Abstract
Genetic experiments have determined that Ku, XRCC4, and ligase IV are required for repair of double-strand breaks by the end-joining pathway. The last two factors form a tight complex in cells. However, ligase IV is only one of three known mammalian ligases and is intrinsically the least active in intermolecular ligation; thus, the biochemical basis for requiring this ligase has been unclear. We demonstrate here a direct physical interaction between the XRCC4-ligase IV complex and Ku. This interaction is stimulated once Ku binds to DNA ends. Since XRCC4-ligase IV alone has very low DNA binding activity, Ku is required for effective recruitment of this ligase to DNA ends. We further show that this recruitment is critical for efficient end-joining activity in vitro. Preformation of a complex containing Ku and XRCC4-ligase IV increases the initial ligation rate 20-fold, indicating that recruitment of the ligase is an important limiting step in intermolecular ligation. Recruitment by Ku also allows XRCC4-ligase IV to use Ku's high affinity for DNA ends to rapidly locate and ligate ends in an excess of unbroken DNA, a necessity for end joining in cells. These properties are conferred only on ligase IV, because Ku does not similarly interact with the other mammalian ligases. We have therefore defined cell-free conditions that reflect the genetic requirement for ligase IV in cellular end joining and consequently can explain in molecular terms why this factor is required.
A double-strand break (DSB) is the most lethal form of cellular DNA damage. Such breaks can be caused by exposure to certain exogenous damaging agents, like ionizing radiation. DSBs are also normal intermediates in several cellular recombination processes, including meiotic recombination, mating type switching in yeast, and antigen receptor gene rearrangement [V(D)J recombination] in vertebrates. Regardless of the source, DSBs are repaired by one of two pathways. End joining (also termed nonhomologous end joining) rejoins broken ends, removing damaged nucleotides as necessary. In contrast, homologous recombination uses an intact copy (homolog or sister chromatid) of the broken chromosome as a template for repair dependent on processive DNA synthesis. End joining is likely to be the more error-prone pathway; nevertheless, in vertebrates it is a major pathway for general repair of DSBs. It is also the only pathway used to resolve intermediates in V(D)J recombination (reviewed, e.g., in reference 16).
Genetic experiments indicate that the Ku heterodimer, XRCC4, and ligase IV are required for efficient end-joining DSB repair in all eukaryotic species examined so far. Mutation of any one of these three factors in both the yeast Saccharomyces cerevisiae (15, 32) and mammalian cells (excepting neurological lineages) (9, 11, 13) have roughly equivalent effects, arguing that the functions of these three factors in the end-joining pathway are interdependent (reviewed in reference 20). Studies of S. cerevisiae also implicate a variety of additional factors in end joining, including the MRE11-RAD50-XRS2 complex. A complex containing orthologs to MRE11 and RAD50 has been identified in vertebrates, and although it is clearly linked in some way to DSB repair, it is not yet certain that the vertebrate version of the MRE11 complex is required for end joining (reference 33 and references therein).
In mammalian cells, end joining also typically requires the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs, or XRCC7). However, an ortholog for this factor has not been found in the fully sequenced genome of S. cerevisiae or the nearly complete genome of Caenorhabditis elegans (29). Even in species with a DNA-PKcs ortholog there are circumstances where its mutation still allows much greater levels of end joining than are observed when Ku, XRCC4, or ligase IV is mutated. Mice completely deficient in DNA-PKcs can join signal end intermediates in V(D)J recombination (2, 10, 30), and embryonic stem cells from such mice possess a normal level of resistance to ionizing radiation (10). The function of DNA-PKcs in end joining therefore may be more dispensable than that of Ku, XRCC4, or ligase IV, depending on the organism, cell type, and molecular context of the ends to be joined.
Ku is perhaps the best biochemically characterized of the factors required for end joining. Ku is a heterodimer of 83- and 70-kDa subunits (encoded by the XRCC5 and XRCC6 genes, respectively) that binds to DNA ends, including forks, single-stranded overhangs, and hairpins, with high specificity and affinity (reviewed in reference 8). Ku can juxtapose DNA ends (5, 24, 25), likely explaining its ability to generally stimulate intermolecular ligation (25).
Ku also recruits DNA-PKcs to DNA ends. DNA-PKcs is a 460-kDa serine/threonine protein kinase that is activated upon binding to DNA ends. Although DNA-PKcs is most efficiently recruited to and activated through formation of a complex on DNA ends with Ku, end binding and kinase activation can also occur in vitro in a Ku-independent manner (14, 34). Kinase activity is required for the role of DNA-PKcs in cellular end joining, but the biologically relevant substrates are not yet known (17).
XRCC4 forms a tight complex with ligase IV in cells (7, 12) (we will subsequently refer to this complex as X4-LIV). By itself, XRCC4 binds weakly to DNA, but this property has no apparent effect on the ligation activity of the X4-LIV complex (23). XRCC4 also interacts weakly with (19) and is phosphorylated by DNA-PK in vitro (7, 19, 23). However, interaction with DNA-PKcs is not essential for XRCC4 function, as mutant XRCC4 lacking these phosphorylation sites is capable of complementing XRCC4-deficient cells (22, 23) and some XRCC4-dependent repair pathways are intact in cells lacking DNA-PKcs (10).
Ligase IV protein is not stable in XRCC4-deficient cells, either in mammals or yeast (4, 15). While interaction of XRCC4 with ligase IV results in a modest stimulation of ligase IV activity in vitro (12, 23), even this complex is less effective at joining DSBs than either of the other two more abundant mammalian ligases (ligase I and ligase III) (25, 27). Nevertheless, both genetic (9, 13) and biochemical (1) experiments argue that it is the primary ligase used for end joining. Moreover, cells defective in ligase IV cannot be complemented by overexpression of the other two ligases (13). Ligase IV (presumably with XRCC4) may thus interact with one or more other DSB repair factors in cells and consequently becomes much more effective than the other ligases at joining DNA ends.
