Abstract
Genetic correlations suggest that the genetic relationship of alcohol use with internalizing psychopathology depends on the measure of alcohol use. Problematic alcohol use (PAU) is positively genetically correlated with internalizing psychopathology, whereas alcohol consumption ranges from not significantly correlated to moderately negatively correlated with internalizing psychopathology. To explore these different genetic relationships of internalizing psychopathology with alcohol use, we performed a multivariate GWAS of four correlated factors (internalizing psychopathology, PAU, quantity of alcohol consumption and frequency of alcohol consumption) and then assessed genome-wide and local genetic covariance between these factors. We identified 14 significant regions of local, largely positive, genetic covariance between PAU and internalizing psychopathology and 12 regions of significant local genetic covariance (including both positive and negative genetic covariance) between consumption factors and internalizing psychopathology. Partitioned genetic covariance among functional annotations suggested that brain tissues contribute significantly to positive genetic covariance between internalizing psychopathology and PAU, but not to the genetic covariance between internalizing psychopathology and quantity or frequency of alcohol consumption. We hypothesize that genome-wide genetic correlations between alcohol use and psychiatric traits may not capture the more complex shared or divergent genetic architectures at the locus or tissue specific level. This study highlights the complexity of genetic architectures of alcohol use and internalizing psychopathology, and the differing shared genetics of internalizing disorders with problematic alcohol use compared to consumption.
Keywords: problematic alcohol use, substance use, internalizing psychopathology, genome-wide association studies, genetic risk
INTRODUCTION
Internalizing disorders and alcohol use disorders (AUDs) have a high rate of comorbidity, co-occurring at a rate two to four times more likely than would be expected if independent (Burns & Teesson, 2002; Himle & Hill, 1991; Kessler et al., 1997; Regier et al., 1990). There is strong evidence that shared genetic factors contribute to this co-occurrence (Enoch et al., 2006; Merikangas et al., 1996; Nurnberger et al., 2002; Torvik et al., 2019). However, the specific underlying genetic mechanisms of this comorbidity are generally unknown.
Complicating this connection is the fact that the relationships of individual internalizing disorders, such as depression and generalized anxiety disorder (GAD), with problematic alcohol use (PAU) are distinct from their relationships with alcohol consumption (Hasin et al., 2007; Himle & Hill, 1991; Kessler et al., 1997; Kushner et al., 1999; Merikangas et al., 1996; Regier et al., 1990). Recent work suggested that when alcohol use, measured with the Alcohol Use Disorders Identification Test (AUDIT) (Saunders et al., 1993), is partitioned into a problem use score (AUDIT-P) and a consumption score (AUDIT-C), these two alcohol use phenotypes are genetically separable, with a genetic correlation (rg) of 0.695, and they have distinct genetic correlations with psychiatric traits (Sanchez-Roige et al., 2019). Importantly, AUDIT-P was significantly positively genetically correlated (rg=0.26) with depression, while AUDIT-C was negatively genetically correlated (rg=−0.24) (Sanchez-Roige et al., 2019).
Such correlations may be impacted by self-reporting biases or from composite AUDIT scores not fully capturing differential impacts of individual items. Xue et al. showed that correcting for self-reporting biases in alcohol use GWAS led to a positive genetic correlation of AUDIT-C with depression (rg=0.11), but this correlation remained lower than the correlation with AUDIT-P (Xue et al., 2021). Alternatively, individual AUDIT items may be disproportionately affecting AUDIT-P and AUDIT-C composite scores (Mallard, Savage, et al., 2020), where equally weighting each AUDIT item does not accurately represent measures of PAU and alcohol consumption. However, after identifying alcohol consumption and problematic use latent factors with Genomic Structural Equation Modeling (Genomic SEM) (Grotzinger et al., 2019), psychopathology phenotypes remained more strongly genetically correlated with PAU than consumption (Mallard, Savage, et al., 2020).
It is worth noting that consumption itself, typically measured as a combination of frequency and quantity of use, may be comprised of genetically differentiable components. Within a Genomic SEM analysis, AUDIT items 2 & 3, which are measures of quantity of consumption, load much more strongly onto a consumption factor than AUDIT item 1, which relates to frequency of alcohol consumption (Mallard, Savage, et al., 2020). This pattern is consistent with other work that observed a more moderate genetic correlation between quantity and frequency of alcohol consumption (rg=0.52) (Polimanti et al., 2019). However, while quantity of alcohol consumption was positively genetically correlated with depression (rg = 0.14), frequency of alcohol consumption was negatively correlated (rg = −0.17) (Polimanti et al., 2019). These findings suggest that measures of alcohol consumption frequency (hereafter referred to as frequency) are distinct from measures of alcohol consumption quantity (hereafter referred to as quantity), which may themselves have separable relationships with internalizing disorders.
Previous studies have examined the individual genetic correlations of PAU or alcohol consumption with either depression or anxiety symptoms (Mallard, Savage, et al., 2020; Sanchez-Roige et al., 2019; H. Zhou et al., 2019). However, a general internalizing psychopathology factor may be a more informative measure when examining these relationships because, 1) the genetic correlation of anxiety and depression is quite high (Kendler et al., 1992, 2007; Levey et al., 2020; Wray et al., 2018), and 2) the relationship between internalizing and alcohol use traits could primarily result from the shared elements of internalizing disorders rather than any specific disorder’s unique components (Krueger, 1999; Kushner et al., 2012). The overlapping genetic architecture between alcohol use traits and internalizing psychopathology has not been well characterized but could lead to a better understanding of their neurobiology and comorbidity of the two.
Of particular interest when evaluating the shared genetics of alcohol use and internalizing psychopathology is where in the genome and in which specific tissues shared biological mechanisms may exert effects. The shared genetic influences of internalizing psychopathology and problematic use have not been localized, but doing so could provide insight into the mechanisms of their comorbidity and the distinct relationships of internalizing traits with PAU versus consumption. This localization includes assessing how genome-wide relationships are partitioned among genes expressed within specific tissues or brain regions to identify key tissues or regions that contribute to their comorbidity, as well as identifying specific loci (SNPs and genomic regions) of traits’ shared genetic architecture. Recent methods have been developed to partition estimates of heritability and genetic covariance along functional annotations and linkage-disequilibrium (LD) blocks (Finucane et al., 2015, 2018; Zhang et al., 2020; Zhu et al., 2018). These partitioned estimates can be used to identify potentially pleiotropic regions of the genome and the specific brain regions involved, as well as facilitate functional follow-up studies (So et al., 2017), for example using animal models (Gusev et al., 2018; Meier et al., 2019; Schumann et al., 2016; Sekar et al., 2016; Y. Zhou et al., 2016) and for drug discovery (Fromer et al., 2016; Nelson et al., 2015; Okada et al., 2014). Relationships at the locus and tissue specific level have the potential to better characterize genetic relationships that may be over-simplified at the genome-wide level.
Here, we examine the shared genetic architecture of multiple aspects of alcohol use (PAU, quantity and frequency) and internalizing psychopathology. First, we evaluate genetic correlations at a genome-wide scale. Second, we analyze the genetic relationships between internalizing psychopathology and alcohol use phenotypes to specify a correlated four-factor model. Third, we localize genomic regions underlying the genetic relationships between these latent factors. Fourth, we apply partitioned genetic covariance analyses to identify specific functional categories involved.
METHODS
Preregistration
We preregistered our analysis (https://osf.io/kuqgf). All preregistered analyses are reported in a publicly available pre-print (Colbert et al., 2020). After performing these preregistered analyses, we expanded the scope of the study to include new summary statistics and used a Genomic SEM framework. We additionally separated alcohol consumption into quantity and frequency measures. As a result, our methods deviated significantly to include latent genetic factors estimated in Genomic SEM. However, the general aim of the project—to compare the genetic architectures of alcohol use and internalizing psychopathology—remained the same. It should also be noted that in the preregistration we designated Bonferonni correction as our method of multiple testing correction; however, in these analyses we also used a False Discovery Rate multiple test correction to follow the procedure used in the original LD score regression (LDSC) method (Bulik-Sullivan et al., 2015).
Study samples and phenotypes
We utilized published, publicly-available summary statistics for depression, GAD, AUD, and drinks per week, and performed GWAS using the UK Biobank for individual AUDIT items and frequency of alcohol intake (Table 1 and S1). To reduce possible confounding from genetic stratification, all summary statistics were derived from analyses of individuals of European ancestry.
Table 1.
Description of univariate genome-wide association summary statistics. Full descriptions of the AUDIT items are available in Supplemental Table 1. SNP-h2 was calculated using LDSC.
| Phenotype | Item Descriptor (if applicable) |
Source | N | SNP-h2 | SNP-h2 SE |
SNP-h2 Z-score |
|---|---|---|---|---|---|---|
| Depression | PGC + UKB (Howard et al., 2019) | 500,199 | 0.053 | 0.0022 | 24.273 | |
| Generalized Anxiety Disorder | MVP (Levey et al., 2020) | 175,163 | 0.047 | 0.0037 | 12.730 | |
| Anxiety meta-analysis | ANGST + UKB + iPSYCH (Purves et al., 2019) | 114,091 | 0.069 | 0.0051 | 13.471 | |
| Alcohol Use Disorder | MVP + PGC (Zhou et al., 2020) | 313,963 | 0.077 | 0.0049 | 15.673 | |
| AUDIT item 1 | Frequency of drinking | UKB | 148,222 | 0.057 | 0.0045 | 12.711 |
| AUDIT item 2 | How many drinks | UKB | 141,749 | 0.040 | 0.0040 | 10.100 |
| AUDIT item 3 | 6+ drinks | UKB | 142,034 | 0.049 | 0.0039 | 12.462 |
| AUDIT item 4 | Not able to stop | UKB | 84,621 | 0.023 | 0.0047 | 4.787 |
| AUDIT item 5 | Failed expectations | UKB | 84,737 | 0.016 | 0.0049 | 3.163 |
| AUDIT item 7 | Guilt/remorse | UKB | 84,675 | 0.030 | 0.0053 | 5.698 |
| AUDIT item 8 | Blackout | UKB | 84,708 | 0.029 | 0.0055 | 5.218 |
| AUDIT item 10 | Friend/family concerned | UKB | 148,185 | 0.020 | 0.0029 | 6.897 |
| Drinks per week | GSCAN (Liu et al., 2019) | 537,349 | 0.045 | 0.0021 | 21.476 | |
| Alcohol intake frequency | UKB | 462,016 | 0.054 | 0.0026 | 20.577 |
We drew internalizing psychopathology summary statistics from three independent studies: 1) A meta-analysis of the published summary statistics of three studies that used DSM-based diagnoses of the five core anxiety disorders (GAD, panic disorder, social phobia, agoraphobia and specific phobia) to define cases (hereafter the ‘anxiety meta-analysis’ dataset) (Purves et al., 2019). 2) A GWAS of the Generalized Anxiety Disorder 2-item scale (GAD-2) using data from the Million Veteran Program (Levey et al., 2020) (hereafter ‘GAD’). 3) A general depression meta-analysis GWAS from the PGC29 cohort and UK Biobank (Howard et al., 2019). We included both depression and anxiety phenotypes, including multiple anxiety case definitions, as there is strong evidence for a shared internalizing genetic factor between the two (Grotzinger et al., 2020; Gustavson et al., 2020; Krueger, 1999).
