Abstract
RORγt, the master transcription factor for cytokine IL-17, is expressed explicitly in Th17 cells, γδT cells, and type 3 innate lymphoid cells in mice and humans. Since dysregulated IL-17 expression is strongly linked to several human inflammatory diseases, the RORγt-IL-17 axis has been the focus of intense research. Recently, several studies have shown that RORγt is modified by multiple post-translational mechanisms, including ubiquitination, acetylation, SUMOylation, and phosphorylation. This review discusses how post-translational modifications modulate RORγt function and its turnover to regulate IL-17-driven inflammation. Broad knowledge of these pathways is crucial for a clear understanding of the pathogenic role of RORγt+IL-17+ cells and for the development of putative therapeutic strategies to target IL-17-driven diseases such as multiple sclerosis, psoriasis, rheumatoid arthritis, systemic lupus erythematosus, and inflammatory bowel disease.
RORγt and autoimmune diseases
Interleukin (IL)-17 is crucial for mammalian host defense against fungal and bacterial infections. However, dysregulated IL-17 expression and its downstream signaling are involved in several inflammatory diseases such as Inflammatory bowel disease (IBD) (see Glossary), multiple sclerosis (MS), psoriasis, systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and asthma [1,2]. IL-17 gained extensive interest after discovering the Th17 subset of helper T cells as the primary source of IL-17 [3]. Additionally, CD8+ T cells, γδT cells, natural killer T (NKT) cells, and Type 3 innate lymphoid cells (ILC3) also produce IL-17 in mice, humans, and non-human primates [1,2].
RORγt is a member of the retinoic acid-related orphan receptor (ROR) family of transcription factors (Box 1) [4,5]. Rorc−/− mice exhibit diminished autoimmune disease, e.g. in experimental autoimmune encephalomyelitis (EAE) [6] and experimental psoriasis [7], and these clinical symptoms have been associated with the absence of tissue-infiltrating Th17 cells [6]. In the last few years, substantial attention has been paid to understanding how RORγt induces IL-17 expression and the differentiation of CD4+ T cells into Th17 cells (Box 2). To induce these processes, RORγt collaborates with several other transcription factors, including IRF4, STAT3, Bob1, and BATF [8,9].
Box 1. Chromatin architecture of RORγt.
The Rorc gene in both humans and mice has two different isoforms denoted as RORγ1 (RORγ) and RORγ2 (popularly known as RORγt) [50]. The Rorc gene is comprised of 11 exons (1–11) in mice and humans [50]. Both isoforms of the Rorc gene use the last nine exons (exons 3–11), but exons 1 and 2 are transcribed differently, leading to two different isoforms [50]. As a result, the mRNA of RORγt is 100 nucleotides smaller than RORγ, which translates into a smaller N-terminal amino acid (aa) sequence (495 aa for RORγt versus 516 aa for RORγ in mice, and 508 aa for RORγt versus 518 aa for RORγ in humans) [50,51]. The two isoforms show a distinct tissue-specific expression pattern and regulate the expression of a different set of genes [50,51]. RORγt is predominantly expressed in Th17 cells and thymocytes, while RORγ1 protein expression is found ubiquitously in mice and humans [6,50,52]. The mechanism underlying the differential expression of the two isoforms remains unclear and needs to be validated rigorously. One study reported two functional Rorc promoters and two distinct forms of 5’-capped Rorc mRNAs in differentiated murine Th17 cells by using the Rapid amplification of cDNA ends (5’-RACE) approach [51]. The authors identified two different transcription start sites (TSS) on the Rorc gene, producing two types of Rorc mRNA and translating into the two isoforms, RORγ1 and RORγt (Figure I A–C) [51]. Sequence alignment of human and mouse promoters showed several putative transcription factor binding sites such as NFAT, AP-1, GATA3, SP1, Rel/NF-κB, and TATA box on each Rorc promoter close to the conserved non-coding sequence (CNS)1 region [51], suggesting alternative promoter usage to form different isoforms.
In addition to differential promoter usage, alternate splicing can also generate protein isoforms [53]. Isoforms of RORα with different N- and C-termini are produced by alternative RNA splicing in humans [53]. Therefore, the possibility of alternative RNA splicing in generating the two isoforms of RORγ also needs to be explored. Another study suggested the existence of a new RORγt isoform, RORγt-Δ (5–8), that does not contain the hinge region encoding exons 5 to 8 in humans [54]. Luciferase assays in human T cells suggested that this RORγt variant represses Il17a transcription [54]. Whether RORγt-Δ (5–8) generation occurs via alternate-splicing or different promoter usage remains unknown. Similarly, how RORγt-Δ (5–8) represses Il17a expression remains to be investigated. Because the DNA binding domain (DBD) of both isoforms are conserved, how the existence or loss of the hinge region encoding exons 5 to 8 can decide whether the isoform serves as a trans-repressor or trans-activator is one area for future investigation.
Figure I in Box 1. Rorc/RORC loci and alternative promoter usage for different isoforms in mice and humans.
(A) Schematic representations of the Rorc locus and two Rorc promoters that generate isoforms of RORγ proteins in mice and humans. The transcription starts site (TSS) for RORγ on Exon 1 is shown in orange, and for RORγt on Exon 3, in purple [50,51]. (B) Schematic representation of the Rorc regulatory region in mice and humans. Three NFAT and NF-κB binding sites on the Rorc promoter are shown (consensus binding sites 4, 6, and 7), while no binding whas been observed at consensus sites 1, 2, and 3 shared between the two promoters [91]. (C) Schematic representation of the domain architecture of RORγ isoforms in mice. The variable A/B domain, DNA binding domain (DBD), ligand-binding domain (LBD), and activation function 2 domain (AF-2) are indicated [51]. The total number of amino acids is shown on the right. This figure was created using BioRender (https://biorender.com/).
Box 2. RORγt and IL-17 transcription.
RORγt binds to the ROR response elements (Rore) within the Il17a promoter in mice and humans [55,56]. The CNS2 of the Il17a gene contains two Rore [57]. Luciferase reporter assays and chromatin conformation capture analysis in murine Th17 cells have shown that RORγt binds to CNS2 and mediates Il17a transcription by controlling chromatin remodeling [57]. Histone modifying enzymes such as p300 and JmjC domain-containing protein (JMJD)3 also bind the CNS2 and mediate permissive histone acetylation, as validated by ChIP analysis in murine Th17 cells [57]. Further, CNS2 associates with the Il17a promoter via a chromatin loop and recruits histone-modifying enzymes associated with CNS2 to the promoter to activate Il17a transcription in murine Th17 cells [8,57]. In a recent report using ATAC seq analysis, CNS6 and CNS9 were demonstrated to be essential for RORγt expression in mouse Th17 cells but not in innate lymphoid cells or γδT cells [58]. IL-6-induced STAT3 binds CNS9, and partially CNS6, whereas TGF-β-induced SMADs predominantly bind to CNS6 to epigenetically activate the Rorc locus and thus, RORγt expression in mouse Th17 cells [58]. Moreover, genetic deletion of CNS9 (Cns9−/− mice) has resulted in complete resistance to EAE, whereas CNS6 deletion (Cns6−/− mice) has led to only partial resistance to EAE, due to a defect in IL-17 expression by Th17 cells [58].
RORγt also cooperates with other transcription factors with known functions in inflammatory signaling and hypoxia responses, for the optimum transcription of Il17a in mouse and human Th17 cells [8]. In murine Th17 cells, ChIP analysis showed that the Runt-related transcription factor (RUNX)1 formed a complex with RORγt, and bound the CNS2 region of the Il17a promoter [59]. A gel-shift assay showed that RORα bound CNS2 along with RORγt in murine Th17 cells [60]. Retrovirus-mediated overexpression of RORα along with RORγt, synergized IL-17 expression in murine Th17 cells [60]. In addition, the nuclear protein inhibitor of kB (IκB)ζ bound CNS2 elements in the Il17a locus, resulting in the recruitment of transcriptional coactivators in mouse Th17 cells [61,62]. Further, ChIP analysis in combination with luciferase reporter assays of mouse Th17 cells showed that HIF-1α directly activated Rorc gene transcription. Additionally, HIF-1α recruited p300 to RORγt transcription complexes on IL-17 promoters and induced Th17 differentiation in mice [63].
