Abstract
Objectives:
Reported associations of the cannabinoid receptor 1 (CNR1) single nucleotide polymorphisms (SNPs) with alcohol dependence (AD) have been inconsistent, prompting a meta-analysis to obtain more precise estimates.
Methods:
A Boolean search of 4 databases (PubMed, Scopus, Google Scholar, and Mednar) sought articles that evaluated the association between CNR1 polymorphisms and risk of AD. We selected the articles with sufficient genotype frequency data to enable calculation of odds ratios (ORs) and 95% confidence intervals (CIs). Using the Population Intervention Comparators Outcome elements, AD patients (P) were compared by genotype data between AD-participants (I) and non-AD-participants (C) in order to determine the risk of AD (O) attributed to the CNR1 SNPs. Analyzing 4 SNPs (rs1049353, rs1535255, rs2023239, and rs806379) using standard genetic models, we examined associations where multiple comparisons were Holm–Bonferroni corrected. The pooled ORs were assessed for aggregate statistical power and robustness (sensitivity analysis). Subgroups were Caucasians and African-Americans.
Results:
From 32 comparisons, 14 were significant indicating increased risk, from which 5 outcomes (P-value for association [Pa] = .003 to <.001) survived the Holm–Bonferroni-correction, which were deemed robust. In the rs1535255 outcomes, the codominant effect (OR = 1.43, 95% CIs = 1.24–1.65, Pa < .001) had greater statistical power than the dominant effect (OR = 1.30, 95% CI = 1.08–1.57, Pa = .006). In contrast, the rs2023239 codominant outcome was underpowered. Significance of both rs806379 Caucasian outcomes (ORs = 1.20–1.43, 95% CIs = 1.07–1.57, Pa = .003) contrasted with the null effects in African-Americans (ORs = 0.98–1.08, 95% CIs = 0.70–1.53).
Conclusions:
Three CNR1 SNPs (rs1535255, rs2023239, and rs806379) were implicated in their associations with development of AD: based on aggregate statistical power, rs1535255 presented greater evidence for associations than rs2023239; rs806379 implicated the Caucasian subgroup. Multiple statistical and meta-analytical features (consistency, robustness, and high significance) underpinned the strengths of these outcomes. Our findings could render the CNR1 polymorphisms useful in the clinical genetics of AD.
Keywords: alcohol dependence, cannabinoid receptor 1 polymorphisms, meta-analysis
1. Introduction
The Diagnostic and Statistical Manual of Mental Disorders (DSM 5), alcohol use disorder including alcohol abuse and alcohol dependence (AD) defined as a psychiatric dysfunction, marked by compulsive drinking, leading to pathological alcohol seeking behavior.[1] Alcohol abuse corrodes the security of health, job, and family.[2,3] Moreover, the morbidity and mortality that result from AD, adversely impacts individuals and society, contributing to the global burden of disease.[4]
Persistence of AD in humans through the millennia may have come from the driving force of positive reinforcement mechanisms that stimulates reward pathways of the brain, which involve the endocannabinoid system (ECS).[5] Studies have shown that the ECS regulates dopamine reward circuits, which play an important role in the reward processes involved in substance dependence[6,7] and facilitate vulnerability to the progression of addiction.[8] The ECS is composed of cannabinoid receptors as well as enzymes that synthesize, degrade, and transport endocannabinoids (endogenous cannabinoids).[9] The cannabinoid receptors, CB1 and CB2 are encoded by cannabinoid receptor 1 (CNR1) and cannabinoid receptor 2 (CNR2) genes, respectively. CNR2 is found at the brain periphery and appears to have an immune function.[10]CNR1 is expressed at high levels in brain regions that act on drug reward and drug memories,[11] which include the hippocampus, striatum, and cerebral cortex.[12] The machinery of drug rewards and memories lead to risks of psychiatric disorders that include substance abuse which involves the ECS.[13] Activating the same reward pathways in the brain are cannabinoids, chemical substances that attach to the cannabinoid receptors of the brain and elicit pharmacological effects similar to marijuana (cannabis).[14] It has been shown that delta-9-tetrahydrocannabinol and alcohol share similar behavioral profiles where at low and high doses, both induce euphoria/motor incoordination and sedation, respectively.[15] delta-9-tetrahydrocannabinol is mediated by CB1, encoded by the CNR1 gene, which maps to chromosome 6q14-q15. CB1/CNR1 displays at least 4 exons that spans 25 kb and produces several transcripts. Common single-nucleotide polymorphisms (SNPs) account for as much as 30% of the variance in AD.[16] Various locations of CNR1 SNPs point to the silent rs1049353 found in the coding region involving a substitution of “G” to “A” at nucleotide position 1359 in codon 435 (threonine). The rs806379 SNP (A/T) is located in intron 2 of the CNR1 gene (–385 to exon 3 alternate transcript initiation site). The rs2023239 and rs1535255 SNPs are non-coding intron variants in the 3 prime untranslated region.
These CNR1 SNPs were found to be associated with polysubstance abuse.[17] A study showed that C allele carriers of rs2023239 had elevated cravings in response to alcohol-associated cues.[18] However, the association of rs1049353 with alcohol and drug dependence elicited variable results from different studies.[10,19–22] A nicotine study found that the T allele on rs806379 reduced mRNA expression resulting in less mRNA activity,[23] which agreed with an earlier haplotype study involving rs2023239, rs806379 and rs1535255.[24] Yet, examinations of these SNPs for their associations with development of AD[25] have produced inconsistent outcomes, that ranged from reduced to increased risks. Hence, using the Population Intervention Comparators Outcome approach, we performed this meta-analysis to realize our objectives of obtaining less ambiguous and better estimates of associations. Here, we examined SNPs that might provide clues of the roles of their proteins in the neuro-metabolic pathways, fostering better understanding of risk biomarkers in AD.
