Piezo1 transduces mechanical forces at the intercellular bridge to coordinate the machinery necessary to split dividing cells.
Abstract
Mechanical forces are exerted throughout cytokinesis, the final step of cell division. Yet, how forces are transduced and affect the signaling dynamics of cytokinetic proteins remains poorly characterized. We now show that the mechanosensitive Piezo1 channel is activated at the intercellular bridge (ICB) connecting daughter cells to regulate abscission. Inhibition of Piezo1 caused multinucleation both in vitro and in vivo. Piezo1 positioning at the ICB during cytokinesis depends on Pacsin3. Pharmacological and genetic inhibition of Piezo1 or Pacsin3 resulted in mislocation of Rab11-family-interacting protein 3 (Rab11-FIP3) endosomes, apoptosis-linked gene 2-interacting protein X (ALIX), and endosomal sorting complex required for transport III (ESCRT-III). Furthermore, we identified FIP3 as the link between Piezo1-generated Ca2+ signals and ALIX delivery to the ICB, where ALIX recruits the ESCRT-III component charged multivesicular body protein 4B, which promotes abscission. These results provide a different view of how mechanical forces participate in cytokinesis and identify Piezo1 as a key modulator of endosome trafficking.
INTRODUCTION
Cytokinetic abscission, the severing of the connecting intercellular bridge (ICB) between nascent daughter cells at the end of mitosis, is a sequential process that requires, among other events, the formation of the midbody at the ICB, delivery of Rab11-family-interacting protein 3 (Rab11-FIP3) endosomes to the ICB, cytoskeletal reorganization, and, lastly, the recruitment of the endosomal sorting complex required for transport III (ESCRT-III) (1–3). Enrolment of the ESCRT-I subunit tumor susceptibility gene-101 (TSG101) and the ESCRT-III targeting factor apoptosis-linked gene 2-interacting protein X (ALIX) at the midbody are prominent events initiated following the accumulation of the centrosomal protein Cep55 at the midbody. Both TSG101 and ALIX, through parallel pathways, recruit the ESCRT-III component charged multivesicular body protein 4B (CHMP4B) to direct abscission (4, 5). Successful cytokinesis also involves mechanical forces (6–11) and intracellular signals such as calcium (12–14), whose impact on the timely and spatially organized ESCRT-III components are unknown. Additional proteins able to bind to the membrane curvatures of the ICB, such as BIN-amphiphysin-Rvs (BAR) domain containing proteins, facilitate cytokinesis by means of anchoring key cytokinetic proteins at the correct location (15, 16). In this study, we aimed to identify how mechanical forces generated during cytokinesis are transduced to regulate the dynamics of key cytokinetic proteins, with a special focus on the activity of calcium-permeable mechanosensitive ion channels.
RESULTS
Localization and activity of Piezo1 during cytokinesis
The presence of functional mechanosensitive transient receptor potential vanilloid 4 (TRPV4) (17), transient receptor potential melastatin 7 (TRPM7) (18, 19), and Piezo1 channels (20) was detected in human microvascular endothelial cells (HMECs) (fig. S1, A to D). Piezo1 inhibition with GsMTx4 (Gs4) (21, 22) or addition of the Ca2+ chelator 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA) significantly triggered the presence of multinucleated cells (Fig. 1, A and B, and fig. S2A). GsMTx4 also prolonged the abscission time and the presence of daughter cells connected with elongated ICB (indicative of defective abscission), without affecting the time for mitotic rounding before metaphase (fig. S2, B to D). Transfection of HMECs with Piezo1 small interfering RNA (siRNA) reduced channel expression and activity (fig. S3, A to E) while increasing the number of nuclei per cell (Fig. 1B) and the length of the ICB (fig. S3F), verifying the involvement of Piezo1 in cytokinesis. To generalize our data showing a key role for Piezo1 in cytokinesis and to test the relevance of such mechanisms in vivo, we examined cytokinesis in zebrafish embryos and human MDA-MB-231-BrM2 adenocarcinoma cells, both expressing functional Piezo1 channel (23, 24). Akin to HMECs, both synchronized MDA-MB-231-BrM2 cells in which Piezo1 was knocked down (fig. S4, A and B) and zebrafish embryos treated with GsMTx4 displayed an increased percentage of multinucleated cells (Fig. 1, C and D, and fig. S4C). These results indicate that Piezo1-mediated signaling pathway during cytokinesis is highly conserved in vertebrates in both synchronized cell cultures and tissue unsynchronized cells.
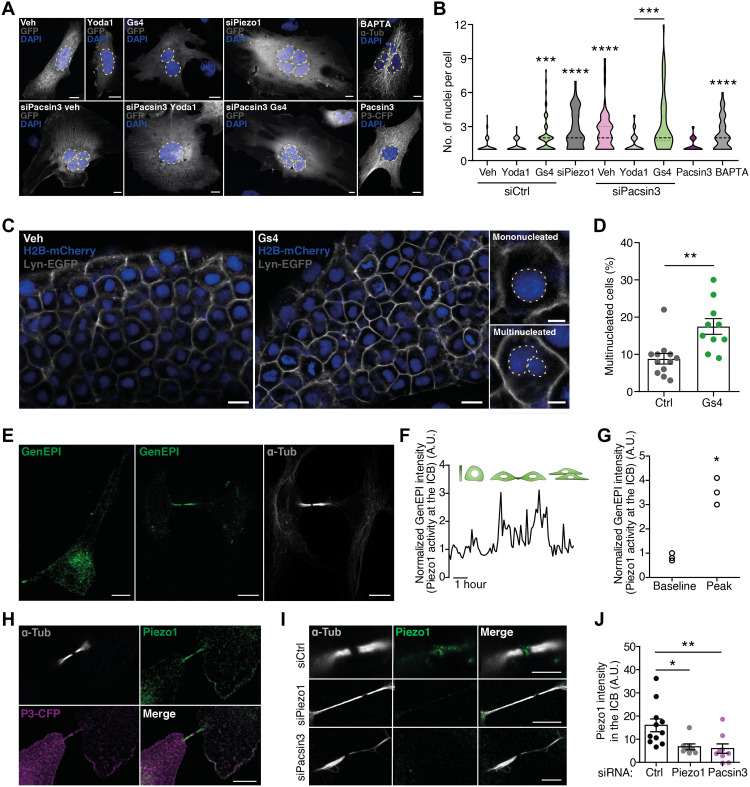
Fig. 1. Piezo1 is required for an effective cytokinesis.
(A) Nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI) in HMECs expressing green fluorescent protein (GFP) and treated with GsMTx4 (1 μM), Yoda1 (5 μM), and BAPTA (1 μM) and/or transfected with siControl (siCtrl), siPiezo1, siPacsin3, or Pacsin3-cyan fluorescent protein (CFP). Nuclei perimeter marked with discontinuous lines. Scale bars, 10 μm. Veh, vehicle; α-Tub, α-tubulin. (B) Violin plot of the number of nuclei/cell counted in HMECs exposed to the conditions indicated. siControl (n = 72), siControl + Yoda1 (n = 47), siControl + GsMTx4 (n = 61), siPiezo1 (n = 47), BAPTA (n = 74), Pacsin3 overexpression (n = 43), siPacsin3 (n = 51), siPacsin3 + Yoda1 (n = 49), and siPacsin3 + GsMTx4 (n = 54). (C) Cell nuclei (H2B-mCherry) and plasma membrane (lyn-GFP) visualization in zebrafish embryos treated with vehicle or 10 μM GsMTx4. Zoom shows representative images of mononucleated and multinucleated cells. Scale bars, 10 μm. (D) Percentage of multinucleated cells in vehicle- or GsMTx4-treated embryos. (E) Piezo1-GCaMP6 (GenEPI) localization in interphase (left) and late mitosis (middle); anti–α-tubulin staining in late mitosis (right). Scale bars, 10 μm. (F) Piezo1-mediated Ca2+ signal, measured at the ICB in GenEPI-transfected HMECs, peaks around 2 hours after release from nocodazole synchronization. A.U., arbitrary units. (G) Peak and baseline GenEPI fluorescence intensity at the ICB. (H) Colocalization of overexpressed Pacsin3-CFP with endogenous Piezo1 and α-tubulin at the ICB. Scale bar, 5 μm. (I) Immunodetection of Piezo1 and α-tubulin in HMECs transfected with control siRNA, siPiezo1, or siPacsin3. Scale bar, 5 μm. (J) Quantification of Piezo1 signal at the ICB in HMECs transfected with siControl, siPiezo1, or siPacsin3. Data are means ± SEM. Number of cells (or experimental repeats) is indicated in each graph. Significance values are respect control condition as determined by Kruskal-Wallis test followed by Dunn’s post hoc test, Mann-Whitney test (D), or paired t test (G).