In this review, we discuss how post-translational modifications such as ubiquitination, SUMOylation, acetylation, and phosphorylation regulate RORγt function, the recruitment of coactivators or corepressors, and the degradation of this transcription factor in mice and humans (Table 1). Broad knowledge of this area is essential for the development of selective and safe modulators of RORγt activity to target the RORγt-IL-17 pathway in an effort to inhibit autoimmune responses and inflammation.
Table 1:
List of post-translational modifications and their effect on RORγt-mediated Il17a expression in mice and humans.
| Modifications | Effects on RORγt | Outcomes | Ref. |
|---|---|---|---|
| E3 ligase ITCH, NDFIP1, and UBR5-mediated ubiquitination | Promotes degradation of RORγt | Inhibits Il17a expression, inhibits colonic inflammation in mouse models and human colon cancer. | [14,16,86] [25] |
| E3 ligase TRAF5-mediated ubiquitination | Stabilizes RORγt | Enhances Il17a expression and increased Traf5 amounts are linked to elevated IL-17 in SLE patients. | [87] |
| USP4-mediated deubiquitination (K48-linked) of RORγt, and USP17-mediated deubiquitination (K48-linked) of RORγt at K360 | Stabilizes RORγt and enhances transcriptional activity. | Promotes Il17a expression in human Th17 cells. | [23,24] |
| DUBA-mediated deubiquitination | Promotes Degradation of RORγt | Inhibits Il17a expression and Inhibits colonic inflammation in mice model. | [25] |
| USP19-mediated deubiquitination (K63-linked) of RORγt at K313 | Disrupts RORγt/SRC3 association and attenuates differentiation of pathogenic Th17 cells. | Inhibits EAE in mouse model. | [26] |
| USP15-mediated deubiquitination of RORγt at K446 | Stimulates RORγt activity by enhancing coactivator SRC1 recruitment | Promotes Il17a expression in murine Th17 cells. | [88] |
| SUMOylation at K187 | Facilitates recruitment of HDAC2 and inhibits transcriptional activity. | Inhibits Il17a expression and colonic inflammation in mouse models and IBD patients. | [28] |
| SUMOylation at K31 | Facilitates recruitment of histone acetyltransferase KAT2A and maintains the binding of SRC1 resulted in enhanced transcriptional activity of RORγt. | Enhances Th17 differentiation and modulates EAE severity in mice model. Affected the development of thymic immature CD8+ single-positive cells in the murine thymus. |
[32] |
| p300-mediated acetylation of RORγt at K81 | Enhances DNA-binding, and the transcriptional activity of RORγt | Enhances Il17a expression in human Th17 cells. | [34] |
| HDAC1-mediated deacetylation | Reverses the effect of p300 | Inhibits Il17a expression in human Th17 cells. | [34] |
| Sirtuin 1-mediated deacetylation | Increases RORγt transcriptional activity | Increases Il17a expression and EAE severity in mouse model. | [89] |
| IKKα-mediated phosphorylation at S376 | Increases RORγt transcriptional activity | Enhances murine Th17 differentiation. | [90] |
| IKKβ-mediated phosphorylation of RORγt at S489 | Promotes RORγt-AHR interaction and Increases RORγt transcriptional activity | Increases disease severity in mouse models of EAE and IL-17 by T cells of SLE patients. | [39] |
RORγt ubiquitination
Ubiquitin (Ub) conjugation plays a crucial regulatory role in immune cells [10–12]. Ubiquitination is a multi-enzymatic biochemical process that is carried out by three classes of enzymes, i.e., Ub activating (E1), Ub conjugating (E2), and Ub ligating (E3) enzymes (Box 3).
Box 3. Biochemical process of protein ubiquitination.
Ubiquitination is a multi-enzymatic post-translational modification that is carried out by three classes of enzymes, i.e., Ub activating (E1), Ub conjugating (E2), and Ub ligating (E3) enzymes [10,12,64]. Ub activating (E1) performs activation of Ub molecule and tis hen transferred to the Ub conjugating (E2) enzyme. The Ub ligating (E3) enzyme then associates with the Ub-conjugated E2 enzyme and the substrate molecule, which in turn forms the iso-peptide bond between the Ub (C-terminus) and the substrate lysine residues [65,66]. Initially, ubiquitin was suggested to provide a ‘kiss of death’, facilitating the degradation of misfolded proteins via the 26S proteasome. However, several lines of evidence show that ubiquitination of target proteins has wide implications for several subcellular processes, including subcellular localization of proteins, protein complex formations, receptor turnover on cell surfaces, and the regulation of gene expression [10–12]. Not surprisingly, dysregulation of the ubiquitin pathway is linked to several inflammatory diseases and malignancies in humans [10].
E3 ligases play a crucial role in the process of ubiquitination as they confer substrate specificity [10,12,64]. E3s are classified depending on domain structure and the mechanisms of transferring the Ub molecule to the target protein: 1) Really interesting new gene (RING) E3 ligases which contains a RING domain, catalyzes the transfer of the E2 Ub molecule to the substrate protein, or 2) Homologous to the E6AP carboxyl terminus (HECT) is a domain type comprising a conserved HECT domain at the C-terminus (responsible for catalysis) and a substrate-binding domain at the N-terminus (responsible for substrate recognition) [10,12,64]. The HECT domain E3 ligases transfer the Ub molecule in a two-step process involving the transfer of the Ub molecule to catalytic cysteine of E3 ligase and finally to substrate protein [10,12,64].
ITCH, an E3 ligase, is comprised of an N-terminal C2 domain (protein kinase C related), four WW domains (WW 1–4), and a C-terminal HECT domain [13]. ITCH inhibits IL-17 production by targeting RORγt for ubiquitination in Th17 cells in mice and humans [14,15]. ITCH protein binds to the conserved PPLY region on RORγt via its WW domain and leads to its degradation, resulting in inhibition of IL-17 expression (Figure 1A–B) [14]. Accordingly, Itch−/− mice spontaneously develop rectal prolapse due to severe colonic inflammation, which has been associated with increased expression of IL-17 by colonic lamina propria lymphocytes (CLPLs) [14]. Consistent with these data, another study reported that mice deficient in NDFIP1 (Ndfip1−/−), a coactivator of ITCH, exhibited increased numbers of Th17 cells in the lung and intestinal mucosae, relative to wild-type controls [16]. NDFIP1 binds to ITCH and promotes its ligase activity in murine CD4+ T cells following TCR ligation [17] via recruitment of ubiquitin-conjugating enzyme E2 (UBCH7) to ITCH [18]. Similar to Itch−/− Th17 cells, Ndfip1−/− Th17 cells have exhibited increased IL-17 expression in lung and intestinal mucosae due to reduced degradation and accumulation of RORγt, relative to wild-type Th17 cells [16]. Moreover, Ndfip1−/− Th17 cells have been reported to be highly colitogenic upon transfer to Rag1−/− mice [16], suggesting a crucial role for the NDFIP1-ITCH pathway in the regulation of IL-17-mediated colonic inflammation, at least in mice.
Figure 1. RORγt ubiquitination by the E3 ligase ITCH in mice.
(A) Cartoon representation of homology-modeled structure of mouse RORγt, binding to the consensus Rore binding sites at the Il17a promoter. DNA binding domain (DBD) and PPLY-motif are shown [19]. The binding of RORγt to Rore sites on the Il17a promoter drives Il17a transcription in mice and humans. (B) Regulation of RORγt by E3 ligase ITCH mediated ubiquitination [14]. Cartoon representation of homology-modeled structure of mouse ITCH, showing HECT and WW domains. The WW domain of ITCH and PPLY domain of RORγt are labeled. The WW domain of ITCH interacts with the PPLY domain of RORγt, which mediates ubiquitination, and proteasomal degradation of RORγt, reducing Il17a transcription in mice [14].
Accordingly, a deficiency in ITCH-mediated RORγt degradation was reported in colorectal cancer (CRC) patients [15]. In this report, CRNDE-h (Colon rectal neoplasia differentially expressed) protein was shown to form a complex with the PPLY region of RORγt in tumor infiltrating Th17 cells [15]. Using fluorescent in situ hybridization (FISH) and RNA pull-down assays, the authors of this study demonstrated that the binding of CRNDE-h to RORγt in patient Th17 cells prevented ubiquitination and proteasomal degradation of RORγt by impeding its association with ITCH [15]. A positive correlation was also observed between CRNDE-h expression and the percentage of Th17 cells among tumor-infiltrating lymphocytes (TILs) from CRC patients [15]. These results are in agreement with previous evidence demonstrating that Itch−/− mice exhibit increased colon cancer growth [15] and further highlight the role of ITCH in Th17-meditated tumor-promoting inflammation in colon cancer.
