Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal types of cancer with an overall 5-year survival rate of less than 10%. The 1-year survival rate of patients with locally advanced or metastatic disease is abysmal. The aggressive nature of cancer cells, hypovascularization, extensive desmoplastic stroma, and immunosuppressive tumor microenvironment (TME) endows PDAC tumors with multiple mechanisms of drug resistance. With no obvious genetic mutation(s) driving tumor progression or metastatic transition, the challenges for understanding the biological mechanism(s) of these processes are paramount. A better understanding of the molecular and cellular mechanisms of these processes could lead to new diagnostic tools for patient management and new targets for therapeutic intervention. microRNAs (miRNAs) are an evolutionarily conserved gene class of short non-coding regulatory RNAs. miRNAs are an extensive regulatory layer that controls gene expression at the posttranscriptional level. This review focuses on preclinical models that functionally dissect miRNA activity in tumor progression or metastatic processes in PDAC. Collectively, these studies suggest an influence of miRNAs and RNA-RNA networks in the processes of epithelial to mesenchymal cell transition and cancer cell stemness. At a cell-type level, some miRNAs mainly influence cancer cell–intrinsic processes and pathways, whereas other miRNAs predominantly act in distinct cellular compartments of the TME to regulate fibroblast and immune cell functions and/or influence other cell types’ function via cell-to-cell communications by transfer of extracellular vesicles. At a molecular level, the influence of miRNA-mediated regulation often converges in core signaling pathways, including TGF-β, JAK/STAT, PI3K/AKT, and NF-κB.
Keywords: microRNA (miR, miRNA), Long non-coding RNAs (lncRNA), Circular non-coding RNA (circRNA), Extracellular vesicles
Introduction
Pancreatic ductal adenocarcinoma is one of the most lethal types of cancer with an overall 5-year survival rate of less than 10% [1]. Virtually all long-term survivors are patients with early stage disease for whom surgical resection and adjuvant systemic therapy provides a potential cure [2, 3]. Due to late clinical presentation, about 30% of patients are diagnosed with locally advanced disease and about 50% of patients with metastatic disease [2]. The 1-year survival rate of these advanced stage cases is abysmal [2, 3]. Modifications in systemic chemotherapy regimens such as FOLFIRINOX (5-fluorouracil, folinic acid, irinotecan, and oxaliplatin) or gemcitabine-containing combination treatments have resulted in a modest improvement of patient outcome in recent years [2]. Targeted therapies and immunotherapies, which have improved outcome in other cancer types, have failed to provide a clear clinical signal in PDAC [2, 4]. The aggressive nature of cancer cells, hypovascularization, extensive desmoplastic stroma, and immunosuppressive tumor microenvironment (TME) endows PDAC tumors with multiple mechanisms of drug resistance [3, 5, 6]. Combination strategies that target cancer cells and elements of the TME may offer new opportunities and hope in PDAC. Myeloid cell-modifying agonist CD40 antibody, stroma-modifying angiotensin-receptor blocker Losartan, CXCR4 antagonist in combination with PD-1 blockade, and connective tissue growth factor inhibitors are showing promising results in on-going clinical trials [2, 7, 8]. In addition, recent studies continue to investigate the clinical value of circulating tumor DNA and other molecular biomarkers for early disease detection [9, 10].
PDAC has a limited number of driver genes, with frequent and predominant mutations in KRAS (> 90%), CDKN2A (~ 90%), TP53 (~ 70%), and SMAD4 (~ 55%) [3, 11]. With no obvious genetic mutation(s) driving tumor progression or metastatic transition, the challenges for understanding the biological mechanism(s) of these processes are paramount. These require a comprehensive and integrative analysis of dynamic changes in gene dosage, transcriptome, epigenome, core signaling, and metabolomic pathways that collectively contribute to tumor evolution and metastatic spread [11–15]. A better understanding of the molecular and cellular mechanisms of these processes could lead to new diagnostic tools for patient management and new targets for therapeutic intervention. microRNAs (miRNAs) are an evolutionarily conserved gene class of short non-coding regulatory RNAs [16–18]. miRNAs are an extensive regulatory layer that controls gene expression at the posttranscriptional level. miRNA expression and function has been linked to different aspects of PDAC biology and disease progression [18–23]. This review focuses on preclinical models that functionally dissect miRNA activity in tumor progression or metastatic processes. Collectively, these studies suggest an influence of miRNAs and RNA-RNA networks in the processes of epithelial to mesenchymal cell transition (EMT), proliferation, and cancer cell stemness. At a cell-type level, some miRNAs mainly influence the processes and pathways in a cancer cell–intrinsic manner, whereas other miRNAs predominantly act in distinct cellular compartments of the TME to regulate fibroblast and immune cell functions and/or to dictate other cell types’ functions via cell-to-cell communication by transfer of extracellular vesicles (EVs). At a molecular level, the influence of miRNA-mediated regulation often converges in core signaling pathways, including TGF-β, JAK/STAT, PI3K/AKT, and NF-κB. In the following sections, we discuss salient examples of these mechanisms of action and crosstalk between cancer cells and other cell types in the TME. We focus our discussion on recent studies in which there is an in vivo demonstration of miRNA activity in tumor progression and/or metastatic processes. This in vivo demonstration may involve the use of genetically engineered mouse models, organoid or cell line xenograft models, and/or patient-derived tumor models.
microRNA biogenesis and dysregulation during PDAC progression
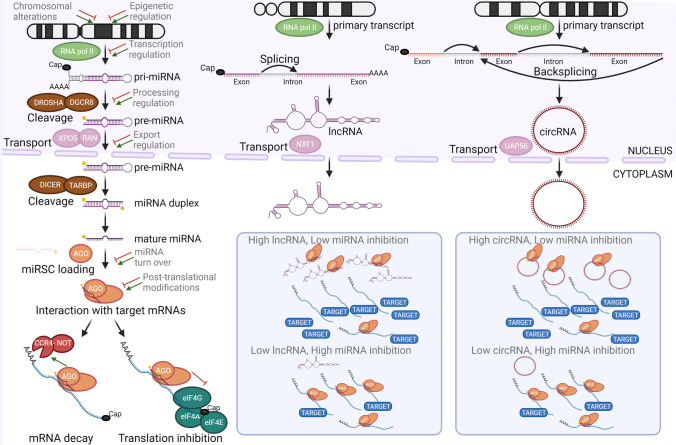
The mature and biological active miRNA consists of a 19–24 nucleotide-long RNA molecule resulting from a stepwise processing of a much longer mRNA-like primary transcript (Fig. 1). The immense majority of the mRNA-like primary transcripts are transcribed by RNA polymerase II [16, 17]. Some miRNAs are arranged in tightly linked gene clusters, and this primary transcript (pri-miRNA) may contain the precursor hairpin RNA (pre-miRNA) for several miRNAs [24]. Transcription of either a single miRNA or a miRNA gene cluster may initiate from a proximal promoter to the miRNA gene or distal promoter driving expression of the host gene [16, 17]. In the nucleus, RNAse III DROSHA-containing RNA microprocessor complex cleaves precursor hairpin RNA that is exported to the cytoplasm by XPO5-facililated pathway [17]. In the cytoplasm, another RNAse III DICER cleaves the final mature RNA molecule that becomes functionally active when loaded into the ARGONAUTE-containing miRNA-induced silencing complex (miRISC). The miRNA serves as a guide to direct miRISC in close proximity to target mRNA by binding to partially complementary sites, typically on the 3′ UTR. The interaction of miRISC with other mRNA-bound protein complexes (e.g., CCR4-NOT, eIF4A-G) leads to mRNA decay, mRNA cleavage, and/or inhibition of translation initiation that ultimately results in a decreased protein output of the target gene(s) [16, 17]. In most cases, miRNA binding to the mRNA is driven by the seed region (2nd to 8th nucleotides of the miRNA) which allows for interactions of a single miRNA with tens or hundreds of mRNAs. The relative abundance of miRNA to target mRNA and number of binding sites and binding affinity determines the extent of protein output reduction for each target gene [18]. A single miRNA can influence cellular programs by profound downregulation of a handful of key targets and/or by more modest but coordinated downregulation of a larger number of target genes. About 2,000 bona fide miRNA genes have been identified in the human genome [24]; it is estimated that all miRNAs expressed in a particular cell can modulate the expression of up to 60% of the protein-encoding genes and correspondingly influence a large number of cellular programs [16, 17, 25].
Fig. 1.
Biogenesis of miRNAs and other non-coding RNAs. Key steps of biogenesis of miRNAs and miRNA-interacting lncRNAs and circRNAs. Several mechanisms are known that affect regulation of miRNA expression and/or activity. Nuclear export is key step for cytoplasmic interaction of miRNAs and these other classes of ncRNAs that sequester miRNAs away from target mRNAs and indirectly increase protein product of the target gene. Abbreviations: AGO, argonaute RISC component; circRNA, circular non-coding RNA; CCR4-NOT, carbon catabolite repression-negative on TATA-less complex; DICER, ribonuclease III Dicer1; DGRCR8, DiGeorge syndrome critical region gene 8, microprocessor complex subunit; DROSHA, ribonuclease III Dicer1; eiF4, eukaryotic translation initiation factor 4; lncRNA, long non-coding RNA; miRISC, miRNA-induced silencing complex; NXF1, nuclear RNA export factor 1; RAN, member of RAS oncogene family, small nuclear GTPase; RNA pol II, RNA polymerase II; miRNA, microRNA; TARBP, TAR (HIV-1) RNA-binding protein 1; UAP56, U2AF65-associated protein 56 also known as DDX39B (DExD-box helicase 39B); XPO5, exportin 5
Dysregulation of miRNA expression and activity in PDAC can occur at different levels from chromosomal alteration (e.g., gain or loss of copy numbers), epigenetic and transcriptional regulation of the pri-miRNA, processing of pri-miRNA, export and processing of pre-miRNA, and/or physical interaction with other non-coding RNAs (Fig. 1). Dysregulation of miRNA expression has been extensively characterized between normal pancreas and PDAC tumor specimens, chronic pancreatitis and PDAC tumor specimens, and PDAC precursor lesions and PDAC tumor specimens at different clinical stages. These tissue correlative studies have led to the identification of specific miRNAs or miRNA signatures that could serve as diagnostic and/or prognostic biomarkers [18, 20, 21]. While there is no complete overlap among different studies, altered expression of miRNAs (e.g., let-7a, miR-10b, miR-21, miR-217) has been frequently associated with disease progression and/or metastatic disease. Guided by these clinical observations, many of these miRNAs have been subjected to a battery of in vitro and in vivo functional assays to dissect the etiological contribution of each of these miRNAs to a specific metastatic process [22, 23]. In most of these studies, in vivo evidence was obtained in different xenograft models in which immunocompromised mouse host is inoculated with human PDAC cells. In xenograft models in which the PDAC cells are inoculated subcutaneously (animal’s flank) or orthotopically (pancreas), the effects of miRNA activity modulation can be interrogated at several stages of disease progression from growth of the primary tumor to increase migration and invasive behavior to intravasation and extravasation and colonization and growth at distant sites. In xenograft models in which the PDAC cells are inoculated intrasplenically (spleen) or intravenously (tail vein), the effects of miRNA activity modulation can be interrogated at more advance stages of disease progression from extravasation to colonization and growth at distant sites. This in vivo evidence suggests that some miRNAs (e.g., miR-200b, miR-323-3p, miR-367, miR-489) appears to exclusively or predominantly influence a specific step of the metastatic process, whereas other miRNAs (e.g., let-7a, miR-10b, miR-21, miR-29b, miR-34a) may influence multiple cellular processes (proliferation, survival, and/or drug resistance) that collectively contribute to tumor progression and metastatic spread.