2. Methods
2.1. Selection of studies
We searched MEDLINE using PubMed, Google Scholar, Scopus, and Mednar for association studies as of March 2, 2021. Using Boolean descriptors, search terms (Table S1, Supplemental Digital Content) were as follows: (CNR1 OR cannabinoid receptor 1 OR endocannabinoid OR CB1) and (polymorphisms, genetic OR gene OR single nucleotide polymorphism OR genome-wide association studies OR GWAS) and (alcoholism OR alcohol dependence OR alcohol abuse) medical subject heading and text, unrestricted by language. The Boolean search started the process of study selection and finalized with full-text examination of the included studies.
This provided the timeline of 2002 to 2010 indicating the years for including the studies. References in the retrieved articles were screened manually to identify additional eligible studies. In cases of duplicates, we selected the one with a later date of publication. The 4 Population Intervention Comparators Outcome elements were applied: Population: AD patients; Intervention: CNR1 gene polymorphisms; Comparators: AD patients versus non-AD patients; and Outcome: AD risk. Inclusion criteria were: case–control design evaluating the association between CNR1 polymorphisms and risk of AD; sufficient genotype frequency (provision of wild-type [wt], variant [var], and heterozygous [wt-var] numbers under a case-control design) data to enable calculation of odds ratios (ORs) and 95% confidence intervals (CIs). Exclusion criteria were: those not involving AD; reviews; functional articles; not about the CNR1 SNPs; those without controls; studies whose genotype controls deviated from the Hardy-Weinberg equilibrium (HWE); and studies whose genotype or allele frequencies were unusable/absent.
2.2. Data extraction and HWE assessment
Two investigators (NP and PC) independently extracted data on March 3 and March 4, respectively. PT adjudicated disagreements, which facilitated consensus. The following information was extracted from each article: first author's name, year of the study, country of origin, ethnicity, age of the subjects, CNR1 SNPs (rs number), comparators, article features needed to tally the Clark–Baudouin (CB) scores, sample sizes, genotype data of AD, and controls and minor allele frequency. HWE was assessed using the application in https://ihg.gsf.de/cgi-bin/hw/hwa1.pl where a P-value for HWE [PHWE] > .05 indicated HW-compliance.
2.3. Statistical power and study quality
Using the G∗Power program,[26] we evaluated aggregate statistical power (ASP). Based on previous single-study results and meta-analysis outcomes,[27–29] we chose 3 OR levels (1.1, 1.2, and 1.5) at a genotypic risk level of α = 0.05 (2-sided) where we considered power adequate at ≥80%. We used the CB scale to evaluate methodological quality of the included studies.[30] In this scale, low, moderate and high have scores of <5, 5 to 6, and ≥7, respectively.
2.4. Meta-analysis protocol
Because the CNR1 genotypes in the included studies were varyingly notated by rs number, we used the generic var and wt notations (Table S2, Supplemental Digital Content). Given the hypothesis of association between CNR1 SNPs and risk of AD, we estimated the ORs with 95% CIs for each study by comparing cases with controls. Pooled ORs with 95% CIs were calculated for the following genetic models: homozygous: (var–var and wt–wt) genotypes compared with wt–wt; recessive: (var–var vs wt–var + wt–wt); dominant: (var–var + wt–var vs wt–wt); and codominant: (var vs wt). We used raw data for frequencies to estimate study specific ORs of AD development. Comparing the effects on the same baseline, we calculated pooled ORs using the Z-test, where we confined our analyses to ≥3 studies. Multiple comparisons were Holm–Bonferroni corrected (HBC).[31] Subgrouping was ethnicity-based (Caucasians and African-Americans [AA]). Heterogeneity[32] was estimated with the chi-squared-based Q test, the significance threshold of which was set at P-value for heterogeneity [Phet] ≤ .10 and quantified with the measure of variability (I2) statistic, which measures variability between studies. Random-effects model[33] was used in the presence of heterogeneity, otherwise, the fixed-effects model was used.[34] Random-effects derived pooled ORs were subjected to outlier treatment, which dichotomized the comparisons into pre- and postoutlier. We used sensitivity analysis to assess robustness of the pooled ORs. Assesment of publication bias was considered for significant outcomes with ≥10 studies.[35] Except for heterogeneity estimation, 2-sided P-values of <.05 were considered significant. Data were analyzed using Review Manager 5.3 (Cochrane Collaboration, Oxford, England) and SIGMASTAT 2.03 (Systat Software, San Jose, CA).
3. Results
3.1. Search results and data extraction
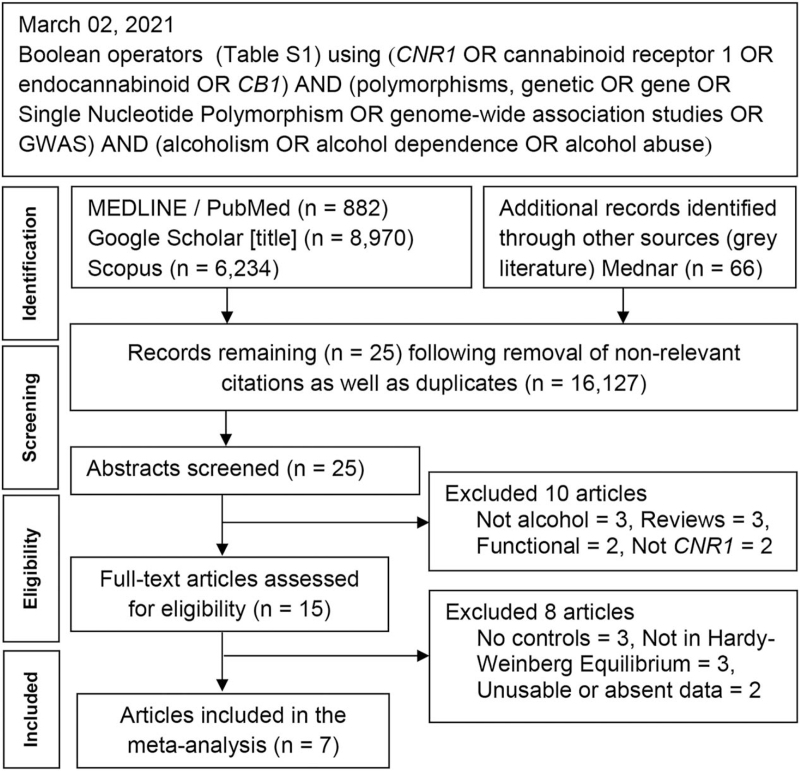
Figure 1 outlines the study selection process following the guidelines in Preferred Reporting Items for Systematic Reviews and Meta-Analyses (Table S3, Supplemental Digital Content). Initial search resulted in 16,152 citations, followed by a series of omissions that yielded 6 genetic association studies (GAS).[20–22,24,36,37] We found 7 GWAS that examined AD[38–44] in our search and 1[38] provided CNR1 data (Table S1, Supplemental Digital Content) warranting inclusion in our analysis. Of the total 7 articles (6 GAS and 1 GWAS), 4[20,21,36,38] examined a single SNP (rs1049353) and the rest examined multiple SNPs (Table 1). Table S2 (Supplemental Digital Content) lists genotype data, sample sizes for each SNP, minor allele frequencies, and control PHWE-values.
