Abstract
The yeast vacuole functions both as a degradative organelle and as a storage depot for small molecules and ions. Vacuoles are dynamic reticular structures that appear to alternately fuse and fragment as a function of growth stage and environment. Vac8p, an armadillo repeat-containing protein, has previously been shown to function both in vacuolar inheritance and in protein targeting from the cytoplasm to the vacuole. Both myristoylation and palmitoylation of Vac8p are required for its efficient localization to the vacuolar membrane (Y.-X. Wang, N. L. Catlett, and L. S. Weisman, J. Cell Biol. 140:1063–1074, 1998). We report that mutants with conditional defects in the rate-limiting enzyme of fatty acid synthesis, acetyl coenzyme A carboxylase (ACC1), display unusually multilobed vacuoles, similar to those observed in vac8 mutant cells. This vacuolar phenotype of acc1 mutant cells was shown biochemically to be accompanied by a reduced acylation of Vac8p which was alleviated by fatty acid supplementation. Consistent with the proposed defect of acc1 mutant cells in acylation of Vac8p, vacuolar membrane localization of Vac8p was impaired upon shifting acc1 mutant cells to nonpermissive condition. The function of Vac8p in protein targeting, on the other hand, was not affected under these conditions. These observations link fatty acid synthesis and availability to direct morphological alterations of an organellar membrane.
In addition to amino acids, nucleotides, and sugars, fatty acids are one of four families of basic cellular components. As such, they serve different biological functions; e.g., they form the hydrophobic core of lipid membranes, serve as an efficient storage form of metabolic energy, and may direct membrane association of otherwise soluble proteins.
Synthesis of fatty acids is catalyzed by a cytosolic multifunctional fatty acid synthetase complex (FAS) which in yeast is encoded by two genes, FAS1 (β subunit [6]) and FAS2 (α subunit [30]), whose products form the active hexameric α6β6 complex. Following the condensation of acetyl coenzyme A (acetyl-CoA) with the sulfhydryl group of the FAS-bound phosphopantetheine prosthetic group, sequential two-carbon unit elongation of the growing acyl chain occurs by condensation with malonyl-CoA, under the concomitant release of CO2. Seven elongation cycles are thus required to produce palmitoyl-CoA (C16:0). FAS elongates acetyl-CoA-primed substrates only (43, 51). Acyl chains of intermediate length are elongated by a different, membrane-bound system that also uses malonyl-CoA for elongation (8, 40, 47). Null mutations in either FAS1 or FAS2 are lethal unless the cells are supplemented with exogenous fatty acids 12 to 18 carbon atoms in length (42).
Malonyl-CoA, required for chain elongation, is supplied by acetyl-CoA carboxylase (Acc1p in yeast; ACC in other organisms), the rate-limiting enzyme of fatty acid synthesis. In prokaryotes, ACC activity requires the functional association of three different proteins, which provide a biotin-carboxylase, biotin-binding site, and transcarboxylase function to the active heteromeric complex. In eukaryotes, on the other hand, ACC activity is encoded by a single, trifunctional protein. Yeast Acc1p has a subunit molecular mass of 250 kDa, is active as a tetramer, and is subject to short-term regulation by phosphorylation (4, 29, 46, 56). ACC1 transcription is regulated positively by Ino2p/Ino4p and negatively by Opi1p and is thus under the general transcriptional control of phospholipid biosynthetic genes (13). In mammalian cells, active ACC forms long filamentous structures (19). Similar structures have been observed in yeast cells that overexpress the protein (40). In wild-type yeast, Acc1p is a cytosolic enzyme that is in association with the endoplasmic reticulum membrane (17).
The first acc1 mutant strains have been isolated in screens for fatty acid auxotrophic cells (28, 35). These early acc1 alleles represent reduced-function rather than loss-of-function mutations, as indicated by the observation that an acc1Δ allele is lethal even when fatty acids are supplemented (4, 13, 40). More recently, a novel temperature-sensitive (ts) allele of ACC1, mtr7 (hereafter referred to as acc1ts), has been isolated in a screen for mutants affected in nuclear mRNA transport (mtr mutants) (18, 40). Analysis of this conditional mutant revealed that the second essential function of Acc1p is to provide malonyl-CoA for elongation of long-chain (C16 to C18) fatty acids to very long chain (C26) fatty acids (40, 41). The synthesis of C26 is essential, as it forms part of the yeast ceramide (7, 38). Acyl chain elongation of C16/C18 to C26 is catalyzed by two microsomal membrane proteins with overlapping substrate specificity, Elo2p and Elo3p (31). The function of Acc1p in C26 synthesis cannot be restored by exogenous supplementation with long-chain and very long chain fatty acids, probably due to limited uptake and/or activation of the very hydrophobic C26 compound (40). The observations that acc1ts, but not the fatty acid auxotrophic acc1 alleles, affects nuclear export of mRNA and that acc1ts mutant cells have an altered morphology of the nuclear membrane have been taken to suggest a requirement of C26 synthesis in nuclear membrane-nuclear pore complex function (40, 41).
The biochemical, morphological, and genetic characterization of a novel cold-sensitive (cs) allele of acetyl-CoA carboxylase, acc1-200cs (hereafter referred to as acc1cs), is reported. This cs allele has been isolated in a screen for mutations that exhibit synthetic lethal interaction with hpr1Δ, a hyperrecombination mutation of Saccharomyces cerevisiae (1–3, 12). The synthetic lethal interaction between hpr1Δ and acc1cs is not allele specific, and we have previously shown that hpr1Δ and acc1cs mutant cells affect nuclear export of polyadenylated RNA (39).
We now report that similar to the ts allele, the cs allele affects the synthesis of the C26 fatty acid and that acc1cs mutant cells have greatly reduced steady-state levels of C26 under permissive conditions. Electron microscopic analysis of the morphology of acc1cs mutant cells revealed a fragmented vacuolar phenotype. This altered vacuolar morphology was rescued by the exogenous addition of fatty acids. We provide biochemical evidence that this multilobed vacuolar phenotype is due to defective acylation of Vac8p, a myristoylated and palmitoylated vacuolar protein whose acylation has previously been shown to be required for vacuolar morphology and inheritance (32, 52).
MATERIALS AND METHODS
Strains, plasmids, and genetic techniques.
Yeast strains and plasmids used in this study are listed in Tables 1 and 2. Media were prepared as described elsewhere (36). Media supplemented with fatty acids contained 1% Tween 40, 0.015% palmitic acid, and 0.015% stearic acid (35). Soraphen A, a kind gift from A. Hinnen, was added to media from a 10-mg/ml stock solution in methanol (48). Yeast cells were grown in liquid YEPD medium at 30°C. Optical density at 600 nm (OD600) was monitored every hour for growth rate determinations. Yeast cells were transformed as described elsewhere (15).
TABLE 1.
Yeast strain genotypes and construction
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| 479-2A | MATα acc1cs leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 | H. Klein |
| 485-13A | MATa acc1cs leu2-3,112 ura3-1 ade2-1 his3-11,15 can1-100 | H. Klein |
| LWY3682 | MATα leu2-3,112 lys2-801 ura3-52 his3-Δ200 suc2-Δ9 trp1-Δ901 vac8::HIS3 pYW40 (URA3) | L. Weisman (52) |
| YB336 | MATa nmt1-181 leu2-3,112 ura3-52 ade2-101 his3Δ200 lys2-801 | Duronio et al. (9) |
| W303d | MATa/MATα leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-1/ade2-1 his3-11,15/his3-11,15 can1-100/can1-100 | R. Rothstein |
| YRXS12 | MATα acc1ts leu2-Δ1 ura3-52 ade2-101 lys2-801 his3-Δ200 | Schneiter et al. (40) |
| 001 | MATa/MATα ACC1/acc1cs leu2-3,112/leu2-3,112 TRP1/trp1-1 ura3-1/ura3-1 ade2-1/ade2-1 his3-11,15/his3-11,15 can1-100/can1-100 | This work |
| 003 | MATa/MATα ACC1/acc1::TRP1 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-1/ade2-1 his3-11,15/his3-11,15 can1-100/can1-100 | This work |
| 009 | MATa/MATα acc1cs/acc1cs leu2-3,112/leu2-3,112 TRP1/trp1-1 ura3-1/ura3-1 ade2-1/ade2-1 his3-11,15/his3-11,15 can1-100/can1-100 | This work |
| 011-2 | MATa/MATα acc1::TRP1/acc1cs leu2-3,112/leu2-3,112 TRP1/trp1-1 ura3-1/ura3-1 ade2-1/ade2-1 his3-11,15/his3-11,15 can1-100/can1-100 | This work |
| YRXS301 | MATa/MATα ACC1/acc1::TRP1 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-1/ade2-1 his3-11,15/his3-11,15 can1-100/can1-100 pRXS53 (URA) pRXS89 (HIS) pCG002::Tn10-LUK#21 (LEU) | This work, strain 003 transformed with pRXS53, pRXS89, and pCG002::Tn10-LUK#21 |
| YRXS301 | MATa/MATα acc1ts/acc1cs leu2-Δ1/leu2-3,112 ura3-52/ura3-1 ade2-101/ade2-1 LYS2/lys2-801 TRP1/trp1-1 his3-Δ200/his3-11,15 CAN1/can1-100 | This work, diploid from cross between YRXS12 and 485-13A |
| YRXS303 | MATa VAC8-GFP-KanMX6 ura3-52 his3Δ200 | This work |
| YRXS304 | MATα acc1cs VAC8-GFP-KanMX6 leu2-3,112 ura3-1 ade2-1 trp1-1 his3-11,15 can1-100 | This work |
| YRXS305 | MAT? acc1cs vac8::HIS3 leu2-3,112 ura3 ade2-1 trp1 his3 pYW40 (URA3) | This work |
| YRXS306 | MAT? acc1cs nmt1-181 leu2-3,112 ura3 ade2 lys2-801 his3 | This work |
TABLE 2.