miRNA-mediated regulation of epithelial to mesenchymal transition
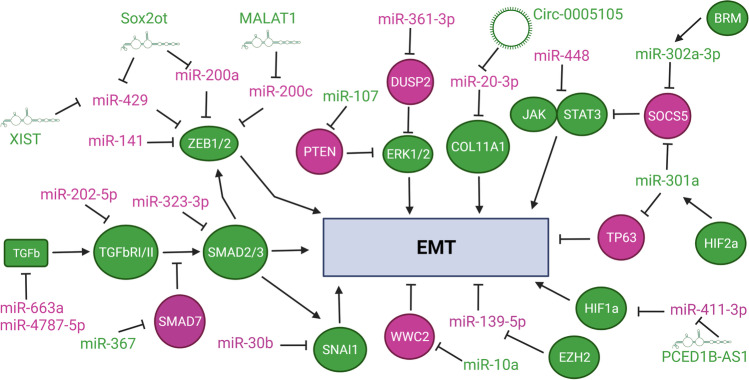
Epithelial to mesenchymal transition (EMT) is a process associated with progression and metastatic spread, by which cancer cells acquire increased motility and invasive behavior [26]. TGF-β and HIF signaling are known inducers and regulators of EMT in PDAC [26]. Ultimately, the integration of these and other input signals leads to upregulation of ZEB1/2, TWIST1, and/or SNAI1/2: transcription factors that promote mesenchymal program and downregulation of epithelial cell adhesion protein, E-cadherin. Several miRNAs have been identified in PDAC with a major or predominant role in regulating EMT (Fig. 2; refs [27–46]). miR-202-5p, -323-3p, -663a, and -4787-5p put a break on the EMT program by inhibiting expression of key components of TGF-β signaling pathway [28–30], whereas miR-367 promotes EMT by inhibiting expression of SMAD7, negative regulator of TGF-β signaling [31]. Similarly, miR-301a promotes EMT by relaying hypoxic HIF2α signal to increase JAK1/STAT3 signaling via inhibition of direct target SOCS5 [32] and to decrease transcription program of direct target TP63 [33]. miR-302a relays pro-metastatic signal of BRM, a catalytic ATPse subunit of the SWI/SNF chromatin remodeling complex, to increase JAK2/STAT3 signaling also via inhibition of direct target SOCS5 [39]. In contrast, miR-448 activity suppresses metastatic spread by downregulating expression of direct target gene JAK1 and dampening pro-EMT JAK/STAT3 signaling [40]. miR-10a decreases Hippo signaling and its negative regulation of EMT and cancer cell stemness via inhibition of direct target WWC2 [46]. Increased activity of WWC2 leads to phosphorylation and activation of LATS1/2, which in turn prevents nuclear translocation and activity YAP/TAZ transcription complex [46]. The miR-200 family members are arranged in two gene clusters (MIR200b ~ MIR200a ~ MIR429 at chromosome 1 and MIR200c ~ MIR141 at chromosome 12) in humans. miR-200 family members are considered safeguards of epithelial cell program and identity by inhibiting expression of direct target genes ZEB1 and ZEB2 [34–36]. Unlike breast and other cancer types, individual activity of these miRNAs leads to different regulation of EMT and tumor suppressive properties in orthotopic Panc-1 xenograft tumor model [34]. Enforced expression of miR-141 or miR-429 results in a significant inhibition of tumor growth in this orthotopic model, whereas that of other miR-200 family members or their combination with miR-141 and miR-429 does not [34]. miR-141 tumor suppressive and anti-metastatic function is also mediated by direct inhibition of MAP4K4 [37] and WIPF1 [38], respectively. There is a differential association between expression changes of individual miR-200 family members and that of ZEB1, ZEB2, and E-cadherin in PDAC clinical samples and patient-derived xenograft tumors [34]. In a cohort of 31 PDAC primary tumors, there is no consistent trend of downregulation for all miR-200 family members, but there is a positive correlation between expression of all miR-200 family and that of E-cadherin [34]. Negative association is only statistically significant for expression of miR-200a and miR-141 in relation to that of ZEB1 and ZEB2 [34]. These results add a level of complexity to the regulation of EMT and perhaps other metastatic processes by each of these related miRNAs and suggest the importance of context, timing, and differential inhibition of key target genes (other than ZEB1/ZEB2).
Fig. 2.
miRNA-mediated regulatory networks influencing EMT program. miRNAs and protein-encoding genes (ovals) depicted in violet maintain or favor epithelial program (anti-EMT, tumor suppressive), whereas miRNAs, lncRNAs, circRNAs, and protein-encoding genes in green favor mesenchymal program (pro-EMT, tumor promoting). In other contexts, these miRNAs may influence other cellular programs and decisions. Abbreviations: BRM, Brahma; COL11A1 collagen 11A1 ; DUSP2, dual specificity phosphatase 2, ERK1/2, extracellular signal-regulated kinases; EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; HIF1a/2a, hypoxia inducible factor 1/2 subunit alpha; JAK, Janus kinase; PTEN, phosphatase and tensin homolog; SMAD2/3/7, similar to gene product of C. elegans Sma (Small) and Drosophila Mad (mothers against decapentaplegic) 2/3/7; SNAI1, snail family transcriptional repressor 1; SOCS5, suppressor of cytokine signaling 5; STAT3, signal transducer and activator of transcription 3; TGFb, tumor growth factor β; TGFbRI/II, TGF-β receptor I/II; TP63, tumor protein 63; ZEB1/2, zinc finger E-box binding homeobox 1/2
miRNA-mediated regulation of extracellular matrix remodeling, invasion, and metastatic spread
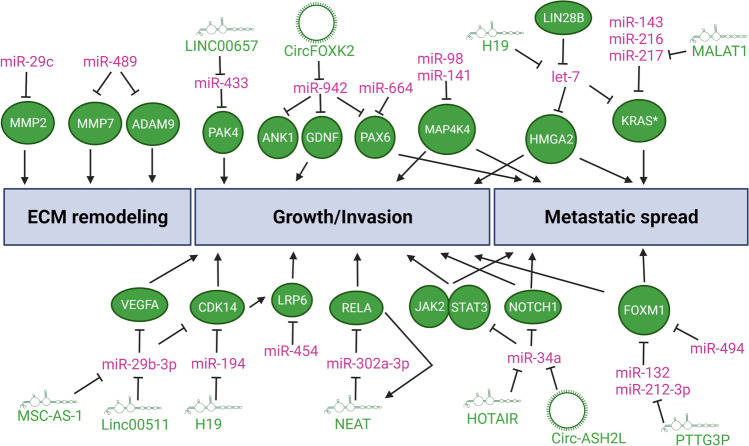
The extracellular matrix (ECM) is composed proteoglycans, proteins, and matricellular associated proteins. ECM remodeling promotes growth, survival, and invasion of PDAC cells at the primary tumor and facilitates colonization at premetastatic niches in the liver and other distant organs [47]. Some miRNAs have a direct role in limiting the tumorigenic remodeling of the ECM (Fig. 3, refs [48, 49]) by modulating the expression of matrix metalloproteinases (MMPs) and disintegrin and metalloproteinases (ADAMs). miR-29c activity inhibits the expression of direct target gene MMP2 [48]. Sequential selection of Hs766t cells with preferential metastasis to the liver shows the requirement of miR-29c to limit metastatic spread [48]. It is not clear if miR-29c is more crucial at escaping the primary tumor site and/or reducing colonization and growth at distant metastatic site. miR-489 activity inhibits the expression of direct target genes MMP7 and ADAM9 and is required for limiting colonization and growth at distant metastatic sites [49]. Other miRNAs influence cellular processes of growth, invasion, and/or metastatic spread that indirectly affect cancer cell-mediated remodeling of the ECM, interaction with ECM elements, and/or mechanosensing of stroma stiffness (Fig. 3, refs [27, 50–65]). miR-664 and miR-942 modulate cancer cell growth and invasion by direct inhibition of common target gene transcription factor PAX6 [53, 54]. Similarly, miR-132, miR-212-3p, and miR-494 modulate invasion and metastatic spread by direct inhibition of common target gene transcription factor FOXM1 [55, 56]. SMAD4 positively regulates expression of miR-494 via canonical TGF-β signaling pathway [56]. Genetic loss of SMAD4 downregulates miR-494 expression and leads to upregulation of FOXM1 along with potentiation of WNT signaling by FOXM1-mediated nuclear translocation of β-catenin [56]. This illustrates the complexity and duality of TFG-β signaling in suppressing tumor growth in early stages of carcinogenesis while promoting tumor growth and metastasis via EMT and immunosuppression (most of the other cases in this review) in later stages. Gene-dosage increases of mutant KRAS alleles drive early stages of carcinogenesis and later metastatic spread [14]. Several miRNAs, including let-7a, miR-143, miR-216, and miR-217, directly bind to the 3′UTR of both wild-type and mutant KRAS mRNAs [27, 57–65]. Overcoming this miRNA-mediated regulation of KRAS expression may also contribute to the mutant KRAS dosage-dependent switch to tumor progression and metastatic spread. let-7a, a founding member of the miRNA gene family, is considered a potent tumor suppressor gene. Other targets of let-7 include potent oncogenic factors such as c-MYC and HMGA2 [27, 64, 65]. By coordinately regulating the expression of KRAS, HMGA2, and c-MYC, let-7 activity restrains cell proliferation, cell cycle progression, invasion, EMT, and metastasis. LIN28B is an oncofetal RNA-binding protein that interferes with the maturation process of let-7 precursor RNA and in so doing increases oncogenic expression of these let-7 key target genes in PDAC [64, 65]. LIN28B is highly expressed in circulating tumor cells of PDAC patients and is a driver of metastatic dissemination [64]. CRISPR-mediated knockout of LIN28B gene, chemical inhibition of LIN28B binding to let-7, or knockdown of HMGA2 significantly diminish the metastatic potential of circulating tumor cells [64]. This indicates the crucial role of let-7 in suppressing LIN28B-depedent metastatic program in PDAC.