Figure 1.
Summary flowchart of the literature search.
Table 1.
Characteristics of the included studies in cannabinoid receptor 1 associations with alcohol dependency.
| First author | Bierut | Schmidt | Preuss | Marcos | Zhang | Herman | Zuo |
| Year | 2010 | 2002 | 2003 | 2012 | 2004 | 2006 | 2007 |
| Countries | USA and Germany | Germany | Germany | Spain | USA | USA | USA |
| Ethnicities | Caucasian (European descent) AA | Caucasian | Caucasian | Caucasian | Caucasian (European Americans)∗ AA∗, Japanese | Caucasian (Non-Hispanic), AA∗ | Caucasian (European Americans) AA |
| Sample sizes (cases / controls) | 1897/1932 | 265/136 | 196/210 | 298/155 (AD + alcohol abuse) | 1161/566 | 769/472 | 550/451 |
| Age of cases in years (mean ± standard deviation) or range | 39.2 ± 9.2 | 39.2 ± 8.6 | 41.1 ± 10.0 | 50.8 ± 10.7 | Not mentioned | 43.8 ± 9.6 | 39.5 ± 18.0 |
| Psychiatric diagnoses of cases | DSM-IV | CIDI†,‡ | ICD-10 DSM-IV | DSM-IV No AD history | DSM-IV | DSM-III-R # DSM-IV-SCID # | DSM-III-R, SADS |
| Controls screening procedure | Screened for AD | Questionnaire | Minnesota Multiphasic Personality Inventory | Examination | Not mentioned | Semi-structured assessment for drug dependence and alcoholism | DSM-III |
| Gender ratio (male: female) | 1:1 | Not mentioned | 3.7:1 | Male only | Not mentioned | 3.4:1 | 2.8:1 |
| Number of SNPs examined in the article | 132 | 1 | 1 | 3 | 5 | 4 | 8 |
| CNR1 polymorphisms | rs1049353 | rs1049353 | rs1049353 | rs1049353 | rs1535255 | rs1535255 | rs1049353 |
| examined in this meta-analysis | rs806379 | rs806379 | rs806379 | ||||
| rs2023239 | rs2023239 | ||||||
| Performed haplotype analysis | No | No | No | Yes | Yes | Yes | Yes |
| Performed linkage disequilibrium analysis | No | No | No | Yes | Yes | Yes | Yes |
| Addressed Hardy–Weinberg Equilibrium | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Clark–Baudouin (CB) score | 9 | 8 | 9 | 8 | 8 | 9 | 8 |
3.2. Study features
Table 1 shows that all 7 articles (2002–2010) had Caucasian subjects; with AA in 4 papers.[22,24,37,38] The Japanese samples in the Zhang et al[24] study were excluded in order to minimize ethnic heterogeneity. The mean and ± standard deviation (42.3 ± 4.5 years) of the age of the cases indicated a middle-aged demographic profile. Controls in most studies were screened for AXIS I or AXIS II psychiatric disorders, such as schizophrenia, with positive findings meriting exclusion from the studies. Phenotypic variation in the subjects was minimized with a battery of tests that involved third edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-III) and fourth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV) guidelines[1] (Table 1). The sex ratio in GWAS[38] was 1:1 whereas in GAS, 3 articles ranged from 2.8 to 3.7 men for every women.[20,22,37] Table S2 (Supplemental Digital Content) lists the 4 CNR1 SNPs used in the meta-analysis (rs1049353, rs1535255, rs2023239, and rs806379) which were Caucasians and AA. The number of studies (n) per SNP ranged from 4 to 6. Combined sample sizes in each of the 4 SNPs ranged from 90 to 1897 for cases and 46 to 1932 for controls. Table 1 shows that in 4 articles,[22,24,36,37] linkage disequilibrium and haplotype analyses were performed. The median (8.0) and interquartile range (8–9) of the CB scores indicated high methodological quality of the studies.
3.3. Meta-analysis outcomes
Table 2 shows 32 outcomes, 28 (88%) of which were fixed-effects and 4 (12%) were random-effects (heterogeneous). Of the 32, 14 were significant (Pa < .05), found in all the genetic models. Of the 14, 6 robust outcomes survived the HBC, which implicated three CNR1 SNPs (rs1535255, rs2023239, and rs806379) in risk for developing AD (Table 3). Publication bias was not performed as our results did not meet the 2 a priori criteria set in the Methods.
Table 2.
