Abstract
Shiga toxigenic Escherichia coli (STEC) strains are a diverse group of organisms capable of causing severe gastrointestinal disease in humans. Within the STEC family, certain strains appear to have greater virulence for humans. STEC strains carrying eae and belonging to serogroup O157 or O111 have been responsible for the vast majority of outbreaks of STEC disease reported to date. Here we describe a STEC O113:H21 strain lacking eae that was responsible for a cluster of three cases of hemolytic-uremic syndrome. This strain produces a single Stx2-related toxin and adheres efficiently to Henle 407 cells.
Shiga toxigenic Escherichia coli (STEC) strains are an important cause of gastrointestinal disease in humans, particularly since such infections may result in life-threatening sequelae such as hemolytic-uremic syndrome (HUS) and thrombotic thrombocytopenic purpura (11, 17, 26). It has been recognized for a number of years that STEC strains causing human disease may belong to a very broad range of O serogroups (11). However, a subset of these (notably O157 and O111) appear to be responsible for the majority of serious cases (those complicated by HUS) (8, 11, 26). These STEC strains have the capacity to produce attaching and effacing lesions on intestinal mucosa, a property mediated by the eae gene product intimin. Production of intimin is not essential for pathogenesis, as a minority of sporadic cases of HUS are caused by STEC strains lacking eae (26). However, all outbreaks of STEC disease complicated by HUS reported to date have been caused by STEC strains carrying eae and belonging to serogroup O157 or O111 (26). We now describe an eae-lacking STEC strain belonging to serotype O113:H21 that was responsible for a cluster of cases of HUS in South Australia.
Examination of fecal culture extracts by multiplex PCR.
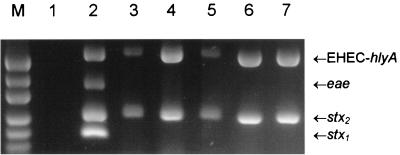
During April 1998, three children with HUS were admitted to the Women’s and Children’s Hospital, North Adelaide, S.A., Australia. Preliminary screening of crude fecal culture extracts by PCR (22) indicated the presence of Shiga toxin gene (stx) sequences. This screening PCR yields 212- and 215-bp amplicons in the presence of genes encoding Shiga toxin type 1 or type 2 (stx1 or stx2), respectively (22). Further characterization was achieved by using two recently described multiplex PCR assays (19). Assay 1 utilizes four PCR primer pairs and detects the presence of stx1, stx2 (including variants of stx2), eae, and enterohemorrhagic E. coli (EHEC) hlyA (which encodes a STEC-specific enterohemolysin), generating amplification products of 180, 255, 384, and 534 bp, respectively. Assay 2 uses two primer pairs specific for portions of the rfb (O-antigen-encoding) regions of E. coli serotypes O157 and O111, generating PCR products of 259 and 406 bp, respectively. The results obtained with assay 1 are shown in Fig. 1. Extracts from all three HUS patients were positive for stx2 and EHEC hlyA only. The fact that the extracts were negative for eae was unexpected, given that all previously reported HUS outbreaks have been caused by eae-carrying STEC. When tested with assay 2, all three samples were negative for rfbO157 or rfbO111 sequences (result not shown), which again was unexpected, as all previously reported outbreaks of HUS have been caused by STEC strains belonging to serogroups O157 or O111.
FIG. 1.
Multiplex PCR analysis of fecal cultures from HUS patients and STEC isolates. Crude DNA extracts of broth cultures from the indicated samples or strains were analyzed by multiplex PCR assay 1, as previously described (19). PCR products were electrophoresed on 2% agarose gels and stained with ethidium bromide. Lanes: M, DNA size markers (pUC19 DNA digested with HpaII; fragment sizes visible are 501/489, 404, 331, 242, 190, and 147 bp); 1, negative control; 2, positive control (O111:H− STEC strain 95NR1, which is positive for stx1, stx2, eae, and EHEC hlyA); 3, fecal culture from HUS patient 1; 4, fecal culture from HUS patient 2; 5, fecal culture from HUS patient 3; 6, STEC isolate 98NK2; 7, STEC isolate 98BN1. The expected mobilities for the various specific PCR products are also indicated.