Fig. 3.
miRNA-mediated regulatory networks influencing metastatic programs. miRNAs depicted in violet act as a tumor suppressive and/or anti-metastatic factor whereas lncRNAs, circRNAs, and protein-encoding genes (ovals) in green as tumor promoting and/or pro-metastatic factors. In other contexts, these miRNAs may influence other cellular programs and decisions. Abbreviations: ADAM9, A disintegrin and metalloproteinase domain 9; ANK1, ankyrin 1; CDK14, cyclin-dependent kinase 14; FOXM1, forkhead box M1; GDNF, glial cell derived neurotrophic factor; HMGA2, high mobility group AT-Hook 2; JAK2, Janus kinase 2; KRAS*, mutant KRAS Proto-Oncogene, GTPase; LIN28B, Lin-28 homolog B; LRP6, LDL receptor related protein 6; MAP4K4, mitogen-activated protein kinase kinase kinase kinase 4; MMP2/9, matrix metallopeptidase 2/9; NOTCH1, notch receptor 1; PAK4, P21 (RAC1) activated kinase 4; PAX6, paired box 6; RELA, RELA Proto-Oncogene, NF-κB Subunit; STAT3, signal transducer and activator of transcription 3; VEGFA, vascular endothelial growth factor A
Interactions between miRNAs and other non-coding RNAs dictate metastatic programs
An emerging layer of regulation in many miRNA/key target pathways in PDAC is that of non-coding RNA dampening miRNA activity. While miRNAs are a well-defined class of short ~ 22 nts RNAs, longer non-coding RNAs are more diverse and complex classes of RNAs (Fig. 1). Long non-coding RNAs (lncRNAs) are a heterogenous class that broadly includes a variety of non-coding RNAs longer than 200 nts [66]. lncRNAs can act as local regulators of transcription by interacting with epigenetic machinery, squelching, or blocking access of transcription factors; provide scaffolding structure for RNA-RNA and RNA–protein interactions; and bind and sequester miRNA molecules [66–68]. Circular non-coding RNAs (circRNAs) are covalently closed loops resulting from backsplicing of mRNAs [67, 69]. The main role of circRNAs is to bind and act as sponges of miRNAs [67, 69]. We describe below a few examples of these miRNA-ncRNA interactions in regulatory pathways pertinent to tumor progression and metastatic processes already highlighted in previous sections (Figs. 2 and 3, refs [27, 35, 36, 54, 70–78]). lncRNA MALAT1 binds to miR-200c and sequesters it away from binding sites on the 3′UTR of ZEB1/ZEB2 mRNAs and other target genes, thereby skewing cellular program towards EMT [36]. Similarly, lncRNA Sox2ot and XIST promote EMT by sequestering other miR-200 family members (miR-200a and miR-429) [35, 72]. This differential interaction and modulation of specific miR-200 family members by these lncRNA may explain, in part, discordant results of in vivo functional analysis of each family member [34]. lncRNA PCED1B-AS1 potentiates HIF-mediated EMT by sequestering miR-411-3p which directly inhibits expression of HIF1α [73]. lncRNA MSC-AS-1 sequesters miR-29b, a family member of EMT-regulating miR-29c, which leads to upregulation of cyclin-dependent kinase CDK14 [74]. lncRNA H19 sequesters miR-194, which also leads to upregulation of CDK14 [75]. CDK14 activates WNT signaling by phosphorylating the LRP5/6 receptors during G2/M phase of the cell cycle. Conversely, miR-454 inhibits expression of LRP6 and dampens WNT signaling [50]. This suggests a converging network of lncRNA-miRNA interactions that modulate cell cycle-dependent WNT signaling. H19 also sequesters let-7a and interferes with its ability to inhibit HMGA2-mediated EMT program [27].
miR-34a is a potent tumor suppressive miRNA that can concomitantly inhibit expression of several proto-oncogenic genes (e.g., BCL2 and c-MET) and signaling pathways (e.g., NOTCH, WNT) [79]. lncRNA HOTAIR sequesters miR-34a and leads to activation of JAK2/STAT3 signaling pathway [77]. By interfering with miR-34a activity, HOTAIR promotes cancer cell stemness, EMT, and metastatic spread of PDAC cells [77]. circRNA Circ-ASH2L sequesters miR-34a what leads to upregulation of miR-34a–direct target gene NOTCH1 [76]. Circ-ASH2L-mediated activation of NOTCH signaling pathway promotes angiogenesis, tumor growth, and tumor invasion [76]. Another circRNA, CircFOXK2, also promotes tumor growth and metastasis by sequestering miR-942 and upregulating expression of its direct target genes ANK1, GDNF, and PAX6 [54]. CircFOXK2 may also influence these processes in a miR-942-independent manner by interacting with proteins involved in mRNA splicing, YBX1 and hnRNPK, which leads to upregulation of NUF2 and PDXK oncogenic expression [54]. There are additional RNA-RNA interactions of metastasis-modulating miRNAs and other ncRNA classes (e.g., lncRNA TUG1-miR-29c, lncRNA DUXAP8-miR-488) in PDAC [80–82]. Although these interactions have been reported in the context of other regulatory pathways and/or cellular processes, they may also contribute to metastatic programs.
microRNA-mediated cellular crosstalk influences metastatic processes
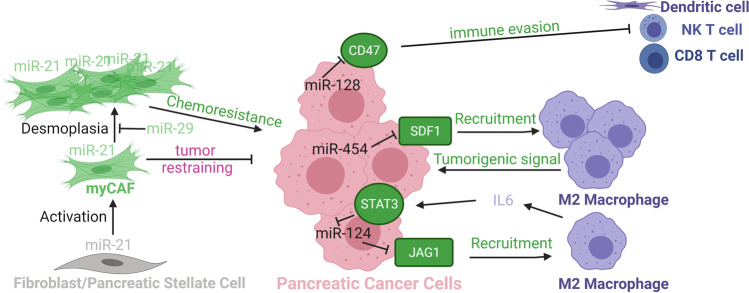
It is well established that cancer cell-stroma interactions in PDAC have an influence on chemoresistance, disease progression, and metastatic spread. Several miRNAs have been implicated in this crosstalk between cancer cells and other cell types in the TME (Fig. 4; refs [83–88]). In some instances, miRNA-mediated regulation in cancer cells leads to a receptor/ligand-mediated response in another cell type of the TME that may then lead to a paracrine signal back to the cancer cells or other cell types in the TME. In other instances, miRNA-mediated regulation in cancer-associated fibroblasts (CAFs) or other cell types in the TME leads to physical interaction and/or paracrine signal that affects cancer cell growth and treatment resistance. miR-124 is engaged in a regulatory feedback loop with NOTCH signaling pathway in cancer cells and crosstalk with tumor-associated macrophages (TAMs). miR-124 inhibits metastatic program by directly targeting JAG1 in cancer cells dampening NOTCH signaling [83]. Increased NOTCH signaling recruits and polarizes TAMs to a tumorigenic M2 phenotype [83]. IL-6 secreted by these M2 TAMs upregulates STAT3 signaling in cancer cells, which downregulates miR-124 expression and promotes EMT program [83]. Similarly, cancer cell–expressed miR-454 inhibits the expression of stromal cell derived factor 1 (SDF1, also known as chemokine CXCL12) [84]. Increased SDF1 levels recruit TAMs via activation of their chemokine receptor CXCR4, creating a pro-tumorigenic TME [84]. In contrast, cancer cell–expressed miR-128 inhibits metastatic program by directly targeting ZEB1, a positive regulator of EMT and CD47-mediated tumor immune evasion [85]. Enforced expression of miR-128 in orthotopic syngeneic Panc02 cell model dampens CD47 expression and thereby increases anti-tumor immunity mediated by dendritic cells, CD8 + , and Natural Killer T cells [85].
Fig. 4.
miRNA-mediated cellular crosstalk influencing metastatic programs. The color of the miRNA indicates the identity of the expressing cell type. Proteins (ovals and round-edge rectangles for ligands) and processes in green depict a pro-metastatic role and process in violet an anti-metastatic role. In other contexts, these miRNAs may influence other cellular programs and decisions. Abbreviations: CD8, T-cell surface glycoprotein CD8 alpha chain; CD47, leukocyte surface antigen CD47; IL6, interleukin 6; JAG1, jagged canonical Notch ligand 1; myCAF, myofibroblastic cancer-associated fibroblast; NK T cell, Natural Killer T cell; SDF1, stroma derived factor 1; STAT3, signal transducer and activator of transcription 3
miR-21 is generally considered an oncogenic miRNA and important mediator of TGF-β-induced EMT via inhibition of PTEN, PDCD4, and/or RECK expression [89]. However, miR-21 also responds to these inputs and regulates these and other direct targets in other cellular elements of the TME, including CAFs and TAMs [18, 90]. New 3D co-culture techniques, single-cell RNAseq, and mechanistic studies in GEMMs have uncovered the existence of distinct CAF subtypes in PDAC [91, 92]. Low or high expression of α-smooth muscle actin (SMA) is a consistent differential feature of CAF subtypes; “myofibroblastic” CAFs (myCAFs) express high levels of SMA, are juxtaposed or in close proximity to cancer cells, and exhibit tumor-restraining function [93–99]. Co-detection of miR-21 and SMA expression demonstrates a predominant upregulation of miR-21 expression in myCAFs in the majority of PDAC tumors from patients and GEMMs [86, 87, 100–107]. myCAF expression of miR-21 provides more robust prognostic information than epithelial expression for predicting treatment response and overall survival of PDAC patients [87, 104, 105]. In vitro and in vivo co-culture assays of cancer cells and fibroblasts/CAFs suggest a CAF-driven role of miR-21 [87, 104, 108]. Inhibition of miR-21 activity in CAFs before co-inoculation with cancer cells enhances efficacy of gemcitabine in a subcutaneous syngeneic Pan02 cell model [87]. Strikingly, global loss of miR-21 activity accelerates tumor development and results in a much shorter overall survival in a mutant K-Ras p53-deleted PDAC GEMM (KPC model) [86]. The absence of myCAFs in precursor and invasive lesions is the most outstanding consequence of miR-21 loss. Cancer cell–specific activity of miR-21 is not required for in vitro or in vivo growth nor affects in vitro chemoresistance to gemcitabine in this KPC model [86]. This study uncovers a pro-fibrotic yet tumor-restraining activity of miR-21 suggestive of a cell-intrinsic myCAF role. Timing, duration, and extent of miR-21 inhibition, mutational landscape, and/or tumor subtype may help explain the discrepancies between studies (Fig. 4). Interestingly, expression of miR-29a and miR-29b is downregulated in pancreatic stellate cells/resting fibroblasts during TGF-β-induced pancreatic stellate cell/CAF activation [88]. miR-29 activity appears to limit ECM protein accumulation and desmoplastic reaction via direct inhibition of collagen (COL1A1, COL3A1) and laminin (LAMC1) mRNAs [88]. Understanding how these miR-21–regulated and miR-29–regulated processes interfere with each other to affect myCAF function and composition of other CAF subtypes under chemotherapy treatment is a key question to answer in order to achieve more effective and durable clinical responses.
