubicin-NIP groups and zero for untreated controls. Similarly, zinc acrylate and vinylphenylboronic acid were used to coat SNPs with a MIP layer against bleomycin, another common anticancer drug, and the 71-80 peptide of human fibroblast growth factor–inducible 14 (Fn14) modified with glucose, to target a specific type of pancreatic cancer cells (170). In vitro imaging showed specific targeting of Fn14-overexpressing cells, while in vivo administration of bleomycin-loaded MINPs inhibited xenografted tumor growth [1.05× increase in size, compared to 1.5× for free bleomycin and 1.6× for bleomycin-loaded nonimprinted NPs (NINPs)].
Polymeric NP imprinting
MINPs can also be prepared to be fully polymer-based. Two different methods have been mainly used in these strategies. One is precipitation polymerization, which happens in aqueous media and uses organic solvents for the subsequent NP precipitation (94, 153, 156). The second is inverse microemulsion polymerization (130). In this technique, a dispersed aqueous phase containing the functional monomers and a continuous organic phase containing surfactants are mixed to form an emulsion. For enameling the imprinting template, it must be modified with a hydrocarbon tail to become amphiphilic, thus positioning itself at the interface of the inverted micelles. Unlike in the former method, this ensures that the template remains at the surface of the forming MINPs after initiating polymerization in the aqueous phase (Fig. 9A) (48, 71). It can therefore be considered a particular case of surface imprinting. This technique also allows a relatively precise control over particle diameter, yielding NP populations with a low polydispersity index.
Fig. 9. Inverse microemulsion MI and representative applications.
(A) Scheme depicting the process of surface imprinting of polymeric NPs by inverse microemulsion polymerization. (B) MINPs against cancer biomarker p32, carrying the chemotherapeutic drug methylene blue, tendentially accumulated in tumoral tissues and drastically reduced tumor growth in mice. Adapted with permission (71). Copyright 2015, Wiley. (C) Tissue culture plastic coverslips coated with MINPs (but not NINPs) can sequester and retain target transforming growth factor–β3 (TGF-β3). Left: AFM images of coated coverslips. Right: fluorescent immunolabeling of TGF-β3. Scale bars, 250 nm (left), 100 mm (right). (D) ASC cell pellets cultured with MINPs (left) grew to larger sizes and produced more collagenous matrix than pellets cultured with NINPs (right). Adapted with permission (48). Copyright 2020, Wiley. Scale bars, 150 mm (lower magnification images), 50 mm (insets).
Adopting this strategy, polymeric MINPs were developed to bind Lpp20, an outer membrane protein of Helicobacter pylori, a bacterium that has been linked to stomach cancer (92). These were able to bind significantly more bacteria than NINPs and NPs imprinted with the unmodified epitope (without the hydrophobic tail), constituting a promising first step for the development of a potential future bactericidal therapy. Following this synthesis concept, another study designed an innovative targeted drug delivery system, by imprinting NPs with a palmitic acid–modified conformational epitope of cancer biomarker p32 (71). These MINPs could be loaded with the drug methylene blue during the preparation process, demonstrated the ability to localize to tumor tissues, and significantly inhibited their growth in mice models (Fig. 9B), while being biocompatible with healthy tissues. As previously mentioned, this imprinting strategy has also been recently used to produce polyacrylamide-based MINPs against TGF-β3 (48). These MINPs were then applied as receptors in adipose tissue–derived stem cell (ASC) culture substrates to sequester endogenously produced TGF-β3 (Fig. 9C). ASCs in 2D cultures showed a marked up-regulation of SOX-9 (a downstream target of TGF-β3 signaling), while in 3D cultures, there was an increased deposition of collagen II–rich matrix (Fig. 9D). These outcomes suggest that MINPs could direct stem cell commitment without exogenous growth factor supplementation, thus constituting cheap, stable alternative growth factor receptors for tissue engineering applications.
Polymeric NPs can also be coupled with fluorescent QDs for tracking and quantification assays, as previously mentioned for silica-based systems (76, 88, 95). These have been used, for example, to quantify albumin in human serum samples (88), or for cell imaging, by targeting specific glycosylation sites (157). In the latter work, two types of fluorescent core-shell particles were prepared: glucuronic acid–imprinted NPs coupled with green QDs and N-acetylneuraminic acid–imprinted NPs coupled with red QDs. This allowed simultaneous recognition of HA and sialylated glycoproteins/glycolipids on keratinocytes. These results helped to demonstrate the applicability of MIPs for saccharide recognition, a fast-expanding research field with increasingly recognized impact in human health and biomedicine (200). In this context, these MINPs might contribute to the emergence of new diagnostic tools, since aberrant glycosylation is known to occur in disease conditions such as several cancer phenotypes (201).
Another relatively recent addition to the MINP toolbox has been reported by the Zhao group (202, 203) and consists of a MI process in micelles. This creative method is based on the use of a novel cross-linkable surfactant monomer that self-assembles into mixed micelles, where the template imprinting takes place. A first round of cross-linking of azide groups yields surface–cross-linked micelles. These can be surface-functionalized in a second click reaction with hydroxyl-rich monomers to ensure that MINPs are highly soluble in water and to facilitate their separation. Last, the micellar core, which encompasses a vinyl-based cross-linker and other vinyl-containing functional monomers (for example, to incorporate electrostatic interactions with charged amino acids), is photocrosslinked. This carefully designed supramolecular construction allows the production of MINPs with sizes close to natural receptors (∼5 nm) that can precisely discriminate between closely related small peptides (204, 205) and, as recently demonstrated, also glycans (39, 40). This ability has been leveraged to achieve impressive biologically relevant results in sequence-selective protection of larger peptides (angiotensins, amyloid-β) from proteolysis (206), selective phosphorylation of peptides and proteins (207), and synthetic hydrolysis of oligo- and polysaccharides (such as cellulose) (208, 209). Although the synthesis of MINPs and its precursors involves nontrivial laborious processes, this is a versatile one-pot method that results in extraordinary templating effects for a variety of different biomolecule epitopes. Thus, we foresee exciting developments in the application of this system in the near future.
Solid-phase imprinting
Several MI studies have been adopting a recently introduced innovative method of MINP synthesis, developed in parallel by two groups from the University of Leicester (UK) and Compiègne University of Technology (France), termed solid-phase imprinting (24, 210, 211). In this technique, the template molecule is tethered to the surface of a solid-phase support (originally, glass microbeads). The functionalized supports are then immersed in a liquid phase containing the prepolymer mixture, of which NIPAAm is a main component to confer thermoresponsiveness to the polymer. At this point, the monomers in solution reorganize around the immobilized template through noncovalent interactions. After starting the polymerization process in the liquid phase, an array of polymeric NPs with different affinities and sizes are formed in suspension.
In the protocol proposed by the Piletsky group, the mixture is then washed several times at or below room temperature to remove weakly bound NPs, which did not form suitable binding sites. When this process is complete, only the particles with a strong affinity for the template remain bound to the solid phase. These are the NPs that were formed by the monomers directly interacting with the immobilized template, acquiring full-fledged imprinted cavities. They are then recovered by eluting with hot water, with enough energy to disrupt those bonds. This is eased by poly-NIPAAm’s thermoresponsive behavior, transitioning from its swollen hydrophilic to its shrunken hydrophobic state above 32°C, thereby changing its spatial arrangement and detaching from the template molecules. This procedure is illustrated in Fig. 10A. On the other hand, works performed by Bui and Haupt’s group (159, 212) have presented a variation of this step, where the polymerization and affinity purification temperatures are switched. In this instance, polymerization occurs above 32°C (in collapsed state), and high-affinity MINPs are eluted below this temperature (in their swollen form). The rationale behind this choice is that when these MINPs are used in physiological conditions (37°C), they will be in the same collapsed state in which they were imprinted, thus ensuring that the conformation of their binding sites is adequate for target binding. Despite the sound logic of this hypothesis and the results achieved in these studies, the fact remains that MINPs produced at room temperature also performed successfully in vivo (73, 169). Future studies comparing both variations could further elucidate this question.
Fig. 10. Solid phase imprinting method.
(A) Solid-phase epitope imprinting procedure. The affinity purification steps allow removal of low-affinity particles and unreacted monomers by eluting at the same temperature as polymerization (blue arrow), followed by recovery of high-affinity MINPs at a different temperature (red arrow). (B) Confocal microscopy images of cancer cell line cultures with low (left) and high (right) expression of epidermal growth factor receptor (EGFR). MINPs against EGFR (green) accumulate significantly only in cells with high expression. Adapted with permission (77). Copyright 2018, American Chemical Society. (C) MINPs against β2 microglobulin, a marker of senescent cells, preferentially accumulate in older mice. Adapted with permission (169). Copyright 2019, The Royal Society of Chemistry. DAPI, 4′,6-diamidino-2-phenylindole.