Interestingly, a DNA extract from a fecal culture from a household contact of patient 3 (an asymptomatic adult) also tested positive by PCR for stx2 and EHEC hlyA. However, extracts from fecal cultures from a sibling of patient 1 and the parents of patient 2 were negative by PCR.
Isolation of the causative STEC strain(s).
In order to isolate the causative STEC strain or strains, fecal samples from each patient were plated on MacConkey agar. Colonies were picked and inoculated into 150 μl of Luria-Bertani (LB) broth (14) in 96-well (U-bottomed) microtiter plates. Cell lysates were prepared, spotted onto nylon filters (Hybond N+; Amersham), and probed with a digoxigenin-labelled stx-specific probe, as described previously (24). Approximately 20% (56 of 288) of the colonies tested from patient 2, which yielded the strongest PCR signal, were stx probe positive. However, no probe-positive isolates were obtained after testing 480 colonies from each of the fecal samples from patient 1 or patient 3. This was not unexpected, as the PCR signal for both of these patients was much less intense than that for patient 2 (Fig. 1, lanes 3 and 5), suggesting the presence of very low numbers of STEC. Interestingly however, 100% of 192 colonies tested from the fecal sample from the household contact of Patient 3 were stx positive. Representative STEC isolates from patient 2 and the contact of patient 3 were designated 98NK2 and 98BN1, respectively. DNA extracts from these two isolates yielded a multiplex PCR profile identical to that obtained for the fecal culture extracts from the three HUS patients (Fig. 1, lanes 6 and 7). During previous characterization of the multiplex PCR assay (19) with a large collection of STEC isolates, we observed this particular profile only in STEC strains belonging to serotype O113:H21. Accordingly, 98NK2 and 98BN1 were tested with O113- and H21-specific typing sera at the Salmonella Reference Laboratory, Institute of Medical and Veterinary Science, Adelaide, S.A., Australia; both isolates were seropositive.
Serological analysis.
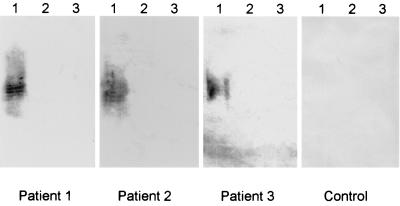
Most HUS patients exhibit a transient serum antibody response to lipopolysaccharide (LPS) of the infecting serotype, and so in cases where STEC have not been isolated from fecal cultures, reliable etiological information can frequently be obtained by serological analysis (3, 5, 26, 32). Accordingly, convalescent-phase sera from all three HUS patients in the present study were analysed by Western blotting for the presence of O113-specific antibodies. Sera were also tested for antibodies to O111 and O157 LPS, as STEC strains belonging to these serogroups are the most common causes of HUS in Australia (26). LPS was purified from STEC O113:H21, O111:H−, and O157:H− according to the method of Westphal and Jann (30), and aliquots were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12) and electrophoretically transferred to nitrocellulose filters, as described by Towbin et al. (28). Replicate filters were then reacted with convalescent-phase patient serum (kindly provided by K. F. Jureidini and P. Henning, Renal Unit, Women’s and Children’s Hospital) or serum from three healthy controls at a dilution of 1:1,000, followed by goat anti-human immunoglobulin G conjugated to alkaline phosphatase (Bio-Rad Laboratories, Hercules, Calif.). Filters were then developed with chromogenic substrate (4-nitroblue tetrazolium and X-phosphate). All three HUS patient sera reacted with O113 LPS but did not label either the O111 or the O157 LPS extracts (Fig. 2). None of the LPS extracts reacted with the control sera (the result for one of these is also shown in Fig. 2). These data, combined with the results of multiplex PCR analysis of fecal cultures described above, are compelling evidence that all three HUS patients were infected with STEC O113.