Next, we evaluated Piezo1 localization and activity in both resting and dividing cells transfected with a genetically encoded fluorescent Ca2+ reporter tagged to Piezo1 (GenEPI) (25). Resting cells showed plasma membrane GenEPI signal reinforcement (Fig. 1E), resembling the immunolocalization of native Piezo1 channel (fig. S3D), while cells in late telophase showed increased GenEPI signal and colocalization with α-tubulin at the ICB (Fig. 1E). GenEPI was sensitive to Piezo1 channel activation and inhibition with Yoda1 and GsMTx4, respectively (fig. S5, A to C). Time-lapse imaging showed transient increases of GenEPI intensity at the ICB after release from cell cycle synchronization (Fig. 1, F and G), indicating a predominant activation of Piezo1 at the ICB of mitotic cells. The use of the intracellular Ca2+ indicator Calbryte further confirmed that mitotic cells show a localized increase in intracellular Ca2+ concentration at the ICB (fig. S5D).
Pacsin3 regulates location and activity of Piezo1
Piezo1 is activated by changes in membrane curvature in response to mechanical stimulation (26). Membrane curvature is also sensed and influenced by BAR domain containing proteins (16), of which Pacsin3 regulates trafficking and function of TRPV4 channel too (27, 28). Therefore, we examined whether Pacsin3 may be responsible for Piezo1 localization at the ICB. In mitotic HMECs, Piezo1 and Pacsin3 signals colocalized with α-tubulin at the ICB (Fig. 1H and fig. S6A). Knockdown of Piezo1 with siRNA significantly decreased Piezo1 signal at the ICB and midbody (Fig. 1, I and J). Similarly, siPacsin3 (fig. S6, B to E) reduced Piezo1 signal at the ICB (Fig. 1, I and J) without affecting Piezo1 expression (fig. S6C). In accordance with the effect of Pacsin3 on Piezo1 localization, knockdown of Pacsin3 incremented ICB length (fig. S3F) and multinucleation (Fig. 1, A and B, and figs. S2A and S6F), an effect that was not modified in the presence of GsMTx4 (Fig. 1, A and B). On the other hand, overexpression of Pacsin3 did not affect multinucleation (Fig. 1, A and B).
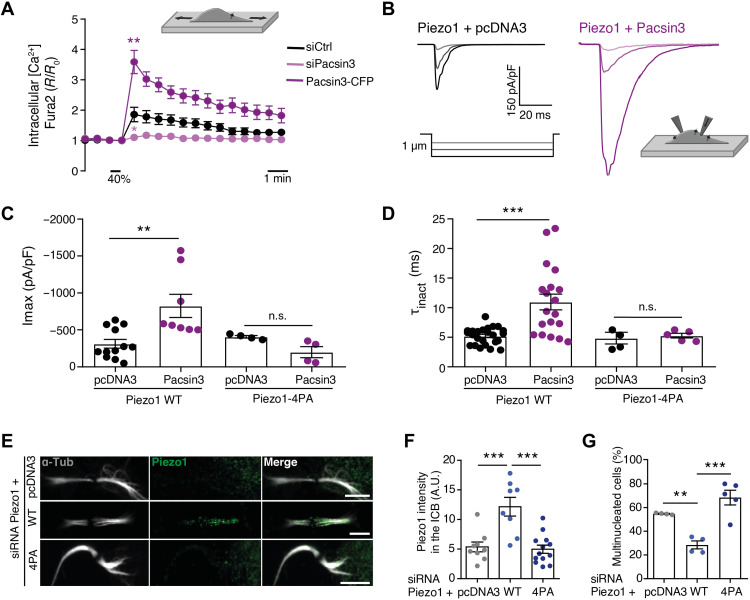
To test whether Pacsin3, in addition to regulating Piezo1 location, also modulates the activity of Piezo1, we imaged changes in intracellular Ca2+ concentration and carried out electrophysiological recordings on mechanically stimulated cells. Knockdown of Pacsin3 reduced mechanically stimulated and Piezo1-mediated Ca2+ signal, whereas overexpression of Pacsin3 increased it (Fig. 2A). Coimmunoprecipitation experiments further confirmed the close interaction between endogenous (fig. S6G) or overexpressed (fig. S6H) Piezo1 and Pacsin3 proteins. Furthermore, and consistent with the Ca2+ imaging experiments in HMECs, we observed that human embryonic kidney (HEK) 293 cells cotransfected with Piezo1 and Pacsin3 and mechanically stimulated with a blunt glass probe or by suction through the recording pipette presented larger whole-cell currents (Fig. 2, B and C, and fig. S7A), and increased single-channel open probability (fig. S7, C and D) than cells overexpressing Piezo1 alone. Piezo1 inactivation, a crucial mechanism determining channel function, was also slowed (higher inactivation time constant τinact) when coexpressed with Pacsin3 (Fig. 2D). Thus, Pacsin3 interaction with Piezo1 regulates both channel activity and location. Next, we tested whether the effect of Pacsin3 knockdown on multinucleation could be overturned in the presence of the potent Piezo1 activator Yoda1 (29). We reasoned that Yoda1 may boost the activity of Piezo1 channels remaining at the ICB of siPacsin3-treated cells. Exposure of cells depleted of Pacsin3 to Yoda1 reverted the presence of multinucleated cells (Fig. 1, A and B), whereas the use of the TRPV4 activator GSK1016790A (30) or the TRPM7 activator naltriben (31) did not reverse the effect of knocking down Pacsin3 on multinucleation (fig. S6, I and J).
Fig. 2. Piezo1 localization and activity is regulated by Pacsin3.
(A) Mean intracellular Ca2+ signals obtained from HMECs loaded with Fura-2 acetoxymethyl ester transfected with siControl (N = 37), siPacsin3 (N = 17), or overexpressing Pacsin3 (N = 15) and exposed to uniaxial stretching (40% of the initial chamber length, inset illustration). (B) Whole-cell traces of mechanically activated Piezo1 currents recorded at a holding potential of −80 mV from HEK293 cells overexpressing Piezo1 (left) or Piezo1 + Pacsin3 (right) following stimulation with a series of mechanical steps of 1 μm (inset illustration). (C) Maximum peak whole-cell currents obtained in HEK293 cells overexpressing pcDNA3, Piezo1–wild type (WT), Piezo1-4PA, and Pacsin3 as indicated. (D) Mean inactivation time constant (t) of whole-cell currents obtained in HEK293 cells overexpressing pcDNA3, Piezo1-WT, Piezo1-4PA, and Pacsin3 as indicated. n.s., not significant. (E) Immunodetection of Piezo1 and α-tubulin in HMECs, in which endogenous Piezo1 was knocked down with an siRNA directed against Piezo1 5′ untranslated region and transfected with the indicated siRNA-resistant Piezo constructs. Scale bars, 5 μm. (F) Quantification of Piezo1 signal at the ICB under the conditions indicated. (G) Percentage of multinucleated HMEC under the conditions indicated. Data are means ± SEM. Number of cells (or experimental repeats) is indicated in each graph. Significance values are respect control condition as determined by analysis of variance (ANOVA) followed by Bonferroni post hoc test or Kruskall-Wallis test followed by Dunn’s post hoc test (A).
We further explored the structural determinants of the interaction between Piezo1 and Pacsin3. Pacsin3 uses the BAR domain for binding to and bending of membranes and its SH3 domain to bind proline-rich domains present in its interacting partners (16). We identified a proline-rich domain in Piezo1 responsible for the interaction with Pacsin3 and the localization of Piezo1 to the ICB. Mutation of Pro1578, Pro1580, Pro1584, and Pro1587 to Ala (Piezo1-4PA) generated a functional channel that lost modulation by Pacsin3, i.e., mutant Piezo1-4PA did not increase peak current or τinact in the presence of overexpressed Pacsin3 (Fig. 2, C and D, and fig. S7B). To test whether the mutant Piezo channel also lacks the correct location and function during cytokinesis, we carried out rescue experiments overexpressing Piezo1–wild type (WT) and Piezo1-4AP in HMECs depleted of endogenous Piezo1 using an siRNA directed against its 5′ untranslated region (fig. S3, A to C). Only Piezo1-WT localized to the cytokinetic bridge (Fig. 2, E and F) and reverted the multinucleation phenotype induced by knocking down Piezo1 (Fig. 2G). Thus, overexpression of a mutant Piezo1 channel that lacks the Pacsin3-interacting domain cannot localize correctly during cytokinesis nor revert the multinucleation phenotype induced by the knockdown of the endogenous channel. Together, these experiments suggested that both Pacsin3 and Piezo1 are in the same regulatory pathway required for efficient cytokinesis and that Pacsin3 modulates localization and activity of Piezo1 at the ICB of mitotic cells.
