Abstract
Two novel assays, a restriction fragment length polymorphism (RFLP) assay and an assay based on the 5′-nuclease activity of Taq DNA polymerase, were developed for screening viral variants in lamivudine-treated patients’ sera containing <1,000 copies of the hepatitis B virus (HBV) genome per ml. Both assays were designed to detect single-nucleotide changes within the HBV DNA polymerase gene that are associated with lamivudine resistance in vitro and have been used to screen a number of patients’ sera for variant virus. Results obtained with these assays and standard sequencing technology were compared with regard to throughput, ability to detect individual virus species present at low concentrations, and ability to detect, distinguish, and quantitate wild-type (wt) and HBV tyrosine methionine552 aspartate aspartate motif variants in mixed viral populations. Unlike DNA sequencing, both assays are amenable to high-throughput screening and were shown to be able to quantitatively detect variant virus in the presence of a background of wt virus. As with DNA sequencing, both new assays incorporate a PCR amplification step and are able to detect the relatively low amounts of virus found in lamivudine-treated patients’ sera. However, these assays are far less labor intensive than the DNA-sequencing techniques presently in use. Overall, the RFLP assay was more sensitive than DNA sequencing in detecting and determining the ratios of wt to variant virus. Furthermore, the RFLP assay and 5′-nuclease assay were equally sensitive in the detection of mixed viral species, but the RFLP assay was superior to the 5′-nuclease assay in the quantitation of mixed viral species. These assays should prove useful for further understanding of virological response to therapy and disease progression.
Hepatitis B virus (HBV) is a blood-borne pathogen and a member of the hepadnavirus family. It is estimated that 350 million individuals are chronically infected with HBV and that 1 to 2% will die each year from complications associated with infection, with the majority of these deaths occurring from cirrhosis of the liver and hepatocellular carcinoma (9, 23, 29). The HBV virion contains a small, circular, partially double-stranded segment of DNA of approximately 3.2 kb in size. Replication of HBV occurs via reverse transcription of an intermediate RNA molecule, called pregenomic RNA, which contains a greater-than-unit-length copy of the genome. The HBV genome contains four overlapping reading frames, the largest of which encodes the viral polymerase. HBV polymerase contains both RNA-dependent DNA polymerase (reverse transcriptase) activity and DNA-dependent DNA polymerase activity. Many current therapeutic approaches for treating HBV infection focus on the DNA polymerase as a target for inhibition of viral replication, including nucleoside analogs that terminate viral DNA synthesis (6, 10, 28).
One such nucleoside antiviral for treatment of HBV infection, lamivudine [(−)2′-deoxy-3′-thiacytidine] has been evaluated in phase III clinical trials for the treatment of patients with chronic hepatitis B (19, 31). In these trials, lamivudine treatment was generally well tolerated and was reported to be effective in rapidly reducing serum HBV DNA levels to below the detection limit of standard commercial assays (19). Similarly, an earlier phase II study of extended lamivudine treatment in patients with chronic hepatitis B showed a substantial decrease in patients’ viral load from baseline values, with a median drop in serum HBV DNA levels of 5 log10 units (12). However, reduced virologic response to lamivudine therapy can occur in a portion of patients after long-term treatment, with treatment-emergent HBV variants containing sequence variations in and near the tyrosine methionine552 aspartate aspartate (YMDD) motif of the HBV polymerase (3, 4, 13, 21, 26, 32). Specifically, codon changes at site 528 (from a leucine to a methionine in the B domain of the polymerase) and at site 552 (from a methionine to a valine or isoleucine in the C domain) are involved in reduced susceptibility of HBV to lamivudine in vitro (1).
To confirm the development of HBV nucleotide sequence markers of potential resistance to lamivudine therapy in a patient, it is important to be able to rapidly and easily detect treatment-emergent genomic variations in HBV with accuracy and sensitivity. Furthermore, for patients at high risk for disease progression, it may be important to detect these variations early during the emergence of viral resistance when viral load in the patient is very low and/or when variant virus represents only a minor fraction of the total viral population. In this case, the method used for detecting sequence variations has to be sensitive enough to detect and discern subpopulations of wild-type (wt) and variant HBV at low levels within the same sample. Initially, lamivudine variations were detected by conventional DNA sequencing. Although DNA sequencing provides sequence information over a span of several nucleotides, it is time consuming, laborious, and not sensitive for detecting and quantifying sites of sequence heterogeneity. Due to these limitations, large-scale, detailed studies of the emergence or kinetics of development of HBV variants during treatment are not practical.
Other molecular genetic techniques that overcome some of the limitations of DNA sequencing are available. This report describes two techniques, a restriction fragment length polymorphism (RFLP) assay and a 5′-nuclease assay, that have been developed for detecting single-nucleotide changes in the HBV polymerase gene known to be responsible for the decrease in sensitivity to lamivudine in vitro. The RFLP assay is a sensitive PCR-based assay that utilizes restriction endonucleases that recognize and cut specific sequences in template DNA. Specific DNA patterns, observed by using polyacrylamide gel electrophoresis after enzyme digestion, are indicative of the presence or absence of a specific sequence of DNA. Therefore, restriction enzyme sites, present naturally in the HBV sequence or introduced by PCR primers, were used to confirm the presence or absence of variations associated with in vitro lamivudine resistance. The RFLP method has been commonly used for identifying known polymorphisms in DNA from many organisms or tissues (25), including a report by Chayama et al. that described an RFLP assay for detecting YMDD motif variations associated with in vitro lamivudine resistance at methionine codon 552 from 20 patients on lamivudine therapy (7). Our RFLP assay, which differs from the assay described by Chayama et al., was developed for codons 528 and 552 and was validated by using conventional DNA sequencing. Results showed that the RFLP assay has certain advantages over DNA sequencing, such as higher throughput capacity for sample analysis, greater sensitivity for the detection of mixed populations of wt and variant HBV, and the ability to quantitate mixtures of wt and variant virus after PCR amplification.
In addition to RFLP, a 5′-nuclease assay has been developed for identifying wt and in vitro lamivudine-resistant variant HBV DNA. This technique has been used successfully for discriminating between closely related DNA sequences for the purpose of identifying species from the same family (5, 14, 24, 30). It has also been used for genotyping DNA with known polymorphisms for the purpose of allelic discrimination, such as identifying DNA samples that contain polymorphisms associated with genetic diseases (8, 20).
The 5′-nuclease assay is a sensitive PCR-based assay that utilizes the 5′→3′-exonuclease activity of Taq DNA polymerase to cleave a hybridization probe labeled at the 5′ end with a fluorescent reporter dye and at the 3′ end with a quencher dye during template amplification (20, 22). Probe cleavage occurs only when it is hybridized to DNA that is complementary to the probe sequence. Probe cleavage occurs during extension from the upstream primer during PCR amplification and generates specific fluorescent signals whose intensity can be quantitated by fluorometry. A probe that does not hybridize to the template DNA due to sequence differences is not cleaved and does not generate a fluorescent signal. Therefore, detection of the unique emission spectra from each fluorescent reporter is used to indicate the presence and sequence of the DNA. In the case of allelic discrimination, two probes with different sequences (i.e., wt and variant sequences) are labeled at the 5′ end with a different fluorescent reporter dye and are used during PCR to identify specific DNA sequences that may differ in only 1 bp. Detection of either fluorescent signal or both indicates the sequence(s) of DNA present in the sample.
The 5′-nuclease assay was compared to the RFLP assay for detecting wt and YMDD-variant HBV DNA. Results showed that the 5′-nuclease assay developed for detecting sequence changes in HBV DNA at polymerase codons 528 and 552 was similar to the RFLP assay in terms of sample throughput and detection of mixed populations. However, the RFLP was better than the 5′-nuclease assay in the detection of HBV DNA at low virus concentrations and for quantitation of mixed viral species.