Solid-phase imprinting holds several advantages over traditional MI protocols. First, removal of the template and isolation of MINPs is easier and straightforward. Second, since the template is immobilized at the solid phase, not only is it not lost during purification but it is also entirely reusable for multiple synthesis cycles, a possibility that is especially attractive for more expensive template molecules. Third, the two-step elution purification process results in the recovery of a more uniform suspension, with similar high-affinity NPs, reminiscent of monoclonal antibodies. Last, it can be scaled up and adapted into a continuous reactor, holding great promise for upscaling in industrial production (213).
Solid-phase imprinting using epitope templates and acrylamide-derived monomers polymerized under mild conditions in PBS has been applied to generate NPs capable of recognizing several distinct human macromolecules (73, 77, 159, 167, 169). For example, MINPs coupled with QDs were produced to recognize human VEGF, achieving a KD of 1.78 nM. When injected in zebrafish models, these NPs were able to successfully accumulate in tumors overexpressing this growth factor, while in a human VEGF–negative model, this ability was completely lost (73). A similar approach was followed to imprint NPs with an epitope of epidermal growth factor receptor (EGFR) (77, 167). In a first study, these MINPs contained a fluorescent monomer, N-fluoresceinyl acrylamide, and were able to localize to the cytoplasm of cells with high EGFR expression but not to cells without the receptor (Fig. 10B). Incorporating fluorescent monomers in the composition of MINPs is an interesting alternative to the encapsulation of QDs for targeted imaging or drug delivery applications, because it simplifies the preparation procedures while avoiding the employment of potentially toxic components (QDs can leak heavy metals into the body, as discussed before) (159, 169). In a subsequent study, EGFR-MINPs produced by applying the double-template imprinting approach could be loaded with doxorubicin, augmenting the efficacy of the drug 1.5 times compared to its administration in free form. The drug’s cytotoxic effects were shown to be restricted to EGFR-expressing cells (77).
Adopting this concept, other groups have developed fluorescent MINPs targeting β2 microglobulin (B2M), an extracellular component of major histocompatibility complex class I (169). Since these proteins are overexpressed in senescent cells, B2M-MINPs could efficiently target them, staining up to 90.3% of target cells compared to 9.7% of control cells. When injected intravenously in mice, they accumulated in older specimens (11 months old), while in young ones (2 months old), no fluorescence was observed 2 hours after the treatment (Fig. 10C). In another work, solid-phase imprinting was used to prepare MINPs tagged with rhodamine B for specific visualization of HA (159). With their relatively small sizes (~70 nm), they were capable of staining intracellular HA, unlike previous MINPs produced by precipitation polymerization.
Last, an innovative cancer therapy was demonstrated based on MINPs targeting the common N-terminal sequence of E-, R-, and N-cadherins (212). These membrane proteins are responsible for cell-cell adhesion, constituting an essential mechanism in tumor resistance to drugs, as well as metastasis. Since these MINPs targeted the protein site directly responsible for adhesion, they were able to inhibit HeLa cell aggregation, disrupt preformed tumor spheroids, and even inhibit cancer cell invasiveness in vitro.
In summary, this production method has quickly asserted itself as an efficient way to realize the promise of artificial antibodies for biomolecules. Since its introduction, it has quickly shown its potential for targeted drug delivery, bioimaging, and biosensing. It has consistently attained subnanomolar KD values, comparable to monoclonal antibodies, which, along with its reproducibility and scalability, make it very attractive for macromolecule targeting. Its main downside thus far seems to be the relatively low production yield per batch, between 0.2 and 0.6 mg/g of glass beads (24, 35). A possible solution could be to use solid-phase supports with smaller diameters, in particular, magnetic microparticles/NPs (214). These types of supports present a greatly increased surface area, thus allowing a higher template density and, consequently, production of larger amounts of MINPs. Furthermore, glass beads should not be stirred during synthesis steps owing to the possibility of surface abrasion and loss of immobilized template. This makes the process good for continuous solvent percolation in large flow reactors, but less than ideal for small batch reactors. Well-dispersed magnetic NPs minimize this hurdle and can be easily washed and separated from solution by magnetic sorting, being thus more versatile. This protocol has recently been expanded to protein imprinting with promising results (35, 215), setting high expectations for future developments in epitope imprinting applying similar systems.
CONCLUSIONS AND PERSPECTIVES
MI has come a long way since its inception, becoming a standard technique for application-oriented research in small-molecule recognition, with commercial MI products being available today (216). Its use in the biological field, however, has historically been hampered by a variety of drawbacks. Despite its relatively recent introduction, the epitope approach has quickly gained traction over the past decade, thanks to its attractive advantages helping to overcome the limitations typically associated with imprinting of entire macromolecules in polymeric materials. Its considerably more versatile and economic production makes this approach particularly interesting for the imprinting of biologically relevant molecules. In this context, the completely synthetic nature of MIPs is especially relevant in an era where ethical, political, and financial concerns regarding animal use increasingly take center stage (13). Along with their potential low cost, scalability, and long-term stability, they present an elegant alternative to biological recognition molecules like antibodies (12).
Nonetheless, as an emerging strategy, it too faces a number of challenges to realize its potential. For example, the parameters analyzed in each study to describe MIP performance differ greatly between them, making comparisons difficult. Harmonizing and adopting standardized testing methods could greatly improve research progress (78, 80). Considering the choice of molecular targets, it is interesting to notice that researchers have only recently begun to introduce complexity into the process and expand to more than proof-of-concept experiments. For years, the important developments of different MI protocols have dominated research focus in this field. Logically, for optimization purposes, the preferential molecular targets remained restricted to affordable proteins that do not provide much of a practical use. However, thanks to the remarkable progress made on synthesis methods, epitope imprinting is expanding to more valuable targets, such as cell surface biomarkers or growth factors. In parallel, the increasingly sought bioinformatic tools allow a more detailed screening of protein structures, thereby improving the epitope selection. Together, this means that more applications can be envisioned (217), such as targeted drug delivery (25, 77, 171), theranostics (218, 219), or tissue engineering (12, 48, 220). Progressively, the use of molecular modeling platforms is also making MIP development faster and more efficient, lowering the time and resources necessary for optimizing the polymer composition. These steady advances, along with the appearance of new companies based on this technology (e.g., MIP Diagnostics Ltd., Semorex), show that we can confidently expect MIPs to become true “plastic antibodies” and serve as far-reaching tools in biomedicine in the not-too-distant future.
Acknowledgments
Schematics were created with BioRender.com.
Funding: This work was supported by Project NORTE-01-0145-FEDER-000021 supported by the Norte Portugal Regional Operational Program (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF); by the European Union Framework Program for Research and Innovation HORIZON 2020, under the Twinning grant agreement no. 810850–Achilles, European Research Council grant agreement no. 772817; and by FCT/MCTES (Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia, e Ensino Superior) through PhD grant PD/BD/143039/2018 for S.P.B.T., financed through the Doctoral Program in Advanced Therapies for Health (PATH) (FSE/POCH/PD/169/2013), project PTDC/NAN-MAT/30595/2017, and individual contract 2020.03410.CEECIND for R.M.A.D. Project PTDC/NAN-MAT/30595/2017 was co-funded by ERDF, through Norte Portugal Regional Operational Program (NORTE-01-0145-FEDER-030595) and by the State Budget, through FCT. N.A.P. acknowledges support from the Cockrell Family Chair Foundation; the Institute for Biomaterials, Drug Delivery, and Regenerative Medicine; and the UT-Portugal Collaborative Research Program.
Author contributions: S.P.B.T. conceived the review framework and conducted literature review. R.M.A.D. was a visiting scientist at the University of Texas at Austin and performed early research related to the conception and writing of this critical review. All authors participated in the discussion and writing of the paper.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper.