The WW domains of HECT type E3 ligases fold to form a twisted triple-stranded antiparallel β-sheet structure (Figure 1B) [19]. The first loop of the antiparallel β-strand is responsible for ligand recognition, and the threonine-hydroxyl group of WW domain forms an H-bond with the ligand and stabilizes the WW-PPLY complex [19]. ITCH contains four WW domains (WW 1–4) [11], but it remains to be investigated which of the four WW domains binds to the PPLY motif. This structural information might lead to a more precise characterization of the conditions that lead to RORγt ubiquitination, which could in turn inform new targeting strategies for fine-tuning ITCH ligase activity and promoting RORγt ubiquitination.
In a group of pediatric patients from an Amish family, a truncating mutation in ITCH resulted in multi-organ inflammatory disorders similar to the phenotype of Itch−/− mice [20]. Sequencing of DNA isolated from the peripheral blood of these patients revealed a single base pair insertion of nucleotide adenine between positions 394–395 of exon 6, resulting in a frame shift-mutation and early termination of the ITCH protein (139 amino acids compared to 903 amino acids of full-length ITCH) [20]. The affected children presented with splenomegaly, hepatomegaly, autoimmune hepatitis, and chronic lung inflammation [20,21]. They also exhibited hypothyroidism, which was associated with autoantibodies directed against thyroglobulin and thyroid peroxidase [20]. It remains to be investigated if a defect in the degradation of RORγt (due to lack of a functional ITCH) might contribute to the observed phenotype of these patients. Analysis of IL-17 expression and RORγt protein amounts in the inflamed lung and liver tissues might provide evidence of a dysregulated RORγt-IL-17 pathway. Additionally, if reconstitution of full-length ITCH in CD4+ T cells obtained from these children reduced IL-17 expression, it could confirm a mechanism by which dysregulated RORγt might contribute, at least in part, to the multi-organ autoimmune syndrome observed in these patients.
Ubiquitination is a reversible process, and the removal of Ub chains bound to protein substrates is mediated by deubiquitinating enzymes (DUBs) [22]. The ubiquitin-specific protease (USP) 4, a DUB, was shown to be highly expressed in mouse and human Th17 cells [23]. Co-immunoprecipitation experiments using human CD4+ T cell lysates demonstrated that USP4 associates with RORγt. Further, it was shown that USP4 co-localizes with RORγt in the nucleus [23]. Cycloheximide (CHX) chase experiments in 293T cells expressing either wild-type or a catalytically inactive mutant of USP4 (C311A) inhibited ubiquitination and promoted the stability of the RORγt protein [23]. Similarly, USP17 was reported to inhibit RORγt degradation by decreasing its polyubiquitination at the Lysine (K)360 residue [24]. In contrast, the deubiquitinating enzyme DUBA (also known as OTUD5) can function as a negative regulator of IL-17 production in mice [25]. Moreover, intraperitoneal injection of anti-CD3 antibodies -- triggering Th17-driven inflammation in the small intestine-- into CD4+ T cell-specific conditional DUBA knockout mice (Dubafl/flCd4Cre mice) resulted in severe intestinal inflammation compared to control mice [25]. Incubation of mouse Th17 cells with CHX showed that RORγt protein stability in Duba−/− cells was extended beyond six hours compared to wild-type mice in which RORγt was stable for one hour [25]. Mass spectrometry coupled to short interfering RNA (siRNA) screening of ubiquitinated proteins in Duba−/− Th17 cells revealed that DUBA directly bound and deubiquitinated UBR5 (Ubiquitin protein ligase E3 component n-recognin 5). The co-immunoprecipitation experiments in murine Th17 cells showed that the UBR5-RORγt interaction led to ubiquitination and proteasomal degradation of RORγt, resulting in decreased IL-17 expression (Figure 2) [25]. In addition, siRNA-mediated knockdown of UBR5 in Th17 cells increased IL-17 production and exacerbated inflammation in the murine small intestine [25]. Since DUBA and UBR5 deficiency resulted in a similar phenotype, the authors of this study speculated that DUBA suppressed RORγt expression indirectly, by stabilizing UBR5. In support of this, treatment of murine Th17 cells with MG-132, a proteasome inhibitor, increased the amount of RORγt protein in wild-type cells but not in Duba−/− cells [25]. Furthermore, Duba−/− Th17 cells contained less UBR5 protein than Duba+/+ Th17 cells, despite similar expression of UBR5 mRNA [25]. Thus, DUBA seems to provide an indirect regulatory mechanism via which IL-17 is produced from Th17 cells, and which involves regulating the stability of RORγt-associated E3 ubiquitin ligase, UBR5 [25].
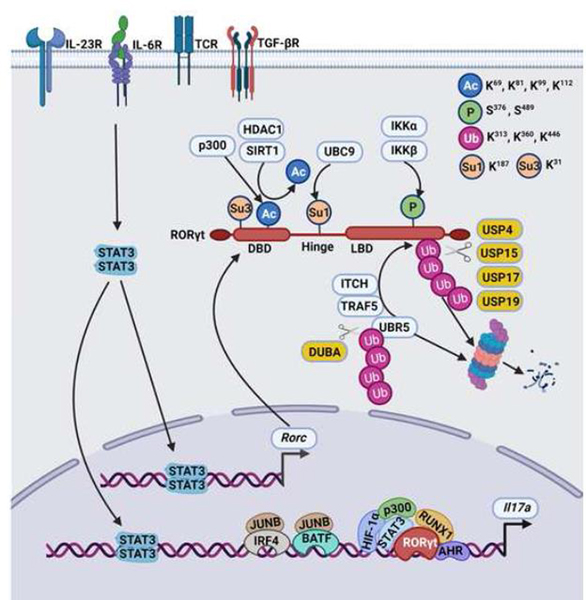
Figure 2. Summary of post-translational modifications of RORγt in mice and humans.
TCR and cytokines signals lead to transcription of the Rorc gene. RORγt-mediated IL-17 expression is regulated by post-translational modifications (PTMs). p300 acetylates RORγt at the DBD and inhibits binding to consensus binding sequence (Rore) at the Il17a promoter in murine Th17 cells [34]. SIRT1, an NAD+-dependent protein deacetylase and histone deacetylase 1 (HDAC1), removes the acetylation and restores the RORγt binding ability to promoter sequences in murine Th17 cells [34,36]. The SUMO E2 ligase, UBC9, causes SUMOylation of RORγt, facilitating the binding of HDAC2 to the Il17a promoter and repressing Il17a transcription in murine Th17 cells [28]. The kinase IKKα phosphorylates RORγt and induces the RORγt-AHR interaction that in turn, activates transcription of the Il17a gene in murine Th17 cells [39]. RORγt is ubiquitinated by the E3 ligase ITCH, TRAF5,as well as UBR5 and undergoes 26S proteasomal degradation [14,25,87]. DUBA is a deubiquitinase of UBR5 in murine Th17 cells [25]. Deubiquitinases such as USP4, USP15, USP17, and USP19 directly act on RORγt in human and murine Th17 cells [23,24,26]. The color code symbol represents a green sphere: phosphorylation; blue sphere: Acetylation; orange sphere: SUMOylation; and red sphere: ubiquitination. The amino acid residues of RORγt that undergo PTMs are labeled. This figure was created using BioRender (https://biorender.com/).
Ubiquitination is often regulated by modulating the expression of enzymes involved in this process [10]. Chromatin immunoprecipitation (ChIP)-sequencing analysis of murine Th17 cells showed that RORγt binds to the promoters of genes involved in the ubiquitination pathway, including E1 and E2 enzymes [26]. Treatment of MOG35–55 peptide-specific murine Th17 cells with PR-619, a deubiquitinase inhibitor, increased EAE severity compared to cells treated with a vehicle control [26]. Further, mass spectrometry and shRNA screening identified USP19 as the most active deubiquitinase in murine pathogenic Th17 cells compared to nonpathogenic Th17 cells (Box 4) [26]. USP19 deubiquitinates the K63-linked ubiquitin chain and disrupts the association of RORγt with steroid receptor coactivator (SRC)3, contributing to suppressing the pathogenicity of murine Th17 cells [26]. This has suggested that different E3 ligases and DUBs can enable RORγt to adapt to various inflammatory signals, and responds to these by recruiting specific cofactors. Collectively, these observations indicate that such a mechanism might help determine the pathogenic versus homeostatic functions of different RORγt+IL-17+ subsets.