MATERIALS AND METHODS
Patients.
Sera were collected from hepatitis B patients enrolled in a phase II Glaxo Wellcome-sponsored clinical trial of lamivudine therapy. Serum samples of varying viral levels containing wt, variant, or mixtures of wt and variant HBV DNA at detectable levels or undetectable levels of HBV DNA, as previously determined by results from the RFLP assay, were utilized in the study. This was to ensure that all HBV DNA samples observed in the clinic during lamivudine therapy were represented in the comparison studies. To obtain this variety, some of the serum samples used for analysis were sequentially collected from individual patients at different times before, during, and after lamivudine therapy.
Extraction of HBV DNA from patient sera.
HBV DNA was extracted from patient sera using either the QIAamp blood kit (Qiagen, Chatsworth, Calif.) or conventional lysis–phenol-chloroform extraction–ethanol precipitation procedures. By using the QIAamp blood kit according to instructions, 200 μl of serum was mixed with 25 μl of Qiagen protease and 200 μl of Qiagen lysis buffer AL and incubated at 70°C for 30 min, followed by an incubation at 95°C for 10 min. All clinical samples were processed at the same time with both negative (water) and positive (wt, variant, and a mixture of wt and variant HBV DNA) control clinical samples. To minimize the risk of cross-contaminating samples, only one tube of serum was opened at a time during the aliquoting-lysis step. Following incubation, all tubes were subjected to brief centrifugation at 6,000 × g to collect liquid from the caps of the tubes. The samples were vigorously mixed with 210 μl of ethanol, subjected to centrifugation briefly at 6,000 × g, transferred to QIAamp spin columns, and centrifuged at 6,000 × g for 1 min to collect nucleic acids onto filters. The filters were washed twice with 500 μl of Qiagen wash buffer AW, with intermediate centrifugation at 6,000 × g for 1 min followed by centrifugation at 16,000 × g for 3 min to remove residual wash buffer. To elute HBV DNA from the columns, each filter was treated with 100 μl of 70°C preheated 10 mM Tris-HCl, pH 8.0. The filter was subsequently incubated at 70°C for 5 min and subjected to centrifugation at 6,000 × g for 1 min. To obtain a higher yield of DNA from each column, the eluate was reapplied to each column, incubated at 70°C for 5 min, and subjected to centrifugation at 6,000 × g for 1 min. During the entire procedure, including sample transfer, all tubes and columns (except the working column) were covered in order to minimize cross-contamination of samples. The DNA was stored at −20°C until further processing.
Using conventional phenol-chloroform methods of DNA extraction, 250 μl of patient serum was lysed in a final volume of 400 μl containing 2.2 mg of proteinase K per ml (Boehringer Mannheim, Indianapolis, Ind.), 5 mM EDTA, pH 8.0, 10 mM Tris-HCl, pH 8.3, and 0.5% sodium dodecyl sulfate and was incubated at 55°C for 2 h. The lysate was transferred to a 1.5-ml microcentrifuge tube containing 300 μl of heavy Phase lock gel (5 prime-3 prime, Inc., Boulder, Colo.) and 400 μl of phenol-chloroform-isoamyl alcohol (25:24:1). The mixture was mixed vigorously for 1 min by vortexing and was subjected to centrifugation for 10 min at 16,000 × g. An additional 400 μl of phenol-chloroform-isoamyl alcohol (25:24:1) was added to the mixture, and the vortexing and centrifugation steps were repeated. The aqueous phase was carefully removed, placed in a 1.5-ml microcentrifuge tube containing 400 μl of chloroform-isoamyl alcohol (24:1), vigorously mixed for 1 min by vortexing, and subjected to centrifugation for 10 min at 16,000 × g. The aqueous phase was removed and placed in a new 1.5-ml microcentrifuge tube to which 50 ng of glycogen was added along with sodium acetate, pH 5.2, to a final concentration of 0.3 M. The DNA was precipitated with 2 volumes of 95% ethanol and was incubated for at least 1 h at −80°C. The DNA was pelleted by centrifugation for 20 min at room temperature at 16,000 × g, washed with 500 μl of cold 80% ethanol, mixed briefly, and subjected to centrifugation for 10 min at 16,000 × g. The DNA pellet was allowed to dry at room temperature for 30 min and subsequently resuspended in either 25 or 250 μl of 10 mM Tris-HCl, pH 8.0. The DNA was stored at −20°C until further processing.
DNA sequencing.
All sequencing reactions and gel assays were performed at the Glaxo Wellcome Sequencing Facility using ABI standard reagents and protocols for the ABI 373 automated DNA Sequencer. HBV DNA samples were prepared for sequencing by amplification with PCR. A total of two forward and reverse primer pairs were used for amplification, 377/840 and 12F/5RC (see Table 1 for sequence of primers). Primers were selected based on conserved regions in the HBV genome, and each pair spanned the genome to include the YMDD motif. HBV DNA extracted from patient sera was amplified by PCR in a final volume of 50 μl containing 20 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 18 mM NaCl, a 0.2 mM concentration of each 2′-deoxynucleoside triphosphate (2′-dNTP) (dATP, dCTP, dTTP, and dGTP), 0.01% gelatin, a 250 nM concentration of each forward and reverse primer, 0.625 U of AmpliTaq polymerase (Perkin Elmer, Foster City Calif.), and 0.14 μg of Taq Start antibody (ClonTech, Palo Alto, Calif.). Thermocycler conditions for amplification were one cycle of 94°C for 5 min, 40 cycles of 94°C for 30 s, 55°C for 15 s, and 72°C for 60 s, and one cycle of 72°C for 5 min followed by a hold at 4°C. PCR products were purified with a QIAquick PCR purification kit (Qiagen) according to instructions and were eluted from the column with 80 μl of molecular-biology-grade distilled deionized water. An estimation of DNA quality and concentration was determined by absorbance measurements at 260 and 280 nm and by gel electrophoresis on a 1% agarose gel. The DNA sequence of the 377-840 PCR product was generated by the 377, 840, and 5RC primers. The DNA sequence of the 12F-5RC PCR product was generated by the 12F, 5RC, and 377 primers.
TABLE 1.
DNA sequence of all primers and probes used for HBV DNA analyses
| Primers and probes | DNA sequence |
|---|---|
| Primers used for conventional DNA sequencing | |
| 377 | 5′-GGATGTGTCTGCGGCGTTT-3′ |
| 840 | 5′-ACCCCATCTTTTTGTTTTGTTAGG-3′ |
| 12F | 5′-AGACTCGTGGTGGACTTCTCT-3′ |
| 5RC | 5′-CAAAAGAAAATTGGTAACAGCGGTA-3′ |
| Primers used for RFLP analysis | |
| F1 | 5′-CACTGTTTGGCTTTCAGTCAT-3′ |
| F3 | 5′-GTGGGCCTCAGTCCGTTTCTC-3′ |
| B2 | 5′-GTTCAAATGTATACCCAAAG-3′ |
| Primers used for amplification for the 5′-nuclease assay for: | |
| Codon 528 | |
| HBVG.F464 | 5′-TGTTGCCCGTTTGTCCTC-3′ |
| HBVG.R711 | 5′-AGCCAAACAGTGGGGGAA-3′ |
| Codon 552 | |
| HBVG.F696A | 5′-TCAGTGGTTCGTAGGGCTTT-3′ |
| HBVG.R840A | 5′-CCATCTCTTTGTTTTGTTAGG-3′ |
| Fluorescent probes used for 5′-nuclease assay for: | |
| Codon 528 | |
| Set I | |
| 528wt | 5′-FAM-CCCGTTTCTCCTGGCTCAGTTTACTAGT-TAMRA 3′ |
| 528mut | 5′-TET-CCCGTTTCTCATGGCTCAGTTTACTAGT-TAMRA 3′ |
| Set II | |
| 528wt | 5′-FAM-TCCGTTTCTCTTGGCTCAGTTTACTAGTGC-TAMRA 3′ |
| 528mut | 5′-TET-TCCGTTTCTCATGGCTCAGTTTACTAGTGC-TAMRA 3′ |
| Codon 552 | |
| Set I | |
| 552wt | 5′-FAM-TGGCTTTCAGTTATATGGATGATGTGG-TAMRA 3′ |
| 552mut1 | 5′-TET-TGGCTTTCAGTTATGTGGATGATGTG-TAMRA 3′ |
| 552mut2 | 5′-HEX-TTGGCTTTCAGTTATATTGATGATGTGG-TAMRA 3′ |
| Set II | |
| 552wt | 5′-FAM-TGGCTTTCAGCTATATGGATGATGTG-TAMRA 3′ |
| 552mut1 | 5′-TET-TGGCTTTCAGCTATGTGGATGATGT-TAMRA 3′ |
| 552mut2 | 5′-HEX-TGGCTTTCAGCTATATTGATGATGTGG-TAMRA 3′ |
Boldface and italics indicate sites of heterogeneity due to subtype differences; boldface and underlining indicate sites of sequence variations in HBV species associated with in vitro lamivudine resistance.