REFERENCES AND NOTES
- 1.Clegg J. R., Peppas N. A., Molecular recognition with soft biomaterials. Soft Matter 16, 856–869 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron R., McCammon J. A., Molecular recognition and ligand association. Annu. Rev. Phys. Chem. 64, 151–175 (2013). [DOI] [PubMed] [Google Scholar]
- 3.P. Quintero-Ronderos, M.-T. Arango, J. Castiblanco, N. E. Correa, G. Montoya-Ortíz., Autoimmunity: From Bench to Bedside (El Rosario University Press, 2013). [PubMed] [Google Scholar]
- 4.Lin W. C., Lien C. C., Yeh H. J., Yu C. M., Hsu S. H., Bacterial cellulose and bacterial cellulose-chitosan membranes for wound dressing applications. Carbohydr. Polym. 94, 603–611 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Au J. L.-S., Yeung B. Z., Wientjes M. G., Lu Z., Wientjes M. G., Delivery of cancer therapeutics to extracellular and intracellular targets: Determinants, barriers, challenges and opportunities. Adv. Drug Deliv. Rev. 97, 280–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shukla A. A., Thömmes J., Recent advances in large-scale production of monoclonal antibodies and related proteins. Trends Biotechnol. 28, 253–261 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Murphy B. M., Swarts S., Mueller B. M., van der Geer P., Manning M. C., Fitchmun M. I., Protein instability following transport or storage on dry ice. Nat. Methods 10, 278–279 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Szenczi Á., Kardos J., Medgyesi G. A., Závodszky P., The effect of solvent environment on the conformation and stability of human polyclonal IgG in solution. Biologicals 34, 5–14 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Zhou J., Rossi J., Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 16, 181–202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang Z., Guan J., Qian J., Zhan C., Peptide ligand-mediated targeted drug delivery of nanomedicines. Biomater. Sci. 7, 461–471 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Bole A. L., Manesiotis P., Advanced materials for the recognition and capture of whole cells and microorganisms. Adv. Mater. 28, 5349–5366 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Teixeira S. P. B., Domingues R. M. A., Shevchuk M., Gomes M. E., Peppas N. A., Reis R. L., Biomaterials for sequestration of growth factors and modulation of cell behavior. Adv. Funct. Mater. 30, 1909011 (2020). [Google Scholar]
- 13.Gray A., Bradbury A. R. M., Knappik A., Plückthun A., Borrebaeck C. A. K., Dübel S., Animal-free alternatives and the antibody iceberg. Nat. Biotechnol. 38, 1234–1239 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Piletsky S., Canfarotta F., Poma A., Bossi A. M., Piletsky S., Molecularly imprinted polymers for cell recognition. Trends Biotechnol. 38, 368–387 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Ahmad O. S., Bedwell T. S., Esen C., Garcia-Cruz A., Piletsky S. A., Molecularly imprinted polymers in electrochemical and optical sensors. Trends Biotechnol. 37, 294–309 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Culver H. R., Peppas N. A., Protein-imprinted polymers: The shape of things to come? Chem. Mater. 29, 5753–5761 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haupt K., Mosbach K., Molecularly imprinted polymers and their use in biomimetic sensors. Chem. Rev. 100, 2495–2504 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Bossi A., Bonini F., Turner A. P. F., Piletsky S. A., Molecularly imprinted polymers for the recognition of proteins: The state of the art. Biosens. Bioelectron. 22, 1131–1137 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Culver H. R., Daily A. M., Khademhosseini A., Peppas N. A., Intelligent recognitive systems in nanomedicine. Curr. Opin. Chem. Eng. 4, 105–113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koetting M. C., Peters J. T., Steichen S. D., Peppas N. A., Stimulus-responsive hydrogels: Theory, modern advances, and applications. Mater. Sci. Eng. R Rep. 93, 1–49 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergmann N. M., Peppas N. A., Molecularly imprinted polymers with specific recognition for macromolecules and proteins. Prog. Polym. Sci. 33, 271–288 (2008). [Google Scholar]
- 22.MIP Database—All database items (available at https://mipdatabase.com/all_items.php).
- 23.F. Canfarotta, A. Cecchini, S. Piletsky, Molecularly Imprinted Polymers for Analytical Chemistry Applications, W. Kutner, P. S. Sharma, Eds. (The Royal Society of Chemistry, 2018), pp. 1–27. [Google Scholar]
- 24.Canfarotta F., Poma A., Guerreiro A., Piletsky S., Solid-phase synthesis of molecularly imprinted nanoparticles. Nat. Protoc. 11, 443–455 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Culver H. R., Clegg J. R., Peppas N. A., Analyte-responsive hydrogels: Intelligent materials for biosensing and drug delivery. Acc. Chem. Res. 50, 170–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheong W. J., Yang S. H., Ali F., Molecular imprinted polymers for separation science: A review of reviews. J. Sep. Sci. 36, 609–628 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Speltini A., Scalabrini A., Maraschi F., Sturini M., Profumo A., Newest applications of molecularly imprinted polymers for extraction of contaminants from environmental and food matrices: A review. Anal. Chim. Acta 974, 1–26 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Zaidi S. A., Molecular imprinted polymers as drug delivery vehicles. Drug Deliv. 23, 2262–2271 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Peppas N. A., Clegg J. R., The challenge to improve the response of biomaterials to the physiological environment. Regen. Biomater. 3, 67–71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S., Cao S., Whitcombe M. J., Piletsky S. A., Size matters: Challenges in imprinting macromolecules. Prog. Polym. Sci. 39, 145–163 (2014). [Google Scholar]
- 31.Kryscio D. R., Peppas N. A., Critical review and perspective of macromolecularly imprinted polymers. Acta Biomater. 8, 461–473 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Liao J. L., Wang Y., Hjertén S., A novel support with artificially created recognition for the selective removal of proteins and for affinity chromatography. Chromatographia 42, 259–262 (1996). [Google Scholar]
- 33.Hjertén S., Liao J. L., Nakazato K., Wang Y., Zamaratskaia G., Zhang H. X., Gels mimicking antibodies in their selective recognition of proteins. Chromatographia 44, 227–234 (1997). [Google Scholar]
- 34.S. R. Marek, M. L. Gran, N. A. Peppas, M. Caldorera-Moore, in Chemoresponsive Materials: Stimulation by Chemical and Biological Signals, H.-J. Schneider, Ed. (Royal Society of Chemistry, ed. 1, 2015), pp. 10–43. [Google Scholar]
- 35.Ashley J., Feng X., Halder A., Zhou T., Sun Y., Dispersive solid-phase imprinting of proteins for the production of plastic antibodies. Chem. Commun. 54, 3355–3358 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Lv Y., Tan T., Svec F., Molecular imprinting of proteins in polymers attached to the surface of nanomaterials for selective recognition of biomacromolecules. Biotechnol. Adv. 31, 1172–1186 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Boysen R. I., Advances in the development of molecularly imprinted polymers for the separation and analysis of proteins with liquid chromatography. J. Sep. Sci. 42, 51–71 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Ariga K., Kunitake T., Molecular recognition at air-water and related interfaces: Complementary hydrogen bonding and multisite interaction. Acc. Chem. Res. 31, 371–378 (1998). [Google Scholar]
- 39.Duan L., Zangiabadi M., Zhao Y., Synthetic lectins for selective binding of glycoproteins in water. Chem. Commun. 56, 10199–10202 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zangiabadi M., Zhao Y., Selective binding of complex glycans and glycoproteins in water by molecularly imprinted nanoparticles. Nano Lett. 20, 5106–5110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oshovsky G. V., Reinhoudt D. N., Verboom W., Supramolecular chemistry in water. Angew. Chemie Int. Ed. 46, 2366–2393 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Li S., Yang K., Deng N., Min Y., Liu L., Zhang L., Zhang Y., Thermoresponsive epitope surface-imprinted nanoparticles for specific capture and release of target protein from human plasma. ACS Appl. Mater. Interfaces 8, 5747–5751 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Rachkov A., Minoura N., Recognition of oxytocin and oxytocin-related peptides in aqueous media using a molecularly imprinted polymer synthesized by the epitope approach. J. Chromatogr. A 889, 111–118 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Clegg J. R., Zhong J. X., Irani A. S., Gu J., Spencer D. S., Peppas N. A., Student award for outstanding research winner in the Ph.D. category for the 2017 society for biomaterials annual meeting and exposition, April 5–8, 2017, Minneapolis, Minnesota: Characterization of protein interactions with molecularly imprinted hydrogels. J. Biomed. Mater. Res. Part A 105, 1565–1574 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Byrne M. E., Hilt J. Z., Peppas N. A., Recognitive biomimetic networks with moiety imprinting for intelligent drug delivery. J. Biomed. Mater. Res. Part A. 84A, 137–147 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Venkataraman A. K., Clegg J. R., Peppas N. A., Polymer composition primarily determines the protein recognition characteristics of molecularly imprinted hydrogels. J. Mater. Chem. B 8, 7685–7695 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Culver H. R., Steichen S. D., Peppas N. A., A closer look at the impact of molecular imprinting on adsorption capacity and selectivity for protein templates. Biomacromolecules 17, 4045–4053 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Teixeira S. P. B., Domingues R. M. A., Babo P. S., Berdecka D., Miranda M. S., Gomes M. E., Peppas N. A., Reis R. L., Epitope-imprinted nanoparticles as transforming growth factor-β3 sequestering ligands to modulate stem cell fate. Adv. Funct. Mater. 31, 2003934 (2021). [Google Scholar]