We included both PAU and alcohol consumption traits. First, we included the reported number of alcoholic drinks an individual consumed per week phenotype of the GWAS & Sequencing Consortium of Alcohol and Nicotine (GSCAN) (Liu et al., 2019). We included GWAS summary statistics of alcohol dependence (H. Zhou et al., 2019) defined by either clinician ratings, semi-structured interviews, or at least one inpatient or two outpatient alcohol-related ICD-9/10 codes. We used individual level AUDIT items to reduce the possibility of any single item disproportionately affecting composite score, following Mallard et al. (2020). AUDIT items 1-3 are considered to be components of alcohol consumption, while AUDIT items 4-10 are treated as PAU measures. In addition to these individual AUDIT items, we also generated GWAS summary statistics for a separate measure of frequency of alcohol intake in the UK Biobank (Table 1). For all UK Biobank traits, we limited our sample to only current drinkers, to reduce misreporting and longitudinal bias. We excluded AUDIT items 6 and 9 as they both had low SNP-heritabilities (Table S2), similar to Mallard, Savage, et al. (2020).
Analyses
Univariate GWAS
We performed GWASs for individual AUDIT items and alcohol intake frequency in the UK Biobank samples using BOLT-LMM (Loh et al., 2015, 2018). To avoid longitudinal bias, we used the UK Biobank drinker status (field ID 20117) to subset our sample to only include current drinkers. Briefly, GWASs were conducted using imputed dosages on individuals of European ancestry based on scores from the first 4 genomic principal components (PCs); SNPs with MAF < 0.001 and INFO score < 0.9 were excluded from analyses. We included as covariates age at time of assessment (field 21003), age-squared, sex (field 31), Townsend Deprivation Index (hereafter, deprivation, field 189), educational attainment (“qualification”, categorical, field 6138), genotyping batch (field 22000), assessment center (field 54), and the first 10 genetic principal components. Principal components were estimated with flashpca (Abraham & Inouye, 2014) applied to MAF- and LD-pruned array markers (plink2 (Chang et al., 2015) command: --maf 0.01 –hwe 1e-8 –indep-pairwise 50 5 0.2), which also (using the same quality control, but without LD-pruning) were used in BOLT-LMM to control for relatedness. We included tbe socioeconomic status (SES) measures of deprivation and educational attainment as covariates because we were primarily interested in genetic effects that act on alcohol use or internalizing traits directly, rather than loci whose effects on these traits are mediated by direct effects on SES variables (Marees et al., 2021). However, we explored the influence of removing these covariates in a sensitivity analysis, described below.
Genome-Wide Genetic Correlations
We used LDSC (Bulik-Sullivan et al., 2015) to estimate genome-wide genetic correlations (rg) among all pairs of alcohol use and internalizing psychopathology summary statistics. Significance of genetic correlations was determined using FDR < 5%.
Genomic structural equation modeling and Multivariate GWAS
We modeled the genetic correlation matrix using a confirmatory factor analysis with four latent factors based on prior knowledge of the alcohol and internalizing phenotypes (K. S. Kendler et al., 2012; Mallard, Savage, et al., 2020; Polimanti et al., 2019; Sanchez-Roige et al., 2019), as well as the patterns identified in the genetic correlation matrix. We chose to use confirmatory factor analysis, as opposed to exploratory factor analysis, to validate our model because of these previous studies supporting this factor structure. The first factor was defined by internalizing phenotypes: depression, GAD and the anxiety meta-analysis. The next three factors corresponded to the distinct blocks of alcohol use phenotypes we observed in our genome-wide genetic correlation analyses: PAU: AUD and AUDIT items 4, 5, 7, 8, and 10; quantity: AUDIT items 2, 3, and drinks per week; and frequency: AUDIT item 1 and alcohol intake frequency. We performed factor analysis in Genomic SEM of this four-factor model. We subsequently assessed fit of three- and two-factor solutions, in which we consolidated frequency and quantity into a consumption factor, and all alcohol use into a single factor, respectively. We assessed model fit with the following statistics: χ2, Akaike information criterion (AIC), comparative fit index (CFI), standardized root mean square residual (SRMR). With large sample sizes such as those typically used with Genomic SEM, the χ2 is typically significant, but CFI > 0.95 and SRMR < 0.08 indicate good fit. Lower values of AIC indicate better it when comparing models, and χ2 difference tests can be used to compare nested models (Hu & Bentler, 1998).
We next performed a multivariate GWAS within Genomic SEM by incorporating SNP effects into the model (Figure S1A). We then estimated the QSNP heterogeneity index (Grotzinger et al., 2019) for each SNP and factor by constructing models in which each SNP predicted three of the factors and the remaining factor’s individual indicators (Figure S1B). Significant QSNPs (p<5e-8), indicating the SNP does not operate solely via the factor, were removed from the latent factor summary statistics used in follow-up analyses as they are not representative of the broader factor. Resulting GWAS summary statistics were uploaded to FUMA, where we identified significant independent lead SNPs and loci using default parameters (Watanabe et al., 2017). We then estimated the effective sample size for each latent factor (Mallard, Linnér, et al., 2020) (Table S3).
Latent Causal Variable
We used latent causal variable analysis (O’Connor & Price, 2018) to test for partial or full genetic causality between factors. While latent causal variable analysis cannot detect simultaneous bidirectional causal relationships, it is less biased by horizontal pleiotropy and sample overlap than traditional Mendelian randomization (MR) methods. Latent causal variable analysis also has the added advantage that while methods such as generalized summary data-based MR (GSMR (Zhu et al., 2018)) only use “top” variants as instruments (SNPs with significant associations with both traits) and thus depend on a large number of genome-wide significant SNPs, latent causal variable analysis uses complete genome-wide data. We did not perform latent causal variable analysis between traits with non-significant genetic covariance (internalizing psychopathology and frequency), as recommended by O’Connor and Price (2018). As an exploratory follow-up, we tested causality using the top ten independent SNPs for each factor in inverse-variance weighted (IVW) MR analysis (Hemani et al., 2018), to see if restricting to top SNPs, rather than using whole genome data, might reduce noise and increase power.
Local Genetic Covariance
Next, we identified specific regions of the genome with local genetic covariances between the four latent factors using SUPERGNOVA, which is robust to sample overlap (Zhang et al., 2020). We estimated local genetic covariances in a total of 1,852 approximately independent LD genomic regions in which there were sufficient numbers of SNPs with GWAS summary statistics. To correct for the number of factor pairs and number of regions tested, we applied a Bonferroni-corrected significance threshold of p<0.05/(6 factor pairs x 1852 regions)<4.5x10−6.
Partitioned Genetic Covariance
Finally, we stratified the genetic covariance between pairs of latent factors into functional categories using GNOVA (Lu, Li, et al., 2017). First, we stratified the covariance by broad tissue type: brain, cardiovascular, epithelium, gastrointestinal, immune, muscular, and other from GenoSkyline-Plus annotations (Lu, Powles, et al., 2017); next, we stratified among annotations of brain region-specific expression derived from GTEx v6 (Lonsdale et al., 2013) using annotations from Finucane et al. (Finucane et al., 2018). Sample overlap correction, implemented within GNOVA, was used on the six factor pairs, as all have substantial sample overlap. We applied Bonferroni correction for 42 tests in the tissue analysis and 78 tests in the brain analysis, resulting in significance thresholds of ptissue<1.19x10−3 and pbrain<6.4x10−4.
SES Sensitivity Analyses
SES measures impact alcohol use and internalizing phenotypes, and are influenced themselves by genetic factors (Braucht, 1983; J. J. Lee et al., 2018; Marees et al., 2020, 2021). Because our focus is primarily on genetic effects on these phenotypes themselves, rather than genetic influences on SES measures that may impact alcohol use or internalizing traits, our primary analysis included SES measures (deprivation and educational attainment) as covariates. Following these analyses, we performed a sensitivity analysis by conducting GWASs of AUDIT items and depression and anxiety within the UK Biobank in which we did not control for deprivation and educational attainment. We identified DSM-V-like major depressive disorder and GAD cases within the UK Biobank using DSM-V criteria. We were not able to test the influence of SES covariates on traits from previously published GWAS summary statistics. We estimated genetic correlations and a similar Genomic SEM confirmatory factor analysis using these GWAS summary statistics and compared the genetic correlations and factor loadings to assess the influence SES GWAS covariates have on our results and conclusions.