Box4: Nonpathogenic and pathogenic Th17 cells.
IL-17 producing Th17 cells that are found at sites of inflammation are intricately involved in autoimmune and inflammatory diseases [1,2]. However, all h17 cells do not contribute to pathogenic inflammation [67]. The Th17 cells that are present naturally in mucosal surfaces, such as the gastrointestinal tract, are essential for maintaining tissue homeostasis [67]. These cells are termed nonpathogenic and are not linked to autoimmune inflammation [68]. The Th17 cells generated in vitro by the addition of TGF-β and IL-6 are poor inducers of EAE in mice [2] and are considered nonpathogenic. While Th17 cells generated by stimulation of CD4+ T cells by the combination of IL-6+IL-1β+IL-23, or IL-6+IL-23+TGF-β3 acquire pathogenicity in mouse models of EAE [69,70]. The gene expression profiling of pathogenic and nonpathogenic Th17 cells has shown distinct regulatory and pathogenic signatures in mouse Th17 cells [70]. Single-cell sequencing of in vitro generated Th17 cells and sorted Th17 cells from mice at the peak of EAE have shown that pathogenic Th17 cells express higher amounts of IL-23R and GM-CSF and lower amounts of CD5L and IL-10 than from healthy control mice [71,72]. The nonpathogenic cells express higher concentrations of IL-10 and lower concentrations of GM-CSF and IL-23R in mouse Th17 cells [71,72]. However, the extracellular signals and intracellular mechanisms that regulate distinct transcriptional profiles and the functional diversity of Th17 cells in vivo remain unclear. IL-23 and IL-1β promote pathogenicity by inducing the expression of Activin-A, a member of the TGF-β superfamily in mice [73]. Consistently, elevated amounts of Activin-A were found in the serum of multiple sclerosis (MS) patients and in mice with EAE [73]. Of note, whole-genome transcriptome analysis of murine CD4+ T cells stimulated with Activin-A, along with IL-6, induced a molecular program of pathogenic Th17 cells [73]. Immunoblotting studies showed that TGF-β but not Activin-A blocked the phosphorylation of ERK that was required for the development of pathogenic Th17 cells, thus suggesting an opposing role for Activin-A and TGF-β1 in controlling the pathogenic and nonpathogenic differentiation of murine Th17 cells [73]. In a mouse model of uveitis, IL-24 was shown to inhibit Th17 pathogenicity in an autocrine manner as a feedback mechanism [74]. In addition, the pathogenic Th17 cells in mice utilize distinct metabolic pathways and have been found to be associated with the arginine and polyamine metabolism of Th17 cells [75,76]. A clear understanding of these pathways could lead to the development of candidate selective inhibitors for pathogenic Th17 cells without affecting homeostatic nonpathogenic Th17 cells.
SUMOylation of RORγt
Similar to ubiquitination, SUMOylation includes a series of enzymatic reactions which involve activating (E1), conjugating (E2), and ligases (E3) enzymes [27]. SUMOylation is carried out by a single E2 enzyme, UBC9, which transfers SUMO to substrate proteins [27]. A mass spectrometry-based approach showed that UBC9 associates with RORγt in murine Th17 cells isolated from the intestinal mucosa [28]. In general, UBC9 recognizes on its target proteins, the consensus binding motif Ψ-K-X-E/D (where Ψ is a hydrophobic residue, K is the lysine conjugated to SUMO, X is any amino acid, and D/E is eitheraspartic acid or glutamic acid respectively) [27]. Sequence alignment of mouse, sheep, orangutan, and human RORγt has demonstrated a highly conserved GKAE consensus motif [28].
Site-directed mutagenesis and lentiviral-mediated reconstitution experiments in murine Th17 cells showed that UBC9 recognizes the GKAE motif and targets K187 within this motif for SUMOylation [28]. Th17 cells, expressing SUMOylation-defective RORγt (RORγt-K187R mutant), led to severe colonic inflammation upon being adoptively transferred into Rag1−/− mice [28]. SUMOylated RORγt assisted in the recruitment of histone deacetylase HDAC2 onto the Il17a promoter, which resulted in reduced transcription of Il17a expression from murine Th17 cells relative to controls (Figure 2) [28]. Accordingly, compared to healthy controls, the expression of UBC9 was significantly decreased in the colonic mucosa of patients with ulcerative colitis (UC) --a form of IBD [29,30]. Promoter methylation analysis of DNA isolated from UC patients was performed via the bisulfite conversion method and showed that reduced UBC9 expression was due to promoter hyper-methylation [30]. Also, a ChIP-assay of murine Th17 cells showed that hypoxia-inducible factor (HIF)-1α bound to a CpG island in the promoter of the Ubc9 gene and enhanced Ubc9 promoter methylation [30]. These data suggested that epigenetic mechanisms could regulate RORγt indirectly by modulating the expression of Ubc9 in mice.
Five different subtypes of SUMO (SUMO1–5) have been reported in mammals [31]. Analysis of SUMO3 deficient mice (Sumo3−/−) showed an impairment of CD4+ T cell differentiation into Th17 cells compared to wild-type cells [32]. K31 was identified as the site of SUMO3-mediated RORγt sumoylation via mass spectrometry [32]. Also, co-immunoprecipitation assays showed that RORγt SUMOylation at K31 recruited histone acetyltransferase KAT2A and stabilized SRC1 binding to enhance the transcriptional activity of RORγt [32]. SUMO1 and SUMO3 facilitate SUMOylation at K187 and K31, respectively, and modulate the transcriptional activity of RORγt by recruiting different cofactors. It is likely that distinct external and intracellular signals lead to RORγt SUMOylation at different sites via selective use of SUMO1 or SUMO3. However, the mechanistic basis for the recruitment of different co-factors onto SUMOylated RORγt at K187 [28] (within the hinge region) and K31 [32] (within the DNA binding domain) remains unknown. Of note, SUMO1 forms mono-SUMOylation at K187 [28], whereas SUMO3 seems to form poly-SUMOylation at K31 [32]. Therefore, it is possible that mono- vs poly-SUMOylation differentially alters the interaction of RORγt with its co-factors, either by masking binding sites, or mediating conformational changes that reveal or block binding sites. Thus, X-ray crystal structures or Cryo-EM studies of unmodified and SUMOylated RORγt can provide additional insight into the molecular consequences of SUMOylation.
RORγt acetylation
Acetylation is another major form of post-translational modification in which the acetyl group is transferred to specific sites on proteins [33]. Co-immunoprecipitation experiments in human Th17 cells showed that p300 (also known as Ep300 or KAT3B), a histone acetyltransferase, interacts with RORγt and acetylates at residue K81 [34]. CHX chase experiments in 293T cells showed that p300-mediated acetylation stabilized RORγt protein [34]. shRNA-mediated knockdown of p300 in human Th17 cells reduced the amount of RORγt and RORγt -mediated IL-17 expression relative to Th17 cells treated with control nonspecific shRNA [34]. JQ1, an inhibitor of p300, impaired p300-mediated RORγt acetylation in human Th17 cells and attenuated Schistosoma-induced fibrosis in mice [35]. Of note, in an overexpression system using 293T cells, HDAC1 reciprocally reversed the effect of p300 and deacetylated RORγt and inhibits Il17a expression in an Il17a promoter-driven luciferase assay [34]. Since acetylation and ubiquitination often happen on the same lysine residues [33], it is reasonable to speculate that direct competition for acetylation or ubiquitination at this lysine residue might inhibit RORγt ubiquitination and degradation. Alternatively, acetylation might induce a conformational change in RORγt, or it might shield it from ubiquitin E3s, following an acetylation-dependent interaction with its partners.