- 49.H. Culver, L. Strong, N. Peppas, Frontiers in Bioengineering and Biotechnology. Conference Abstract: 10th World Biomaterials Congress (2016). [Google Scholar]
- 50.Sette A., Fikes J., Epitope-based vaccines: An update on epitope identification, vaccine design and delivery. Curr. Opin. Immunol. 15, 461–470 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Hart B. R., Shea K. J., Molecular imprinting for the recognition of N-terminal histidine peptides in aqueous solution. Macromolecules 35, 6192–6201 (2002). [Google Scholar]
- 52.Titirici M. M., Hall A. J., Sellergren B., Hierarchical imprinting using crude solid phase peptide synthesis products as templates. Chem. Mater. 15, 822–824 (2003). [Google Scholar]
- 53.Titirici M. M., Sellergren B., Peptide recognition via hierarchical imprinting. Anal. Bioanal. Chem. 378, 1913–1921 (2004). [DOI] [PubMed] [Google Scholar]
- 54.Rachkov A., Hu M., Bulgarevich E., Matsumoto T., Minoura N., Molecularly imprinted polymers prepared in aqueous solution selective for [Sar1,Ala8]angiotensin II. Anal. Chim. Acta 504, 191–197 (2004). [Google Scholar]
- 55.Tai D. F., Lin C. Y., Wu T. Z., Chen L. K., Recognition of dengue virus protein using epitope-mediated molecularly imprinted film. Anal. Chem. 77, 5140–5143 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Nishino H., Huang C. S., Shea K. J., Selective protein capture by epitope imprinting. Angew. Chemie - Int. Ed. 45, 2393–2396 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Özcan A. A., Say R., Denizli A., Ersöz A., L-histidine imprinted synthetic receptor for biochromatography applications. Anal. Chem. 78, 7253–7258 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Brown M. E., Puleo D. A., Protein binding to peptide-imprinted porous silica scaffolds. Chem. Eng. J. 137, 97–101 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rechichi A., Cristallini C., Vitale U., Ciardelli G., Barbani N., Vozzi G., Giusti P., New biomedical devices with selective peptide recognition properties. Part 1: Characterization and cytotoxicity of molecularly imprinted polymers. J. Cell. Mol. Med. 11, 1367–1376 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosellini E., Barbani N., Giusti P., Ciardelli G., Cristallini C., Molecularly imprinted nanoparticles with recognition properties towards a laminin H–Tyr–Ile–Gly–Ser–Arg–OH sequence for tissue engineering applications. Biomed. Mater. 5, 065007 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Hoshino Y., Urakami T., Kodama T., Koide H., Oku N., Okahata Y., Shea K. J., Design of synthetic polymer nanoparticles that capture and neutralize a toxic peptide. Small 5, 1562–1568 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoshino Y., Koide H., Urakami T., Kanazawa H., Kodama T., Oku N., Shea K. J., Recognition, neutralization, and clearance of target peptides in the bloodstream of living mice by molecularly imprinted polymer nanoparticles: A plastic antibody. J. Am. Chem. Soc. 132, 6644–6645 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang H., Molecularly imprinted nanoparticles for biomedical applications. Adv. Mater. 32, 1806328 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Patel K. D., Kim H., Knowles J. C., Poma A., Molecularly imprinted polymers and electrospinning: Manufacturing convergence for next-level applications. Adv. Funct. Mater. 30, 2001955 (2020). [Google Scholar]
- 65.BelBruno J. J., Molecularly imprinted polymers. Chem. Rev. 119, 94–119 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Haupt K., Medina Rangel P. X., Bui B. T. S., Molecularly imprinted polymers: Antibody mimics for bioimaging and therapy. Chem. Rev. 120, 9554–9582 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Bagán H., Zhou T., Eriksson N. L., Bülow L., Ye L., Synthesis and characterization of epitope-imprinted polymers for purification of human hemoglobin. RSC Adv. 7, 41705–41712 (2017). [Google Scholar]
- 68.Li H., Li D., Preparation of a pipette tip-based molecularly imprinted solid-phase microextraction monolith by epitope approach and its application for determination of enkephalins in human cerebrospinal fluid. J. Pharm. Biomed. Anal. 115, 330–338 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Pan G., Shinde S., Yeung S. Y., Jakštaitė M., Li Q., Wingren A. G., Sellergren B., An epitope-imprinted biointerface with dynamic bioactivity for modulating cell–biomaterial interactions. Angew. Chemie Int. Ed. 56, 15959–15963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang G., Jiang L., Zhou J., Hu L., Feng S., Epitope-imprinted mesoporous silica nanoparticles for specific recognition of tyrosine phosphorylation. Chem. Commun. 55, 9927–9930 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y., Deng C., Liu S., Wu J., Chen Z., Li C., Lu W., Active targeting of tumors through conformational epitope imprinting. Angew. Chemie Int. Ed. 54, 5157–5160 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Palladino P., Minunni M., Scarano S., Cardiac troponin T capture and detection in real-time via epitope-imprinted polymer and optical biosensing. Biosens. Bioelectron. 106, 93–98 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Cecchini A., Raffa V., Canfarotta F., Signore G., Piletsky S., Macdonald M. P., Cuschieri A., In vivo recognition of human vascular endothelial growth factor by molecularly imprinted polymers. Nano Lett. 17, 2307–2312 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Ji X., Li D., Li H., Preparation and application of a novel molecularly imprinted solid-phase microextraction monolith for selective enrichment of cholecystokinin neuropeptides in human cerebrospinal fluid. Biomed. Chromatogr. 29, 1280–1289 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Yang Y. Q., He X. W., Wang Y. Z., Li W. Y., Zhang Y. K., Epitope imprinted polymer coating CdTe quantum dots for specific recognition and direct fluorescent quantification of the target protein bovine serum albumin. Biosens. Bioelectron. 54, 266–272 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Li D. Y., Zhang X. M., Yan Y. J., He X. W., Li W. Y., Zhang Y. K., Thermo-sensitive imprinted polymer embedded carbon dots using epitope approach. Biosens. Bioelectron. 79, 187–192 (2016). [DOI] [PubMed] [Google Scholar]
- 77.Canfarotta F., Lezina L., Guerreiro A., Czulak J., Petukhov A., Daks A., Smolinska-Kempisty K., Poma A., Piletsky S., Barlev N. A., Specific drug delivery to cancer cells with double-imprinted nanoparticles against epidermal growth factor receptor. Nano Lett. 18, 4641–4646 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Castell O. K., Barrow D. A., Kamarudin A. R., Allender C. J., Current practices for describing the performance of molecularly imprinted polymers can be misleading and may be hampering the development of the field. J. Mol. Recognit. 24, 1115–1122 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Rushton G. T., Karns C. L., Shimizu K. D., A critical examination of the use of the Freundlich isotherm in characterizing molecularly imprinted polymers (MIPs). Anal. Chim. Acta 528, 107–113 (2005). [Google Scholar]
- 80.R. J. Ansell, in Advances in Biochemical Engineering/Biotechnology (Springer Science and Business Media Deutschland GmbH, 2015), vol. 150, pp. 51–93. [Google Scholar]
- 81.Qin Y.-P., Jia C., He X.-W., Li W.-Y., Zhang Y.-K., Thermosensitive metal chelation dual-template epitope imprinting polymer using distillation–precipitation polymerization for simultaneous recognition of human serum albumin and transferrin. ACS Appl. Mater. Interfaces 10, 9060–9068 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Qin Y. P., Li D. Y., He X. W., Li W. Y., Zhang Y. K., Preparation of high-efficiency cytochrome c-imprinted polymer on the surface of magnetic carbon nanotubes by epitope approach via metal chelation and six-membered ring. ACS Appl. Mater. Interfaces 8, 10155–10163 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Zhou J., Gan N., Li T., Hu F., Li X., Wang L., Zheng L., A cost-effective sandwich electrochemiluminescence immunosensor for ultrasensitive detection of HIV-1 antibody using magnetic molecularly imprinted polymers as capture probes. Biosens. Bioelectron. 54, 199–206 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Dong Y., Li W., Gu Z., Xing R., Ma Y., Zhang Q., Liu Z., Inhibition of HER2-positive breast cancer growth by blocking the HER2 signaling pathway with HER2-glycan-imprinted nanoparticles. Angew. Chemie Int. Ed. 58, 10621–10625 (2019). [DOI] [PubMed] [Google Scholar]
- 85.Shinde S., El-Schich Z., Malakpour A., Wan W., Dizeyi N., Mohammadi R., Rurack K., Gjörloff Wingren A., Sellergren B., Sialic acid-imprinted fluorescent core–shell particles for selective labeling of cell surface glycans. J. Am. Chem. Soc. 137, 13908–13912 (2015). [DOI] [PubMed] [Google Scholar]