Plasmid construction
| Designation | Markers | Comments | Source |
|---|---|---|---|
| pCG001 and pCG002 | CEN LEU2 ACC1 | ACC1-containing plasmid isolated from the pBS32 genomic library | Guerra et al. (12) |
| pYW40 | CEN URA3 vac8-3 | Palmitoylation-deficient mutant of VAC8 | L. Weisman (52) |
| pYW10 | 2μm URA3 VAC8 | VAC8 high-copy-number plasmid | L. Weisman (52) |
| pCG005 | CEN URA3 ACC1 | 8-kb SacI fragment containing ACC1 cloned into pRS316 | This work |
| pCG006 | CEN URA3 acc1::TRP1 | 5.9-kb BglII fragment of ACC1 replaced by a 1.6-kb BglII fragment containing TRP1 | This work |
| pCG002::Tn10-LUK#21 | CEN LEU2 lacZ acc1::Tn10-LUK | C-terminally truncated ACC1, obtained after Tn10-LUK mutagenesis of pCG002 | This work |
| pRXS53 | CEN URA3 ACC1 | SphI fragment containing ACC1 into SphI site of pFL38 | Schneiter et al. (40) |
| pRXS89 | CEN HIS3 acc1K735R | Biotinylation-deficient allele of ACC1 | Schneiter et al. (40) |
| pGG2 | CEN HIS3 acc1ts | ts allele cloned into pRXS89 | This work |
Plasmids pCG001 and pCG002 have previously been described (12). Plasmid pCG005 consists of the 8-kb SacI restriction fragment containing the ACC1 gene from pCG001, subcloned into pRS316 (44). Plasmid pCG006 contains the ACC1 gene with most of the coding region (a 5.9-kb BglII restriction fragment) replaced by the TRP1 gene (contained on a 1.6-kb BglII restriction fragment). Strain 003 was derived from strain W303 by one-step gene disruption (37), using a 3.6-kb SacI restriction fragment from pCG006. Strain 011-2 is a 5-fluoro-orotic acid-resistant derivative from a diploid strain that was obtained by crossing strain 479-2A with a MATa acc1::TRP1 strain carrying the ACC1 plasmid pCG005.
To identify the DNA lesion in the two conditional acc1 alleles, we amplified the mutant and wild-type alleles by PCR and sequenced the products. The ts allele was cloned, following amplification of a 5,438-bp fragment from acc1ts (YRXS12) genomic DNA, using the primers ACC1-P05 (5′-ATCGTTGCGCCCGTTAAAAT-3′) and ACC1-P06 (5′-AGGCAACCATACCAATAGCGT-3′). The amplified fragment was cut with EagI and BamHI, gel purified, and cloned into EagI/NarI/BamHI-cut pRXS89 (40), giving rise to pGG2. pGG2 was tested for conferring temperature-dependent growth to a haploid acc1::TRP1 strain.
The carboxy-terminal fusion of green fluorescent protein (GFP) to Vac8p resulted from the genomic integration of a PCR-amplified GFP-KanMX6 cassette (50) immediately 5′ of the stop codon of VAC8, using primers pVAC8-1 (5′-GCAAGTTTGG AATTGTATAA TATTACTCAA CAGATTTTAC AATTTTTACA TGGAGCAGGT GCTGGTGCTG GTGCTGGAGC A-3′) and pVAC8-2 (5′-CGAAGATATA GATGTTATCT AGAATTGGTT TTTGTATGTA GCCCTTCTCT CCTTCATCGA TGAATTCGAG CTCGTTTAAA C-3′).
Tn10-LUK mutagenesis.
Plasmid pCG002 was mutagenized by Tn10-LUK transposon insertion (16). The transposon insertion site in the target plasmid was determined by restriction enzyme analysis and in some cases by sequencing the region upstream of the IS10-lacZ region of the transposon. DNA sequencing was performed with a Sequenase version 2.0 kit (U.S. Biochemical Corp. Cleveland, Ohio) and the oligonucleotide 5′-TGTAAAACGACGGGATC-3′ as primer. Plasmids carrying different transpositions were used to transform an acc1cs strain (479-2A). Transformants were scored for the cold-sensitive phenotype on YEPD medium at 20°C and for β-galactosidase activity on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) indicator medium at 30°C. β-Galactosidase assays were performed as described elsewhere (36).
Fatty acid analyses.
Glycerophospholipids and sphingolipids were extracted as previously described (14). After alkaline hydrolysis of lipids, fatty acids were converted to methyl esters by BF3-catalyzed methanolysis and separated by gas-liquid chromatography on a Hewlett-Packard Ultra 2 capillary column (5% phenyl dimethyl silicone) with a temperature gradient (20 min at 200°C, 10°C/min to 280°C, 15 min at 280°C). Fatty acids were identified by comparison to commercially available methyl ester standards (NuCheck, Inc., Elysian, Minn.).
Subcellular fractionation and Western blot analysis.
To determine the membrane association of Vac8p, exponentially growing cells were lysed by vigorous agitation with 0.5-mm-diameter glass beads in lysis buffer (0.3 M sorbitol, 10 mM Tris [pH 7.5], 0.1 M NaCl, 1 mM MgCl2, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) containing protease cocktail (57). Extracts were precleared by centrifugation at 500 × g for 10 min and separated into pellet (P13) and supernatant (S13) fractions by centrifugation at 13,000 × g for 10 min. The S13 fraction was then further separated into pellet (P100) and supernatant (S100) fractions by centrifugation at 100,000 × g for 30 min as previously described (52).
Protein concentration was determined according to Lowry et al. (24), using bovine serum albumin as a standard. Proteins were separated by one-dimensional sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (22), using 4% stacking and 10% separating gels. Proteins were transferred to nitrocellulose sheets (Hybond C; Amersham) and stained with Ponceau S to assess transfer efficiency. Membranes were incubated overnight in blocking solution containing 2% bovine serum albumin in Tris-buffered saline (TBS; 50 mM Tris-HCl, 150 mM NaCl [pH 8.0]). After two washes with TTBS (0.1% Tween 20 in TBS), membranes were incubated with one of the following rabbit primary antibodies: anti-Vac8p (1:1,000) (52), anti-Acc1p (1:700) (17), anti-Fas1/2p (1:600 and 1:5,000), and anti-aminopeptidase I (API) (1:5,000) (20). Peroxidase-conjugated ExtrAvidin (1:2,000; Sigma, St. Louis, Mo.) was diluted in TBS. The secondary antibody used was peroxidase-conjugated anti-rabbit immunoglobulin G (Sigma), and signal detection was carried out as instructed by the manufacturer, using the SuperSignal chemiluminescence-horseradish peroxidase substrate system (Pierce, Rockford, Ill.). Densitometric scanning of X-ray films and quantification of signals was carried out using NIH Image 1.54 (National Institutes of Health, Bethesda, Md.).
Enzyme activity measurements.
Cytosol used for ACC and FAS enzyme activity assays was prepared as follows. Cells were harvested at 1,200 × g for 10 min, washed with 0.1 M K2HPO4-KH2PO4 buffer (pH 6.5), mixed with breaking buffer (50 mM Tris-HCl, 100 mM NaF, 1 mM EDTA, 10 mM β-mercaptoethanol, 0.25 M sucrose, 1 mM phenylmethylsulfonyl fluoride [pH 7.5]) and glass beads (0.30-mm diameter) in a ratio of 1:1:1 (wt/vol/wt), and disrupted by four 1-min bursts in a Braun-Melsungen homogenizer under CO2 cooling. Glass beads were collected by centrifugation at 5,000 rpm for 5 min; the supernatant was centrifuged at 20,000 × g for 20 min and then centrifuged again at 195,000 × g for 80 min. Saturated (NH4)2SO4 was added in three portions within 20 min to 50% saturation, and samples were stirred for an additional 30 min. The precipitate was collected by centrifugation at 15,000 × g, dissolved in a HEPES buffer (50 mM HEPES, 1 mM EDTA, 0.02% sodium azide, 50% glycerol [pH 7.0]), and stored frozen at −20°C. Enzymatic activities of Acc1p and Fas1/2p were stable over a period of 3 weeks after freezing of samples. All steps were carried out at 4°C.
Activity of acetyl-CoA carboxylase was determined using a photometric assay in a coupled enzymatic reaction as described elsewhere (26), and FAS activity was measured by an established procedure (25). All enzyme measurements were carried out at 23°C.
Fluorescence microscopy and vital staining.