Summary outcomes for cannabinoid receptor 1 associations with alcohol dependency.
| Test of association | Test of heterogeneity | |||||||
| n | OR | 95% CI | P a | P het | I2 (%) | Analysis model | ||
| rs1049353 overall | ||||||||
| 1 | Homozygous | 6 | 1.24 | 0.92–1.67 | .15 | .13 | 42 | Fixed |
| 2 | Recessive | 6 | 1.14 | 0.85–1.51 | .38 | .33 | 13 | Fixed |
| 3 | Dominant | 6 | 1.49 | 0.86–2.60 | .16 | <.001 | 92 | Random |
| 4 | Dominant∗ | 5 | 1.13 | 0.95–1.33 | .16 | .25 | 26 | Fixed |
| 5 | Codominant | 6 | 1.08 | 0.96–1.23 | .20 | .16 | 36 | Fixed |
| rs1049353 Caucasian-Mixed | ||||||||
| 6 | Homozygous | 4 | 1.31 | 0.94–1.82 | .11 | .13 | 46 | Fixed |
| 7 | Recessive | 4 | 1.24 | 0.90–1.70 | .18 | .26 | 25 | Fixed |
| 8 | Dominant | 4 | 1.14 | 0.94–1.37 | .08 | .15 | 44 | Fixed |
| 9 | Codominant | 4 | 1.13 | 0.98–1.31 | .09 | .10 | 54 | Fixed |
| rs1535255 overall | ||||||||
| 10 | Homozygous | 4 | 1.54 | 1.05–2.26 | .03 | .65 | 0 | Fixed |
| 11 | Recessive | 4 | 1.30 | 0.90–1.88 | .17 | .65 | 0 | Fixed |
| 12 | Dominant | 4 | 1.08 | 0.77–1.52 | .64 | .02 | 71 | Random |
| 13 | Dominant∗ | 3 | 1.30 | 1.08–1.57 | .006 † | .40 | 0 | Fixed |
| 14 | Codominant | 4 | 1.43 | 1.24–1.65 | <.001 † | .21 | 33 | Fixed |
| rs2023239 overall | ||||||||
| 15 | Homozygous | 4 | 1.36 | 0.94–1.97 | .10 | .19 | 36 | Fixed |
| 16 | Recessive | 4 | 0.75 | 0.14–4.10 | .74 | <.001 | 91 | Random |
| 17 | Recessive∗ | 3 | 1.61 | 1.03–2.52 | .04 | .86 | 0 | Fixed |
| 18 | Dominant | 4 | 1.24 | 1.04–1.48 | .01 | .15 | 44 | Fixed |
| 19 | Codominant | 4 | 1.21 | 0.97–1.51 | .10 | .08 | 56 | Random |
| 20 | Codominant∗ | 3 | 1.33 | 1.13–1.56 | <.001 † | .40 | 0 | Fixed |
| rs806379 overall | ||||||||
| 21 | Homozygous | 6 | 1.31 | 1.07–1.60 | .009 | .22 | 29 | Fixed |
| 22 | Recessive | 6 | 1.23 | 1.03–1.46 | .02 | .40 | 3 | Fixed |
| 23 | Dominant | 6 | 1.18 | 1.01–1.38 | .04 | .36 | 9 | Fixed |
| 24 | Codominant | 6 | 1.15 | 1.04–1.27 | .007 † | .18 | 34 | Fixed |
| rs806379 Caucasian-Mixed | ||||||||
| 25 | Homozygous | 3 | 1.43 | 1.13–1.81 | .003 † | .17 | 43 | Fixed |
| 26 | Recessive | 3 | 1.28 | 1.05–1.57 | .01 | .32 | 11 | Fixed |
| 27 | Dominant | 3 | 1.26 | 1.05–1.52 | .01 | .26 | 26 | Fixed |
| 28 | Codominant | 3 | 1.20 | 1.07–1.35 | .003 † | .15 | 47 | Fixed |
| rs806379 African-American | ||||||||
| 29 | Homozygous | 3 | 1.03 | 0.70–1.53 | .88 | .43 | 0 | Fixed |
| 30 | Recessive | 3 | 1.08 | 0.78–1.51 | .64 | .34 | 0 | Fixed |
| 31 | Dominant | 3 | 0.98 | 0.71–1.33 | .88 | .65 | 0 | Fixed |
| 32 | Codominant | 3 | 1.02 | 0.84–1.24 | .85 | .39 | 0 | Fixed |
Table 3.
Power and sensitivity analyses for the main outcomes.
| Sample sizes | ASP (%) α = 0.05 | Test of association | Test of heterogeneity | ||||||||||||
| Comparison rs number | Genetic model | Outlier status | n | Cases | Controls | OR 1.5 | OR 1.2 | OR 1.1 | OR | 95% CI | P a | P het | I2 (%) | Analysis model | Sensitivity treatment outcome |
| Overall | |||||||||||||||
| rs1535255 | Dominant | Post- | 3 | 784 | 896 | 98.3 | 68.9 | 53.3 | 1.30 | 1.08–1.57 | .006 | .40 | 0 | Fixed | Robust |
| rs1535255 | Codominant | Pre- | 4 | 1135 | 1,086 | 99.7 | 80.7 | 65.3 | 1.43 | 1.24–1.65 | <.001 | .21 | 33 | Fixed | Robust |
| rs2023239 | Codominant | Post- | 3 | 566 | 287 | 78.7 | 38.0 | 28.1 | 1.33 | 1.13–1.56 | <.001 | .40 | 0 | Fixed | Robust |
| rs806379 | Codominant | Pre- | 6 | 1497 | 1,843 | 99.9 | 93.2 | 81.9 | 1.15 | 1.04–1.27 | .007 | .18 | 34 | Fixed | Robust |
| Caucasian-Mixed | |||||||||||||||
| rs806379 | Homozygous | Pre- | 3 | 1045 | 1,316 | 99.8 | 82.5 | 67.5 | 1.43 | 1.13–1.81 | .003 | .17 | 43 | Fixed | Robust |
| rs806379 | Codominant | Pre- | 3 | 1045 | 1,316 | 99.8 | 82.5 | 67.5 | 1.20 | 1.07–1.35 | .003 | .15 | 47 | Fixed | Robust |
3.3.1. rs1049353
This SNP had the highest aggregate sample sizes (3862 cases and 2863 controls) (Table S2, Supplemental Digital Content). However, none of the 9 rs1049353 outcomes were significant (ORs = 1.13–1.49, 95% CIs = 0.85–2.60, Pa = .15–.38), not even when confined to Caucasian (ORs = 1.13–1.31, 95% CIs = 0.90–1.82, Pa = .08–.18) (Table 2).