- 86.Kunath S., Panagiotopoulou M., Maximilien J., Marchyk N., Sänger J., Haupt K., Cell and tissue imaging with molecularly imprinted polymers as plastic antibody mimics. Adv. Healthc. Mater. 4, 1322–1326 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Ertürk G., Mattiasson B., Molecular imprinting techniques used for the preparation of biosensors. Sensors 17, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Y. Z., Li D. Y., He X. W., Li W. Y., Zhang Y. K., Epitope imprinted polymer nanoparticles containing fluorescent quantum dots for specific recognition of human serum albumin. Microchim. Acta. 182, 1465–1472 (2015). [Google Scholar]
- 89.Drzazgowska J., Schmid B., Süssmuth R. D., Altintas Z., Self-assembled monolayer epitope bridges for molecular imprinting and cancer biomarker sensing. Anal. Chem. 92, 4798–4806 (2020). [DOI] [PubMed] [Google Scholar]
- 90.Urraca J. L., Aureliano C. S. A., Schillinger E., Esselmann H., Wiltfang J., Sellergren B., Polymeric complements to the Alzheimer’s disease biomarker β-amyloid isoforms Aβ1–40 and Aβ1–42 for blood serum analysis under denaturing conditions. J. Am. Chem. Soc. 133, 9220–9223 (2011). [DOI] [PubMed] [Google Scholar]
- 91.Tchinda R., Tutsch A., Schmid B., Süssmuth R. D., Altintas Z., Recognition of protein biomarkers using epitope-mediated molecularly imprinted films: Histidine or cysteine modified epitopes? Biosens. Bioelectron. 123, 260–268 (2019). [DOI] [PubMed] [Google Scholar]
- 92.Han J., Sun Y., Hou J., Wang Y., Liu Y., Xie C., Lu W., Pan J., Preliminary investigations into surface molecularly imprinted nanoparticles for Helicobacter pylori eradication. Acta Pharm. Sin. B. 5, 577–582 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tan L., Yu Z., Zhou X., Xing D., Luo X., Peng R., Tang Y., Antibody-free ultra-high performance liquid chromatography/tandem mass spectrometry measurement of angiotensin I and II using magnetic epitope-imprinted polymers. J. Chromatogr. A 1411, 69–76 (2015). [DOI] [PubMed] [Google Scholar]
- 94.Liu S., Bi Q., Long Y., Li Z., Bhattacharyya S., Li C., Inducible epitope imprinting: “Generating” the required binding site in membrane receptors for targeted drug delivery. Nanoscale 9, 5394–5397 (2017). [DOI] [PubMed] [Google Scholar]
- 95.Zhang Y., Li S., Ma X. T., He X. W., Li W. Y., Zhang Y. K., Carbon dots-embedded epitope imprinted polymer for targeted fluorescence imaging of cervical cancer via recognition of epidermal growth factor receptor. Microchim. Acta 187, 228 (2020). [DOI] [PubMed] [Google Scholar]
- 96.Qin Y.-T., Feng Y.-S., Ma Y.-J., He X.-W., Li W.-Y., Zhang Y., Tumor-sensitive biodegradable nanoparticles of molecularly imprinted polymer-stabilized fluorescent zeolitic imidazolate framework-8 for targeted imaging and drug delivery. ACS Appl. Mater. Interfaces 12, 24585–24598 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Qin Y. P., Wang H. Y., He X. W., Li W. Y., Zhang Y. K., Metal chelation dual-template epitope imprinting polymer via distillation-precipitation polymerization for recognition of porcine serum albumin. Talanta 185, 620–627 (2018). [DOI] [PubMed] [Google Scholar]
- 98.Tai D. F., Jhang M. H., Chen G. Y., Wang S. C., Lu K. H., Der Lee Y., Liu H. T., Epitope-cavities generated by molecularly imprinted films measure the coincident response to anthrax protective antigen and its segments. Anal. Chem. 82, 2290–2293 (2010). [DOI] [PubMed] [Google Scholar]
- 99.Lee M. H., Thomas J. L., Liao C. L., Jurcevic S., Crnogorac-Jurcevic T., Lin H. Y., Epitope recognition of peptide-imprinted polymers for Regenerating protein 1 (REG1). Sep. Purif. Technol. 192, 213–219 (2018). [Google Scholar]
- 100.Li Z., Guan P., Hu X., Ding S., Tian Y., Xu Y., Qian L., Preparation of molecularly imprinted mesoporous materials for highly enhancing adsorption performance of cytochrome C. Polymers 10, 298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang X., Zhang N., Du C., Guan P., Gao X., Wang C., Du Y., Ding S., Hu X., Preparation of magnetic epitope imprinted polymer microspheres using cyclodextrin-based ionic liquids as functional monomer for highly selective and effective enrichment of cytochrome c. Chem. Eng. J. 317, 988–998 (2017). [Google Scholar]
- 102.Ma X. T., He X. W., Li W. Y., Zhang Y. K., Epitope molecularly imprinted polymer coated quartz crystal microbalance sensor for the determination of human serum albumin. Sens. Actuators B 246, 879–886 (2017). [Google Scholar]
- 103.Li M. X., Wang X. H., Zhang L. M., Wei X. P., A high sensitive epitope imprinted electrochemical sensor for bovine serum albumin based on enzyme amplifying. Anal. Biochem. 530, 68–74 (2017). [DOI] [PubMed] [Google Scholar]
- 104.Lee M. H., Thomas J. L., Liao C. L., Jurcevic S., Crnogorac-Jurcevic T., Lin H. Y., Polymers imprinted with three REG1B peptides for electrochemical determination of regenerating protein 1B, a urinary biomarker for pancreatic ductal adenocarcinoma. Microchim. Acta. 184, 1773–1780 (2017). [Google Scholar]
- 105.Tan L., He R., Li Y., Liang Y., Li H., Tang Y., Fabrication of a biomimetic adsorbent imprinted with a common specificity determinant for the removal of α- and β-amanitin from plasma. J. Chromatogr. A 1459, 1–8 (2016). [DOI] [PubMed] [Google Scholar]
- 106.Schwark S., Sun W., Stute J., Lütkemeyer D., Ulbricht M., Sellergren B., Monoclonal antibody capture from cell culture supernatants using epitope imprinted macroporous membranes. RSC Adv. 6, 53162–53169 (2016). [Google Scholar]
- 107.Lin C. Y., Tsai S. H., Tai D. F., Detection of oxytocin, atrial natriuretic peptide, and brain natriuretic peptide using novel imprinted polymers produced with amphiphilic monomers. J. Pept. Sci. 25, e3150 (2019). [DOI] [PubMed] [Google Scholar]
- 108.Lu C. H., Zhang Y., Tang S. F., Bin Fang Z., Yang H. H., Chen X., Chen G. N., Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens. Bioelectron. 31, 439–444 (2012). [DOI] [PubMed] [Google Scholar]
- 109.Kushwaha A., Srivastava J., Singh A. K., Anand R., Raghuwanshi R., Rai T., Singh M., Epitope imprinting of Mycobacterium leprae bacteria via molecularly imprinted nanoparticles using multiple monomers approach. Biosens. Bioelectron. 145, 111698 (2019). [DOI] [PubMed] [Google Scholar]
- 110.Martins M. V. S. B., da S. Guimarães M. M., Spencer J. S., Hacker M. A. V. B., Costa L. S., Carvalho F. M., Geluk A., van der Ploeg-van Schip J. J., Pontes M. A. A., Gonçalves H. S., de Morais J. P., Bandeira T. J. P. G., Pessolani M. C. V., Brennan P. J., Pereira G. M. B., Pathogen-specific epitopes as epidemiological tools for defining the magnitude of Mycobacterium leprae transmission in areas endemic for leprosy. PLOS Negl. Trop. Dis. 6, e1616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gupta N., Shah K., Singh M., An epitope-imprinted piezoelectric diagnostic tool for Neisseria meningitidis detection. J. Mol. Recognit. 29, 572–579 (2016). [DOI] [PubMed] [Google Scholar]
- 112.Gupta N., Singh R. S., Shah K., Prasad R., Singh M., Epitope imprinting of iron binding protein of Neisseria meningitidis bacteria through multiple monomers imprinting approach. J. Mol. Recognit. 31, e2709 (2018). [DOI] [PubMed] [Google Scholar]
- 113.Altintas Z., Takiden A., Utesch T., Mroginski M. A., Schmid B., Scheller F. W., Süssmuth R. D., Integrated approaches toward high-affinity artificial protein binders obtained via computationally simulated epitopes for protein recognition. Adv. Funct. Mater. 29, 1807332 (2019). [Google Scholar]
- 114.Liang Y., Guttman M., Davenport T. M., Hu S. L., Lee K. K., Probing the impact of local structural dynamics of conformational epitopes on antibody recognition. Biochemistry 55, 2197–2213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.C. J. Burrell, C. R. Howard, F. A. Murphy, Fenner and White’s Medical Virology 5th Edition, C. J. Burrell, C. R. Howard, F. A. Murphy, Eds. (Academic Press, ed. 5, 2016), pp. 65–76. [Google Scholar]
- 116.Cenci L., Guella G., Andreetto E., Ambrosi E., Anesi A., Bossi A. M., Guided folding takes a start from the molecular imprinting of structured epitopes. Nanoscale 8, 15665–15670 (2016). [DOI] [PubMed] [Google Scholar]
- 117.Xu J., Merlier F., Avalle B., Vieillard V., Debré P., Haupt K., Tse B., Bui S., Molecularly imprinted polymer nanoparticles as potential synthetic antibodies for immunoprotection against HIV. ACS Appl. Mater. Interf. 11, 9824–9831 (2019). [DOI] [PubMed] [Google Scholar]
- 118.Hashemi-Moghaddam H., Zavareh S., Karimpour S., Madanchi H., Evaluation of molecularly imprinted polymer based on HER2 epitope for targeted drug delivery in ovarian cancer mouse model. React. Funct. Polym. 121, 82–90 (2017). [Google Scholar]
- 119.Vita R., Mahajan S., Overton J. A., Dhanda S. K., Martini S., Cantrell J. R., Wheeler D. K., Sette A., Peters B., The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 47, D339–D343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.The Immune Epitope Database (IEDB); available at www.iedb.org.
- 121.The PyMOL Molecular Graphics System, Schrödinger, LLC.
- 122.Ponomarenko J., Bui H., Li W., Fusseder N., Bourne P. E., Sette A., Peters B., ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics 9, 514–521 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.ElliPro: Antibody Epitope Prediction, available at http://tools.iedb.org/ellipro/.
