Abstract
The transcription factors PAX5, IKZF1 and EBF1 are frequently mutated in B cell acute lymphoblastic leukemia (B-ALL). We demonstrate that compound heterozygous loss of multiple genes critical for B and T cell development drives transformation, including Pax5+/−xEbf1+/−, Pax5+/−xIkzf1+/− and Ebf1+/−xIkzf1+/− mice for B-ALL, or Tcf7+/−xIkzf1+/− mice for T-ALL. To identify genetic defects that cooperate with Pax5 and Ebf1 compound heterozygosity to initiate leukemia, we performed a Sleeping Beauty (SB) transposon screen that identified cooperating partners including gain-of-function mutations in Stat5b (~65%) and Jak1 (~68%), or loss-of-function mutations in Cblb (61%) and Myb (32%). These findings underscore the role of JAK/STAT5B signaling in B cell transformation and demonstrate roles for loss-of-function mutations in Cblb and Myb in transformation. RNA-Seq studies demonstrated upregulation of a PDK1>SGK3>MYC pathway; treatment of Pax5+/−xEbf1+/− leukemia cells with PDK1 inhibitors blocked proliferation in vitro. In addition, we identified a conserved transcriptional gene signature between human and murine leukemias characterized by upregulation of myeloid genes, most notably involving the GM-CSF pathway, that resemble a B cell/myeloid mixed-lineage leukemia. Thus, our findings identify multiple mechanisms that cooperate with defects in B cell transcription factors to generate either progenitor B cell or mixed B/myeloid-like leukemias.
Introduction
Loss-of-function mutations in B cell transcription factors are a common feature of B cell acute lymphoblastic leukemia (ALL)[1]. This is clearly evident for three transcription factors - EBF1, PAX5 and IKZF1[1, 2]. Interestingly, alterations involving these transcription factors commonly occur together[1, 3]. This is particularly pronounced in BCR-ABL+ leukemia in which 50% of leukemias with IKZF1 deletions also have mutations affecting Pax5 expression or function[4]. Therefore, an important question is whether compound haploinsufficiency for these transcription factors drives transformation and which combinations of transcription factors promote transformation. Finally, the genetic alterations that cooperate with haploinsufficiency for these transcription factors to drive transformation have also not been comprehensively elucidated.
To address the above questions, we generated a set of mice that exhibited compound haploinsufficiency for various combinations of Ebf1, Pax5, Ikzf1, Cebpa, and Tcf7. Herein, we demonstrate that Pax5+/−xEbf1+/−, Pax5+/−xIkzf1+/−, and Ebf1+/−xIkzf1+/− mice generated B cell leukemia, while Tcf7+/−xIkzf1+/− mice generated T cell leukemia. We used a SB Transposon screen to identify mutations that cooperate with Pax5+/−xEbf1+/− compound haploinsufficiency to promote transformation. This analysis yielded two primary types of leukemia – one subset that resembled classical progenitor B cells and a second subset with a striking mixed B cell/myeloid gene signature, characterized by upregulation of Csf2 (which encodes GM-CSF) and Csf2r. The most common alterations in all leukemia subtypes were gain-of-function mutations in Stat5b and Jak1, and loss-of-function mutations in Clbb. Our findings document the key role that compound haploinsufficiency for critical transcription factors plays in transformation and identify mutations that cooperate with such alterations to initiate transformation.
MATERIALS AND METHODS
Mice and Cells
All mice were previously described [5-10]; the University of Minnesota IACUC approved all animal experiments. Mice were monitored for up to 400 days for leukemia. Spleen, lymph nodes, and bone marrow were isolated from tumor-bearing mice and used for further experiments.
Sleeping Beauty Mutagenesis
Pax5+/−xEbf1+/−xCd79a-cre mice were crossed to mice with a concatemer of mutagenic transposon vectors (T2/Onc) on chromosome 1 or 15 and a Cre-inducible SB transposase gene (Rosa26LSL-SB11) (T2/OncxRosa26LSL-SB11 combination, referred to as SB hereafter) [11]. The Rosa26LSL-SB11 transposase transgene contains a Gfp cDNA that is removed upon CRE-mediated recombination and allows identification of cells in which CRE recombinase is active and the SB11 transposase is expressed. We generated 34 leukemic mice; 12 mice from the Chromosome 1 system and 19 mice from the Chromosome 15 system.
Fusions generated by transposon insertion [12] were identified in the RNA-Seq dataset as described [13]. GCESS is a method to systematically identify similar gene clusters across multiple complex transcriptional datasets [13-15]. Transcriptional profile datasets are separately log transformed and mean centered, filtered for highly variant genes, and clustered using average linkage hierarchical clustering using Pearson correlation as the metric. The resulting trees are parsed to identify gene clusters with node correlation greater than a given value and greater than a given number of genes. The identified cluster members are then statistically compared across datasets to identify clusters, that are enriched for common genes present greater than expected by random chance using the Fisher exact test. This method allows facile identification of conserved transcriptional patterns observed across complex datasets without prior biological knowledge.
Accession Numbers
RNA-Seq data was deposited with GEO - GEO Series accession number GSE148680.
Supplemental methods
Supplemental Methods includes detailed protocols of cell lines and culture conditions, NGS, flow cytometry, qPCR, western blotting, RNA-Seq Analysis, Gene Set Enrichment Analysis, WGCNA analysis, SB Mutagenesis, Transposon Insertion Analysis, Reverse Phase Proteomics and Inhibitor Assay.
RESULTS
Reduced expression of transcription factors critical for lymphocyte development promotes transformation
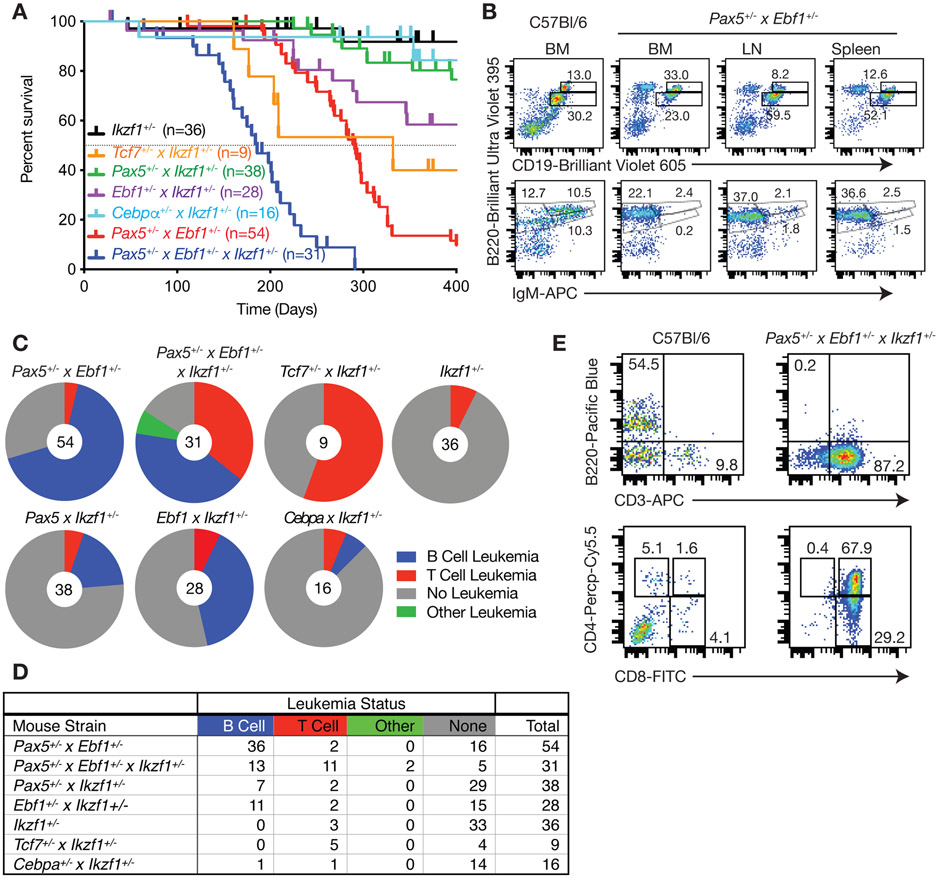
To explore whether compound haploinsufficiency for Ebf1 and Pax5 leads to B cell transformation, we generated Pax5+/−xEbf1+/− mice. Pax5+/−xEbf1+/− mice develop leukemia with a mean survival of 296 days (Fig. 1A). Flow cytometry analysis from bone marrow, lymph nodes and spleens of these mice demonstrated that leukemias resemble progenitor-B cells with a B220+CD19+IgM− phenotype (Fig. 1B), and express pre-BCR, CD43, IL7R, TSLPR, c-KIT, AA4.1 and CD25 confirming their progenitor-B cell like phenotype (Sup Fig. 1). Although both male and female mice developed leukemia in this model, female mice exhibited greater penetrance (97% versus 71% at 400 days) and reduced median survival (265 days vs 298 days, p= 0.005; Sup Fig. 2). Similar results were observed in Pax5+/−xIkzf1+/− and Ebf1+/−xIkzf1+/− mice (Fig. 1A). Compound haploinsufficiency for all three transcription factors (Pax5+/−xEbf1+/−xIkzf1+/− mice) resulted in highly penetrant leukemia and shorter mean survival (202 days). Neither Pax5+/− or Ebf1+/− mice develop leukemia[16]. In contrast, Ikzf1+/− mice develop leukemia with low penetrance (Fig. 1A)[17, 18]; however, these were always T cell leukemias (Fig. 1C,D). Deleting one copy of Pax5 and Ebf1 increased the frequency of B cell leukemias in Ikzf1+/− mice (none to ~40%), and resulted in a dramatic increase in T cell leukemias (~5% in Ikzf1+/− mice versus ~35% in Pax5+/−xEbf1+/−xIkzf1+/− mice) (Fig. 1C-E). Thus, although PAX5 and EBF1 are only expressed in B cells, reduced expression of these two transcription factors paradoxically also promoted T cell leukemia.