3.3.2. rs1535255
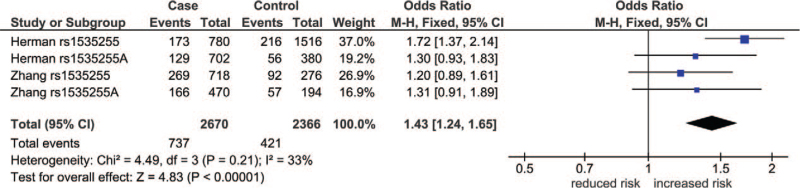
Aggregate sample sizes for this SNP were 1135 cases and 1086 controls (Table S2, Supplemental Digital Content). This SNP generated 5 comparisons, 3 of which were significant. Of the 3, 2 survived the HBC. In the first, the dominant outcome was postoutlier (ORs = 1.30, 95% CIs = 1.08–1.57, Pa = .006) with an ASP of 98.3% (OR 1.5). In the second, the codominant outcome was preoutlier (ORs = 1.43, 95% CIs = 1.24–1.65) with ASPs of 99.7% and 80.7% (ORs 1.5 and 1.2), respectively. Table 3 and Table S2 (Supplemental Digital Content) tabulate the data for which rs1535255 and Fig. 2 shows the forest plot for this outcome. In this figure, 3 study-specific ORs were non-significant (ORs = 1.20–1.31, 95% CIs = 0.91–1.89) and one was (OR = 1.72, 95% CI = 1.37–3.14), which resulted in a significant pooled OR (Pa < .001).
Figure 2.
Preoutlier forest plot in the codominant model of rs1533255. ∗: Caucasian; +: African-American; CI = confidence interval; CNR1 = cannabinoid receptor 1 gene; df = degree of freedom; I2 = measure of variability; Pa = P-value for association; Phet = P-value for heterogeneity.
3.3.3. rs2023239
This SNP had aggregate sample sizes of 704 cases and 681 controls (Table S2, Supplemental Digital Content). Of the 6 comparisons, 3 outcomes were significant, 1 of which survived the HBC. This postoutlier effect in the codominant model (OR = 1.33, 95% CI = 1.13–1.56, Pa < .001) was underpowered (Table 3).
3.3.4. rs806379
With aggregate sample sizes of 1497 cases and 1843 controls (Table S2, Supplemental Digital Content), this SNP generated 12 outcomes, 4 and 8 for the overall analysis and subgroups, respectively (Table 2). The 4 overall outcomes in all genetic models were significant (ORs = 1.15–1.31, 95% CIs = 1.01–1.60, Pa = .007–.04) one of which survived HBC. This was the codominant pooled effect (OR 1.15, 95% CI = 1.04–1.27, Pa = .007), which was statistically powered at all 3 levels of ORs (Table 3). Of the 4 significant outcomes in the Caucasian subgroup, 2 (powered at OR 1.5 and 1.2) survived HBC in the homozygous and codominant models (ORs = 1.20–1.43, 95% CIs = 1.07–1.81, Pa = .003). In contrast, all 4 AA outcomes were null (ORs = 0.98–1.08, Pa = .64–.88) (Table 2).
4. Discussion
4.1. Summary of the findings
Screened for false positives (HBC), the 14 significant outcomes were reduced to 6 pooled ORs implicating 3 SNPs (rs1535255, rs2023239, and rs806379). These 6 main outcomes showed up to 1.4-fold increased risks, 67% (4/6) which had a codominant effect. This effect in rs1535255, rs2023239, and rs806379 indicated consistency of associations. The rs806379 SNP identified Caucasians as a susceptible subgroup, but not the AA. Differential outcomes between the ethnicities could have been due to a number of confounding factors that involved sex or variations in phenotype definition. Our use of HBC, power analysis, subgrouping, outlier, and sensitivity approaches were instrumental in generating strong evidence for association, delineating subgroup effects and identifying CNR1 SNP associations with AD development. By design, such features are not present in the component single-study outcomes. Conflicting outcomes between primary studies may be attributed to small sample sizes, hence, lack of power.[30]
4.2. Gene–gene, gene–environment interactions
In spite of the evidence for associations, complexity of AD development involves interactions between genetic and non-genetic factors allowing for the strong likelihood of environmental and behavioral involvement. Although all the included articles focused on CNR1, gene–gene and gene–environment interactions have been reported to influence associations of CNR1 SNPs with development of AD. All 6 GAS studies addressed haplotypes, with 4[22,24,36,37] performing haplotype analysis.
4.3. Related meta-analysis
To our knowledge, this is the first meta-analysis to focus on the associations between CNR1 SNPs and susceptibility to AD development. We have used the multi-SNP approach of GWAS (1 CNR1 SNP + SNPs in other genes) and GAS (several SNPs in CNR1) in order to unmask associations of CNR1 SNPs with risk of developing AD. A previous meta-analysis[17] examined addictive disorders that included AD, with detailed AD results only for rs1048353. Hence, we compare outcomes of the A allele of Benyamina (2011) with ours in the codominant model of the overall and Caucasian analyses. Thus, both meta-analyses showed non-significant associations for developing AD in rs1048353 (ORs = 1.14–1.16, P = .14–.19 vs ORs = 1.13–1.14, Pa = .07–.09). However, in the broader scheme of substance dependence (AD included), the Benyamina et al[17] findings for rs806379 were null (ORs = 1.01–1.03, P = .40–.48) while ours were significant (ORs = 1.12–1.20, Pa = .02–.003). Moreover, AA outcomes were non-significant (P = .20) 1.1-fold association while ours were null (OR = 1.02, Pa = .85).