In a parallel study, mass-spectrometric analysis of 293T cells showed that RORγt was acetylated at K69, K81, K99, and K112 [36]. SIRT1 (sirtuin 1), a deacetylase, interacted with RORγt in mouse Th17 cells and deacetylated the protein at three of these residues (K69, K81, and K99) [36]. Additionally, genetic deletion of Sirt1 (using Sirt1fl/flRorcCre mice) in Th17 cells attenuated IL-17 expression and protected mice from EAE [36]. This finding contrasts an earlier report showing that acetylation promoted RORγt stability and enhanced IL-17 expression [34]. The exact reason for the discrepancy between these two studies remains unclear; however, SIRT1 deacetylates RORγt at K69, and K99 (in addition to K81) [36]. Thus, it is possible that acetylation at different sites on RORγt might have opposing functions, and hence, relying on SIRT1 deletion may not be conclusive. Moreover, in a recent report, pharmacological activation of SIRT1 using metformin, SRT1720, and resveratrol, blunted IL-17 expression in mouse and human Th17 cells [37]. Reduced IL-17 expression via SIRT1 activation was mediated by STAT3 deacetylation in mouse Th17 cells [37]. Thus, we posit that further detailed studies involving knock-in mice in which K69, K81, and K99 are individually mutated to arginine (R) at respective positions might provide clear evidence for the regulation of Th17-mediated inflammation via RORγt acetylation.
RORγt phosphorylation
Phosphorylation of RORγt itself, as well as modulators of RORγt, can enhance as well as inhibit the transcriptional activity of RORγt and Il17a expression, depending on the site of phosphorylation. SRC1, which functions as a coactivator of RORγt, was demonstrated to be phosphorylated at S1271 and S1272 by protein kinase C theta (PKC-θ) in murine Th17 cells [38]. Co-immunoprecipitation experiments using wild-type and Pkc-θ−/− Th17 cells, demonstrated that T cell receptor (TCR) signaling-induced PKC-θ activation was crucial for the association of RORγt with SRC1, as well as for RORγt binding to the Il17a promoter [38]. Moreover, SRC1 phosphorylation led to the recruitment of CARM1 (coactivator of arginine methyltransferase 1), resulting in progressive di-methylation of H3R17, while suppressing inhibitory H3K9 trimethylation, further modifying the open chromatin accessibility region of the Il17a locus for efficient IL-17 expression in murine Th17 cells [38]. Furthermore, Forkhead box protein P3 (FOXP3)-- a CD4+ regulatory T cell key transcription factor -- was displaced upon binding of phospho-SRC1 to RORγt, resulting in proteasomal degradation of FOXP3 and removal of its inhibitory effect on RORγt activity [38]. This implied that phosphorylated SRC1 could override FOXP3-mediated inhibition of RORγt transcriptional activity to induce Th17 differentiation, at least in mice.
Similarly, using confocal microscopy and immunoblotting in murine Th17 cells, germinal center kinase-like kinase (GLK) (also known as MAP4K3) was reported to phosphorylate aryl hydrocarbon receptor (AHR) at serine (S)36 and to promote AHR nuclear translocation [39]. This was further validated in Th17 cells of Lck-GLK transgenic mice (full-length human GLK inserted downstream of the Lck promoter) [39]. In addition, experiments of co-transfection of kinases, and GST-pull down assays showed that IKKβ-mediated phosphorylation of RORγt at S489, induce an RORγt-AHR interaction [39]. Altogether, these data suggested that IL-17 production by Th17 cells was dependent on TCR triggering-induced phosphorylation of the RORγt-AHR complex. It is possible that this mechanism might serve as a means for preventing IL-17 expression from Th17 cells in the absence of foreign antigens, and for preventing autoimmunity (Figure 2). Accordingly, an increased percentage of GLK+IL-17A+ T cells was detected among peripheral blood CD4+ and CD8+ T cells taken from SLE patients, compared with that of healthy controls [40]. Indeed, CD4+ and CD8+ T cells from SLE patients exhibited increased phosphorylation of RORγt as well as RORγt-AHR associations relative to controls, suggesting an important role for the GLK-RORγt-IL-17 pathway in SLE pathology [41]. Of note, verteporfin, a small-molecule inhibitor, reduced IL-17 production from mouse Th17 cells and total T cells isolated from SLE and RA patients [41]. Collectively, these studies suggest that inhibitors of GLK or the RORγt-AHR complex might be considered as candidate IL-17A blocking agents in the potential treatment of IL-17-mediated autoimmune diseases.
Targeting the RORγt-IL-17 axis
Given the significant role of IL-17 in autoimmune and inflammatory diseases, the RORγt-IL-17 axis is thus an attractive therapeutic target. Digoxin has been recognized as the first RORγt inhibitor [42]; since then, a number of new RORγt inhibitors have been actively developed for the potential treatment of autoimmune diseases. As a result, several molecules have been assessed in clinical trials, such as ABBV-553, GSK-2981278, JTE-151, VTP-43742, JNJ-3534, AZD-0284, and TAK-828, but were either suspended or discontinued due to either a lack of efficacy, or safety concerns in humans [43]. Most of these inhibitors bind to the ligand-binding domain (LBD) [43,44], which is identical in both RORγt and RORγ in mice and humans. Thus, it is reasonable to speculate that these inhibitors might also inhibit RORγ, which is expressed ubiquitously. In addition, RORγt is essential for thymocyte development as well as lymphoid organ development in mice [43,44]. Therefore, the long-term effects of RORγt inhibitors might result in off-target adverse effects. New targeting strategies to specifically inhibit RORγt at the site of inflammation without affecting RORγ would overcome these limitations. Advancement in our knowledge of the post-translational modifications of RORγt could also open novel alternative avenues for targeting RORγt for drug discovery in the possible treatment of autoimmune diseases.
The intramolecular interaction between the different domains of the ITCH protein inhibits its E3 ligase activity [45,46]. Following TCR stimulation, JNK1 phosphorylates ITCH at S199, threonine (T) 222, and S232 residues, followed by a change in conformation, resulting in disrupted intramolecular interactions, subsequently enabling the E3 ligase activity [47]. Therefore, small molecules that block intramolecular interactions might potentially activate ITCH and degrade RORγt. Further, the specificity of degradation might be enhanced by using a proteolysis-targeting chimera (PROTAC). We envision that a putative ITCH-modulating small molecule could be attached to a flexible chemical linker and an RORγt binding molecule. This binding and formation of RORγt-PROTAC-ITCH E3 ubiquitin ligase complex would be expected to degrade RORγt, but this remains to be rigorously tested. Thus, with careful design and by combining precision chemistry with cellular protein degradation machinery, it may be possible to promote RORγt degradation at sites of inflammation.
Finally, it is currently unknown if TCR-induced phosphorylation of RORγt causes any conformational changes on the protein. In such a scenario, highly specific RORγt inhibitors might be developed such that TCR-induced events would be expected to be specific for RORγt but not for RORγ considering that TCR signaling is T-cell specific. Therefore, we posit that a detailed understanding of the post-translational regulation of RORγt is an essential component to efficiently targeting IL-17- driven inflammatory diseases.
Concluding remarks
The RORγt-IL-17 axis has emerged as a crucial pathogenic factor in several autoimmune diseases such as psoriasis, SLE, RA, and IBD. Despite the remarkable efficacy of antibodies against IL-17A and IL-17Rs in treating psoriasis [2], concerns remain for the potential risk of systemic inactivation of IL-17 activity, which provides essential host defense and proper barrier function at mucosal surfaces [2,48,49]. During the last few years, substantial advancements have been made in our understanding of the regulation of RORγt, which could help in developing strategies to inhibit selective aspects of IL-17-mediated inflammation in a site-specific manner.
The necessity for such diverse and non-redundant post-translational mechanisms to promote or turn off RORγt -mediated IL-17 expression remains unclear (see Outstanding Questions). It is possible that these pathways may not be individually sufficient, but a coordinated effort is necessary at multiple levels. The rate of stimulation or inhibition of each regulatory molecule may act uniquely to module RORγt at different stages of signaling to fine-tune Th17-mediated inflammation. In addition, it is likely that these pathways may differentially regulate nonpathogenic and pathogenic Th17 cells (see Outstanding Questions). Further, the binding of different natural ligands of RORγt (Box 5) may trigger distinct post-translational modifications of RORγt. Detailed mechanistic knowledge of these issues will be advantageous in developing safe and selective drug targets for managing IL-17-driven diseases, and we posit that it certainly merits future attention.
Outstanding questions.
Pathogenic and non-pathogenic Th17 cells express distinct phenotypic and transcriptional profiles. Do different post-translational modifications molecularly regulate the development of pathogenic and non-pathogenic Th17 cells?
