Abstract
Purpose:
The ClinGen Variant Curation Expert Panels (VCEPs) provide disease-specific rules for accurate variant interpretation. Using the hearing loss-specific ACMG/AMP guidelines, the Hearing Loss VCEP (HL VCEP) illustrates the utility of expert specifications in variant interpretation.
Methods:
A total of 157 variants across nine HL genes, previously submitted to ClinVar, were curated by the HL VCEP. The curation process involved collecting published and unpublished data for each variant by biocurators, followed by bi-monthly meetings of an expert curation subgroup that reviewed all evidence and applied the HL-specific ACMG/AMP guidelines to reach a final classification.
Results:
Before expert curation, 75% (117/157) of variants had single or multiple VUS submissions (17/157) or had conflicting interpretations in ClinVar (100/157). After applying the HL-specific ACMG/AMP guidelines, 24% (4/17) of VUS and 69% (69/100) of discordant variants were resolved into Benign (B), Likely Benign (LB), Likely Pathogenic (LP), or Pathogenic (P). Overall, 70% (109/157) variants had unambiguous classifications (B, LB, LP, P). We quantify the contribution of the HL-specified ACMG/AMP codes to variant classification.
Conclusion:
Expert specification and application of the HL-specific ACMG/AMP guidelines effectively resolved discordant interpretations in ClinVar. This study highlights the utility of ClinGen VCEPs in supporting more consistent clinical variant interpretation.
Introduction
Hearing loss (HL) is the most common congenital sensory condition, with approximately 50% of affected individuals having an identifiable genetic etiology1. HL is a heterogeneous condition with more than 100 genes underlying nonsyndromic HL, and over 400 genes implicated in syndromic forms of deafness2,3. A clinical genetics evaluation is recommended as part of a standard of care diagnostic work-up, and results from genetic testing can often inform clinical management, especially if a genetic syndrome is identified before the onset of additional clinical manifestations1,4. As an expert panel of the Clinical Genome Resource (ClinGen), a National Institute of Health-funded resource focused on defining the clinical validity of gene and variant contributions to disease, the Hearing Loss Clinical Domain Working Group (HL-CDWG) (https://www.clinicalgenome.org/working-groups/clinical-domain/hearing-loss/) has worked to evaluate gene-disease relationships and standardize variant interpretation in hereditary HL5,6,7.
Interpretation of variants’ clinical significance is a rigorous process that involves collating and analyzing available literature and evidence, followed by a formal classification based on this evidence. The ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) archives and aggregates variant interpretations from various submitters and indicates whether the submitted interpretations are concordant or discordant. Data sharing through ClinVar provides an invaluable opportunity to identify classification differences and to collaborate with submitters to resolve these discrepancies. A pilot study conducted by four clinical laboratories showed that 87.2% (211/242) of discordant variants were resolved when the variant was reassessed with current criteria and/or through internal data sharing8. The resolution of conflicting interpretations helps provide crucial diagnostic information to clinicians and patients.
Here we demonstrate the successful implementation of the HL-specific ACMG/AMP guidelines5 by the ClinGen HL Variant Curation Expert Panel (HL VCEP) to resolve discrepancies in variant interpretations.
Methods
HL-CDWG Organizational Structure
The HL-CDWG has worked to create a standardized, thorough set of genes and variants that are associated with syndromic and nonsyndromic HL5,9. The members include otolaryngologists, clinical geneticists, molecular geneticists, ClinGen biocurators, clinical researchers, and genetic counselors from 11 different countries including China, France, Israel, Japan, Netherlands, Singapore, South Korea, Spain, Tunisia, the United Arab Emirates, and the United States, representing 26 institutions. In order to accomplish this goal, the HL-CDWG formed two other groups defined by ClinGen as a Gene Curation Expert Panel (GCEP) and a Variant Curation Expert Panel (VCEP).
HL VCEP Rule Specification and Curation Process
A smaller task team of the HL VCEP worked to provide expert guidance for standardized variant interpretation and adapted the ACMG/AMP guidelines for the interpretation of sequence variants in nine HL genes, specifically USH2A, SLC26A4, GJB2, MYO7A, CDH23, TECTA, COCH, KCNQ4, and MYO65. Those genes represent the major causes of hereditary hearing loss encompassing dominant and recessive inheritance models, different disease mechanisms, penetrance and expressivities.