- 124.U.S. National Library of Medicine, National Center for Biotechnology Information, BLASTP.
- 125.Li C., Pazgier M., Liu M., Lu W.-Y., Lu W., Apamin as a template for structure-based rational design of potent peptide activators of p53. Angew. Chemie Int. Ed. 48, 8712–8715 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E., The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.RCSB Protein Data Bank, available at www.rcsb.org.
- 128.Lamiable A., Thévenet P., Rey J., Vavrusa M., Derreumaux P., Tufféry P., PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 44, W449–W454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.PEP-FOLD 3. De novo peptide structure prediction, available at https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/. [DOI] [PMC free article] [PubMed]
- 130.Zeng Z., Hoshino Y., Rodriguez A., Yoo H., Shea K. J., Synthetic polymer nanoparticles with antibody-like affinity for a hydrophilic peptide. ACS Nano 4, 199–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bossi A. M., Sharma P. S., Montana L., Zoccatelli G., Laub O., Levi R., Fingerprint-imprinted polymer: Rational selection of peptide epitope templates for the determination of proteins by molecularly imprinted polymers. Anal. Chem. 84, 4036–4041 (2012). [DOI] [PubMed] [Google Scholar]
- 132.NCBI Protein, available at https://www.ncbi.nlm.nih.gov/protein/.
- 133.UniProt, available at https://www.uniprot.org/.
- 134.E. Gasteiger, C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, A. Bairoch, The Proteomics Protocols Handbook. Springer Protocols Handbooks, John M. Walker, Ed. (Humana Press, 2005), pp. 571–607. [Google Scholar]
- 135.ExPASy PeptideCutter, available at https://web.expasy.org/peptide_cutter/.
- 136.Mol* Viewer, available at https://molstar.org/viewer/.
- 137.Xing R., Wang S., Bie Z., He H., Liu Z., Preparation of molecularly imprinted polymers specific to glycoproteins, glycans and monosaccharides via boronate affinity controllable–oriented surface imprinting. Nat. Protoc. 12, 964–987 (2017). [DOI] [PubMed] [Google Scholar]
- 138.V. W. Rodwell, D. A. Bender, K. M. Botham, P. J. Kennelly, P. A. Weil, Harper’s Illustrated Biochemistry (McGraw-Hill Education, ed. 31, 2018). [Google Scholar]
- 139.Bie Z., Chen Y., Ye J., Wang S., Liu Z., Boronate-affinity glycan-oriented surface imprinting: A new strategy to mimic lectins for the recognition of an intact glycoprotein and its characteristic fragments. Angew. Chemie Int. Ed. 54, 10211–10215 (2015). [DOI] [PubMed] [Google Scholar]
- 140.Qader A. A., Urraca J., Torsetnes S. B., Tønnesen F., Reubsaet L., Sellergren B., Peptide imprinted receptors for the determination of the small cell lung cancer associated biomarker progastrin releasing peptide. J. Chromatogr. A 1370, 56–62 (2014). [DOI] [PubMed] [Google Scholar]
- 141.Rossetti C., Abdel Qader A., Halvorsen T. G., Sellergren B., Reubsaet L., Antibody-free biomarker determination: Exploring molecularly imprinted polymers for pro-gastrin releasing peptide. Anal. Chem. 86, 12291–12298 (2014). [DOI] [PubMed] [Google Scholar]
- 142.Çulha S., Armutcu C., Uzun L., Şenel S., Denizli A., Synthesis of L-lysine imprinted cryogels for immunoglobulin G adsorption. Mater. Sci. Eng. C 52, 315–324 (2015). [DOI] [PubMed] [Google Scholar]
- 143.Emgenbroich M., Borrelli C., Shinde S., Lazraq I., Vilela F., Hall A. J., Oxelbark J., De Lorenzi E., Courtois J., Simanova A., Verhage J., Irgum K., Karim K., Sellergren B., A phosphotyrosine-imprinted polymer receptor for the recognition of tyrosine phosphorylated peptides. Chem. A Eur. J. 14, 9516–9529 (2008). [DOI] [PubMed] [Google Scholar]
- 144.Helling S., Shinde S., Brosseron F., Schnabel A., Müller T., Meyer H. E., Marcus K., Sellergren B., Ultratrace enrichment of tyrosine phosphorylated peptides on an imprinted polymer. Anal. Chem. 83, 1862–1865 (2011). [DOI] [PubMed] [Google Scholar]
- 145.Chen J., Shinde S., Subedi P., Wierzbicka C., Sellergren B., Helling S., Marcus K., Validation of molecularly imprinted polymers for side chain selective phosphopeptide enrichment. J. Chromatogr. A 1471, 45–50 (2016). [DOI] [PubMed] [Google Scholar]
- 146.Bllaci L., Torsetnes S. B., Wierzbicka C., Shinde S., Sellergren B., Rogowska-Wrzesinska A., Jensen O. N., Phosphotyrosine biased enrichment of tryptic peptides from cancer cells by combining pY-MIP and TiO2 affinity resins. Anal. Chem. 89, 11332–11340 (2017). [DOI] [PubMed] [Google Scholar]
- 147.Wan W., Descalzo A. B., Shinde S., Weißhoff H., Orellana G., Sellergren B., Rurack K., Ratiometric fluorescence detection of phosphorylated amino acids through excited-state proton transfer by using molecularly imprinted polymer (MIP) recognition nanolayers. Chem. - A Eur. J. 23, 15974–15983 (2017). [DOI] [PubMed] [Google Scholar]
- 148.Wierzbicka C., Torsetnes S. B., Jensen O. N., Shinde S., Sellergren B., Hierarchically templated beads with tailored pore structure for phosphopeptide capture and phosphoproteomics. RSC Adv. 7, 17154–17163 (2017). [Google Scholar]
- 149.Wierzbicka C., Liu M., Bauer D., Irgum K., Sellergren B., Cationic pTyr/pSer imprinted polymers based on a bis-imidazolium host monomer: Phosphopeptide recognition in aqueous buffers demonstrated by μ-liquid chromatography and monolithic columns. J. Mater. Chem. B 5, 953–960 (2017). [DOI] [PubMed] [Google Scholar]
- 150.Liu M., Tran T. M., Abbas Elhaj A. A., Bøen Torsetnes S., Jensen O. N., Sellergren B., Irgum K., Molecularly imprinted porous monolithic materials from melamine-formaldehyde for selective trapping of phosphopeptides. Anal. Chem. 89, 9491–9501 (2017). [DOI] [PubMed] [Google Scholar]
- 151.Liu M., Torsetnes S. B., Wierzbicka C., Jensen O. N., Sellergren B., Irgum K., Selective enrichment of phosphorylated peptides by monolithic polymers surface imprinted with bis-imidazolium moieties by UV-initiated cryopolymerization. Anal. Chem. 91, 10188–10196 (2019). [DOI] [PubMed] [Google Scholar]
- 152.Incel A., Arribas Díez I., Wierzbicka C., Gajoch K., Jensen O. N., Sellergren B., Selective enrichment of histidine phosphorylated peptides using molecularly imprinted polymers. Anal. Chem. 93, 3857–3866 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xu L., Hu Y., Shen F., Li Q., Ren X., Specific recognition of tyrosine-phosphorylated peptides by epitope imprinting of phenylphosphonic acid. J. Chromatogr. A 1293, 85–91 (2013). [DOI] [PubMed] [Google Scholar]
- 154.Li Q., Shen F., Zhang X., Hu Y., Zhang Q., Xu L., Ren X., One-pot synthesis of phenylphosphonic acid imprinted polymers for tyrosine phosphopeptides recognition in aqueous phase. Anal. Chim. Acta 795, 82–87 (2013). [DOI] [PubMed] [Google Scholar]
- 155.Li D. Y., Qin Y. P., Li H. Y., He X. W., Li W. Y., Zhang Y. K., A “turn-on” fluorescent receptor for detecting tyrosine phosphopeptide using the surface imprinting procedure and the epitope approach. Biosens. Bioelectron. 66, 224–230 (2015). [DOI] [PubMed] [Google Scholar]
- 156.Yang X., Xia Y., Selective enrichment and separation of phosphotyrosine peptides by thermosensitive molecularly imprinted polymers. J. Sep. Sci. 39, 419–426 (2016). [DOI] [PubMed] [Google Scholar]
- 157.Panagiotopoulou M., Salinas Y., Beyazit S., Kunath S., Duma L., Prost E., Mayes A. G., Resmini M., Bui B. T. S., Haupt K., Molecularly imprinted polymer coated quantum dots for multiplexed cell targeting and imaging. Angew. Chemie Int. Ed. 55, 8244–8248 (2016). [DOI] [PubMed] [Google Scholar]