Vital staining of vacuoles with the lipophilic styryl dye N-(3-triethylammoniumpropyl)-4-(p-diethylaminophenyl-hexatrienyl) pyridinium dibromide (FM4-64; Molecular Probes, Eugene, Oreg.) was performed as described elsewhere (49). In vivo localization of the Vac8p-GFP fusion and microscopic analysis of FM4-64-stained samples was performed using a Leica TCS 4d confocal microscope with a PL APO 100×/1.40 objective. Figures were composed using Adobe Photoshop 5.0 (Adobe Systems, San Jose, Calif.) on a Macintosh PowerPC (Apple Computer Co., Cupertino, Calif.) and printed on an HP8500 color laser printer (Hewlett-Packard, Palo Alto, Calif.). Corresponding pictures were recorded using identical pin hole openings and amplification settings.
Electron microscopy.
For ultrastructural investigations, cells were fixed in 4% paraformaldehyde–5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.0)–1 mM CaCl2 for 90 min at room temperature, then washed in buffer with 1 mM CaCl2 for 1 h, and incubated for 1 h with a 2% (aqueous) solution of KMnO4. Fixed cells were washed in distilled water for 30 min and incubated in 1% sodium metaperiodate for 20 min. Then samples were rinsed in distilled water for 15 min and postfixed for 2 h in 2% OsO4 buffered with 0.1 M cacodylate at pH 7.0. After another wash with buffer for 30 min, the samples were dehydrated in a graded series of ethanol (50 to 100%, with en bloc staining in 2% uranyl acetate in 70% ethanol overnight) and embedded in Spurr's resin. Ultrathin sections were stained with lead citrate and viewed with a Philips CM10 transmission electron microscope.
RESULTS
Growth of the acc1cs mutant is not fully rescued by fatty acid supplementation.
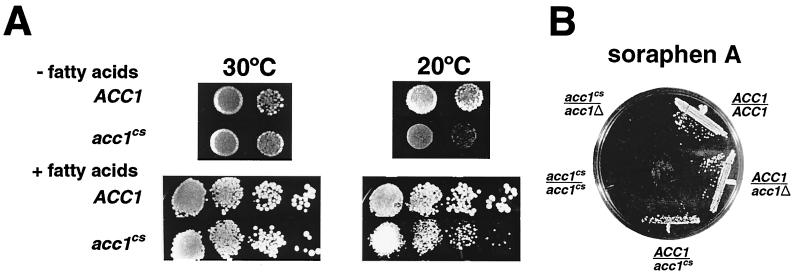
The acc1cs allele, acc1-200cs, was recovered in a screen for mutants that displayed synthetic lethal interaction with a null allele of HPR1 (12). The acc1cs strain showed a slight decrease in mitotic growth rate at the permissive temperature but grew poorly at the nonpermissive temperature of 20°C. Addition of fatty acids to the YEPD medium improved growth at 20°C but did not restore wild-type growth rates (Fig. 1A).
FIG. 1.
Effects of fatty acids and of soraphen A on growth of the acc1cs mutant. (A) Haploid yeast colonies of the indicated genotype were resuspended in water, and aliquots of dilutions were plated on medium with and without fatty acid supplementation; 10 μl containing ca. 104, 103, 102, or 101 cells was spotted onto plates, which were incubated for 3 days at 30°C or for 7 days at 20°C. (B) The soraphen A-sensitive phenotype of acc1cs is semidominant. Diploid strains of indicated genotypes were streaked onto YEPD medium containing 0.25 μg of soraphen A per ml, and the plate was incubated for 2 days at 30°C.
Soraphen A is a potent inhibitor of ACC activity in yeast (48). The acc1cs allele has previously been found to confer hypersensitivity to soraphen A at the permissive temperature (39). This hypersensitivity of acc1cs toward soraphen A was suppressed by the addition of fatty acids, consistent with the idea that inhibition of ACC activity leads to reduced fatty acid synthesis in vivo. Interestingly, the soraphen A-sensitive phenotype of acc1cs is semidominant (Fig. 1B), suggesting that the acc1cs product can form heteromeric complexes with wild-type Acc1p protein and thereby inhibit its activity. This mechanism has previously been proposed to explain the semidominant nature of other soraphen A-resistant acc1 alleles (48).
acc1cs is defective in malonyl-CoA-dependent acyl chain elongation and the synthesis of C26.
Supplementation with long-chain fatty acids is not sufficient to fully complement the loss of Acc1p function but is sufficient to rescue fas null alleles (13, 40, 42).
Fatty acid analysis of the acc1cs mutant strain shifted to nonpermissive conditions for 4 h revealed a dramatic increase in the level of C14:1 and a concomitant approximate twofold reduction of steady-state levels of C26 (Table 3). Levels of C16:0 and C18:0 were reduced approximately twofold, while those of C16:1 were slightly increased. These changes in the fatty acid profile of the cs mutant were comparable to what we previously observed for acc1ts (40), suggesting that the cs allele also affects malonyl-CoA-dependent acyl chain elongation. C26 synthesis requires both malonyl-CoA and long-chain acyl-CoA; long-chain fatty acid supplementation alone thus is not sufficient to relieve a malonyl-CoA-dependent block in C26 synthesis. The poor rescue of the cold sensitivity of the mutant by long-chain fatty acid supplementation is thus likely to be due to limiting levels of C26.
TABLE 3.
Fatty acid composition of wild-type and acc1cs mutant cellsa
| Fatty acid | Fatty acid composition (%)
|
|||
|---|---|---|---|---|
| 30°C
|
17°C
|
|||
| Wild type | acc1cs | Wild type | acc1cs | |
| C14:0 | 2.1 ± 0.2 | 4.4 ± 0.7 | 2.1 ± 0.3 | 4.8 ± 0.4 |
| C14:1 | 0.5 ± 0.1 | 1.3 ± 0.5 | 1.1 ± 0.1 | 7.5 ± 1.2 |
| C16:0 | 13.2 ± 1.6 | 15.3 ± 1.3 | 13.5 ± 0.9 | 7.9 ± 1.1 |
| C16:1 | 40.7 ± 2.2 | 50.1 ± 2.4 | 49.2 ± 2.2 | 60.9 ± 3.1 |
| C18:0 | 3.7 ± 0.4 | 2.5 ± 1.0 | 2.9 ± 0.3 | 1.1 ± 0.3 |
| C18:1 | 36.5 ± 0.9 | 25.8 ± 1.4 | 30.2 ± 1.6 | 17.2 ± 1.5 |
| C26:0 | 1.0 ± 0.1 | 0.6 ± 0.1 | 1.0 ± 0.2 | 0.5 ± 0.1 |
Cells were grown in liquid YEPD medium at 30°C to an OD600 of 0.3. The cultures were split; one fraction was maintained at 30°C, and the other was shifted to nonpermissive conditions for 4 h. Total lipids were extracted and subjected to alkaline hydrolysis, and fatty acids were converted to methyl esters by BF3-catalyzed methanolysis. Fatty acid methyl esters were separated by gas-liquid chromatography as described in Materials and Methods. Values represent means and standard deviations of three experiments.
Fate of Acc1p mutant proteins.
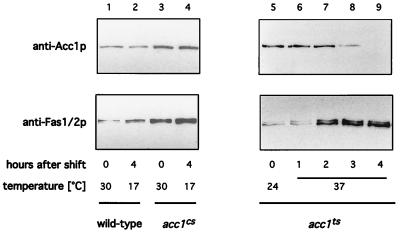
Previous analysis of the fate of ts mutant cells revealed that a shift to a nonpermissive temperature results in a rapid and irreversible loss of cell viability (40). Shifting the cs mutant strain to nonpermissive conditions, on the other hand, resulted in a reversible growth inhibition but did not affect cell viability (data not shown). To understand the difference in behavior of the two conditional alleles, the fate of the two mutant proteins was investigated in more detail. First, steady-state levels of Acc1p upon shifting cells to nonpermissive conditions were investigated by Western blot analysis. Levels of Acc1p in the cs strain did not visibly change upon a 4-h shift to nonpermissive conditions. Those in the ts mutant, however, declined to nondetectable levels within the same period of time at 37°C. This block in synthesis and/or increased turnover of Acc1p appeared to be specific, as steady-state levels of Fas1/2p displayed an apparently compensatory increase rather than decrease in acc1ts (Fig. 2).
FIG. 2.
Steady-state levels of Acc1p in conditional acc1 mutants, determined by Western blotting to detect Acc1p and Fas1/2p expression in wild-type (W303; lane 1 and 2) acc1cs (479-2A; lane 3 and 4), and acc1ts (YRXS12; lanes 5 to 9) strains at the indicated times at permissive and nonpermissive conditions.
Since neither steady-state levels nor the subcellular distribution of the cs mutant protein appeared to be affected in acc1cs cells (data not shown), we determined whether ACC enzymatic activity was conditionally affected. At permissive conditions, ACC activity was approximately sevenfold lower in the mutants than in the wild type. This activity further declined to nondetectable levels upon shifting cells to nonpermissive conditions. FAS activity, on the other hand, was not affected (Table 4).
TABLE 4.