The HL VCEP group then continued to curate variants in these nine genes according to the new HL specific standards with an emphasis on those that have conflicting interpretations in ClinVar (Figure 1). The HL VCEP utilizes monthly meetings in addition to email correspondence to present, review, and reach consensus on the classification of these variants. Each variant was assigned to a single trained ClinGen biocurator who utilized various literature search engines and databases including PubMed, the Deafness Variation Database (DVD)10, Human Gene Mutation Database (HGMD)11, Google Scholar, Leiden Open Variation Database (LOVD)12, and LitVar13 to find publications associated with these variants. HL VCEP members also contributed case observations and phenotypic information from their laboratories or clinics. Additionally, ClinVar submitters were contacted via email to provide internal genotype and phenotype data for their submissions so that biocurators could aggregate all the relevant data. Curators utilized ClinGen’s Variant Curation Interface (https://curation.clinicalgenome.org/) to assess and document the applicable rules for each variant. Provisional classifications made by the curators were comprehensively presented to the chairs and members of the HL VCEP during the monthly meetings. Members provided verbal feedback during these calls to help modify and approve the various ACMG/AMP codes as necessary. Monthly calls between the biocurators and the chairs of the group were utilized to finalize the classification of variants and to discuss the language of the classification summary text. The average curation time for an individual variant was 40 minutes, with a range from 10 minutes to 2 hours. The expert classifications of these variants were submitted to ClinVar on a quarterly basis (Figure 1).
Figure 1.
Workflow highlighting the steps in the Expert curation process to resolve conflicting interpretations or variants of uncertain significance. Refer to Methods for a detailed description of the curation process. HL, Hearing Loss; ACMG, American College of Medical Genetics; AMP, Association for Molecular Pathology; VCEP, Variant Curation Expert Panel.
Results
Curated Variants
Since the specification and promulgation of the ACMG/AMP variant rules by the HL VCEP (Oza 2018; Abou Tayoun 2018), 157 variants – including the 51 variants used to pilot the newly specified rules – across nine HL genes (Supplementary Figure 1, Supplementary Table 1) underwent an expert review and classification process as shown in Figure 1. The majority of variants (79%) were missense (n=124), while the remaining (21%) were in essential (+/−1,2) splice sites (n=4), frameshift (n=7), synonymous (n=5), non-essential splice regions (n=10), nonsense (n=3), inframe deletion (n=1), exon deletion (n=1), start loss (n=1), or UTR (n=1) variants (Supplementary Table 1). As of January 8, 2021, 937 classifications have been submitted from 70 submitters for these 157 variants. Submitters were mainly affiliated with academic or commercial diagnostic laboratories, while a few were from academic research institutions.
Prior to expert curation, 100 variants had conflicting interpretations, 17 variants had VUS classifications, and 40 variants had no conflicts in ClinVar. Of the 100 variants with conflicting interpretations, 76 variants had medically significant conflicting classifications (P/LP vs. VUS/LB/B as defined by Harrison et al 20178) and 24 variants had VUS vs LB/B conflicts. Only three variants had conflicting classifications of P/LP vs LB/B. For the 40 variants with no conflicting classifications, 33 variants were supported by multiple submissions, five variants had only one submission, and two variants had no previous submissions. Of these 40 variants, some were chosen to pilot the initial specifications of the rules while others were considered clinically significant variants or common pathogenic variants in a specific subpopulation5 (Supplementary Figure 1).
