Abstract
Background:
Suppressor 3 of cytokine signaling (SOCS3) hypermethylation has been reported to participate in hepatocellular carcinoma (HCC) development and progression, but conflicting results were published. This study aimed to analyze the clinical effects of SOCS3 hypermethylation in HCC and the effects of sex and age on SOCS3 hypermethylation in HCC.
Methods:
Databases were searched for relevant case-control and cohort studies on SOCS3 hypermethylation in HBV-related HCC. In vitro and in vivo studies and studies of patients with serious comorbidities were excluded. Review Manager 5.2 was used to estimate the effects of the results among the selected studies. Forest plots, sensitivity analysis, and bias analysis for the included studies were also conducted.
Results:
Finally, 8 relevant studies met the inclusion criteria. A significant difference in SOCS3 hypermethylation in HCC was found between tumor and nontumor groups (the odds ratio [OR] = 2.01, 95% confidence interval [CI]: 1.48–2.73, P < .00001; P for heterogeneity = .39, I2 = 5%). The meta-analysis suggested no significant difference in the effect of sex (OR = 1.00, 95% CI: 0.76–1.31, P = .76; P for heterogeneity = .44, I2 = 0%) and age on SOCS3 hypermethylation in HCC (OR = 1.11, 100% CI: 0.78–1.29, P = .03; P for heterogeneity = .14, I2 = 36%). Limited publication bias was observed in this study.
Conclusion:
SOCS3 hypermethylation is associated with HBV-related HCC. Sex and age do not affect the association between SOCS3 hypermethylation and HCC. SOCS3 might be a treatment target for HCC.
Keywords: age, hypermethylation, meta-analysis, sex, suppressor 3 of cytokine signaling
1. Introduction
Hepatocellular carcinoma (HCC) is a common malignant tumor worldwide, with rapid progression and high short-term mortality. The occurrence of HCC is closely related to the liver tissue in chronic diseases caused by hepatitis B virus (HBV) or hepatitis C virus infection, toxin exposure, excessive alcohol consumption, and/or other environmental or genetic factors.[1,2]
In the liver, the continuous progression of cancer is a multistep process involving genetic and epigenetic changes that lead to the activation of oncogenes and the inactivation of tumor suppressor genes.[3,4] DNA methylation is the first epigenetic regulation and has one of the most important molecular mechanisms considered to be the cause of liver cancer.[4]
Many studies showed that HCC tumors exhibited specific DNA methylation characteristics, which were related to the main risk factors and tumor progression.[5] However, the methylation status of tumor suppressor genes is not fully understood in patients with liver cancer and different viral infections, especially HBV infection.[6]
The members of the suppressors of cytokine signaling (SOCS) gene family suppress tumor progression by inhibiting Janus-activated kinase signal transducers and activators of transcription (JAK/STAT) and NF-κB signaling pathways and promoting the p53 signaling pathway. In the SOCS family, SOCS3 is reported to be hypermethylated in a variety of cancers, including head and neck cancer, lung cancer, prostate cancer, Barrett esophagus cancer, and colorectal cancer associated with ulcerative colitis.[7,8] The hypermethylated SOCS3 gene cannot be translated, leading to activated JAK/STAT and SOCS3 NF-κB signaling pathways, which promote tumor cell growth and invasion, and repressed p53 signaling pathway, decreasing apoptosis.[9–12] Studies showed that down-regulated SOCS3 was associated with poorer survival and increased invasion,[13,14] silenced SOCS3 increased liver tumor formation and growth,[15] and loss of SOCS3 increased the aggressiveness of liver tumors.[16] Still, a previous study reported no excessive SOCS3 methylation in HBV-related HCC tissues.[8] These conflicting results might be due to the complex etiological mechanisms of HCC, in which SOCS3 is only one of many actors and different loci detected in the SOCS3 promoter region.[17,18] Therefore, it was imperative to explore the detailed methylation status of methyl methacrylate and the relationship between SOCS3 and its methylation in HCC with different viral infection backgrounds and clinicopathology.