The RFLP assay for DNA sequence determination at codons 528 and 552 in the HBV polymerase gene associated with lamivudine resistance in vitro.
A rapid method using RFLP technology for detection of lamivudine-resistant variations in HBV has been developed. To examine the presence or absence of a methionine codon at site 552 or the presence or absence of a leucine codon at site 528 in the HBV polymerase gene, HBV DNA extracted from patient sera was amplified by PCR in a final volume of 50 μl containing 20 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 18 mM NaCl, a 0.2 mM concentration of each 2′-dNTP (dATP, dCTP, dTTP, and dGTP), 0.01% gelatin, a 500 nM concentration of each primer, 1.5 U of AmpliTaq polymerase, 0.34 μg of Taq Start antibody, and 10 μl of DNA. The starting concentration of HBV DNA amplified ranged from 25 to 100 pg/ml based on values previously determined by the conventional solution hybridization assay (Abbott Diagnostics, North Chicago, Ill.). Those samples containing greater than 100 pg/ml were diluted to the appropriate range with 10 mM Tris-HCl, pH 8.0. Thermocycler conditions for amplification were 40 cycles of 92°C for 30 s, 55°C for 45 s, and 72°C for 30 s, followed by a hold at 8°C.
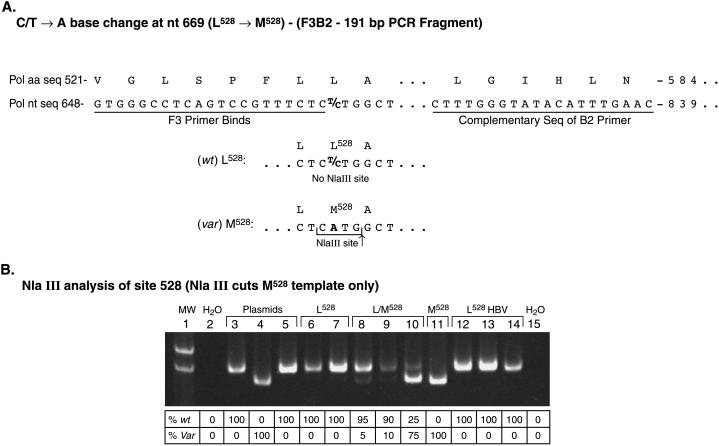
To examine the 528 codon, primer pairs F3 and B2 were used for PCR amplification. Primer F3 anneals just upstream from the leucine 528 codon in the polymerase. The variation at the 528 codon, associated with lamivudine resistance in vitro, from leucine (C/TTG) to methonine (ATG) created an NlaIII restriction site (Fig. 1).
FIG. 1.
Design and gel image of an RFLP assay for rapid detection of variant HBV at codon 528. (A) PCR primers designed to amplify the region of codon 528 (primers F3 and B2) have been used to detect the presence of the methionine codon at 528. A methionine at codon 528 creates an NlaIII restriction site (CATG↓), whereas the NlaIII site is absent in the wt sequence. (B) Result of sample analysis at codon 528 after NlaIII enzyme digestion. HBV DNA containing either the wt (Leu), variant (Met), or both sequences at codon 528 can be determined by PCR amplification with primers F3 and B2, digestion of PCR products with NlaIII, and resolution of DNA band size on a 6% polyacrylamide gel. The faster migrating band indicates the variant sequence, whereas the slower migrating band indicates the wt sequence. Lane 1, molecular weight markers; lane 2, water (negative control); lane 3, wt pCMVHBV plasmid; lane 4, L528M/M552V pCMVHBV plasmid; lane 5, M552I pCMVHBV plasmid; lanes 6 to 14, HBV DNAs from sera of different patients on lamivudine therapy that are either wt at codon 528, L528M, or both; lane 15, water. The amount of each DNA present in lanes (expressed as a percentage) is shown below the image and was quantitated as described in Materials and Methods. Var, variant.
To examine the 552 codon by RFLP analysis, primer pairs F1 and B2 were used for PCR amplification. Primer F1 anneals just upstream from the methionine codon of the YMDD motif of the HBV polymerase and changes the tyrosine codon from TAT to CAT. This nucleotide change created an NdeI (CA↓TATG) site when the HBV DNA template was of the wt sequence (methionine or ATG at codon 552) (Fig. 2). If the HBV DNA template had a variation at codon 552, either a valine (GTG) or isoleucine (ATT) associated with lamivudine resistance, then the NdeI restriction site was absent from the amplified DNA. However, the presence of the valine variation at 552 created a new restriction site, NlaIII (CATG↓), allowing the determination of the presence of either a valine or isoleucine residue (or both) in the variant template by digestion with the NlaIII restriction enzyme.
FIG. 2.
Design and gel image of an RFLP assay for rapid detection of HBV variant at codon 552. (A) For detecting variations at codon 552 in the HBV polymerase, the YMDD nucleotide sequence TATATGGATGAT was used, and a 5′-PCR primer (primer F1) was designed that contains a CAT sequence at the 3′ end (changing the first tyrosine codon from a TAT to a CAT) in order to introduce an NdeI restriction site (CA↓TATG) upon amplification of part of the HBV polymerase gene with the 5′-PCR primer and a 3′-PCR primer located 120 bases downstream. The creation of an NdeI site occurs only if the template HBV has the wt DNA sequence and is absent if the template HBV is mutated, indicating the loss of the NdeI site only. (B) Result of sample analysis at codon 552 after NdeI enzyme digestion. Resolution of PCR products on a polyacrylamide gel after amplification of template HBV DNA using primers F1 and B2 with subsequent digestion using NdeI indicates either wt sequence (faster moving band) or variant sequence (slower moving band). Lane 1, molecular weight markers; lane 2, water (negative control); lane 3, wt pCMVHBV plasmid; lane 4, L528M/M552V pCMVHBV plasmid; lane 5, M552I pCMVHBV plasmid; lanes 6 to 11, HBV DNA from sera of different patients undergoing lamivudine therapy that are either wt at codon 552, M552V, or both; lanes 12 to 14, HBV DNA from sera of different patients on lamivudine therapy that are wt at codon 552, M552I, or both; lane 15, water. The amount of each DNA present in lanes (expressed as a percentage) is shown below the image and was quantitated as described in Materials and Methods. var, variant. (C) After determining the sequence (wt or variant) of a sample from results of the NdeI analysis shown in Fig. 2B, the sequence of the variant template (Val or Ile) can be determined by NlaIII enzyme digestion of the PCR products. Resolution of the PCR products on a polyacrylamide gel after amplification of template HBV DNA using primers F1 and B2 with subsequent digestion using NlaIII indicates either a valine sequence (faster moving band) or methionine or isoleucine sequence (slower moving band) at codon 552. Lane 1, molecular weight markers; lane 2, water (negative control); lane 3, wt pCMVHBV plasmid; lane 4, L528M/M552V pCMVHBV plasmid; lane 5, M552I pCMVHBV plasmid; lanes 6 to 8, HBV DNA from sera of different patients undergoing lamivudine therapy that are either wt at codon 552, M552V, or both; lanes 9 to 11, HBV DNA from sera of different patients undergoing lamivudine therapy that are wt at codon 552, M552I, or both; lane 15, water. The amount of each DNA present in lanes (expressed as a percentage) is shown below the image and was quantitated as described in Materials and Methods.