Expert Curation Outcomes
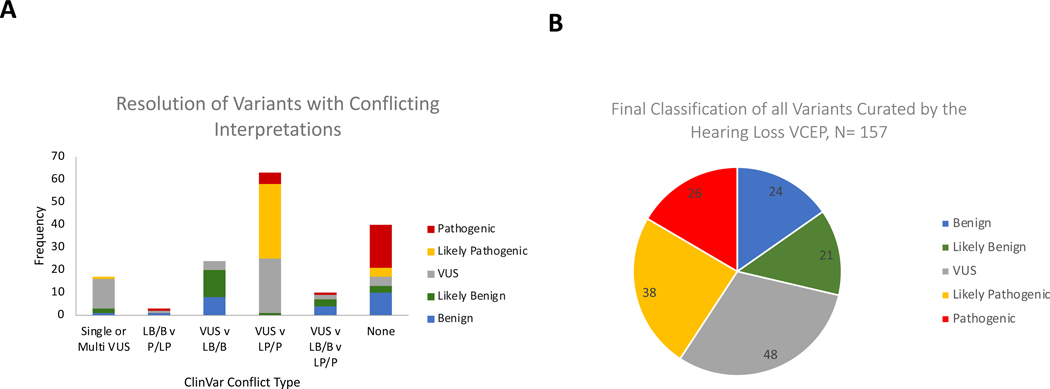
After applying the HL-specific ACMG/AMP variant rules, 73 out of the 117 variants (63%) with either conflicting or VUS classifications, were classified as B, LB, LP, or P, while 44 variants (37%) were classified as VUS (Figure 2). A significant proportion of the resolved classifications belonged to the VUS vs LB/B conflicting category where 20 out of those 24 variants (83%) were classified as B or LB and the remaining as VUS. On the other hand, 38 out of the 63 variants (60.3%) with VUS vs LP/P conflicting interpretations were classified as P or LP, one was classified as LB, and the remaining to VUS. Four out of 17 variants (24%) with single or multiple VUS submissions in ClinVar were classified as B (n=1) or LB (n=2) or LP (n=1) (Figure 2A). Of the 40 non-VUS variants without conflicts, 13 (32.5%) were classified as B or LB, while 23 (57.5%) were classified as P or LP (Figure 2A). Overall, 109 out of the 157 variants (69%) were assigned non-VUS classifications (B, LB, LP, P) after applying the HL-specific ACMG/AMP modified variant interpretation guidelines by the expert group (Figure 2B).
Figure 2.
Outcomes of expert variant interpretation. A, Resolution of variants with different conflict types into Pathogenic (P), Likely Pathogenic (LP), Variant of Uncertain Significance (VUS), Likely Benign (LB), or Benign (B) by the ClinGen HL EP. Y-axis represents the number of variants; X-axis represents the conflict type in ClinVar. ‘Single’ and ‘multi’ refer to single and multiple submitters in ClinVar, respectively. B, Final classifications of all variants curated by the Hearing Loss VCEP (N=157).
Criteria contribution to the final classification
In classifying the 157 variants, the most commonly applied criteria codes were PM2 (absence or rare occurrence in population databases) at two different strength levels (PM2_Supporting and PM2) and PM3 (allelic observations in recessive cases) at four different strength levels (PM3_Supporting, PM3, PM3_Strong, PM3_Very Strong), as specified by the HL VCEP (Oza et al., 2018). Both the PM2 and PM3 codes were used 80 times and were applied to 77% (49/64) and 88% (56/64) of the variants classified as P/LP, respectively (Supplementary Figure 2). The in silico prediction code PP3, specified using REVEL5,14, was applied 64 times as the second most commonly used code, and was applied to 59% (38/64) of P/LP variants. The BA1, BS1, and BS1_Supporting allele frequency codes, as specified by the expert panel, were applied 59 times and to 91% (41/45) of the B/LB classifications. The phenotype code PP4 was used 47 times and was applied to 58% (37/64) of P/LP variants. The specified segregation code PP1 at three different strength levels (PP1, PP1_Moderate, PP1_Strong) was used 31 times and was applied to 41% (26/64) of P/LP variants. Finally, the specified functional codes (BS3_P, PS3_P, PS3_M, and PS3) (Oza et al., 2018) were used 31 times, 48% (15/31) of which were towards P/LP classifications. Several other codes were utilized as specified by the HL VCEP and as shown in Supplementary Figure 2.