In recent years, the distortion of SOCS3 in human malignant tumors has been noted,[7] but the detailed methylation status of SOCS3 in HCC with different viral infection backgrounds has not been fully understood. It was hypothesized that SOCS3 hypermethylation was associated with HBV-related HCC. This meta-analysis was conducted to confirm the relationship between SOCS3 methylation and HCC and the association of the degree of SOCS3 methylation with sex and age. Confirming the involvement of SOCS3 hypermethylation could indicate new treatment targets for the treatment of HCC.
2. Materials and methods
2.1. Literature search strategy
The study was designed according to the population, intervention, comparison, outcome principle. Relevant literature was retrieved from PubMed, Embase, Cochrane Library, and China National Knowledge Infrastructure databases using a combination of the following keywords: “SOCS3” or “inhibitors of the cytokine signaling” and “hepatocellular carcinoma” or “liver cancer” or “HCC,” and “methylation”, followed by screening based on the eligibility criteria. All relevant studies were published from January 2005 to May 2020, without language restrictions. Literature searches were conducted independently by 2 authors (HZ and YY) using a standardized approach. As this study used published data, the ethics committee deemed ethical approval unnecessary.
2.2. Study selection
The inclusion criteria were as follows:
Study design: case-control or cohort studies; patients: diagnosed with HCC, cirrhosis, and chronic hepatitis B; exposure: HBV identified by the detection of HBV surface antigen; hypermethylation status determination: the methods suitable for methylation detection limited to MethyLight arrays, pyrosequencing, and methylation-specific polymerase chain reaction; reported the frequency of SOCS3 methylation evaluated in liver cancer tissues; and when authors from the same institution published multiple studies using overlapping sample data, only recent studies or studies with complete information were selected.
The exclusion criteria were as follows: duplicate studies in different databases; patients with other serious diseases, such as other malignancies, heart failure, or kidney failure; patients receiving a combination of other treatments; cell or animal studies; and reviews, letters, case reports, or meeting abstracts.
2.3. Comparison
The methylation status of SOCS3 was examined between HCC and non-cancer tissues.
2.4. Data extraction and quality assessment
The full texts of the studies were read carefully, and the characteristics of each study were extracted by two authors (HZ and YY) using a predetermined form. Discrepancies in the extraction were solved by discussion. The data extracted from these studies included the first author's name, year of publication, country, sex, and sample size and range. For case-control or cohort studies, the Newcastle-Ottawa Scale (NOS) quality evaluation tool was often used, including 8 aspects, with a total of 9 points. Generally, NOS score ≥6 was regarded as high quality.
2.5. Statistical analysis
Review Manager 5.2 software (The Cochrane Collaboration, 2011) was used to estimate the differences in the overall outcomes between methylation and unmethylation groups in this study. The NOS table was used to assess the quality of the included studies. For binary data, the odds ratio (OR) with 95% confidence intervals (CIs) were calculated to estimate the relationship between SOCS3 methylation and HCC and also the effect of sex and age on the relationship. Heterogeneity was assessed using the inconsistency index (I2) statistic in this study; the value of the I2 statistic also reflected the heterogeneity level. If I2 was >50%, the trials were considered heterogeneous, and the random-effects model was adopted. Otherwise, a fixed-effects model was chosen. For all comparisons, a P value <.05 was considered statistically significant. In addition, bias analyses of the studies were conducted with Stata 10.0 software (Stata Corporation, TX) to examine the quality of studies. Funnel plots were used to estimate publication bias.
3. Results
3.1. Search process
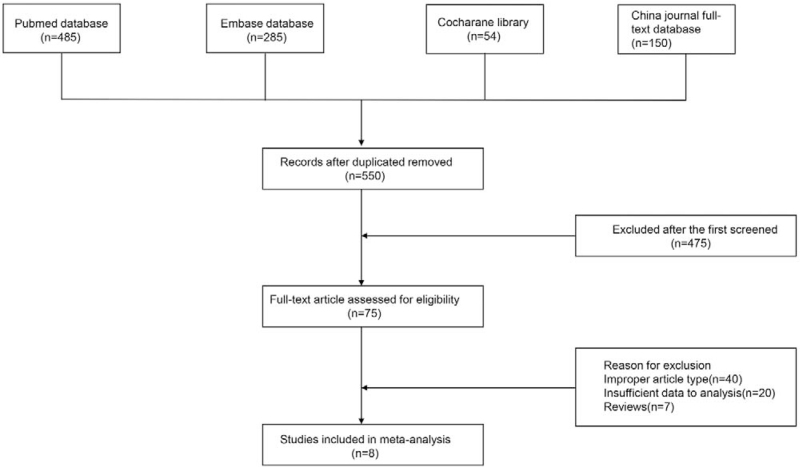
The electronic search ended with a total number of 550 studies. After a thorough reading, 75 studies met the preliminary criteria. In further screening, 67 studies were excluded because of the study design, insufficient data, and type of studies. Finally, 8 studies[19–26] were selected for analysis. Figure 1 shows a flowchart of identification, inclusion, and exclusion of studies, reflecting the search and the reasons for exclusion.