4.4. Complex phenotype of AD
An overriding issue in the genetics of AD development is recognizing that the AD phenotype is complex, involving other dependent-prone substances.[45] These include dependency on nicotine, marijuana (cannabis), and the hard drugs (cocaine, heroin) which involve various CNR1 SNPs. Significant influence of rs1049353 with heroin among Caucasians indicated that the AA and GG genotypes conferred protection and contributed susceptibility, respectively.[46] In a polysubstance abuse study, minor allele frequency involving rs1535255 was significantly higher in cases.[24] A significantly statistical association of impulsivity was found in several polymorphisms, which included rs1049353, rs1535255, rs2023239.[47] A significant association was found between polysubstance abuse in rs1535255, rs2023239, and rs806379.[24] The association of rs2023239 with alcohol, cannabis, and tobacco use resulted in greater activation in the nucleus accumbens and ventral medial prefrontal cortex, as shown with neuroimaging.[48] In the development of AD, C allele carriers of rs2023239 were found to modulate the effect of olanzapine,[24] but these findings were not replicated in a later study.[37] In contrast with these lack of effects, our findings for rs2023239 showed a highly significant codominant post-outlier-derived association, underpinned by robustness. More studies may be needed to confirm this finding. A 3-marker haplotype involving rs806379 had significant allelic frequency differences between various sample populations[24] but was not replicated in subsequent studies.[10,22,37]
4.5. Strengths and limitations
Limitations of our study include: first, insufficient number of studies precluded investigation of CNR2; second, most of the component studies were underpowered; third, our conclusions were limited to Caucasians and AAs. More studies may be needed to examine other ethnicities; and fourth, the studies that presented data on polysubstance abuse subtracted from the precision of the AD phenotype. The use of AD development as the sole phenotypic end point has been proposed to be a limitation.[49] Focusing on a narrow phenotype (AD) generated limitation fourth. Broadening the phenotype (e.g., polysubstance abuse) as a countermeasure would have altered our objectives. Having defined our objectives, we were constrained at countering limitations first to third. On the other hand, strengths of this study include: first, quality (CB scores) of component studies was high; second, restricted to HW-compliant studies reduced the risk for genotyping errors and possible selection bias; third, outlier treatment was key to generating significance and eliminating heterogeneity; fourth, applying HBC minimized the risk of a Type 1 error; fifth, power analysis outcomes strengthened the evidence for associations and minimized the risk for Type 2 error.[50]
In conclusion, this meta-analysis identified 3 CNR1 SNPs that may increase the risk for developing AD. Caucasian susceptibility differed from the lack of associations in AAs. Additional well-designed studies exploring other parameters would confirm or modify our results and add to the extant knowledge about the association of the CNR1 polymorphism and susceptibility to the development of AD.
Author contributions
Conceptualization: Noel Pabalan.
Data curation: Noel Pabalan, Phanthip Chaweeborisuit.
Formal analysis: Noel Pabalan, Phanthip Chaweeborisuit, Hamdi Jarjanazi.
Funding acquisition: Noel Pabalan.
Investigation: Adis Tasanarong, Pairath Tapanadechopone.
Methodology: Noel Pabalan, Hamdi Jarjanazi, Phanthip Chaweeborisuit.
Project administration: Adis Tasanarong.
Resources: Adis Tasanarong, Hamdi Jarjanaz.
Software: Noel Pabalan, Hamdi Jarjanazi.
Supervision: Pairath Tapanadechopone.
Validation: Adis Tasanarong, Thanee Eiamsitrakoon, Pairath Tapanadechopone.
Visualization: Phanthip Chaweeborisuit.
Writing – original draft: Noel Pabalan, Phanthip Chaweeborisuit, Phuntila Tharabenjasin.
Writing – review & editing: Noel Pabalan, Phanthip Chaweeborisuit, Phuntila Tharabenjasin.
Supplementary Material
Supplementary Material
Supplementary Material
Footnotes
Abbreviations: AA = African-American, AD = alcohol dependence, ASP = aggregate statistical power, CB = Clark–Baudouin, CB1 = cannabinoid receptor 1, CI = confidence interval, CIDI = Composite International Diagnostic Interview, CNR1 = cannabinoid receptor 1 gene, CNR1 = cannabinoid receptor 1 protein, CNR2 = cannabinoid receptor 2 gene, DSM-III, IV, and V = Third, fourth, and fifth editions of the Diagnostic and Statistical Manual of Mental Disorders, ECS = endocannabinoid system, GAS = genetic association studies, GWAS = genome-wide association studies; HBC = Holm–Bonferroni-correction, HWE = Hardy Weinberg equilibrium, I2 = measure of variability, n = number of studies, OR = odds ratio, Pa = P-value for association, Phet = P-value for heterogeneity, PHWE = P-value for HWE, SNP = single nucleotide polymorphism, var = variant, wt = wild-type.
How to cite this article: Pabalan N, Chaweeborisuit P, Tharabenjasin P, Tasanarong A, Jarjanazi H, Eiamsitrakoon T, Tapanadechopone P. Associations of CB1 cannabinoid receptor (CNR1) gene polymorphisms with risk for alcohol dependence: evidence from meta-analyses of genetic and genome-wide association studies. Medicine. 2021;100:43(e27343).
Ethics approval: An ethics committee was not necessary for this study.