Figure 1.
Compound haploinsufficiency for transcription factor genes drives B cell or T cell leukemia. A Kaplan-Meier survival analysis of mice of the indicated genotype. Tick marks represent censored mice. B Flow cytometric analysis of control C57Bl/6 bone marrow (BM) cells or bone marrow, lymph node (LN), and spleen cells from Pax5+/−xEbf1+/− leukemic mice. Representative flow cytometric analysis of B220, CD19, and IgM expression is shown. Doublets were gated out and a lymphocyte gate was set based on side and forward scatter properties. All gates shown are based on bone marrow isolated from control C57Bl/6 mice. This mouse was 267 days old. C Pie charts showing the number of leukemias from each genotype that were either of the B cell (Blue), T cell (Red) or other (Green) phenotype; grey represents mice that failed to develop leukemia or were censored. D The table provides the numbers of mice from the pie charts included in each group and is color-coded to match the pie charts. E Flow cytometric analysis of bone marrow cells from control C57Bl/6 and Pax5+/−xEbf1+/−xIkzf1+/− leukemic mice. Representative flow cytometric analysis of B220, CD3, CD4, and CD8 expression on bone marrow cells is shown. Doublets were gated out and a lymphocyte gate was set based on side and forward scatter properties. All gates shown are based on bone marrow isolated from control C57Bl/6 mice.
We next examined whether compound haploinsufficiency for lineage-determining transcription factors was a general mechanism that could promote transformation of multiple cell lineages. To this end, we generated Tcf7+/−xIkzf1+/− mice, as Tcf7, which encodes TCF1, and Ikzf1, are both required for T cell development[8, 9]. We also generated Cebpa+/−xIkzf1+/− mice, as Cebpa and Ikzf1 are both involved in myeloid cell development[19]. Cebpa+/−xIkzf1+/− mice did not develop myeloid leukemia and the rate of T cell leukemia in Cebpa+/−xIkzf1+/− mice was no higher than that observed for Ikzf1+/− mice. Thus, not all combinations of transcription factor haploinsufficiency promote transformation. However, Tcf7+/−xIkzf1+/− mice developed T cell leukemias with penetrance comparable to that for B cell leukemias in Pax5+/−xEbf1+/− mice (Fig. 1A,C). Thus, compound haploinsufficiency for lineage defining transcription factors promotes transformation in multiple cell lineages and may underlie many types of leukemias.
The role of DNA repair and Pax5 and Ebf1 heterozygosity in B-ALL induction
Previous reports suggested that Ebf1+/− mice have defects in DNA repair with decreased expression of Rad51 and increased γH2AX foci[20]. These studies further claimed that defects in DNA repair resulted in increased mutation rates in Pax5+/−xEbf1+/− leukemias and that this accounts, in part, for progenitor B cell transformation in those mice[20]. This suggestion is difficult to reconcile with the low frequency of somatic mutations reported in human B-ALL[21]. We re-examined this issue using Ebf1+/− mice in our colony. We found no difference in Rad51, Rad51AP or γH2AX expression when examining two independent RNA-Seq experiments (Sup. Fig. 3A). In fact, the low level of variation paralleled that observed for a panel of housekeeping genes (B2m, Hprt, and Actb; Sup. Fig. 3A). Since the previous studies compared progenitor-B cells from C57Bl/6 and Ebf1+/− mice that had been cultured extensively in vitro we examined γH2AX expression in long-term cultured progenitor-B cells from C57Bl/6 and Ebf1+/− mice; no significant difference was observed (Sup. Fig. 3B). Further, we examined γH2AX expression by flow cytometry in progenitor-B cells directly from the bone marrow of C57Bl/6 and Ebf1+/− mice. Again, we found no significant difference in expression (Sup. Fig. 3C). Finally, we also examined progenitor B cells from Pax5+/− mice and Pax5+/−x Ebf1+/− pre-leukemic mice, as well as Pax5+/− x Ebf1+/− leukemias and found no difference in Rad51, Rad51AP or γH2AX expression (Sup. Fig. 3D and Sup. Table 1). Next, we examined whether other genes involved in DNA repair were enriched in Ebf1+/− cells by Gene Set Enrichment Analysis (GSEA) using our RNA-Seq data. We saw a significant enrichment for DNA repair genes (Sup. Fig. 3E-G, Sup. Table 2), although it is unclear whether this reflects a direct effect of EBF1 on genes involved in DNA repair or just a relative increase in cells stuck at a stage undergoing VDJ recombination, as there is significant overlap between genes involved in DNA damage response and VDJ recombination. This suggests other mechanisms may be involved in transformation of progenitor B cells harboring loss of one allele of Pax5 and Ebf1.
Sleeping Beauty mutagenesis screen identifies genes that cooperate with Pax5 x Ebf1 heterozygosity to induce leukemia.
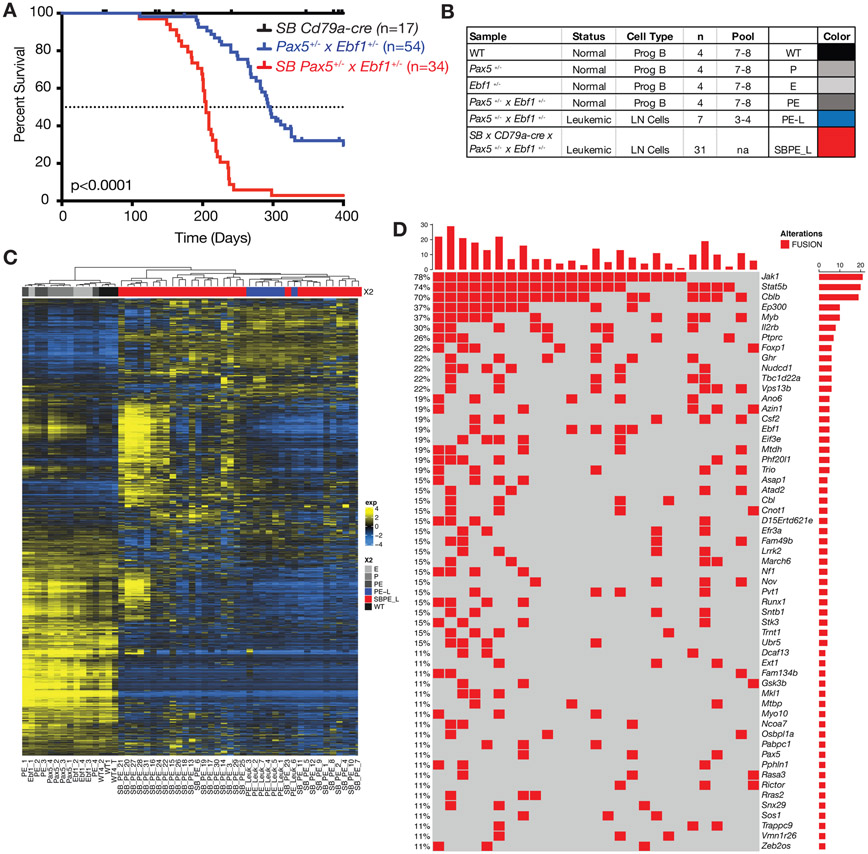
To identify mutations that cooperate with Pax5+/−xEbf1+/− heterozygosity to initiate leukemia we crossed Pax5+/−xEbf1+/−xCd79a-cre mice to mice expressing the mutagenic transposon SB in a Cd79a-Cre-dependent, and hence B cell-specific, manner. We generated 34 mice that were heterozygous for both Ebf1 and Pax5 and expressed the mutagenic transposon. Mice were housed for up to 400 days to allow them to develop leukemia. We included single heterozygous combinations (Pax5+/−xCd79a-CrexSB and Ebf1+/−xCd79a-CrexSB) but neither of these cohorts developed leukemia (data not shown). As seen in figure 2A, all Pax5+/−xEbf1+/−xCd79a-CrexSB mice developed leukemia. Thus, the presence of the sleeping beauty transposon increased penetrance of leukemia to 100% and decreased the median age of death from 296 to 205 days. Thus, other genes mobilized or silenced by SB transposition, which are described in this manuscript (see Fig. 2d), clearly cooperate with Pax5 and Ebf1 heterozygosity to initiate transformation.
Figure 2.