Comparison of Acc1p and Fas1/2p activities in wild-type, acc1cs, and acc1ts mutantsa
| Enzyme | Enzyme activity (mU/mg)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild-type
|
acc1cs
|
acc1ts
|
||||||
| 17°C | 23°C | 30°C | 37°C | 17°C | 30°C | 23°C | 37°C | |
| Acc1p | ||||||||
| 0 h | 47.1 | 58.7 | 47.1 | 58.7 | 7.7 | 7.7 | 7.1 | 7.1 |
| 3 h | 41.3 | 48.4 | 45.0 | 41.4 | ND | 10.3 | 9.0 | ND |
| 4 h | 52.9 | 50.2 | 47.7 | 24.6 | ND | 16.8 | 5.2 | ND |
| Fas1/2p | ||||||||
| 0 h | 181.9 | 203.8 | 218.2 | 89.0 | 366.4 | 298.2 | 99.3 | 112.2 |
| 3 h | 200.0 | 170.3 | 234.8 | 107.1 | 406.4 | 334.1 | 100.6 | 92.9 |
| 4 h | 187.1 | 183.2 | 187.2 | 183.2 | 287.7 | 287.7 | 82.6 | 82.6 |
Cells were grown in YEPD medium at 30°C (acc1cs) or 23°C (acc1ts) to an OD600 of 0.3. The cultures were split; one fraction was maintained at the permissive temperature, and the other was shifted to the nonpermissive conditions for 3 or 4 h, respectively. Cells were harvested, and cytosol was prepared as described in Materials and Methods. Enzyme activities were determined as described elsewhere (25, 26). The standard deviation of four experiments was below 10%. ND, not detectable.
Interallelic complementation of different acc1 alleles.
To map the mutation responsible for the Cs phenotype, transposon insertion mutagenesis was performed. During the course of this work, an insertion within the coding region of ACC1 that complemented the Cs− phenotype of acc1cs was recovered. DNA sequence analysis of the insertion junction indicated that this insertion would create a carboxy-terminally truncated ACC1 allele (acc1C-term) that lacked the 574 amino acids after position 1772, including the CoA-binding site of the intact protein. Since the CoA-binding site is essential for the transcarboxylase activity of Acc1p, the truncated subunit would be expected to be enzymatically inactive (4, 23). Accordingly, a plasmid carrying this truncated acc1 allele failed to complement the lethality of an acc1Δ allele or the Ts phenotype of acc1ts.
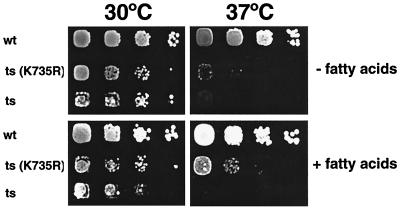
Since the ts and cs alleles appeared to affect different domains of the trifunctional enzyme, one would predict that they complement each other. To test this hypothesis, diploids from a cross of the two conditional mutants were examined for growth at 17 and 37°C. The acc1cs/acc1ts diploid (YRXS302) grew at all temperatures tested, indicating that the two alleles fully complemented each other. To further define which domain of the trifunctional enzyme is affected in the conditional mutants, the ts and cs mutant strains were transformed with a nonbiotinylatable point mutant allele of ACC1 in which the biotin-carrying lysine at position 735 had been replaced by an arginine residue (40). Surprisingly, this acc1K735R allele fully rescued the growth phenotype of the acc1cs strain at 17°C. It also rescued the acc1ts strain at the semipermissive temperature of 35°C and partially rescued growth of this strain at 37°C on fatty acid-supplemented plates (Fig. 3). ACC enzymatic activity of the different heteroallelic combinations are consistent with the observed growth phenotypes and revealed that the activity of the cs mutant protein is rescued to wild-type levels by either the carboxy-terminally truncated allele or the biotinylation-deficient allele of ACC1. The activity of the ts mutant allele, on the other hand, was increased only approximately twofold when combined with acc1C-term or acc1K735R but was fully restored in combination with acc1cs. These results on the interallelic complementation of different acc1 alleles are summarized in Table 5.
FIG. 3.
A biotinylation-deficient allele of ACC1, acc1K735R, partially rescues the acc1ts allele. Wild-type (wt; W303), the acc1ts mutant strain (ts; YRXS12), and the acc1ts mutant harboring a plasmid encoding the biotinylation deficient allele of ACC1 [ts(K735R); pRXS89] were serially diluted 10-fold and spotted onto YEPD and YEPD plates supplemented with fatty acids. Plates were incubated for 3 days at 30 or 37°C.
TABLE 5.
Interallelic complementation and ACC activity of different combinations of acc1 allelesa
| Allele combination | Growth (ACC activity [mU/mg]) at:
|
||
|---|---|---|---|
| 17°C | 30°C | 37°C | |
| None (wild type) | + | + (51.6) | + |
| acc1cs/acc1ts | + | + (48.2) | + |
| acc1cs/acc1C-term | + | + (47.4) | + |
| acc1cs/acc1K735R | + | + (45.1) | + |
| acc1ts/acc1C-term | + | + (12.3) | − |
| acc1ts/acc1K735R | + | + (14.4) | −b |
Cells containing the indicated combination of acc1 alleles were serially diluted 10-fold and replica plated on YEPD and YEPD plates supplemented with fatty acids. Plates were incubated at the indicated growth temperatures. Growth at 30 and 37°C was assessed after 3 days; growth at 17°C was determined after 7 days. ACC enzyme activity of strains bearing the indicated combination of acc1 alleles was determined from cytosolic extracts of strains grown in liquid minimal medium at 30°C. +, growth; −, no growth.
Partial rescue on fatty acid-supplemented plates.
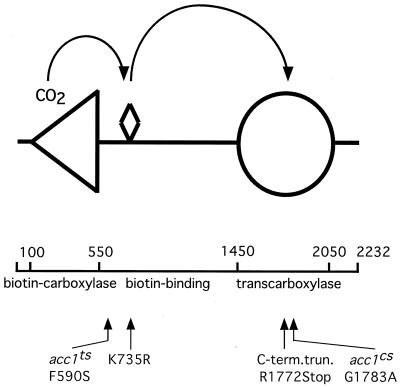
To gain a better understanding of how the mutations impaired ACC activity, the sequence alterations responsible for the conditional growth phenotype were identified. The alteration that conferred the Ts phenotype was identified as a mutation of codon 590 of the published sequence (4) from TTC to TCC, which resulted in a phenylalanine-to-serine change (F590S). Position 590 maps carboxy terminally to the biotin-carboxylase domain but lies before the biotin-binding domain of the enzyme. The mutation in the cs allele was identified as a GGT-to-GCT change of codon 1783, which resulted in a replacement of glycine by alanine (G1783A). Position 1783 maps within the transcarboxylase domain of the enzyme (Fig. 4).
FIG. 4.
Map (drawn to scale) summarizing the positions of mutations in different acc1 alleles. The biotin prosthetic group is represented by a diamond-shaped symbol, the biotin-carboxylase domain is represented by a triangle (positions 100-550), and the transcarboxylase domain is denoted by a circle (positions 1450 to 2050). Lesions associated with different alleles are indicated.
Interallelic complementation by stabilization of the heteroallelic enzymatic complex.
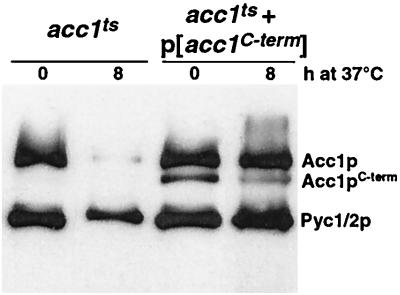
The fact that the cs mutation fell into the transcarboxylase domain but was nevertheless complemented by the carboxy-terminally truncated version of the enzyme suggested that in this case, interallelic complementation was not due solely to the functional substitution of one mutant protein domain by the corresponding wild-type domain of the complementing partner. Instead, the complementation map was consistent with the idea that interallelic complementation between different alleles could also be due to a stabilization of the enzymatically active heteroallelic complex. To test this hypothesis, we followed the fate of the thermolabile Acc1tsp mutant in the presence or absence of the carboxy-terminally truncated version of the protein. Western blot analysis of acc1ts mutant cells that coexpressed Acc1C-termp revealed a marked stabilization of the full-length thermolabile enzyme upon a shift to nonpermissive conditions (Fig. 5).
FIG. 5.
Acc1C-termp stabilizes the thermolabile enzymatic complex formed by Acc1tsp. For Western blot analysis of acc1ts (YRXS12) transformed with an empty vector or with pCG002::Tn10-LUK#21 encoding the C-terminally truncated version of Acc1p, cells were grown at permissive conditions in synthetic medium lacking leucine and shifted to nonpermissive conditions for 8 h. Protein extracts were prepared and separated by SDS-PAGE, and blots were probed with peroxidase-conjugated ExtrAvidin to detect biotinylated Acc1p and the two isoforms of pyruvate carboxylase (Pyc1/2p).
Conditional acc1 alleles affect vacuolar morphology.