RESULTS
Genome-Wide Genetic Correlations
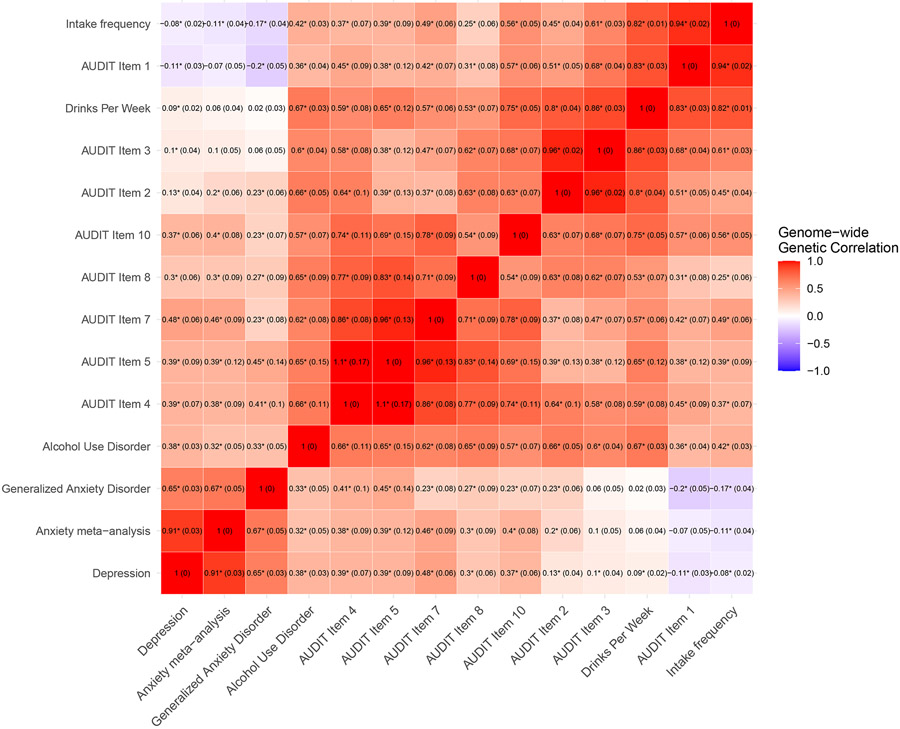
We observed significant, positive genome-wide LDSC-based genetic correlations between all pairs of three internalizing psychopathology phenotypes, and, separately, between all pairs of 11 alcohol use traits (Figure 1 and Table S4). Among pairs of alcohol use traits, items related to PAU, frequency and quantity were somewhat differentiated. PAU-related items (AUD and AUDIT items 4, 5, 7, 8, and 10) were all strongly correlated with one another (rg=0.57-1.10). Consumption-related items (drinks per week, AUDIT items 1-3, and intake frequency), were strongly positively genetically correlated (rg=0.60-0.97). In general, genetic correlations of PAU items were stronger with quantity measures (drinks per week, AUDIT items 2-3) than frequency measures (AUDIT item 1 and intake frequency), suggesting a differentiation between quantity and frequency measures and three alcohol use trait groupings.
Figure 1. Genome-wide genetic correlations (and standard errors) amongst 14 traits using LDSC.
Asterisks indicate significant (FDR<5%) genetic correlations. See Table S4 for complete numerical results.
PAU, quantity, and frequency traits were differentially genetically correlated with internalizing phenotypes (Figure 1). Internalizing phenotypes had moderately positive genetic correlations with PAU traits (rg=0.25-0.50), but weak and only sometimes significant positive correlations with quantity items (rg=0.02-0.21). In contrast, the internalizing phenotypes were either uncorrelated or weakly negatively genetically correlated with frequency phenotypes.
Genomic structural equation modeling of internalizing disorders and alcohol use
The four-factor confirmatory factor analysis model using Genomic SEM fit the data well (χ2(71)=638.155; AIC=706.155; CFI=0.9741; SRMR=0.081). Results indicated strong, significant loadings of all individual indicators, as well as significant correlations between several pairs of latent factors (Figure 2). Unsurprisingly, all alcohol use factors correlated strongly with each other (rg=0.60-0.82), with the weakest genetic correlation between PAU and Frequency (rg=0.60, SE = 0.04, p = 8.59x10−52). The Internalizing Psychopathology factor was correlated with PAU (rg=0.48, SE = 0.04, p = 1.28x10−39), but only weakly with Quantity (rg=0.09, SE = 0.03, p = 3.54x10−4) and Frequency (rg=−0.06, SE = 0.03, p = 2.02x10−2).
Figure 2. Genetic Relationships between internalizing disorders and alcohol use phenotypes.
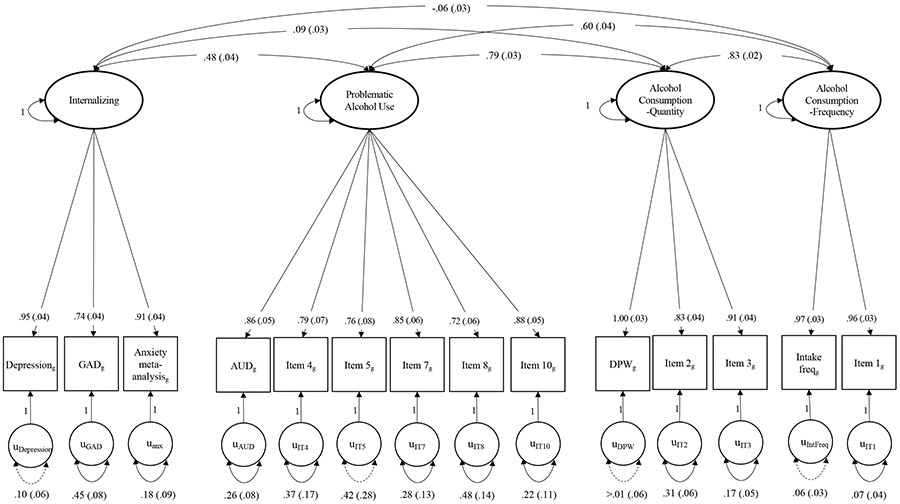
Path diagram of the final model structure. Parameter estimates are standardized with standard errors in parentheses. Dashed lines represent non-significant parameter estimates (p>0.05). Unit variance identification was used to present the model without SNP effects. Generalized anxiety disorder (GAD), alcohol use disorder (AUD), drinks per week (DPW).
We compared this model to a three-factor model (Figure S2; χ2(74)=1240.436; AIC=1302.436; CFI=0.9459; SRMR=0.091) and a two-factor model (Figure S3; χ2(76)=2074.206; AIC=2132.206; CFI=0.9073; SRMR=0.1670) in which the alcohol factors were collapsed. Both fit significantly worse than the four-factor model according to both chi-square difference tests and other fit metrics. Better fit of the four-factor model indicates that partitioning measures of alcohol use by PAU, frequency, and quantity better models the genetics of alcohol use than either not partitioning at all (Figure S3) or only partitioning into PAU and consumption factors (Figure S2).
Inclusion of SES variables as covariates in the GWAS did not noticeably impact the correlations or the factor structure, and loadings remained nearly identical (Figures S4-5). Therefore, below we present analyses which use the SES-controlled GWASs as they represent more trait-specific measures of genetic variance (Marees et al., 2021).
Multivariate GWAS of internalizing disorders and alcohol use
We observed no loci with heterogeneous effects for the Internalizing Psychopathology and PAU factors (all QSNP p>5e-8). We identified two significant independent QSNP loci (each containing one independent lead SNP per locus) for the Frequency factor (Table S5) and six independent lead SNPs in 4 independent QSNP loci for the Quantity factor (Table S6). We removed these SNPs from subsequent analyses. We identified 76 independent lead SNPs in 63 independent genomic loci for Internalizing Psychopathology (Figure S6A, Table S7), 22 independent lead SNPs in 20 genomic risk loci for PAU (Figure S6B, Table S8), 40 independent lead SNPs in 33 genomic risk loci for Quantity (Figure S6C, Table S9), and 46 independent lead SNPs in 40 genomic risk loci for Frequency (Figure S6D, Table S10).
Latent Causal Variable
Using the latent causal variable method, we did not detect a genetically causal relationship, either full or partial, in either direction, between any factors (Table S11). Results of exploratory follow-up analysis using IVW MR were similar and non-significant.
Local Genetic Covariance
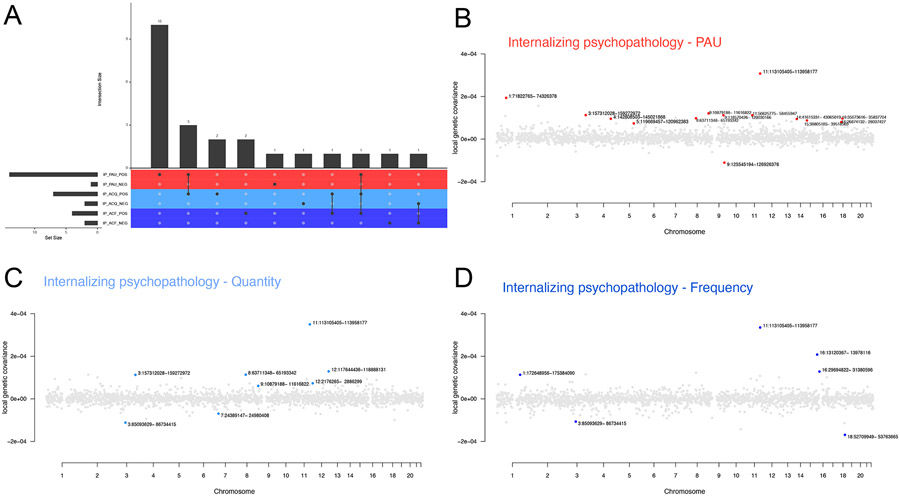
Among all factor pairs, we identified a total of 90 significant (p<4.5x10−6) local genetic covariance estimates between pairs of factors across 58 independent genomic loci out of 1852 throughout the genome (Tables S12-17). Of the significant covariances, 42 were between pairs of alcohol use factors (Figure S7, Tables S15-17).