HBV DNA samples extracted from patient sera were subjected to PCR amplification with primer pair F1 and B2 (for analysis of the 552 site) or F3 and B2 (for analysis of the 528 site). A 5-μl aliquot of each PCR product was digested with NdeI or NlaIII for 1 h at 37°C with enzyme replenishment after the initial 30-min incubation. The resulting DNA fragments were resolved by electrophoresis on a 6% nondenaturing polyacrylamide gel. Since no other NdeI or NlaIII sites were present in the amplified regions, the differences between undigested PCR products and digested PCR products were 20 (NdeI) and 24 (NlaIII) bp. DNA bands were detected by exposure to UV illumination after staining with 0.01% SYBR Green (FMC, Rockland Maine) for 10 min. Gel images were captured by using a Bio-Rad Gel Doc 1000 system (Bio-Rad Laboratories, Inc., Hercules, Calif.). Band intensities in lanes with both wt and variant sequences, as indicated by the presence of digested and undigested PCR products, were analyzed with Molecular Analyst version 2.1.1 software (Bio-Rad).
A plasmid (pCMVHBV) (11) containing a copy of the wt HBV genome (subtype ayw) was used to generate the variant HBV constructs L528M/M552V and M552I as described by Allen et al. (1). Each plasmid was linearized by XhoI restriction enzyme digestion and used at 25 pg/ml as a control for amplification and enzyme digestion during RFLP analysis. Prior to amplification with the F1 and B2 primers for codon 552 analysis only, the control wt plasmid was diluted and used at the following concentrations: 1010, 109, 108, 107, 104, 103, 500, 250, 125, 62.5, and 31.25 copies/ml. The resulting standard curve was used for every RFLP analysis to ensure that the PCR products were completely digested within this range of DNA concentrations. The wt plasmid control was also used to establish a lower limit of HBV DNA detection for each RFLP analysis at codon 552. In addition, the wt plasmid was used to establish the lower limit of detection by the RFLP assay for codon 528 analysis using the same concentrations of plasmid as described for codon 552 analysis.
The wt and L528M/M552V plasmids were used to determine the capacity of the RFLP assay to detect wt HBV and HBV variant at codon 552 in a single sample. An estimation of DNA quality and concentration was determined by absorbance measurements at 260 and 280 nm at several dilutions of each plasmid for accuracy. The two control plasmids were diluted to 50,000, 25,000, 10,000, 5,000, 2,500, 1,250, and 625 copies/ml and were mixed at 0:100, 10:90, 25:75, 50:50, 75:25, 90:10, and 100:0 wt-to-variant ratios for each concentration. RFLP analysis for the 552 codon was performed in replicates of six for each ratio at each concentration, using the same conditions described above. The data was compiled using a logistic-regression curve of the log of copies per milliliter versus the probability of detecting the response, and from logistic regression, an inverse prediction was performed to obtain an estimate of the copies per milliliter associated with a 95% confidence level by JMP statistical software (SAS, Cary, N.C.).
The 5′-nuclease PCR assay for detecting single-base polymorphisms at codons 528 and 552.
A fluorogenic 5′-nuclease assay was developed and evaluated for its ability to specifically detect single nucleotide polymorphisms at codons 528 and 552, characteristic of variant HBV with genotypic lamivudine resistance. Reagents for the 5′-nuclease assay were purchased from Perkin-Elmer. The fluorescently labeled oligonucleotide probes (Table 1) were synthesized by either Genset Corp (Paris, France) or Synthetic Genetics (San Diego, Calif.). The oligonucleotides contained one of the following reporter dyes: 6-carboxyfluorescein (FAM), 6-carboxy-4,7,2′,7′-tetrachlorofluorescein (TET), or hexachlorofluorescein (HEX) phosphoramidites at the 5′ end and the quencher dye 6-carboxytetramethylrhodamine (TAMRA) with a phosphate linked to the 3′ end of each probe to prevent probe extension during amplification. All primer and probe sequences were designed with Oligo 5.0 software (National Biosciences, Plymouth, Minn.). Due to the sequence heterogeneity that naturally exists between the different subtypes of HBV (see below) in the region where the probes hybridize, two sets of probes (wt and variant) were designed for each virus type, to take into account those sequence variations that are not due to variants associated with genotypic lamivudine resistance. This was necessary since sequence determination of HBV by this technique uses probe hybridization to distinguish a single nucleotide difference between wt and lamivudine-variant templates.
The site 528 wt sequences were as follows: set I (subtypes ayw, ayr, and adr) (nucleotides 659 to 685), CCCGTTTCTCCTGGCTCAGTTTACTAG, and set II (subtypes adw2 and adw), TCCGTTTCTCTTGGCTCAGTTTACTAG. The site 552 wt sequences were as follows: set I (subtypes ayw and adr) (nucleotides 726 to 753), TTGGCTTTCAGTTATATGGATGATGTGG, and set II (subtypes adw2, adw, and ayr), TTGGCTTTCAGCTATATGGATGATGTGG. Boldface nucleotides indicate HBV DNA sequence differences due to subtype heterogeneity. Underlined nucleotides indicate sites where differences in the DNA sequence occur between wt and genotypic lamivudine-resistant variant HBV.
All reactions were performed in Perkin-Elmer 96-well optical plates with optical caps by the Perkin-Elmer ABI7700 thermocycler-fluorescent plate reader. Replicates of eight wells containing either wt or variant XhoI-linearized pCMVHBV plasmid or water were used as control standards for amplification and allelic discrimination. ABI7700 Sequence Detector version 1.6.3 software was used for sample analysis to determine the final fluorescent measurements of each well after each cycle during amplification and for allelic discrimination.
All reactions for site 552 containing set I or II probes were performed under similar conditions. Each reaction mixture contained a final concentration of 1× Taqman buffer, 4.5 or 4.75 mM MgCl2, 200 μM (each) dNTP, 5% glycerol, 0.01 U of uracil-N-glycosylase, 0.04 U of AmpliTaq Gold, 200 nM (each) wt and one variant (Val or Ile) probe, and 800 nM (each) forward and reverse primer in a final volume of 50 μl. Thermocycling conditions were one cycle at 50°C for 2 min, one cycle at 95°C for 10 min, 40 cycles at 95°C for 15 s, and 65°C for 60 s, followed by a hold at 4°C.