We demonstrate here that X4-LIV directly interacts with Ku in vitro. Ku specifically recruits X4-LIV to DNA ends, and together these proteins are capable of efficient end joining under conditions where Ku and the other mammalian ligases are not. Ku, XRCC4, and ligase IV thus cooperate to form a complex that greatly facilitates intermolecular ligation, consistent with the coordinate requirement for all three components for efficient end joining in eukaryotic species from yeast to mammals.
MATERIALS AND METHODS
DNA constructs.
Full-length ligase IV (911 amino acids [12]) was amplified from a cDNA (the gift of T. Lindahl) and inserted into pFASTBAC1 (Life Technologies, Bethesda, Md.). The coding sequence was modified to introduce a short DNA encoding the hexahistidine tag-containing sequence LEIEGRHHHHHH immediately before the stop codon. The 70-kDa subunit of Ku was also amplified from a cloned cDNA (the gift of D. Capra), inserted into pFASTBAC1, and modified to include the same tag sequence at the C terminus. The 83-kDa subunit of Ku was transferred from a cDNA to pFASTBAC1 without an affinity tag sequence. XRCC4 was amplified from pMMX4 (the gift of M. Modesti), and the hexahistidine tag already present in this construct was removed prior to insertion into pFASTBAC1. Each of these constructs was verified by sequencing the entire coding region.
Protein purification.
Viral isolates for insect cell expression of ligase IV, XRCC4, Ku70, and Ku80 were prepared from pFASTBAC1 constructs according to the manufacturer's instructions. SF-9 cells were coinfected with either XRCC4 and ligase IV virus at a 1:1 ratio or with Ku70 and Ku80 virus at a 1:2 ratio. Extracts were made in 50 mM sodium phosphate (pH 8.0), 1 M KCl, 10% glycerol, 0.25% Triton X-100 (Sigma), and 7 mM β-mercaptoethanol. Under these conditions, the amount of tagged protein (Ku70 for the Ku70-Ku80 infection or ligase IV for the X4-LIV infection) was limiting for both coinfections (see Fig. 1A, lane 1).
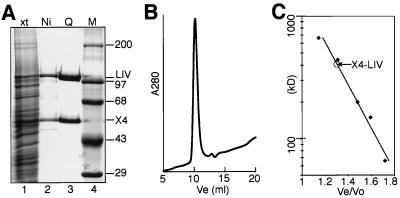
FIG. 1.
Purification of X4-LIV. (A) SDS-PAGE gel of protein fractions stained with Coomassie blue. Lane 1, total soluble extract (xt); lane 2, peak fractions from Ni column; lane 3, peak fractions from Mono Q column; lane 4, molecular weight marker (M). X4, XRCC4, LIV, ligase IV. (B) Gel filtration of purified X4-LIV. A280, UV absorbance at 280 nm; Ve, volume eluted after injection of sample. (C) Elution plot of X4-LIV relative to mass standards (in kilodaltons). Vo, excluded volume for this column. The Ve/Vo ratios for five molecular mass standards are marked by diamonds. The determined Ve/Vo ratio for X4-LIV is noted relative to a line derived from regression of the five standards.
Extracts were supplemented with imidazole to 10 mM, loaded onto a Ni-NTA Superflow (Qiagen) column, and eluted with extraction buffer plus 350 mM imidazole. Ku- or X4-LIV-containing fractions were dialyzed against buffer A (25 mM Tris [pH 8.0], 150 mM KCl, 10% glycerol, 0.05% Triton X-100, and 2 mM dithiothreitol [DTT] [Calbiochem]), loaded onto a Mono Q HR5/5 column (Pharmacia) preequilibrated in buffer A, and eluted with a linear gradient to 400 mM KCl over 20 column volumes. Ku was further purified by chromatography over a native DNA cellulose column as previously described (25). XRCC4 and ligases I and III were expressed in bacteria and purified as previously described (23, 25).
A filter binding assay was used to test for the presence of contaminating nucleic acids in our Ku and X4-LIV preparations. No nucleic acids were detected in 5-μg samples of each protein preparation, while parallel spotting with control DNA-containing samples indicated that this assay had a detection limit of 10 ng. We conclude that nucleic acid contamination of our protein preparations is less than 1:500, by mass, compared with the amount of protein and thus could not significantly impact our results.
Ku (purified as described in reference 6) and ligase IV (the gift of J. Turchi, Wright State University) from HeLa cells formed a complex equivalent to that shown with recombinant Ku and X4-LIV by the electrophoretic mobility shift assay (EMSA) (unpublished data). In addition, a similar complex was detected in whole-cell extracts from HeLa cells (see Fig. 4). This complex is therefore not a consequence of the presence of hexahistidine tags or the recombinant source. All proteins were frozen in small aliquots on liquid nitrogen and stored at −80°C. Protein concentrations were estimated by a modified Bradford assay (Coomassie Plus; Pierce) and comparison to a standard curve generated using serial dilutions of bovine serum albumin (BSA). Molar concentrations were calculated assuming a heterodimer for Ku (150 kDa), monomers for ligase I (125 kDa) and ligase III (100 kDa), and a heterotetramer with two molecules each for X4-LIV (280 kDa; our gel filtration data suggest this is likely a lower limit for the size of X4-LIV in solution). Proteins were diluted in buffer A plus 50 μg of BSA (New England Biolabs)/ml for use in all assays.
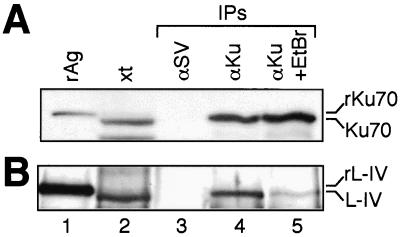
FIG. 4.
Immunoprecipitation of Ku from HeLa cell extracts. A 20-ng aliquot of the appropriate recombinant antigen (rAg; recombinant 70-kDa subunit of Ku or recombinant ligase IV), a 20-μg aliquot of the input extract (xt), and material recovered from immunoprecipitations (IPs) were electrophoresed and blotted onto a nitrocellulose membrane. αKu, immunoprecipitation with a monoclonal antibody to Ku; αSV, immunoprecipitation with an isotype-matched monoclonal antibody to SV40 T antigen; +EtBr, immunoprecipitation supplemented with 50 μg of ethidium bromide/ml. Recombinant antigens migrate slightly slower than the native antigens from HeLa cells due to the presence of C-terminal hexahistidine tags. Immunoblotting with the appropriate polyclonal antisera was used to detect Ku70 (A) and ligase IV (B).
