Abstract
This study investigated the effects of inulin on rumen fermentation parameters, ruminal microbiome and metabolites, as well as lactation performance and serum indexes in dairy cows. Sixteen Holstein dairy cows with similar body conditions were randomly divided into 2 groups (n = 8 per group), with inulin addition at 0 and 200 g/d per cow. The experiment lasted for 6 weeks, including a 1-week adaptation period and a 5-week treatment period. At the end of the experimental period, the milk, serum and rumen fluid were sampled and analyzed. The microbiome and metabolome in the rumen fluid were analyzed via 16S rRNA sequencing and untargeted metabolomics, respectively. The results showed that supplementation with inulin (200 g/d per cow) increased the milk yield (P = 0.001), milk protein (P = 0.032), lactose rate (P = 0.004) and proportion of saturated fatty acids (SFA) in milk (P < 0.001), but decreased the proportion of unsaturated fatty acids (USFA) (P = 0.041). Rumen pH (P = 0.040) and the concentration of NH3–N (P = 0.024) were decreased; however, acetate (P < 0.001), propionate (P = 0.003), butyrate (P < 0.001) and lactic acid (LA) (P = 0.043) were increased. The total cholesterol (TC) (P = 0.008) and triglycerides (TG) (P = 0.01) in serum were also reduced. Additionally, inulin addition elevated the relative abundance of several beneficial symbiotic and short-chain fatty acid (SCFA)-producing bacteria, such as Muribaculaceae (false discovery rate [FDR]-adjusted P < 0.01), Acetitomaculum (FDR-adjusted P = 0.043), and Butyrivibrio (FDR-adjusted P = 0.036), while elevating the levels of L-lysine (FDR-adjusted P = 4.24 × 10−3), L-proline (FDR-adjusted P = 0.0158), and L-phenylalanine (FDR-adjusted P = 0.027). In contrast, several pathogens and ruminal bacteria abundant in high-fat diets, such as Escherichia-Shigella (FDR-adjusted P = 0.022), Erysipelotrichaceae__UCG-004 (FDR-adjusted P < 0.01) and RF39 (FDR-adjusted P = 0.042) were decreased along with the reduction of lysophosphatidylcholine (LysoPC) (18:1 (9Z)) (FDR-adjusted P = 1.03 × 10−3), LysoPC (16:0) (FDR-adjusted P = 0.0108), LysoPC (18:2 (9Z, 12Z)) (FDR-adjusted P = 1.65 × 10−3) and 8-methylnonenoate. In conclusion, dietary inulin supplementation could increase the relative abundance of commensal microbiota and SCFA-producing bacteria, upregulate amino acidmetabolism and downregulate lipid metabolism in the rumen of dairy cows, which might further improve lactation performance and the level of serum lipids.
Keywords: Inulin, Ruminal microbiota, Metabolomics, Milk quality, Serum lipid, Dairy cow
1. Introduction
Inulin is a reserve polysaccharide in plants (Roberfroid, 2007). Statistically, inulin is contained in approximately 36,000 plants, of which Jerusalem artichoke (Helianthus tuberosus L.) tubers are the most abundant (Saengthongpinit and Sajjaanantakul, 2005). As a kind of fructo-oligosaccharide (FOS), inulin is composed mainly of β-D-fructofuranose and glucopyranose residues connected by β-(1, 2) glucosidic bonds (Roberfroid, 2007). There is a mild smell in inulin with the characteristics of high sweetness and low calorific value. Although inulin has high stability in water and tends to agglomerate when dispersed in water (Niness, 1999), Ronkart et al. (2006) reported that inulin could be slowly degraded into fructose and glucose in solutions with pH < 4 and a proper temperature for a period of time. Due to the existence of a β-(1, 2) glycosidic bond structure, inulin cannot be digested and decomposed in the mouth, stomach and small intestine, and can only be partially fermented by Bifidobacteria in the colon (Roberfroid, 2007). Therefore, inulin is scientifically proven to be an edible nondigestible polysaccharide that meets the definition of prebiotics (Roberfroid, 2007).