Figure 1.
Flow diagram of study selection.
3.2. Characteristics of included studies
Detailed characteristics of the included studies are presented in Table 1. All these studies were published from 2005 to 2020. The sample size ranged from 37 to 269. The methylation and unmethylation groups comprised 886 and 409 patients, respectively. Of the included studies, 7 analyzed the difference in methylation between tumor and nontumor groups, 5 analyzed the difference in the effect of sex on hypermethylation, and 4 evaluated the difference in the age on hypermethylation.
Table 1.
Characteristics of studies included in the meta-analysis.
| Study | Year | Language | Country | Age range (mean) | Groups | n | Years of onset |
| Calvis[11] | 2006 | English | America | 53.2 ± 11.8 | Male | 80 | 2000–2005 |
| Female | 0 | ||||||
| Jiang[12] | 2017 | English | China | 56 ± 7.5 | Male | 181 | 2010–2014 |
| Female | 88 | ||||||
| Ogata[13] | 2006 | English | Japan | 63.4 ± 7.5 | Male | 25 | 2000–2004 |
| Female | 12 | ||||||
| Wei[14] | 2020 | English | China | 58 ± 8.5 | Male | 87 | 2015–2018 |
| Female | 9 | ||||||
| Wei[15] | 2017 | English | China | 47.4 ± 6.5 | Male | 355 | 2008–2015 |
| Female | 68 | ||||||
| Yang[16] | 2020 | English | China | 52 ± 8.9 | Male | 56 | 2014–2017 |
| Female | 6 | ||||||
| Zhang[17] | 2015 | English | China | 57 ± 18 | Male | 174 | 2008–2014 |
| Female | 80 | ||||||
| Zhao[18] | 2017 | English | China | 60.5 ± 7.5 | Male | 77 | 2009–2011 |
| Female | 20 |
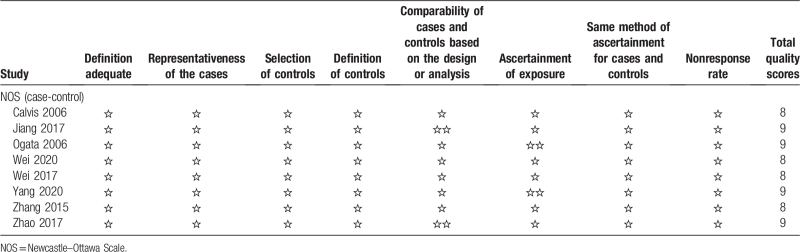
3.3. Results of quality assessment
The quality of the included studies is shown in Table 2. The NOS table was used to evaluate the risk of patient selection in 8 studies. Of these 8 studies, 4 had 9 stars, and the other 4 had 8 stars, indicating that the included studies were of good quality.
Table 2.
Quality assessment using the Newcastle–Ottawa Scale.