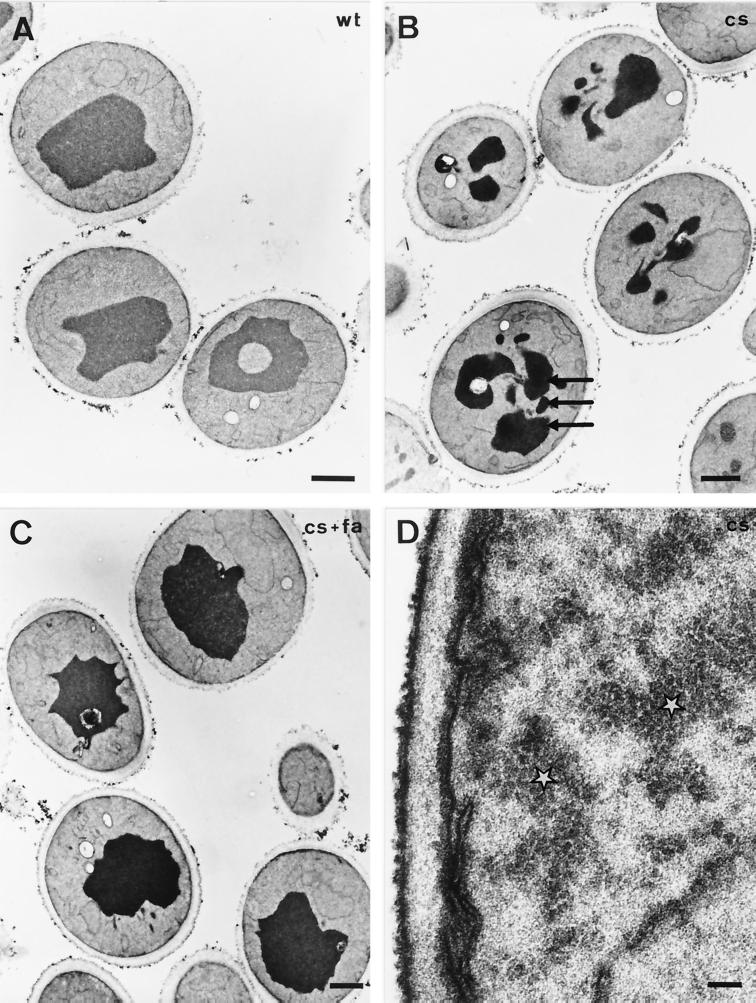
Our previous analysis of the morphological alterations observed in acc1ts at nonpermissive conditions revealed a striking alteration of the nuclear membrane. The two nuclear membranes were frequently separated from each other, and the newly formed intermembrane space contained vesicle-like structures (40, 41). To determine whether the cs allele exhibited a similar phenotype, cells were fixed and prepared for thin-section electron microscopy. As shown in Fig. 6B, acc1cs mutant cells shifted to nonpermissive conditions for 4 h displayed a fragmented vacuole, but no alteration of the nuclear envelope was observed. The vacuolar phenotype was exhibited by the majority of cells and was not observed in similarly treated wild-type cells (Fig. 6A). Vacuolar fragmentation appeared to be dependent on long-chain fatty acid synthesis, as it was rescued in cells supplemented with long-chain fatty acids (Fig. 6C). In addition, some of the acc1cs mutant cells contained electron-dense, granular cytosolic material, reminiscent of the structures that we previously observed in cells overexpressing Acc1p (40), suggesting that the cs mutant protein forms cytosolic aggregates at nonpermissive conditions (Fig. 6D).
FIG. 6.
Morphological analysis of acc1cs mutant cells. Transmission electron micrographs show wild-type (wt; A) and acc1cs mutant cells (B to D) shifted to nonpermissive conditions for 4 h in the absence (B and D) or presence (C) of supplemented fatty acids (fa). Fragmented vacuoles are indicated by arrows in panel B. Electron-dense granular structures reminiscent of Acc1p filaments are indicated by white stars in panel D. Bars: A to C, 1 μm; D, 0.1 μm.
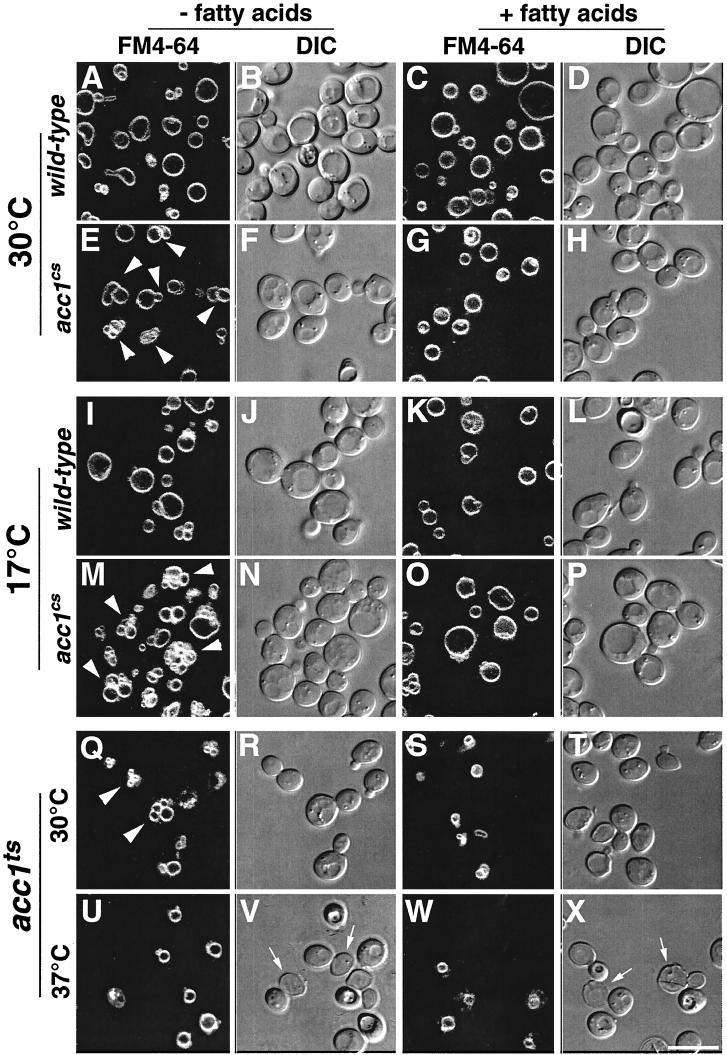
The appearance of the fragmented vacuole, a feature of several yeast mutants defective in endocytic trafficking (33), prompted us to investigate whether the endocytic marker FM4-64 (49) was properly delivered to the vacuolar compartment in conditional acc1 mutants. In wild-type cells, the vacuole of the mother cell was typically seen as a large round structure composed of an average of one to three lobes. Daughter cells normally inherit vacuoles from their mothers. Soon after bud formation, vacuoles from the maternal cell become organized into a so-called segregation structure that moves through the bud neck and gives rise to the vacuole of the daughter (11, 34, 55). As shown in Fig. 7, FM4-64 staining of conditional acc1 mutant cells revealed strong fluorescence of the large vacuolar compartment, suggesting that the dye is efficiently internalized and delivered to the vacuolar membrane. Unlike the vacuoles of wild-type cells, however, the vacuoles in the two conditional acc1 mutants were multilobed at permissive conditions (Fig. 7E and Q). This phenotype was even more pronounced in cs mutant cells shifted to nonpermissive conditions (Fig. 7M). Confirming the results obtained by electron microscopy, the multilobed vacuolar phenotype seen in the cs mutant was rescued by fatty acid supplementation (Fig. 7O).
FIG. 7.
Vacuolar morphology in wild-type, acc1cs, and acc1ts cells stained with FM4-64 Wild-type, acc1cs, and acc1ts cells were cultivated in YEPD or YEPD supplemented with fatty acids to early logarithmic growth phase at 30°C. Cells were then incubated with 30 μM FM4-64 for 30 min, followed by a chase for 1 h in YEPD or YEPD supplemented with fatty acids at 30°C. They were then incubated at the permissive temperature (30°C) or shifted to nonpermissive conditions (17°C for 3 h or 37°C for 30 min) and examined by confocal microscopy. Multilobed vacuoles are indicated by arrowheads in panels E, M, and Q. Dead cells in panels V and X are indicated by arrows. DIC (differential interference contrast) pictures of the visual fields are shown to the right of the fluorescence images. Bar, 10 μm.
A quantitative analysis of the number of vacuolar lobes observed upon FM4-64 staining of wild-type cells and the two conditional acc1 mutants revealed an average of 1.5 vacuolar lobes in wild-type cells, with 96% of buds containing FM4-64 stained vacuolar membranes. In the conditional acc1cs alleles, on the other hand, an average of 7.3 vacuolar lobes were observed, and 96% of daughters contained a stained vacuolar membrane, indicating that vacuolar inheritance was not affected in this mutant. In the acc1ts mutant cells, an average of 5.4 vacuolar lobes were observed. In these cells, however, only 59% of buds contained an FM4-64 stained vacuolar membrane, indicating that vacuolar inheritance was also impaired.
Acylation and vacuolar membrane association of Vac8p is impaired in acc1cs mutants.
Several vacuolar inheritance mutants, isolated through selection with a fluorescence-activated cell sorter, have previously been divided into three classes based on vacuolar morphology (53). Among these, the class I vac8 mutant appears to arrest early in vacuolar inheritance and displays multilobed vacuoles, similar to what we observed in the conditional acc1 alleles (53). Interestingly, Vac8p/Yeb3p/Yel013p is an armadillo repeat-containing protein that requires both myristoylation and palmitoylation for localization to the vacuolar membrane (10, 32, 52). Moreover, a palmitoylation-deficient allele of VAC8, vac8-3, displays a multilobed vacuolar phenotype similar to that of a vac8Δ allele, indicating that palmitoylation is essential for a wild-type vacuolar morphology (52).