As a soluble dietary fiber, inulin has been widely studied in humans and monogastric animals. Inulin can promote the growth of probiotics, such as Bifidobacteria and Lactobacilli, and inhibit the proliferation of pathogenic and putrid bacteria, such as Clostridium pneumoniae, Enterococcus and mold (Roberfroid, 2007), and the changes in intestinal flora caused by inulin fermentation can regulate the immune function of intestinal lymphoid tissue (Schley and Field, 2002). Chen et al. (2017) found that inulin could delay the occurrence of immune diseases such as acute pancreatitis by regulating the homeostasis of the intestinal flora. However, inulin can induce metabolic changes. Inulin can promote the production of the endogenous active substance cathelicidin related antimicrobial peptide. The effect depends on the production of short-chain fatty acids (SCFA). In addition, a large number of SCFA and lactic acid (LA) produced by inulin fermentation in the colon can not only stimulate intestinal peristalsis and prevent constipation (Roberfroid, 2007), but also lower the pH value of the intestine and facilitate mineral absorption (Lopez et al., 2000). Additionally, inulin has been shown to have the ability to inhibit the degradation of ingested fat by lipolytic enzymes, thereby lowering blood lipids (Williams, 1999). Further, the lipid-lowering effect may also be related to the regulation of cholesterol and triglycerides (TG) in the blood by SCFA produced by inulin fermentation (Causey et al., 2000). In summary, inulin can improve the intestinal environment and body health in humans and monogastric animals by changing the microbial population and metabolic processes.
However, studies on inulin in ruminants, especially in dairy cows, are quite limited. Previous studies supported that inulin could not exert a prebiotic role in ruminants because of the existence of the rumen, in which inulin would be strongly degraded (Biggs and Hancock, 1998; Zhao et al., 2016). The fermentation process of inulin in the colon of monogastric animals is basically analogous to the process that occurs in the rumen of ruminants (Biggs and Hancock, 1998). An in vitro rumen fermentation test in sheep showed that adding 1.5 g DM inulin to incubation bottles containing 1 g basic diet could significantly reduce the concentration of ammoniacal nitrogen (NH3–N), which showed the potential of inulin to improve rumen nitrogen utilization (Gndz, 2009). Zhao et al. (2016) investigated the effects of inulin on rumen fermentation and microbial growth in goats by rumen simulation technology. The results showed that inulin treatment decreased the concentration of acetate, the ratio of acetate to propionate (A/P) and methane production but increased the concentration of butyrate. Furthermore, the abundances of Fibrobacter succinogenes and Ruminococcus flavefaciens were reduced, which indicated that inulin may inhibit the growth of some rumen cellulose decomposition bacteria. Nevertheless, Umucalilar et al. (2010) reported that inulin could only play a limited modifier role in regulating rumen fermentation parameters in vitro. Another study on finishing beef showed that the supplementation of inulin in the diet diverted the rumen fermentation pattern from acetate to propionate and butyrate; moreover, the α-diversity index of rumen bacteria was increased, in which the abundance of Bacteroides and Firmicutes were significantly increased, as well as weight and feed utilization (Tian et al., 2019). Most of the previous studies on the effects of inulin on ruminants have focused mainly on in vitro tests, which may have restricted the knowledge and in-depth study of the effects of inulin on the actual physiological regulation and production performance in ruminants.
Intestinal microbes are the most important microbial group in monogastric animals, and ruminant microbes have the same status in ruminants (Young et al., 2015). The successful application of inulin in monogastric animals to improve the intestinal environment has aroused great interest in its potential roles in rumen function, and physiology and production performance in ruminants. To date, however, the effects of inulin on lactation performance, immune capacity, ruminal microorganism composition and metabolite activity in dairy cows have received little research attention. We hypothesized that dietary supplementation with inulin may affect lactation performance and several lipid metabolites in the blood via regulating the rumen inner-environment of dairy cows. In this study, 16S rRNA sequencing and untargeted metabolomics technology were applied to investigate the effects of inulin on rumen microbial profile and metabolite composition, which was helpful to evaluate the feasibility of using inulin as a feed additive in dairy cows.