Sleeping Beauty mutagenesis screen to identify genes that cooperate with Pax5 and Ebf1 heterozygosity to induce leukemia. A Kaplan-Meier survival analysis of mice comparing Pax5+/−xEbf1+/− leukemic mice (n=54) and SB Pax5+/−xEbf1+/− (n=34) leukemic mice to control mice SBxCd79a-Cre (n=17). P-value compares Pax5+/−xEbf1+/− versus SB Pax5+/−xEbf1+/− mice. B Table indicating all of the samples used in RNA-Seq analysis. The table indicates the status, type and number of samples utilized for the RNA-Seq. Control samples represent progenitor B cell pools from 7-8 C57Bl/6 (WT), Pax5+/−, Ebf1+/−, and Pax5+/−xEbf1+/− pre-leukemic mice. Three to four leukemic mouse samples were pooled using the online tool from Graphpad Quickcalcs Randomizer. Colors are added for interpretation of subsequent figures. B220+CD19+Lambda−Kappa− cells were used for all control samples. C Hierarchical clustering of all leukemic and control samples. D Fusions identified from RNA-Seq analysis of our sleeping beauty mutagenesis screen. This chart identifies recurrent insertions found in 27 of the 31 samples tested. Each column represents a different leukemic mouse and each row different targeted genes (red indicates an SB insertion was found ).
We performed RNA-Seq analysis on 31 SB induced leukemias, to identify genes targeted by the transposon. The SB transposon contains a unique 5’ leader sequence with a splice donor site that allows for splicing into transcripts and a splice acceptor and SV40 polyA tail that allows for splicing of upstream exons to the SV40 polyA sequence. These unique 5’ SB sequences and 3’ SV40 polyA signal sequences can be identified by RNA-Seq as novel fusion proteins. We carried out RNA-Seq analysis on progenitor B cells (CD19+B220+Igκ−/λ−) from C57Bl/6, Ebf1+/−, Pax5+/−, and Pax5+/− Ebf1+/− pre-leukemic mice (~6-12 weeks of age), as well as spontaneous Pax5+/−xEbf1+/− leukemias (Fig. 2B). Differential gene expression analysis showed that C57Bl/6 and pre-leukemic samples all clustered distinctly from SB-induced and spontaneous leukemias. The spontaneous Pax5+/−xEbf1+/− leukemias were interspersed among the SB-induced leukemias demonstrating that the SB-induced leukemias shared gene expression signatures with the spontaneous leukemias. Finally, the leukemias were clearly heterogenous with distinct subsets harboring unique gene signatures (Fig. 2C).
We next identified fusion transcripts from the RNA-Seq data. Figure 2D lists all the recurrent RNA fusions identified in our screen. Consistent with the heterogeneity of the gene expression profiles in distinct B-ALL subsets (Fig 2C), many of the targeted genes were only found in a fraction of the leukemias (Fig. 2D). The notable exception was that almost all leukemias had SB insertions involving Jak1 or Stat5b (Fig. 2D). Thus, targeting the JAK/STAT5b pathway appears to be required for transformation.
JAK/STAT signaling cooperates with Pax5/Ebf1 haploinsufficiency to induce leukemia
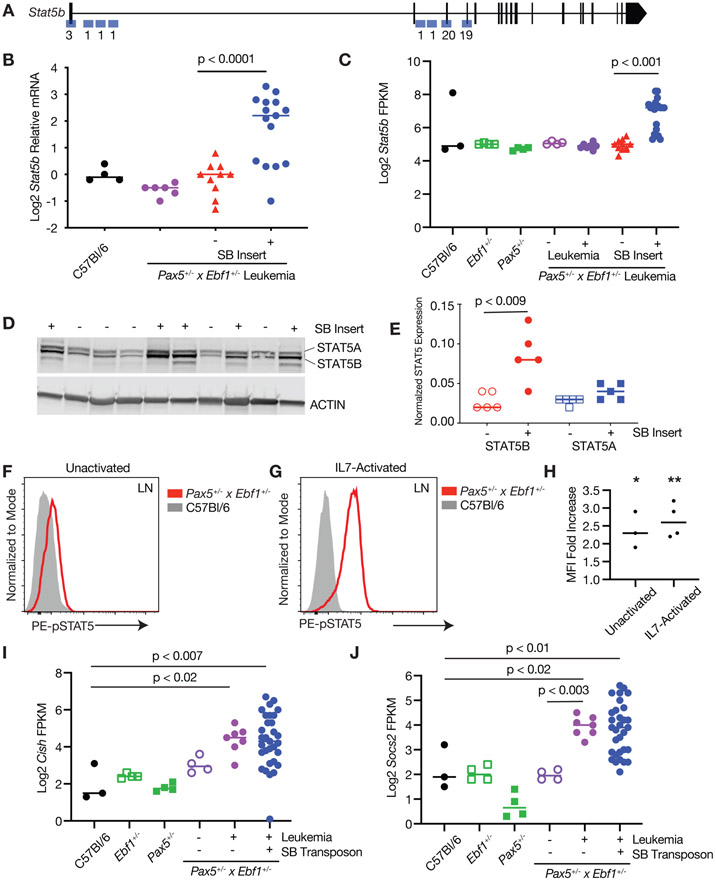
Our SB RNA fusion analysis demonstrated that the SB 5’ leader UTR sequence typically fused to the first 1-4 coding exons of Stat5b or Jak1 genes (Fig. 3A, Sup. 4A). This suggested that a full-length or nearly full-length coding transcript would be generated for both Jak1 and Stat5b. Stat5b mRNA was expressed at significantly higher levels in leukemic samples harboring a SB transposon insertion (Fig. 3B,C). STAT5B, but not STAT5A, protein was increased in samples with an SB insertion in the Stat5b gene (Fig. 3D,E). The spontaneous Pax5+/−xEbf1+/− leukemias did not exhibit significant increases in Stat5b expression (Fig. 3B,C). However, levels of phosphorylated STAT5 (Tyr694/699) in Pax5+/−xEbf1+/− leukemic cells expressed significantly higher levels of phosphorylated STAT5 than C57Bl/6 control mice, either directly ex vivo (Fig. 3F,H) or following in vitro stimulation with IL7 (Fig. 3G,H). We next looked at known targets of STAT5 - Cish and Socs2. Both Cish and Socs2 expression levels were elevated in Pax5+/−xEbf1+/− and Pax5+/−xEbf1+/−xCd79a-CrexSB leukemias, suggesting that STAT5 is active in leukemic cells (Fig. 3I,J). Similar expression results were seen for Jak1, with a significant increase in Jak1 mRNA in mice harboring insertions in the Jak1 locus (Sup. Fig. 4B,C), which correlated with increased expression of JAK1 protein in leukemic samples with an SB transposon insertion in the Jak1 gene locus (Sup. Fig. 4D,E). Our findings are consistent with the high rate of STAT5 activation observed in both human and murine B-ALL[16, 22] and underscore the critical role of JAK/STAT5 signaling in B cell leukemia – particularly those with reduced expression of Pax5 and Ebf1.
Figure 3. Increased Expression of Stat5b in leukemia.
A Map of common insertion sites in the Stat5b gene; numbers refer to number of insertions at a particular site. B Quantitative Real Time PCR (qRT-PCR) for Stat5b normalized to Actin in progenitor B cells isolated from the bone marrow of C57Bl/6 (black, n=4) mice, and leukemic cells isolated from the lymph nodes of Pax5+/−xEbf1+/− (purple, n=6) and SB Pax5+/−xEbf1+/− mice. The samples from the SB Pax5+/−xEbf1+/− mice were split between those with (blue, n=15) or without (red, n=10) an insertion in the Stat5b locus. The normalized values were log2 transformed and an ordinary one-way ANOVA with Holm-Sidak’s multiple comparison test was used to determine significance. The line represents the median value. C Log2 transformed fragments per kilobase of exon model per million reads mapped (FPKM) values from C57Bl/6 (black filled, n=3), Ebf1+/−(green open, n=4), Pax5+/−(green filled, n=4), Pax5+/−xEbf1+/− pre-leukemic (purple open, n=4), Pax5+/−xEbf1+/− leukemic (purple filled, n=7), and SB Pax5+/−xEbf1+/− leukemic samples with (blue filled, n=20) or without (red filled, n=11) a transposon insertion in Stat5b locus. A Kruskal-Wallis test with Dunn’s multiple comparison test was used to test for significance. The line represents the median value. D Western blot analysis showing expression of total STAT5A and STAT5B. The + or − indicates the presence or absence of a SB transposon insertion in each representative sample. E Plotted ratio of STAT5A or STAT5B to actin from the western blot. Shown are STAT5B samples without (clear red circles; n=5) or with a transposon insert (solid red circles; n=5) and STAT5A samples without (clear blue squares; n=5) or with a transposon insert (solid blue squares; (n=5). A Kruskal-Wallis test with Dunn’s multiple comparison test was used to test for significance. The line represents the median value. F Flow cytometric analysis of bone marrow cells from Pax5+/−xEbf1+/− leukemic mice and C57BL/6 progenitor B cells. Representative flow cytometric analysis of pSTAT5 expression in cells where doublets were gated out, a lymphocyte gate was applied, and cells were further gated on B220 and AA4.1. This is representative of at least 3 independent experiments. G Flow cytometric analysis of leukemic B cells from Pax5+/−xEbf1+/− leukemic mice and C57BL/6 progenitor B cells. Lymph node cells from leukemic mice or bone marrow cells from C57Bl/6 mice were activated with IL-7 for 30 minutes and subjected to flow cytometric analysis for pSTAT5 expression. Doublets were gated out, a lymphocyte gate was applied, and cells were further gated on B220 and AA4.1. This is a representative plot of 4 independent experiments. H Plotted ratio of pSTAT5 mean fluorescence intensity (MFI) in bone marrow cells from Pax5+/−xEbf1+/− leukemic cells mice divided by progenitor B cells from C57Bl/6 mice. This was done for either cells activated with IL7 (IL7-activated) or cells left untreated (unactivated). These results represent at least 3 independent experiments with 1 mouse of each genotype in each experiment. The line represents the median. A one sample t test was performed to determine significance. *Unactivated p<0.01 and **IL7-activated p<0.002. I Log2 transformed FPKM values for Cish from C57Bl/6 (black filled, n=3), Ebf1+/−(green open, n=4), Pax5+/−(green filled, n=4) Pax5+/−xEbf1+/− pre-leukemic (purple open, n=4), Pax5+/−xEbf1+/− leukemic (purple filled, n=7), and SB Pax5+/−xEbf1+/− leukemic (blue filled, n=31) samples. An ordinary one-way ANOVA with multiple comparisons was used to test for significance. The line represents the median value. J Log2 transformed FPKM values for Socs2 from C57Bl/6 (black filled, n=3), Ebf1+/−(green filled, n=4), Pax5+/−(green open, n=4), Pax5+/−xEbf1+/− pre-leukemic (purple open, n=4), Pax5+/−xEbf1+/− leukemic (purple filled, n=7), and SB Pax5+/−xEbf1+/− leukemic (blue filled, n=31) samples. An ordinary one-way ANOVA with Holm-Sidak’s test for multiple comparisons was used to test for significance. The line represents the median value.