Piezo1 controls localization of ESCRT-III components
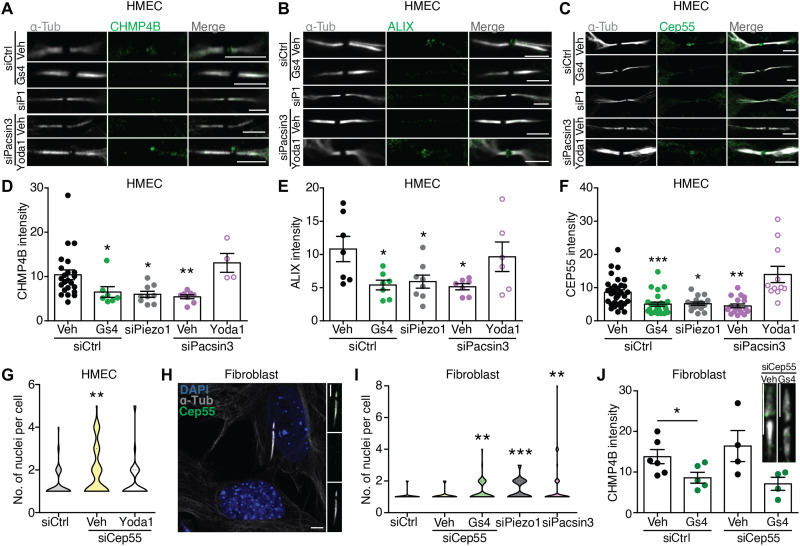
The presence of multinucleated cells and longer ICB following pharmacological or genetic inhibition of Piezo1 suggested a defect in abscission, the cutting of the narrow ICB, which depends on the ESCRT-III machinery (1). Consistent with the cytokinetic phenotype, endogenous CHMP4B was recruited to late cytokinetic bridges in control conditions but was absent in HMECs depleted of Piezo1, Pacsin3, or treated with GsMTx4, whereas pharmacological activation of Piezo1 with Yoda1 reverted the siPacsin3-induced mislocation of CHMP4B (Fig. 3, A and D). The CHMP4B-recruiting factor ALIX (Fig. 3, B and E) and its upstream regulator Cep55 (Fig. 3, C and F) also disappeared from the ICB, indicating that Piezo1 controls the recruitment of the ALIX-CHMP4B pathway to the midbody of late cytokinetic bridges. The role of Cep55 as a regulator of abscission has been recently disputed (32, 33). To test the relevance of Cep55 in this process, we treated HMECs with siCep55 and observed an increase in the number of nuclei/cell that was reverted in the presence of Yoda1 (Fig. 3G). Similarly, in Cep55-lacking human primary skin fibroblasts, siCep55 (Fig. 3H), unlike siPiezo1, siPacsin3, or GsMTx4, did not modify the number of nuclei/cell (Fig. 3I) nor the effect of GsMTx4 on CHMP4B recruitment to the midbody (Fig. 3J). Together, these results indicate that Piezo1 involvement in cytokinetic abscission is independent of Cep55.
Fig. 3. Piezo1 regulates localization of ESCRT-III components to the late cytokinetic bridge.
Immunolocalization of α-tubulin and CHMP4B (A), ALIX (B), and Cep55 (C) at the midbody of siControl-, siPiezo1-, GsMTx4-, siPacsin3-, and siPacsin3 + Yoda1–treated HMECs. Scale bars, 5 μm. Quantification of CHMP4B (D), ALIX (E), and Cep55 (F) immunofluorescence intensities at the midbody. (G) Quantification of the number of nuclei per cell in HMECs transfected with control siRNA (n = 57) or siCep55 (vehicle = 73 and Yoda1 = 64) in the presence/absence of Yoda1. (H) Immunolocalization of Cep55 and α-tubulin in primary human dermal fibroblasts. Nuclear staining with DAPI. Scale bars, 5 μm. (I) Quantification of the number of nuclei per cell in primary human dermal fibroblasts transfected with control siRNA (n = 143), siCep55 (vehicle = 33 and GsMTx4 = 55), siPiezo1 (n = 61), or siPacsin3 (n = 57). (J) CHMP4B immunofluorescence intensities at the midbody of primary human dermal fibroblasts transfected with control siRNA or siCep55 in the presence or absence of GsMTx4. Scale bar, 5 μm. Data are means ± SEM. Number of cells (or experimental repeats) is indicated in each graph. Significance values are respect control condition as determined by Kruskal-Wallis test followed by Dunn’s post hoc test or ANOVA followed by Dunnett’s post hoc test (E).
Piezo1 activity is linked to Rab11-FIP3 location at the cytokinetic bridge
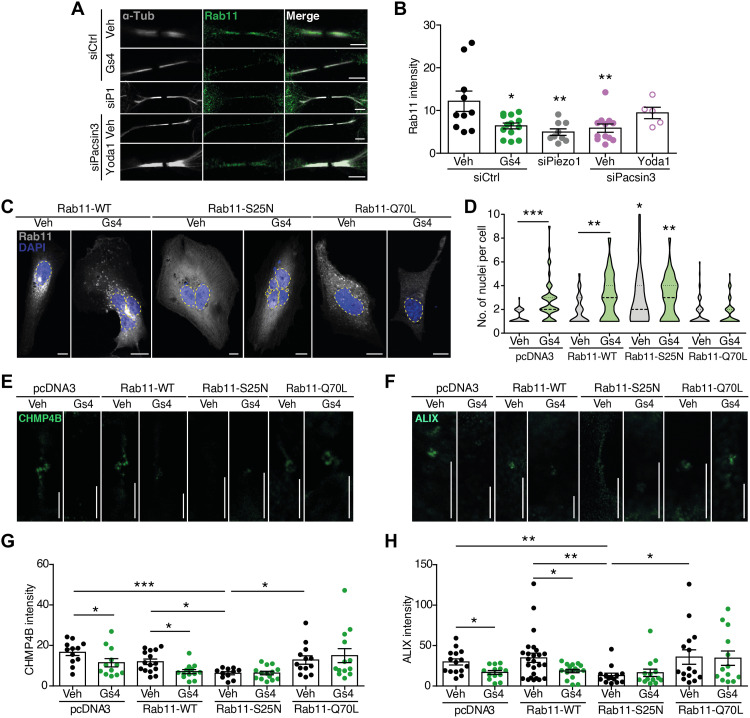
ALIX is a direct recruiter of CHMP4B and participates in the formation of secondary ingression during late cytokinesis (3). Midbody location of ALIX is affected by genetic or pharmacological inhibition of Piezo1. Therefore, we focused on how Piezo1 may control the correct location of ALIX, independently of Cep55. ALIX is present in endosomes (34), where it is capable of binding the ESCRT-III component CHMP4B (35). Thus, we tested the impact of Piezo1 on endosome trafficking and delivery of key molecules to the ICB. The small guanosine triphosphatase (GTPase) Rab11 and its effector FIP3 are present in endosomes and control endosome targeting to the ICB (3, 36). Interference of Piezo1 or Pacsin3 reduced Rab11 presence at the ICB (Fig. 4, A and B), consistent with the reduced ALIX signal. Piezo1 inhibition with GsMTx4 mimicked the effect of siPiezo1, whereas siPacsin3 effect was reverted with Yoda1 (Fig. 4, A and B). However, Yoda1 did not revert the effect of knocking down Piezo1 on Rab11 localization (fig. S8, A and B), confirming the specificity of Yoda1. To further explore the link between Rab11-containing endosomes and Piezo1-mediated signaling, we overexpressed WT (Rab11-WT), dominant-negative (Rab11-S25N), and dominant-positive (Rab11-Q70L) Rab11 and checked multinucleation (Fig. 4, C and D) as well as ICB localization of CHMP4B (Fig. 4, E and G) and ALIX (Fig. 4, F and H). Overexpression of Rab11-WT did not alter the number of nuclei, the intensity of the CHMP4B, and ALIX signals at the ICB or the effect of GsMTx4. Rab11-S25N overexpression significantly increased multinucleation and reduced CHMP4B and ALIX signals, effects that were not further increased in the presence of GsMTx4. On the other hand, Rab11-Q70L prevented the effect of GsMTx4 on any parameter tested, an effect not observed in cells overexpressing a dominant-positive Rab35 (fig. S8, C and D) (37). Other molecules relevant to cytokinesis and/or calcium signaling such as apoptosis-linked gene (ALG-2) and Beclin-1 (38, 39) were tested and showed no changes in their location at the ICB (fig. S9, A to D). Piezo channels have been linked to the activation of Rho GTPase and the downstream target myosin (22, 24). Considering that Rho-dependent regulation of the actomyosin cytoskeleton participates in different steps of cytokinesis (40), we tested whether the effect of Piezo1 on cytokinesis was related to the activation of Rho signaling. For that purpose, we used the Rho kinase inhibitor III, Rockout (41), and counted nuclei in the presence or absence of Yoda1. Figure S9E shows increased number of nuclei in Rockout-treated cells, an effect reverted in the presence of Yoda1, suggesting that Piezo1 participates in cytokinesis abscission independently of Rho signaling. We also noticed that inhibition of Piezo1 increased the presence of tubulin in the ICB (fig. S9F), most likely as the result of a defective recruitment of the microtubule-depolymerizing enzyme spastin by ESCRT-III (42).