FIG. 2.
Western immunoblot detection of anti-O113 LPS. Aliquots of LPS purified from E. coli serogroups O113 (lanes 1), O111 (lanes 2), and O157 (lanes 3) were separated by SDS-PAGE, transferred to nitrocellulose filters, and reacted with convalescent-phase sera from the three HUS patients or serum from a healthy control.
Genetic characterization of STEC isolates.
Although the STEC O113:H21 isolates were negative by PCR for eae, it was considered possible that such strains carry a variant eae gene with sufficient sequence differences in the primer-specific region to interfere with the efficiency of PCR. This was nonetheless thought to be unlikely, since sequence variation in eae from diverse classes of pathogenic E. coli occurs principally in the 3′ portion of the gene (13, 29), and the PCR primers used in the present study anneal to conserved sequences in the 5′ region. As additional confirmation, chromosomal DNA from 98NK2 and 98BN1 was subjected to Southern hybridization analysis with a 3-kb probe prepared by PCR amplification of the complete eae gene from STEC 95SF2 (O157:H−) in the presence of digoxigenin-11-dUTP. No hybridization was detected, even under low-stringency conditions (result not shown).
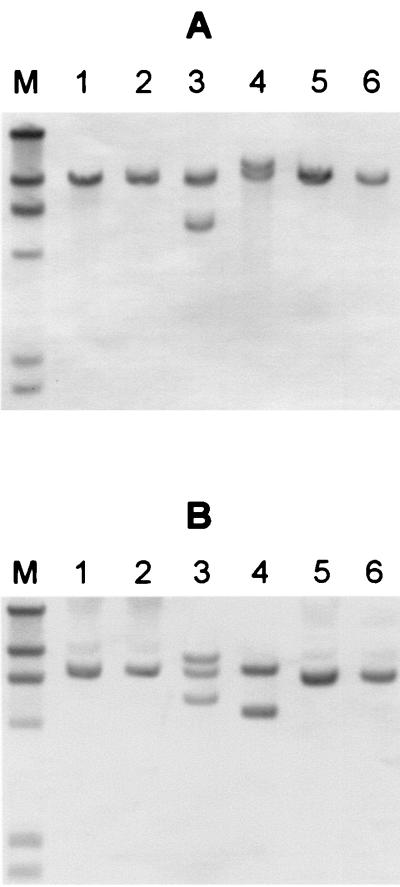
To examine the relationship between the two STEC O113:H21 isolates 98NK2 and 98BN1, chromosomal DNA from these as well as other STEC O113 strains in our collection was restricted with EcoRI or SphI and subjected to restriction fragment length polymorphism (RFLP) analysis with an stx2-specific probe, as described previously (24) (Fig. 3). 98NK2 and 98BN1 both contained single stx2 genes on a 9.4-kb EcoRI or 7.3-kb SphI fragment. The other STEC O113 strains tested had distinct RFLP patterns. DNA from 97MW1, which was isolated in Adelaide, S.A., Australia, in 1997 from a patient with microangiopathic hemolytic anemia and thrombocytopenia, contained two stx2-reactive EcoRI fragments (9.4 and 5.7 kb). However, three reactive fragments were detected in SphI digests (8.7, 7.3, and 5.5 kb), suggesting that this strain contains three stx2-related genes. Strain MW10, which was isolated from fermented sausage in Adelaide in 1995, contains two stx2-related genes located on 11.7- and 10.0-kb EcoRI fragments (7.7- and 4.9-kb SphI fragments). Strains 1183 and 3848, clinical isolates from New Zealand (kindly provided by Jenny Bennet), contained single stx2-related genes located on 10.0- and 9.4-kb EcoRI fragments.
FIG. 3.