Discussion
Of the 28 ACMG/AMP criteria, 25 were specifically tailored to hereditary HL5,9. The HL VCEP included an international group of scientists, clinicians, geneticists, genetic counselors, and laboratory directors. This panel leveraged the collective members’ expertise and knowledge, along with internal unpublished datasets accessible to the group, to characterize several HL attributes such as prevalence, penetrance, genetic and allelic heterogeneity, and phenotypic presentations (onset, severity, frequencies affected, symmetry and audiometric profile) specific to several HL-associated genes. This information was then used to define the criteria for variant classification in hearing loss5.
Our panel implemented these HL-specific guidelines to classify 157 variants. One hundred and nine variants (69%) were classified into non-VUS categories. Interestingly, the commonly used classification criteria codes were either specified for HL (e.g. PM2, BA1, BS1, BS1_Supporting, PP3, PP4) or, in collaboration with the ClinGen Sequence Variant Interpretation (SVI) workgroup, specified more broadly for recessive disorders (e.g. PM3) or any disorder (e.g. PP1). Application of criteria without disease-specific guidance is a large contributor of conflicting classifications, and emphasizes the importance of expert guidance and specification in usage of the ACMG/AMP rules. Several variants classified as P and LP also had BA1 and BS1 codes that were applied to them. Given that many of these variants were founder variants in specific subpopulations or had other evidence such as case-control data and case counts, the group decided to exclude the conflicting benign population frequency codes from impacting the overall clinical interpretation of the variant.
Sharing of internal clinical laboratory case data among members of the HL VCEP has also served as an invaluable resource for resolving variants with conflicting interpretations. For example, the c.1708G>A (p.Val570Ile) variant in SLC26A4 previously had a conflicting classification and a medically significant difference of VUS vs LP. The HL VCEP resolved this conflict by counting 2 compound heterozygous internal cases from the Laboratory of Molecular Medicine, which led to the application of PM3_Strong, PP4, and a LP classification. Without these internal cases, the variant would have been classified as VUS due to a lack of published case-level evidence and a highly-specific phenotype.
Our curation process included bimonthly calls where biocurators first present all evidence used for each variant classification to the experts. Based on the feedback, biocurators often reached out to ClinVar submitters, authors of certain publications, and/or any of the experts to gather additional unpublished data required for final variant interpretation. The group also relied on feedback from expert clinicians regarding any genotype-phenotype correlations. The final evidence is then discussed on intervening monthly calls with the HL VCEP chairs, and if needed on subsequent calls with the larger group, to reach a final classification, including a text-based variant evidence summary documenting all applied evidence to be submitted to ClinVar and publicly accessible through ClinGen’s Evidence Repository (https://erepo.clinicalgenome.org/evrepo/). This process has been, and continues to be, optimized for more efficient expert interpretations while still leveraging the diverse expertise within the group. It is important to note that this initial effort was mostly focused on variants with conflicting or uncertain significance to highlight the utility of the specified rules. In this process, a significant effort was dedicated to contacting ClinVar submitters and obtaining additional unpublished evidence to resolve interpretation discrepancies. We therefore expect this process to be faster for novel variants or ones without conflicting evidence. Ultimately, this process can be optimized the process to strike a balance between efficiency and leveraging the expertise of any disease group
We have focused this pilot project on variants in nine HL-associated genes, USH2A, SLC26A4, GJB2, MYO7A, CDH23, TECTA, COCH, KCNQ4, and MYO6, for which ACMG/AMP specifications were made. However, most of those specified rules are equally applicable to other HL-associated genes. As such, the population frequency (BA1, BS1, PM2), allelic (PM3, BP2), de novo (PS2, PM6), segregation (PP1), predicted effects (PVS1, PM4, PP3, BP3, BP4, BP7), case-control (PS4), and their varied strength levels as specified by the expert group, are currently being extended to other genes. The group will also continue to prioritize curation of variants in HL-genes with medically significant differences and those of uncertain significance.
In summary, our study shows that expert curation using disease-specific modifications of ACMG/AMP guidelines resolves discrepancies in variant classification, leading to more consistent results for patients in need of accurate diagnoses and management decisions.
Data Availability
All variants were submitted to ClinVar by the HL-EP. Refer to Supplementary Table 1 for a list of all variants and their ClinVar IDs.