Most ROR-γt post-translational modifications are studied in Th17 cells. Do similar regulations occur in ILC3, γδT cells, NK cells, or other cells?
What is the role of other post-translational modifications such as deSUMOylation, methylation, glycosylation, and palmitoylation in the regulation of ROR-γt and IL-17-driven inflammation?
Post-translational modifications are generally triggered by extracellular or intracellular signals. Which external/internal signals drive ROR-γt modifications? Are these signals different for different post-translational modifications?
Is there any crosstalk between different types of post-translational modifications of ROR-γt? In particular, can phosphorylation or acetylation positively or negatively regulate ROR-γt ubiquitination?
Can ITCH or other E3 ligases be targeted using PROTAC approaches to ideally treat psoriasis, IBD, or other IL-17-associated diseases?
Box 5: The natural ligands of RORγt.
The nuclear receptor (NR) ligands in metazoans are mostly hydrophobic molecules that are mainly derivatives of cholesterol, fatty acids, retinoids, vitamins, and lipophilic hormones [77]. In an insect cell-based, RORγ reporter assay using Drosophila melanogaster cells, supplementing the lipid-free medium with cholesterol and its derivatives restored luciferase activity [78]. Overexpression of cholesterol biosynthetic enzymes in 293T cells co-transfected with an RORγ reporter plasmid could regulate the transcriptional activity of RORγ [78]. Ketoconazole inhibition of CYP51, the cholesterol biosynthetic enzyme causing demethylation of the canonical sterol in the nucleus, reduced IL-17 production in mouse and human Th17 cells and reduced skin inflammation in a mouse imiquimod-induced psoriasis model, relative to controls [79]. These studies established the functional role of cholesterol biosynthesis intermediates as natural ligands of RORγt. Similarly, a surface plasmon resonance (SPR) binding assay demonstrated that 7β, 27-OHC, a naturally occurring sterol, acted as a ligand for RORγt in mice and humans [80]. Incubation of mouse and human CD4+ Th17 cells with 7β, 27-OHC enhanced IL-17 production by Th17 cells in vitro [80]. It remains to be investigated if endogenous ligands bind the LBD domain of RORγt to induce IL-17 expression during an inflammatory response.
X-ray crystallography and biochemical analysis of other nuclear receptors (NR) such as peroxisome proliferator-activated receptor (PPAR)α, PPARγ, and constitutive androstane receptor (CAR) have shown that ligand binding to NRs is dependent on their post-translational modifications [81–85]. Therefore, an interesting area of research will be to explore the role of post-translational modifications that modulating ligand binding of RORγt. Further, exciting areas for future study include the characterization off different cell types that can produce IL-17 (e.g., ILCs and Th17 cells), and whether these utilize distinct ligands of the transcription factor based on cellular metabolic and activation states. Additionally, It remains to be assessed whether Th17 cells respond to external or internal signals by modulating the availability and type of natural ligands when regulating the RORγt-mediated transcriptional activity that can impart pathogenic vs protective Th17 functions.
Highlights.
ROR-γt is a member of the ROR family of transcription factors, and is selectively expressed in Th17 cells, γδT cells, and innate lymphoid cells in mice and humans. ROR-γt cooperates with STAT3, IRF4, and BATF to induce Il17a transcription in human and murine Th17 cells.
ROR-γt protein turnover is regulated by the interplay between its ubiquitination and deubiquitination. The ubiquitin E3 ligase ITCH, UBR5, and TRAF5 facilitate ROR-γt ubiquitination in mouse Th17 cells, whereas USP4, USP17, and mouse Th17 cells USP19 promote its deubiquitination in human Th17 cells. DUBA indirectly promotes ROR-γt degradation by deubiquitinating UBR5 in murine Th17 cells, which promotes degradation of ROR-γt.
SUMOylation of ROR-γt at K-187 facilitates the recruitment of histone deacetylase HDAC2 and inhibits Il17a expression in mouse Th17 cells and IBD patients. In contrast, SUMOylation at K-31 facilitates the recruitment of histone acetyltransferase KAT2A and enhances ROR-γt transcription activity in murine Th17 cells.
DNA-binding and the transcriptional activity of ROR-γt are regulated via p300-mediated acetylation in mouse Th17 cells. HDAC1 inhibits Il17a transcription by reversing the effect of p300, whereas Sirtuin-1 enhances Il17a transcription.
ACKNOWLEDGEMENTS
We thank Dr. Ezra Burstein for the helpful discussions and Dr. Wayne Lancaster for the critical reading of the manuscript. This work was supported by funds from the National Institutes of Health (R01-DK115668, R01-AI155786) and Cancer Prevention Research Institute of Texas (RP160577, RP190527), and a translational pilot project grant from the Harold C. Simmons Comprehensive Cancer Center, UT Southwestern Medical Center (23001045) to K. Venuprasad and R01- DK117001 to K. Venuprasad and Arianne Theiss.
Glossary:
- Inflammatory bowel disease
category of intestinal disorders characterized by chronic inflammation of the gastrointestinal tract. Ulcerative colitis (UC) and Crohn’s disease (CD) are two major forms of IBD.
- Rag1−/− mice
Recombination activating gene1 (RAG1) is involved in antibody and T-cell receptor V(D)J recombination. Deletion of Rag1 (Rag1−/− mice) impairs T cell and B cell development and deletes mature T and B cells from the immune system.
- Colon rectal neoplasia differentially expressed-h
CRNDE-h is a 1kb long noncoding RNA, transcribed from chromosome 16q12.2. It is a proto-oncogene secreted from the exosomes of colorectal cancer patients.
- Aryl hydrocarbon receptor
basic-helix-loop-helix transcription factor encoded by the AHR gene; ligand-activated factor that integrates environmental, dietary, microbial, and metabolic cues to control complex transcriptional programs in a ligand-, cell-type-, and context-specific manner
- Ligand-binding domain
consists of three-layer fold of 11–12 alpha helices and 2–3 β-strands to which ligands binds on orthosteric as well allosteric sites and can inhibit or activate the function of a protein.
- Proteolysis Targeting Chimera (PROTAC)
hetero-bifunctional small molecules that contain two active domains connected by a linker- one domain binds to the target protein, and the other to the E3 ligase. The ternary complex of E3 ligase, PROTAC, and the target protein brings the ubiquitination machinery in close proximity and facilitates ubiquitination and degradation of the target protein.
- Conserved Non-coding Sequence (CNS) region
regions on the genome that do not code for proteins and are evolutionary conserved; involved in gene regulation, including transcription factor binding sites, and cis-regulatory elements.
- Alternative promoter usage
can influence gene expression and generates transcriptome diversity within the genome; in turn, provides a framework for differential subcellular specificity and functions of proteins.
- Alternative pre-mRNA splicing
mechanism by which introns are removed, and exons or portions of exons are assembled to construct different isoforms of mature mRNA from a single pre-mRNA.
- Chromatin remodeling
mechanism of gene regulation in which chromatin undergoes rearrangement from a condensed state to an open state (transcriptionally active), allowing binding of transcription factors and gene regulatory elements.
- Imiquimod (IMQ)-induced dermatitis
most commonly used experimental model to study dermatitis in rodents. Topical application of 5% IMQ cream induces localized scaly skin lesions similar to human psoriasis.