Fig. 4. Piezo1 controls delivery of Rab11-FIP3 endosomes to the cytokinetic ring.
(A) Immunolocalization of Rab11 at the midbody of siControl-, siPiezo1-, GsMTx4-, siPacsin3-, and siPacsin3 + Yoda1–treated HMECs. Immunolocalization of α-tubulin in white. Scale bars, 5 μm. (B) Quantification of Rab11 intensity at the ICB under the conditions is shown in (A). (C) Nuclear staining with DAPI (blue) in HMECs transfected with Rab11-WT-GFP, dominant-negative Rab11-S25N-GFP, or dominant-positive Rab11-Q70L–GFP in the presence/absence of GsMTx4. Scale bars, 10 μm. (D) Quantification of nuclei in HMECs overexpressing pcDNA3 (vehicle = 41 and GsMTx4 = 44), Rab11-WT (vehicle = 62 and GsMtx4 = 57), Rab11-S25N (vehicle = 50 and GsMTx4 = 38), or Rab11-Q70L (vehicle = 59 and GsMTx4 = 56) in the presence or absence of GsMTx4. CHMP4B (E) and ALIX (F) immunofluorescence intensities at the midbody of HMECs overexpressing pcDNA3, Rab11-WT, Rab11-S25N, and Rab11-Q70L in the presence or absence of GsMTx4. Scale bars, 5 μm. Quantification of CHMP4B (G) and ALIX (H) immunofluorescence intensities in HMECs under the conditions shown. Data are means ± SEM. Number of cells (or experimental repeats) is indicated in each graph. Significance values are respect control condition as determined by Kruskal-Wallis test followed by Dunn’s post hoc test or ANOVA followed by Dunnett’s post hoc test (B).
The role of TRPV4- and TRPM7-mechanosensitive channels in the recruitment of ALIX was also studied. The lack of an effect of TRPV4 and TRPM7 activators on siPacsin3-induced multinucleation (fig. S6I) did not support a key role for these channels in abscission. We confirmed this observation by localizing overexpressed TRPV4–green fluorescent protein (GFP) (43) and TRPM7–yellow fluorescent protein (YFP) (19, 44) and by studying the effect of knocking down these two channels on the location of ALIX at the ICB. Neither TRPV4-GFP nor TRPM7-YFP localized to the ICB (fig. S10A), and knockdown of TRPV4 or TRPM7 (fig. S10, B and C) did not alter the location of ALIX at the ICB (fig. S10, D and E). Together, these experiments pointed to Piezo1 as the key mechanosensitive channel participating in abscission.
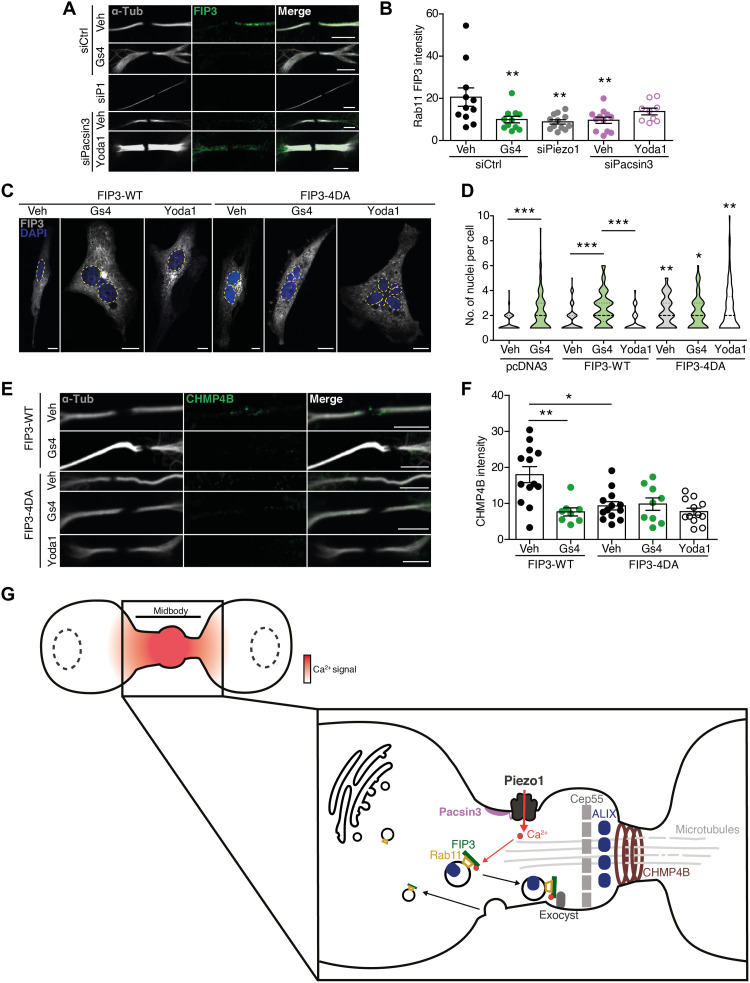
Next, we searched for the target of the Piezo1-mediated Ca2+ signal that controls the trafficking of Rab11 endosomes to the ICB. Rab11 has no Ca2+-binding sites identified, whereas its effector, FIP3, presents two Ca2+-binding EF-hand domains (www.uniprot.org/uniprot/O75154). First, we localized FIP3 at the ICB and showed that FIP3 signal, like Rab11 signal, decreased in HMECs treated with GsMTx4, siPiezo1, and siPacsin3 but not in cells treated with siPacsin3 in the presence of Yoda1 (Fig. 5, A and B). Second, we transfected HMECs with FIP3-WT or FIP3-4DA, in which four Asp (D215, D217, D219, and D247) present in the EF-hand domains that are required for Ca2+ binding were mutated to Ala, and checked for the appearance of extra nuclei and CHMP4B at the ICB. Overexpression of FIP3-WT did not induce multinucleation or CHMP4B localization nor altered the response to GsMTx4, whereas overexpression of FIP3-4DA significantly increased the number of nuclei and reduced the CHMP4B signal, effects that were not further modified in the presence of GsMTx4 (Fig. 5, C to F). Similar effects to those obtained with FIP3-4DA were obtained when cells were transfected with a mutated FIP3 (FIP3-I738E) that does not interact with Rab11 (fig. S11, A and B) (45). Together, these results pointed to Rab11-FIP3 as a target of Piezo1-mediated signaling during cytokinesis.
Fig. 5. FIP3 links Piezo1-generated Ca2+ signals to the recruitment of the abscission machinery.
(A) Immunolocalization of FIP3 at the midbody of siControl-, siPiezo1-, GsMTx4-, siPacsin3-, and siPacsin3 + Yoda1–treated HMECs. Immunolocalization of α-tubulin in white. Scale bars, 5 μm. (B) Quantification of FIP3 signal at the ICB under the conditions shown in (A). (C) DAPI staining of the nuclei in HMECs overexpressing FIP3-WT and FIP3-4DA in the presence and absence of GsMTx4 or Yoda1. Scale bars, 10 μm. (D) Quantification of nuclei in HMECs overexpressing pcDNA3 (vehicle = 97 and GsMTx4 = 104), FIP3-WT (vehicle = 114, GsMTx4 = 65, and Yoda1 = 71), and FIP3-4DA (vehicle = 75, GsMTx4 = 67, and Yoda1 = 89). (E) CHMP4B immunofluorescence of HMECs overexpressing FIP3-WT and FIP3-4DA in the presence and absence of GsMTx4 or Yoda1. (F) Quantification of CHMP4B immunofluorescence intensity in HMECs under the conditions shown. Data are means ± SEM. When required, number of cells (or experimental repeats) is indicated in each graph. Significance values are respect control condition as determined by Kruskal-Wallis test followed by Dunn’s post hoc test or ANOVA followed by Dunnett’s post hoc test (B). (G) Cartoon model of cytokinesis regulation by the Piezo1 channel. Mechanical forces exerted at the ICB activate Piezo1 channel generating a marked and localized increase in intracellular calcium (red shading). The increase in intracellular Ca2+ concentration is sensed by FIP3 to direct the transport of ALIX-containing Rab11-FIP3 endosomes to the ICB where ALIX recruits CHMP4B to complete abscission.