microRNA-mediated pro-metastatic cell-to-cell communication via extracellular vesicles
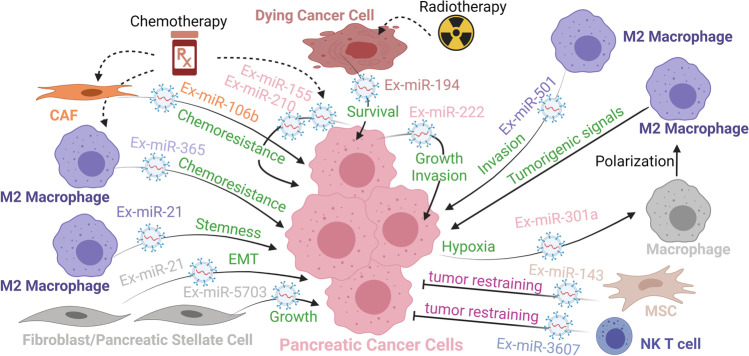
miRNAs themselves can be the regulatory signal from one cell type to another via transfer of extracellular vesicles (EVs). There are a few classes of membrane-bound EVs depending on their origin and size that mediate in cell-to-cell communications: microparticles, shed microvesicles, or ectosomes are directly formed by budding of the cell plasma membrane and range in size 10 nm to 3 μm; exosomes are secreted via multivesicular-body endocytic process and are about 100 nm in size (40–150 nm range). Because there are technical and methodological challenges in the purification of exosomes from a mixed population of other classes of secreted vesicles [23, 109–111], we will use the more general term of EV in accordance with the International Society of Extracellular Vesicles [112] even though the original research study may have used the term exosome. We will refer to the miRNA loaded and transferred in EVs from one cell to another as Ex-miRNA. In some instance, the Ex-miRNA can act as a hormone-like signal and interact with RNA sensing receptors such as Toll-like receptor 8 [113], but generally the transferred Ex-miRNA is loaded to miRISC and controls gene expression at the posttranscriptional level in the recipient cells [23, 109, 111]. Ex-miRNAs can be secreted from cancer cells in response to treatment, hypoxia, or other inputs in the TME to affect cellular processes at a distance; recipient cells may be other cancer cells or other cell types in the TME (Fig. 5, refs [114–117]). Similarly, ex-miRNAs can be secreted from CAFs, TAMs, and other cell types in the TME to affect a variety of cellular processes in the recipient cancer cells (Fig. 5, [61, 118–125]). Ex-miR-222 secreted from more aggressive and metastasis-prone cancer cells can stimulate proliferative and invasion program in neighboring cancer cells by downregulating the expression and activity of cyclin-dependent kinase inhibitor p27 [114]. miR-222 directly binds and inhibits p27 mRNA and indirectly increases AKT-mediated phosphorylation and cytoplasmic translocation of p27 via inhibition of serine/threonine-protein phosphate PPP2R2A expression [114]. Radiotherapy induces the secretion of Ex-miR-194-5p from dying cancer cells [115]. Ex-miR-194-5p potentiates the survival of neighbor cancer cells, especially those with stem cell–like and tumor repopulating properties, by inducing a temporary G1/S arrest and upregulation of DNA damage response via direct inhibition of E2F3 and HMGA2 [115]. Latent release of prostaglandin E2 from dying cells stimulates proliferation of Ex-miR-194-5p–protected cancer cells. Combining radiotherapy with low-dose aspirin enhances treatment efficacy by reducing both the amount of Ex-miR-194-5 pm and prostaglandin E2 released from dying cells [115]. Similarly, chemotherapy treatment with gemcitabine induces the secretion of Ex-miR-155 and Ex-miR-210 from chemosensitive cancer cells, which leads to increase chemoresistance in recipient cancer cells [116, 117]. Higher levels of miR-210 in recipient cells activate mTOR pathway, though the direct key target genes of miR-210 leading to this activation have not been identified. Gemcitabine also induces the secretion of Ex-miR-106b from CAFs. In recipient cancer cells, miR-106b enhances chemoresistance via direct inhibition of TP3INP1 expression [119], a stress-induced p53-target gene with anti-proliferative and pro-apoptotic activity. M2-polarized TAMs secrete Ex-miR365 that contributes to gemcitabine chemoresistance in cancer cells by augmenting the gemcitabine-competing triphospho-nucleotide pool and upregulating gemcitabine-inactivating cytidine deaminase [120]. Direct target genes that miR-365 regulates in the process have not been identified.
Fig. 5.
microRNA-mediated cell-to-cell communication via extracellular vesicles. The color of the Ex-miRNA indicates the identity of the cell type secreting the EV-bound miRNA. Processes in violet depict an anti-metastatic role whereas those in green a pro-metastatic role. In other contexts, these Ex-miRNAs may influence other cellular programs and decisions. Abbreviations: CAF, cancer-associated fibroblast; MSC, mesenchymal stem cell; NK T cell, Natural Killer T cell
Hypoxic TME induces HIF-dependent release of Ex-miR-301a from cancer cells to TAMs [121]. In TAMs, miR-301a upregulates PI3Kγ signaling pathway via direct inhibition of PTEN expression, which favors polarization to tumorigenic M2 phenotype [121]. These M2 TAMs secrete TGF-β, IL-10, and arginase that promote cancer cell metastasis and potentiate miR-301a-mediated EMT program as described above (Fig. 2). While it is not clear if in direct response to Ex-miR-301a transfer, M2 TAMs can secrete Ex-miR-21 and Ex-miR-501 that are taken up by cancer cells. In cancer cells, miR-501-3p downregulates expression of TGFβR3, whose shedded ectodomain curtails TGF-β signaling, to favor migration and invasive behavior [122]. In cancer cells, miR-21 downregulates the expression of transcription factor KLF3 to promote NANOG/OCT4-dependent cancer cell stemness program [123]. Pancreatic stellate cells secrete Ex-miR-21 which in recipient cancer cells activates PI3K/AKT pathway and promotes EMT program and MMP2/9-mediated ECM remodeling [124]. Pancreatic stellate cells also secrete Ex-miR-5703, which in recipient cancer cells activates PI3/AKT pathway and promotes cell proliferation via downregulation of CMTM4 [125]. While beyond the scope of this review, there is growing evidence of cell-to-cell communication via secreted lncRNAs and circRNAs that inhibit miRNA activity in recipient cells in PDAC tumors [35, 126]. This adds another exciting and complex regulatory layer to the field of RNA-RNA interactions and endogenous competing RNA hypothesis.
Pharmacological interventions to modulate miRNA activity
Several therapeutics strategies to modulate the activity of a specific miRNA with synthetic RNA analogs and different delivery technologies have been tested in preclinical models and some are being investigated in clinical trials [18, 127, 128]. An attractive feature of miRNAs as drug target is that activity modulation of a single miRNA could have a broad influence in several direct targets and processes downstream of these regulatory interactions. A potential advantage of a miRNA drug is that it could be more directed to a particular cell type and/or have a more restricted effect on selected cellular processes than small molecule inhibitors targeting upstream signaling pathways such TGF-β, JAK/STAT, or PI3K/AKT. There are several technical considerations for the chemical modifications and delivery approaches when replenishing the activity of tumor suppressive and/or anti-metastatic miRNA vs. inhibiting the activity of a tumor promoting and/or pro-metastatic miRNA [18, 127, 128]. miRNA mimics to replenish miRNA activity are typically double-stranded modified RNA analogs that need to be taken up by cells, released in an intact form in the cytoplasm for DICER processing, miRISC loading and binding to target mRNAs, to be functionally active. In contrast, anti-miRNA inhibitors to inhibit miRNA activity are typically single-stranded modified RNA analogs that need to be taken up by cells and then released in the cytoplasm to bind to perfectly complementary miRNA molecules. Thus, the design and delivery of a miRNA mimic is more challenging than that of an anti-miRNA inhibitor [18, 128]. miRNA mimics require encapsulation in liposomal nanoparticles or complexing with polymers or other coated nanoparticles [18, 128]. A double-stranded miR-217 mimic encapsulated in a PEGylated lipid nanoparticle, decorated with iRDG tumor penetrating–peptide, is effective at downregulating KRAS expression in vitro cell assays, but it may not be as effective as short interfering RNAs (siRNAs) against KRAS in an in vivo subcutaneous PDAC tumor model [129]. miR-34a is an excellent candidate for replenishing therapy in PDAC [130]. A miR-34a-expressing DNA vector encapsulated in liposomal nanoparticles (nanovector) is effective at reducing tumor growth in subcutaneous and orthotopic xenograft models, and more so than similar nanovector strategy with KRAS-targeting miR-143 and miR-145 [130]. Forced expression of miR-34a significantly reduces tumor expression of direct target genes (CD44, ALDH) involved in cancer cell stemness [130]. miR-34 mimetic drug, MRX34, was the first miRNA replacement therapy to enter clinical trials for treatment of primary liver tumors or cases with liver metastasis [18]. Unfortunately, on-target adverse immunological effects caused termination of these clinical trials [131]. Re-formulation of the liposomal nanoparticle or use of other platforms that may preferentially deliver the miR-34 mimic to cancer cells could bypass this immune toxicity. A poly (D,L-lactide-co-glycolide) (PLGA)-based nanoparticle formulation has a high encapsulation capacity for and effectively can delivery miRNA mimics to PDAC cell lines in vitro [132]. While this nanoparticle formulation was only tested for a miR-150 mimic and its interaction with direct target MUC4 [132], it may be a viable and low toxicity option for systemic delivery of a miR-34 mimic. Natural or synthetic exosomes may also be used as an encapsulation and delivery vehicle to modulate miRNA activity at primary or metastatic tumor sites in PDAC [133–137]. In comparison to other nanoparticle formulation (e.g., liposomes, micelles, inorganic nanoparticles), exosomes can have a greater drug delivery potential due to their lower clearance rate, deeper tissue penetration, and enhanced biocompatibility [138, 139]. Ultrasound-assisted loading of miR-34a mimic molecules in 293 T-derived exosomes provides a promising delivery platform to inhibit cancer cell growth in an in vivo xenograft PDAC model [140]. In this study, miR-34-loaded exosomes effectively downregulate expression of anti-apoptotic BCL2 in the xenograft tumors [140]. This or a similar strategy could be considered for investigating miR-34 replacement therapy in PDAC patients.
Anti-miRNA inhibitors can tolerate more chemical modifications since they do not require processing by the cellular machinery. Anti-miRNA inhibitors can be delivered systemically without the need of encapsulation, conjugation, or complexing, though these can offer intrinsic imaging and/or more targeted delivery capabilities. Administration of an unconjugated heavily modified single-stranded anti-miR-21 antisense oligonucleotide in the K-Ras-driven p53-mutated KPC model at an early age is remarkably effective at preventing tumor progression from pancreatic intraductal precursor lesion to malignant invasive carcinoma in this aggressive GEMM [141]. As we described above, miR-21 is a multifaceted miRNA with functional activity in different cell types. An important consideration for clinical evaluation of anti-miR-21-based cancer interception strategy will be to understand if a specific cell type or multiple ones are driving malignancy. A similar anti-miR-21 inhibitor encapsulated in PEGylated lipid nanoparticle, decorated with iRDG tumor penetrating–peptide and/or transportan cell-penetrating–peptide, reduces tumor growth in patient-derived and organoid-derived xenograft models [129, 142]. This therapy appears to be effective only in cancer cell–derived models with high levels of miR-21 at baseline, suggesting an intertumoral heterogeneity in terms of which miR-21-expressing cell types contribute to malignancy. Expression and function of miR-10 family members, miR-10a and miR-10b, have been associated with pro-metastatic programs in PDAC [46, 143–148]. In glioblastoma and breast cancer models, several encapsulation and nanoparticle strategies, including dextran-coated iron oxide nanoparticles, have been successful at delivering anti-miR-10b modified oligonucleotides to primary and metastatic tumor sites and causing miR-10b-dependent growth inhibition [18]. A similar dextran-coated iron oxide nanoparticle platform was recently used to deliver siRNAs against immune checkpoint PD-L1 in an orthotopic Pan02 syngeneic model of PDAC [149]. Thus, this nanoparticle platform with intrinsic magnetic resonance imaging capability could be applied to investigate the therapeutic efficacy of anti-miR-10b in vivo models of PDAC.