3.4. Results of meta-analysis
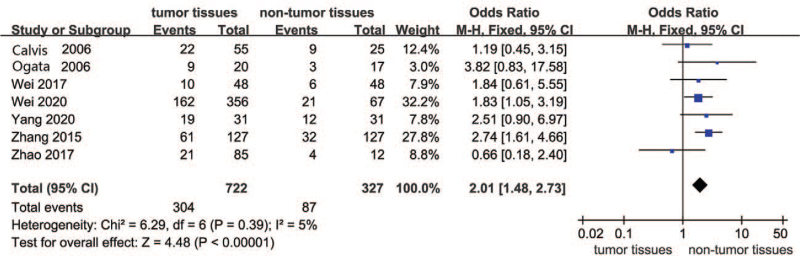
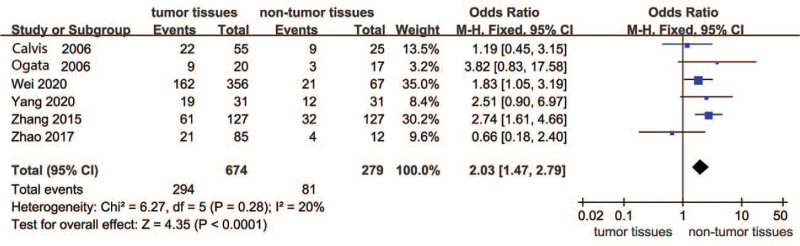
Figure 2 presents the forest plot for the comparison of SOCS3 hypermethylation in HCC between the tumor and nontumor groups. Seven studies were involved in this comparison. The overall results suggested that SOCS3 hypermethylation was much higher in tumor tissues than in nontumor tissues. Therefore, in the included studies, SOCS3 hypermethylation had a significant effect on HCC tumor tissues (OR = 2.01, 95% CI: 1.48–2.73, P < .00001; P for heterogeneity = .39, I2 = 5%).
Figure 2.
Forest plots of SOCS3 hypermethylation in HCC between tumor and nontumor groups. HCC = hepatocellular carcinoma, SOCS3 = suppressor 3 of cytokine signaling.
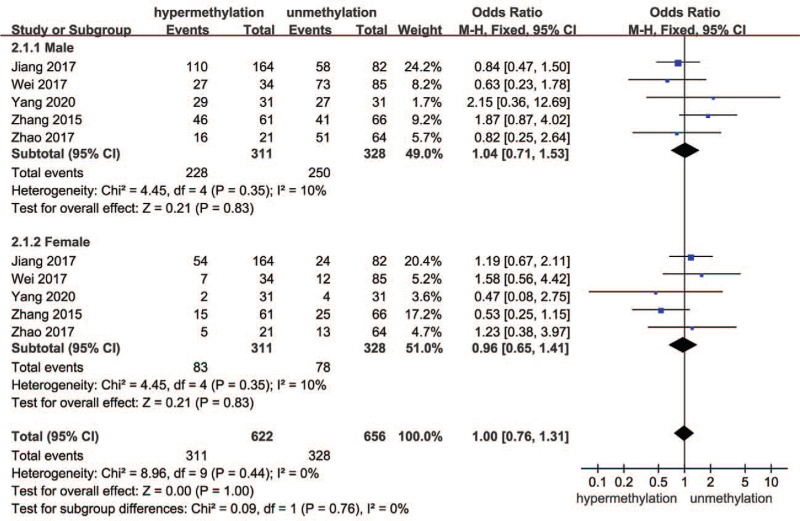
The forest plot for the effect of sex on SOCS3 hypermethylation in HCC is shown in Fig. 3. Since I2 was <50%, the fixed-effects model was adopted. The results of subgroup analysis suggested that the effect of both male and female sex in hypermethylation and unhypermethylation groups had no difference, indicating no effect of sex on SOCS3 hypermethylation in HCC (OR = 1.00, 95% CI 0.76–1.31, P = .76; P for heterogeneity = .44, I2 = 0%).
Figure 3.
Forest plots of the effect of sex on SOCS3 hypermethylation and unhypermethylation groups. SOCS3 = suppressor 3 of cytokine signaling.
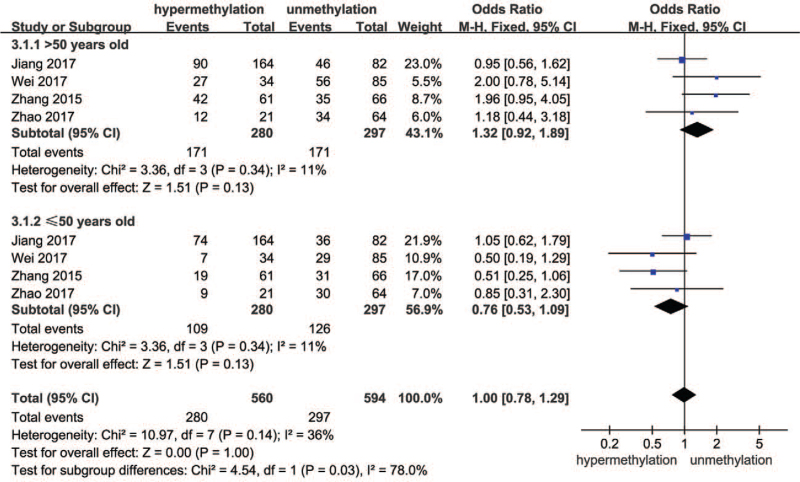
Four of the 8 studies included the effect of age on SOCS3 hypermethylation in HCC. The forest plot is shown in Fig. 4. These 4 studies displayed a difference in patients aged ≤50 and >50 years between hypermethylation and unhypermethylation groups. The combined results suggested that age had no effect on SOCS3 hypermethylation in HCC (OR = 1.00, 95% CI: 0.78–1.29, P = .03; P for heterogeneity = .14, I2 = 36%).