Footnotes
Competing interests
The authors declare that they do not have any competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- 1.McGeachy MJ et al. (2019) The IL-17 Family of Cytokines in Health and Disease. Immunity 50, 892–906. 10.1016/j.immuni.2019.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel DD and Kuchroo VK (2015) Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity 43, 1040–1051. 10.1016/j.immuni.2015.12.003 [DOI] [PubMed] [Google Scholar]
- 3.Harrington LE et al. (2005) Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 6, 1123–1132. 10.1038/ni1254 [DOI] [PubMed] [Google Scholar]
- 4.Robinson-Rechavi M et al. (2003) The nuclear receptor superfamily. J Cell Sci 116, 585–586. 10.1242/jcs.00247 [DOI] [PubMed] [Google Scholar]
- 5.Rutz S et al. (2016) Post-translational regulation of RORgammat-A therapeutic target for the modulation of interleukin-17-mediated responses in autoimmune diseases. Cytokine Growth Factor Rev 30, 1–17. 10.1016/j.cytogfr.2016.07.004 [DOI] [PubMed] [Google Scholar]
- 6.Ivanov II et al. (2006) The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133. S0092–8674(06)01105–6 [pii] 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- 7.Xue X et al. (2016) Pharmacologic modulation of RORgammat translates to efficacy in preclinical and translational models of psoriasis and inflammatory arthritis. Sci Rep 6, 37977. 10.1038/srep37977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capone A and Volpe E (2020) Transcriptional Regulators of T Helper 17 Cell Differentiation in Health and Autoimmune Diseases. Front Immunol 11, 348. 10.3389/fimmu.2020.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikegami I et al. (2019) Bob1 enhances RORgammat-mediated IL-17A expression in Th17cells through interaction with RORgammat. Biochem Biophys Res Commun 514, 1167–1171. 10.1016/j.bbrc.2019.05.057 [DOI] [PubMed] [Google Scholar]
- 10.Venuprasad K et al. (2006) Immune regulation by ubiquitin conjugation. Adv Exp Med Biol 584, 207–217. 10.1007/0-387-34132-3_15 [DOI] [PubMed] [Google Scholar]
- 11.Venuprasad K et al. (2015) Multifaceted role of the ubiquitin ligase Itch in immune regulation. Immunol Cell Biol 93, 452–460. 10.1038/icb.2014.118 [DOI] [PubMed] [Google Scholar]
- 12.Venuprasad K (2010) Cbl-b and itch: key regulators of peripheral T-cell tolerance. Cancer Res 70, 3009–3012. 10.1158/0008-5472.CAN-09-4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu K et al. (2017) Allosteric auto-inhibition and activation of the Nedd4 family E3 ligase Itch. EMBO Rep 18, 1618–1630. 10.15252/embr.201744454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kathania M et al. (2016) Itch inhibits IL-17-mediated colon inflammation and tumorigenesis by ROR-gammat ubiquitination. Nat Immunol 17, 997–1004. 10.1038/ni.3488 [DOI] [PubMed] [Google Scholar]
- 15.Sun J et al. (2021) Tumor exosome promotes Th17 cell differentiation by transmitting the lncRNA CRNDE-h in colorectal cancer. Cell Death Dis 12, 123. 10.1038/s41419-020-03376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Layman AAK et al. (2017) Ndfip1 restricts Th17 cell potency by limiting lineage stability and proinflammatory cytokine production. Sci Rep 7, 39649. 10.1038/srep39649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver PM et al. (2006) Ndfip1 protein promotes the function of itch ubiquitin ligase to prevent T cell activation and T helper 2 cell-mediated inflammation. Immunity 25, 929–940. 10.1016/j.immuni.2006.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kathania M et al. (2015) Ndfip1 Regulates Itch Ligase Activity and Airway Inflammation via UbcH7. J Immunol. 10.4049/jimmunol.1402742 [DOI] [PubMed] [Google Scholar]
- 19.Lorenz S (2018) Structural mechanisms of HECT-type ubiquitin ligases. Biol Chem 399, 127–145. 10.1515/hsz-2017-0184 [DOI] [PubMed] [Google Scholar]
- 20.Lohr NJ et al. (2010) Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Hum Genet 86, 447–453. 10.1016/j.ajhg.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleine-Eggebrecht N et al. (2019) Mutation in ITCH Gene Can Cause Syndromic Multisystem Autoimmune Disease With Acute Liver Failure. Pediatrics 143. 10.1542/peds.2018-1554 [DOI] [PubMed] [Google Scholar]
- 22.Massoumi R (2010) Ubiquitin chain cleavage: CYLD at work. Trends in biochemical sciences 35, 392–399. 10.1016/j.tibs.2010.02.007 [DOI] [PubMed] [Google Scholar]
- 23.Yang J et al. (2015) Cutting edge: Ubiquitin-specific protease 4 promotes Th17 cell function under inflammation by deubiquitinating and stabilizing RORgammat. J Immunol 194, 4094–4097. 10.4049/jimmunol.1401451 [DOI] [PubMed] [Google Scholar]
- 24.Han L et al. (2014) The E3 deubiquitinase USP17 is a positive regulator of retinoic acid-related orphan nuclear receptor gammat (RORgammat) in Th17 cells. J Biol Chem 289, 25546–25555. 10.1074/jbc.M114.565291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rutz S et al. (2015) Deubiquitinase DUBA is a post-translational brake on interleukin-17 production in T cells. Nature 518, 417–421. 10.1038/nature13979 [DOI] [PubMed] [Google Scholar]
- 26.Zhang J et al. (2021) USP19 Suppresses Th17-Driven Pathogenesis in Autoimmunity. J Immunol. 10.4049/jimmunol.2100205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hickey CM et al. (2012) Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13, 755–766. 10.1038/nrm3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh AK et al. (2018) SUMOylation of ROR-gammat inhibits IL-17 expression and inflammation via HDAC2. Nat Commun 9, 4515. 10.1038/s41467-018-06924-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mustfa SA et al. (2017) SUMOylation pathway alteration coupled with downregulation of SUMO E2 enzyme at mucosal epithelium modulates inflammation in inflammatory bowel disease. Open Biol 7. 10.1098/rsob.170024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar R et al. (2021) Cutting Edge: Hypoxia-Induced Ubc9 Promoter Hypermethylation Regulates IL-17 Expression in Ulcerative Colitis. J Immunol 206, 936–940. 10.4049/jimmunol.2000015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Celen AB and Sahin U (2020) Sumoylation on its 25th anniversary: mechanisms, pathology, and emerging concepts. FEBS J 287, 3110–3140. 10.1111/febs.15319 [DOI] [PubMed] [Google Scholar]
- 32.He Z et al. (2018) Sumoylation of RORgammat regulates TH17 differentiation and thymocyte development. Nat Commun 9, 4870. 10.1038/s41467-018-07203-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choudhary C et al. (2014) The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol 15, 536–550. 10.1038/nrm3841 [DOI] [PubMed] [Google Scholar]
- 34.Wu Q et al. (2015) Reciprocal regulation of RORgammat acetylation and function by p300 and HDAC1. Sci Rep 5, 16355. 10.1038/srep16355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X et al. (2020) JQ1, a bromodomain inhibitor, suppresses Th17 effectors by blocking p300-mediated acetylation of RORgammat. Br J Pharmacol 177, 2959–2973. 10.1111/bph.15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim HW et al. (2015) SIRT1 deacetylates RORgammat and enhances Th17 cell generation. J Exp Med 212, 607–617. 10.1084/jem.20132378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Limagne E et al. (2017) Sirtuin-1 Activation Controls Tumor Growth by Impeding Th17 Differentiation via STAT3 Deacetylation. Cell Rep 19, 746–759. 10.1016/j.celrep.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 38.Sen S et al. (2018) SRC1 promotes Th17 differentiation by overriding Foxp3 suppression to stimulate RORgammat activity in a PKC-theta-dependent manner. Proc Natl Acad Sci U S A 115, E458–E467. 10.1073/pnas.1717789115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chuang HC et al. (2018) GLK-IKKbeta signaling induces dimerization and translocation of the AhR-RORgammat complex in IL-17A induction and autoimmune disease. Sci Adv 4, eaat5401. 10.1126/sciadv.aat5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chuang HC et al. (2011) The kinase GLK controls autoimmunity and NF-kappaB signaling by activating the kinase PKC-theta in T cells. Nat Immunol 12, 1113–1118. 10.1038/ni.2121 [DOI] [PubMed] [Google Scholar]
- 41.Chuang HC et al. (2019) AhR-ROR-gammat complex is a therapeutic target for MAP4K3/GLK(high)IL-17A(high) subpopulation of systemic lupus erythematosus. FASEB J 33, 11469–11480. 10.1096/fj.201900105RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huh JR et al. (2011) Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature 472, 486–490. 10.1038/nature09978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun N et al. (2019) Retinoic acid receptor-related orphan receptor gamma-t (RORgammat) inhibitors in clinical development for the treatment of autoimmune diseases: a patent review (2016-present). Expert Opin Ther Pat 29, 663–674. 10.1080/13543776.2019.1655541 [DOI] [PubMed] [Google Scholar]