RFLP analysis of STEC O113 isolates. Genomic DNA purified from the indicated STEC strains was digested with EcoRI (A) or SphI (B), electrophoresed, and subjected to Southern hybridization analysis with an stx2-specific DNA probe, as described previously (24). Lanes: 1, 98NK2; 2, 98BN1; 3, 97MW1; 4, MW10; 5, 1183; 6, 3848. Lane M contains DNA size markers of 23.1, 9.4, 6.6, 4.4, 2.3, and 2.0 kb.
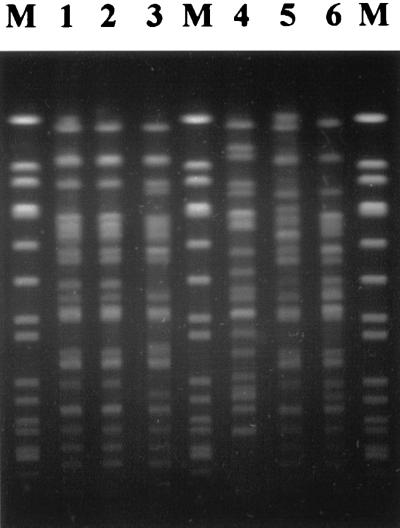
The genetic relationships between the various STEC O113 strains were also examined by pulsed-field gel electrophoresis (PFGE), as described previously (24) (Fig. 4). 98NK2 and 98BN1 had indistinguishable PFGE patterns. The overall PFGE pattern for the other Adelaide clinical isolate, 97MW1, was similar but not identical; there were differences in at least four DNA fragments. The PFGE patterns for the food isolate MW10 and the two clinical isolates from New Zealand were all quite distinct. These findings are consistent with the RFLP analysis and collectively support the conclusion that 98NK2 and 98BN1 are representatives of the same STEC O113:H21 clone.
FIG. 4.
Pulsed-field gel electrophoresis of XbaI-digested genomic DNA of STEC O113. Lanes 1 to 6 are labelled as for Fig. 3. The lanes marked M contain marker DNA (SmaI-digested genomic DNA of Staphylococcus aureus NCTC8325).
Analysis of the O113:H21 gene encoding Stx2.
In order to characterize the Stx2 protein produced by 98NK2, the complete stx2 operon was amplified by PCR with previously described primers (23). Amplification was carried out using the Expand High Fidelity PCR System (Boehringer Mannheim) under conditions recommended by the manufacturer. The resultant 1,495-bp PCR product was cloned into pBluescript (Stratagene, La Jolla, Calif.) and transformed into E. coli JM109 (33). Both strands of the insert were then sequenced by using dye terminator chemistry and custom-made primers on an ABI model 373A automated DNA sequencer. The deduced amino acid sequences of the A and B subunits of Stx2 produced by 98NK2 (designated Stx2O113) aligned with those for classical Stx2 (10) and the subtypes Stx2c (27) and Stx2d (9) are shown in Fig. 5. Although Stx2O113 has three amino acid differences in the A subunit with respect to classical Stx2, it clearly belongs to this subtype, as it lacks the specific A subunit amino acid substitutions at the C terminus (S291 and E297) that are associated with Stx2d (15, 16). The B subunit of Stx2O113 differs from that of classical Stx2 by a single residue in the signal peptide. It does not contain the specific D16→N substitution associated with the Stx2c and Stx2d subtypes.
FIG. 5.
Deduced amino acid sequences of the A and B subunits of Stx2O113 aligned with published sequences for Stx2 (10), Stx2c (27), and Stx2d (9). Dots denote residues identical to Stx2O113; the dash denotes an absent residue. Arrows indicate the first residue of the mature polypeptide for each subunit.
Epithelial cell adherence.