Gel filtration analysis was performed on a 1- by 30-cm Superdex 200 column (Pharmacia) using a 100-μl aliquot of X4-LIV at 0.3 mg/ml, both in buffer A and in buffer A with 1 M KCl. Thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), and BSA (66 kDa) were used as standards, and the excluded volume (7.89 ml) for this column was determined by using blue dextran (2,000 kDa; Sigma).
EMSA.
A radiolabeled 60-bp double-stranded (ds) DNA substrate was made by 5′ 32P end labeling DAR166 (5′-CAGCTGGGAATTCCATATGAGTACTGCAGATGCACTTGCTCGATAGATCTAACATGAGCC-3′) and annealing the labeled DNA to DAR167 (5′-GTAGGGCTCATGTTAGATCTATCGAGCAAGTGCATCTGCAGTACTCATATGGAATTCCCAGCTGAG-3′). The DNA was incubated with the various proteins for 30 min on ice in a volume of 10 μl in our standard reaction buffer (standard buffer; 25 mM Tris [pH 7.5], 100 mM NaCl, 30 mM KCl, 0.1 mM EDTA, 0.05% Triton X-100, 50 μg of BSA/ml, 2% glycerol, and 2 mM DTT). Samples were subjected to electrophoresis for 25 min at 18 V/cm using a 3.5% polyacrylamide gel in a buffer containing 45 mM Tris-acetate (pH 8.0) and 1 mM EDTA. Antibody supershifts were performed by adding 1 μl of antibodies to each reaction mixture after the 30-min complex formation step and incubating the reaction mixtures for a further 10 min before electrophoresis. Sera were used after a 1:25 dilution, while monoclonal-antibody-expressing hybridoma supernatants were used undiluted. Electrophoresis of antibody-containing complexes was performed as described above, except that 90 mM Tris-borate (pH 8.2) and 1 mM EDTA were used as the electrophoresis buffer. Antibodies used included an anti-XRCC4 rabbit serum (Serotec), normal rabbit serum (Sigma), a monoclonal antibody to Ku (antibody 162; the gift of W. Reeves) and a monoclonal antibody to simian virus 40 (SV40) T antigen (antibody 101; the gift of W. Reeves).
Immunoprecipitations.
For the initial immunoprecipitation assay using purified protein, a sample of X4-LIV was deadenylated by treatment with sodium pyrophosphate, dialyzed, and then radiolabeled by incubation on ice for 30 min in buffer A plus 5 mM Mg2+ and approximately 1 μM [α-32P]ATP (3,000 Ci/mmol; Amersham). Radiolabeled X4-LIV was incubated with Ku and/or DNA in a volume of 500 μl of standard buffer for 30 min on ice. Aliquots of antibody-protein A-Sepharose complexes (made with 50 μl of protein A-Sepharose [Sigma] and 50 μl of hybridoma supernatant) for each reaction were prepared and washed twice in standard buffer. The reaction mixtures were added directly to antibody-protein A-Sepharose pellets and incubated for an additional 20 min at 4°C with constant mixing. Immune complexes were washed four times in standard buffer, pelleted by brief centrifugation, released from the beads by incubation at 95°C for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and electrophoresed on an SDS–8% PAGE gel. The gel was stained with Coomassie blue, photographed, and dried, and radiolabeled proteins were detected with a PhosphorImager (Molecular Dynamics).
Immunoprecipitation assays were also performed using whole-cell extracts from HeLa cells harvested from a 70% confluent 15-cm-diameter dish. The cells were washed twice with cold phosphate-buffered saline, and the washed cell pellet was resuspended in 0.5 ml of a buffer containing 25 mM Tris, 500 mM NaCl, 150 mM KCl, 0.5 mM EDTA, 0.25% Triton X-100, 2 mM DTT, and a mixture of protease inhibitors (Complete protease inhibitors; Boehringer Mannheim) and lysed by sonication. The extract was then diluted with 2 ml of a buffer containing 25 mM Tris (pH 7.5) and 2 mM DTT and clarified by centrifugation at 15,000 × g for 10 min. This produced a whole-cell extract with soluble protein at a concentration of approximately 1.5 mg/ml in a buffer roughly equivalent to the previously described standard buffer. Aliquots (0.5 ml) of extract were then precleared with protein A-Sepharose for 30 min before being added to antibody-protein A-Sepharose pellets prepared as described above. Where indicated, ethidium bromide was also added to a final concentration of 50 μg/ml; this concentration completely disrupted the independent association of Ku, Oct2, and a 110-kDa protein with DNA in a similar immunoprecipitation experiment (18). Immune complexes were purified as described above, but after a 1-h incubation with rotation at 4°C. Seventy percent of the material from each immunoprecipitation, as well as a 20-ng aliquot of recombinant X4-LIV complex and a 20-μg aliquot of the input extract, was electrophoresed on an SDS–6% PAGE gel, transferred to a nitrocellulose membrane using a semidry transfer apparatus (Hoeffer), and probed with a polyclonal rabbit antiserum raised against purified X4-LIV complex (Cocalico Biologicals). This antiserum did not cross-react with recombinant ligase I or III (unpublished data). The remainder of the material (30%) from each immunoprecipitation, as well as a 20-ng aliquot of recombinant Ku and a 20-μg aliquot of input extract, was separately electrophoresed, blotted, and probed with a polyclonal rabbit antiserum raised against purified Ku70 (Serotec). Both Ku and ligase IV were then detected with horseradish peroxidase-conjugated anti-rabbit immunoglobulin and Lumiglo peroxidase substrate (New England Biolabs).
Ligation assays.