We identified 28 regions with significant local genetic covariances between Internalizing Psychopathology and alcohol use factors (Figure 3, Tables S12-14). Fourteen genomic regions showed significant positive local genetic covariance between the PAU and Internalizing Psychopathology factors (Figure 3A-B, Table S12), 10 of which showed no significant genetic covariance between the Internalizing factor and Quantity or Frequency (Figure 3C-D, Table S13-14). We identified a total of eight and six regions with significant covariances between the Internalizing factor and Quantity and Frequency, respectively.
Figure 3. Local genetic covariances between internalizing psychopathology and alcohol use factors.
(A) Number of specific genomic regions with significant local genetic covariance between internalizing psychopathology and alcohol use traits. Intersect size indicates the number of correlated loci which overlap among pairs of internalizing psychopathology (IP) and alcohol use traits. Pairs are also separated to show positive and negative covariances. Colors represent the three alcohol use factors: problematic alcohol use (PAU; red), quantity of alcohol consumption (ACQ; light blue) and frequency of alcohol consumption (ACF; dark blue). (B-D) Manhattan-style plots showing local genetic covariance estimates between (B) internalizing psychopathology and PAU, (C) internalizing psychopathology and quantity of alcohol consumption, (D) internalizing psychopathology and frequency of alcohol consumption. Red, light blue and dark blue points indicate genomic regions with significant local genetic covariances between internalizing psychopathology and PAU, internalizing psychopathology and quantity of alcohol consumption and internalizing psychopathology and frequency of alcohol consumption, respectively. A p-value of 4.5x10−6 was used to determine significance.
Consistent with the strong positive genome-wide genetic correlation between Internalizing Psychopathology and PAU, we observed mostly positive local genetic covariances, identifying only one region (chr9:125,545,194–126,926,376) of negative genetic covariance (Figure 3A-B, Table S12). In contrast, identified regions between alcohol consumption factors and Internalizing Psychopathology contained both positive and negative covariances (two of eight and two of six were negative for Quantity and Frequency, respectively; Figure 3C-D, Tables S13-14), indicating that the direction of effect of this relationship was more variable (positive or negative) throughout the genome than that of the Internalizing factor-PAU relationship. Only one region (chr11:113,105,405–113,958,177) had statistically significant (and positive) local covariance for all pairs of Internalizing Psychopathology and alcohol use factors (Figure 3).
Genetic Covariance Stratified by Functional Annotations
For most tissues, the genetic covariance among pairs of factors partitioned by genes specifically expressed within those tissues was not significant (Table S18). However, genetic covariance annotated to genes specifically expressed within the brain was positive among pairs of alcohol use factors, as was genetic covariance of quantity and frequency at loci annotated to immune tissue specific expression (p<0.00119; Figure S8, Table S18). We found no significant tissue-specific genetic covariance between any pairs of internalizing psychopathology and alcohol use factors (Figure S8, Table S18).
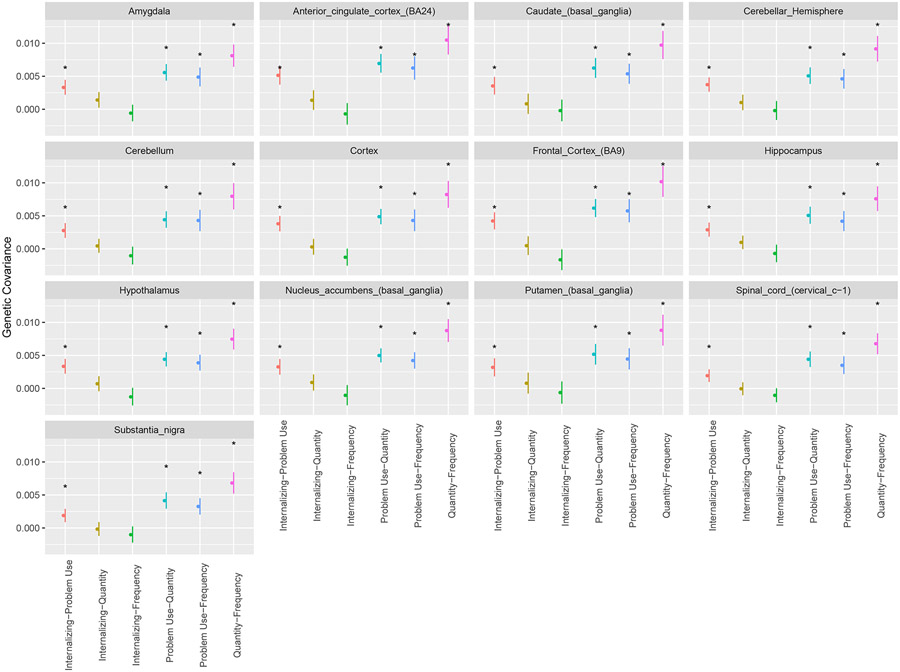
We found significant (p<0.00064), positive genetic covariance due to variants in genes specifically expressed in individual brain regions across all brain region annotations for pairs of alcohol use factors (Figure 4, Table S19). Interestingly, while the genetic covariance between internalizing psychopathology and PAU was not significant in the overall brain tissue annotation (Figure S8, Table S18), there was significant positive genetic covariance of internalizing psychopathology and PAU among all specific brain region annotations (all p<0.00064; Figure 4, Table S19), suggesting that nuanced differences in brain region expression are lost when brain tissue is treated as homogeneous. We found no significant genetic covariance in specific brain regions amongst pairs of internalizing psychopathology and quantity or frequency (Figure 4, Table S19).
Figure 4. Estimates of genetic covariance in specific brain regions.
Bars show 95% confidence intervals. Colors represent different pairs of the four factors: Internalizing-Problem Use (red: internalizing psychopathology and problematic alcohol use), Internalizing-Quantity (ochre: internalizing psychopathology and quantity of alcohol consumption), Internalizing-Frequency (green: internalizing psychopathology and frequency of alcohol consumption), Problem Use-Quantity (light blue: problematic alcohol use and quantity of alcohol consumption), Problem Use -Frequency (blue: problematic alcohol use and frequency of alcohol consumption), Quantity-Frequency (pink: quantity of alcohol consumption and frequency of alcohol consumption). Asterisks indicate significant (p<0.00064) stratified genetic covariance estimates.
DISCUSSION
We characterized the shared genetic architecture of internalizing psychopathology and alcohol use phenotypes, differentiating between problematic use and consumption, using multivariate analysis of four correlated latent factors. While problematic use showed a strong, positive genome-wide genetic correlation with internalizing psychopathology, quantity and frequency largely did not. At a genome-wide level, this pattern is consistent with previous reports (Sanchez-Roige et al., 2019; H. Zhou et al., 2020) and supports the hypothesis that alleles that increase genetic risk of internalizing psychopathology also increase risk of PAU, but not necessarily other aspects of alcohol use.
At the tissue level, analyses of partitioned genetic covariance demonstrated further evidence of distinct relationships of internalizing psychopathology with PAU and alcohol consumption. Our analyses revealed significant covariances across all 13 specific brain region annotations between the internalizing psychopathology and PAU factor, but not internalizing psychopathology and alcohol consumption. Notably, the covariance between internalizing psychopathology and PAU is not concentrated across any broad tissue categories, including the overall brain annotation. Gene expression among specific brain regions can vary greatly, and therefore a “whole brain” annotation aggregating all regions together may not be representative of important region-specific effects (Lonsdale et al., 2013). We also consider that the comparability of these annotations is limited, as the whole brain tissue annotation was derived from GenoSkyline and the specific brain tissue annotations were derived from GTEx. While all individual brain regions examined contributed to the PAU-Internalizing Psychopathology genetic covariance, we are currently unable to identify which among these may be particularly influential. We expect that growing reference panels of expression data (e.g., GTEx (Lonsdale et al., 2013) and PsychENCODE (Akbarian et al., 2015)) will improve the resolution of such analyses in the future. Our results concur with prior reports of individual brain regions contributing to both internalizing psychopathology and alcohol use (Becker et al., 2017; Gilpin et al., 2015; Hägele et al., 2015; Neupane, 2016; Ray et al., 2018), while adding evidence that these relationships could be unique to PAU.
While significant genome-wide and brain-specific genetic covariance of internalizing psychopathology with alcohol use were found only with PAU and not consumption, at the individual genomic region level we found significant local genetic covariances of internalizing psychopathology with all three alcohol use factors, though the specific regions of covariance and their directions of effect differed among alcohol use factors. This local genomic region scale of analysis localized differences that may underlie the distinct genetic relationships between internalizing psychopathology and the different dimensions of alcohol use. It also highlights the fact that fine-scale analyses can clarify complex relationships between traits, even if traits show no genome-wide correlation. Our analyses cannot distinguish between direct pleiotropic effects of loci on both traits, whether the causal effect of a locus on one trait is mediated by effects on the other, or if a region represents physically linked loci with separate direct effects. However, these findings do suggest specific regions and genes for functional follow-up analyses, for example, internalizing psychopathology was significantly negatively correlated with both quantity and frequency, but not PAU, within a chromosome 3 locus (85,093,629–86,734,415) which only contains one protein coding gene, CADM2 (cell adhesion molecule 2), signifying that CADM2 may influence internalizing psychopathology and alcohol consumption, but in different directions. CADM2 has been associated with various behavioral traits (Day et al., 2016; Ibrahim-Verbaas et al., 2016; Karlsson Linnér et al., 2019; Morris et al., 2019) including alcohol consumption (Clarke et al., 2017; Sanchez-Roige et al., 2019), however not with PAU (Kranzler et al., 2019; Sanchez-Roige et al., 2019; Walters et al., 2018; H. Zhou et al., 2020), consistent with our results. Internalizing psychopathology was also significantly negatively correlated with frequency at a chromosome 18 locus (52,709,949-53,763,665) and the only protein coding gene in this region, DCC (Deleted in Colorectal Carcinoma gene), is implicated in a variety of cognitive (Reynolds et al., 2018; Savage et al., 2018) and psychiatric traits (P. H. Lee et al., 2019; Manitt et al., 2013; Turley et al., 2018; Wray et al., 2018). Therefore, genetic components in this region, and possibly the DCC gene, might be implicated in the puzzling negative genetic relationship observed between internalizing psychopathology and frequency.