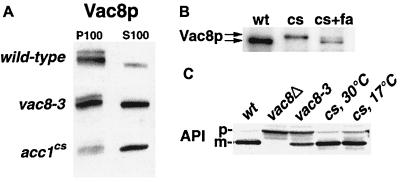
We thus investigated whether acylation and hence vacuolar localization of Vac8p are affected in acc1cs mutants. Subcellular fractionation followed by Western blot analysis with an antibody against Vac8p revealed that the protein was enriched in the soluble cytosolic fraction from acc1cs mutant cells but pelleted with the membrane fraction of wild-type cells. In the palmitoylation-deficient vac8-3 mutant, the protein partitioned approximately equally between the particulate and the soluble fraction (Fig. 8A). These results are consistent with the idea that acylation of Vac8p was impaired in acc1cs and that a reduced level of palmitoylated Vac8p may result in the multilobed vacuolar phenotype observed in the mutant. A reduced capacity to acylate Vac8p in cs mutant cells is furthermore indicated by the reduced mobility of Vac8p upon SDS-PAGE of whole cell extracts from cells shifted to nonpermissive conditions (Fig. 8B). It has previously been shown for acylation-defective point mutant alleles of VAC8 that this difference in mobility is attributable to the lack of posttranslational acylation of the protein (52). Consistent with this idea, we find that wild-type mobility of Vac8p is restored in cs mutant cells supplemented with fatty acids (Fig. 8B).
FIG. 8.
Membrane association and acylation of Vac8p, but not processing of API, is affected in acc1cs mutant cells. (A) Membranes isolated from exponentially growing wild-type, vac8-3 (expressing a nonpalmitoylatable allele of Vac8p) (52), and acc1cs cells were subfractionated into P100 and S100 fractions, and membrane association of Vac8p was determined by immunoblot analysis with an anti-Vac8p serum. (B) Whole cell extracts of wild-type (wt) and cs mutant cells shifted to nonpermissive conditions for 4 h in the presence (+fa) or absence of exogenously added fatty acids were separated by SDS-PAGE, and the relative mobility of Vac8p was assessed by immunoblot analysis with an anti-Vac8p serum. (C) API processing in wild-type (wt), vac8Δ, vac8-3, and acc1cs mutant cells (cs) incubated at either permissive (30°C) or nonpermissive (17°C) conditions for 4 h was determined by immunoblot analysis with an anti-API serum. The positions of the precursor (p) and mature (m) forms of API are indicated.
Vac8p not only is required for vacuolar morphology and inheritance but also affects the cytoplasm-to-vacuole-targeting (Cvt) pathway (21), as indicated by the observation that a vac8Δ allele accumulates the precursor form of API (52, 53). In contrast to the function of Vac8p in vacuolar morphology and inheritance, the function of Vac8p in protein targeting is independent of its acylation (52). To determine whether this function of Vac8p is affected in the cs mutant, maturation of API was analyzed by Western blotting. As shown in Fig. 8C, maturation of API was not impaired in acc1cs at either permissive or nonpermissive conditions.
acc1cs affects the subcellular localization of a Vac8p-GFP fusion.
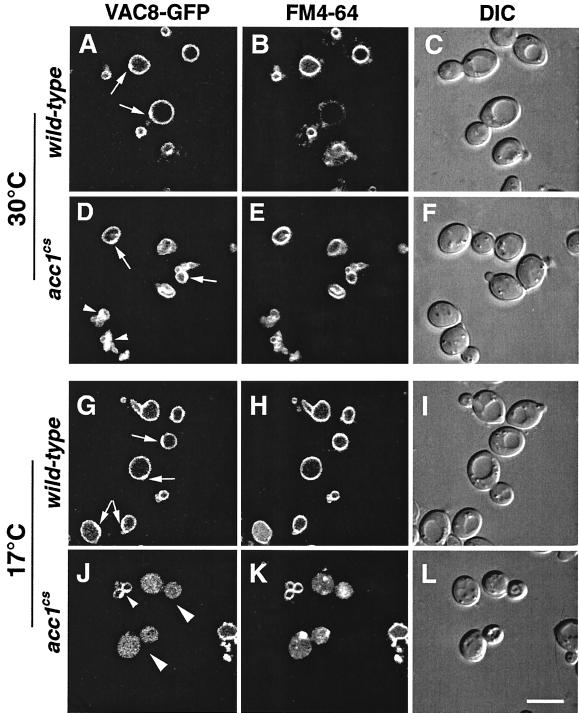
To determine the in vivo localization of Vac8p in acc1cs mutant cells, GFP was fused to the carboxy terminus of Vac8p by genomic integration of a GFP-KanMX6 module just prior to the stop codon of VAC8 (50). The resulting Vac8p-GFP fusion was functional in both API maturation and vacuolar morphology and inheritance (data not shown). Remarkably, subcellular localization of this fusion protein by confocal microscopy revealed a somewhat polarized distribution of Vac8p-GFP on the vacuolar membrane of wild-type cells, compared to the more uniform signal obtained from FM4-64 staining of the membrane. On relaxed vacuolar membranes of wild-type cells, the more intense Vac8p-GFP signal was always located on the site of the vacuole that faced the nucleus and the bud site (Fig. 9A, D, and G). In acc1cs mutant cells at permissive conditions, Vac8p-GFP colocalized with the multilobed vacuole. Under nonpermissive conditions, however, a significant fraction of the cells displayed aberrant, uniform labeling of Vac8p-GFP throughout the cell (Fig. 9J). This mislocalization of Vac8p-GFP was not observed in acc1cs mutant cells supplemented with fatty acids (data not shown).
FIG. 9.
Subcellular distribution of Vac8p fused to GFP in wild-type and acc1cs mutant cells. Wild-type and acc1cs mutant cells expressing a functional Vac8p-GFP fusion protein were grown to early logarithmic growth phase at 30°C. Cells were then incubated with 30 μM FM4-64 for 30 min, followed by a chase for 1 h in YEPD. The cultures were split and incubated at either permissive (30°C) or nonpermissive (17°C) conditions for 4 h. Vacuolar morphology and the subcellular distribution of Vac8p-GFP was then analyzed by confocal microscopy. The polarized distribution of Vac8p-GFP on the relaxed vacuolar membrane of wild-type cells is indicated by arrows in panels A, D, and G. Small arrowheads in panels D and J point to a polarized distribution of Vac8p-GFP on multilobed vacuolar structures. Aberrant distribution of Vac8p-GFP in acc1cs mutant cells at nonpermissive conditions is indicated by larger arrowheads in panel J. DIC (differential interference contrast) pictures of the visual fields are shown to the right of the fluorescence images. Bar, 5 μm.
DISCUSSION
Acc1p is a cytoplasmic enzyme that catalyzes the rate-limiting step of fatty acid synthesis. The gene is essential, and we have shown in this report that synthesis of the gene product is rate limiting. The inability to fully complement the cold-sensitive growth of the acc1cs mutant by exogenous long-chain fatty acids reflects deficiencies in other essential malonyl-CoA-dependent processes, i.e., the microsomal fatty acid elongation systems (8, 31, 40). Consistent with such a defect in acyl chain elongation, the chain length profile of fatty acids in the acc1cs mutant was shifted toward shorter chains. Most importantly, steady-state levels of C26 were reduced approximately twofold in the mutant, suggesting that the malonyl-CoA pool in acc1cs is limiting at permissive conditions and becomes depleted at nonpermissive conditions.
In an attempt to map the cold-sensitive lesion, a transposon insertion in the coding sequence of the ACC1 gene was recovered. This carboxy-terminally truncated allele complemented the cold-sensitive growth of the acc1cs mutant but did not complement the temperature-sensitive growth of the acc1ts mutant, suggesting that the cs allele affected a different domain of the enzyme than the ts allele. Consistent with this hypothesis, an acc1cs/acc1ts diploid strain was wild type for growth at all temperatures tested, indicating that the two alleles fully complemented each other's defect by interallelic complementation (58). The fact that the enzymatically inactive carboxy-terminally truncated version complemented the cs allele, despite bearing a defect in the same domain of the protein, suggested that in this case, complementation was due to a stabilizing effect of the truncated protein on the heteroallelic enzyme complex. In cs mutant cells, cytosolic structures resembling the filamentous form of Acc1p were apparent by electron microscopy, suggesting that the mutant enzyme aggregated at the nonpermissive temperature. In vitro sedimentation experiments, however, did not allow us to recapitulate this possible aggregation of the cs mutant protein, thus precluding a direct assay of the stabilizing effect of the truncated allele on the cs mutant protein (data not shown). A stabilizing effect of Acc1C-termp on a heteroallelic enzymatic complex, however, could be demonstrated with the heat-labile enzyme, indicating that stabilization of an otherwise labile enzymatic complex is one possible mechanism to account for the complementation between conditional alleles and the nonfunctional alleles of ACC1.