Substrate L is a pair of dsDNA molecules: a radiolabeled 60-bp duplex, described above for use in the EMSA (DAR166 annealed to DAR167) as well as a 30-bp duplex made by annealing 5′-phosphorylated DAR 165 (5′-ACTGCAGTTCTAGACCTCATCGAGGGATTA-3′) to DAR 164 (5′-GATGTAATCCCTCGATGAGGTCTAGAACTGCAGTCT-3′). Substrate S is a second pair of dsDNA molecules, including 5′ 32P-labeled DAR296 (5′-CAGC TGGGAAT TCCATATGAG TAC TGCAGATGCAC T TGC TCGATAGATCTAACATGAG-3′) annealed to DAR281 (5′-GTAGGGCTCATGTTAGATCTATCGAGCAAGTGCATCTGCAGTACTCATATGGAATTCCCAGCTGGA-3′) and 5′-phosphorylated DAR 165 (also used to make substrate L) annealed to DAR297 (5′-GATGTAATCCCTCGATGAGGTCTAGAACTGCAGTTC-3′). Preincubations were performed in standard buffer supplemented with 10% (wt/vol) polyethylene glycol (molecular mass, greater than 8,000 kDa), and ligation was initiated by the addition of Mg2+ and ATP to a final concentration of 5 and 0.1 mM, respectively. The reactions were stopped by addition of an equal volume of 20 mM Tris (pH 8.0), 100 mM NaCl, 5 mM EDTA, and 0.2% SDS, incubated at 80°C for 5 min, extracted with 2 volumes of chloroform, and electrophoresed on a denaturing 8% polyacrylamide gel. Radiolabeled DNA was detected and quantified using a PhosphorImager and ImagequaNT software (Molecular Dynamics).
RESULTS
XRCC4 and ligase IV form a complex with Ku on DNA ends.
As might be predicted from the instability of ligase IV protein in XRCC4-deficient cells (4, 15), overexpression of recombinant ligase IV without XRCC4 resulted in low solubility and relatively inactive protein. We therefore generated a highly purified preparation of the X4-LIV complex by coexpressing human ligase IV and human XRCC4 in insect cells (Fig. 1). Although XRCC4 was observed in excess over ligase IV to varying degrees in extracts from different preparations, purification on the basis of an affinity tag in ligase IV always recovered the two proteins at an approximately 1:1 ratio (e.g., Fig. 1A, lane 3). Gel filtration chromatography and comparison to a series of globular protein standards indicates that XRCC4 and ligase IV form a homogeneous oligomer with an apparent molecular mass of 400 kDa, suggesting it possesses at least two molecules of each of the two proteins (Fig. 1B and C). This species was very stable, eluting as a single peak (without significant aggregation or the presence of free XRCC4 or ligase IV) upon gel filtration in 150 mM KCl (Fig. 1B) or 1 M KCl (unpublished data). Taken together, these data are suggestive of a very strong association and a highly favored architecture. Recombinant human Ku heterodimer was also purified from insect cells, as previously described (25).
We initially examined interaction between X4-LIV and Ku using an EMSA. Ku, X4-LIV, or combinations of the two factors were incubated with a 32P-labeled 60-bp DNA duplex for 30 min on ice. We detected DNA binding upon addition of Ku alone (Fig. 2A, lane 1, species I) but not X4-LIV alone (Fig. 2A, lane 2). However, addition of both the Ku heterodimer and an excess of X4-LIV eliminated the Ku-plus-DNA species and produced a species of even slower mobility (a supershift [Fig. 2A, lane 3, species II]).
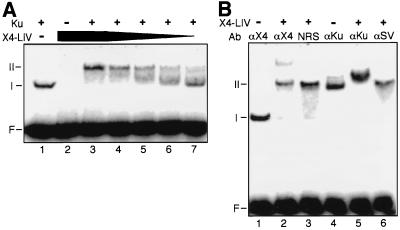
FIG. 2.
EMSA of Ku and X4-LIV. 5′ 32P-labeled 60-bp DNA duplex (50 nM) was present in all reactions. F, free DNA probe. Species I and II are DNA-protein complexes. (A) Formation of DNA-protein complexes with Ku and X4-LIV. The concentration of Ku, when present (+), was 5 nM. X4-LIV was added to a concentration of 50 (lanes 2 and 3), 25 (lane 4), 12.5 (lane 5), 5 (lane 6), and 2.5 nM (lane 7). (B) Characterization of DNA-protein complexes with antibodies. Ku (5 nM) was present in all reactions. The concentration of X4-LIV when present was 25 nM. αX4, polyclonal antiserum to XRCC4; NRS, normal rabbit serum; αKu, monoclonal antibody to Ku; αSV, monoclonal antibody to SV40 T antigen.
The presence of both X4-LIV and Ku in species II was verified by using antibodies. Addition of polyclonal antiserum against XRCC4 retarded some of species II (Fig. 2B, lane 2) but not species I (Fig. 2B, lane 1); higher concentrations of this antiserum shifted all of species II, but the immune complex no longer entered the gel (unpublished data). Association of XRCC4 and ligase IV is stable under these conditions; therefore, we conclude that the X4-LIV complex is present in species II. The participation of Ku in these DNA-protein complexes was verified by using a monoclonal antibody to the Ku heterodimer, which retarded species I (containing only Ku and DNA [Fig. 2B, lane 4]) but produced an even slower migrating species when added to reactions with Ku, X4-LIV, and DNA (Fig. 2B, compare lane 4 to lane 5). Control experiments with normal rabbit serum (Fig. 2B, lane 3) or an isotype-matched monoclonal antibody (Fig. 2B, lane 6) demonstrated that both the anti-XRCC4 and anti-Ku reagents, respectively, were specific for these antigens. Species II therefore represents a DNA-protein complex containing both Ku and X4-LIV.
Taken together, these results suggest that X4-LIV binds stably to DNA only in the context of a ternary complex including Ku. This Ku-X4-LIV complex could form efficiently even at low concentrations of X4-LIV (5 nM; equimolar to Ku [Fig. 2A, lane 6]). At concentrations lower than 25 nM, X4-LIV apparently progressively disassociates from Ku-bound DNA during electrophoresis, resulting in more heterogeneous retardation of the Ku-plus-DNA species. Thus, while X4-LIV was recruited to Ku-bound DNA when present at as low as 5 nM, X4-LIV alone was unable to bind DNA, even in reactions where X4-LIV was present at a 20-fold-higher concentration (Fig. 2A, compare lane 2 to lane 7). Moreover, in reactions containing equimolar amounts of both Ku and X4-LIV but 10-fold more molecules of DNA, X4-LIV supershifts Ku-bound DNA and does not bind to free DNA (Fig. 2A, lane 6). These results are not consistent with independent or coincident binding of X4-LIV and Ku to the same DNA molecule. We therefore conclude that X4-LIV has high affinity only for Ku-bound DNA, presumably because its recruitment to the DNA molecule has been facilitated by a direct protein-protein interaction between X4-LIV and Ku.