A locus at chr11:113,105,405–113,958,177 was the only region to show significant positive covariances between internalizing psychopathology and all three alcohol use factors. This locus contains variants associated with worry and neuroticism (Nagel et al., 2018), depression (Wray et al., 2018), PAU (H. Zhou et al., 2019), and AUDIT scores (Sanchez-Roige et al., 2019). Within this locus is DRD2, a gene which researchers hypothesize may moderate stress-induced ethanol consumption in mice (Chuang et al., 2020; Delis et al., 2013). Substances such as alcohol produce surges of the neurotransmitter dopamine, causing changes in neural connectivity that reinforce and reward the repeated use of the substance (Spanagel & Weiss, 1999). In addition to DRD2, this region also houses the genes NCAM1 (Neural Cell Adhesion Molecule 1), TTC12 (Tetratricopeptide Repeat Domain 12) and ANKK1 (Ankyrin Repeat And Kinase Domain Containing 1). Together, these genes make up the NTAD gene cluster, which may contribute to various psychiatric disorders as well as the comorbidity of psychiatric disorders (Mota et al., 2015; Yang et al., 2008). Variants associated with the NTAD gene cluster may therefore represent key genetic risk factors between internalizing psychopathology and alcohol use.
Finally, we isolated nine loci with significant positive covariances between internalizing psychopathology and PAU, but not quantity or frequency, contributing to the distinct genome-wide genetic correlations between internalizing psychopathology and alcohol use traits. In particular, one region (chr1:71,822,765-74,326,378) contains NEGR1 (Neuronal growth regulator 1), a gene with a regulatory region containing genetic variants associated with depression (Turley et al., 2018; Wang et al., 2020; Wray et al., 2018), PAU (H. Zhou et al., 2020) and several other psychiatric disorders (P. H. Lee et al., 2019). NEGR1, as well as DRD2 and DCC, all play roles in neural development (Finci et al., 2015; Singh et al., 2018; Todd, 1992), hinting that genetic components involved in neural development processes may be important to the multivariate genetic architectures of internalizing psychopathology and alcohol use traits. As sample sizes increase, we expect power to detect pleiotropic loci and causal effects to also improve. Our conclusions are also restricted by the lack of diverse ancestry in available summary statistics; this limits the generalizability of these findings to populations of European ancestry. We emphasize here, however, that the magnitude of these local pleiotropic effects is small, consistent with the polygenic nature of these traits, and that as larger and more diverse samples become available, the local structure of pleiotropic effects throughout the genome will become clearer.
Direct causal relationships are hypothesized to explain the comorbidity of AUDs and alcohol consumption with internalizing disorders, including self-medication and substance-induced anxiety or depression (Castillo-Carniglia et al., 2019; Polimanti et al., 2019; Stewart & Conrod, 2007). For example, some individuals with anxiety or depression use alcohol to alleviate symptoms that subsequently lead to AUDs, while in other cases, withdrawal after alcohol use leads to symptoms of depression and anxiety. These processes are not mutually exclusive and can lead to a self-reinforcing loop (i.e., through simultaneous bi-directional causal paths Smith & Randall, 2012). Risk factors for both AUDs and internalizing disorders include genetic and environmental causes. Our results revealed much stronger and positive genome-wide and local genetic covariances of internalizing psychopathology with PAU compared to consumption. This finding confirms that genetic variation contributes to the differential relationships between consumption and PAU and their relationships with internalizing traits, and hints at distinct causal relationships. However, our MR and latent causal variable analyses did not identify full or partial causality among any pairs of traits, including consumption metrics and PAU. LCV is conservative and limited in detecting simultaneous bi-directional causality (O’Connor & Price, 2018); therefore, we cannot rule out causal genetic relationships between these factors, and larger sample sizes may be needed for more powerful tests (Zhu et al., 2018) of causality.
While we cannot conclude with confidence causal relationships from these analyses, measures of consumption like quantity and frequency ultimately contribute to PAU, as PAU is unlikely to occur without heavy alcohol use. However, heavy use may not always lead to PAU, and should be interpreted within the context of cultural norms. Individuals demonstrating higher levels of these consumption behaviors may be perfectly in line with normative drinking and may not develop AUDs, or they may be drinking in contexts that are protective against disordered drinking. For example, having a glass of wine with dinner every night is not considered abnormal in western culture, and while this behavior would result in high frequency of drinking, it is not alone indicative of problematic alcohol use. Similarly, loneliness is positively genetically correlated with depression and alcohol dependence, but negatively genetically correlated with alcohol consumption (Abdellaoui et al., 2019), while increased participation in sports or social clubs is positively genetically correlated with alcohol intake (Day et al., 2018). In this context, we interpret the weak or null genome-wide and both positive and negative local genetic covariances between internalizing disorders and consumption as reflecting multiple, relationships of internalizing disorders with both normative and disordered drinking. On the contrary, the stronger and positive genome-wide and local genetic covariances of internalizing disorders and PAU reflect a stronger, potentially causal relationship among the disorders.
In summary, we identified shared genetic factors of internalizing psychopathology and alcohol use, distinguishing among PAU, quantity, and frequency, while also presenting a framework for examining comorbid relationships at multiple scales, from genome-wide patterns to functional annotations to individual genomic loci. The genetic components shared between internalizing psychopathology and PAU and internalizing psychopathology and alcohol consumption factors are separable at all of these scales. Future work will further evaluate directionality of causation in the alcohol-internalizing relationship and focus on functional characterization of identified genomic loci.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Matthew C. Keller, Naomi Wray, and Peter Visscher for helpful discussion. This work was supported by NIH MH100141 (PI: M.C. Keller), R01 AG046938 (PI: C.A. Reynolds & S. J Wadsworth), T32MH016880 (PI: John Hewitt), F32AA027435 (PI: E.C. Johnson), R01 DA042742 (PI: R.H.C. Palmer), R01 NS86933 (PI: C. A. Hoeffer), NIH U01 DA046413 (PIs: S.I Vrieze & N. P. Friedman), Linda Crnic Foundation, Le Jeune Foundation, and the Institute for Behavioral Genetics. This work utilized the Summit supercomputer, which is supported by the National Science Foundation (awards ACI-1532235 and ACI-1532236), the University of Colorado Boulder, and Colorado State University. The Summit supercomputer is a joint effort of the University of Colorado Boulder and Colorado State University.
This research has been conducted using the UK Biobank Resource (application number 1665). We thank the UK Biobank and the participants of the UK Biobank.
This work was conducted using the summary statistics from the Psychiatric Genomics Consortium’s Substance Use Disorders Working group. The Psychiatric Genomics Consortium's Substance Use Disorders (PGC-SUD) working group is supported by MH109532 with funding from NIMH and NIDA. We gratefully acknowledge prior support from NIAAA and thank all our contributing investigators and study participants who make this research possible.
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal.
Funding information:
NIH MH100141 (PI: M.C. Keller), R01 AG046938 (PI: C.A. Reynolds & S. J Wadsworth), T32MH016880 (PI: John Hewitt), F32AA027435 (PI: E.C. Johnson), R01 DA042742 (PI: R.H.C. Palmer), R01 NS86933 (PI: C. A. Hoeffer), NIH U01 DA046413 (PIs: S.I Vrieze & N. P. Friedman), Linda Crnic Foundation, Le Jeune Foundation, Institute for Behavioral Genetics
Footnotes
The authors declare that there is no conflict of interest.
DATA AVAILABILITY STATEMENT
All previously published summary statistics used in these analyses are publicly available. The AUDIT data used to conduct GWAS were derived from the UK Biobank (https://www.ukbiobank.ac.uk/) under application number 1665.