Electron microscopic analysis of the morphology of acc1cs mutant cells revealed a fragmented vacuole. This vacuolar phenotype was also observed by fluorescence microscopy of cells stained with FM4-64. The phenotype was not specific for the cs allele but was also observed in ts mutant cells. Addition of fatty acids fully rescued the vacuolar fragmentation of acc1cs mutant cells as determined by electron microscopy and FM4-64 staining of viable cells. Acidification of the vacuolar compartment, however, was not affected in the conditional acc1 mutants, as revealed by quinacrine staining (data not shown) (54).
There were remarkable differences between the two conditional acc1 alleles with respect to the way in which they affected vacuolar morphology and inheritance: (i) in contrast to the ts mutant allele, the cs mutant cells displayed no defect in vacuolar inheritance; (ii) the multilobed vacuolar morphology of the ts mutant was not fully rescued by fatty acid addition; and (iii) shifting acc1ts to nonpermissive conditions rapidly resulted in the relaxation of the vacuolar membrane. The signals that affect vacuolar structure may be manifold, and acylation of Vac8p is probably only one of these. The observation that the presence of fatty acids is not sufficient to fully relax the vacuolar membrane in the ts mutant suggests that additional signals required for vacuolar relaxation are also impaired in this mutant.
This apparent discrepancy between the two conditional acc1 alleles may be explained by recalling that the ts allele is the stronger of the two alleles, as indicated by the decline of steady-state levels of Acc1tsp at 37°C and the fact that ts mutant cells shifted to nonpermissive conditions rapidly die. The acyl-CoA pool in this mutant thus appears to become depleted more efficiently than in the cs mutant cells, which require a shift to low temperature, a condition that by itself reduces metabolic turnover. The same argument may also explain why the nuclear envelope alteration is observed only in ts mutant cells (40, 41). Other phenotypes, such as the synthetic lethal interaction with hpr1Δ (39), however, are shared between the two alleles, suggesting that different levels of depletion of malonyl-CoA may result in distinguishable phenotypic consequences.
The multilobed vacuolar phenotype observed in the conditional acc1 mutants is a hallmark of the class I vacuolar inheritance (vac) mutation, vac8 (53). Vac8p is an armadillo repeat-containing protein whose function is required both for the Cvt pathway and for vacuolar morphology and inheritance (52, 53). Interestingly, Vac8p is both myristoylated and palmitoylated, and acylation of the protein is required for its localization to the vacuolar membrane but not for its function in protein targeting (32, 52). Characterization of the membrane association of Vac8p and its mobility in SDS-gels revealed that Vac8p was mostly soluble and not fully acylated in the acc1cs mutant, suggesting that a reduced level of acylation of Vac8p may result in the observed multilobed vacuolar phenotype of acc1cs mutant cells. Consistent with this proposed role of ACC1 in vacuolar morphology, API processing was not affected in acc1cs mutant cells.
In contrast to a myristoylation-deficient mutant, a palmitoylation-deficient point mutant allele of VAC8, vac8-3, renders the mutant completely defective in vacuolar inheritance (52). Given that vacuolar inheritance is affected in the ts but not the cs mutant strain, one might suggest that the ts allele affects palmitoylation of Vac8p more strongly than does the cs mutation at 30°C.
Analysis of the subcellular distribution of a GFP-tagged functional version of Vac8p revealed a polarized distribution of Vac8p on the relaxed vacuolar membrane of wild-type cells, where it appeared to be concentrated on those parts of the membrane that faced the nucleus and the growing bud site. The subcellular localization of a carboxy-terminal fusion of Vac8p with GFP has previously been analyzed (32). In this study, the Vac8p fusion was observed to be concentrated ∼5 to 7-fold in bands located between clustered vacuoles. While we see a similar clustering of Vac8p-GFP on multilobed membranes (Fig. 9D and J), our analysis extends this observation of the polarized distribution of Vac8 on relaxed vacuolar membranes. Consistently, previous localization of Vac8p by immunogold electron microscopy revealed that Vac8p appeared to be enriched on the vacuolar membrane at sites of membrane-membrane contact (10). The functional significance of this remarkably polarized localization of Vac8p remains to be determined. However, the fact that Vac8p is required for polarization of the vacuole to the presumptive bud site and specifically associates with actin filaments in vitro has been taken to suggest that Vac8p may mediate vacuole membrane-actin interactions (52).
In the cs mutant, mislocalization of Vac8p-GFP throughout the cell was observed at nonpermissive conditions. This mislocalization was not observed when the cells were supplemented with fatty acids (data not shown). The microscopic assay for membrane association of Vac8p, however, appears less sensitive than the biochemical fractionation, since minor amounts of soluble Vac8p will escape microscopic detection due to dilution of the protein in the cytosol, but these minor amounts will be revealed quantitatively by the immunoblot analysis. The mutant cells that displayed mislocalized Vac8p-GFP appeared to be more generally impaired, as indicated by the aberrant FM4-64 staining of the vacuolar membrane.
The relationship between fatty acid synthesis and Vac8p function was further investigated by epistatic analysis. The vacuolar morphology of an acc1cs vac8-3 double mutant was strongly multilobed already at 30°C, indicating that vac8-3 is downstream of acc1cs. Similarly, transformation of acc1cs with a VAC8-harboring high-copy-number plasmid did not rescue the vacuolar phenotype of the cs mutant (data not shown). These results are consistent with the proposed function of ACC1 in acylation of Vac8p. Moreover, analysis of the vacuolar morphology of a conditional N-myristoylation-deficient (nmt1-181) strain (9) also revealed a multilobed vacuolar phenotype. Consistent with the myristic acid auxotrophy of this mutant strain, the vacuolar phenotype of nmt1-181 was fully rescued by supplementation with C14:0 but not C16:0 fatty acids (our unpublished observations). Thus, like the ts mutant acc1 allele, the vacuolar membrane of nmt1-181 did not relax upon fatty acid (C16:0) supplementation. Finally, analysis of an acc1cs nmt1-181 double mutant revealed a vacuolar morphology characteristic of nmt1-181, indicating that myristoylation is downstream of acc1cs. Taken together, these genetic data are consistent with the proposed function of ACC1 in affecting vacuolar morphology through acylation of Vac8p.
Unlike myristoylation, palmitoylation appears to be reversible and thus may be regulated in a dynamic manner (27, 45). Protein acylation is generally thought of as a mechanism to shuttle a protein on and off a membrane. In case of Vac8p, however, acylation may be important for functions in addition to membrane targeting. The observed polarized localization of Vac8p is intriguing and might possibly stand in relation to the acyl anchor of Vac8p and the proposed properties of saturated acyl chains to form structurally and functionally defined membrane domains (5).
ACKNOWLEDGMENTS
We thank G. Daum for kind support and helpful discussions, A. Hinnen for the gift of soraphen A. G. Gogg for fatty acid analysis, L. Weisman for generously providing vac8 mutant strains, anti-Vac8p serum, and the VAC8 high-copy-number plasmid, D. Goldfarb for providing a Vac8p-GFP fusion plasmid, J. Gordon for providing the conditional N-myristoyltransferase mutant, and D. Klionsky and M. Thumm for making the anti-API serum available to us. Critical reading of the manuscript by A. Leber and K. Athenstaedt is gratefully acknowledged.
This work was supported by the National Institutes of Health (grant GM30439 to H.L.K.), the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich (project 11731 to S.D.K.; M00304 and 13767 to R.S.), the Austrian Nationalbank (P7273 to S.D.K.), and the Swiss National Science Foundation (823A-046702 to R.S.).