Loss of Cblb cooperates with Pax5/Ebf1 haploinsufficiency to induce leukemia
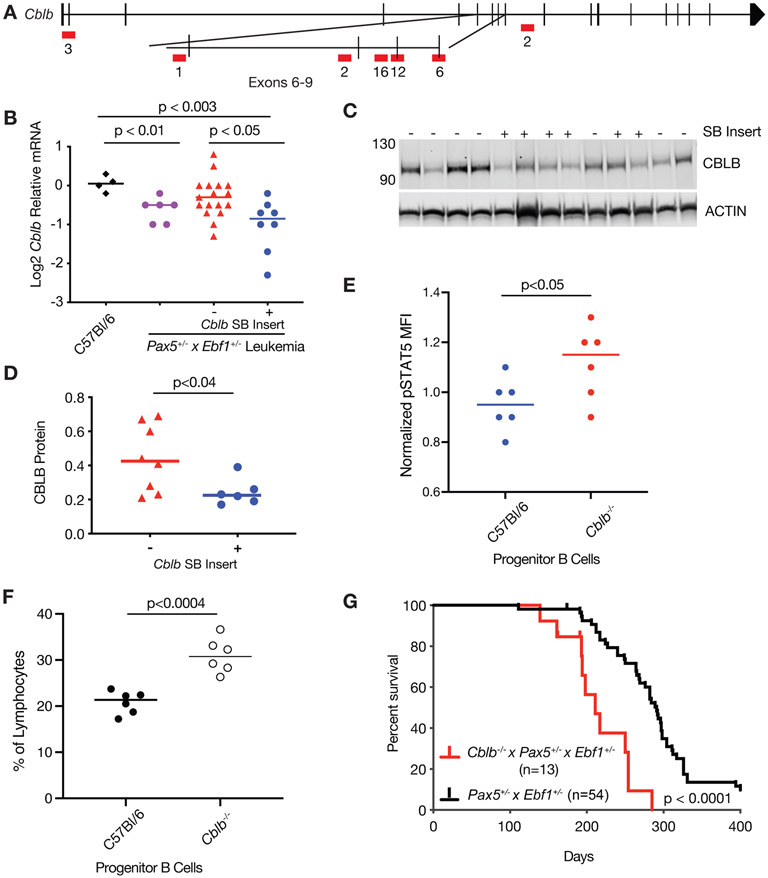
Another top hit in our mutagenesis screen was Cblb (Fig. 4A). SB-induced Pax5+/−x Ebf1+/− leukemias with an SB insertion in Cblb, showed significantly reduced Cblb mRNA (4.6-fold) and protein (1.8-fold) expression compared to C57Bl/6 controls (Fig. 4B-D). Cblb levels were also reduced in spontaneous Pax5+/−xEbf1+/− leukemias (Fig. 4B). To determine whether CBL-B affects STAT5 activation, we stained progenitor B cells from C57BL/6 and Cblb−/− mice with antibodies to pSTAT5(Tyr694/699). Loss of CBL-B resulted in a significant increase in STAT5 phosphorylation (Fig. 4E). Cblb−/− mice also showed an enrichment in progenitor-B cells (B220int, Intracellular IgM+) relative to C57Bl/6 controls (Fig. 4F). To confirm that CBL-B deficiency accelerates leukemogenesis, we generated Cblb−/−xPax5+/−xEbf1+/− mice; such mice developed B-ALL and died significantly faster than Pax5+/−xEbf1+/− mice, demonstrating that Cblb acts as a tumor suppressor in progenitor B cells (Fig. 4G).
Figure 4. Loss of Cblb accelerates the onset of B cell ALL.
A Map of common insertion sites in the Cblb gene; numbers represent number of insertions at a specific site. B qRT-PCR for Cblb normalized to Actin in progenitor B cells isolated from the bone marrow of C57Bl/6 (black, n=4) mice, and leukemic cells isolated from the lymph nodes of Pax5+/−xEbf1+/− (purple, n=6) and SB Pax5+/−xEbf1+/− mice. The samples from the SB Pax5+/−xEbf1+/− mice were split between those without (red, n=17) or with (blue, n=8) an insertion in the Cblb locus. The normalized values were log2 transformed and significance was determined using a Kruskal-Wallis test with Dunn’s multiple comparison test. The line represents the median value. C Western blot analysis showing expression of CBLB. The + or − indicates the presence or absence of a SB transposon insertion in each representative sample. D Plotted ratio of CBLB to actin from the western blot. Samples without a transposon insert in Cblb are red (n=6) and those with a transposon insert in Cblb are blue (n=8). Significance was determined using an unpaired student t-test and the line represents the median. E Plotted are the mean fluorescence intensities (MFI) of phospho-STAT5 intracellular staining in progenitor cells (B220intCD19int) normalized to the average MFI from the C57BL/6 samples. These results are from two independent experiments with three mice per experiment. Significance was determined using an unpaired student t-test and the line represents the median. F Plotted are the percent of lymphocytes for progenitor cells (B220intIgMint) from either C57BL/6 or Cblb−/− mice. These results are from two independent experiments with three mice per experiment. Significance was determined using an unpaired student t-test and the line represents the median. G Kaplan-Meier survival analysis of mice comparing Pax5+/−xEbf1+/− leukemic mice (n=54) and Cblb−/−x Pax5+/−xEbf1+/− (n=13) leukemic mice.
Reduced levels of Myb cooperate with Pax5/Ebf1 haploinsufficiency to induce leukemia
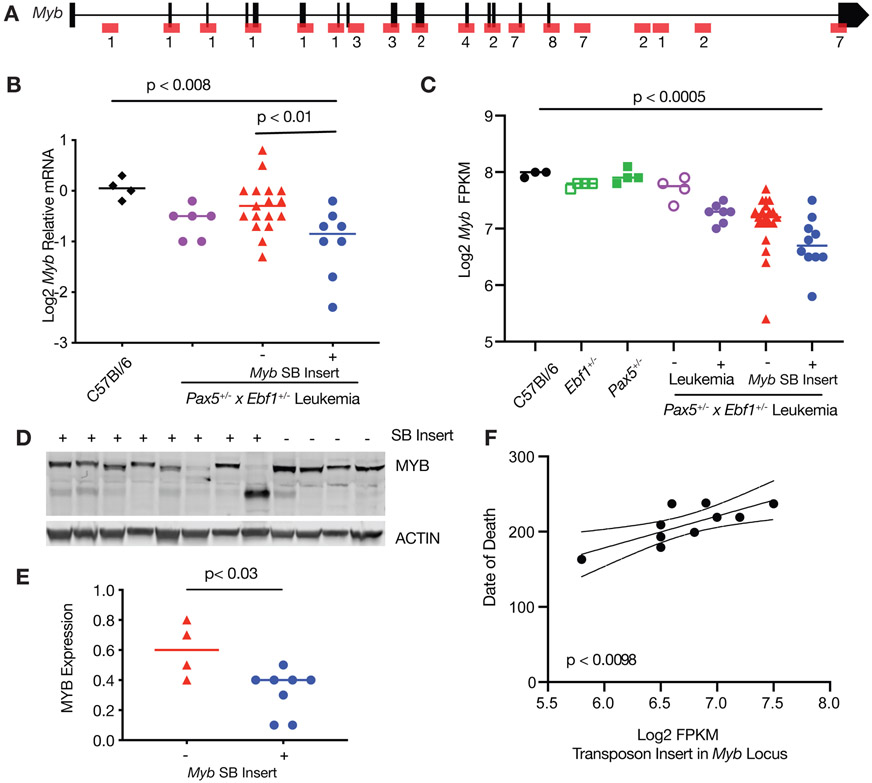
Myb was another target of our mutagenesis screen. SB transposon insertions were scattered throughout the Myb gene locus (Fig. 5A). Spontaneous Pax5+/−xEbf1+/− leukemias showed a clear trend towards reduced Myb expression. Likewise, SB-induced Pax5+/−xEbf1+/− leukemias exhibited decreased Myb expression in leukemias without an SB insertion in Myb (1.5-fold, Fig. 5B), and an additional significant decrease in leukemias with an SB insertion in Myb (2.3-fold, Fig. 5C). Thus, downregulation of Myb appears to be a general feature of Pax5+/−x Ebf1+/− leukemias. In SB-induced Pax5+/−xEbf1+/− leukemias with insertions in the Myb gene, there was also a significant reduction (2.8-fold, Fig. 5D,E) at the protein level; notably many leukemias with SB insertions in Myb also appear to express truncated MYB proteins. Importantly, we found that Myb expression as assessed by RNA-Seq correlated with age of death - leukemias with less Myb were more aggressive resulting in earlier lethality (Fig. 5F).