The adherence of 98NK2 and the O113 food isolate MW10 to human intestinal epithelial (Henle 407) cells was compared with that of 95NR1, an eae-carrying STEC O111:H− strain responsible for a large food-borne outbreak of HUS in Adelaide in 1995 (24), as previously described (25). E. coli cells were grown overnight at 37°C in LB broth and diluted to a density of 104 CFU/ml (confirmed by viable count) in Dulbecco’s modified Eagle’s medium buffered with 20 mM HEPES and supplemented with 10% fetal calf serum and 2 mM l-glutamine. Washed Henle 407 monolayers in 24-well tissue culture plates were then infected with 1-ml aliquots of bacterial suspension. After incubation at 37°C for 3 h, the culture medium was removed and the monolayers were washed four times with phosphate-buffered saline to remove nonadherent bacteria. The cell monolayers were then detached from the plate by treatment with 100 μl of 0.25% trypsin–0.02% EDTA. Cells were then lysed by addition of 400 μl of 0.025% Triton X-100, and 50-μl aliquots (and serial 10-fold dilutions thereof) were plated on LB agar to determine the total number of adherent bacteria. Under these conditions, total adherence (mean ± standard error of quadruplicate assays) of 95NR1 was (8.85 ± 0.46) × 104 CFU per well. Total adherence for 98NK2 was (7.32 ± 0.20) × 104 CFU per well, which is 83% of that for the STEC O111 strain. In contrast, adherence of the food isolate MW10 was markedly lower, at only (3.45 ± 0.49) × 104 CFU per well (39 and 47% of that for 95NR1 and 98NK2, respectively).
Discussion and conclusions.
Although a wide variety of STEC serogroups have been associated with human disease, O157 strains are the predominant causes of disease in many parts of the world. The dominance of these strains is even more marked when one considers those responsible for outbreaks of STEC disease which include cases complicated by HUS. Indeed, only a small number of HUS outbreaks due to non-O157 STEC have been reported to date, all of which were caused principally by STEC strains belonging to serogroup O111 (1, 4, 5, 24). Like STEC O157, these strains are positive for eae. An eae-lacking STEC strain belonging to serotype O104:H21 was responsible for an outbreak of diarrheal disease in Montana in 1994, but none of the infected persons developed HUS (6).
In the present study, we have provided molecular microbiological evidence that an eae-lacking STEC strain belonging to serogroup O113 was responsible for a cluster of three cases of HUS. Multiplex PCR analysis of fecal culture extracts from the three HUS patients yielded identical patterns (stx1 negative, stx2 positive, eae negative, EHEC hlyA positive, rfbO111 negative, rfbO157 negative), which also agreed with those obtained when DNA from the STEC O113:H21 isolates from patient 2 and a household contact of patient 3 were tested. Furthermore, all three HUS patients had serological evidence of infection due to E. coli O113. The possibility that these patients had been coinfected with an eae-carrying STEC strain is extremely remote, given the fact that none of the primary fecal cultures were positive for eae by PCR. Moreover, these fecal cultures were also negative for rfbO111 and rfbO157 by PCR, and O111- or O157-specific antibodies were not detected in convalescent-phase sera.
We recently cloned and sequenced the entire rfb locus of E. coli O113 (20), and we have used this information to design a PCR assay specific for this serogroup (21). In an attempt to obtain additional confirmation that all three HUS patients described in the present study were infected with a STEC O113 strain, we tested crude DNA extracts from the original fecal cultures. Even though these had been stored at 4°C for approximately 1 year, an O113-specific PCR product was observed in extracts from patient 1 and patient 2 (21); unfortunately, the extract from patient 3 was negative. However, the latter extract was also negative when retested for stx2 and EHEC hlyA, suggesting that it had deteriorated during prolonged storage.
The inability to isolate STEC O113 from HUS patients 1 and 3 is undoubtedly a consequence of the fact that the numbers of STEC organisms in the gut often decrease rapidly as disease progresses (26). The fecal samples from these two patients were collected after HUS had developed. On the other hand, the fecal sample from HUS patient 2 which yielded 98NK2 was collected during the diarrheal prodrome, about 5 days before the onset of HUS symptoms.