Figure 4.
Forest plots of the effect of age on SOCS3 hypermethylation and unhypermethylation groups. SOCS3 = suppressor 3 of cytokine signaling.
3.5. Results of sensitivity analysis and publication bias
According to the meta-analysis, the heterogeneity of SOCS3 hypermethylation and HCC was low (I2 = 4%). As shown in Fig. 5, the heterogeneity of SOCS3 hypermethylation and HCC might be attributed to the different results of each study. When the article of Wei in 2017 was excluded, I2 changed to 20%. This indicated that the result in this article was robust.
Figure 5.
Funnel plot of publication bias.
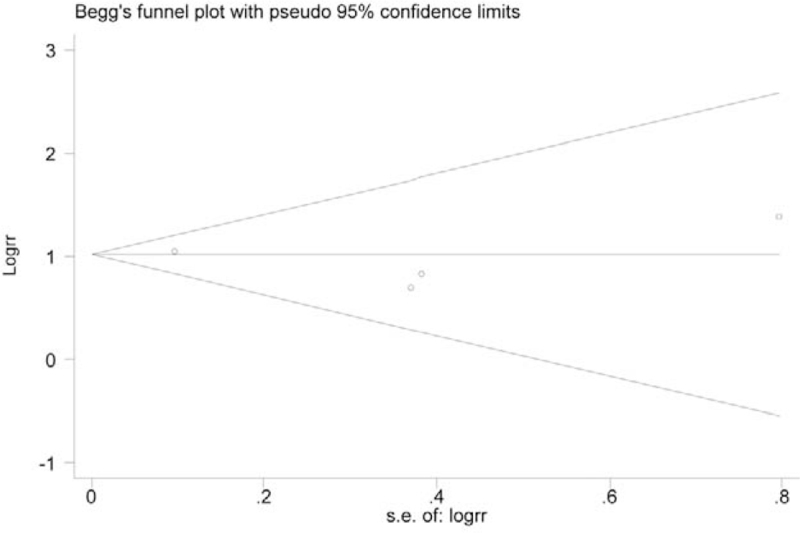
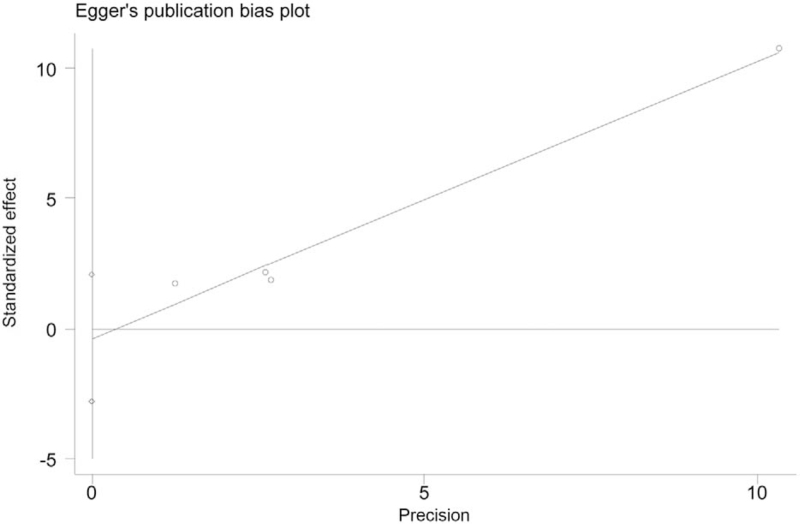
A funnel plot for SOCS3 hypermethylation in HCC tumor tissues was performed. All 8 studies were included in the plot. To some extent, the result indicated no publication bias because the symmetrical characteristic of the funnel plot was good (Fig. 5). The result of Begg test suggested no significant evidence of potential publication bias (z = 1.35, P = .125, Fig. 6). The Egger test also suggested no significant evidence of potential publication bias (t = 1.21, P = .201, Fig. 7).