Figure 5.
Loss of MYB expression results in worse outcome in ALL. A Map of common insertion sites in the Myb gene; numbers refer to number of insertions at a specific site. B qRT-PCR for Myb normalized to Actin in progenitor B cells isolated from the bone marrow of C57Bl/6 (black, n=4) mice, and leukemic cells isolated from the lymph nodes of Pax5+/−xEbf1+/− (purple, n=6) and SB Pax5+/−xEbf1+/− mice. The samples from the SB Pax5+/−xEbf1+/− mice were split between those without (red, n=17) or with (blue, n=8) an insertion in the Myb locus. Significance was tested using an ordinary one-way ANOVA with Holm-Sidak’s multiple comparison test and the lines represent median. C Log2 transformed fragments per kilobase of exon model per million reads mapped (FPKM) values from C57Bl/6 (black filled, n=3), Ebf1+/−(green open, n=4), Pax5+/−(green filled, n=4), Pax5+/−xEbf1+/− pre-leukemic (purple open, n=4), Pax5+/−x Ebf1+/− leukemic (purple filled, n=7), and SB Pax5+/−xEbf1+/− leukemic samples without (red filled, n=21) or with (blue filled, n=10) a transposon insertion. Significance was tested using a Kruskal-Wallis test with Dunn’s multiple comparison test and the line represents median. D Western blot analysis showing decreased expression of MYB in SB leukemia samples harboring a transposon insertion. The + or − indicates the presence or absence of a SB transposon insertion in each representative sample. E Plotted ratio of MYB to actin from the western blot in panel d. Samples without a transposon insert are red (n=4) and those with a transposon insert are blue (n=8). Significance was determined using an unpaired student t-test and the line represents the median. F Linear regression analysis comparing date of death versus FPKM value for leukemic samples harboring a transposon insertion. The dashed lines represent 95% confidence bands.
PDK1-signaling pathway is deregulated in Pax5+/− x Ebf1+/− leukemias
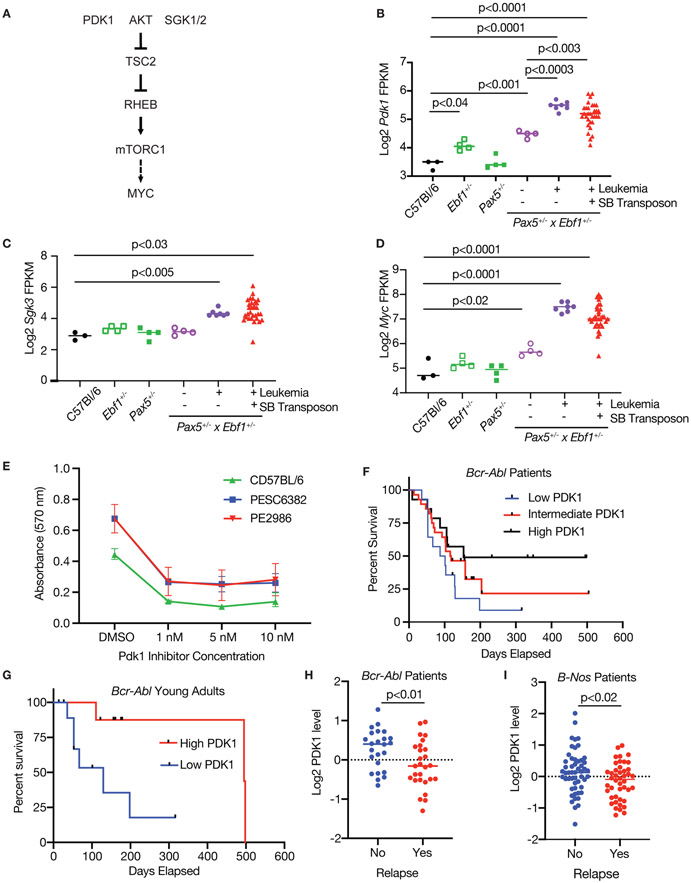
In addition to gene alterations directly targeted by SB transposition, we identified other genes whose expression was significantly altered in Pax5+/−xEbf1+/− leukemias relative to non-transformed controls. These included genes such as the tumor suppressor Bach2, which was significantly reduced (Sup. Fig. 5A). Other genes were strikingly upregulated, including Pdk1 (3.0-fold) and its downstream targets Sgk3, and Rhebl1 (Fig. 6A-C, Sup. Fig. 5C). Conversely Tsc2, which inhibits this pathway, was downregulated (Sup. Fig. 5B); this pathway has been previously shown to enhance mTORC1 function and ultimately Myc expression[23, 24]. Consistent with this concept, Myc expression was significantly increased in both spontaneous and SB-induced Pax5+/−xEbf1+/− leukemias (Fig. 6D). To determine whether PDK1 plays a critical role in maintaining viability of Pax5+/−xEbf1+/− leukemias, we treated two cell lines generated from Pax5+/−xEbf1+/− leukemias along with progenitor B cells from C57Bl/6 mice in vitro with either vehicle control or the PDK1 inhibitor (GSK2334750)[25]. PDK1 inhibitor treatment resulted in a dose-dependent decrease in the survival of these leukemic cell lines as well as progenitor B cells from C57BL/6 mice (Fig. 6E). Thus, while treatment with PDK1 inhibitors might be an effective treatment for B-ALL subsets with reduced Pax5 and Ebf1 expression, like many other therapeutic approaches (Rituximab, CAR-T cells, broad-based chemotherapeutics, etc.), it does affect non-transformed B cells as well.
Figure 6.
Inhibition of PDK1 blocks leukemic proliferation. A Schematic of PDK1 signaling pathway. B Log2 transformed FPKM values for Pdk1 from the RNA-Seq datasets. Significance was determined by an ordinary one-way ANOVA with Holm-Sidak’s multiple comparison test. C Log2 transformed FPKM values for Sgk3 from the RNA-Seq datasets. Significance was tested using a Kruskal-Wallis test with Dunn’s multiple comparison test. D Log2 transformed FPKM values for Myc from the RNA-Seq datasets. Significance was determined by an ordinary one-way ANOVA with Holm-Sidak’s multiple comparison test. E PDK1 inhibitor blocks growth. MTT assay was performed on two Pax5+/−xEbf1+/− leukemic cell lines generated from lymph node cells from leukemic mice and C57BL/6 progenitor B cells. The cells were subjected to differing concentrations of GSK2334470. Each dot represents the average of two biological replicates - each biological replicate represents the mean of triplicate technical replicates within an experiment. Error bars represent the range between experiments. F Overall survival of Bcr-Abl patients. Bcr-Abl patients were separated into three equal groups representing higher (black line, n=14), intermediate (red line, n=28) and lower (blue line, n=14) levels of PDK1. Patients with lower levels of PDK1 did significantly worse than patients with higher PDK1 (p=0.04, Log Rank test for trend). G Overall survival of Bcr-Abl young adult patients. Young adult patients were separated into two equal groups representing higher (red line, n=9) and lower (blue line, n=9) levels of PDK1. Patients with lower levels of PDK1 did significantly worse than patients with higher PDK1 (p=0.02, Log Rank (Mantel-Cox) Test). H PDK1 expression of Bcr-Abl patients split by relapse status. Bcr-Abl patients were separated into two groups representing no relapse (blue dots, n=14), and relapse (red dots, n=14) levels of PDK1 were graphed. Patients with lower levels of PDK1 did significantly worse than patients with higher PDK1 (p<0.01, unpaired T-test). I PDK1 expression of B-Nos patients split by relapse status. Patients were separated into two groups representing no relapse (blue dots, n=14), and relapse (red dots, n=14) and levels of PDK1 were graphed. Patients with lower levels of PDK1 did significantly worse than patients with higher PDK1 (p<0.02, unpaired T-test).