DISCUSSION
Cytokinetic abscission, the severing of the connecting ICB between nascent daughter cells at the end of mitosis, is a complex process involving mechanical forces (6–11), intracellular signals (13, 14, 46), endosome trafficking, and, lastly, the recruitment of the ESCRT-III component CHMP4B via ALIX and/or TSG101 to complete abscission (1–5). The participation of mechanical forces throughout cytokinesis is generally accepted, although how these forces control cytokinesis, particularly its late steps, has been largely unexplored. There are also claims suggesting a differential impact of mechanical forces on dividing cells depending on cell geometry or cell density (7, 11), which complicate the understanding of how mechanical forces integrate into the signaling generated by a dividing cell. Our findings reveal that Piezo1-mediated transduction of mechanical forces (generated by cell crawling and elongation, by cell interaction with extracellular matrix, or by contraction of cytokinetic ring and protrusion of the cell poles) is key to the control of cytokinetic abscission both in isolated cells or confluent cell cultures and in vivo zebrafish embryos, thereby strengthening the central role of mechanotransduction in cytokinesis.
Generation of the actomyosin ring and furrow ingression is a cytokinetic step in which both mechanical forces and Ca2+ signaling may be coupled via the Ca2+-dependent regulation of the contraction process (14), although the source of Ca2+ during the furrow ingression appears to originate at the endoplasmic reticulum. The presence of long ICB, typical of abscission defects, following inhibition of Piezo1 points toward a defect in abscission rather than alterations in the furrow ingression. Activation of the mechanosensitive Piezo1 channel at the ICB generates local and transient increases in intracellular [Ca2+] that are sensed by FIP3 to target Rab11-FIP3 endosomes containing ALIX (or an ALIX-recruiting element) to the ICB. The fact that a diffusible signal such as Ca2+ participates in the recruitment of abscission elements to the midbody may facilitate coordination of different cytokinetic events without the need of a physical/sequential interaction.
Despite the numerous experimental approaches used for the demonstration of the involvement of Piezo1 in the cytokinesis, our study is not exempt of limitations. One is the technical difficulty to directly record channel activity in the ICB with a recording pipette attached to the cell membrane under different mechanical stress regimes. Additional limitations of our study are the following: (i) Although we clearly demonstrated the pathway that links Piezo1 to ESCRT-III recruitment via Rab11-FIP3 endosomes, we cannot completely discard the participation of other Piezo1- and/or mechanosensitive Ca2+-dependent events in the abscission process; and (ii) the phenotype observed in zebrafish embryos by using the most specific Piezo1 blocker, GsMTx4 (21), strongly supports the role of Piezo1 in vivo model, although we cannot completely discard that GsMTx4 may also affect other ion pathways in zebrafish.
Far-reaching implications of our work are the following: (i) the identification of the BAR domain containing protein Pacsin3 as a Piezo1-binding protein that regulates channel location and activity, being particularly interesting the regulation of channel inactivation, a parameter that is at the core of Piezo1-linked pathologies (47, 48); (ii) the role of force-induced Ca2+-dependent regulation of the endosomal pathway and how it may affect other mechanically regulated physiological functions such as cargo targeting and release of exosomes (35, 49–51); (iii) the role of mechanically induced nuclear envelope repair of confined migrating cells that also depends on recruitment of ESCRT-III (52); and (iv) the presence of cytokinesis failures may result in altered proliferation and chromosomal instability that affect adaptation and fitness of tumors (53). In this regard, analysis of several cancer databases, including Human Protein Atlas and The Cancer Genome Atlas, has shown an association of Piezo1 with different cancers and that Piezo1 may be a valuable prognostic tool (54–57). In addition, in view of our results, the mechanism leading to embryonic lethality in Piezo1 knockout animal models (58, 59) and human phenotypes generated by loss-of-function mutations in Piezo1 (60) should be revised. Most probably, there is not a single mechanism but a more complex scenario. The current view is that embryonic lethality, or human disease, in the absence of functional Piezo1 is due to the loss of vascular development because endothelial cells are unable to sense shear stress induced by blood flow. However, our results clearly show the key role of Piezo1 in cytokinesis, a basic cellular routine required for embryonic development and vasculogenesis.
MATERIALS AND METHODS
Cell lines and constructs
HEK293 (catalog number 85120602) and HeLa (catalog number 93021013) cells were purchased from the European Collection of Authenticated Cell Cultures. HEK293 and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (D6046, Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 U/ml). HMEC-1 were purchased from the American Type Culture Collection (ATCC) (Crl-3243). HMECs were maintained in MCDB131 (without l-glutamine; Life Technologies catalog number 10372019) supplemented with epidermal growth factor (10 ng/ml; Thermo Fisher Scientific catalog number PHG0314), hydrocortisone (1 μg/ml; Sigma-Aldrich catalog number H0396), 10 mM glutamine (ATCC 30-2214), and 10% FBS (ATCC 30-2020). HEK293 cells were used to transfect Piezo1 and perform electrophysiological studies and coimmunoprecipitation assays because of the high amount of expression and protein obtained from transfected cells. MDA-MB-231-BrM2 cells (61) were provided by J. Massagué, Memorial Sloan Kettering Cancer Center, NY. Knockdown of Piezo1 in MDA-MB-231BrM2 cells were obtained using lentivirus as previously described (24). HEK293 were transfected with 2 μg of pMD2-G + 4 μg of pSPAX2 and 5 μg of pLKO.1-shPIEZO1 or pLKO.1-scramble (table S1) using Lipofectamine 3000 (Invitrogen). After collection, centrifugation, and titration, lentiviral supernatants were used to infect MDA-MB-231BrM2 cells. MDA-MB-231BrM2–shPIEZO1 and MB-231BrM2–scramble stable cell lines were generated by selecting with puromycin (1 μg/ml). MDA-MB-231-BrM2 cells were synchronized with a double nocodazole block procedure (100 nM; 16, 8, and 16 hours) and released with normal medium 2 hours before experiments were carried out.
FIP3 Asp residues in EF-1 (D215/D217/D219) and EF-2 (D247) coordinating Ca2+ binding were mutated to Ala (FIP3-4DA) on the pEGFP-C1-FIP3 plasmid. Mutations were introduced by site-directed mutagenesis using the QuikChange kit (Stratagene). All materials used in this study are shown in table S1.
Electrophysiological recordings
Stretch-activated currents were recorded on HEK293 cells 2 days after transfection. For whole-cell recordings, the bath solution contained 140 mM NaCl, 2.5 mM KCl, 1.2 mM CaCl2, 0.5 mM MgCl2, 5 mM glucose, and 10 mM Hepes (adjusted to pH 7.45 with NaOH). Borosilicate glass patch pipettes had a tip resistance of 1 to 5 megohms and contained 140 mM CsCl, 0.3 mM Na3GTP, 4 mM Na2ATP, 1 mM EGTA, and 10 mM Hepes (adjusted to pH 7.3 with tris). The membrane potential inside the patch was held at −80 mV using an Axopatch 200 A (Molecular Devices, USA). Piezo1 activity was assessed by applying a mechanical stimulation generated with a fire-polished glass probe driven by a piezoelectric controller (E-665, Physik Instrumente). The probe had an upward motion consisting of 1-μm increment, 200-ms duration, and a spaced interval of 10 s. Currents were sampled at 10 kHz and filtered at 1 kHz using pCLAMP 10.5 software (Axon Instruments).
For cell-attached configuration, borosilicate glass patch pipettes (2 to 3 megohms) were filled with a solution containing 130 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 10 mM tetraethylammonium-Cl, and 10 mM Hepes (adjusted to pH 7.3 with NaOH). Piezo1 activity was also triggered under this recording condition using the Piezo activator Yoda (20 μM) (29), added to the pipette solution. The bath solution contained 140 mM KCl, 1 mM MgCl2, 10 mM glucose, and 10 mM Hepes (adjusted to pH 7.3 with tris). This extracellular potassium concentration was used to zero the membrane potential, allowing better control of the voltage (−80 mV) using an EPC10-USB patch-clamp amplifier (HEKA Elektronik). Stimulation of the Piezo1 channel was performed using a High Speed Pressure Clamp (HSPC-1, ALA Scientific Instruments) applying a prepulse of −8 mmHg. Experiments were run at room temperature (22° to 26°C). Data were analyzed using Igor Pro 8 (WaveMetrics) and MATLAB. Because of the biphasic kinetic of the currents, the time constant of inactivation (τ) was obtained by fitting the results to a double exponential curve between the peak current and the end of the stimulus according to the next equation
We only used the value of τ1 for comparison of the time constants of inactivation, as τ1 represents the main current decay due to inactivation of Piezo1 [~5 ms according to (62)] and τ2 represents the slow residual current at the end of the mechanical stimulation.