Figure 6.
Begg funnel plot with pseudo 95% confidence limits.
Figure 7.
Egger publication bias plot.
4. Discussion
The results showed a difference in SOCS3 hypermethylation between the tumor and nontumor groups, indicating that SOCS3 hypermethylation was vital in HCC, as previously described.[27] Villanueva et al[28] showed that the SOCS3 gene was hypermethylated in endometrial cancer, prostate cancer, Barrett esophageal cancer, and ulcerative colorectal cancer. In vitro, SOCS3 methylation promoted the growth of pancreatic cancer cell lines.[27] The frequency of methylation of SOCS3 in HCC was significantly higher than that in nontumor and benign liver tissues, but it had no effect on the prognosis of patients with HCC.[29] The methylation status of SOCS3 was closely related to transarterial chemoembolization response and prognosis, suggesting that SOCS3 methylation status could be used as a biomarker to predict transarterial chemoembolization efficacy in patients with liver cancer. Still, in contrast with previous studies, the present study only included studies of HBV-related HCC. These results imply that determining the SOCS3 hypermethylation status might have prognostic value and help tailor the treatments.
DNA methylation is the addition of a methyl group to cytosine in CpG dinucleotide catalyzed by DNA methyltransferase. This epigenetic mechanism is used by cells to control gene expression. Hypermethylation associated with SOCS protein silencing has been found in a variety of cancers, including myeloma, melanoma, bladder cancer, liver cancer, gastric cancer, and colorectal cancer.[30] The effect of risk factors on DNA methylation in HCC needed to be analyzed. Therefore, this study analyzed the effects of sex on the relationship between DNA methylation and HCC. The results showed no impact of sex. The incidence rate of HCC in the population gradually increased with age. However, a recent study found that patients with HCC were younger, and the incidence rate of HCC was significantly higher in men than in women. The incidence rate of HCC in men was 4 to 8 times higher than that in women. Wu et al[30] reported that the methylation of the SOCS3 gene was high in HCC and had no significant correlation with the patient's sex, age, tumor size, and differentiation degree. Li et al[31] showed that the expression of SOCS3 positively correlated with the prevalence of HCC, but not with the age and sex of patients with HCC.
The methylation status of SOCS3 in circulating DNA was consistent with that in tissue DNA and could be used as an effective noninvasive molecular marker to screen high-risk patients with HCC and predict the poor prognosis of patients with partial hepatectomy. Meanwhile, the methylation of the SOCS3 gene promoter in liver cancer tissue and plasma significantly correlated with AFP 400, tumor size, tumor differentiation, cirrhosis, metastasis, and recurrence.[28,32]
The strength of this study is that it only included data about HBV-related HCC, eliminating the possible confounding effect of various etiologies. Still, this study had some limitations. First, more indicators evaluating other risk factors should be included in future studies. Second, more studies from various areas could be included in future analyses. Third, data on the correlation between SOCS3 and clinicopathological features and prognosis were limited in published studies. Hence, these correlations should be analyzed in the future.
5. Conclusion
In conclusion, SOCS3 hypermethylation is associated with HBV-related HCC. Sex and age do not affect the association between SOCS3 hypermethylation and HCC. The SOCS3 axis might be a treatment target for HCC, which requires additional studies. Future studies should also examine whether this association remains true in HCC from other etiologies.
Author contributions
Conceptualization: Hairu Zheng.
Data curation: Hairu Zheng, Jiajia Cheng, Yong Wang.
Formal analysis: Shuyong Yu, Yong Wang.
Methodology: Yanggang Yan.
Resources: Yanggang Yan.
Supervision: Jiajia Cheng.
Writing – original draft: Hairu Zheng.
Writing – review & editing: Shuyong Yu, Yong Wang.