Conclusions
The level of evidence and biological effects of these discussed miRNAs and RNA-RNA networks varies depending on experimental design, model used, and clinical validation. By design, most studies focus on a particular miRNA-mediated process, but all these miRNA-regulated processes may be occurring at once in a PDAC tumor. What the cumulative effects of these miRNAs and RNA-RNA networks are and which of these miRNA-regulated processes may have a more impactful contribution to tumor progression and metastatic spread and in what cell types are still important remaining questions. Answering these questions should guide prioritizing of clinical development of miRNA targeting strategies that would be most beneficial for PDAC patients.
Author contribution
LS conceptualized and wrote manuscript, curated data, and created figures; KP edited manuscript, curated data, and edited figures; JR, AB, and TS edited manuscript and curated data. All authors reviewed and approved final version of the article.
Funding
This work was supported in part by the National Cancer Institute R21 CA226579 (LS), 2020 American Society for Investigative Pathology Summer Research Opportunity Program in Pathology, and 2021 American Society for Investigative Gotlieb Undergraduate Student in Pathobiology Travel Award (KP). Figures were created with BioRender.com.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA: A Cancer Journal for Clinicians. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395:2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 3.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV, Neale RE, Tempero M, Tuveson DA, Hruban RH, Neoptolemos JP. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022. doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- 4.Conway JR, Herrmann D, Evans TJ, Morton JP, Timpson P. Combating pancreatic cancer with PI3K pathway inhibitors in the era of personalised medicine. Gut. 2019;68:742–758. doi: 10.1136/gutjnl-2018-316822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lafaro KJ, Melstrom LG. The paradoxical web of pancreatic cancer tumor microenvironment. American Journal of Pathology. 2019;189:44–57. doi: 10.1016/j.ajpath.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collisson EA, Bailey P, Chang DK, Biankin AV. Molecular subtypes of pancreatic cancer. Nature Reviews. Gastroenterology & Hepatology. 2019;16:207–220. doi: 10.1038/s41575-019-0109-y. [DOI] [PubMed] [Google Scholar]
- 7.Bockorny B, Semenisty V, Macarulla T, Borazanci E, Wolpin BM, Stemmer SM, Golan T, Geva R, Borad MJ, Pedersen KS, Park JO, Ramirez RA, Abad DG, Feliu J, Munoz A, Ponz-Sarvise M, Peled A, Lustig TM, Bohana-Kashtan O, et al. BL-8040, a CXCR4 antagonist, in combination with pembrolizumab and chemotherapy for pancreatic cancer: The COMBAT trial. Nature Medicine. 2020;26:878–885. doi: 10.1038/s41591-020-0880-x. [DOI] [PubMed] [Google Scholar]
- 8.Picozzi, V., Alseidi, A., Winter, J., Pishvaian, M., Mody, K., Glaspy, J., Larson, T., Matrana, M., Carney, M., Porter, S., Kouchakji, E., Rocha, F., and Carrier, E. (2020). Gemcitabine/nab-paclitaxel with pamrevlumab: A novel drug combination and trial design for the treatment of locally advanced pancreatic cancer. ESMO Open 5. [DOI] [PMC free article] [PubMed]
- 9.Yang, J., Xu, R., Wang, C., Qiu, J., Ren, B., and You, L. (2021). Early screening and diagnosis strategies of pancreatic cancer: A comprehensive review. Cancer Commun (Lond). [DOI] [PMC free article] [PubMed]
- 10.Affolter KE, Hellwig S, Nix DA, Bronner MP, Thomas A, Fuertes CL, Hamil CL, Garrido-Laguna I, Scaife CL, Mulvihill SJ, Underhill HR. Detection of circulating tumor DNA without a tumor-informed search using next-generation sequencing is a prognostic biomarker in pancreatic ductal adenocarcinoma. Neoplasia. 2021;23:859–869. doi: 10.1016/j.neo.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyabayashi K, Nakagawa H, Koike K. Molecular and phenotypic profiling for precision medicine in pancreatic cancer: Current advances and future perspectives. Front Oncol. 2021;11:682872. doi: 10.3389/fonc.2021.682872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whittle MC, Izeradjene K, Rani PG, Feng L, Carlson MA, DelGiorno KE, Wood LD, Goggins M, Hruban RH, Chang AE, Calses P, Thorsen SM, Hingorani SR. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell. 2015;161:1345–1360. doi: 10.1016/j.cell.2015.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher T, Konukiewitz B, Ollinger R, Zwiebel M, Strong A, Yen HY, Banerjee R, Louzada S, Fu B, Seidler B, Gotzfried J, Schuck K, Hassan Z, Arbeiter A, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554:62–68. doi: 10.1038/nature25459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung EC, DeNicola GM, Nixon C, Blyth K, Labuschagne CF, Tuveson DA, Vousden KH. Dynamic ROS control by TIGAR regulates the initiation and progression of pancreatic cancer. Cancer Cell. 2020;37:168–182 e164. doi: 10.1016/j.ccell.2019.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nature Reviews Molecular Cell Biology. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sempere, L.F., Azmi, A.S., and Moore, A. (2021). microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip Rev RNA, e1662. [DOI] [PMC free article] [PubMed]
- 19.Lotfi Z, Najjary S, Lotfi F, Amini M, Baghbanzadeh A, Rashid DJ, Asl ER, Baradaran B, Mokhtarzadeh A. Crosstalk between miRNAs and signaling pathways involved in pancreatic cancer and pancreatic ductal adenocarcinoma. Eur J Pharmacol. 2021;901:174006. doi: 10.1016/j.ejphar.2021.174006. [DOI] [PubMed] [Google Scholar]
- 20.Rawat, M., Kadian, K., Gupta, Y., Kumar, A., Chain, P.S.G., Kovbasnjuk, O., Kumar, S., and Parasher, G. (2019). MicroRNA in pancreatic cancer: From biology to therapeutic potential. Genes (Basel) 10. [DOI] [PMC free article] [PubMed]
- 21.Tesfaye AA, Azmi AS, Philip PA. miRNA and gene expression in pancreatic ductal adenocarcinoma. American Journal of Pathology. 2019;189:58–70. doi: 10.1016/j.ajpath.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weidle UH, Birzele F, Nopora A. Pancreatic ductal adenocarcinoma: MicroRNAs affecting tumor growth and metastasis in preclinical in vivo models. Cancer Genomics & Proteomics. 2019;16:451–464. doi: 10.21873/cgp.20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uddin, M.H., Al-Hallak, M.N., Philip, P.A., Mohammad, R.M., Viola, N., Wagner, K.U., and Azmi, A.S. (2021). Exosomal microRNA in pancreatic cancer diagnosis, prognosis, and treatment: From bench to bedside. Cancers (Basel) 13. [DOI] [PMC free article] [PubMed]
- 24.Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human micrornAome. Annual Review of Genetics. 2015;49:213–242. doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomson DW, Dinger ME. Endogenous microRNA sponges: Evidence and controversy. Nature Reviews Genetics. 2016;17:272–283. doi: 10.1038/nrg.2016.20. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Aznar, E., Wiesmuller, L., Sainz, B., Jr., and Hermann, P.C. (2019). EMT and stemness-key players in pancreatic cancer stem cells. Cancers (Basel) 11. [DOI] [PMC free article] [PubMed]
- 27.Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan Z, Ai K. H19 promotes pancreatic cancer metastasis by derepressing let-7's suppression on its target HMGA2-mediated EMT. Tumour Biology. 2014;35:9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 28.Mody HR, Hung SW, Pathak RK, Griffin J, Cruz-Monserrate Z, Govindarajan R. miR-202 diminishes TGFbeta receptors and attenuates TGFbeta1-induced EMT in pancreatic cancer. Molecular Cancer Research. 2017;15:1029–1039. doi: 10.1158/1541-7786.MCR-16-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, Liu P, Wu H, Cui P, Li Y, Liu Y, Liu Z, Gou S. MicroRNA-323-3p inhibits cell invasion and metastasis in pancreatic ductal adenocarcinoma via direct suppression of SMAD2 and SMAD3. Oncotarget. 2016;7:14912–14924. doi: 10.18632/oncotarget.7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mody HR, Hung SW, AlSaggar M, Griffin J, Govindarajan R. Inhibition of S-adenosylmethionine-dependent methyltransferase attenuates TGFbeta1-induced EMT and metastasis in pancreatic cancer: Putative roles of miR-663a and miR-4787-5p. Molecular Cancer Research. 2016;14:1124–1135. doi: 10.1158/1541-7786.MCR-16-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z, Xu Y, Zhao J, Liu Q, Feng W, Fan J, Wang P. miR-367 promotes epithelial-to-mesenchymal transition and invasion of pancreatic ductal adenocarcinoma cells by targeting the Smad7-TGF-beta signalling pathway. British Journal of Cancer. 2015;112:1367–1375. doi: 10.1038/bjc.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu H, Zhang Q, Chen W, Wu T, Liu S, Li X, Luo B, Zhang T, Yan G, Lu H, Lu Z. MicroRNA-301a promotes pancreatic cancer invasion and metastasis through the JAK/STAT3 signaling pathway by targeting SOCS5. Carcinogenesis. 2020;41:502–514. doi: 10.1093/carcin/bgz121. [DOI] [PubMed] [Google Scholar]
- 33.Zhang KD, Hu B, Cen G, Yang YH, Chen WW, Guo ZY, Wang XF, Zhao Q, Qiu ZJ. MiR-301a transcriptionally activated by HIF-2alpha promotes hypoxia-induced epithelial-mesenchymal transition by targeting TP63 in pancreatic cancer. World Journal of Gastroenterology. 2020;26:2349–2373. doi: 10.3748/wjg.v26.i19.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diaz-Riascos ZV, Ginesta MM, Fabregat J, Serrano T, Busquets J, Buscail L, Cordelier P, Capella G. Expression and role of MicroRNAs from the miR-200 family in the tumor formation and metastatic propensity of pancreatic cancer. Mol Ther Nucleic Acids. 2019;17:491–503. doi: 10.1016/j.omtn.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang X, Chen K, Zhang Y, Liu H, Gan L, Bi H, Zhen P, Zhu J, Li X. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene. 2018;37:3822–3838. doi: 10.1038/s41388-018-0237-9. [DOI] [PubMed] [Google Scholar]
