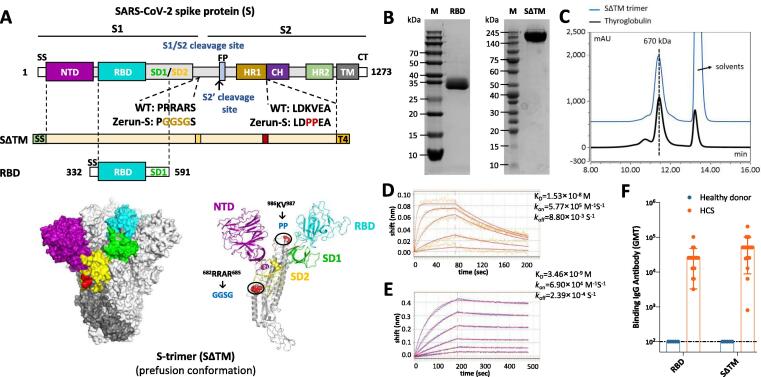
Fig. 1.
Molecular design and characterization of SΔTM and RBD. (A) Domain architecture of the SARS-CoV-2 S protein. SS, signal sequence; NTD, N-terminal domain; RBD, receptor-binding domain; SD1, subdomain 1; SD2, subdomain 2; S1/S2, S1/S2 protease cleavage site; S2′, S2′ protease cleavage site; FP, fusion peptide; HR1, heptad repeat 1; CH, central helix; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane domain; CT, cytoplasmic tail. Two recombinant SARS-CoV-2 spike antigens were designed: SΔTM (the prefusion S ectodomain with proline substitutions at residues 986 and 987 to retain S2 in the prefusion conformation, a “GGSG” substitution at the furin cleavage site, a C-terminal T4 fibritin trimerization motif), and 260-mer RBD (RBD-SD1). The structure model of S-trimer was generated by the SWISS-MODEL using homology modelling techniques (http://swissmodel.expasy.org/), and the 3D structure figures were prepared using PyMOL (www.pymol.org). (B) SDS-PAGE analysis of purified SΔTM and RBD. Molecular weight standards are indicated at the left in kDa. (C) Size-Exclusion HPLC chromatogram of purified SΔTM (shown as blue line) and a 670 kDa molecular weight standard (shown as black line). (D) and (E) Binding profiles of SΔTM and RBD to human ACE2 measured by BLI in GatorPrime. The data are shown as blue and orange lines for SΔTM and RBD, respectively, and the best fit of the data to a 1:1 binding model is shown in red. (F) Antigenicity of SΔTM and RBD measured by serially diluted HCS. HCS, human convalescent sera.