In the present study, the O113:H21 isolates 98NK2 and 98BN1 were genetically indistinguishable, as judged by RFLP and PFGE analysis. This is not a consequence of lack of diversity among STEC O113 clones, as the RFLP and PFGE patterns for other such strains in our collection were different, both from 98NK2/98BN1 and from each other. Thus, it appears that patient 2 and the contact of patient 3 were infected with the same STEC clone. It is reasonable to conclude by inference that patient 3 was also infected with this O113:H21 strain. However, notwithstanding the clustering of the three HUS cases (dates of presentation fell within a 20-day period) and the convincing molecular and serological evidence that a STEC O113 strain was responsible, the possibility that patient 1 represented a sporadic case caused by a distinct STEC O113 strain cannot be eliminated, as no isolate was obtained for RFLP or PFGE analysis. Such a coincidence would be highly improbable, given that until the present cluster of HUS cases, there had been only one culture-proven case of human infection with a STEC O113 strain in South Australia (strain 97MW1 was isolated from this patient). An extensive epidemiological investigation did not identify a clear link between any of the HUS cases, and testing of a range of foods from the homes of the patients by stx-specific PCR also failed to detect any positive samples.
STEC strains which carry eae are generally considered to have higher virulence for humans than those which lack eae (2, 31). However, a minority of sporadic cases of HUS are caused by eae-lacking STEC strains, and the findings of the present study indicate that such strains are capable of causing HUS outbreaks. Melton-Celsa and colleagues (15, 16) have demonstrated that certain eae-lacking STEC strains produce a variant form of Stx2 (designated Stx2d) which, unlike other members of the Stx family, is activated by intestinal mucus. This property was found to be associated with specific amino acids at the C terminus of the A subunit of Stx2d, notably S291 and E297. Those authors suggested that production of an activatable toxin might compensate for the lack of eae, as strains producing Stx2d were extremely virulent for streptomycin-treated mice. However, the STEC O113:H21 strain isolated in the present study has a single toxin gene encoding a member of the classical Stx2 subtype. Interestingly, Stx2O113 is identical to the Stx2-related toxin produced by an eae-lacking STEC O48:H21 strain, 94CR, which we isolated previously from a sporadic case of HUS (18). Although the activatable Stx2d toxins have so far been detected only in eae-lacking STEC strains (16), it is now clear that this property is not essential for such strains to cause life-threatening human disease.
Although production of Stx is a sine qua non of virulence, capacity to adhere to the intestinal epithelium and colonize the gut undoubtedly plays an important role in the pathogenesis of human STEC disease (17, 26). We have previously shown that STEC isolates from HUS cases have an enhanced capacity to adhere to Henle 407 cells relative to isolates from nonhuman sources (25). Similarly, in the present study, 98NK2 exhibited twice the level of adherence to this cell line than did the STEC O113 strain (MW10) which was isolated from a food source. Although not directly compared in the present study, 98NK2 and the O48:H21 strain 94CR (25) exhibit similar degrees of adherence relative to a reference O111:H− STEC strain. In spite of the absence of eae, and by inference the inability to adhere intimately to enterocytes, the STEC O113:H21 strain associated with this cluster of HUS cases is clearly capable of efficient colonization of the human gut. This is demonstrated by the fact that the fecal sample from the asymptomatic family contact of HUS patient 3 yielded a virtually pure culture of 98BN1 on MacConkey agar. However, the precise bacterial factor(s) mediating adherence of eae-lacking STEC is yet to be characterized. Dytoc et al. (7) have previously studied the adhesion phenotype of an eae-lacking STEC O113:H21 strain. This strain was reported to be piliated, and it adhered to rabbit ileal brush border membranes and to both Hep-2 and Henle 407 cells in a diffuse pattern. Interestingly, it was capable of microvillus effacement in vivo, although it did not cause the cytoskeletal rearrangements and intimate attaching and effacing lesions typical of STEC strains carrying eae.