- 158.Panagiotopoulou M., Kunath S., Medina-Rangel P. X., Haupt K., Bui B. T. S., Fluorescent molecularly imprinted polymers as plastic antibodies for selective labeling and imaging of hyaluronan and sialic acid on fixed and living cells. Biosens. Bioelectron. 88, 85–93 (2017). [DOI] [PubMed] [Google Scholar]
- 159.Medina Rangel P. X., Laclef S., Xu J., Panagiotopoulou M., Kovensky J., Bui B. T. S., Haupt K., Solid-phase synthesis of molecularly imprinted polymer nanolabels: Affinity tools for cellular bioimaging of glycans. Sci. Rep. 9, 3923 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chen K., He R., Luo X., Qin P., Tan L., Tang Y., Yang Z., A fluorescent glycosyl-imprinted polymer for pH and temperature regulated sensing of target glycopeptide antibiotic. Biosens. Bioelectron. 94, 609–615 (2017). [DOI] [PubMed] [Google Scholar]
- 161.Tan L., Li Y., Pan X., Marina M. L., Jiang Z., Boronate affinity glycosyl molecularly imprinted polymer microspheres for the determination of teicoplanin using ultra-high performance liquid chromatography coupled with tandem mass spectrometry. J. Chromatogr. A 1615, 460776 (2020). [DOI] [PubMed] [Google Scholar]
- 162.Bedwell T. S., Anjum N., Ma Y., Czulak J., Poma A., Piletska E., Whitcombe M. J., Piletsky S. A., New protocol for optimisation of polymer composition for imprinting of peptides and proteins. RSC Adv. 9, 27849–27855 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Singh L. K., Singh M., Singh M., Biopolymeric receptor for peptide recognition by molecular imprinting approach‑—Synthesis, characterization and application. Mater. Sci. Eng. C 45, 383–394 (2014). [DOI] [PubMed] [Google Scholar]
- 164.Wendler K., Thar J., Zahn S., Kirchner B., Estimating the hydrogen bond energy. J. Phys. Chem. A 114, 9529–9536 (2010). [DOI] [PubMed] [Google Scholar]
- 165.T. Ohara, T. Sato, N. Shimizu, G. Prescher, H. Schwind, O. Weiberg, K. Marten, H. Greim, Ullmann’s Encyclopedia of Industrial Chemistry (Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, 2011). [Google Scholar]
- 166.Hoshino Y., Kodama T., Okahata Y., Shea K. J., Peptide imprinted polymer nanoparticles: A plastic antibody. J. Am. Chem. Soc. 130, 15242–15243 (2008). [DOI] [PubMed] [Google Scholar]
- 167.Canfarotta F., Czulak J., Betlem K., Sachdeva A., Eersels K., Van Grinsven B., Cleij T. J., Peeters M., A novel thermal detection method based on molecularly imprinted nanoparticles as recognition elements. Nanoscale 10, 2081–2089 (2018). [DOI] [PubMed] [Google Scholar]
- 168.Moczko E., Guerreiro A., Cáceres C., Piletska E., Sellergren B., Piletsky S. A., Epitope approach in molecular imprinting of antibodies. J. Chromatogr. B 1124, 1–6 (2019). [DOI] [PubMed] [Google Scholar]
- 169.Ekpenyong-Akiba A. E., Canfarotta F., Abd H B., Poblocka M., Casulleras M., Castilla-Vallmanya L., Kocsis-Fodor G., Kelly M. E., Janus J., Althubiti M., Piletska E., Piletsky S., Macip S., Detecting and targeting senescent cells using molecularly imprinted nanoparticles. Nanoscale Horiz. 4, 757–768 (2019). [Google Scholar]
- 170.Jia C., Zhang M., Zhang Y., Ma Z. B., Xiao N. N., He X. W., Li W. Y., Zhang Y. K., Preparation of dual-template epitope imprinted polymers for targeted fluorescence imaging and targeted drug delivery to pancreatic cancer BxPC-3 cells. ACS Appl. Mater. Interfaces 11, 32431–32440 (2019). [DOI] [PubMed] [Google Scholar]
- 171.Wang H. Y., Cao P. P., He Z. Y., He X. W., Li W. Y., Li Y. H., Zhang Y. K., Targeted imaging and targeted therapy of breast cancer cells: Via fluorescent double template-imprinted polymer coated silicon nanoparticles by an epitope approach. Nanoscale 11, 17018–17030 (2019). [DOI] [PubMed] [Google Scholar]
- 172.Xing R., Ma Y., Wang Y., Wen Y., Liu Z., Specific recognition of proteins and peptides via controllable oriented surface imprinting of boronate affinity-anchored epitopes. Chem. Sci. 10, 1831–1835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Li T., Shi S., Goel S., Shen X., Xie X., Chen Z., Zhang H., Li S., Qin X., Yang H., Wu C., Liu Y., Recent advancements in mesoporous silica nanoparticles towards therapeutic applications for cancer. Acta Biomater. 89, 1–13 (2019). [DOI] [PubMed] [Google Scholar]
- 174.Zhao Z., Fang R., Rong Q., Liu M., Bioinspired nanocomposite hydrogels with highly ordered structures. Adv. Mater. 29, 1703045 (2017). [DOI] [PubMed] [Google Scholar]
- 175.Clegg J. R., Wechsler M. E., Peppas N. A., Vision for functionally decorated and molecularly imprinted polymers in regenerative engineering. Regen. Eng. Transl. Med. 3, 166–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Altintas Z., Abdin M. J., Tothill A. M., Karim K., Tothill I. E., Ultrasensitive detection of endotoxins using computationally designed nanoMIPs. Anal. Chim. Acta 935, 239–248 (2016). [DOI] [PubMed] [Google Scholar]
- 177.Bates F., Cela-Pérez M. C., Karim K., Piletsky S., López-Vilariño J. M., Virtual screening of receptor sites for molecularly imprinted polymers. Macromol. Biosci. 16, 1170–1174 (2016). [DOI] [PubMed] [Google Scholar]
- 178.Bates F., Busato M., Piletska E., Whitcombe M. J., Karim K., Guerreiro A., del Valle M., Giorgetti A., Piletsky S., Computational design of molecularly imprinted polymer for direct detection of melamine in milk. Sep. Sci. Technol. 52, 1441–1453 (2017). [Google Scholar]
- 179.Busato M., Distefano R., Bates F., Karim K., Bossi A. M., López Vilariño J. M., Piletsky S., Bombieri N., Giorgetti A., MIRATE: MIps RAtional dEsign science gateway. J. Integr. Bioinform. 15, 10.1515/jib-2017-0075, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Karim K., Cowen T., Guerreiro A., Piletska E., Whitcombe M., A protocol for the computational design of high affinity molecularly imprinted polymer synthetic receptors. Glob. J. Biotechnol. Biomater. Sci. 3, 001–007 (2017). [Google Scholar]
- 181.Wren S. P., Piletsky S. A., Karim K., Gascoine P., Lacey R., Sun T., Grattan K. T. V., Computational design and fabrication of optical fibre fluorescent chemical probes for the detection of cocaine. J. Light. Technol. 33, 2572–2579 (2015). [Google Scholar]
- 182.Viveiros R., Karim K., Piletsky S. A., Heggie W., Casimiro T., Development of a molecularly imprinted polymer for a pharmaceutical impurity in supercritical CO2: Rational design using computational approach. J. Clean. Prod. 168, 1025–1031 (2017). [Google Scholar]
- 183.Boroznjak R., Reut J., Tretjakov A., Lomaka A., Öpik A., Syritski V., A computational approach to study functional monomer-protein molecular interactions to optimize protein molecular imprinting. J. Mol. Recognit. 30, e2635 (2017). [DOI] [PubMed] [Google Scholar]
- 184.T. Cowen, K. Karim, S. A. Piletsky, Smart Polymer Catalysts and Tunable Catalysis (Elsevier, 2019), pp. 51–75. [Google Scholar]
- 185.Iskierko Z., Sharma P. S., Noworyta K. R., Borowicz P., Cieplak M., Kutner W., Bossi A. M., Selective PQQPFPQQ gluten epitope chemical sensor with a molecularly imprinted polymer recognition unit and an extended-gate field-effect transistor transduction unit. Anal. Chem. 91, 4537–4543 (2019). [DOI] [PubMed] [Google Scholar]
- 186.M. F. Brigatti, A. Mottana, Layered Mineral Structures and their Application in Advanced Technologies (European Mineralogical Union and the Mineralogical Society of Great Britain and Ireland, London, UK, 2011).