For site 528, all reactions were performed using the Taqman Universal PCR master mix (Perkin-Elmer). For set I, each reaction mixture contained the components of 1× PCR master mix (buffer, MgCl2, dNTPs, glycerol, uracil-N-glycosylase, and AmpliTaq Gold), 300 nM forward and 900 nM reverse primers, and 200 nM wt and 50 nM variant probes in a final volume of 50 μl. Thermocycling conditions were identical to those conditions used for site 552, except the annealing step was performed at 62°C. For set II, components of the PCR mixture were identical to set I except the variant probe concentration was 100 nM, and the annealing temperature was 64°C. The reason for the differences in probe concentrations between set I and II reactions was due to fluorescent variations in the probes themselves. Probes were optimized based on their initial fluorescence according to Perkin-Elmer protocol number 4303267.
RESULTS
A more sensitive and rapid method than DNA sequencing was needed to detect and quantitate HBV wt and YMDD-variant species in the sera of patients undergoing lamivudine therapy, in order to study the kinetics of emergence of these viral variants in large numbers of patients. An RFLP assay was developed for detecting, in HBV DNA amplified from patients’ sera, viral DNA markers associated with lamivudine resistance in vitro. To determine the sensitivity of the RFLP assay, lower limits of detecting HBV DNA were established for codons 528 and 552 of 300 and 500 copies/ml (95% confidence), respectively. The capacity of the RFLP assay to detect wt and variant (L528M/M552V) HBV in a single sample at codon 552 was determined by mixing different ratios of wt and variant plasmids and diluting over a range of concentrations. The concentrations of HBV DNA (lower limit) at the 95% confidence level for detecting samples containing mixed populations of virus for each wt-to-variant ratio are shown in Table 2.
TABLE 2.
Lower limit of HBV DNA concentrations for the detection of mixed populations of wt and M552V sequences by RFLP assay
| Ratio [wt(M552): variant(V552)] | Total HBV DNA in log10 units (approx copies/ml) |
|---|---|
| 100:0 | <2.80 (<625) |
| 10:90 | 3.67 (5,000) |
| 25:75 | 3.29 (2,000) |
| 50:50 | 2.83 (700) |
| 75:25 | 3.54 (3,500) |
| 90:10 | 3.49 (3,100) |
| 0:100 | <2.80 (<625) |
To compare sensitivity and validity of results, both conventional DNA sequencing and RFLP analyses were performed on 50 serum samples harvested from patients before, during, or after lamivudine therapy. Results obtained by these methods for identifying defined single-nucleotide changes within codons 528 and 552 are presented in Table 3. Results from samples analyzed with both the RFLP method and the DNA-sequencing procedure were in good agreement, regardless of virus type (wt or variant). Using the RFLP analysis, we were able to quantitate the mixtures of wt and variant virus of samples, whereas DNA sequencing revealed only the presence or absence of an absorbance peak for each base and was not quantitative. Identical results were achieved by each method for codon 528 in 38 of 50 (76%) samples, whereas 36 of 50 (72%) samples had identical results after both methods were used at codon 552. In 7 of 50 (14%) (at codon 528) and 10 of 50 (20%) (at codon 552) samples, the RFLP method was more sensitive than DNA sequencing for detecting samples with mixed populations of wt and variant sequences. In those samples, DNA sequencing was unable to detect any DNA or was able to detect only one population and was unable to detect a mixture of viruses. For analysis of both codons, DNA sequencing produced results in only 3 of 50 (6%) samples where RFLP could not. Results for two samples at codon 528 and one sample at codon 552 could not be obtained by either method.
TABLE 3.
Comparison of results obtained by DNA sequencing versus RFLP at codons 528 and 552 for 50 HBV DNA samplesa
| Codon 528
|
Codon 552
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Results (no. of samples) | Amino acid(s) detected (no. of samples) | Results of:
|
Summary of viral load | Results (no. of samples) | Amino acid(s) detected (no. of samples) | Results of:
|
Summary of viral load | ||
| DNA sequencing | RFLP | DNA sequencing | RFLP | ||||||
| RFLP equals DNA sequencing (38) | wt-L528 (27) Variant-M528 (7) wt/variant mixes (4) | L M (L)/M (L)/M L/M L/M | L M L(75)/M(25) L(65)/M(35) L/(M) L(80)/M(20) | 17 samples had HBV DNA that was detectable by the SH or bDNA assay. Of the 14 samples undetectable by the SH assay, 10 contained greater than 10,000 copies of DNA per ml by the in-house PCR quantitative assay, three had greater than 1,000 copies of DNA ml, and only one sample had less than 1,000 copies of DNA per ml. Of the seven samples that were NA by commercial assays, six had greater than 10,000 copies of DNA ml by PCR assay, and one sample was NA. | RFLP equals DNA sequencing (36) | wt-M552 (15) Variant I-V552 (8) Variant II-I552 (5) wt/variant mixes (8) | M V I (M)/V (M)/V M/V M/V (M)/V M/I M/I M/I | M V I M(45)/V(55) M(15)/V(85) M(55)/V(45) M(80)/V(20) M(40)/V(60) (M)/I M(90)/I(10) M(35)/I(65) | 17 samples had HBV DNA that was detectable by the SH or bDNA assay. Of the 11 samples undetectable by the SH assay, eight contained greater than 10,000 copies of DNA per ml by the PCR assay, two had greater than 1,000 copies of DNA per ml, and only one had less than 1,000 copies of DNA per ml. Of the eight samples that were NA by commercial assays, six had greater than 10,000 copies of DNA per ml by the PCR assay, and two samples were NA. |
| RFLP greater than DNA sequencing (7) | L L M M M M 0 | L/(M) L(15)/M(85) (L)/M L(10)/M(90) L(10)/M(90) L(50)/M(50) L | Only three samples had HBV DNA that was detectable by the SH or bDNA assay. Of the three samples undetected by the SH assay, two contained greater than 10,000 and one contained less than 1,000 copies of DNA per ml by the PCR assay. Only one sample was NA by both assays. | RFLP greater than DNA sequencing (10) | M M M M M M M V 0 0 | M(85)/V(15) M(85)/V(15) M(90)/V(10) M(80)/V(20) M(85)/V(15) M/(V) M(25)/I(75) M(25)/V(75) (M) M(10)/I(90) | Only three samples had HBV DNA that was detectable by the SH or bDNA assay. Of the six samples undetected by the SH assay, four contained greater than 10,000, one contained greater than 1,000, and one contained less than 1,000 copies of DNA per ml by the PCR assay. One sample was NA but was less than 1,000 copies of DNA per ml by the PCR assay. | ||
| DNA sequencing greater than RFLP (3) | L L L/(M) | 0 0 0 | All samples contained HBV DNA below the detection limit of the SH assay and were less than 1,000 copies of DNA per ml by the PCR assay. | DNA sequencing greater than RFLP (3) | M M/V/I M | 0 0 0 | All samples contained HBV DNA below the detection limit of the SH assay and were less than 1,000 copies of DNA per ml by the PCR assay. | ||
| Undetermined (2) | 0 0 | 0 0 | One sample contained HBV below the detection limit of the SH assay and was less than 1,000 copies of DNA per ml by the PCR assay; one sample was NA by both commercial assays and was less than 1,000 copies of DNA per ml by the PCR assay. | Undetermined (1) | 0 | 0 | Sample was below the detection limit of the SH assay and was less than 1,000 copies of DNA per ml by the PCR assay. | ||
HBV DNA was extracted from 50 different serum samples from individual patients undergoing lamivudine therapy and was analyzed for variations associated with lamivudine resistance at codon 528 and 552 in HBV polymerase by DNA sequencing or by the RFLP method described above. Results are indicated by the one-letter amino acid code. More than one population in a single sample is indicated by more than one letter and letters are separated by a forward slash. The numbers shown in parentheses indicate the percentage of HBV DNA of a particular viral species. The letters in parentheses indicate that the HBV DNA species, representing <5% of the entire sample, could be visualized on a polyacrylamide gel after electrophoresis, but not quantitated by the software. 0 indicates that a result could not be obtained. SH, solution hybridization; NA, data not available for that sample.