- 36.Zhuo M, Yuan C, Han T, Cui J, Jiao F, Wang L. A novel feedback loop between high MALAT-1 and low miR-200c-3p promotes cell migration and invasion in pancreatic ductal adenocarcinoma and is predictive of poor prognosis. BMC Cancer. 2018;18:1032. doi: 10.1186/s12885-018-4954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao G, Wang B, Liu Y, Zhang JG, Deng SC, Qin Q, Tian K, Li X, Zhu S, Niu Y, Gong Q, Wang CY. miRNA-141, downregulated in pancreatic cancer, inhibits cell proliferation and invasion by directly targeting MAP4K4. Molecular Cancer Therapeutics. 2013;12:2569–2580. doi: 10.1158/1535-7163.MCT-13-0296. [DOI] [PubMed] [Google Scholar]
- 38.Pan Y, Lu F, Xiong P, Pan M, Zhang Z, Lin X, Pan M, Huang H. WIPF1 antagonizes the tumor suppressive effect of miR-141/200c and is associated with poor survival in patients with PDAC. Journal of Experimental & Clinical Cancer Research. 2018;37:167. doi: 10.1186/s13046-018-0848-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Z, Li J, Guo H, Wang F, Ma L, Du C, Wang Y, Wang Q, Kornmann M, Tian X, Yang Y. BRM transcriptionally regulates miR-302a-3p to target SOCS5/STAT3 signaling axis to potentiate pancreatic cancer metastasis. Cancer Letters. 2019;449:215–225. doi: 10.1016/j.canlet.2019.02.031. [DOI] [PubMed] [Google Scholar]
- 40.Yu DL, Zhang T, Wu K, Li Y, Wang J, Chen J, Li XQ, Peng XG, Wang JN, Tan LG. MicroRNA-448 suppresses metastasis of pancreatic ductal adenocarcinoma through targeting JAK1/STAT3 pathway. Oncology Reports. 2017;38:1075–1082. doi: 10.3892/or.2017.5781. [DOI] [PubMed] [Google Scholar]
- 41.Xiong Y, Wang Y, Wang L, Huang Y, Xu Y, Xu L, Guo Y, Lu J, Li X, Zhu M, Qian H. MicroRNA-30b targets snail to impede epithelial-mesenchymal transition in pancreatic cancer stem cells. Journal of Cancer. 2018;9:2147–2159. doi: 10.7150/jca.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong J, Wang D, Wei A, Lu H, Tan C, Li A, Tang J, Wang Y, He S, Liu X, Hu W. Deregulated expression of miR-107 inhibits metastasis of PDAC through inhibition PI3K/Akt signaling via caveolin-1 and PTEN. Experimental Cell Research. 2017;361:316–323. doi: 10.1016/j.yexcr.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 43.Hu J, Li L, Chen H, Zhang G, Liu H, Kong R, Chen H, Wang Y, Li Y, Tian F, Lv X, Li G, Sun B. MiR-361-3p regulates ERK1/2-induced EMT via DUSP2 mRNA degradation in pancreatic ductal adenocarcinoma. Cell Death & Disease. 2018;9:807. doi: 10.1038/s41419-018-0839-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma G, Li G, Fan W, Xu Y, Song S, Guo K, Liu Z. Circ-0005105 activates COL11A1 by targeting miR-20a-3p to promote pancreatic ductal adenocarcinoma progression. Cell Death & Disease. 2021;12:656. doi: 10.1038/s41419-021-03938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma J, Zhang J, Weng YC, Wang JC. EZH2-mediated microRNA-139-5p regulates epithelial-mesenchymal transition and lymph node metastasis of pancreatic cancer. Molecules and Cells. 2018;41:868–880. doi: 10.14348/molcells.2018.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C, Yin W, Liu H. MicroRNA-10a promotes epithelial-to-mesenchymal transition and stemness maintenance of pancreatic cancer stem cells via upregulating the Hippo signaling pathway through WWC2 inhibition. Journal of Cellular Biochemistry. 2020;121:4505–4521. doi: 10.1002/jcb.29716. [DOI] [PubMed] [Google Scholar]
- 47.Kai F, Drain AP, Weaver VM. The extracellular matrix modulates the metastatic journey. Developmental Cell. 2019;49:332–346. doi: 10.1016/j.devcel.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou Y, Li J, Chen Z, Li X, Zheng S, Yi D, Zhong A, Chen J. miR-29c suppresses pancreatic cancer liver metastasis in an orthotopic implantation model in nude mice and affects survival in pancreatic cancer patients. Carcinogenesis. 2015;36:676–684. doi: 10.1093/carcin/bgv027. [DOI] [PubMed] [Google Scholar]
- 49.Yuan P, He XH, Rong YF, Cao J, Li Y, Hu YP, Liu Y, Li D, Lou W, Liu MF. KRAS/NF-kappaB/YY1/miR-489 signaling axis controls pancreatic cancer metastasis. Cancer Research. 2017;77:100–111. doi: 10.1158/0008-5472.CAN-16-1898. [DOI] [PubMed] [Google Scholar]
- 50.Fan Y, Shi C, Li T, Kuang T. microRNA-454 shows anti-angiogenic and anti-metastatic activity in pancreatic ductal adenocarcinoma by targeting LRP6. American Journal of Cancer Research. 2017;7:139–147. [PMC free article] [PubMed] [Google Scholar]
- 51.Fu Y, Liu X, Chen Q, Liu T, Lu C, Yu J, Miao Y, Wei J. Downregulated miR-98-5p promotes PDAC proliferation and metastasis by reversely regulating MAP4K4. Journal of Experimental & Clinical Cancer Research. 2018;37:130. doi: 10.1186/s13046-018-0807-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu D, Zheng S, Fang C, Guo X, Han D, Tang M, Fu H, Jiang M, Xie N, Nie Y, Yao X, Chen Y. Dysbindin promotes pancreatic ductal adenocarcinoma metastasis by activating NF-kappaB/MDM2 via miR-342-3p. Cancer Letters. 2020;477:107–121. doi: 10.1016/j.canlet.2020.02.033. [DOI] [PubMed] [Google Scholar]
- 53.Wang Q, Wang J, Niu S, Wang S, Liu Y, Wang X. MicroRNA-664 targets paired box protein 6 to inhibit the oncogenicity of pancreatic ductal adenocarcinoma. International Journal of Oncology. 2019;54:1884–1896. doi: 10.3892/ijo.2019.4759. [DOI] [PubMed] [Google Scholar]
- 54.Wong CH, Lou UK, Li Y, Chan SL, Tong JH, To KF, Chen Y. CircFOXK2 promotes growth and metastasis of pancreatic ductal adenocarcinoma by complexing with RNA-binding proteins and sponging MiR-942. Cancer Research. 2020;80:2138–2149. doi: 10.1158/0008-5472.CAN-19-3268. [DOI] [PubMed] [Google Scholar]
- 55.Liu W, Tang J, Zhang H, Kong F, Zhu H, Li P, Li Z, Kong X, Wang K. A novel lncRNA PTTG3P/miR-132/212-3p/FoxM1 feedback loop facilitates tumorigenesis and metastasis of pancreatic cancer. Cell Death Discov. 2020;6:136. doi: 10.1038/s41420-020-00360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W, Cui J, Du Y, Wei D, Huang S, Xie K. Down-regulation of microRNA-494 via loss of SMAD4 increases FOXM1 and beta-catenin signaling in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147:485–497 e418. doi: 10.1053/j.gastro.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 57.Azevedo-Pouly AC, Sutaria DS, Jiang J, Elgamal OA, Amari F, Allard D, Grippo PJ, Coppola V, Schmittgen TD. miR-216 and miR-217 expression is reduced in transgenic mouse models of pancreatic adenocarcinoma, knockout of miR-216/miR-217 host gene is embryonic lethal. Functional & Integrative Genomics. 2017;17:203–212. doi: 10.1007/s10142-016-0512-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu P, Yang H, Zhang J, Peng X, Lu Z, Tong W, Chen J. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Science and Reports. 2017;7:5186. doi: 10.1038/s41598-017-05274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sutaria DS, Jiang J, Azevedo-Pouly AC, Wright L, Bray JA, Fredenburg K, Liu X, Lu J, Torres C, Mancinelli G, Grippo PJ, Coppola V, Schmittgen TD. Knockout of acinar enriched microRNAs in mice promote duct formation but not pancreatic cancer. Science and Reports. 2019;9:11147. doi: 10.1038/s41598-019-47566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM, Chen J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis. 2010;31:1726–1733. doi: 10.1093/carcin/bgq160. [DOI] [PubMed] [Google Scholar]
- 61.Wang B, Xu Y, Wei Y, Lv L, Liu N, Lin R, Wang X, Shi B. Human mesenchymal stem cell-derived exosomal microRNA-143 promotes apoptosis and suppresses cell growth in pancreatic cancer via target gene regulation. Front Genet. 2021;12:581694. doi: 10.3389/fgene.2021.581694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie F, Li C, Zhang X, Peng W, Wen T. MiR-143–3p suppresses tumorigenesis in pancreatic ductal adenocarcinoma by targeting KRAS. Biomed Pharmacother. 2019;119:109424. doi: 10.1016/j.biopha.2019.109424. [DOI] [PubMed] [Google Scholar]
- 63.Hu Y, Ou Y, Wu K, Chen Y, Sun W. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumour Biology. 2012;33:1863–1870. doi: 10.1007/s13277-012-0446-8. [DOI] [PubMed] [Google Scholar]
- 64.Franses JW, Philipp J, Missios P, Bhan I, Liu A, Yashaswini C, Tai E, Zhu H, Ligorio M, Nicholson B, Tassoni EM, Desai N, Kulkarni AS, Szabolcs A, Hong TS, Liss AS, Fernandez-Del Castillo C, Ryan DP, Maheswaran S, et al. Pancreatic circulating tumor cell profiling identifies LIN28B as a metastasis driver and drug target. Nature Communications. 2020;11:3303. doi: 10.1038/s41467-020-17150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Li J, Guo S, Ouyang Y, Yin L, Liu S, Zhao Z, Yang J, Huang W, Qin H, Zhao X, Ni B, Wang H. Lin28B facilitates the progression and metastasis of pancreatic ductal adenocarcinoma. Oncotarget. 2017;8:60414–60428. doi: 10.18632/oncotarget.19578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nature Reviews Molecular Cell Biology. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anfossi S, Babayan A, Pantel K, Calin GA. Clinical utility of circulating non-coding RNAs - An update. Nature Reviews. Clinical Oncology. 2018;15:541–563. doi: 10.1038/s41571-018-0035-x. [DOI] [PubMed] [Google Scholar]
- 68.Machiela, E., Popkie, A., and Sempere, L. (2015). Individual noncoding RNA variations: Their role in shaping and maintaining the epigenetic landscape. In Personalized Epigenetics, Trygve Tollefsbol, ed. Waltham, Massachusetts: Academic Press, 2015, Chapter 4 pages: 84–114. ISBN-13: 978–0124201354.