Cell stretching and calcium imaging
To measure cytosolic calcium increases in response to stretch, cells were cultured in a silicone chamber (STB-CH-04, STRETX Inc.) to a 50% confluence and loaded with 5 μM fura- 2 AM (Thermo Fisher Scientific) as previously described (17). Isotonic bath solutions contained 140 mM NaCl, 2.5 mM KCl, 1.2 mM CaCl2, 0.5 mM MgCl2, 5 mM glucose, 300 mOsm (adjusted with d-mannitol), and 10 mM Hepes (adjusted to pH 7.3 with tris base). For mechanical stimulation of Piezo1, two steps of 0.5-s uniaxial stretching (40 and 80% of the initial chamber length) were applied with a delay of 8 min, which is the time necessary to recover basal calcium levels after the first stimulus. The fluorescence ratio (F340/F380) was acquired with an imaging processing software (HCImage, Hamamatsu Photonics). Signals were normalized to the F340/F380 measured before cell stimulation. Intracellular [Ca2+] was also measured in cells loaded with the single-wavelength Ca2+ dye Calbryte (23). Signals were normalized to the fluorescence intensity measured before the application of different stimuli.
Immunofluorescence and time-lapse microscopy
HMEC and MDA-MB-231-BrM2 cells were washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 20 min at 37°C. Cells were then permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) in PBS for 5 min and blocked in 5% bovine serum albumin (BSA) in PBS for 1 hour at room temperature. Primary antibodies (table S1) were incubated overnight at 4°C diluted in 1% BSA in PBS. Secondary antibodies (table S1) were diluted in PBS containing 1% BSA and incubated for 1 hour at room temperature. For the identification and quantification of nuclei, cells were incubated for 10 min with 4′,6-diamidino-2-phenylindole (DAPI) (table S1) diluted in PBS containing 1% BSA. To visualize cell plasma membrane, a pretreatment of 20 min with concanavalin A (1:50; in ice-cooled PBS) was carried out before cell fixation. Coverslips were mounted with Fluoromount (0100-01, Southern Biotech), and cells were examined with a Leica TCS-SP8 confocal microscope with a 63× 1.40 immersion oil objective and using maximal microscope resolution (95 × 95 nm). For time-lapse experiments using GenEPI, transfected HMECs plated in 35-mm glass-bottom dishes were synchronized in G2/M with 100 nM nocodazole in complete culture medium for 16 to 20 hours and released 2 hours before image acquisition. Experiments were performed 48 hours after transfection in a Leica TCS-SP8 confocal microscope with a 40× 1.40 immersion oil objective using a resonant scanner and a hybrid detector. For time length measurements during cell cycle, HMECs at 70 to 90% confluence were synchronized in G2/M with 100 nM nocodazole in complete culture medium. Following release from synchronization, cells were exposed to either vehicle or the indicated drugs, and image acquisition was started. Cytokinesis start time and the abscission time were manually calculated from the videos obtained. A time-lapse series was acquired using a fully motorized Zeiss Cell Observer HS microscope, 10× objective. Temperature was controlled at 37°C using the built-in microscope incubator providing a humidified atmosphere with 5% CO2. Imaging was performed for 24 hours with a lapse time of 10 or 15 min. ImageJ software was used for image processing and immunofluorescence quantification at the ICB and midbody (Plot Profile tool).
Cytokinesis in zebrafish embryos
Zebrafish (Danio rerio) embryos were obtained by mating adult WT fish using standard methods. All protocols were approved by the Institutional Animal Care and Use Ethic Committee (Comitè D’Etica en l’Experimentació Animal, Barcelona Biomedical Research Park) and the Generalitat of Catalonia (Departament de Territori i Sostenibilitat) and were implemented according to European regulations for the handling of animals in research. Experiments were carried out in accordance with the principles of the 3Rs (refinement, reduction, and replacement). All embryos had passed the mid blastula transition phase and were all at the same stage. No cell cycle pharmacological synchronization was carried out on zebrafish embryos. To quantify multinucleated cells in zebrafish embryos, cell nuclei and plasma membranes were visualized by injection of mRNAs encoding for H2B-mCherry and lyn-GFP fusion proteins, respectively. mRNAs were synthetized from pCS2 plasmids using the mMESSAGE mMACHINE kit (Thermo Fisher Scientific). WT embryos were injected at the one-cell stage with 2 nl of a mix containing 50 ng/μl of each mRNA and either 10 μM GsMTx4 or vehicle (H2O). Injected embryos were developed until 30% epiboly stage, moment in which they were in vivo imaged. Embryos were mounted in 0.7% low-melting point agarose in glass-bottom petri dishes (MatTek), and image acquisition was performed on a Leica Sp8 confocal microscope using a 20× objective. Multinucleation was quantified by the mean percentage of cells with >2 nuclei from two different Z stacks per embryo with a minimum of 200 cells per embryo.
Coimmunoprecipitation and Western blot
Coimmunoprecipitation assay was performed as previously described (63). In brief, cell extracts were obtained by incubation in 1% Triton X-100 lysis buffer supplemented with 10% of protease inhibitor cocktail (Thermo Fisher Scientific) for 30 min at 4°C. Cell lysates were centrifuged at 10.000g for 5 min at 4°C, and the soluble fraction was incubated with equilibrated GFP-Trap beads (gta-20, Chromotek) for 2 hours at 4°C. For Western blotting, cells were harvested 72 hours after transfection and solubilized in radioimmunoprecipitation assay buffer [10 mM tris buffer, 1 mM EDTA, 140 mM NaCl, 0.1% sodium deoxycholate, 0.1% SDS, and 1% Triton X-100 (pH 7.2)] supplemented with EDTA-free protease inhibitor cocktail tablets (Sigma-Aldrich), 1 mM phenylmethylsulfonyl fluoride, 2 mM tris(2-carboxyethyl)phosphine, and 1 mM N-ethylmaleimide for 10 to 20 min on a rotating wheel at 4°C. Complexes or cell lysates were centrifuged and washed four times with PBS buffer, denatured with SDS–polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (NP0007, Invitrogen) for 10 min at 70°C, and separated by SDS-PAGE in tris-acetate 3 to 8% precast gels (EA0375BOX, Invitrogen). The immunoprecipitated proteins or cell lysates cleared by centrifugation were analyzed by Western blotting, and soluble fractions from cell lysis were used as input. Membranes were blocked by 5% nonfat dry milk in tween-tris-buffered saline and incubated in the same blocking buffer with anti-Myc, anti-GFP, anti-Pacsin3, or anti-Piezo1 (table S1) overnight at 4°C. Secondary antibodies peroxidase-conjugated anti-rabbit immunoglobulin G (IgG) or anti-mouse IgG were used in blocking buffer and incubated for 1 hour at room temperature. Detection was performed using the enhanced chemiluminescence detection kit (1705061; Clarity, Bio-Rad). For coimmunoprecipitation of endogenous proteins in HMECs, cell extracts were centrifuged at 14.000g at 4°C for 10 min to remove aggregates. Solubilized proteins were incubated overnight with an anti-Piezo1 antibody. Immunocomplexes were then incubated with 50 μl of protein G beads for 2 hours at 4°C.
Real-time polymerase chain reaction
Total RNA was isolated from cells using a NucleoSpin RNA isolation kit (740955, Macherey-Nagel) or an NZY Total RNA Isolation kit (MB13402, NZYTech). SuperScript III reverse transcriptase system (18080044, Thermo Fisher Scientific) was used for complementary DNA synthesis. Quantitative real-time polymerase chain reaction (PCR) was performed using SYBR Green (4367659, Applied Biosystems) in a QuantStudio 12K Flex system (Applied Biosystems) with the specific human primers shown in table S1. Glyceraldehyde-3-phosphate dehydrogenase or hypoxanthine-guanine phosphoribosyltransferase were used as a housekeeping gene for the quantification of relative gene expression using 2DDCt.
Statistical analysis
All data are represented as means ± SEM. Statistics and graphics were performed using GraphPad Prism. Before checking statistical significance, a Shapiro-Wilk omnibus normality test was applied in all cases. For the data that followed normal distributions, a Student’s t test was applied between two groups and one-way analysis of variance (ANOVA) followed by Bonferroni or Dunnett post hoc tests across multiple groups. For the data that did not assume Gaussian distributions, a Mann-Whitney’s U test was used for comparing two groups, while Kruskal-Wallis followed by Dunn’s post hoc test was applied for multiple groups. Significant difference was considered when a final value of *P < 0.05 was reached. **P < 0.01; ***P < 0.001; and ****P < 0.0001.