Of the patients’ serum samples that gave results with either method for each codon, the majority had high HBV DNA levels as determined by the conventional solution hybridization assay, the Chiron bDNA Quantiplex assay (Chiron Corporation, Emeryville, Calif.), or by a quantitative PCR-based assay. The solution hybridization assay and the Chiron bDNA assay have relatively high lower limits of HBV DNA detection (1.6 pg/ml; equivalent to roughly 1 × 106 copies of HBV DNA/ml and 7 × 105 copies/ml, respectively) (16). A quantitative and sensitive PCR assay for HBV DNA has been developed with the much lower limit of detection of approximately 750 copies/ml, and it is an adaptation of a viral quantitation assay described in Jansen et al. (15). This assay was used for determining viral load for most of the samples that had a solution hybridization assay reading of <1.6 pg/ml. The ability to obtain a result by the RFLP assay or DNA sequencing correlated with the amount of virus present in the serum sample or viral level. The majority of samples that yielded results by either one or both methods had viral level measurements of at least 10,000 (38 samples [76%]) or 1,000 (3 samples [6%]) copies of HBV DNA/ml (Table 3). Only 10% (codon 528) or 12% (codon 552) of the samples that could be identified by either one or both methods had low viral levels below 1,000 copies/ml. Samples that could not be detected by either method had viral levels that were below the detection limit of both the solution hybridization and PCR quantitative assays.
The results from these experiments demonstrate that the RFLP assay has some advantages over DNA sequencing, including higher throughput of sample analysis and the ability to more precisely detect and quantitate mixed populations of wt and variant viruses. Since each method is based on PCR amplification, sequence information could be obtained from samples with relatively low viral levels. Overall, the results obtained by RFLP analysis were confirmed and were comparable to results obtained by DNA sequencing.
RFLP analysis and a 5′-nuclease assay were performed on 40 HBV DNA samples isolated from the sera of patients on lamivudine therapy to compare the sensitivity and validity of each assay. Results from using these methods are presented in Table 4. Results from all samples analyzed by the RFLP assay and by the 5′-nuclease assay were in agreement. Both methods were sensitive in their ability to detect viral DNA and were able to detect the presence of both wt and variant viruses in a single sample. The ability of the 5′-nuclease assay to detect mixtures of virus was examined by mixing known ratios of plasmids containing wt and variant HBV DNA, and results indicated that samples containing at least 20% of the minor species could consistently be detected (data not shown).
TABLE 4.
Comparison of results obtained by RFLP versus 5′-nuclease assay at codons 528 and 552 for 40 HBV DNA samplesa
| Codon 528
|
Codon 552
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Results (no. of samples) | Amino acid(s) detected (no. of samples) | Results of:
|
Summary of viral load | Results (no. of samples) | Amino acid(s) detected (no. of samples) | Results of:
|
Summary of viral load | ||
| RFLP | 5′-NA | RFLP | 5′-NA | ||||||
| 5′-NA equals RFLP (18) | wt-L528 (15) Variant-M528 (3) wt/variant mixes (0) | L M | L M | 17 samples had HBV DNA that was detectable by the SH assay. One sample was undetectable by the SH assay and contained greater than 1,000 copies of DNA per ml by the PCR quantitative assay. | 5′-NA equals RFLP (23) | wt-M552 (18) Variant I-V552 (3) Variant II-I552 (1) wt/variant mixes (1) | M V I M(45)/V(55) | M V I M/V | 19 samples had HBV DNA that was detectable by the SH or bDNA assay. Three samples were undetectable by the SH assay and had greater than 10,000 copies of DNA per ml by the PCR quantitative assay. One sample was NA by both assays. |
| 5′-NA greater than RFLP (6) | 6 | L | L/M | All samples had HBV DNA that was undetectable by the SH assay and contained greater than 10,000 copies of DNA per ml by the PCR assay. | 5′-NA greater than RFLP (1) | 1 | M | M/V | Sample had HBV DNA that was detectable by the SH assay. |
| RFLP greater than 5′-NA (8) | 1 5 1 1 | L/M L (L) L(75)/M(25) | M 0 0 0 | Four samples contained HBV DNA that was detectable by the SH assay. Of the two that were undetectable by the SH assay, one contained greater than 10,000 copies of DNA per ml, and one contained less than 1,000 copies of DNA per ml by the PCR assay. Two samples were NA by both assays. | RFLP greater than 5′-NA (8) | 2 1 1 3 1 | M/(V) M(85)/V(15) M(55)/V(45) M V | M M V 0 0 | One sample was detectable by the SH assay. Six samples contained HBV DNA below the detection limit of the SH assay; four were greater than 10,000 copies of DNA per ml, one was greater than 1,000 copies of DNA per ml, and one was less than 1,000 copies of DNA per ml by the PCR assay. One sample was NA by both assays. |
| Undetermined (8) | 8 | 0 | 0 | One sample contained HBV DNA that was detectable by the SH assay. Six samples contained HBV DNA below the detection limit of the SH assay; all samples contained less than 1,000 copies of DNA per ml by the PCR assay. One sample was NA by the SH assay but had less than 1,000 copies of DNA per ml by the PCR assay. | Undetermined (8) | 8 | 0 | 0 | One sample was detectable by the SH assay. Six samples contained HBV DNA below the detection limit of the SH assay and contained less than 1,000 copies of DNA per ml by the PCR assay. One sample was NA by the SH assay and contained less than 1,000 copies of DNA per ml by the PCR assay. |
HBV DNA was extracted by conventional phenol-chloroform methods (described above) from 40 different serum samples from individual patients undergoing lamivudine therapy and was analyzed for variants associated with lamivudine resistance at codons 528 and 552 in the HBV polymerase by the RFLP method or by the 5′-nuclease assay described above. Results are indicated by the one-letter amino acid code. More than one population in a single sample is indicated by more than one letter and letters are separated by a forward slash. The numbers shown in parentheses indicate the percentage of HBV DNA of a particular virus type. The letters in parentheses indicate that the HBV DNA species, representing <5% of the entire sample, could be visualized on a polyacrylamide gel but not quantitated by the software. 0 indicates that a result could not be obtained. 5′-NA, 5′-nuclease assay; SH, solution hybridization; NA, data not available for that sample.
Identical results were achieved by the RFLP and 5′-nuclease methods for codon 528 in 18 of 40 (45%) samples, whereas 23 of 40 (57.5%) samples had identical results with both methods at codon 552. For the analysis of both codons, the RFLP assay detected virus or more than one virus in 8 of 40 (20%) samples that were not detected by the 5′-nuclease assay. In 6 of 40 (15%) (at codon 528) and 1 of 40 (2.5%) (at codon 552) samples, the 5′-nuclease assay was more sensitive than RFLP for detecting samples with mixed populations of wt and variant sequences. Results could not be obtained with eight samples (20%) by either method.
Viral level measurements of most of these samples were determined by either the solution hybridization assay or the in-house quantitative PCR assay. The majority of the samples that could be identified by either one or both methods had viral level measurements of at least 10,000 (29 samples) or 1,000 (one sample) copies of HBV DNA/ml (Table 4). Only one sample that was identified by either one or both methods had a viral level of <1,000 copies/ml. The majority of samples that could not be identified by either one or both methods had viral levels that were less than 1,000 copies/ml (seven samples).