Supplementary Material
Acknowledgements
The authors would like to acknowledge Larry Babb for his help in compiling data from ClinVar to conduct some of these analyses. Research reported in this publication was supported by the National Human Genome Research Institute (NHGRI) under award number U41HG006834. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
APPENDIX
Hearing Loss Clinical Domain Working Group (non-author contributors)
Sonia Abdelhak, PhD1, John Alexander, PhD2, Zippora Brownstein, PhD3, Rachel Burt, PhD4, Byung Yoon Choi, PhD5, Lilian Downie, MBBS6, Thomas Friedman, PhD7, Anne Giersch, PhD8, John Greinwald, MD9, Jeffrey Holt, PhD, CGC10, Makoto Hosoya, MD, PhD11, Un-Kyung Kim, PhD12, Ian Krantz, MD13,14, Suzanne Leal, PhD15, Saber Masmoudi, PhD16, Tatsuo Matsunaga, MD, PhD17,18, Matías Morín, PhD19, Cynthia Morton, PhD20,21,22, Hideki Mutai, PhD11, Arti Pandya, MD, MBA23, Richard Smith, MD24, Mustafa Tekin, MD25, Shin-Ichi Usami, MD, PhD26, 27, Guy Van Camp, PhD28, Kazuki Yamazawa, MD, PhD29, Hui-Jun Yuan, PhD30, Elizabeth Black-Zeigelbein31, and Kejian Zhang, MD, MBA32,33
1 Biomedical Genomics and Oncogenetics, Institut Pasteur de Tunis, Tunis, Tunisia
2 EGL Genetics / Department of Human Genetics, Emory University School of Medicine, Atlanta, GA, USA (Current affiliations: ConsulGene, LLC, Jacksonville, FL, USA and Department of Ophthalmology, University of Alabama at Birmingham School of Medicine, Birmingham, AL, USA)
3 Department of Human Molecular Genetics and Biochemistry, Sackler Faculty of Medicine and Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel
4 Molecular Hearing Laboratory, Murdoch Children’s Research Institute, Parkville, Australia
5 Department of Otorhinolaryngology-Head and Neck Surgery, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, South Korea
6 Murdoch Children’s Research Institute, Melbourne, Australia
7 Laboratory of Molecular Genetics1, National Institute on Deafness and Other Communication Disorders, National Institutes of Health, Rockville, Maryland, USA
8 Department of Pathology, Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts
9 Department of Otolaryngology–Head and Neck Surgery, University of Cincinnati College of Medicine, Cincinnati, Ohio, USA
10 Indiana University School of Medicine, Indianapolis, Indiana
11 National Hospital Organization Tokyo Medical Center, National Institute of Sensory Organs, Tokyo, Japan
12 Department of Biology, College of Natural Sciences, Kyungpook National University, Daegu, South Korea
13 Department of Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA
14 Division of Human Genetics, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA
15 Laboratory of Statistical Genetics, Rockefeller University, New York, NY, USA
16 Laboratoire Procédés de Criblage Moléculaire et Cellulaire, Centre de Biotechnologie de Sfax, Université de Sfax, Sfax, Tunisia
17 Division of Hearing and Balance Research, National Hospital Organization Tokyo Medical Center, Tokyo, Japan
18 Medical Genetics Center, National Institute of Sensory Organs, National Hospital Organization Tokyo Medical Center, Tokyo, Japan
19 Servicio de Genética, Ramón y Cajal Institute of Health Research (IRYCIS) and Biomedical Network Research Centre on Rare Diseases (CIBERER), Madrid, Spain
20 Departments of Obstetrics and Gynecology and of Pathology, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA
21Manchester Centre for Audiology and Deafness, University of Manchester, Manchester Academic Health Science Centre, Manchester, UK
22 The Broad Institute, Cambridge, MA, USA
23 Division of Pediatric Genetics and Metabolism, Department of Pediatrics, The University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA
24 Department of Internal Medicine and Otolaryngology, University of Iowa Carver College of Medicine, Iowa City, Iowa, USA
25 John P. Hussman Institute for Human Genomics, University of Miami, Miami, FL, USA
26 Department of Otolaryngology, Shinshu University School of Medicine, Matsumoto, Japan
27 Department of Hearing Implant Sciences, Shinshu University School of Medicine, Matsumoto, Japan
28 Department of Medical Genetics, University of Antwerp, Universiteitsplein 1, Antwerp, Belgium
29 Medical Genetics Center, National Hospital Organization Tokyo Medical Center, 2–5–1 Higashigaoka, Meguro, Tokyo, Japan
30Southwest Hospital in China, Chongqing Shi, China
31 Molecular Otolaryngology and Renal Research Laboratories, Department of Otolaryngology-Head and Neck Surgery, Carver College of Medicine, University of Iowa, Iowa City, IA, USA
32 Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA
33Department of Pediatrics, University of Cincinnati School of Medicine, Cincinnati, OH, USA
Footnotes
Disclosure Statement
Conflict of Interest Statement
The authors declare no conflict of interest.