REFERENCES
- 1.Aguilera A, Klein H L. Genetic and molecular analysis of recombination events in Saccharomyces cerevisiae occurring in the presence of the hyper-recombination mutation hpr1. Genetics. 1989;122:503–517. doi: 10.1093/genetics/122.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilera A, Klein H L. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera A, Klein H L. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol Cell Biol. 1990;10:1439–1451. doi: 10.1128/mcb.10.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Feel W, Chirala S S, Wakil S J. Cloning of the yeast FAS3 gene and primary structure of yeast acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1992;89:4534–4538. doi: 10.1073/pnas.89.10.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown D A, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 6.Chirala S S, Kuziora M A, Spector D M, Wakil S J. Complementation of mutations and nucleotide sequence of FAS1 gene encoding β- subunit of yeast fatty acid synthase. J Biol Chem. 1987;262:4231–4240. [PubMed] [Google Scholar]
- 7.Dickson R C. Sphingolipid functions in Saccharomyces cerevisiae: comparison to mammals. Annu Rev Biochem. 1998;67:27–48. doi: 10.1146/annurev.biochem.67.1.27. [DOI] [PubMed] [Google Scholar]
- 8.Dittrich F, Zajone D, Hühne K, Hoja U, Ekici A, Greiner E, Klein H, Hofmann J, Bessoule J J, Sperling P, Schweizer E. Fatty acid elongation in yeast: biochemical characteristics of the enzyme system and isolation of elongation-defective mutants. Eur J Biochem. 1998;252:477–485. doi: 10.1046/j.1432-1327.1998.2520477.x. [DOI] [PubMed] [Google Scholar]
- 9.Duronio R J, Rudnick D A, Johnson R L, Johnson D R, Gordon J I. Myristic acid auxotrophy caused by mutation of S. cerevisiae myristoyl-CoA:protein N-myristoyltransferase. J Cell Biol. 1991;113:1313–1330. doi: 10.1083/jcb.113.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleckenstein D, Rohde M, Klionsky D J, Rüdiger M. Ye1013p (Vac8p), an armadillo repeat protein related to plakoglobin and importin alpha is associated with the yeast vacuole membrane. J Cell Sci. 1998;111:3109–3118. doi: 10.1242/jcs.111.20.3109. [DOI] [PubMed] [Google Scholar]
- 11.Gomes de Mesquita D S, ten Hoopen R, Woldringh C L. Vacuolar segregation to the bud of Saccharomyces cerevisiae: an analysis of morphology and timing in the cell cycle. J Gen Microbiol. 1991;137:2447–2454. doi: 10.1099/00221287-137-10-2447. [DOI] [PubMed] [Google Scholar]
- 12.Guerra C E, Klein H L. Mapping of the ACC1/FAS3 gene to the right arm of chromosome XIV of Saccharomyces cerevisiae. Yeast. 1995;11:697–700. doi: 10.1002/yea.320110711. [DOI] [PubMed] [Google Scholar]
- 13.Hasslacher M, Ivessa A S, Paltauf F, Kohlwein S D. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 14.Hechtberger P, Daum G. Intracellular transport of inositol-containing sphingolipids in the yeast, Saccharomyces cerevisiae. FEBS Lett. 1995;367:201–204. doi: 10.1016/0014-5793(95)00567-s. [DOI] [PubMed] [Google Scholar]
- 15.Hill J, Ian K A, Donald G, Griffiths D E. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huisman O, Raymond W, Froehlich K, Errada P, Kleckner N, Botstein D, Hoyt M A. A Tn10-lacZ-kanR-URA3 gene fusion transposon for insertion mutagenesis and fusion analysis of yeast and bacterial genes. Genetics. 1987;116:191–199. doi: 10.1093/genetics/116.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivessa A S, Schneiter R, Kohlwein S D. Yeast acetyl-CoA carboxylase is associated with the cytoplasmic surface of the endoplasmic reticulum. Eur J Cell Biol. 1997;74:399–406. [PubMed] [Google Scholar]
- 18.Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff A M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleinschmidt A K, Moss J, Lane M D. Acetyl coenzyme A carboxylase: filamentous nature of the animal enzymes. Science. 1969;166:1276–1278. doi: 10.1126/science.166.3910.1276. [DOI] [PubMed] [Google Scholar]
- 20.Klionsky D J, Cueva R, Yaver D S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky D J, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Cronan J E J. The genes encoding the two carboxyltransferase subunits of Escherichia coli acetyl-CoA carboxylase. J Biol Chem. 1992;267:16841–16847. [PubMed] [Google Scholar]
- 24.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 25.Lynen F. Yeast fatty acid synthase. Methods Enzymol. 1969;14:17–33. [Google Scholar]
- 26.Matsuhashi M. Acetyl-CoA carboxylase from yeast. Methods Enzymol. 1969;14:3–8. [Google Scholar]
- 27.Milligan G, Parenti M, Magee A I. The dynamic role of palmitoylation in signal transduction. Trends Biochem Sci. 1995;20:181–186. doi: 10.1016/s0968-0004(00)89004-0. [DOI] [PubMed] [Google Scholar]
- 28.Mishina M, Roggenkamp R, Schweizer E. Yeast mutants defective in acetyl-coenzyme A carboxylase and biotin: apocarboxylase ligase. Eur J Biochem. 1980;111:79–87. doi: 10.1111/j.1432-1033.1980.tb06077.x. [DOI] [PubMed] [Google Scholar]
- 29.Mitchelhill K I, Stapleton D, Gao G, House C, Michell B, Katsis F, Witters L A, Kemp B E. Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J Biol Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- 30.Mohamed A H, Chirala S S, Mody N H, Huang W-Y, Wakil S J. Primary structure of the multifunctional α subunit protein of yeast fatty acid synthase derived from FAS2 gene sequence. J Biol Chem. 1988;263:12315–12325. [PubMed] [Google Scholar]
- 31.Oh C S, Toke D A, Mandala S, Martin C E. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 32.Pan X Z, Goldfarb D S. YEB3/VAC8 encodes a myristylated armadillo protein of the Saccharomyces cerevisiae vacuolar membrane that functions in vacuole fusion and inheritance. J Cell Sci. 1998;111:2137–2147. doi: 10.1242/jcs.111.15.2137. [DOI] [PubMed] [Google Scholar]
- 33.Raymond C K, Howald-Stevenson I, Vater C A, Stevens T H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raymond C K, Roberts C J, Moore K E, Howald I, Stevens T H. Biogenesis of the vacuole in Saccharomyces cerevisiae. Int Rev Cytol. 1992;139:59–120. doi: 10.1016/s0074-7696(08)61410-2. [DOI] [PubMed] [Google Scholar]
- 35.Roggenkamp R, Numa S, Schweizer E. Fatty acid-requiring mutant of Saccharomyces cerevisiae defective in acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1980;77:1814–1817. doi: 10.1073/pnas.77.4.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose M D, Winston F, Hieter F. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 37.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 38.Schneiter R. Brave little yeast, please guide us to Thebes: sphingolipid function in S. cerevisiae. Bioessays. 1999;21:1004–1010. doi: 10.1002/(SICI)1521-1878(199912)22:1<1004::AID-BIES4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 39.Schneiter R, Guerra C E, Lampl M, Gogg G, Kohlwein S D, Klein H L. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Δ is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol Cell Biol. 1999;19:3415–3422. doi: 10.1128/mcb.19.5.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneiter R, Hitomi M, Ivessa A S, Fasch E-V, Kohlwein S D, Tartakoff A M. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneiter R, Kohlwein S D. Organelle structure, function, and inheritance in yeast: a role for fatty acid synthesis? Cell. 1997;88:431–434. doi: 10.1016/s0092-8674(00)81882-6. [DOI] [PubMed] [Google Scholar]
- 42.Schüller H-J, Förtsch B, Rautenstrauss B, Wolf D H, Schweizer E. Differential proteolytic sensitivity of yeast fatty acid synthase subunits α and β contributing to a balanced ratio of both fatty acid synthase components. Eur J Biochem. 1992;203:607–614. doi: 10.1111/j.1432-1033.1992.tb16590.x. [DOI] [PubMed] [Google Scholar]
- 43.Schweizer E. Genetics of fatty acid biosynthesis in yeast. New Compr Biochem. 1984;7:59–83. [Google Scholar]
- 44.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song J, Dohlman H G. Partial constitutive activation of pheromone receptors by a palmitoylation-site mutant of a G protein alpha subunit in yeast. Biochemistry. 1996;35:14806–14817. doi: 10.1021/bi961846b. [DOI] [PubMed] [Google Scholar]
- 46.Sumper M, Riepertinger C. Structural relationship of biotin-containing enzymes. Acetyl-CoA carboxylase and pyruvate carboxylase from yeast. Eur J Biochem. 1972;29:237–248. doi: 10.1111/j.1432-1033.1972.tb01980.x. [DOI] [PubMed] [Google Scholar]
- 47.Toke D A, Martin C E. Isolation and characterization of a gene affecting fatty acid elongation in Saccharomyces cerevisiae. J Biol Chem. 1996;271:18413–18422. doi: 10.1074/jbc.271.31.18413. [DOI] [PubMed] [Google Scholar]
- 48.Vahlensieck H F, Pridzun L, Reichenbach H, Hinnen A. Identification of the yeast ACC1 gene product (acetyl-CoA carboxylase) as the target of the polyketide fungicide soraphen A. Curr Genet. 1994;25:95–100. doi: 10.1007/BF00309532. [DOI] [PubMed] [Google Scholar]
- 49.Vida T A, Emr S D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 51.Wakil S J. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry. 1989;28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y-X, Catlett N L, Weisman L S. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y-X, Zhao H, Harding T M, Gomes de Mesquita D S, Woldringh C L, Klionsky D J, Munn A L, Weisman L S. Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol Biol Cell. 1996;7:1375–1389. doi: 10.1091/mbc.7.9.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weisman L S, Bacallao R, Wickner W. Multiple methods of visualizing the yeast vacuole permit evaluation of its morphology and inheritance during the cell cycle. J Cell Biol. 1987;105:1539–1547. doi: 10.1083/jcb.105.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weisman L S, Wickner W. Intervacuole exchange in the yeast zygote: a new pathway in organelle communication. Science. 1988;241:589–591. doi: 10.1126/science.3041591. [DOI] [PubMed] [Google Scholar]
- 56.Witters L A, Watts T D. Yeast acetyl-CoA carboxylase: in vitro phosphorylation by mammalian and yeast protein kinases. Biochem Biophys Res Commun. 1990;169:369–376. doi: 10.1016/0006-291x(90)90341-j. [DOI] [PubMed] [Google Scholar]
- 57.Xu Z, Wickner W. Thioredoxin is required for vacuole inheritance in Saccharomyces cerevisiae. J Cell Biol. 1996;132:787–94. doi: 10.1083/jcb.132.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zabin I, Villarejo M R. Protein complementation. Annu Rev Biochem. 1975;44:295–313. doi: 10.1146/annurev.bi.44.070175.001455. [DOI] [PubMed] [Google Scholar]