2. Materials and methods
2.1. Animal ethics statement
The experimental animals, designs and animal management in the present study were approved by the Animal Ethics Committee of the Chinese Academy of Agricultural Sciences (Beijing, China) (approval number: IAS-2019-9) and were in accordance with the recommendations of the academy's guidelines for animal research.
2.2. Inulin
The inulin (>85% purity, other sugars including fructose, sucrose and glucose <15%) added in this study was in powder form and extracted from Jerusalem artichoke (H. tuberosus L.) tubers, which were provided by Langfang Academy of Agriculture and Forestry (Hebei, China).
2.3. Experimental animals, diets and design
The experiment was carried out on a well-managed and large-scale dairy farm in the suburbs of Beijing, China. On this dairy farm, there were approximately 560 adult lactating cows. Sponge rubber mats and sterilized rice hulls were laid on the cow bedding. The cowshed was equipped with electric manure scrapers and electric cow body brushes, providing a clean and comfortable environment for the cows. The experiment lasted 6 weeks and was composed of 1 week of adaptation and 5 weeks of treatments from mid-September to late October. Sixteen Holstein dairy cows with similar initial parity (1.83 ± 0.91), body weight (BW) (553 ± 17.3 kg), days in milk (DIM) (166 ± 48.8 d), milk yield (33.7 ± 2.64 kg/d) and milk somatic cell count (SCC) (338 ± 64.5 × 103 cells/mL) were randomly assigned to 2 groups, including the control and inulin groups (n = 8). Cows were housed in individual tie stalls and fed 3 times a day at 07:00, 13:00, and 19:00, respectively, with the same total mixed rations (TMR; concentrate-to-forage ratio of 40:60). Cows were fed TMR and drank freely. Daily orts were collected for feed intake calculation. The TMR ingredients and nutrient components are listed in Appendix Table 1. The chemical composition (DM, CP, EE and ash) of the TMR was analyzed based on the Association of Official Analytical Chemists (AOAC) methods (Helrich, 1990). The NDF and ADF contents were analyzed using an ANKOM A2000i fiber analyzer (ANKOM Technology, New York). The concentrations of Ca and P were analyzed by inductively coupled plasma-atomic emission spectrometry (ICP-AES) (iCAP6300, Thermo Fisher, New York). The net energy for lactation (NEL) was calculated according to NRC (2001).
Based on the basal diet, the inulin addition dose was 0 and 200 g/d per cow in the control and inulin groups, respectively. The addition level of inulin in the current study was determined according to an independent in vitro test through a rumen simulation technique. The results indicated that 0.8% DM inulin in the TMR had a significant effect on rumen fluid incubation performance in dairy cows. Therefore, the dosage in the current study was calculated as 0.8% of the average dry matter intake (25 kg/d per cow), resulting in 200 g/d per cow. Due to the hygroscopicity, the inulin was weighed (200 g/d per cow) and rolled into pills with the same shape and size. A feeder (Boehringer-Ingelheim, Biblach, Germany) was used to dose the pills through the mouth during the morning feeding.
2.4. Sample collection and analyses
2.4.1. Milk sample collection and body weight measurement
The cows were taken to the milking parlor and milked 3 times a day at 06:00, 12:00 and 18:00, using automatic milking systems (Afimilk, Israel). In the last 3 consecutive days of the 6th week, milk samples were collected and the milk yield was recorded. The average milk yield in these 3 d was used as representative for the experimental period. A 50-mL milk sample was collected from each cow, immediately placed in a foam box with ice bags and taken back to the laboratory. The milk samples from the 3 times per day were mixed at the ratio of 4:3:3 (Wang et al., 2017). A part of the mixed milk sample was combined with 0.6 mg/mL potassium dichromate as a preservative and stored at 4 °C for the analysis of milk composition. The other part of the mixed milk sample was stored at −20 °C for the analysis of milk fatty acids (FA).