DISCUSSION
HBV variants with point mutations in the viral polymerase gene have been detected in patients on prolonged lamivudine therapy, and such variants have been associated with reduced susceptibility to lamivudine in vitro (1, 18). The most common variations are located in or near the YMDD motif of the HBV polymerase at codon 528 (L528M) and 552 (M552V/I) (1, 4, 21, 32). These variations are detectable in a portion of patients after a year of therapy (phase III average of 24% of patients) (2) and usually appear after at least 6 to 8 months of therapy (12, 19, 31). However, to obtain more extensive information regarding the kinetics of emergence of these HBV variants, sensitive, quantitative, high-throughput assays are required that can detect small amounts of variant HBV at low viral titers in the presence of wt HBV in a single serum sample. This report describes the development of two assays, an RFLP and a 5′-nuclease assay, for detecting HBV viral populations in patient sera with variations in the HBV DNA polymerase gene associated with reduced susceptibility to lamivudine.
The RFLP assay was used for rapid analysis of potential lamivudine-resistant variants using 50 patient serum samples and was found to produce results comparable to those obtained by conventional DNA sequencing. Variations in results were due to better sensitivity of one method over the other for a given sample, and no discrepancies were observed for the results obtained by either method. The RFLP assay was able to detect more serum samples containing a mixture of wt and variant virus than was the DNA-sequencing assay. Similarly, an RFLP assay designed to detect a mutation resulting in a stop codon in the precore region of HBV was shown to be more sensitive than DNA sequencing for samples containing a mixture of wt and mutant viruses (25).
In addition to identifying HBV variants present in serum samples, we were able to use the RFLP technique to quantitatively determine the ratio of wt to variant HBV within a serum sample. In some cases, DNA sequencing was able to detect samples with mixed virus populations. However, this method could not be used to determine the proportion of each virus present in the sample. Furthermore, results from more samples in a shorter amount of time could be obtained by the RFLP assay than by DNA sequencing. The RFLP assay described in this paper differs from that developed by Chayama et al. (7). Our assay detected variants associated with in vitro lamivudine resistance at codons 528 and 552, utilized different restriction enzyme sites, required a single round of nonnested PCR, and was quantitative.
A 5′-nuclease assay was used to identify wt and variant HBV in 40 patient serum samples, and results were compared to those obtained with the RFLP assay. With both assays, it was possible to detect wt and variant HBV DNA on the basis of a single nucleotide polymorphism. For the 5′-nuclease assay, more than one probe was required for each codon because of the natural variations in different serological subtypes of HBV and the stringent requirement for probe hybridization to the exact DNA sequence. The 5′-nuclease assay has potential to have higher throughput than the RFLP assay due to the absence of post-PCR manipulation. However, the requirement for custom probes due to subtype sequence differences resulted in a decrease in assay throughput, since some samples needed to be reanalyzed using the alternate set of probes. As a result, the throughput of the 5′-nuclease assay was approximately equal to the RFLP assay.
Interestingly, the RFLP assay was able to detect more samples (4 of 40 [10%]) with a mixture of wt and virus variant at codon 552, which is the principal site identified in patients with decreased sensitivity to lamivudine. The 5′-nuclease assay was able to detect more samples (6 of 40 [15%]) with a mixture of wt and virus variant at codon 528, which is a variance detected in some patients with variances at codon 552. Because variations at the polymerase codon 552 are the predominant sequence changes found in HBV DNA from patients exhibiting a loss of sensitivity to lamivudine, accurate DNA typing at this site is critical. It is difficult to determine why this difference in detection of variances at the two sites occurred, but it could be due to different assay conditions optimized for each codon (i.e., primer and probe sequences, etc.).
The RFLP and 5′-nuclease procedures are PCR-based assays with low limits of detecting viral DNA, which is advantageous for analyzing viral DNA present at low concentrations as is typical for patients undergoing lamivudine therapy. This sensitivity is important because serum viral levels of many patients undergoing lamivudine therapy can require PCR amplification for detection. Since both of these methods rely on amplifying DNA by PCR, the lower limit of HBV DNA detection in serum is much lower than that for standard commercial HBV DNA detection assays. The sensitivity limit of the RFLP assay was defined for the PCR conditions described (Table 2), but even lower limits could be achieved by increasing the overall amount of input HBV DNA (i.e., increasing the amount of serum for DNA extraction, increasing the amount of DNA in PCR mixtures, etc.).
Any differences in results observed among the three assays stem from these techniques relying upon a set of PCR primers that are required for the amplification of all species of DNA present. Since the primers for each technique are different, they may bind differently to each sample, especially if there is variation in the sequence at the site where the primers bind. This could account for some of the differences in results predominantly seen when serum samples contained low amounts of viral DNA. This may also account for the observation that some samples with low HBV DNA concentrations were not typeable. However, the RFLP assay was able to provide a more accurate typing from more samples (14 to 20%) than DNA sequencing due to the ability of the RFLP assay to detect low levels of variant virus in samples containing mixed viral populations that DNA sequencing could not distinguish.
Even though the RFLP assay was more sensitive in identifying HBV viral populations, one advantage of DNA sequencing over the RFLP and the 5′-nuclease assay is that it provides DNA sequence information at sites other than at specific codons (i.e., codons 528 and 552) and continues to be useful in the detection of sequence variations at other sites for detecting quasispecies.
All assays were able to generate DNA sequence information from almost all of the samples with high viral titers, and where there were no results, the samples had very low viral titers (<1,000 copies/ml). The extraction methods that were used have been evaluated in the literature for PCR. Although the QIAamp blood kit is reputed to be as efficient as the conventional phenol-chloroform extraction method for extracting HBV DNA from low-titer serum samples (17), we have found that phenol-chloroform extractions are 2 to 25 times more efficient for DNA extraction. This is probably due to the removal of proteins and other cellular contaminants which could inhibit PCR amplification by protein digestion and heat denaturation prior to the ethanol precipitation-extraction methods used herein (27).
Through the use of sensitive, quantitative assays developed and reported here, the kinetics of emergence of YMDD-variant HBV may be more extensively evaluated and correlated with clinical aspects of response to therapy (e.g., changes in necroinflammatory findings in liver biopsies, changes in serum alanine aminotransferase levels, propensity for hepatitis B antigen seroconversion, etc.). Such studies could address the potential differences in long-term efficacy outcomes between patients who develop detectable YMDD variants and patients who maintain only (low-level) wt HBV viremia or who achieve persistently undetectable HBV DNA levels.
ACKNOWLEDGMENTS
This work was partially supported by Glaxo Wellcome, Inc., and by Propetto Finalizzato CNR, grant number 97.01256.PF49.