This study was funded by the Chulabhorn International College of Medicine, Thammasat University Recipient: Noel Pabalan (2019–2020).
The authors have no conflicts of interest to disclose.
Data availability statement: All data generated or analyzed during this study are included in this article and its supplementary information files.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Supplemental digital content is available for this article.
AA = African-American, AD = alcohol dependency, CNR1 = cannabinoid receptor 1 gene, DSM = Diagnostic and Statistical Manual of Mental Disorders, ICD = International Classification of Diseases, SADS = schedule for affective disorders and schizophrenia, SCID = structured clinical interview for DSM, USA = United States of America.
Polysubstance cases with data for alcohol dependency.
CIDI = Composite International Diagnostic Interview.
Cases were subgrouped for clinical homogeneity.
CI = confidence interval, I2 = measure of variability, n = number of studies, OR = odds ratio, Pa = P-value for association, Phet = P-value for heterogeneity, bold values indicate significant associations.
Postoutlier.
Survived the Holm–Bonferroni correction.
Values in bold indicate significant association and in italics, statistically powered comparisons.
ASP = aggregate statistical power, CI = confidence interval, I2 = measure of variability, n = number of studies, OR = odds ratio, Pa = P-value for association, Phet = P-value for heterogeneity.
References
- [1].Association AP. Diagnostic and Statistical Manual of Mental Disorders 5 (DSM-5). Washington, DC: American Psychiatric Press; 2013. [Google Scholar]
- [2].Sher KJ, Grekin ER, Williams NA. The development of alcohol use disorders. Annu Rev Clin Psychol 2005;1:493–523. [DOI] [PubMed] [Google Scholar]
- [3].Rehm J. The risks associated with alcohol use and alcoholism. Alcohol Res Health 2011;34:135–43. [PMC free article] [PubMed] [Google Scholar]
- [4].Rehm J, Mathers C, Popova S, Thavorncharoensap M, Teerawattananon Y, Patra J. Global burden of disease and injury and economic cost attributable to alcohol use and alcohol-use disorders. Lancet 2009;373:2223–33. [DOI] [PubMed] [Google Scholar]
- [5].Augustin SM, Lovinger DM. Functional relevance of endocannabinoid-dependent synaptic plasticity in the central nervous system. ACS Chem Neurosci 2018;9:2146–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kunos G. Interactions between alcohol and the endocannabinoid system. Alcohol Clin Exp Res 2020;44:790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lavanco G, Castelli V, Brancato A, Tringali G, Plescia F, Cannizzaro C. The endocannabinoid-alcohol crosstalk: recent advances on a bi-faceted target. Clin Exp Pharmacol Physiol 2018. [DOI] [PubMed] [Google Scholar]
- [8].Manzanares J, Uriguen L, Rubio G, Palomo T. Role of endocannabinoid system in mental diseases. Neurotox Res 2004;6:213–24. [DOI] [PubMed] [Google Scholar]
- [9].Mechoulam R, Parker LA. The endocannabinoid system and the brain. Annu Rev Psychol 2013;64:21–47. [DOI] [PubMed] [Google Scholar]
- [10].Hartman CA, Hopfer CJ, Haberstick B, et al. The association between cannabinoid receptor 1 gene (CNR1) and cannabis dependence symptoms in adolescents and young adults. Drug Alcohol Depend 2009;104:11–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Childers SR, Breivogel CS. Cannabis and endogenous cannabinoid systems. Drug Alcohol Depend 1998;51:173–87. [DOI] [PubMed] [Google Scholar]
- [12].Herkenham M. Cannabinoid receptor localization in brain: relationship to motor and reward systems. Ann N Y Acad Sci 1992;654:19–32. [DOI] [PubMed] [Google Scholar]
- [13].van der Stelt M, Di Marzo V. The endocannabinoid system in the basal ganglia and in the mesolimbic reward system: implications for neurological and psychiatric disorders. Eur J Pharmacol 2003;480:133–50. [DOI] [PubMed] [Google Scholar]
- [14].Ameri A. The effects of cannabinoids on the brain. Prog Neurobiol 1999;58:315–48. [DOI] [PubMed] [Google Scholar]
- [15].Mechoulam R, Parker L. Cannabis and alcohol--a close friendship. Trends Pharmacol Sci 2003;24:266–8. [DOI] [PubMed] [Google Scholar]
- [16].Palmer RHCea. Shared additive genetic influences on DSM-IV criteria for alcohol dependence in subjects of European ancestry. Addiction 2015;110:1922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Benyamina A, Kebir O, Blecha L, Reynaud M, Krebs MO. CNR1 gene polymorphisms in addictive disorders: a systematic review and a meta-analysis. Addict Biol 2011;16:01–6. [DOI] [PubMed] [Google Scholar]
- [18].van den Wildenberg E, Janssen RG, Hutchison KE, van Breukelen GJ, Wiers RW. Polymorphisms of the dopamine D4 receptor gene (DRD4 VNTR) and cannabinoid CB1 receptor gene (CNR1) are not strongly related to cue-reactivity after alcohol exposure. Addict Biol 2007;12:210–20. [DOI] [PubMed] [Google Scholar]
- [19].Heller D, Schneider U, Seifert J, Cimander KF, Stuhrmann M. The cannabinoid receptor gene (CNR1) is not affected in German i.v. drug users. Addict Biol 2001;6:183–7. [DOI] [PubMed] [Google Scholar]
- [20].Preuss UW, Koller G, Zill P, Bondy B, Soyka M. Alcoholism-related phenotypes and genetic variants of the CB1 receptor. Eur Arch Psychiatry Clin Neurosci 2003;253:275–80. [DOI] [PubMed] [Google Scholar]
- [21].Schmidt LG, Samochowiec J, Finckh U, et al. Association of a CB1 cannabinoid receptor gene (CNR1) polymorphism with severe alcohol dependence. Drug Alcohol Depend 2002;65:221–4. [DOI] [PubMed] [Google Scholar]
- [22].Zuo L, Kranzler HR, Luo X, Covault J, Gelernter J. CNR1 variation modulates risk for drug and alcohol dependence. Biol Psychiatry 2007;62:616–26. [DOI] [PubMed] [Google Scholar]