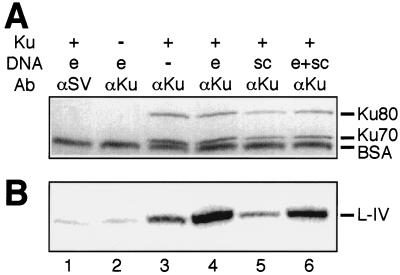
We used immunoprecipitation to assess the relative contributions of protein-protein interactions and DNA-protein interactions in formation of the Ku-X4-LIV complex. For this experiment, we radiolabeled X4-LIV by adenylation of the ligase with [α-32P]ATP and also confirmed that neither the Ku nor the X4-LIV preparations contained significant amounts of contaminating nucleic acids (see Materials and Methods). An antibody to Ku was able to coimmunoprecipitate X4-LIV in the absence of added DNA (Fig. 3B, lane 3), indicating that interaction of these two factors is not completely dependent on DNA and is mediated at least in part by protein-protein contacts. However, inclusion of DNA ends, but not supercoiled DNA, stimulated X4-LIV's ability to interact with Ku over fivefold (Fig. 3B, compare lane 3 to lane 4 or lane 5 to lane 6). Controls performed in the absence of Ku or using an isotype-matched control antibody (against SV40 T antigen) failed to recover significant amounts of either Ku or X4-LIV (Fig. 3A and B, lanes 1 and 2).
FIG. 3.
Detection of protein complexes by immunoprecipitation. All reactions contained 25 nM 32P-labeled X4-LIV. When present (+), Ku was at 5 nM. DNA species included in the reactions were e, a 60-bp duplex at 25 nM (0.5 μg), and sc, 0.5 μg of a supercoiled plasmid. Ab, antibody. Protein A-Sepharose and a monoclonal antibody to Ku (αKu) or an isotype-matched control antibody to SV40 T antigen (αSV; lane 1) were used to precipitate DNA-protein complexes. (A) Coomassie blue-stained gel of precipitated complexes. Ku80, 83-kDa subunit of Ku; Ku70, 70-kDa subunit of Ku. BSA was present due to its inclusion in the wash buffer. (B) Phosphorimage of dried gel. L-IV, ligase IV.
To determine if this interaction could be detected in a more physiological context, we repeated the immunoprecipitation, using whole-cell extracts from HeLa cells instead of purified proteins. An antibody to Ku, but not the control antibody, recovered both ligase IV (Fig. 4, lanes 3 and 4) and XRCC4 (unpublished data). We wished to once again address the role of DNA in the interaction between Ku and X4-LIV, but whole-cell extracts are typically already rich in nucleic acids. We therefore added ethidium bromide to an immunoprecipitation at a concentration that has previously been shown to disrupt DNA-protein interactions (18). Addition of ethidium bromide reduced, but did not eliminate, recovery of X4-LIV with the antibody to Ku (Fig. 4, compare lane 4 to lane 5). This is consistent with the previous in vitro immunoprecipitation result showing that while interaction between Ku and X4-LIV is stimulated by DNA, DNA is not required (Fig. 3, lanes 3 and 4).
Formation of the Ku-plus-X4-LIV complex facilitates end joining.
To determine whether the Ku-X4-LIV complex was functional, we devised a competition assay to see if preloading Ku and X4-LIV on a ligation substrate committed X4-LIV to joining that substrate (Fig. 5A). However, Ku is generally required for ligation activity (25) (see Fig. 7) irrespective of its possible role in the recruitment of X4-LIV. To allow us to distinguish the possible functional effects of complex formation on ligation activity from general effects of Ku on ligation activity, it was therefore necessary to preform the Ku-X4-LIV complex on one substrate and compare X4-LIV activity on that substrate to activity on a competitor substrate also preloaded with Ku.
FIG. 5.
Functional test for formation of Ku-X4-LIV complexes. (A) Competition assay design. Substrate L is a pair of dsDNA molecules. One dsDNA is 5′ 32P labeled on a 60-nucleotide (nt) strand, which can be ligated only to the 36-nt strand of a second dsDNA due to complementary 2-bp 3′ overhangs. Substrate S is another pair of dsDNA molecules. One dsDNA is 5′ 32P labeled on a 58-nt strand that can be ligated only to a 36-nt strand of a second dsDNA due to a different pair of complementary 2-bp 3′ overhangs. Asterisks show the locations of a 5′ 32P label. Step 1, substrates are preincubated on ice for 20 min, with or without Ku and/or X4-LIV, varied for each of the three reactions as noted. Step 2, preincubations are mixed together. In reaction I only, Ku is added after mixing. All three reactions are now equivalent and contain a final concentration of 5 nM for each of the four DNA molecules, 2.5 nM X4-LIV, and 10 nM Ku. Step 3, ligation is initiated by addition of 5 mM Mg2+ and 0.1 mM ATP and incubation at 37°C. Step 4, aliquots of each reaction are taken at successive time points and analyzed by denaturing gel electrophoresis. (B) Results of competition assay. Reactions I, II, and III were assembled as described above. Time is noted in minutes after the start of the reaction. The positions of the labeled strands of the substrates and products for both L and S pairs are marked. (C) Effect of preincubation on reaction kinetics. Reaction mixtures were preincubated with substrate L (described above; 5 nM each duplex) and (i) 5 nM Ku, and 2.5 nM X4-LIV, (ii) 2.5 nM X4-LIV only, (iii) 5 nM Ku only, or (iv) no protein (No Pre.) for 20 min on ice. Ku and/or X4-LIV was then added so that all four reaction mixtures were equivalent, and the reactions were initiated by the addition of Mg2+. Aliquots of each reaction mixture were taken at the indicated times (in minutes), and the ligation product was quantified as a percentage of the total starting substrate.