- 69.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 70.Zhao X, Liu Y, Li Z, Zheng S, Wang Z, Li W, Bi Z, Li L, Jiang Y, Luo Y, Lin Q, Fu Z, Rufu C. Linc00511 acts as a competing endogenous RNA to regulate VEGFA expression through sponging hsa-miR-29b-3p in pancreatic ductal adenocarcinoma. Journal of Cellular and Molecular Medicine. 2018;22:655–667. doi: 10.1111/jcmm.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luo Z, Yi ZJ, Ou ZL, Han T, Wan T, Tang YC, Wang ZC, Huang FZ. RELA/NEAT1/miR-302a-3p/RELA feedback loop modulates pancreatic ductal adenocarcinoma cell proliferation and migration. Journal of Cellular Physiology. 2019;234:3583–3597. doi: 10.1002/jcp.27039. [DOI] [PubMed] [Google Scholar]
- 72.Shen J, Hong L, Yu D, Cao T, Zhou Z, He S. LncRNA XIST promotes pancreatic cancer migration, invasion and EMT by sponging miR-429 to modulate ZEB1 expression. International Journal of Biochemistry & Cell Biology. 2019;113:17–26. doi: 10.1016/j.biocel.2019.05.021. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, Y., Ma, H., and Chen, C. (2021). Long noncoding RNA PCED1BAS1 promotes pancreatic ductal adenocarcinoma progression by regulating the miR4113p/HIF1alpha axis. Oncol Rep 46. [DOI] [PMC free article] [PubMed]
- 74.Sun Y, Wang P, Yang W, Shan Y, Zhang Q, Wu H. The role of lncRNA MSC-AS1/miR-29b-3p axis-mediated CDK14 modulation in pancreatic cancer proliferation and Gemcitabine-induced apoptosis. Cancer Biology & Therapy. 2019;20:729–739. doi: 10.1080/15384047.2018.1529121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun Y, Zhu Q, Yang W, Shan Y, Yu Z, Zhang Q, Wu H. LncRNA H19/miR-194/PFTK1 axis modulates the cell proliferation and migration of pancreatic cancer. Journal of Cellular Biochemistry. 2019;120:3874–3886. doi: 10.1002/jcb.27669. [DOI] [PubMed] [Google Scholar]
- 76.Chen Y, Li Z, Zhang M, Wang B, Ye J, Zhang Y, Tang D, Ma D, Jin W, Li X, Wang S. Circ-ASH2L promotes tumor progression by sponging miR-34a to regulate Notch1 in pancreatic ductal adenocarcinoma. Journal of Experimental & Clinical Cancer Research. 2019;38:466. doi: 10.1186/s13046-019-1436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deng S, Wang J, Zhang L, Li J, Jin Y. LncRNA HOTAIR promotes cancer stem-like cells properties by sponging miR-34a to activate the JAK2/STAT3 pathway in pancreatic ductal adenocarcinoma. Oncotargets and Therapy. 2021;14:1883–1893. doi: 10.2147/OTT.S286666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bi S, Wang Y, Feng H, Li Q. Long noncoding RNA LINC00657 enhances the malignancy of pancreatic ductal adenocarcinoma by acting as a competing endogenous RNA on microRNA-433 to increase PAK4 expression. Cell Cycle. 2020;19:801–816. doi: 10.1080/15384101.2020.1731645. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Li WJ, Wang Y, Liu R, Kasinski AL, Shen H, Slack FJ, Tang DG. MicroRNA-34a: potent tumor suppressor, cancer stem cell inhibitor, and potential anticancer therapeutic. Front Cell Dev Biol. 2021;9:640587. doi: 10.3389/fcell.2021.640587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu Y, Tang L, Zhang Z, Li S, Liang S, Ji L, Yang B, Liu Y, Wei W. Long noncoding RNA TUG1/miR-29c axis affects cell proliferation, invasion, and migration in human pancreatic cancer. Disease Markers. 2018;2018:6857042. doi: 10.1155/2018/6857042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li JR, Liu L, Luo H, Chen ZG, Wang JH, Li NF. Long noncoding RNA DUXAP8 promotes pancreatic carcinoma cell migration and invasion via pathway by miR-448/WTAP/Fak signaling axis. Pancreas. 2021;50:317–326. doi: 10.1097/MPA.0000000000001751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiong G, Liu C, Yang G, Feng M, Xu J, Zhao F, You L, Zhou L, Zheng L, Hu Y, Wang X, Zhang T, Zhao Y. Long noncoding RNA GSTM3TV2 upregulates LAT2 and OLR1 by competitively sponging let-7 to promote gemcitabine resistance in pancreatic cancer. Journal of Hematology & Oncology. 2019;12:97. doi: 10.1186/s13045-019-0777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Geng Y, Fan J, Chen L, Zhang C, Qu C, Qian L, Chen K, Meng Z, Chen Z, Wang P. A Notch-dependent inflammatory feedback circuit between macrophages and cancer cells regulates pancreatic cancer metastasis. Cancer Research. 2021;81:64–76. doi: 10.1158/0008-5472.CAN-20-0256. [DOI] [PubMed] [Google Scholar]
- 84.Fan Y, Xu LL, Shi CY, Wei W, Wang DS, Cai DF. MicroRNA-454 regulates stromal cell derived factor-1 in the control of the growth of pancreatic ductal adenocarcinoma. Science and Reports. 2016;6:22793. doi: 10.1038/srep22793. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Xi Q, Chen Y, Yang GZ, Zhang JY, Zhang LJ, Guo XD, Zhao JY, Xue ZY, Li Y, Zhang R. miR-128 regulates tumor cell CD47 expression and promotes anti-tumor immunity in pancreatic cancer. Frontiers in Immunology. 2020;11:890. doi: 10.3389/fimmu.2020.00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schipper J, Westerhuis JJ, Beddows I, Madaj Z, Monsma D, Hostetter G, Kiupel M, Conejo-Garcia JR, Sempere LF. Loss of microRNA-21 leads to profound stromal remodeling and short survival in K-Ras-driven mouse models of pancreatic cancer. International Journal of Cancer. 2020;147:2265–2278. doi: 10.1002/ijc.33041. [DOI] [PubMed] [Google Scholar]
- 87.Zhang L, Yao J, Li W, Zhang C. Micro-RNA-21 regulates cancer-associated fibroblast-mediated drug resistance in pancreatic cancer. Oncology Research. 2018;26:827–835. doi: 10.3727/096504017X14934840662335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kwon JJ, Nabinger SC, Vega Z, Sahu SS, Alluri RK, Abdul-Sater Z, Yu Z, Gore J, Nalepa G, Saxena R, Korc M, Kota J. Pathophysiological role of microRNA-29 in pancreatic cancer stroma. Science and Reports. 2015;5:11450. doi: 10.1038/srep11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bautista-Sanchez D, Arriaga-Canon C, Pedroza-Torres A, De La Rosa-Velazquez IA, Gonzalez-Barrios R, Contreras-Espinosa L, Montiel-Manriquez R, Castro-Hernandez C, Fragoso-Ontiveros V, Alvarez-Gomez RM, Herrera LA. The promising role of miR-21 as a cancer biomarker and its importance in RNA-based therapeutics. Mol Ther Nucleic Acids. 2020;20:409–420. doi: 10.1016/j.omtn.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sheedy FJ. Turning 21: Induction of miR-21 as a key switch in the inflammatory response. Frontiers in Immunology. 2015;6:19. doi: 10.3389/fimmu.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Biffi G, Tuveson DA. Diversity and biology of cancer-associated fibroblasts. Physiological Reviews. 2021;101:147–176. doi: 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Helms E, Onate MK, Sherman MH. Fibroblast heterogeneity in the pancreatic tumor microenvironment. Cancer Discovery. 2020;10:648–656. doi: 10.1158/2159-8290.CD-19-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, Corbo V, Oni TE, Hearn SA, Lee EJ, Chio II, Hwang CI, Tiriac H, Baker LA, Engle DD, Feig C, Kultti A, Egeblad M, Fearon DT, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. Journal of Experimental Medicine. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Biffi G, Oni TE, Spielman B, Hao Y, Elyada E, Park Y, Preall J, Tuveson DA. IL1-induced JAK/STAT signaling is antagonized by TGFbeta to shape CAF Heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discovery. 2019;9:282–301. doi: 10.1158/2159-8290.CD-18-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, Teinor JA, Belleau P, Biffi G, Lucito MS, Sivajothi S, Armstrong TD, Engle DD, Yu KH, Hao Y, Wolfgang CL, Park Y, Preall J, Jaffee EM, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discovery. 2019;9:1102–1123. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Steele, N.G., Biffi, G., Kemp, S.B., Zhang, Y., Drouillard, D., Syu, L., Hao, Y., Oni, T.E., Brosnan, E., Elyada, E., Doshi, A., Hansma, C., Espinoza, C., Abbas, A., The, S., Irizarry-Negron, V., Halbrook, C.J., Franks, N.E., Hoffman, M.T., et al. (2021). Inhibition of hedgehog signaling alters fibroblast composition in pancreatic cancer. Clin Cancer Res. [DOI] [PMC free article] [PubMed]
- 97.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, Laklai H, Sugimoto H, Kahlert C, Novitskiy SV, De Jesus-Acosta A, Sharma P, Heidari P, Mahmood U, Chin L, Moses HL, Weaver VM, Maitra A, Allison JP, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, Dekleva EN, Saunders T, Becerra CP, Tattersall IW, Westphalen CB, Kitajewski J, Fernandez-Barrena MG, Fernandez-Zapico ME, Iacobuzio-Donahue C, Olive KP, Stanger BZ. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25:735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S, Fitamant J, Jones PD, Ghanta KS, Kawano S, Nagle JM, Deshpande V, Boucher Y, Kato T, Chen JK, Willmann JK, Bardeesy N, Beachy PA. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111:E3091–3100. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sempere LF, Preis M, Yezefski T, Ouyang H, Suriawinata AA, Silahtaroglu A, Conejo-Garcia JR, Kauppinen S, Wells W, Korc M. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clinical Cancer Research. 2010;16:4246–4255. doi: 10.1158/1078-0432.CCR-10-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Preis, M., Gardner, T.B., Gordon, S.R., Pipas, M.J., Mackenzie, T.A., Klein, E.E., Longnecker, D.S., Gutmann, E.J., Sempere, L.F., and Korc, M. (2011). microRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clin.Cancer Res. [DOI] [PMC free article] [PubMed]
- 102.Sempere LF, Korc M. A method for conducting highly sensitive microRNA in situ hybridization and immunohistochemical analysis in pancreatic cancer. Methods in Molecular Biology. 2013;980:43–59. doi: 10.1007/978-1-62703-287-2_4. [DOI] [PubMed] [Google Scholar]
- 103.Sempere LF, Zaluzec E, Kenyon E, Kiupel M, Moore A. Automated five-color multiplex co-detection of Microrna and protein expression in fixed tissue specimens. Methods in Molecular Biology. 2020;2148:257–276. doi: 10.1007/978-1-0716-0623-0_17. [DOI] [PubMed] [Google Scholar]
- 104.Kadera BE, Li L, Toste PA, Wu N, Adams C, Dawson DW, Donahue TR. MicroRNA-21 in pancreatic ductal adenocarcinoma tumor-associated fibroblasts promotes metastasis. PLoS ONE. 2013;8:e71978. doi: 10.1371/journal.pone.0071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Donahue TR, Nguyen AH, Moughan J, Li L, Tatishchev S, Toste P, Farrell JJ. Stromal microRNA-21 levels predict response to 5-fluorouracil in patients with pancreatic cancer. Journal of Surgical Oncology. 2014;110:952–959. doi: 10.1002/jso.23750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ensink E, Sinha J, Sinha A, Tang H, Calderone HM, Hostetter G, Winter J, Cherba D, Brand RE, Allen PJ, Sempere LF, Haab BB. Segment and fit thresholding: A new method for image analysis applied to microarray and immunofluorescence data. Analytical Chemistry. 2015;87:9715–9721. doi: 10.1021/acs.analchem.5b03159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.du Rieu MC, Torrisani J, Selves J, Al ST, Souque A, Dufresne M, Tsongalis GJ, Suriawinata AA, Carrere N, Buscail L, Cordelier P. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clinical Chemistry. 2010;56:603–612. doi: 10.1373/clinchem.2009.137364. [DOI] [PubMed] [Google Scholar]