- 187.Wren S. P., Piletsky S. A., Karim K., Gascoine P., Lacey R., Sun T., Grattan K. T. V., Design and synthesis of a fluorescent molecular imprinted polymer for use in an optical fibre-based cocaine sensor. 23rd Int. Conf. Opt. Fibre Sens. 9157, 91574W (2014). [Google Scholar]
- 188.Piletska E. V., Pink D., Karim K., Piletsky S. A., Development of a computationally-designed polymeric adsorbent specific for mycotoxin patulin. Analyst 142, 4678–4683 (2017). [DOI] [PubMed] [Google Scholar]
- 189.Zadok I., Srebnik S., Coarse-grained simulation of protein-imprinted hydrogels. J. Phys. Chem. B 122, 7091–7101 (2018). [DOI] [PubMed] [Google Scholar]
- 190.Cenci L., Bertolla M., Anesi A., Ambrosi E., Guella G., Bossi A. M., Micro- versus nano-sized molecularly imprinted polymers in MALDI-TOF mass spectrometry analysis of peptides. Anal. Bioanal. Chem. 409, 6253–6261 (2017). [DOI] [PubMed] [Google Scholar]
- 191.Mujahid A., Iqbal N., Afzal A., Bioimprinting strategies: From soft lithography to biomimetic sensors and beyond. Biotechnol. Adv. 31, 1435–1447 (2013). [DOI] [PubMed] [Google Scholar]
- 192.Yang K., Li S., Liu L., Chen Y., Zhou W., Pei J., Liang Z., Zhang L., Zhang Y., Epitope imprinting technology: Progress, applications, and perspectives toward artificial antibodies. Adv. Mater. 31, 1902048 (2019). [DOI] [PubMed] [Google Scholar]
- 193.Bossi A., Piletsky S. A., Piletska E. V., Righetti P. G., Turner A. P. F., Surface-grafted molecularly imprinted polymers for protein recognition. Anal. Chem. 73, 5281–5286 (2001). [DOI] [PubMed] [Google Scholar]
- 194.Yilmaz E., Haupt K., Mosbach K., The use of immobilized templates—A new approach in molecular imprinting. Angew. Chemie Int. Ed. 39, 2115–2118 (2000). [DOI] [PubMed] [Google Scholar]
- 195.Shi H., Tsal W. B., Garrison M. D., Ferrari S., Ratner B. D., Template-imprinted nanostructured surfaces for protein recognition. Nature 398, 593–597 (1999). [DOI] [PubMed] [Google Scholar]
- 196.Schulz A., Jankowski J., Zidek W., Jankowski V., Absolute quantification of endogenous angiotensin II levels in human plasma using ESI-LC-MS/MS. Clin. Proteomics 11, 37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zhao C. J., Ma X. H., Li J. P., An insulin molecularly imprinted electrochemical sensor based on epitope imprinting. Chinese J. Anal. Chem. 45, 1360–1366 (2017). [Google Scholar]
- 198.Qin Y.-T., Peng H., He X.-W., Li W.-Y., Zhang Y.-K., Highly effective drug delivery and cell imaging using fluorescent double-imprinted nanoparticles by targeting recognition of the epitope of membrane protein. Anal. Chem. 91, 12696–12703 (2019). [DOI] [PubMed] [Google Scholar]
- 199.Peng H., Qin Y.-T., He X.-W., Li W.-Y., Zhang Y.-K., Epitope molecularly imprinted polymer nanoparticles for chemo-/photodynamic synergistic cancer therapy guided by targeted fluorescence imaging. ACS Appl. Mater. Interfaces 12, 13360–13370 (2020). [DOI] [PubMed] [Google Scholar]
- 200.Costa A. F., Campos D., Reis C. A., Gomes C., Targeting glycosylation: A new road for cancer drug discovery. Trends Cancer. 6, 757–766 (2020). [DOI] [PubMed] [Google Scholar]
- 201.Mereiter S., Balmaña M., Campos D., Gomes J., Reis C. A., Glycosylation in the era of cancer-targeted therapy: Where are we heading? Cancer Cell 36, 6–16 (2019). [DOI] [PubMed] [Google Scholar]
- 202.Fa S., Zhao Y., Water-soluble nanoparticle receptors supramolecularly coded for acidic peptides. Chem. A Eur. J. 24, 150–158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Awino J. K., Gunasekara R. W., Zhao Y., Sequence-selective binding of oligopeptides in water through hydrophobic coding. J. Am. Chem. Soc. 139, 2188–2191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Fa S., Zhao Y., General method for peptide recognition in water through bioinspired complementarity. Chem. Mater. 31, 4889–4896 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zangiabadi M., Zhao Y., Molecularly imprinted polymeric receptors with interfacial hydrogen bonds for peptide recognition in water. ACS Appl. Polym. Mater. 2, 3171–3180 (2020). [Google Scholar]
- 206.Li X., Chen K., Zhao Y., Sequence-selective protection of peptides from proteolysis. Angew. Chemie Int. Ed. 60, 11092–11097 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Li X., Palhano Zanela T. M., Underbakke E. S., Zhao Y., Controlling kinase activities by selective inhibition of peptide substrates. J. Am. Chem. Soc. 143, 639–643 (2021). [DOI] [PubMed] [Google Scholar]
- 208.Li X., Zhao Y., Synthetic glycosidases for the precise hydrolysis of oligosaccharides and polysaccharides. Chem. Sci. 12, 374–383 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Li X., Zangiabadi M., Zhao Y., Molecularly imprinted synthetic glucosidase for the hydrolysis of cellulose in aqueous and nonaqueous solutions. J. Am. Chem. Soc. 143, 5172–5181 (2021). [DOI] [PubMed] [Google Scholar]
- 210.Poma A., Guerreiro A., Whitcombe M. J., Piletska E. V., Turner A. P. F., Piletsky S. A., Solid-phase synthesis of molecularly imprinted polymer nanoparticles with a reusable template—“Plastic antibodies”. Adv. Funct. Mater. 23, 2821–2827 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ambrosini S., Beyazit S., Haupt K., Bui B. T. S., Solid-phase synthesis of molecularly imprinted nanoparticles for protein recognition. Chem. Commun. 49, 6746 (2013). [DOI] [PubMed] [Google Scholar]
- 212.Medina Rangel P. X., Moroni E., Merlier F., Gheber L. A., Vago R., Bui B. T. S., Haupt K., Chemical antibody mimics inhibit cadherin-mediated cell–cell adhesion: A promising strategy for cancer therapy. Angew. Chemie Int. Ed. 59, 2816–2822 (2020). [DOI] [PubMed] [Google Scholar]
- 213.Poma A., Guerreiro A., Caygill S., Moczko E., Piletsky S., Automatic reactor for solid-phase synthesis of molecularly imprinted polymeric nanoparticles (MIP NPs) in water. RSC Adv. 4, 4203–4206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Berghaus M., Mohammadi R., Sellergren B., Productive encounter: Molecularly imprinted nanoparticles prepared using magnetic templates. Chem. Commun. 50, 8993–8996 (2014). [DOI] [PubMed] [Google Scholar]
- 215.Mahajan R., Rouhi M., Shinde S., Bedwell T., Incel A., Mavliutova L., Piletsky S., Nicholls I. A., Sellergren B., Highly efficient synthesis and assay of protein-imprinted nanogels by using magnetic templates. Angew. Chemie Int. Ed. 58, 727–730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.L. Ye, Molecular Imprinting: Principles and Applications of Micro- and Nanostructure Polymers (CRC Press, 2013). [Google Scholar]
- 217.Clegg J. R., Irani A. S., Ander E. W., Ludolph C. M., Venkataraman A. K., Zhong J. X., Peppas N. A., Synthetic networks with tunable responsiveness, biodegradation, and molecular recognition for precision medicine applications. Sci. Adv. 5, eaax7946 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Singh M., Gupta N., Raghuwanshi R., Epitope imprinting approach to monitor diseases. J. Mol. Genet. Med. 11, 2–7 (2017). [Google Scholar]
- 219.M. Panagiotopoulou, S. Kunath, K. Haupt, B. T. S. Bui, Synthetic Antibodies: Methods and Protocols, T. Tiller, Ed. (Springer Science+Business Media LLC, 2017), vol. 1575, pp. 399–415. [Google Scholar]
- 220.Neves M. I., Wechsler M. E., Gomes M. E., Reis R. L., Granja P. L., Peppas N. A., Molecularly imprinted intelligent scaffolds for tissue engineering applications. Tissue Eng. Part B Rev. 23, 27–43 (2017). [DOI] [PubMed] [Google Scholar]
- 221.Turan E., His-tag-epitope imprinted thermoresponsive magnetic nanoparticles for recognition and separation thyroid peroxidase antigens from whole blood samples. ChemistrySelect 3, 11963–11969 (2018). [Google Scholar]
- 222.Ma X., He X., Li W., Zhang Y., Oriented surface epitope imprinted polymer-based quartz crystal microbalance sensor for cytochrome c. Talanta 191, 222–228 (2019). [DOI] [PubMed] [Google Scholar]
- 223.Zhang W., Zhang T., Chen Y., Simultaneous quantification of Cyt c interactions with HSP27 and Bcl-xL using molecularly imprinted polymers (MIPs) coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based targeted proteomics. J. Proteomics 192, 188–195 (2019). [DOI] [PubMed] [Google Scholar]
- 224.Cenci L., Tatti R., Tognato R., Ambrosi E., Piotto C., Bossi A. M., Synthesis and characterization of peptide-imprinted nanogels of controllable size and affinity. Eur. Polym. J. 109, 453–459 (2018). [Google Scholar]
- 225.Gómez-Arribas L. N., Urraca J. L., Benito-Penìa E., Moreno-Bondi M. C., Tag-specific affinity purification of recombinant proteins by using molecularly imprinted polymers. Anal. Chem. 91, 4100–4106 (2019). [DOI] [PubMed] [Google Scholar]