Ethics Declaration
All curated variants were obtained from ClinVar, and were therefore, publicly available. As per ClinVar policy, variant submitters are expected to have obtained appropriate consent for data submission and sharing.
References
- 1.Alford RL, Arnos KS, Fox M, Lin JW, Palmer CG, Pandya A, et al. American College of Medical Genetics and Genomics guideline for the clinical evaluation and etiologic diagnosis of hearing loss. Genet Med.16, 347–355 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Abou Tayoun AN, Al Turki SH, Oza AM, Bowser MJ, Hernandez AL, Funke BH, et al. Improving hearing loss gene testing: a systematic review of gene evidence toward more efficient next-generation sequencing-based diagnostic testing and interpretation. Genet Med.18, 545–553 (2016). [DOI] [PubMed] [Google Scholar]
- 3.DiStefano MT, Hughes MY, Patel MJ, Wilcox EH, Oza AM Expert interpretation of genes and variants in hereditary hearing loss. Medizinische Genetik. 32:109–115 (2020). [Google Scholar]
- 4.Shearer AE, Shen J, Amr S, Morton CC, Smith RJ, et al. A proposal for comprehensive newborn hearing screening to improve identification of deaf and hard-of-hearing children. Genet Med.21, 2614–2630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oza AM, DiStefano MT, Hemphill SE, Cushman BJ, Grant AR, Siegert RK, et al. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat. 39, 1593–1613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen J, Oza AM, del Castillo I, Duzkale H, Matsunaga T, Pandya A,, et al. Consensus interpretation of the p.Met34Thr and p.Val37Ile variants in GJB2 by the ClinGen Hearing Loss Expert panel. Genet Med. 21, 2442–2452 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiStefano MT, Hemphill SE, Oza AM, Siegert RK, Grant AR, Hughes MY, et al. ClinGen expert clinical validity curation of 164 hearing loss gene-disease pairs. Genet Med. 21, 2239–2247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison SM, Dolinsky JS, Knight Johnson AE, Pesaran T, Azzariti DR, Bale S, et al. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet Med. 19, 1096–1104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abou Tayoun AN, Pesaran T, DiStefano MT, Oza A, Rehm HL, Biesecker LG, et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 39, 1517–1524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azaiez H, Booth KT, Ephraim SS, Crone B, Black-Ziegelbein EA, Marini RJ, et al. Genomic Landscape and Mutational Signatures of Deafness-Associated Genes. Am J Hum Genet. 103, 484–497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenson PD, Mort M, Ball EV. Shaw K, Phillips A, Cooper DN The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomics medicine. Hum. Genet 133, 1–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT LOVD v2.0: the next generation in gene variant databases. Hum. Mutat 32, 557–63 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Allot A, Peng Y, Wei C-H, Lee K, Phan L, Lu Z. LitVar: a semantic search engine for linking genomic variant data in PubMed and PMC. Nucleic Acids Res. 46, W530–W536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am J Hum Genet. 99, 877–885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All variants were submitted to ClinVar by the HL-EP. Refer to Supplementary Table 1 for a list of all variants and their ClinVar IDs.