REFERENCES
- 1.Allen M I, DesLauriers M, Andrews C W, Tipples G A, Walters K-A, Tyrrell D L J, Brown N, Condreay L D. Identification and characterization of mutations in hepatitis B virus resistant to lamivudine. Hepatology. 1998;27:1670–1677. doi: 10.1002/hep.510270628. [DOI] [PubMed] [Google Scholar]
- 2.Atkins M, Hunt C M, Brown N, Gray F, Sanathanan L, Woessner M, Lai C L, Dusheiko G, Dienstag J, Wright T, Barnard J, Bourne E, Condreay L. Clinical significance of YMDD mutant hepatitis B virus (HBV) in a large cohort of lamivudine-treated hepatitis B patients, abstr. 625. Hepatology. 1998;28:319. [Google Scholar]
- 3.Bartholomeusz A, Locarnini S A. Mutations in the hepatitis B virus polymerase gene that are associated with resistance to famciclovir and lamivudine. Int Antiviral News. 1997;5:123–124. [Google Scholar]
- 4.Bartholomew M M, Jansen R W, Jeffers L J, Reddy K R, Johnson L C, Bunzendahl H, Condreay L D, Tzakis A G, Schiff E R, Brown N A. Hepatitis B virus resistant to lamivudine given for recurrent infection after orthotopic liver transplantation. Lancet. 1997;349:20–22. doi: 10.1016/S0140-6736(96)02266-0. [DOI] [PubMed] [Google Scholar]
- 5.Bassler H A, Flood S J A, Livak K J, Marmaro J, Knorr R, Batt C A. Use of a fluorogenic probe in a PCR-based assay for the detection of Listeria monocytogenes. Appl Environ Microbiol. 1995;61:3724–3728. doi: 10.1128/aem.61.10.3724-3728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowden S. Hepatitis B: treatment for the 21st century. Antivir Chem Chemother. 1997;8:77–82. [Google Scholar]
- 7.Chayama K, Suzuki Y, Kobayashi M, Kobayashi M, Tsubota A, Hashimoto M, Miyano Y, Koike H, Kobayashi M, Koida I, Arase Y, Saitoh S, Murashima N, Ikeda K, Kumada H. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild-type after cessation of therapy. Hepatology. 1998;27:1711–1716. doi: 10.1002/hep.510270634. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Zehnbauer B, Gnirke A, Kwok P-Y. Fluorescence energy transfer detection as a homogeneous DNA diagnostic method. Proc Natl Acad Sci USA. 1997;94:10756–10761. doi: 10.1073/pnas.94.20.10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey S. State of the world’s vaccines and immunization. 1996. [Google Scholar]
- 10.Doong S-L, Tsai C-H, Schinazi R F, Liotta D C, Cheng Y-C. Inhibition of the replication of hepatitis B virus In Vitro by 2′,3′-dideoxy-3′-thiacytidine and related analogues. Proc Natl Acad Sci USA. 1991;88:8495–8499. doi: 10.1073/pnas.88.19.8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fallows D A, Goff S P. Mutations in the epsilon sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J Virol. 1995;69:3067–3073. doi: 10.1128/jvi.69.5.3067-3073.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gauthier, J., E. J. Bourne, M. W. Lutz, L. M. Crowther, J. L. Dienstag, N. A. Brown, and L. D. Condreay. Quantitative assessment of HBV viremia and emergence of YMDD variants in chronic hepatitis B patients treated with lamivudine. J. Infect. Dis., in press. [DOI] [PubMed]
- 13.Honkoop P, Niesters H G M, de Man R A M, Osterhaus A D M E, Schalm S W. Lamivudine resistance in immunocompetent chronic hepatitis B. J Hepatol. 1997;26:1393–1395. doi: 10.1016/s0168-8278(97)80476-x. [DOI] [PubMed] [Google Scholar]
- 14.Ibrahim M S, Esposito J J, Jahrling P B, Lofts R S. The potential of 5′ nuclease PCR for detecting a single-base polymorphism in Orthopoxvirus. Mol Cell Probes. 1997;11:143–147. doi: 10.1006/mcpr.1996.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen R W, Johnson L C, Averett D R. High-capacity in vitro assessment of anti-hepatitis B virus compound selectivity by a virion-specific PCR assay. Antimicrob Agents Chemother. 1993;37:441–447. doi: 10.1128/aac.37.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapke G F, Watson G, Sheffler S, Hunt D, Frederick C. Comparison of the chiron quantiplex branched DNA (bDNA) assay and the Abbott genostics solution hybridization assay for quantification of hepatitis B viral DNA. J Viral Hepatitis. 1997;4:67–75. doi: 10.1046/j.1365-2893.1997.00127.x. [DOI] [PubMed] [Google Scholar]
- 17.Kramvis A, Bukofzer S, Kew M C. Comparison of hepatitis B virus DNA extractions from serum by the QIAamp blood kit, GeneReleaser, and the phenol-chloroform method. J Clin Microbiol. 1996;34:2731–2733. doi: 10.1128/jcm.34.11.2731-2733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ladner S K, Miller T J, King R W. The M539V polymerase variant of human hepatitis B virus demonstrates resistance to 2′-deoxy-3′-thiacytidine and a reduced ability to synthesize viral DNA. Antimicrob Agents Chemother. 1998;42:2128–2131. doi: 10.1128/aac.42.8.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai C L, Chien R N, Leung N W, Chang T T, Guan R, Tai D I, Ng K Y, Wu P C, Dent J C, Barber J, Stephenson S L, Gray D F. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group [see comments] N Engl J Med. 1998;339:61–68. doi: 10.1056/NEJM199807093390201. [DOI] [PubMed] [Google Scholar]
- 20.Lee L G, Connell C R, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ling R, Mutimer D, Ahmed M, Boxall E H, Elias E, Dusheiko G M, Harrison T J. Selection of mutations in the hepatitis B virus polymerase during therapy of transplant recipients with lamivudine. Hepatology. 1996;24:711–713. doi: 10.1002/hep.510240339. [DOI] [PubMed] [Google Scholar]
- 22.Livak K J, Flood S J A, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 23.Mast E, Alter M. Epidemiology of viral hepatitis: an overview. Semin Virol. 1993;4:273–283. [Google Scholar]
- 24.McGoldrick A, Lowings J P, Ibata G, Sands J J, Belak S, Paton D J. A novel approach to the detection of classical swine fever virus by RT-PCR with a fluorogenic probe (Taqman) J Virol Methods. 1998;72:125–135. doi: 10.1016/s0166-0934(97)00208-5. [DOI] [PubMed] [Google Scholar]
- 25.Nakahori S, Yokosuka O, Ehata T, Chuang W L, Imazeki F, Ito Y, Ohto M. Detection of hepatitis B virus precore stop codon mutants by selective amplification method: frequent detection of precore mutants in hepatitis B e antigen positive healthy carriers. J Gastroenterol Hepatol. 1995;10:419–425. doi: 10.1111/j.1440-1746.1995.tb01594.x. [DOI] [PubMed] [Google Scholar]
- 26.Niesters H G, Honkoop P, Haagsma E B, de Man R A, Schalm S W, Osterhaus A D. Identification of more than one mutation in the hepatitis B virus polymerase gene arising during prolonged lamivudine treatment. J Infect Dis. 1998;177:1382–1385. doi: 10.1086/517819. [DOI] [PubMed] [Google Scholar]
- 27.Ratnamohan V M, Cunningham A L, Rawlinson W D. Removal of inhibitors of CSF-PCR to improve diagnosis of herpesviral encephalitis. J Virol Methods. 1998;72:59–65. doi: 10.1016/s0166-0934(98)00020-2. [DOI] [PubMed] [Google Scholar]
- 28.Schalm S W, de Man R A, Heijtink R A, Niesters H G M. New nucleoside analogues for chronic hepatitis B. J Hepatol. 1995;22:52–56. [PubMed] [Google Scholar]
- 29.Sherlock S, Dooley J. Diseases of the liver and biliary system. 10th ed. Oxford, England: Blackwell Science; 1997. [Google Scholar]
- 30.Swan D C, Tucker R A, Holloway B P, Icenogle J P. A sensitive, type-specific, fluorogenic probe assay for detection of human papillomavirus DNA. J Clin Microbiol. 1997;35:886–891. doi: 10.1128/jcm.35.4.886-891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tassopoulos N C, Volpes R, Pastore G, Heathcote J, Buti M, Goldin R D, Hawley S, Barber J, Condreay L, Gray D F the Lamivudine Pre-core Mutant Study Group. Efficacy of lamivudine in patients with hepatitis B e antigen-negative/hepatitis B virus DNA-positive (precore mutant) chronic hepatitis B. Hepatology. 1999;29:889–896. doi: 10.1002/hep.510290321. [DOI] [PubMed] [Google Scholar]
- 32.Tipples G A, Ma M M, Fischer K P, Bain V G, Kneteman N M, Tyrrell D L J. Mutations in HBV RNA-Dependent DNA polymerase confers resistance to lamivudine in vivo. Hepatology. 1996;24:714–717. doi: 10.1002/hep.510240340. [DOI] [PubMed] [Google Scholar]