To examine PDK1 expression in human ALL, we examined ALL patient samples using a reverse phase proteomics approach[26]. PDK1 was expressed in five subsets of B-ALL but expression varied widely (Sup. Fig. 5D, Sup. Table 3). We examined PDK1 expression in the two largest cohorts - B-NOS and BCR-ABL+ leukemias. PDK1 levels did not correlate with overall survival in B-NOS patients (data not shown). In contrast, BCR-ABL+ patients with the highest levels of PDK1 expression survived significantly longer than those with lower PDK1 expression (Fig. 6F). The difference in overall survival is driven most strongly by young adults (Fig. 6G). Finally, we examined PDK1 expression in patients with BCR-ABL and B-NOS leukemia based on relapse status. In both subsets of leukemia, lower levels of PDK1 correlated with relapse (Fig. 6H,I). Thus, PDK1 appears to play an important role in B-ALL survival or proliferation, but patients with the highest level of PDK1 expression respond better to therapy.
Murine Pax5 x Ebf1 leukemias show common transcriptional variation patterns with human leukemias
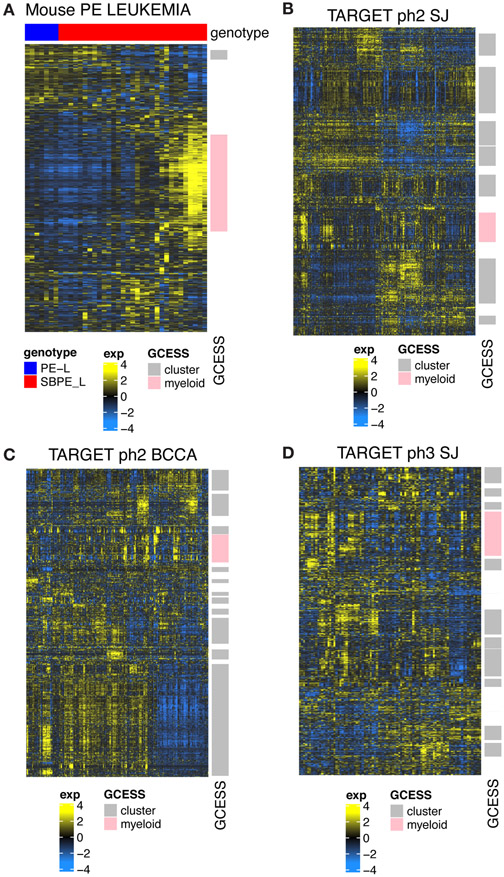
To determine if the murine leukemias that developed in our sleeping beauty screen are similar to human B-ALL, we quantified inter-leukemia transcriptional variation using our newly developed gene cluster expression summary score (GCESS)[14]. The goal of GCESS is to systematically identify similar clusters of genes across multiple transcriptional datasets. This approach has been used to determine common heterogeneity present across related human and murine tumor datasets [13-15]. Analyses restricted to only the mouse tumors identified two distinct transcriptional GCESS (Fig. 7A). We used Fishers exact test to compare the genes found in these clusters to genes found in clusters observed in three independent human B-ALL tumor datasets using an identical GCESS approach. There was conserved heterogeneity between each of the human B-ALL subsets indicating that common variation is present. Visual inspection of the resulting heatmaps show that greater variation (more independent clusters) was present in the human dataset than in our mouse dataset (Fig. 7B-D). Importantly, the variation observed in mouse cluster 1 (myeloid) is clearly present within the variation of each of the human datasets (Fisher exact test, p= 4.3e-07). Thus, the leukemias that developed in our SB system are similar to human leukemias.
Figure 7.
Transcriptome profiles from leukemic progenitor B cells show common inter-leukemic transcriptional variation across human and mouse samples. Heatmaps showing GCESS clusters were generated to allow direct visualization of gene expression heterogeneity in RNA-SEQ from mouse leukemias (A) generated in Pax5+/−xEbf1+/− mice with and without SB accelerated tumorigenesis and (B,C,D) human ALL datasets obtained from the TARGET database. Gene transcript values derived from samples were log transformed and mean centered within each of the 4 datasets independently. Invariant (low SD) genes were removed prior to unsupervised average linkage clustering. GCESS based conservation was apparent in all human datasets despite the fact that the SJ set was summarized as gene symbols while the BCCA set was summarized to ENSEMBL ids. Transcripts with increased levels are shown in yellow, while transcripts with decreased levels are shown in blue. Gene cluster overlap analyses comparing clusters derived from human and mouse tumors show that the variation present in our mouse dataset represents one of the many GCESS clusters conserved in all of the human samples. This cluster is labeled in pink in each of the datasets in the row annotation and is labeled “myeloid”. Gene lists for the conserved pink cluster from each dataset are provided in Supplementary Table 4.
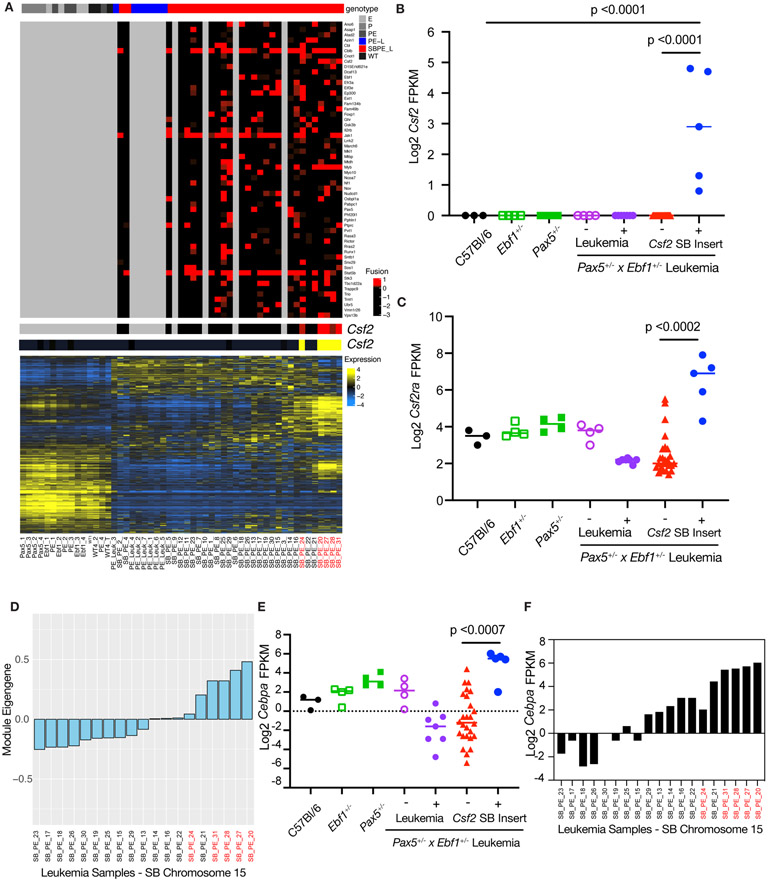
To further characterize these leukemias, we analyzed the mouse GCESS list using ENRICHR (Sup. Table 4)[27, 28]. Consistent with other findings in this study, we found our dataset was enriched for cytokines and cytokine receptor genes, as well as genes involved in JAK/STAT5 signaling. In addition, NFκB signaling pathways were targeted, which is consistent with work from multiple groups on NFκB in B cell development and leukemia [22, 29, 30]. We generated a list of the overlapping genes from the GCESSS analysis using the online Venny tool and used ENRICHR to determine pathways conserved between human and murine ALL (Sup. Table 4) [27, 31]. A surprising observation was a strong myeloid gene signature in the human and murine leukemias (Sup. Table 4). Further analysis identified SB insertions in Csf2 in this GCESS subset of genes in our leukemic mice (Fig. 8A). These mice had significantly enriched expression of both Csf2 and its receptor Csf2ra (Fig. 8B,C). We also identified this set of myeloid-like murine leukemias using an independent approach called weighted gene co-expression network analysis (WGCNA) [32, 33]. Using WGCNA we identified several distinct gene modules. These groups of genes have similar expression patterns which can be transformed into an eigengene value that represents all genes in a module. Hypergeometric testing of genes in each module for GO Term gene enrichment showed significant enrichment of genes in the “myeloid leukocyte activation” GO term list. This WGCNA “myeloid leukocyte activation” module eigengene value had high relative expression in the same subset of myeloid-like leukemias that were identified in our GCESS analysis (Fig 8D). Expression of the myeloid transcription factor Cebpa in our SB Pax5+/−xEbf1+/− leukemia samples was also increased in mice harboring a Csf2 transposon insertion (Fig. 8E). Further, expression of Cebpa was highest in leukemias with the strongest myeloid eigengene values (Fig. 8F). Thus, a subset of Pax5+/−xEbf1+/− leukemias exhibits both B cell and myeloid cell characteristics.
Figure 8.