FIG. 7.
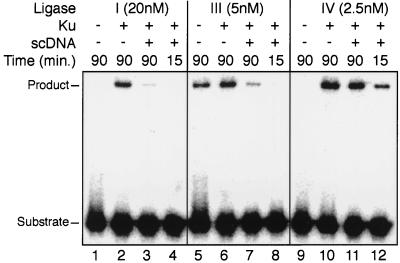
Effect of unbroken competitor DNA on activities of different ligases. Substrate L (Fig. 5A) was present in all reactions at 5 nM for each of the two duplex DNAs. Ku, when present (+), was added to a concentration of 10 nM and preincubated with the ligation substrate for 15 min at 25°C. Two micrograms of supercoiled DNA was added when noted, and ligation was initiated by the subsequent addition of 20 nM ligase I (lanes 1 to 4), 5 nM ligase III (lanes 5 to 8), or 2.5 nM X4-LIV (lanes 9 to 12). Reaction mixtures were incubated for the indicated times (in minutes) at 37°C, and product formation was assessed by denaturing gel electrophoresis.
Formation of a complex was promoted during a preincubation step where X4-LIV was added to a Ku-bound substrate under conditions that do not permit ligation (0.1 mM EDTA and no Mg2+ or ATP [Fig. 5A, step 1]). Potential ternary (Ku–X4-LIV-plus-substrate) complexes were then mixed (Fig. 5A, step 2) with a similarly preincubated competitor lacking only X4-LIV before ligation was initiated by the addition of Mg2+, ATP, and incubation at 37°C (Fig. 5A, step 3). We used two labeled ligation substrates, S and L, that differ such that (i) labeled strands of the two substrates and their ligation products are distinguishable by a 2-bp difference in size and (ii) they can form only one ligation product each due to differences in sequence of 2-bp overhangs (Fig. 5A). This allowed us to compare within the same reaction the extent to which recruitment of X4-LIV to a substrate committed X4-LIV to acting on that substrate.
In reaction II (Fig. 5A), we prepared two preincubations: one contained Ku, X4-LIV, and ligation substrate S, while the competitor reaction contained only Ku and substrate L. Complex formation in reaction II was detected by a strong preference for ligation of substrate S, preincubated with both X4-LIV and Ku, at the expense of substrate L, preincubated with Ku only (Fig. 5B; compare product S to product L for reaction II). This was most evident early in the reaction, presumably due to a progressive redistribution of X4-LIV to both substrates over time. Preferential ligation was not due to the subtle difference between the two substrates, as preincubation of X4-LIV with substrate L led to the expected reciprocal result (Fig. 5B; compare reaction II to reaction III). The preference in ligation was also not due to recruitment of X4-LIV to the substrate during preincubation independent of Ku, as no significant substrate preference was observed in a reaction where X4-LIV was included in a preincubation and Ku was added only after mixing (Fig. 5B, reaction I). This control argues that although XRCC4 may have a low intrinsic DNA binding potential, it is not sufficient to commit X4-LIV to a DNA substrate.
Failure to form the ternary complex in the preincubation also generally reduced the initial rate of the reaction. This was examined in greater detail by comparing reactions without preincubation to reactions with only Ku, only X4-LIV, or with both components included in the preincubation. Each reaction was then made equivalent by addition of the missing components, and ligation was initiated by addition of Mg2+. As expected, no significant difference was observed between reactions with no preincubation and reactions where only X4-LIV was included (Fig. 5C). A modest (approximately fourfold) increase in early product formation was observed when only Ku was included, possibly reflecting the time required for Ku to bind DNA ends. However, addition of both components to the preincubation increased the initial rate approximately 20-fold compared to that of a control with no preincubation.
Ku specifically recruits ligase IV to DNA ends.
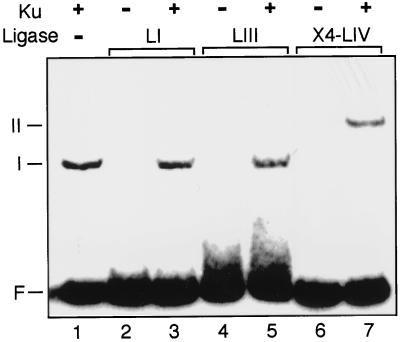
The ability of a preincubation step with Ku and X4-LIV to increase the initial reaction rate is in contrast to a similar experiment using ligase I, where preincubation with Ku did not have this effect (25). We therefore used the EMSA to determine if the ability to form a complex between Ku and a mammalian DNA ligase was specific to X4-LIV. Neither ligase I (Fig. 6, lane 3) nor ligase III (Fig. 6, lane 5) was capable of supershifting Ku-bound DNA, despite being tested under the same conditions and at a 10-fold-higher concentration than that required to observe complex formation with X4-LIV. Similar negative results were observed when association of ligase I to Ku-bound DNA was tested using the pull-down assay described in the legend to Fig. 3 (unpublished data).
FIG. 6.
Specificity of complex formation. A 5′ 32P-labeled 60-bp DNA duplex was present in all reactions at a concentration of 50 nM. F, free DNA probe. The mobilities of DNA-protein complexes are noted for species I and II. When present (+), Ku was added to a concentration of 5 nM. LI, 50 nM ligase I; LIII, 50 nM ligase III; X4-LIV, 25 nM XRCC4 plus ligase IV.
A significant problem for end joining in a cellular context is the need to rapidly locate and repair rare DNA DSBs in the presence of a large, mostly unbroken DNA genome. Since X4-LIV has only low intrinsic affinity for DNA ends, we supposed its participation in the complex could make use of Ku's high affinity and specificity for DNA ends to become much more effective than the other ligases in solving this problem. We therefore prebound Ku to a labeled ligation substrate. The activities of similar amounts of the three mammalian ligases added directly to this substrate were then compared to the activities when they were added following the addition of a large excess of unbroken DNA. Addition of unbroken DNA reduced the activity of ligase I by a factor of 10 (Fig. 7, compare lane 2 to lane 3) and reduced that of ligase III by a factor of 5 (compare lane 6 to lane 7). X4-LIV activity was hardly affected (reduced by one-third; compare lane 10 to lane 11). Moreover, only X4-LIV displayed significant levels of activity shortly after addition (compare lanes 4 and 8 to lane 12). The ability of X4-LIV to form a complex with Ku thus confers an ability to quickly join ends in an excess of unbroken DNA on this ligase alone.