There have been a number of reports of sporadic cases of HUS being caused by O113:H21 and other eae-lacking STEC strains (11, 26). However, to our knowledge, this is the first report of a cluster of HUS cases caused by such a strain. Although historically less common, the severity of disease caused by these strains may be no less than that caused by recognized “enterohemorrhagic” STEC serogroups such as O157 and O111. More widespread use of PCR- or enzyme-linked immunosorbent assay-based screening tests for the presence of STEC of any serogroup in fecal samples will undoubtedly result in increased detection of similar non-O157 outbreaks in the future. This will provide more accurate data on the etiology of human STEC disease.
Acknowledgments
We are grateful to Milka Karna-Marelj for serotyping of E. coli isolates and to Rolf Wise for assistance with imaging of PFGE gels. We are particularly grateful to Rod Givney for providing epidemiological information and to Fred Jureidini and Paul Henning for providing convalescent-phase sera. We also thank Jenny Bennett for providing New Zealand STEC O113 isolates.
This work was supported by a grant from the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Banatvala N, Debeukelaer M M, Griffin P M, Barrett T J, Greene K D, Green J H, Wells J G. Shiga-like toxin-producing Escherichia coli O111 and associated hemolytic-uremic syndrome: a family outbreak. Pediatr Infect Dis J. 1996;15:1008–1011. doi: 10.1097/00006454-199611000-00015. [DOI] [PubMed] [Google Scholar]
- 2.Barrett T J, Kaper J B, Jerse A E, Wachsmuth I K. Virulence factors in Shiga-like toxin-producing Escherichia coli isolated from humans and cattle. J Infect Dis. 1992;165:979–980. doi: 10.1093/infdis/165.5.979. [DOI] [PubMed] [Google Scholar]
- 3.Bitzan M, Karch H. Indirect hemagglutination assay for diagnosis of Escherichia coli O157 infection in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1174–1178. doi: 10.1128/jcm.30.5.1174-1178.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boudailliez B, Berquin P, Mariani-Kurkdjian P, Ilef D, Cuvelier B, Capek I, Tribout B, Bingen E, Piussan C. Possible person-to-person transmission of Escherichia coli O111-associated hemolytic uremic syndrome. Pediatr Nephrol. 1997;11:36–39. doi: 10.1007/s004670050229. [DOI] [PubMed] [Google Scholar]
- 5.Caprioli A, Luzzi I, Rosmini F, Resti C, Edefonti A, Perfumo F, Farina C, Goglio A, Gianviti A, Rizzone G. Communitywide outbreak of hemolytic uremic syndrome associated with non-O157 Verocytotoxin-producing Escherichia coli. J Infect Dis. 1994;169:208–211. doi: 10.1093/infdis/169.1.208. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Outbreak of acute gastroenteritis attributable to Escherichia coli serotype O104:H21—Helena, Montana, 1994. Morbid Mortal Weekly Rep. 1995;44:501–503. [PubMed] [Google Scholar]
- 7.Dytoc M T, Ismaili A, Philpott D J, Soni R, Brunton J L, Sherman P M. Distinct binding properties of eaeA-negative verocytotoxin-producing Escherichia coli of serotype O113:H21. Infect Immun. 1994;62:3494–3505. doi: 10.1128/iai.62.8.3494-3505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin P M. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R I, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 739–761. [Google Scholar]
- 9.Ito H, Terai A, Kurazono H, Takeda Y, Nishibuchi M. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb Pathog. 1990;8:47–60. doi: 10.1016/0882-4010(90)90007-d. [DOI] [PubMed] [Google Scholar]
- 10.Jackson M P, Neill R J, O’Brien A D, Holmes R K, Newland J W. Nucleotide sequence analysis and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli. FEMS Microbiol Lett. 1987;44:109–114. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 11.Karmali M A. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Louie M, De-Azavedo J, Clarke R, Borczyk A, Lior H, Richter M, Brunton J. Sequence heterogeneity of the eae gene and detection of verotoxin-producing Escherichia coli using serotype-specific primers. Epidemiol Infect. 1994;112:449–461. doi: 10.1017/s0950268800051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 15.Melton-Celsa A R, Darnell S C, O’Brien A D. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect Immun. 1996;64:1569–1576. doi: 10.1128/iai.64.5.1569-1576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melton-Celsa A R, O’Brien A D. Structure, biology, and relative toxicity of Shiga toxin family members for cells and animals. In: Kaper J B, O’Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C: American Society for Microbiology; 1998. pp. 121–128. [Google Scholar]