Leukemias with a Myeloid Gene Signature. A The genotype of the sample is shown in the top panel with SB accelerated tumors shown in red and the PE leukemias shown in blue. The second panel shows each recurrent SB fusion based on the number of reads that support the fusion. Four SB accelerated samples did not show any recurrent fusions. The third and fourth panel highlight fusions in CSF2 (red) and the corresponding transcript level (yellow). The final panel shows the overall RNA-Seq landscape, and the samples are ordered based on hierarchical clustering of transcript profiles. Samples with the Csf2 insertion are labeled in red. B Log2 transformed fragments per kilobase of exon model per million reads mapped (FPKM) values for Csf2 from C57Bl/6 (black filled, n=3), Ebf1+/−(green open, n=4), Pax5+/−(green filled, n=4), Pax5+/−xEbf1+/− pre-leukemic (purple open, n=4), Pax5+/−xEbf1+/− leukemic (purple filled, n=7), and SB Pax5+/−xEbf1+/− leukemic samples without (red filled, n=26) or with (blue filled, n=5) a transposon insertion. Significance was tested using a Kruskal-Wallis test with Dunn’s multiple comparison test and the line represents median. C Log2 transformed fragments per kilobase of exon model per million reads mapped (FPKM) values for Csf2ra from C57Bl/6 (black filled, n=3), Ebf1+/−(green open, n=4), Pax5+/−(green filled, n=4), Pax5+/−xEbf1+/− pre-leukemic (purple open, n=4), Pax5+/−xEbf1+/− leukemic (purple filled, n=7), and SB Pax5+/− x Ebf1+/− leukemic samples without (red filled, n=26) or with (blue filled, n=5) a transposon insertion for Csf2. Significance was tested using a Kruskal-Wallis test with Dunn’s multiple comparison test and the line represents median. D Weighted Gene co-expression network analysis (WGCNA) of SB Chromosome 15 RNA-Seq samples. Module eigengene values corresponding to each SB leukemia are plotted. E Log2 transformed fragments per kilobase of exon model per million reads mapped (FPKM) values for Cebpa from C57Bl/6 (black filled, n=3), Ebf1+/−(green open, n=4), Pax5+/−(green filled, n=4), Pax5+/−xEbf1+/− pre-leukemic (purple open, n=4), Pax5+/−xEbf1+/− leukemic (purple filled, n=7), and SB Pax5+/−xEbf1+/− leukemic samples without (red filled, n=26) or with (blue filled, n=5) a Csf2 transposon insertion. Significance was tested using a Kruskal-Wallis test with Dunn’s multiple comparison test and the line represents median. Samples with the Csf2 insertion are labeled in red. F Log2 transformed fragments per kilobase of exon model per million reads mapped (FPKM) values for Cebpa from SB Leukemia Samples that were generated from Chromosome 15 transposon harboring mice. The order of samples on the x-axis is identical to that shown in panel D for comparison. Samples with the Csf2 insertion are labeled in red.
Discussion
The mechanism by which compound haploinsufficiency promotes transformation remains unclear. Previous studies suggested that this may be due to defective DNA repair upon reduced Ebf1 expression. We were unable to validate defects in Rad51, Rad51ap expression or increased γH2AX expression in pre-B cells from Ebf1+/− mice. Moreover, B cell leukemias tend to have relatively low numbers of non-synonymous mutations relative to other cancers[21], making defects in DNA repair a less likely mode of transformation. To better understand the molecular alterations that cooperate with Pax5 and Ebf1 haploinsufficiency to promote transformation we carried out an unbiased SB transposon screen in Pax5+/−xEbf1+/− mice. These studies identified two major pathways that cooperate with Pax5 and Ebf1 haploinsufficiency to drive transformation. First, we found gain-of-function mutations for Jak1 or Stat5b in almost all of our leukemias in this screen. This finding underscores in an unbiased way the critical role of JAK/STAT5B signaling in B cell transformation. An additional important feature we observed was the exclusive targeting of Stat5b versus Stat5a. These two genes are located 10 kb apart in the genome, share a common DNA binding motif, and appear to be highly functionally redundant. Our results suggest that Stat5b either exhibits unique functions required for B cell transformation, or has features that allow it to be more easily transcribed in B cells. This observation is consistent with a recent report showing that loss of STAT5B had a greater impact on reducing the transforming potential of BCR-ABL, than loss of STAT5A [34]. It is also constructive to compare this SB screen with a previous one we carried out in which mice expressing a constitutively active form of STAT5b were crossed to SB mice. In both cases we targeted the Jak1 allele, indicating that both JAK and STAT5B activation appear to be critical for optimal transformation, and that a rather high amount of STAT5 activation is required. SB mice with a STAT5 predisposing mutation (Stat5b-CA) induced more rapid leukemia onset than SB mice with only Pax5+/−xEbf1+/− predisposing mutations (average onset of leukemia ~72 versus 205 days, respectively) [35]. These findings do not allow us to definitively determine the relative order in which JAK/STAT5B related mutations versus loss-of-function mutations in Pax5/Ebf1/Ikzf1 occur, but they do suggest that STAT5B activation is the rate limiting step in transformation. This is consistent with the observation of human germline mutations in PAX5 and IKZF1 that precede the development of leukemia [36, 37]. In contrast, no germline activating mutations in the STAT5 pathway that predispose to leukemia have been reported to date. Taken together, these findings suggest that alterations in B cell transcription factors likely precede gain-of-function mutations in the JAK/STAT5b pathway.
The second major pathway we identified involves CBL-B, and to a lesser extent the related family member CBL. The mechanism by which Cblb loss-of-function affects transformation is unclear. A recent study found that MDM2 and CBL-B compete for binding to STAT5 and that MDM2 stabilizes, while CBL-B degrades, STAT5 protein [38]. Consistent with this hypothesis, we found that pSTAT5 levels were higher in Cblb−/− versus C57Bl/6 progenitor B cells. This finding may explain why mutations in ARF, which sequesters MDM2, are more common than mutations in TP53 in B-ALL, as ARF loss-of-function mutations would simultaneously target the p53 and STAT5 pathway. We also observed an increase in the percentage of progenitor B cells (B220int, intracellular IgM+) in Cblb−/− mice. While this increase mimics effects seen in Stat5b-CA transgenic mice [39], we suspect that CBL-B also plays other roles in transformation (perhaps impinging on pre-BCR signals) as most SB leukemias with SB insertions in Cblb also have SB insertions in Jak1 and Stat5b. Consistent with this idea, a previous study found that loss of CBLB perturbed homing to the bone marrow of chronic myeloid leukemic cells [40]. Thus, our SB screen suggest that human B-ALL samples should be examined more carefully for the presence of CBLB mutations.
A number of other target genes were identified in our SB screen, although none were as prevalent as the mutations in Jak1, Stat5b or Cblb. These include SB insertions in several cytokine/receptor genes that have previously been shown to be involved in transformation including Il2rb, Gh, Csf2, as well as the transcription factor Myb. Mutations in IL2RB have been previously identified in human leukemias [41]. Surprisingly, the mutations in Myb appear to be loss-of-function mutations resulting in reduced or truncated Myb expression. This leads to the somewhat unusual observation that Myb acts operationally as a tumor suppressor in this context and parallels our previous observation that NFκB also acts as a functional tumor suppressor in progenitor B cells[22, 42]. MYB appears to be required for both survival and differentiation of progenitor B cells[43]. The pro-survival functions of MYB are presumably coopted by strong STAT5 activation, while its role in differentiation may explain why Myb was targeted in our screen.