Footnotes
Abbreviations: CIs = confidence intervals, HBV = hepatitis B virus, HCC = hepatocellular carcinoma, NOS = Newcastle–Ottawa Scale, OR = odds ratio, SOCS = suppressors of cytokine signaling, SOCS3 = suppressor 3 of cytokine signaling.
How to cite this article: Zheng H, Yan Y, Cheng J, Yu S, Wang Y. Association between SOCS3 hypermethylation and HBV-related hepatocellular carcinoma and effect of sex and age: a meta-analysis. Medicine. 2021;100:43(e27604).
This work was supported by a Research start-up fund for talent introduction of the Second Affiliated Hospital of Hainan Medical University, Key Research and Development Program of Hainan Province (No. ZDYF2019132), National Natural Science Foundation of China (No. 81960331).
Ethical Experimentation: Ethical approval and informed consent is not applicable in this study.
The authors have no conflicts of interest to disclose.
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
References
- [1].Yuan XD, Wang JW, Fang Y, et al. Methylation status of the T-cadherin gene promotor in peripheral blood mononuclear cells is associated with HBV-related hepatocellular carcinoma progression. Pathol Res Pract 2020;216:152914. [DOI] [PubMed] [Google Scholar]
- [2].Yan X, Wu T, Tang M, et al. Methylation of the ataxia telangiectasia mutated gene (ATM) promoter as a radiotherapy outcome biomarker in patients with hepatocellular carcinoma. Medicine (Baltimore) 2020;99:e18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sun FK, Fan YC, Zhao J, et al. Detection of TFPI2 methylation in the serum of hepatocellular carcinoma patients. Dig Dis Sci 2013;58:1010–5. [DOI] [PubMed] [Google Scholar]
- [4].Liu XY, Fan YC, Gao S, et al. Methylation of SOX1 and VIM promoters in serum as potential biomarkers for hepatocellular carcinoma. Neoplasma 2017;64:745–53. [DOI] [PubMed] [Google Scholar]
- [5].Sun FK, Sun Q, Fan YC, et al. Methylation of tissue factor pathway inhibitor 2 as a prognostic biomarker for hepatocellular carcinoma after hepatectomy. J Gastroenterol Hepatol 2016;31:484–92. [DOI] [PubMed] [Google Scholar]
- [6].Wang YY, Jiang JX, Ma H, et al. Role of ZIC1 methylation in hepatocellular carcinoma and its clinical significance. Tumour Biol 2014;35:7429–33. [DOI] [PubMed] [Google Scholar]
- [7].Zhang T, Guan G, Chen T, et al. Methylation of PCDH19 predicts poor prognosis of hepatocellular carcinoma. Asia Pac J Clin Oncol 2018;14:e352–8. [DOI] [PubMed] [Google Scholar]
- [8].Zhang C, Ge S, Wang J, et al. Epigenomic profiling of DNA methylation for hepatocellular carcinoma diagnosis and prognosis prediction. J Gastroenterol Hepatol 2019;34:1869–77. [DOI] [PubMed] [Google Scholar]
- [9].Dhar K, Rakesh K, Pankajakshan D, Agrawal DK. SOCS3 promotor hypermethylation and STAT3-NF-kappaB interaction downregulate SOCS3 expression in human coronary artery smooth muscle cells. Am J Physiol Heart Circ Physiol 2013;304:H776–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Iwahori K, Serada S, Fujimoto M, et al. Overexpression of SOCS3 exhibits preclinical antitumor activity against malignant pleural mesothelioma. Int J Cancer 2011;129:1005–17. [DOI] [PubMed] [Google Scholar]
- [11].Ogata H, Chinen T, Yoshida T, et al. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta1 production. Oncogene 2006;25:2520–30. [DOI] [PubMed] [Google Scholar]
- [12].Zhang L, Badgwell DB, Bevers JJ, 3rd, et al. IL-6 signaling via the STAT3/SOCS3 pathway: functional analysis of the conserved STAT3 N-domain. Mol Cell Biochem 2006;288:179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Deng J, Jiao X, Liu H, et al. Lymph node metastasis is mediated by suppressor of cytokine signaling-3 in gastric cancer. Tumour Biol 2013;34:3627–36. [DOI] [PubMed] [Google Scholar]