- [23].Evans DE, Sutton SK, Jentink KG, Lin HY, Park JY, Drobes DJ. Cannabinoid receptor 1 (CNR1) gene variant moderates neural index of cognitive disruption during nicotine withdrawal. Genes Brain Behav 2016;15:621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang PW, Ishiguro H, Ohtsuki T, et al. Human cannabinoid receptor 1: 5’ exons, candidate regulatory regions, polymorphisms, haplotypes and association with polysubstance abuse. Mol Psychiatry 2004;9:916–31. [DOI] [PubMed] [Google Scholar]
- [25].Buhler KM, Huertas E, Echeverry-Alzate V, et al. Risky alcohol consumption in young people is associated with the fatty acid amide hydrolase gene polymorphism C385A and affective rating of drug pictures. Mol Genet Genomics 2014;289:279–89. [DOI] [PubMed] [Google Scholar]
- [26].Faul F, Erdfelder E, Lang AG, Buchner A. G∗Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Method 2007;39:175–91. [DOI] [PubMed] [Google Scholar]
- [27].Pabalan N, Bapat B, Sung L, Jarjanazi H, Francisco-Pabalan O, Ozcelik H. Cyclin D1 Pro241Pro (CCND1-G870A) polymorphism is associated with increased cancer risk in human populations: a meta-analysis. Cancer Epidemiol Biomarkers Prev 2008;17:2773–81. [DOI] [PubMed] [Google Scholar]
- [28].Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. Human genome epidemiology network. PLoS Med 2009;6:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dong J, Dai J, Zhang M, Hu Z, Shen H. Potentially functional COX-2-1195G>A polymorphism increases the risk of digestive system cancers: a meta-analysis. J Gastroenterol Hepatol 2010;25:1042–50. [DOI] [PubMed] [Google Scholar]
- [30].Clark MF, Baudouin SV. A systematic review of the quality of genetic association studies in human sepsis. Intensive Care Med 2006;32:1706–12. [DOI] [PubMed] [Google Scholar]
- [31].Eichstaedt KE, Kovatch K, Maroof DA. A less conservative method to adjust for familywise error rate in neuropsychological research: the Holm's sequential Bonferroni procedure. NeuroRehabilitation 2013;32:693–6. [DOI] [PubMed] [Google Scholar]
- [32].Higgins JP. Commentary: heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol 2008;37:1158–60. [DOI] [PubMed] [Google Scholar]
- [33].DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [34].Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719–48. [PubMed] [Google Scholar]
- [35].Ioannidis JP, Trikalinos TA. The appropriateness of asymmetry tests for publication bias in meta-analyses: a large survey. CMAJ 2007;176:1091–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Marcos M, Pastor I, de la Calle C, Barrio-Real L, Laso FJ, Gonzalez-Sarmiento R. Cannabinoid receptor 1 gene is associated with alcohol dependence. Alcohol Clin Exp Res 2012;36:267–71. [DOI] [PubMed] [Google Scholar]
- [37].Herman AI, Kranzler HR, Cubells JF, Gelernter J, Covault J. Association study of the CNR1 gene exon 3 alternative promoter region polymorphisms and substance dependence. Am J Med Genet B Neuropsychiatr Genet 2006;141B:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bierut L. A genome-wide association study of alcohol dependence. PNAS 2010;107:5082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kranzler HR, Zhou H, Kember RL, et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun 2019;10:1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Walters RK, Polimanti R, Johnson EC, et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci 2018;21:1656–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Edenberg HJ, Koller DL, Xuei X, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res 2010;34:840–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gelernter J, Kranzler HR, Sherva R, et al. Genome-wide association study of alcohol dependence: significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 2014;19:41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lai D, Wetherill L, Bertelsen S, et al. Genome-wide association studies of alcohol dependence, DSM-IV criterion count and individual criteria. Genes Brain Behav 2019;18:e12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang KS, Liu X, Zhang Q, Pan Y, Aragam N, Zeng M. A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence. J Psychiatr Res 2011;45:1419–25. [DOI] [PubMed] [Google Scholar]
- [45].Huang SY, Lin WW, Ko HC, et al. Possible interaction of alcohol dehydrogenase and aldehyde dehydrogenase genes with the dopamine D2 receptor gene in anxiety-depressive alcohol dependence. Alcohol Clin Exp Res 2004;28:374–84. [DOI] [PubMed] [Google Scholar]
- [46].Proudnikov D, Kroslak T, Sipe JC, et al. Association of polymorphisms of the cannabinoid receptor (CNR1) and fatty acid amide hydrolase (FAAH) genes with heroin addiction: impact of long repeats of CNR1. Pharmacogenomics J 2010;10:232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ehlers CL, Slutske WS, Lind PA, Wilhelmsen KC. Association between single nucleotide polymorphisms in the cannabinoid receptor gene (CNR1) and impulsivity in southwest California Indians. Twin Res Hum Genet 2007;10:805–11. [DOI] [PubMed] [Google Scholar]
- [48].Hutchison K. Cue-elicited craving and acute responses to alcohol: influence of the dopamine, opioid, and cannabinoid receptor genes. Alcohol Clin Exp Res 2006;30: (suppl): 253A.16441274 [Google Scholar]
- [49].Hutchison KE, Haughey H, Niculescu M, et al. The incentive salience of alcohol: translating the effects of genetic variant in CNR1. Arch Gen Psychiatry 2008;65:841–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Button KS, Ioannidis JP, Mokrysz C, et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci 2013;14:365–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.