References
- Abdellaoui A, Sanchez-Roige S, Sealock J, Treur JL, Dennis J, Fontanillas P, Elson S, Nivard MG, Ip HF, Van Der Zee M, Baselmans BML, Hottenga JJ, Willemsen G, Mosing M, Lu Y, Pedersen NL, Denys D, Amin N, M Van Duijn C, … Boomsma DI (2019). Phenome-wide investigation of health outcomes associated with genetic predisposition to loneliness. Human Molecular Genetics, 28(22). 10.1093/hmg/ddz219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham G, & Inouye M (2014). Fast principal component analysis of large-scale genome-wide data. PLoS ONE, 9(4), e93766. 10.1371/journal.pone.0093766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH, Mill J, Nairn AC, Abyzov A, Pochareddy S, Prabhakar S, Weissman S, Sullivan PF, State MW, Weng Z, … Sestan N (2015). The PsychENCODE project. In Nature Neuroscience (Vol. 18, Issue 12). 10.1038/nn.4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfimova MV, Kondratyev NV, Tomyshev AS, Lebedeva IS, Lezheiko TV, Kaleda VG, Abramova LI, & Golimbet VE (2019). Effects of a GWAS-Supported Schizophrenia Variant in the DRD2 Locus on Disease Risk, Anhedonia, and Prefrontal Cortical Thickness. Journal of Molecular Neuroscience. 10.1007/s12031-019-01324-w [DOI] [PubMed] [Google Scholar]
- Becker A, Gerchen MF, Kirsch M, Ubl B, Subramaniapillai S, Diener C, Kuehner C, Kiefer F, & Kirsch P (2017). Frontostriatal connectivity during reward anticipation: A neurobiological mechanism cutting across alcohol use disorder and depression? Zeitschrift Fur Psychologie / Journal of Psychology. 10.1027/2151-2604/a000307 [DOI] [Google Scholar]
- Braucht GN (1983). How environments and persons combine to influence problem drinking. Current research issues. In Recent developments in alcoholism :an official publication of the American Medical Society on Alcoholism, the Research Society on Alcoholism, and the National Council on Alcoholism (Vol. 1). 10.1007/978-1-4613-3617-4_6 [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Duncan L, Perry JRB, Patterson N, Robinson EB, Daly MJ, Price AL, & Neale BM (2015). An atlas of genetic correlations across human diseases and traits. Nature Genetics, 47(11), 1236–1241. 10.1038/ng.3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns L, & Teesson M (2002). Alcohol use disorders comorbid with anxiety, depression and drug use disorders. Drug and Alcohol Dependence, 68(3), 299–307. 10.1016/s0376-8716(02)00220-x [DOI] [PubMed] [Google Scholar]
- Castillo-Carniglia A, Keyes KM, Hasin DS, & Cerdá M (2019). Psychiatric comorbidities in alcohol use disorder. In The Lancet Psychiatry (Vol. 6, Issue 12). 10.1016/S2215-0366(19)30222-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, & Lee JJ (2015). Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience, 4(1), s13742–015. 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HG, Aziz NNHA, Wong JH, Mustapha M, Abdullah JM, Idris Z, Abdullah Z, Alrafiah A, & Muthuraju S (2020). Role of toll-like receptor 4 antagonist Lipopolysaccharide-Rhodobacter sphaeroides on acute stress-induced voluntary ethanol preference and drinking behaviour: In vivo Swiss Albino mouse model. European Neuropsychopharmacology, 1–14. 10.1016/j.euroneuro.2019.12.121 [DOI] [PubMed] [Google Scholar]
- Clarke TK, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, Murray AD, Smith BH, Campbell A, Hayward C, Porteous DJ, Deary IJ, & McIntosh AM (2017). Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK biobank (N=112117). Molecular Psychiatry, 22(10), 1376–1384. 10.1038/mp.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert SMC, Funkhouser SA, Johnson EC, Hoeffer C, Ehringer MA, Evans LM, Colbert S, & Evans L (2020). Differential shared genetic influences on anxiety with problematic alcohol use compared to alcohol consumption. 10.1101/2020.08.21.20179374 [DOI] [Google Scholar]
- Day FR, Helgason H, Chasman DI, Rose LM, Loh PR, Scott RA, Helgason A, Kong A, Masson G, Magnusson OT, Gudbjartsson D, Thorsteinsdottir U, Buring JE, Ridker PM, Sulem P, Stefansson K, Ong KK, & Perry JRB (2016). Physical and neurobehavioral determinants of reproductive onset and success. Nature Genetics. 10.1038/ng.3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Ong KK, & Perry JRB (2018). Elucidating the genetic basis of social interaction and isolation. Nature Communications, 9(1). 10.1038/s41467-018-04930-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delis F, Thanos PK, Rombola C, Rosko L, Grandy D, Wang GJ, & Volkow ND (2013). Chronic mild stress increases alcohol intake in mice with low dopamine D2 receptor levels. Behavioral Neuroscience, 127(1), 95–105. 10.1037/a0030750 [DOI] [PubMed] [Google Scholar]
- Enoch MA, Schwartz L, Albaugh B, Virkkunen M, & Goldman D (2006). Dimensional anxiety mediates linkage of GABRA2 haplotypes with alcoholism. American Journal of Medical Genetics, Part B: Neuropsychiatric Genetics, 141(6), 599–607. 10.1002/ajmg.b.30336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finci L, Zhang Y, Meijers R, & Wang JH (2015). Signaling mechanism of the netrin-1 receptor DCC in axon guidance. In Progress in Biophysics and Molecular Biology. 10.1016/j.pbiomolbio.2015.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, Anttila V, Xu H, Zang C, Farh K, Ripke S, Day FR, Purcell S, Stahl E, Lindstrom S, Perry JRB, Okada Y, Raychaudhuri S, Daly MJ, … Price AL (2015). Partitioning heritability by functional annotation using genome-wide association summary statistics. Nature Genetics, 47(11), 1228–1235. 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, Gazal S, Loh PR, Lareau C, Shoresh N, Genovese G, Saunders A, Macosko E, Pollack S, Perry JRB, Buenrostro JD, Bernstein BE, Raychaudhuri S, McCarroll S, … Price AL (2018). Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nature Genetics, 50(4), 621–629. 10.1038/s41588-018-0081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, Klei LL, Kramer R, Pinto D, Gümüş ZH, Cicek AE, Dang KK, Browne A, Lu C, Xie L, … Sklar P (2016). Gene expression elucidates functional impact of polygenic risk for schizophrenia. In Nature Neuroscience (Vol. 19, Issue 11). 10.1038/nn.4399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin NW, Herman MA, & Roberto M (2015). The Central Amygdala as an Integrative Hub for Anxiety and Alcohol Use Disorders. Biological Psychiatry, 77(10), 859–869. 10.1016/j.biopsych.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzinger AD, Mallard TT, Akingbuwa WA, Ip HF, Adams MJ, Lewis CM, McIntosh AM, Grove J, Dalsgaard S, Peter-Lesch K, Strom N, Meier SM, Mattheisen M, Børglum AD, Mors O, Breen G, iPSYCH, PGC, T. S. and O. C. D. W. G. of the, PGC, B. D. W. G. of the, … Nivard MG (2020). Genetic Architecture of 11 Major Psychiatric Disorders at Biobehavioral, Functional Genomic, and Molecular Genetic Levels of Analysis. MedRxiv. 10.1101/2020.09.22.20196089v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzinger AD, Rhemtulla M, de Vlaming R, Ritchie SJ, Mallard TT, Hill WD, Ip HF, Marioni RE, McIntosh AM, Deary IJ, Koellinger PD, Harden KP, Nivard MG, & Tucker-Drob EM (2019). Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nature Human Behaviour. 10.1038/s41562-019-0566-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Mancuso N, Won H, Kousi M, Finucane HK, Reshef Y, Song L, Safi A, McCarroll S, Neale BM, Ophoff RA, O’Donovan MC, Crawford GE, Geschwind DH, Katsanis N, Sullivan PF, Pasaniuc B, & Price AL (2018). Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nature Genetics. 10.1038/s41588-018-0092-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavson DE, Friedman NP, Fontanillas P, Elson SL, Palmer AA, & Sanchez-Roige S (2020). The Latent Genetic Structure of Impulsivity and Its Relation to Internalizing Psychopathology. Psychological Science, 31(8), 1025–1035. 10.1177/0956797620938160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hägele C, Schlagenhauf F, Rapp M, Sterzer P, Beck A, Bermpohl F, Stoy M, Ströhle A, Wittchen HU, Dolan RJ, & Heinz A (2015). Dimensional psychiatry: Reward dysfunction and depressive mood across psychiatric disorders. Psychopharmacology. 10.1007/s00213-014-3662-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, Stinson FS, Ogburn E, & Grant BF (2007). Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: Results from the national epidemiologic survey on alcohol and related conditions. Archives of General Psychiatry, 64(7), 830–842. 10.1001/archpsyc.64.7.830 [DOI] [PubMed] [Google Scholar]
- Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, & Haycock PC (2018). The MR-base platform supports systematic causal inference across the human phenome. ELife, 7. 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himle JA, & Hill EM (1991). Alcohol abuse and the anxiety disorders: Evidence from the epidemiologic catchment area survey. Journal of Anxiety Disorders, 5(3), 237–245. 10.1016/0887-6185(91)90004-D [DOI] [Google Scholar]
- Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M, Coleman JRI, Hagenaars SP, Ward J, Wigmore EM, Alloza C, Shen X, Barbu MC, Xu EY, Whalley HC, Marioni RE, Porteous DJ, Davies G, Deary IJ, … McIntosh AM (2019). Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nature Neuroscience. 10.1038/s41593-018-0326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LT, & Bentler PM (1998). Fit Indices in Covariance Structure Modeling: Sensitivity to Underparameterized Model Misspecification. Psychological Methods, 3(4). 10.1037/1082-989X.3.4.424 [DOI] [Google Scholar]
- Ibrahim-Verbaas CA, Bressler J, Debette S, Schuur M, Smith AV, Bis JC, Davies G, Trompet S, Smith JA, Wolf C, Chibnik LB, Liu Y, Vitart V, Kirin M, Petrovic K, Polasek O, Zgaga L, Fawns-Ritchie C, Hoffmann P, … Mosley TH (2016). GWAS for executive function and processing speed suggests involvement of the CADM2 gene. Molecular Psychiatry. 10.1038/mp.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaalund SS, Newburn EN, Ye T, Tao R, Li C, Deep-Soboslay A, Herman MM, Hyde TM, Weinberger DR, Lipska BK, & Kleinman JE (2014). Contrasting changes in DRD1 and DRD2 splice variant expression in schizophrenia and affective disorders, and associations with SNPs in postmortem brain. Molecular Psychiatry. 10.1038/mp.2013.165 [DOI] [PubMed] [Google Scholar]
- Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, Lebreton M, Tino SP, Abdellaoui A, Hammerschlag AR, Nivard MG, Okbay A, Rietveld CA, Timshel PN, Trzaskowski M, Vlaming R. de, Zünd CL, Bao Y, Buzdugan L, … Wagner GG (2019). Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nature Genetics. 10.1038/s41588-018-0309-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Aggen SH, Prescott CA, Crabbe J, & Neale MC (2012). Evidence for multiple genetic factors underlying the DSM-IV criteria for alcohol dependence. Molecular Psychiatry, 17(12). 10.1038/mp.2011.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler, Kenneth S, Gardner CO, Gatz M, & Pedersen NL (2007). The sources of co-morbidity between major depression and generalized anxiety disorder in a Swedish national twin sample. Psychological Medicine, 37(3). 10.1017/S0033291706009135 [DOI] [PubMed] [Google Scholar]
- Kendler, Kenneth S, Neale MC, Kessler RC, Heath AC, & Eaves LJ (1992). Major Depression and Generalized Anxiety Disorder: Same Genes, (Partly) Different Environments? Archives of General Psychiatry, 49(9). 10.1001/archpsyc.1992.01820090044008 [DOI] [PubMed] [Google Scholar]
- Kessler RC, Crum RM, Warner LA, Nelson CB, Schulenberg J, & Anthony JC (1997). Lifetime co-occurrence of DSM-III-R alcohol abuse and dependence with other psychiatric disorders in the national comorbidity survey. Archives of General Psychiatry, 54(4), 313–321. 10.1001/archpsyc.1997.01830160031005 [DOI] [PubMed] [Google Scholar]
- Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, Tsao PS, Klarin D, Baras A, Reid J, Overton J, Rader DJ, Cheng Z, Tate JP, Becker WC, Concato J, Xu K, Polimanti R, Zhao H, & Gelernter J (2019). Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nature Communications, 10(1), 1–11. 10.1038/s41467-019-09480-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger RF (1999). The structure of common mental disorders. Archives of General Psychiatry. 10.1001/archpsyc.56.10.921 [DOI] [PubMed] [Google Scholar]
- Kushner MG, Sher KJ, & Erickson DJ (1999). Prospective analysis of the relation between DSM-III anxiety disorders and alcohol use disorders. American Journal of Psychiatry, 156(5), 723–732. 10.1176/ajp.156.5.723 [DOI] [PubMed] [Google Scholar]
- Kushner MG, Wall MM, Krueger RF, Sher KJ, Maurer E, Thuras P, & Lee S (2012). Alcohol Dependence is Related to Overall Internalizing Psychopathology Load Rather than to Particular Internalizing Disorders: Evidence from a National Sample. Alcoholism: Clinical and Experimental Research, 36(2), 325–331. 10.1111/j.1530-0277.2011.01604.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, Nguyen-Viet TA, Bowers P, Sidorenko J, Karlsson Linnér R, Fontana MA, Kundu T, Lee C, Li H, Li R, Royer R, Timshel PN, Walters RK, Willoughby EA, … Turley P (2018). Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nature Genetics. 10.1038/s41588-018-0147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, Anttila V, Won H, Feng YCA, Rosenthal J, Zhu Z, Tucker-Drob EM, Nivard MG, Grotzinger AD, Posthuma D, Wang MMJ, Yu D, Stahl EA, Walters RK, Anney RJL, Duncan LE, Ge T, Adolfsson R, Banaschewski T, … Smoller JW (2019). Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell, 179(7), 1469–1482.e11. 10.1016/j.cell.2019.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey DF, Gelernter J, Polimanti R, Zhou H, Cheng Z, Aslan M, Quaden R, Concato J, Radhakrishnan K, Bryois J, Sullivan PF, & Stein MB (2020). Reproducible Genetic Risk Loci for Anxiety: Results From ~200,000 Participants in the Million Veteran Program. The American Journal of Psychiatry. 10.1176/appi.ajp.2019.19030256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan X, Agee M, Alipanahi B, Auton A, Bell RK, Bryc K, Elson SL, Fontanillas P, Furlotte NA, … Vrieze S (2019). Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nature Genetics, 51, 237–244. 10.1038/s41588-018-0307-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P-R, Kichaev G, Gazal S, Schoech AP, & Price AL (2018). Mixed-model association for biobank-scale datasets. Nature Genetics, 50(7), 906–908. 10.1038/s41588-018-0144-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P-R, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ, Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B, Patterson N, & Price AL (2015). Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nature Genetics, 47, 284–290. 10.1038/ng.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, Hasz R, Walters G, Garcia F, Young N, Foster B, Moser M, Karasik E, Gillard B, Ramsey K, Sullivan S, Bridge J, Magazine H, Syron J, … Moore HF (2013). The Genotype-Tissue Expression (GTEx) project. Nature Genetics, 45, 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Li B, Ou D, Erlendsdottir M, Powles RL, Jiang T, Hu Y, Chang D, Jin C, Dai W, He Q, Liu Z, Mukherjee S, Crane PK, & Zhao H (2017). A Powerful Approach to Estimating Annotation-Stratified Genetic Covariance via GWAS Summary Statistics. American Journal of Human Genetics, 101(6), 939–964. 10.1016/j.ajhg.2017.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Powles RL, Abdallah S, Ou D, Wang Q, Hu Y, Lu Y, Liu W, Li B, Mukherjee S, Crane PK, & Zhao H (2017). Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease. PLoS Genetics, 13(7), 1–24. 10.1371/journal.pgen.1006933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard TT, Linnér RK, Grotzinger AD, Sanchez-roige S, Okbay A, De Vlaming R, Meddens SFW Working, B. D., Consortium, G., Palmer AA, & Davis LK (2020). Multivariate GWAS of psychiatric disorders and their cardinal symptoms reveal two dimensions of cross-cutting genetic liabilities. 1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard TT, Savage JE, Johnson EC, Huang Y, Edwards AC, Hottenga JJ, Grotzinger AD, Gustavson DE, Jennings MV, Anokhin A, Dick DM, Edenberg HJ, Kramer JR, Lai D, Meyers JL, Pandey AK, Harden KP, Nivard MG, de Geus EJC, … Sanchez-Roige S (2020). Multivariate GWAS elucidates the genetic architecture of alcohol consumption and misuse, corrects biases, and reveals novel associations with disease. BioRxiv. [Google Scholar]
- Manitt C, Eng C, Pokinko M, Ryan RT, Torres-Berrío A, Lopez JP, Yogendran SV, Daubaras MJJ, Grant A, Schmidt ERE, Tronche F, Krimpenfort P, Cooper HM, Pasterkamp RJ, Kolb B, Turecki G, Wong TP, Nestler EJ, Giros B, & Flores C (2013). Dcc orchestrates the development of the prefrontal cortex during adolescence and is altered in psychiatric patients. Translational Psychiatry. 10.1038/tp.2013.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marees AT, Smit DJA, Abdellaoui A, Nivard MG, van den Brink W, Denys D, Galama TJ, Verweij KJH, & Derks EM (2021). Genetic correlates of socio-economic status influence the pattern of shared heritability across mental health traits. Nature Human Behaviour. 10.1038/s41562-021-01053-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marees AT, Smit DJA, Ong JS, Macgregor S, An J, Denys D, Vorspan F, Van Den Brink W, & Derks EM (2020). Potential influence of socioeconomic status on genetic correlations between alcohol consumption measures and mental health. Psychological Medicine, 50(3). 10.1017/S0033291719000357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier SM, Trontti K, Purves KL, Als TD, Grove J, Laine M, Pedersen MG, Bybjerg-Grauholm J, Bækved-Hansen M, Sokolowska E, Mortensen PB, Hougaard DM, Werge T, Nordentoft M, Breen G, Børglum AD, Eley TC, Hovatta I, Mattheisen M, & Mors O (2019). Genetic Variants Associated with Anxiety and Stress-Related Disorders: A Genome-Wide Association Study and Mouse-Model Study. JAMA Psychiatry. 10.1001/jamapsychiatry.2019.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, Stevens D, & Fenton B (1996). Comorbidity of alcoholism and anxiety disorders the role of family studies. Alcohol Research and Health, 20(2), 100–106. [PMC free article] [PubMed] [Google Scholar]
- Morris J, Bailey MES, Baldassarre D, Cullen B, de Faire U, Ferguson A, Gigante B, Giral P, Goel A, Graham N, Hamsten A, Humphries SE, Johnston KJA, Lyall DM, Lyall LM, Sennblad B, Silveira A, Smit AJ, Tremoli E, … Strawbridge RJ (2019). Genetic variation in CADM2 as a link between psychological traits and obesity. Scientific Reports. 10.1038/s41598-019-43861-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota NR, Rovaris DL, Kappel DB, Picon FA, Vitola ES, Salgado CAI, Karam RG, Rohde LA, Grevet EH, & Bau CHD (2015). NCAM1-TTC12-ANKK1-DRD2 gene cluster and the clinical and genetic heterogeneity of adults with ADHD. American Journal of Medical Genetics, Part B: Neuropsychiatric Genetics, 168(6), 433–444. 10.1002/ajmg.b.32317 [DOI] [PubMed] [Google Scholar]
- Nagel M, Jansen PR, Stringer S, Watanabe K, De Leeuw CA, Bryois J, Savage JE, Hammerschlag AR, Skene NG, Muñoz-Manchado AB, Agee M, Alipanahi B, Auton A, Bell RK, Bryc K, Elson SL, Fontanillas P, Furlotte NA, Hinds DA, … Posthuma D (2018). Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nature Genetics, 50(7), 920–927. 10.1038/s41588-018-0151-7 [DOI] [PubMed] [Google Scholar]
- Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y, Floratos A, Sham PC, Li MJ, Wang J, Cardon LR, Whittaker JC, & Sanseau P (2015). The support of human genetic evidence for approved drug indications. Nature Genetics, 47(8). 10.1038/ng.3314 [DOI] [PubMed] [Google Scholar]
- Neupane SP (2016). Neuroimmune interface in the comorbidity between alcohol use disorder and major depression. In Frontiers in Immunology. 10.3389/fimmu.2016.00655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI, Foroud T, Flury L, Meyer ET, & Wiegand R (2002). Is there a genetic relationship between alcoholism and depression? Alcohol Research and Health. [PMC free article] [PubMed] [Google Scholar]