Acknowledgments
We thank members of the laboratories for insightful discussions.
Funding: This study was supported by the Spanish Ministry of Science, Education, and Universities through grants RTI2018-099718-B-100 to M.A.V. and PGC2018-095663-B100 to C.P., an institutional “María de Maeztu” Programme for Units of Excellence in R&D, and FEDER funds. C.P. is the recipient of an ICREA Academia award.
Author contributions: M.A.V. and J.C.-G. designed the research. J.C.-G. performed electrophysiological, calcium imaging and confocal microscopy experiments. S.A.S.: electrophysiology. F.R.-M.: Calcium imaging and generation of cell lines and plasmids. V.H.-F.: MDA231-BrM2 experiments and confocal microscopy. M.V.-G.: PCR experiments. P.D.-M.: Coimmunoprecipitation experiments. C.F.H.: Zebrafish experiments. C.P.: Supervision of zebrafish experiments and funding. M.A.V.: Funding, supervision of the project, and manuscript writing. All authors edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Table S1
REFERENCES AND NOTES
- 1.Carlton J. G., Martin-Serrano J., Parallels between cytokinesis and retroviral budding: A role for the ESCRT machinery. Science 316, 1908–1912 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Green R. A., Paluch E., Oegema K., Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28, 29–58 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Schiel J. A., Simon G. C., Zaharris C., Weisz J., Castle D., Wu C. C., Prekeris R., FIP3-endosome-dependent formation of the secondary ingression mediates ESCRT-III recruitment during cytokinesis. Nat. Cell Biol. 14, 1068–1078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morita E., Sandrin V., Chung H.-Y., Morham S. G., Gygi S. P., Rodesch C. K., Sundquist W. I., Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26, 4215–4227 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vietri M., Radulovic M., Stenmark H., The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21, 25–42 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Burton K., Taylor D. L., Traction forces of cytokinesis measured with optically modified elastic substrata. Nature 385, 450–454 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Lafaurie-Janvore J., Maiuri P., Wang I., Pinot M., Manneville J.-B., Betz T., Balland M., Piel M., ESCRT-III assembly and cytokinetic abscission are induced by tension release in the intercellular bridge. Science 339, 1625–1629 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Gupta D. K., Du J., Kamranvar S. A., Johansson S., Tension-induced cytokinetic abscission in human fibroblasts. Oncotarget 9, 8999–9009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srivastava V., Robinson D. N., Mechanical stress and network structure drive protein dynamics during cytokinesis. Curr. Biol. 25, 663–670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam S., Chaudhuri O., Mitotic cells generate protrusive extracellular forces to divide in three-dimensional microenvironments. Nat. Phys. 14, 621–628 (2018). [Google Scholar]
- 11.Uroz M., Garcia-Puig A., Tekeli I., Elosegui-Artola A., Abenza J. F., Marín-Llauradó A., Pujals S., Conte V., Albertazzi L., Roca-Cusachs P., Raya Á., Trepat X., Traction forces at the cytokinetic ring regulate cell division and polyploidy in the migrating zebrafish epicardium. Nat. Mater. 18, 1015–1023 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Chang D. C., Meng C., A localized elevation of cytosolic free calcium is associated with cytokinesis in the zebrafish embryo. J. Cell Biol. 131, 1539–1545 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boucrot E., Kirchhausen T., Endosomal recycling controls plasma membrane area during mitosis. Proc. Natl. Acad. Sci. U.S.A. 104, 7939–7944 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webb S. E., Miller A. L., Ca2+ signalling and membrane dynamics during cytokinesis in animal cells. Adv. Exp. Med. Biol. 981, 389–412 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Smith C. M., Chircop M., Clathrin-mediated endocytic proteins are involved in regulating mitotic progression and completion. Traffic 13, 1628–1641 (2012). [DOI] [PubMed] [Google Scholar]
- 16.McDonald N. A., Gould K. L., Linking up at the BAR: Oligomerization and F-BAR protein function. Cell Cycle 15, 1977–1985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandes J., Lorenzo I. M., Andrade Y. N., Garcia-Elias A., Serra S. A., Fernandez-Fernandez J. M., Valverde M. A., IP3 sensitizes TRPV4 channel to the mechano- and osmotransducing messenger 5′-6′-epoxyeicosatrienoic acid. J. Cell Biol. 181, 143–155 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei C., Wang X., Chen M., Ouyang K., Song L.-S., Cheng H., Calcium flickers steer cell migration. Nature 457, 901–905 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao R., Afthinos A., Zhu T., Mistriotis P., Li Y., Serra S. A., Zhang Y., Yankaskas C. L., He S., Valverde M. A., Sun S. X., Konstantopoulos K., Cell sensing and decision-making in confinement: The role of TRPM7 in a tug of war between hydraulic pressure and cross-sectional area. Sci. Adv. 5, eaaw7243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coste B., Mathur J., Schmidt M., Earley T. J., Ranade S., Petrus M. J., Dubin A. E., Patapoutian A., Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 330, 55–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bae C., Sachs F., Gottlieb P. A., The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry 50, 6295–6300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung W. C., Yang J. R., Yankaskas C. L., Wong B. S., Wu P. H., Pardo-Pastor C., Serra S. A., Chiang M. J., Gu Z., Wirtz D., Valverde M. A., Yang J. T., Zhang J., Konstantopoulos K., Confinement sensing and signal optimization via Piezo1/PKA and myosin II pathways. Cell Rep. 15, 1430–1441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venturini V., Pezzano F., Castro F. C., Häkkinen H. M., Jiménez-Delgado S., Colomer-Rosell M., Marro M., Tolosa-Ramon Q., Paz-López S., Valverde M. A., Weghuber J., Loza-Alvarez P., Krieg M., Wieser S., Ruprecht V., The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science 370, eba2644 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Pardo-Pastor C., Rubio-Moscardo F., Vogel-González M., Serra S. A., Afthinos A., Mrkonjic S., Destaing O., Abenza J. F., Fernández-Fernández J. M., Trepat X., Albiges-Rizo C., Konstantopoulos K., Valverde M. A., Piezo2 channel regulates RhoA and actin cytoskeleton to promote cell mechanobiological responses. Proc. Natl. Acad. Sci. U.S.A. 115, 1925–1930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaganoglu S., Helassa N., Gaub B. M., Welling M., Shi J., Müller D. J., Török K., Pantazis P., GenEPi: Piezo1-based fluorescent reporter for visualizing mechanical stimuli with high spatiotemporal resolution. bioRxiv 702423 (2019). [Google Scholar]
- 26.Lin Y. C., Guo Y. R., Miyagi A., Levring J., MacKinnon R., Scheuring S., Force-induced conformational changes in PIEZO1. Nature 573, 230–234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuajungco M. P., Grimm C., Oshima K., D’hoedt D., Nilius B., Mensenkamp A. R., Bindels R. J., Plomann M., Heller S., PACSINs bind to the TRPV4 cation channel. PACSIN 3 modulates the subcellular localization of TRPV4. J. Biol. Chem. 281, 18753–18762 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Elias A., Mrkonjic S., Pardo-Pastor C., Inada H., Hellmich U. A., Rubio-Moscardó F., Plata C., Gaudet R., Vicente R., Valverde M. A., Phosphatidylinositol-4,5-biphosphate-dependent rearrangement of TRPV4 cytosolic tails enables channel activation by physiological stimuli. Proc. Natl. Acad. Sci. U.S.A. 110, 9553–9558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Syeda R., Xu J., Dubin A. E., Coste B., Mathur J., Huynh T., Matzen J., Lao J., Tully D. C., Engels I. H., Petrassi H. M., Schumacher A. M., Montal M., Bandell M., Patapoutian A., Chemical activation of the mechanotransduction channel Piezo1. eLife 4, e07369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorneloe K. S., Sulpizio A. C., Lin Z., Figueroa D. J., Clouse A. K., McCafferty G. P., Chendrimada T. P., Lashinger E. S. R., Gordon E., Evans L., Misajet B. A., Demarini D. J., Nation J. H., Casillas L. N., Marquis R. W., Votta B. J., Sheardown S. A., Xu X., Brooks D. P., Laping N. J., Westfall T. D., N-((1S)-1-{[4-((2S)-2-{[(2,4-dichlorophenyl)sulfonyl]amino}-3-hydroxypropanoyl)-1-piperazinyl]carbonyl}-3-methylbutyl)-1-benzothiophene-2-carboxamide (GSK1016790A), a novel and potent transient receptor potential vanilloid 4 channel agonist induces urinary bladder contraction and hyperactivity: Part I. J. Pharmacol. Exp. Ther. 326, 432–442 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Hofmann T., Schäfer S., Linseisen M., Sytik L., Gudermann T., Chubanov V., Activation of TRPM7 channels by small molecules under physiological conditions. Pflügers Arch 466, 2177–2189 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Lie-Jensen A., Ivanauskiene K., Malerød L., Jain A., Tan K. W., Laerdahl J. K., Liestøl K., Stenmark H., Haglund K., Centralspindlin recruits ALIX to the midbody during cytokinetic abscission in Drosophila via a mechanism analogous to virus budding. Curr. Biol. 29, 3538–3548.