DISCUSSION
We have shown that Ku physically interacts with X4-LIV and specifically recruits this ligase to DNA ends. The interaction could be detected in cell extracts (Fig. 4) as well as in several assays (EMSA, immunoprecipitation assay. and activity assay) using purified components at low concentrations of each component (less than 10 nM Ku, X4-LIV, and DNA). The advantages the Ku-X4-LIV complex confers upon ligation in vitro can therefore explain why these factors, and in particular ligase IV, are required for cellular end joining: ligation is fast and efficient, even at low enzyme and substrate concentrations and in the presence of large amounts of unbroken DNA. Ligase I and ligase III are much less capable than X4-LIV by these criteria, as they are both much slower at performing intermolecular ligation and more sensitive to unbroken competitor. This also may explain why previous work argued that ligase I was the most effective ligase for end joining (25, 26), as these experiments were performed for extended periods of time and in the absence of excess unbroken DNA. Just as Ku targets X4-LIV to DSBs, the involvement of ligases I and III in DSB repair is likely further precluded in cells by specific targeting of these ligases to replication forks or other types of DNA damage by factors specific to these processes (reviewed in reference 31).
XRCC4 alone possesses a weak DNA binding activity, raising the possibility that ligase IV could be recruited to DNA ends independently of Ku (23). However, modification of XRCC4 such that it no longer bound DNA had no effect on its ability to stimulate ligase IV activity (23). Consistent with this, we demonstrate here that preincubation of a DNA substrate and the X4-LIV complex does not commit X4-LIV to joining that substrate (Fig. 5B, reaction I), nor does such a preincubation increase the rate of substrate ligation (Fig. 5C). We therefore argue that Ku is required for functionally effective recruitment of the X4-LIV complex to DNA ends. This does not exclude the possibility that XRCC4's DNA binding activity may help stabilize the ternary (Ku-X4-LIV-DNA) complex. The extra stability provided by this DNA-protein contact, in addition to the already high affinity that Ku has for DNA, could explain why DNA stimulates the interaction between X4-LIV and Ku. However, it must be emphasized that the interaction between X4-LIV and Ku is not a simple consequence of independent or coincident binding to the same DNA molecule. We cannot detect binding of X4-LIV to DNA independently of Ku in our assays, even when the concentration of X4-LIV added is 20-fold higher than that needed to form the ternary complex with Ku (Fig. 2A), nor is the ability to detect an interaction between X4-LIV and Ku wholly dependent on the presence of DNA (Fig. 3 and 4).
Ku thus has at least three distinct and separable functions in end-joining DSB repair that can be identified in vitro. It generally facilitates end joining by aligning DNA ends (25), and it specifically recruits both X4-LIV (as described here) and DNA-PKcs (29) to DNA ends. Recruitment of both DNA-PKcs and X4-LIV is stimulated by the prior binding of Ku to DNA ends. This suggests that Ku might act very early in DSB repair, possibly as a first step by sensing the presence of a DSB and subsequently recruiting other DSB repair factors. Initial recognition of DNA ends by Ku could thus be important in nucleating formation of an active DSB repair complex, much as similar damage recognition steps trigger formation of higher-order complexes in other DNA repair pathways (e.g., nucleotide excision repair [28]). Consistent with this argument, the activity of terminal deoxynucleotidyl transferase (TdT), a factor involved in the joining of coding-end DSB intermediates in V(D)J recombination, is dependent on the presence of Ku (3). Direct association of TdT with Ku in cells has also recently been demonstrated (21).
Interaction between X4-LIV and Ku links the end recognition and alignment functions of Ku to ligase activity. This potentially reversible arrangement may provide important advantages over the alternative—a single factor possessing all of these activities—for the following reason. Joining of ends that need modification prior to ligation (e.g., mismatched ends, hairpins, and ends with damaged nucleotides) requires the temporary removal or relocation of the ligase to allow modifying factors access to the ends. The ability to uncouple ligase activity from Ku allows Ku to remain associated with the pair of DNA ends and to align them, possibly also recruiting different modifying factors, until the ends are a substrate for ligation.
This model argues that a “remodeling” activity presumably must be present to remove or temporarily relocate X4-LIV from Ku-bound ends when the ends need to be processed. The differential effects of DNA-PKcs mutation on joining different types of ends in cells (2, 3, 10, 30) suggest this factor may be important in remodeling. The ability of DNA-PKcs to phosphorylate XRCC4 and consequently modify XRCC4's DNA binding activity further suggests a specific mechanism by which DNA-PKcs could perform this function. In vitro experiments testing for modification of the stability and activity of the Ku-X4-LIV complex at DNA ends can now be used to investigate this possibility, as well as the roles of other factors involved in this pathway.
Both genetic and biochemical evidence indicates several factors in addition to Ku, XRCC4, and ligase IV will be required for the full reconstitution of end joining as it occurs in cells (1, 16, 29). Ku and X4-LIV alone join blunt ends less efficiently than ends with complementary overhangs (25; unpublished data), suggesting that the alignment of blunt ends may require additional stabilization over that already provided by Ku. As previously discussed, processing factors will also be required for the joining of ends with more complex structures than those used in the experiments described here.
A ligation step is nevertheless common to all cellular end-joining reactions, regardless of the structure or context of the broken DNA. Our work establishes in vitro conditions that specifically require Ku, XRCC4, and ligase IV to cooperate for efficient ligation of DSBs. Once the complex is formed, ligation is extremely fast, reaching half-maximal levels in less than 1 min (Fig. 5C). We therefore suggest that these factors together constitute a core complex that is essential for efficient end joining and alone may be sufficient for the repair of a significant proportion of cellular DSBs (i.e., broken ends with matching overlaps, similar to the substrates used in the experiments described here). This is consistent with the genetic requirement for Ku, XRCC4, and ligase IV for all types of end joining reactions in both yeast and mammalian cells.