- 108.Chen S, Chen X, Shan T, Ma J, Lin W, Li W, Kang Y. MiR-21-mediated metabolic alteration of cancer-associated fibroblasts and its effect on pancreatic cancer cell behavior. International Journal of Biological Sciences. 2018;14:100–110. doi: 10.7150/ijbs.22555. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 109.Sempere, L.F., Keto, J., and Fabbri, M. (2017). Exosomal MicroRNAs in breast cancer towards diagnostic and therapeutic applications. Cancers (Basel) 9. [DOI] [PMC free article] [PubMed]
- 110.Kalluri R. The biology and function of exosomes in cancer. The Journal of Clinical Investigation. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hussain, Z., Nigri, J., and Tomasini, R. (2021). The cellular and biological impact of extracellular vesicles in pancreatic cancer. Cancers (Basel) 13. [DOI] [PMC free article] [PubMed]
- 112.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pardini, B., and Calin, G.A. (2019). MicroRNAs and long non-coding RNAs and their hormone-like activities in cancer. Cancers (Basel) 11. [DOI] [PMC free article] [PubMed]
- 114.Li Z, Tao Y, Wang X, Jiang P, Li J, Peng M, Zhang X, Chen K, Liu H, Zhen P, Zhu J, Liu X, Liu X. Tumor-secreted exosomal miR-222 promotes tumor progression via regulating P27 Expression and re-localization in pancreatic cancer. Cellular Physiology and Biochemistry. 2018;51:610–629. doi: 10.1159/000495281. [DOI] [PubMed] [Google Scholar]
- 115.Jiang MJ, Chen YY, Dai JJ, Gu DN, Mei Z, Liu FR, Huang Q, Tian L. Dying tumor cell-derived exosomal miR-194-5p potentiates survival and repopulation of tumor repopulating cells upon radiotherapy in pancreatic cancer. Molecular Cancer. 2020;19:68. doi: 10.1186/s12943-020-01178-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yang Z, Zhao N, Cui J, Wu H, Xiong J, Peng T. Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210. Cellular Oncology (Dordrecht) 2020;43:123–136. doi: 10.1007/s13402-019-00476-6. [DOI] [PubMed] [Google Scholar]
- 117.Mikamori M, Yamada D, Eguchi H, Hasegawa S, Kishimoto T, Tomimaru Y, Asaoka T, Noda T, Wada H, Kawamoto K, Gotoh K, Takeda Y, Tanemura M, Mori M, Doki Y. MicroRNA-155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Science and Reports. 2017;7:42339. doi: 10.1038/srep42339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sun H, Shi K, Qi K, Kong H, Zhang J, Dai S, Ye W, Deng T, He Q, Zhou M. Natural Killer cell-derived exosomal miR-3607-3p inhibits pancreatic cancer progression by targeting IL-26. Frontiers in Immunology. 2019;10:2819. doi: 10.3389/fimmu.2019.02819. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 119.Fang Y, Zhou W, Rong Y, Kuang T, Xu X, Wu W, Wang D, Lou W. Exosomal miRNA-106b from cancer-associated fibroblast promotes gemcitabine resistance in pancreatic cancer. Exp Cell Res. 2019;383:111543. doi: 10.1016/j.yexcr.2019.111543. [DOI] [PubMed] [Google Scholar]
- 120.Binenbaum Y, Fridman E, Yaari Z, Milman N, Schroeder A, Ben David G, Shlomi T, Gil Z. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Research. 2018;78:5287–5299. doi: 10.1158/0008-5472.CAN-18-0124. [DOI] [PubMed] [Google Scholar]
- 121.Wang X, Luo G, Zhang K, Cao J, Huang C, Jiang T, Liu B, Su L, Qiu Z. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kgamma to promote pancreatic cancer metastasis. Cancer Research. 2018;78:4586–4598. doi: 10.1158/0008-5472.CAN-17-3841. [DOI] [PubMed] [Google Scholar]
- 122.Yin Z, Ma T, Huang B, Lin L, Zhou Y, Yan J, Zou Y, Chen S. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-beta signaling pathway. Journal of Experimental & Clinical Cancer Research. 2019;38:310. doi: 10.1186/s13046-019-1313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chang, J., Li, H., Zhu, Z., Mei, P., Hu, W., Xiong, X., and Tao, J. (2021). microRNA-21–5p from M2 macrophage-derived extracellular vesicles promotes the differentiation and activity of pancreatic cancer stem cells by mediating KLF3. Cell Biol Toxicol. [DOI] [PMC free article] [PubMed]
- 124.Ma Q, Wu H, Xiao Y, Liang Z, Liu T. Upregulation of exosomal microRNA21 in pancreatic stellate cells promotes pancreatic cancer cell migration and enhances Ras/ERK pathway activity. International Journal of Oncology. 2020;56:1025–1033. doi: 10.3892/ijo.2020.4986. [DOI] [PubMed] [Google Scholar]
- 125.Li M, Guo H, Wang Q, Chen K, Marko K, Tian X, Yang Y. Pancreatic stellate cells derived exosomal miR-5703 promotes pancreatic cancer by downregulating CMTM4 and activating PI3K/Akt pathway. Cancer Letters. 2020;490:20–30. doi: 10.1016/j.canlet.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 126.Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, Zhao X, Zhang Y, Zhen P, Zhu J, Li X. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Letters. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 127.Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nature Reviews. Drug Discovery. 2020;19:673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Forterre, A., Komuro, H., Aminova, S., and Harada, M. (2020). A comprehensive review of cancer MicroRNA therapeutic delivery strategies. Cancers (Basel) 12. [DOI] [PMC free article] [PubMed]
- 129.Gilles ME, Hao L, Brown K, Lim J, Bhatia SN, Slack FJ. Tumor penetrating nanomedicine targeting both an oncomiR and an oncogene in pancreatic cancer. Oncotarget. 2019;10:5349–5358. doi: 10.18632/oncotarget.27160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, Maitra A. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Molecular Cancer Therapeutics. 2011;10:1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, Shin S, Becerra CR, Falchook G, Stoudemire J, Martin D, Kelnar K, Peltier H, Bonato V, Bader AG, et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. British Journal of Cancer. 2020;122:1630–1637. doi: 10.1038/s41416-020-0802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Arora S, Swaminathan SK, Kirtane A, Srivastava SK, Bhardwaj A, Singh S, Panyam J, Singh AP. Synthesis, characterization, and evaluation of poly (D, L-lactide-co-glycolide)-based nanoformulation of miRNA-150: Potential implications for pancreatic cancer therapy. International Journal of Nanomedicine. 2014;9:2933–2942. doi: 10.2147/IJN.S61949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ding Y, Cao F, Sun H, Wang Y, Liu S, Wu Y, Cui Q, Mei W, Li F. Exosomes derived from human umbilical cord mesenchymal stromal cells deliver exogenous miR-145-5p to inhibit pancreatic ductal adenocarcinoma progression. Cancer Letters. 2019;442:351–361. doi: 10.1016/j.canlet.2018.10.039. [DOI] [PubMed] [Google Scholar]
- 134.Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xie Y, Hang Y, Wang Y, Sleightholm R, Prajapati DR, Bader J, Yu A, Tang W, Jaramillo L, Li J, Singh RK, Oupicky D. Stromal modulation and treatment of metastatic pancreatic cancer with local intraperitoneal triple miRNA/siRNA nanotherapy. ACS Nano. 2020;14:255–271. doi: 10.1021/acsnano.9b03978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shang S, Wang J, Chen S, Tian R, Zeng H, Wang L, Xia M, Zhu H, Zuo C. Exosomal miRNA-1231 derived from bone marrow mesenchymal stem cells inhibits the activity of pancreatic cancer. Cancer Medicine. 2019;8:7728–7740. doi: 10.1002/cam4.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu DM, Wen X, Han XR, Wang S, Wang YJ, Shen M, Fan SH, Zhang ZF, Shan Q, Li MQ, Hu B, Lu J, Chen GQ, Zheng YL. Bone marrow mesenchymal stem cell-derived exosomal MicroRNA-126-3p inhibits pancreatic cancer development by targeting ADAM9. Mol Ther Nucleic Acids. 2019;16:229–245. doi: 10.1016/j.omtn.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 138.Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacologica Sinica. 2017;38:754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Elsharkasy OM, Nordin JZ, Hagey DW, de Jong OG, Schiffelers RM, Andaloussi SE, Vader P. Extracellular vesicles as drug delivery systems: Why and how? Advanced Drug Delivery Reviews. 2020;159:332–343. doi: 10.1016/j.addr.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 140.Zuo L, Tao H, Xu H, Li C, Qiao G, Guo M, Cao S, Liu M, Lin X. Exosomes-coated miR-34a displays potent antitumor activity in pancreatic cancer both in vitro and in vivo. Drug Des Devel Ther. 2020;14:3495–3507. doi: 10.2147/DDDT.S265423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chu NJ, Anders RA, Fertig EJ, Cao M, Hopkins AC, Keenan BP, Popovic A, Armstrong TD, Jaffee EM, Zimmerman JW. Inhibition of miR-21 regulates mutant KRAS effector pathways and intercepts pancreatic ductal adenocarcinoma development. Cancer Prevention Research (Philadelphia, Pa.) 2020;13:569–582. doi: 10.1158/1940-6207.CAPR-20-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gilles ME, Hao L, Huang L, Rupaimoole R, Lopez-Casas PP, Pulver E, Jeong JC, Muthuswamy SK, Hidalgo M, Bhatia SN, Slack FJ. Personalized RNA Medicine for Pancreatic Cancer. Clinical Cancer Research. 2018;24:1734–1747. doi: 10.1158/1078-0432.CCR-17-2733. [DOI] [PubMed] [Google Scholar]
- 143.Fei X, Jin HY, Gao Y, Kong LM, Tan XD. Hsa-miR-10a-5p promotes pancreatic cancer growth by BDNF/SEMA4C pathway. Journal of Biological Regulators and Homeostatic Agents. 2020;34:927–934. doi: 10.23812/20-61-A-47. [DOI] [PubMed] [Google Scholar]
- 144.Kong F, Li L, Wang G, Deng X, Li Z, Kong X. VDR signaling inhibits cancer-associated-fibroblasts' release of exosomal miR-10a-5p and limits their supportive effects on pancreatic cancer cells. Gut. 2019;68:950–951. doi: 10.1136/gutjnl-2018-316627. [DOI] [PubMed] [Google Scholar]
- 145.Xiong G, Huang H, Feng M, Yang G, Zheng S, You L, Zheng L, Hu Y, Zhang T, Zhao Y. MiR-10a-5p targets TFAP2C to promote gemcitabine resistance in pancreatic ductal adenocarcinoma. Journal of Experimental & Clinical Cancer Research. 2018;37:76. doi: 10.1186/s13046-018-0739-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sheedy P, Medarova Z. The fundamental role of miR-10b in metastatic cancer. American Journal of Cancer Research. 2018;8:1674–1688. [PMC free article] [PubMed] [Google Scholar]
- 147.Xu C, Qi X. MiR-10b inhibits migration and invasion of pancreatic ductal adenocarcinoma via regulating E2F7. J Clin Lab Anal. 2020;34:e23442. doi: 10.1002/jcla.23442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yoo B, Greninger P, Stein GT, Egan RK, McClanaghan J, Moore A, Benes CH, Medarova Z. Potent and selective effect of the mir-10b inhibitor MN-anti-mir10b in human cancer cells of diverse primary disease origin. PLoS One. 2018;13:e0201046. doi: 10.1371/journal.pone.0201046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yoo B, Jordan VC, Sheedy P, Billig AM, Ross A, Pantazopoulos P, Medarova Z. RNAi-mediated PD-L1 inhibition for pancreatic cancer immunotherapy. Science and Reports. 2019;9:4712. doi: 10.1038/s41598-019-41251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]