- [14].Stofas A, Levidou G, Piperi C, et al. The role of CXC-chemokine receptor CXCR2 and suppressor of cytokine signaling-3 (SOCS-3) in renal cell carcinoma. BMC Cancer 2014;14:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Baltayiannis G, Baltayiannis N, Tsianos EV. Suppressors of cytokine signaling as tumor repressors. Silencing of SOCS3 facilitates tumor formation and growth in lung and liver. J BUON 2008;13:263–5. [PubMed] [Google Scholar]
- [16].Wu WY, Kim H, Zhang CL, Meng XL, Wu ZS. Loss of suppressors of cytokine signaling 3 promotes aggressiveness in hepatocellular carcinoma. J Invest Surg 2014;27:197–204. [DOI] [PubMed] [Google Scholar]
- [17].Um TH, Kim H, Oh BK, et al. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J Hepatol 2011;54:939–47. [DOI] [PubMed] [Google Scholar]
- [18].Revill K, Wang T, Lachenmayer A, et al. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology 2013;145:1424.e1–35.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Calvisi DF, Ladu S, Gorden A, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology 2006;130:1117–28. [DOI] [PubMed] [Google Scholar]
- [20].Jiang BG, Wang N, Huang J, et al. Tumor SOCS3 methylation status predicts the treatment response to TACE and prognosis in HCC patients. Oncotarget 2017;8:28621–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ogata H, Kobayashi T, Chinen T, et al. Deletion of the SOCS3 gene in liver parenchymal cells promotes hepatitis-induced hepatocarcinogenesis. Gastroenterology 2006;131:179–93. [DOI] [PubMed] [Google Scholar]
- [22].Wei L, Huang Y, Zhao R, et al. Detection of promoter methylation status of suppressor of cytokine signaling 3 (SOCS3) in tissue and plasma from Chinese patients with different hepatic diseases. Clin Exp Med 2018;18:79–87. [DOI] [PubMed] [Google Scholar]
- [23].Wei L, Liu Q, Huang Y, et al. Knockdown of CTCF reduces the binding of EZH2 and affects the methylation of the SOCS3 promoter in hepatocellular carcinoma. Int J Biochem Cell Biol 2020;120:105685. [DOI] [PubMed] [Google Scholar]
- [24].Yang Z, Zhu H, Zhang L, et al. DNA methylation of SOCS1/2/3 predicts hepatocellular carcinoma recurrence after liver transplantation. Mol Biol Rep 2020;47:1773–82. [DOI] [PubMed] [Google Scholar]
- [25].Zhang X, You Q, Zhang X, Chen X. SOCS3 methylation predicts a poor prognosis in HBV infection-related hepatocellular carcinoma. Int J Mol Sci 2015;16:22662–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhao R, Chen K, Zhou J, et al. The prognostic role of BORIS and SOCS3 in human hepatocellular carcinoma. Medicine (Baltimore) 2017;96:e6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Quan H, Zhou F, Nie D, et al. Hepatitis C virus core protein epigenetically silences SFRP1 and enhances HCC aggressiveness by inducing epithelial-mesenchymal transition. Oncogene 2014;33:2826–35. [DOI] [PubMed] [Google Scholar]
- [28].Villanueva A, Portela A, Sayols S, et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology 2015;61:1945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lv X, Ye G, Zhang X, Huang T. p16 Methylation was associated with the development, age, hepatic viruses infection of hepatocellular carcinoma, and p16 expression had a poor survival: a systematic meta-analysis (PRISMA). Medicine (Baltimore) 2017;96:e8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wu B, Jiang Y, Zhu F, Sun D, Huang H. Demethylation effects of elemene on the GSTP1 gene in HCC cell line QGY7703. Oncol Lett 2016;11:2545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li CC, Yu Z, Cui LH, Piao JM, Liu M. Role of P14 and MGMT gene methylation in hepatocellular carcinomas: a meta-analysis. Asian Pac J Cancer Prev 2014;15:6591–6. [DOI] [PubMed] [Google Scholar]
- [32].Jiang LH, Hao YL, Zhu JW. Expression and prognostic value of HER-2/neu, STAT3 and SOCS3 in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol 2019;43:282–91. [DOI] [PubMed] [Google Scholar]