Perhaps the most interesting finding was a subset of leukemias that contained SB insertions in Csf2, which encodes the myeloid growth factor GM-CSF, a cytokine not normally expressed by progenitor B cells. These leukemias exhibited a clear myeloid gene signature, including expression of the GM-CSF receptor (Csf2ra), which allows them to respond to GM-CSF (presumably enhancing JAK/STAT5 signaling). There are two potential reasons for this. First, these leukemias could be infiltrated with myeloid cells. Alternatively, the leukemias could have lost lineage fidelity and begun to express myeloid genes. Since PAX5, EBF1 and IKZF1 all play key roles in enforcing B cell lineage fidelity, and our murine B cell leukemias were relatively pure populations of leukemic blasts, we favor this latter possibility. It is likely that expression of GM-CSF also alters the leukemia microenvironment. This could promote leukemia progression by a number of mechanisms including expanding myeloid-derived suppressor cells. The myeloid-like gene signature may also allow these leukemias to escape therapies that target B cells by allowing for differentiation into a more myeloid-like state. Such lineage plasticity would normally be blocked by high level expression of PAX5, EBF1 and IKZF1, but may occur in our leukemias due to reduced expression of these transcription factors. Such loss of lineage fidelity is not only reflected in mixed B cell/myeloid leukemias but also seen in T-ALL. Specifically, we found that compound haploinsufficiency for Pax5 and Ebf1 promotes increased penetrance of B cell and T cell leukemia on an Ikzf1+/− background. Thus, decreased expression of transcription factors that are only expressed in B cells (PAX5 and EBF1) paradoxically enhances the development of T cell leukemia in Ikzf1+/− mice, suggesting that in some cases T-ALL may emerge from a B cell progenitor. This may have implications for how such leukemias develop resistance to therapy if the key progenitor is more closely linked to B cell rather than T cell development. Thus, our studies suggest that reduced B cell lineage fidelity entrained by loss of EBF1, PAX5 and/or IKZF1 could be an important part of the transformation process giving rise to both mixed lineage B/myeloid and B/T-ALL leukemias.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Rost, for technical assistance with mouse breeding; the University of Minnesota’s Supercomputing Institute for providing computing and bioinformatic resources; Dr. Meinrad Busslinger (Pax5+/−), Dr. Rudolf Grosschedl (Ebf1−/−), Dr. Peter Johnson (Cebpa−/−), Dr. Andrew Wells (Ikzf1−/−) and Dr. David Largaespada (Rosa26LSL-SB11T2/OncxRosa26LSL-SB11) for providing the indicated mouse strains. The results published here are in part based upon data generated by the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative, phs000218, managed by the NCI. This work was supported by a Cancer Research Institute Investigator award, a Leukemia and Lymphoma Society Scholar award, funding from the UMN Masonic Cancer center and grants from the NIH (RO1 CA232317) to MAF. TKS was supported by grants from the Randy Shaver Cancer Research and Community Fund, NIH NCI (R21 CA216652), and the Masonic Cancer Center. ALS was supported by NCI (CA211249) and Masonic Cancer Center Support Grant (CA077598). SMK was supported by CPRIT MIRA RP 160693 and NIH/NCI P50 CA100632-09.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007; 446: 758–764. [DOI] [PubMed] [Google Scholar]
- 2.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009; 360: 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mullighan CG. The genomic landscape of acute lymphoblastic leukemia in children and young adults. Hematology Am Soc Hematol Educ Program 2014; 2014: 174–180. [DOI] [PubMed] [Google Scholar]
- 4.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature 2008; 453: 110–114. [DOI] [PubMed] [Google Scholar]
- 5.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 1995; 376: 263–267. [DOI] [PubMed] [Google Scholar]
- 6.Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 1994; 79: 901–912. [DOI] [PubMed] [Google Scholar]
- 7.Chiang YJ, Kole HK, Brown K, Naramura M, Fukuhara S, Hu RJ et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nature 2000; 403: 216–220. [DOI] [PubMed] [Google Scholar]
- 8.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M et al. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity 1996; 5: 537–549. [DOI] [PubMed] [Google Scholar]
- 9.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M et al. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 1995; 374: 70–74. [DOI] [PubMed] [Google Scholar]
- 10.Lee YH, Sauer B, Johnson PF, Gonzalez FJ. Disruption of the c/ebp alpha gene in adult mouse liver. Mol Cell Biol 1997; 17: 6014–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupuy AJ, Rogers LM, Kim J, Nannapaneni K, Starr TK, Liu P et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer research 2009; 69: 8150–8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Temiz NA, Moriarity BS, Wolf NK, Riordan JD, Dupuy AJ, Largaespada DA et al. RNA sequencing of Sleeping Beauty transposon-induced tumors detects transposon-RNA fusions in forward genetic cancer screens. Genome research 2016; 26: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beckmann PJ, Larson JD, Larsson AT, Ostergaard JP, Wagner S, Rahrmann EP et al. Sleeping Beauty Insertional Mutagenesis Reveals Important Genetic Drivers of Central Nervous System Embryonal Tumors. Cancer research 2019; 79: 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott MC, Temiz NA, Sarver AE, LaRue RS, Rathe SK, Varshney J et al. Comparative Transcriptome Analysis Quantifies Immune Cell Transcript Levels, Metastatic Progression, and Survival in Osteosarcoma. Cancer research 2018; 78: 326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarver AL, Xie C, Riddle MJ, Forster CL, Wang X, Lu H et al. Retinoblastoma tumor cell proliferation is negatively associated with an immune gene expression signature and increased immune cells. Lab Invest 2021; 101: 701–718. [DOI] [PubMed] [Google Scholar]
- 16.Heltemes-Harris LM, Willette MJ, Ramsey LB, Qiu YH, Neeley ES, Zhang N et al. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. The Journal of experimental medicine 2011; 208: 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell 1995; 83: 289–299. [DOI] [PubMed] [Google Scholar]
- 18.Papathanasiou P, Perkins AC, Cobb BS, Ferrini R, Sridharan R, Hoyne GF et al. Widespread failure of hematolymphoid differentiation caused by a recessive niche-filling allele of the Ikaros transcription factor. Immunity 2003; 19: 131–144. [DOI] [PubMed] [Google Scholar]
- 19.Heath V, Suh HC, Holman M, Renn K, Gooya JM, Parkin S et al. C/EBPalpha deficiency results in hyperproliferation of hematopoietic progenitor cells and disrupts macrophage development in vitro and in vivo. Blood 2004; 104: 1639–1647. [DOI] [PubMed] [Google Scholar]
- 20.Prasad MA, Ungerback J, Ahsberg J, Somasundaram R, Strid T, Larsson M et al. Ebf1 heterozygosity results in increased DNA damage in pro-B cells and their synergistic transformation by Pax5 haploinsufficiency. Blood 2015; 125: 4052–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katerndahl CDS, Heltemes-Harris LM, Willette MJL, Henzler CM, Frietze S, Yang R et al. Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival. Nature immunology 2017; 18: 694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castel P, Ellis H, Bago R, Toska E, Razavi P, Carmona FJ et al. PDK1-SGK1 Signaling Sustains AKT-Independent mTORC1 Activation and Confers Resistance to PI3Kalpha Inhibition. Cancer Cell 2016; 30: 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan J, Li Z, Lee PL, Guan P, Aau MY, Lee ST et al. PDK1 signaling toward PLK1-MYC activation confers oncogenic transformation, tumor-initiating cell activation, and resistance to mTOR-targeted therapy. Cancer Discov 2013; 3: 1156–1171. [DOI] [PubMed] [Google Scholar]
- 25.Najafov A, Sommer EM, Axten JM, Deyoung MP, Alessi DR. Characterization of GSK2334470, a novel and highly specific inhibitor of PDK1. The Biochemical journal 2011; 433: 357–369. [DOI] [PubMed] [Google Scholar]
- 26.Kornblau SM, Tibes R, Qiu YH, Chen W, Kantarjian HM, Andreeff M et al. Functional proteomic profiling of AML predicts response and survival. Blood 2009; 113: 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013; 14: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016; 44: W90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao X, Yang G, Bai P, Gui S, Nyuyen TM, Mercado-Uribe I et al. Inhibition of nuclear factor-kappa B enhances the tumor growth of ovarian cancer cell line derived from a low-grade papillary serous carcinoma in p53-independent pathway. BMC Cancer 2016; 16: 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen F, Castranova V. Nuclear factor-kappaB, an unappreciated tumor suppressor. Cancer research 2007; 67: 11093–11098. [DOI] [PubMed] [Google Scholar]
- 31.Oliveros JC. Venny. An interactive tool for comparing lists with Venn's diagrams, 2007-2015. [Google Scholar]
- 32.van der Weyden L, Giotopoulos G, Wong K, Rust AG, Robles-Espinoza CD, Osaki H et al. Somatic drivers of B-ALL in a model of ETV6-RUNX1; Pax5(+/−) leukemia. BMC Cancer 2015; 15: 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008; 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kollmann S, Grundschober E, Maurer B, Warsch W, Grausenburger R, Edlinger L et al. Twins with different personalities: STAT5B-but not STAT5A-has a key role in BCR/ABL-induced leukemia. Leukemia 2019; 33: 1583–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heltemes-Harris LM, Larson JD, Starr TK, Hubbard GK, Sarver AL, Largaespada DA et al. Sleeping Beauty transposon screen identifies signaling modules that cooperate with STAT5 activation to induce B-cell acute lymphoblastic leukemia. Oncogene 2016; 35: 3454–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah S, Schrader KA, Waanders E, Timms AE, Vijai J, Miething C et al. A recurrent germline PAX5 mutation confers susceptibility to pre-B cell acute lymphoblastic leukemia. Nat Genet 2013; 45: 1226–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Churchman ML, Qian M, Te Kronnie G, Zhang R, Yang W, Zhang H et al. Germline Genetic IKZF1 Variation and Predisposition to Childhood Acute Lymphoblastic Leukemia. Cancer Cell 2018; 33: 937–948 e938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J, Kryczek I, Li S, Li X, Aguilar A, Wei S et al. The ubiquitin ligase MDM2 sustains STAT5 stability to control T cell-mediated antitumor immunity. Nature immunology 2021; 22: 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goetz CA, Harmon IR, O'Neil JJ, Burchill MA, Farrar MA. STAT5 activation underlies IL7 receptor-dependent B cell development. J Immunol 2004; 172: 4770–4778. [DOI] [PubMed] [Google Scholar]
- 40.Badger-Brown KM, Gillis LC, Bailey ML, Penninger JM, Barber DL. CBL-B is required for leukemogenesis mediated by BCR-ABL through negative regulation of bone marrow homing. Leukemia 2013; 27: 1146–1154. [DOI] [PubMed] [Google Scholar]
- 41.Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 2014; 371: 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia Y, Shen S, Verma IM. NF-kappaB, an active player in human cancers. Cancer Immunol Res 2014; 2: 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fahl SP, Crittenden RB, Allman D, Bender TP. c-Myb is required for pro-B cell differentiation. J Immunol 2009; 183: 5582–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.