- 17.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paton A W, Bourne A J, Manning P A, Paton J C. Comparative toxicity and virulence of Escherichia coli clones expressing variant and chimeric Shiga-like toxin type II operons. Infect Immun. 1995;63:2450–2458. doi: 10.1128/iai.63.7.2450-2458.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paton A W, Paton J C. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602. doi: 10.1128/jcm.36.2.598-602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paton, A. W., and J. C. Paton. Molecular characterization of the locus encoding biosynthesis of the lipopolysaccharide O-antigen of Escherichia coli serotype O113. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 21.Paton A W, Paton J C. Direct detection of Shiga toxigenic Escherichia coli strains belonging to serogroups O111, O157, and O113 by multiplex PCR. J Clin Microbiol. 1999;37:3362–3365. doi: 10.1128/jcm.37.10.3362-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton A W, Paton J C, Goldwater P N, Manning P A. Direct detection of Escherichia coli Shiga-like toxin genes in primary fecal cultures using the polymerase chain reaction. J Clin Microbiol. 1993;31:3063–3067. doi: 10.1128/jcm.31.11.3063-3067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paton A W, Paton J C, Manning P A. Polymerase chain reaction amplification, cloning and sequencing of variant Escherichia coli Shiga-like toxin type II operons. Microb Pathog. 1993;15:77–82. doi: 10.1006/mpat.1993.1058. [DOI] [PubMed] [Google Scholar]
- 24.Paton A W, Ratcliff R, Doyle R M, Seymour-Murray J, Davos D, Lanser J A, Paton J C. Molecular microbiological investigation of an outbreak of hemolytic-uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J Clin Microbiol. 1996;34:1622–1627. doi: 10.1128/jcm.34.7.1622-1627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paton A W, Voss E, Manning P A, Paton J C. Shiga toxin-producing Escherichia coli isolates from cases of human disease show enhanced adherence to intestinal epithelial (Henle 407) cells. Infect Immun. 1997;65:3799–3805. doi: 10.1128/iai.65.9.3799-3805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paton J C, Paton A W. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998;11:450–479. doi: 10.1128/cmr.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt C K, McKee M L, O’Brien A D. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect Immun. 1991;59:1065–1073. doi: 10.1128/iai.59.3.1065-1073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voss E, Paton A W, Manning P A, Paton J C. Molecular analysis of Shiga toxigenic Escherichia coli O111:H− proteins which react with sera from patients with hemolytic-uremic syndrome. Infect Immun. 1998;66:1467–1472. doi: 10.1128/iai.66.4.1467-1472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westphal O, Jann K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 31.Willshaw G A, Scotland S M, Smith H R, Cheasty T, Thomas A, Rowe B. Hybridization of strains of Escherichia coli O157 with probes derived from the eaeA gene of enteropathogenic E. coli and the eaeA homolog from a Vero cytotoxin-producing strain of E. coli O157. J Clin Microbiol. 1994;32:897–902. doi: 10.1128/jcm.32.4.897-902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada S, Kai A, Kudoh Y. Serodiagnosis by passive hemagglutination test and verotoxin enzyme-linked immunosorbent assay of toxin-producing Escherichia coli infections in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1994;32:955–959. doi: 10.1128/jcm.32.4.955-959.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]