ACKNOWLEDGMENTS
We thank John Turchi for the gift of HeLa cell ligase IV, W. Reeves for the gifts of monoclonal antibodies, and T. Lindahl, D. Capra, and M. Modesti for the gifts of the various cDNAs. We thank K. Griffin and R. Johnson for expert technical assistance, the members of B. Mitchell's lab for helpful discussion, and M. Gellert, L. Mechanic, and K. Hiom for critical reading of the manuscript.
This work was supported by Public Health Service grant CA84442-01 and a Searle Scholar award to D.A.R.
REFERENCES
- 1.Baumann P, West S C. DNA end-joining catalyzed by human cell-free extracts. Proc Natl Acad Sci USA. 1998;95:14066–14070. doi: 10.1073/pnas.95.24.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogue M A, Jhappan C, Roth D B. Analysis of variable (diversity) joining recombination in DNA-dependent protein kinase (DNA-PK)-deficient mice reveals DNA-PK-independent pathways for both signal and coding joint formation. Proc Natl Acad Sci USA. 1998;95:15559–15564. doi: 10.1073/pnas.95.26.15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogue M A, Wang C, Zhu C, Roth D B. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. doi: 10.1016/s1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- 4.Bryans M, Valenzano M C, Stamato T D. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat Res. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- 5.Cary R B, Peterson S R, Wang J, Bear D G, Bradbury E M, Chen D J. DNA looping by Ku and the DNA-dependent protein kinase. Proc Natl Acad Sci USA. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan D W, Mody C H, Ting N S, Lees-Miller S P. Purification and characterization of the double-stranded DNA-activated protein kinase, DNA-PK, from human placenta. Biochem Cell Biol. 1996;74:67–73. doi: 10.1139/o96-007. [DOI] [PubMed] [Google Scholar]
- 7.Critchlow S E, Bowater R P, Jackson S P. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 8.Dynan W S, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank K M, Sekiguchi J M, Seidl K J, Swat W, Rathbun G A, Cheng H L, Davidson L, Kangaloo L, Alt F W. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y, Chaudhuri J, Zhu C, Davidson L, Weaver D T, Alt F W. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Sun Y, Frank K M, Dikkes P, Fujiwara Y, Seidl K J, Sekiguchi J M, Rathbun G A, Swat W, Wang J, Bronson R T, Malynn B A, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin S H, Greenberg M E, Alt F W. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 12.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson T E, Mann M, Lieber M R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 13.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber M R. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 14.Hammarsten O, Chu G. DNA-dependent protein kinase: DNA binding and activation in the absence of Ku. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrmann G, Lindahl T, Schar P. Saccharomyces cerevisiae LIF1: a function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 1998;17:4188–4198. doi: 10.1093/emboj/17.14.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanaar R, Hoeijmakers J H, van Gent D C. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–489. doi: 10.1016/s0962-8924(98)01383-x. [DOI] [PubMed] [Google Scholar]
- 17.Kurimasa A, Kumano S, Boubnov N V, Story M D, Tung C S, Peterson S R, Chen D J. Requirement for the kinase activity of human DNA-dependent protein kinase catalytic subunit in DNA strand break rejoining. Mol Cell Biol. 1999;19:3877–3884. doi: 10.1128/mcb.19.5.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leber R, Wise T W, Mizuta R, Meek K. The XRCC4 gene product is a target for and interacts with the DNA-dependent protein kinase. J Biol Chem. 1998;273:1794–1801. doi: 10.1074/jbc.273.3.1794. [DOI] [PubMed] [Google Scholar]
- 20.Lieber M R. The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells. 1999;4:77–85. doi: 10.1046/j.1365-2443.1999.00245.x. [DOI] [PubMed] [Google Scholar]
- 21.Mahajan K N, Gangi-Peterson L, Sorscher D H, Wang J, Gathy K N, Mahajan N P, Reeves W H, Mitchell B S. Association of terminal deoxynucleotidyl transferase with Ku. Proc Natl Acad Sci USA. 1999;96:13926–13931. doi: 10.1073/pnas.96.24.13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizuta R, Cheng H-L, Gao Y, Alt F W. Molecular genetic characterization of XRCC4 function. Int Immunol. 1997;9:1607–1613. doi: 10.1093/intimm/9.10.1607. [DOI] [PubMed] [Google Scholar]
- 23.Modesti M, Hesse J E, Gellert M. DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J. 1999;18:2008–2018. doi: 10.1093/emboj/18.7.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang D, Yoo S, Dynan W S, Jung M, Dritschilo A. Ku proteins join DNA fragments as shown by atomic force microscopy. Cancer Res. 1997;57:1412–1415. [PubMed] [Google Scholar]
- 25.Ramsden D A, Gellert M. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsden D A, Paull T, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- 27.Robins P, Lindahl T. DNA ligase IV from HeLa cell nuclei. J Biol Chem. 1996;271:24257–24261. doi: 10.1074/jbc.271.39.24257. [DOI] [PubMed] [Google Scholar]
- 28.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 29.Smith G C, Jackson S P. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 30.Taccioli G E, Amatucci A G, Beamish H J, Gell D, Xiang X H, Torres Arzayus M I, Priestley A, Jackson S P, Marshak Rothstein A, Jeggo P A, Herrera V L. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 31.Tomkinson A E, Mackey Z B. Structure and function of mammalian DNA ligases. Mutat Res. 1998;407:1–9. doi: 10.1016/s0921-8777(97)00050-5. [DOI] [PubMed] [Google Scholar]
- 32.Wilson T E, Grawunder U, Lieber M R. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi-Iwai Y, Sonoda E, Sasaki M S, Morrison C, Haraguchi T, Hiraoka Y, Yamashita Y M, Yagi T, Takata M, Price C, Kakazu N, Takeda S. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 1999;18:6619–6629. doi: 10.1093/emboj/18.23.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yaneva M, Kowalewski T, Lieber M R. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy studies. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]