- O’Connor LJ, & Price AL (2018). Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nature Genetics. 10.1038/s41588-018-0255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Yoshida S, Graham RR, Manoharan A, Ortmann W, Bhangale T, Denny JC, Carroll RJ, Eyler AE, Greenberg JD, Kremer JM, … Plenge RM (2014). Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature, 506(7488). 10.1038/nature12873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polimanti R, Peterson RE, Ong JS, MacGregor S, Edwards AC, Clarke TK, Frank J, Gerring Z, Gillespie NA, Lind PA, Maes HH, Martin NG, Mbarek H, Medland SE, Streit F, Agrawal A, Edenberg HJ, Kendler KS, Lewis CM, … Derks EM (2019). Evidence of causal effect of major depression on alcohol dependence: Findings from the psychiatric genomics consortium. Psychological Medicine, 49(7), 1218–1226. 10.1017/S0033291719000667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves KL, Coleman JRI, Meier SM, Rayner C, Davis KAS, Cheesman R, Bækvad-Hansen M, Børglum AD, Wan Cho S, Jürgen Deckert J, Gaspar HA, Bybjerg-Grauholm J, Hettema JM, Hotopf M, Hougaard D, Hübel C, Kan C, McIntosh AM, Mors O, … Eley TC (2019). A major role for common genetic variation in anxiety disorders. Molecular Psychiatry, 1–12. 10.1038/s41380-019-0559-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray MH, Hanlon E, & McDannald MA (2018). Lateral orbitofrontal cortex partitions mechanisms for fear regulation and alcohol consumption. PLoS ONE, 13(6), e0198043. 10.1371/journal.pone.0198043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier DA, Farmer ME, Rae DS, Locke BZ, Keith SJ, Judd LL, & Goodwin FK (1990). Comorbidity of Mental Disorders With Alcohol and Other Drug Abuse: Results From the Epidemiologic Catchment Area (ECA) Study. JAMA: The Journal of the American Medical Association, 264(19), 2511–2518. 10.1001/jama.1990.03450190043026 [DOI] [PubMed] [Google Scholar]
- Reynolds LM, Pokinko M, Torres-Berrío A, Cuesta S, Lambert LC, Del Cid Pellitero E, Wodzinski M, Manitt C, Krimpenfort P, Kolb B, & Flores C (2018). DCC Receptors Drive Prefrontal Cortex Maturation by Determining Dopamine Axon Targeting in Adolescence. Biological Psychiatry. 10.1016/j.biopsych.2017.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, Adams MJ, Howard DM, Edenberg HJ, Davies G, Crist RC, Deary IJ, McIntosh AM, Clarke TK, Elson L, Fontanillas P, Furlotte NA, Hinds DA, Huber KE, Kleinman A, Litterman NK, … Wilson CH (2019). Genome-wide association study meta-analysis of the alcohol use disorders identification test (AUDIT) in two population-based cohorts. American Journal of Psychiatry, 176(2), 107–118. 10.1176/appi.ajp.2018.18040369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders JB, Aasland OG, Babor TF, de la Fuente JR, & Grant M (1993). Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on Early Detection of Persons with Harmful Alcohol Consumption—II. Addiction, 791–804. 10.1163/9789004377912_003 [DOI] [PubMed] [Google Scholar]
- Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, De Leeuw CA, Nagel M, Awasthi S, Barr PB, Coleman JRI, Grasby KL, Hammerschlag AR, Kaminski JA, Karlsson R, Krapohl E, Lam M, Nygaard M, Reynolds CA, Trampush JW, … Posthuma D (2018). Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nature Genetics. 10.1038/s41588-018-0152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, Ruggeri B, Amin N, Jia T, Preis S, Lepe MS, Akira S, Barbieri C, Baumeister S, Cauchi S, Clarke TK, Enroth S, Fischer K, Hällfors J, … Elliott P (2016). KLB is associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. Proceedings of the National Academy of Sciences of the United States of America. 10.1073/pnas.1611243113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, De Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, Rose SA, Handsaker RE, Daly MJ, Carroll MC, Stevens B, & McCarroll SA (2016). Schizophrenia risk from complex variation of complement component 4. Nature. 10.1038/nature16549 [DOI] [PubMed] [Google Scholar]
- Selvaggi P, Pergola G, Gelao B, Di Carlo P, Nettis MA, Amico G, Fazio L, Rampino A, Sambataro F, Blasi G, & Bertolino A (2019). Genetic variation of a DRD2 co-expression network is associated with changes in prefrontal function after D2 receptors stimulation. Cerebral Cortex. 10.1093/cercor/bhy022 [DOI] [PubMed] [Google Scholar]
- Singh K, Loreth D, Pöttker B, Hefti K, Innos J, Schwald K, Hengstler H, Menzel L, Sommer CJ, Radyushkin K, Kretz O, Philips MA, Haas CA, Frauenknecht K, Lilleväli K, Heimrich B, Vasar E, & Schäfer MKE (2018). Neuronal growth and behavioral alterations in mice deficient for the psychiatric disease-associated negr1 gene. Frontiers in Molecular Neuroscience. 10.3389/fnmol.2018.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JP, & Randall CL (2012). Anxiety and alcohol use disorders: Comorbidity and treatment considerations. Alcohol Research: Current Reviews, 34(4). [PMC free article] [PubMed] [Google Scholar]
- So HC, Chau CKL, Chiu WT, Ho KS, Lo CP, Yim SHY, & Sham PC (2017). Analysis of genome-wide association data highlights candidates for drug repositioning in psychiatry. Nature Neuroscience. 10.1038/nn.4618 [DOI] [PubMed] [Google Scholar]
- Spanagel R, & Weiss F (1999). The dopamine hypothesis of reward: Past and current status. In Trends in Neurosciences (Vol. 22, Issue 11). 10.1016/S0166-2236(99)01447-2 [DOI] [PubMed] [Google Scholar]
- Stewart SH, & Conrod PJ (2007). Anxiety Disorder and Substance Use Disorder Co-Morbidity: Common Themes and Future Directions. In Anxiety and Substance Use Disorders. 10.1007/978-0-387-74290-8_13 [DOI] [Google Scholar]
- Todd RD (1992). Neural development is regulated by classical neurotransmitters: Dopamine D2 receptor stimulation enhances neurite outgrowth. Biological Psychiatry. 10.1016/0006-3223(92)90311-M [DOI] [PubMed] [Google Scholar]
- Torvik FA, Rosenström TH, Gustavson K, Ystrom E, Kendler KS, Bramness JG, Czajkowski N, & Reichborn-Kjennerud T (2019). Explaining the association between anxiety disorders and alcohol use disorder: A twin study. Depression and Anxiety, 36(6), 522–532. 10.1002/da.22886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, Nguyen-Viet TA, Wedow R, Zacher M, Furlotte NA, Magnusson P, Oskarsson S, Johannesson M, Visscher PM, Laibson D, Cesarini D, Neale BM, Benjamin DJ, Agee M, … Pitts SJ (2018). Multi-trait analysis of genome-wide association summary statistics using MTAG. Nature Genetics. 10.1038/s41588-017-0009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, Aliev F, Bacanu SA, Batzler A, Bertelsen S, Biernacka JM, Bigdeli TB, Chen LS, Clarke TK, Chou YL, Degenhardt F, Docherty AR, Edwards AC, Fontanillas P, … Agrawal A (2018). Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nature Neuroscience, 21(12), 1656–1669. 10.1038/s41593-018-0275-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cheng W, Zhu J, Yin H, Chang S, Yue W, & Yu H (2020). Integrating genome-wide association study and expression quantitative trait loci data identifies NEGR1 as a causal risk gene of major depression disorder. Journal of Affective Disorders. 10.1016/j.jad.2019.11.116 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Taskesen E, Van Bochoven A, & Posthuma D (2017). Functional mapping and annotation of genetic associations with FUMA. Nature Communications. 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, Adams MJ, Agerbo E, Air TM, Andlauer TMF, Bacanu SA, Bækvad-Hansen M, Beekman AFT, Bigdeli TB, Binder EB, Blackwood DRH, Bryois J, Buttenschøn HN, Bybjerg-Grauholm J, … Sullivan PF (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature Genetics, 50(5), 668–681. 10.1038/s41588-018-0090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue A, Jiang L, Zhu Z, Wray NR, Visscher PM, Zeng J, & Yang J (2021). Genome-wide analyses of behavioural traits are subject to bias by misreports and longitudinal changes. Nature Communications. 10.1038/s41467-020-20237-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, & Gelernter J (2008). Haplotypic variants in DRD2, ANKK1, TTC12, and NCAM1 are associated with comorbid alcohol and drug dependence. Alcoholism: Clinical and Experimental Research, 32(12), 2117–2127. 10.1111/j.1530-0277.2008.00800.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu Q, Ye Y, Huang K, Liu W, Wu Y, Zhong X, Li B, Yu Z, Travers BG, Werling DM, Li JJ, & Zhao H (2020). Local genetic correlation analysis reveals heterogeneous etiologic sharing of complex traits. Bio Rxiv, 1–33. 10.1101/2020.05.08.084475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Sealock JM, Sanchez-Roige S, Clarke T-K, Levey D, Cheng Z, Li B, Polimanti R, Kember RL, Smith RV, Thygesen JH, Morgan MY, Atkinson SR, Thursz MR, Nyegaard M, Mattheisen M, Børglum AD, Johnson EC, Program, the V. M. V., … Gelernter J (2019). Meta-analysis of problematic alcohol use in 435,563 individuals identifies 29 risk variants and yields insights into biology, pleiotropy and causality. Bio Rxiv. 10.1101/738088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Sealock JM, Sanchez-Roige S, Clarke TK, Levey DF, Cheng Z, Li B, Polimanti R, Kember RL, Smith RV, Thygesen JH, Morgan MY, Atkinson SR, Thursz MR, Nyegaard M, Mattheisen M, Børglum AD, Johnson EC, Justice AC, … Gelernter J (2020). Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nature Neuroscience. 10.1038/s41593-020-0643-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Kaiser T, Monteiro P, Zhang X, Van der Goes MS, Wang D, Barak B, Zeng M, Li C, Lu C, Wells M, Amaya A, Nguyen S, Lewis M, Sanjana N, Zhou Y, Zhang M, Zhang F, Fu Z, & Feng G (2016). Mice with Shank3 Mutations Associated with ASD and Schizophrenia Display Both Shared and Distinct Defects. Neuron. 10.1016/j.neuron.2015.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, Robinson MR, McGrath JJ, Visscher PM, Wray NR, & Yang J (2018). Causal associations between risk factors and common diseases inferred from GWAS summary data. Nature Communications, 9(1), 1–12. 10.1038/s41467-017-02317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All previously published summary statistics used in these analyses are publicly available. The AUDIT data used to conduct GWAS were derived from the UK Biobank (https://www.ukbiobank.ac.uk/) under application number 1665.