- 44.Huh JR and Littman DR (2012) Small molecule inhibitors of RORgammat: targeting Th17 cells and other applications. Eur J Immunol 42, 2232–2237. 10.1002/eji.201242740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang L et al. (2006) The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell 124, 601–613. 10.1016/j.cell.2006.01.021 [DOI] [PubMed] [Google Scholar]
- 46.Gao M et al. (2004) Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306, 271–275. 10.1126/science.1099414 [DOI] [PubMed] [Google Scholar]
- 47.Gallagher E et al. (2006) Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci U S A 103, 1717–1722. 10.1073/pnas.0510664103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ju J et al. (2020) Crohn’s disease exacerbated by IL-17 inhibitors in patients with psoriasis: a case report. BMC Gastroenterol 20, 340. 10.1186/s12876-020-01474-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Philipose J et al. (2018) Severe de novo Ulcerative Colitis following Ixekizumab Therapy. Case Rep Gastroenterol 12, 617–621. 10.1159/000493922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He YW et al. (1998) RORgamma t, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity 9, 797–806. 10.1016/s1074-7613(00)80645-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruan Q et al. (2011) The Th17 immune response is controlled by the Rel-RORgamma-RORgamma T transcriptional axis. J Exp Med 208, 2321–2333. 10.1084/jem.20110462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Z et al. (2000) Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science 288, 2369–2373 [DOI] [PubMed] [Google Scholar]
- 53.Giguere V et al. (1994) Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev 8, 538–553. 10.1101/gad.8.5.538 [DOI] [PubMed] [Google Scholar]
- 54.Rauen T et al. (2012) A novel isoform of the orphan receptor RORgammat suppresses IL-17 production in human T cells. Genes Immun 13, 346–350. 10.1038/gene.2011.85 [DOI] [PubMed] [Google Scholar]
- 55.Jetten AM et al. (2001) The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes. Prog Nucleic Acid Res Mol Biol 69, 205–247. 10.1016/s0079-6603(01)69048-2 [DOI] [PubMed] [Google Scholar]
- 56.Xiao S et al. (2014) Small-molecule RORgammat antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity 40, 477–489. 10.1016/j.immuni.2014.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X et al. (2012) Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity 36, 23–31. 10.1016/j.immuni.2011.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang D et al. (2020) The Conserved Non-coding Sequences CNS6 and CNS9 Control Cytokine-Induced Rorc Transcription during T Helper 17 Cell Differentiation. Immunity 53, 614–626 e614. 10.1016/j.immuni.2020.07.012 [DOI] [PubMed] [Google Scholar]
- 59.Zhang F et al. (2008) Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol 9, 1297–1306. 10.1038/ni.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang XO et al. (2008) T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity 28, 29–39. 10.1016/j.immuni.2007.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okamoto K et al. (2010) IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature 464, 1381–1385. 10.1038/nature08922 [DOI] [PubMed] [Google Scholar]
- 62.Yamazaki S et al. (2001) A novel IkappaB protein, IkappaB-zeta, induced by proinflammatory stimuli, negatively regulates nuclear factor-kappaB in the nuclei. J Biol Chem 276, 27657–27662. 10.1074/jbc.M103426200 [DOI] [PubMed] [Google Scholar]
- 63.Dang EV et al. (2011) Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell 146, 772–784. 10.1016/j.cell.2011.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venuprasad K et al. (2015) Multifaceted role of the ubiquitin ligase Itch in immune regulation. Immunol Cell Biol. 10.1038/icb.2014.118 [DOI] [PubMed] [Google Scholar]
- 65.Pickart CM (2004) Back to the future with ubiquitin. Cell 116, 181–190. S0092867403010742 [pii] [DOI] [PubMed] [Google Scholar]
- 66.Ciechanover A (2005) Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ 12, 1178–1190. 4401692 [pii] 10.1038/sj.cdd.4401692 [DOI] [PubMed] [Google Scholar]
- 67.Yasuda K et al. (2019) The pathogenicity of Th17 cells in autoimmune diseases. Semin Immunopathol 41, 283–297. 10.1007/s00281-019-00733-8 [DOI] [PubMed] [Google Scholar]
- 68.Omenetti S et al. (2019) The Intestine Harbors Functionally Distinct Homeostatic Tissue-Resident and Inflammatory Th17 Cells. Immunity 51, 77–89 e76. 10.1016/j.immuni.2019.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghoreschi K et al. (2010) Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 467, 967–971. 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee Y et al. (2012) Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 13, 991–999. 10.1038/ni.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang C et al. (2015) CD5L/AIM Regulates Lipid Biosynthesis and Restrains Th17 Cell Pathogenicity. Cell 163, 1413–1427. 10.1016/j.cell.2015.10.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stockinger B and Omenetti S (2017) The dichotomous nature of T helper 17 cells. Nat Rev Immunol 17, 535–544. 10.1038/nri.2017.50 [DOI] [PubMed] [Google Scholar]
- 73.Wu B et al. (2021) The TGF-beta superfamily cytokine Activin-A is induced during autoimmune neuroinflammation and drives pathogenic Th17 cell differentiation. Immunity 54, 308–323 e306. 10.1016/j.immuni.2020.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chong WP et al. (2020) The Cytokine IL-17A Limits Th17 Pathogenicity via a Negative Feedback Loop Driven by Autocrine Induction of IL-24. Immunity 53, 384–397 e385. 10.1016/j.immuni.2020.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagner A et al. (2021) Metabolic modeling of single Th17 cells reveals regulators of autoimmunity. Cell. 10.1016/j.cell.2021.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu L et al. (2020) Niche-Selective Inhibition of Pathogenic Th17 Cells by Targeting Metabolic Redundancy. Cell 182, 641–654 e620. 10.1016/j.cell.2020.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sladek FM (2011) What are nuclear receptor ligands? Mol Cell Endocrinol 334, 3–13. 10.1016/j.mce.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Santori FR et al. (2015) Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell Metab 21, 286–298. 10.1016/j.cmet.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu X et al. (2015) Sterol metabolism controls T(H)17 differentiation by generating endogenous RORgamma agonists. Nat Chem Biol 11, 141–147. 10.1038/nchembio.1714 [DOI] [PubMed] [Google Scholar]
- 80.Soroosh P et al. (2014) Oxysterols are agonist ligands of RORgammat and drive Th17 cell differentiation. Proc Natl Acad Sci U S A 111, 12163–12168. 10.1073/pnas.1322807111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shizu R et al. (2017) Phosphorylated Nuclear Receptor CAR Forms a Homodimer To Repress Its Constitutive Activity for Ligand Activation. Mol Cell Biol 37. 10.1128/MCB.00649-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shao D et al. (1998) Interdomain communication regulating ligand binding by PPARgamma. Nature 396, 377–380. 10.1038/24634 [DOI] [PubMed] [Google Scholar]
- 83.Grimaldi B et al. (2010) PER2 controls lipid metabolism by direct regulation of PPARgamma. Cell Metab 12, 509–520. 10.1016/j.cmet.2010.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tian L et al. (2014) Acetylation-defective mutant of Ppargamma is associated with decreased lipid synthesis in breast cancer cells. Oncotarget 5, 7303–7315. 10.18632/oncotarget.2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brunmeir R and Xu F (2018) Functional Regulation of PPARs through Post-Translational Modifications. Int J Mol Sci 19. 10.3390/ijms19061738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramon HE et al. (2012) The E3 ubiquitin ligase adaptor Ndfip1 regulates Th17 differentiation by limiting the production of proinflammatory cytokines. J Immunol 188, 4023–4031. 10.4049/jimmunol.1102779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X et al. (2015) TRAF5-mediated Lys-63-linked Polyubiquitination Plays an Essential Role in Positive Regulation of RORgammat in Promoting IL-17A Expression. J Biol Chem 290, 29086–29094. 10.1074/jbc.M115.664573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He Z et al. (2016) Ubiquitination of RORgammat at Lysine 446 Limits Th17 Differentiation by Controlling Coactivator Recruitment. J Immunol 197, 1148–1158. 10.4049/jimmunol.1600548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lim HW et al. (2015) SIRT1 deacetylates RORgammat and enhances Th17 cell generation. J Exp Med 212, 973. 10.1084/jem.2013237805062015c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.He Z et al. (2017) Regulation of Th17 Differentiation by IKKalpha-Dependent and - Independent Phosphorylation of RORgammat. J Immunol 199, 955–964. 10.4049/jimmunol.1700457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yahia-Cherbal H et al. (2019) NFAT primes the human RORC locus for RORgammat expression in CD4(+) T cells. Nat Commun 10, 4698. 10.1038/s41467-019-12680-x [DOI] [PMC free article] [PubMed] [Google Scholar]