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Tedeschi A., Almagro J., Renshaw M. J., Messal H. A., Behrens A., Petronczki M., Cep55 promotes cytokinesis of neural progenitors but is dispensable for most mammalian cell divisions. Nat. Commun. 11, 1746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bissig C., Lenoir M., Velluz M. C., Kufareva I., Abagyan R., Overduin M., Gruenberg J., Viral infection controlled by a calcium-dependent lipid-binding module in ALIX. Dev. Cell 25, 364–373 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larios J., Mercier V., Roux A., Gruenberg J., ALIX- And ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 219, e201904113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horgan C. P., Walsh M., Zurawski T. H., McCaffrey M. W., Rab11-FIP3 localises to a Rab11-positive pericentrosomal compartment during interphase and to the cleavage furrow during cytokinesis. Biochem. Biophys. Res. Commun. 319, 83–94 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Dambournet D., MacHicoane M., Chesneau L., Sachse M., Rocancourt M., El Marjou A., Formstecher E., Salomon R., Goud B., Echard A., Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat. Cell Biol. 13, 981–988 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Scheffer L. L., Sreetama S. C., Sharma N., Medikayala S., Brown K. J., Defour A., Jaiswal J. K., Mechanism of Ca2+-triggered ESCRT assembly and regulation of cell membrane repair. Nat. Commun. 5, 5646 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You S. Y., Park Y. S., Jeon H.-J., Cho D.-H., Bae Jeon H., Kim S. H., Chang J. W., Kim J.-S., Oh J. S., Beclin-1 knockdown shows abscission failure but not autophagy defect during oocyte meiotic maturation. Cell Cycle 16, 1611–1619 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollard T. D., O’Shaughnessy B., Molecular mechanism of cytokinesis. Annu. Rev. Biochem. 88, 661–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yarrow J. C., Totsukawa G., Charras G. T., Mitchison T. J., Screening for cell migration inhibitors via automated microscopy reveals a rho-kinase inhibitor. Chem. Biol. 12, 385–395 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Elia N., Sougrat R., Spurlin T. A., Hurley J. H., Lippincott-Schwartz J., Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. U.S.A. 108, 4846–4851 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berna-Erro A., Izquierdo-Serra M., Sepúlveda R. V., Rubio-Moscardo F., Doñate-Macián P., Serra S. A., Carrillo-Garcia J., Perálvarez-Marín A., González-Nilo F., Fernández-Fernández J. M., Valverde M. A., Structural determinants of 5′,6′-epoxyeicosatrienoic acid binding to and activation of TRPV4 channel. Sci. Rep. 7, 10522 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chubanov V., Waldegger S., M. Mederos y Schnitzler, Vitzthum H., Sassen M. C., Seyberth H. W., Konrad M., Gudermann T., Disruption of TRPM6/TRPM7 complex formation by a mutation in the TRPM6 gene causes hypomagnesemia with secondary hypocalcemia. Proc. Natl. Acad. Sci. U.S.A. 101, 2894–2899 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouchet J., del Río-Iñiguez I., Vázquez-Chávez E., Lasserre R., Agüera-González S., Cuche C., McCaffrey M. W., Di Bartolo V., Alcover A., Rab11-FIP3 regulation of Lck endosomal traffic controls TCR signal transduction. J. Immunol. 198, 2967–2978 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Fededa J. P., Gerlich D. W., Molecular control of animal cell cytokinesis. Nat. Cell Biol. 14, 440–447 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Zarychanski R., Schulz V. P., Houston B. L., Maksimova Y., Houston D. S., Smith B., Rinehart J., Gallagher P. G., Mutations in the mechanotransduction protein PIEZO1 are associated with hereditary xerocytosis. Blood 120, 1908–1915 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bae C., Gnanasambandam R., Nicolai C., Sachs F., Gottlieb P. A., Xerocytosis is caused by mutations that alter the kinetics of the mechanosensitive channel PIEZO1. Proc. Natl. Acad. Sci. U.S.A. 110, E1162–E1168 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tytell M., Lasek R. J., Gainer H., Axonal maintenance, glia, exosomes, and heat shock proteins. F1000Res. 5, F1000 Faculty Rev-205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y., Goliwas K. F., Severino P. E., Hough K. P., Van Vessem D., Wang H., Tousif S., Koomullil R. P., Frost A. R., Ponnazhagan S., Berry J. L., Deshane J. S., Mechanical strain induces phenotypic changes in breast cancer cells and promotes immunosuppression in the tumor microenvironment. Lab. Invest. 100, 1503–1516 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savina A., Fader C. M., Damiani M. T., Colombo M. I., Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 6, 131–143 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Denais C. M., Gilbert R. M., Isermann P., McGregor A. L., te Lindert M., Weigelin B., Davidson P. M., Friedl P., Wolf K., Lammerding J., Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lens S. M. A., Medema R. H., Cytokinesis defects and cancer. Nat. Rev. Cancer 19, 32–45 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Li C., Rezania S., Kammerer S., Sokolowski A., Devaney T., Gorischek A., Jahn S., Hackl H., Groschner K., Windpassinger C., Malle E., Bauernhofer T., Schreibmayer W., Piezo1 forms mechanosensitive ion channels in the human MCF-7 breast cancer cell line. Sci. Rep. 5, 8364 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X., Wanggou S., Bodalia A., Zhu M., Dong W., Fan J. J., Yin W. C., Min H. K., Hu M., Draghici D., Dou W., Li F., Coutinho F. J., Whetstone H., Kushida M. M., Dirks P. B., Song Y., Hui C.-C., Sun Y., Wang L.-Y., Li X., Huang X., A feedforward mechanism mediated by mechanosensitive ion channel PIEZO1 and tissue mechanics promotes glioma aggression. Neuron 100, 799, –815.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Zhou W., Liu X., van Wijnbergen J. W. M., Yuan L., Liu Y., Zhang C., Jia W., Identification of PIEZO1 as a potential prognostic marker in gliomas. Sci. Rep. 10, 16121 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu H., Chen Z., Li C., The prognostic value of Piezo1 in breast cancer patients with various clinicopathological features. Anticancer Drugs 32, 448–455 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Ranade S. S., Qiu Z., Woo S. H., Hur S. S., Murthy S. E., Cahalan S. M., Xu J., Mathur J., Bandell M., Coste B., Li Y. S. J., Chien S., Patapoutian A., Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proc. Natl. Acad. Sci. U.S.A. 111, 10347–10352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J., Hou B., Tumova S., Muraki K., Bruns A., Ludlow M. J., Sedo A., Hyman A. J., McKeown L., Young R. S., Yuldasheva N. Y., Majeed Y., Wilson L. A., Rode B., Bailey M. A., Kim H. R., Fu Z., Carter D. A. L., Bilton J., Imrie H., Ajuh P., Dear T. N., Cubbon R. M., Kearney M. T., Prasad R. K., Evans P. C., Ainscough J. F. X., Beech D. J., Piezo1 integration of vascular architecture with physiological force. Nature 515, 279–282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin-Almedina S., Mansour S., Ostergaard P., Human phenotypes caused by PIEZO1 mutations; one gene, two overlapping phenotypes? J. Physiol. 596, 985–992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valiente M., Obenauf A. C., Jin X., Chen Q., Zhang X. H.-F., Lee D. J., Chaft J. E., Kris M. G., Huse J. T., Brogi E., Massagué J., Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 156, 1002–1016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lewis A. H., Grandl J., Inactivation kinetics and mechanical gating of Piezo1 ion channels depend on subdomains within the cap. Cell Rep. 30, 870–880.e2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doñate-Macián P., Jungfleisch J., Pérez-Vilaró G., Rubio-Moscardo F., Perálvarez-Marín A., Diez J., Valverde M. A., The TRPV4 channel links calcium influx to DDX3X activity and viral infectivity. Nat. Commun. 9, 2307 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Table S1