2.4.2. Milk composition and fatty acid analysis
Milk protein, fat, lactose, urea nitrogen (MUN) and SCC were subsequently measured at the Beijing Dairy Cattle Center (Beijing, China) by a milk composition analyzer (Lactoscan SP, Funke Gerber, Berlin, Germany). Milk FA was detected by the acetyl chloride-methanolmethyl esterification method. After extraction with toluene, a gas chromatograph (Agilent 8860 GC, CA, USA) was used for separation and detection and quantified by an external standard method. The milk sample pretreatment was performed according to Wang et al. (2006). In addition, 4.5 mL of toluene was added to 0.5 mL FA triglycerides and mixed thoroughly for use as a standard measurement solution. The chromatographic reference conditions were as follows: Dicyanopropyl polysiloxane column (100 m × 0.25 mm, 0.20 μm) was used; The temperature of the column oven was 140 °C for 5 min, then increased to 240 °C at 4 °C/min for 5 min; The injector and detector temperatures were 260 and 280 °C, respectively (Wang et al., 2006). FA standard measurement solution (1.0 μL) and methylated milk sample (1.0 μL) were pipetted into sample vials for chromatographic analysis. Parallel determination was conducted at least twice, and the chromatographic peak area was used for quantification. In the chromatographic data workstation, the first derivative method was used to determine the starting point, end point and vertex of the chromatographic peak. The area of chromatographic peaks was obtained by the integral method, and the retention time was determined by the positive and negative changes of the first derivative values (Wang et al., 2006).
2.4.3. Blood sampling and analysis
Blood samples were collected from each cow through the caudal vein into procoagulant inert separation tubes (Saiao An Weiye Technology Development Co., Ltd., Beijing, China) approximately 1 h before morning feeding on the last day of the 6th week. Two tubes of blood samples were collected from each cow, with each tube containing 5 mL. The blood samples were placed in a foam box with ice packs and centrifuged at 3,000 × g for 15 min after being brought back to the laboratory. The separated serum from each cow was collected and stored at −20 °C for further analysis of biochemical indicators. The serum TG and total cholesterol (TC) concentrations were analyzed using a serum triglyceride determination kit (TR0100, Sigma–Aldrich, MO, USA) and cholesterol quantitation kit (MAK043, Sigma–Aldrich, MO, USA), respectively. The concentration of total protein (TP) was defined through the biuret-specific absorbance colorimetric method using a total protein detection kit (Bai Olaibo Technology Co., Ltd., Beijing, China). The serum albumin (ALB) concentration was detected by the bromocresol green (BCG) method using a BCG albumin assay kit (MAK124, Sigma–Aldrich, MO, USA). The concentration of serum globulin (GLO) was calculated by the difference between the concentration of TP and ALB. The blood urea nitrogen (BUN) concentration was determined by a urea nitrogen kit (MS1812, Leigen Biotechnology Co., Ltd., Beijing, China). An ultraviolet and visible spectrophotometer (UV5Nano, Mettler-Toledo, Zurich, Switzerland) was used to quantify the above serum indicators.
2.4.4. Rumen fluid sampling
Rumen fluid samples were collected on the last day of this experiment. At 1 h before morning feeding, a gastric tube-type rumen fluid sampler (MDW15, Colebo Equipment Co., Ltd., Wuhan, China) and a 200-mL syringe were used to collect rumen fluid samples from each cow. The first 2 tubes of rumen fluid were discarded to avoid saliva contamination, and a 100-mL rumen fluid sample was collected from each cow (Liu et al., 2019). The pH value of rumen fluid samples was immediately measured after collection by a portable pH meter (Bell Analytical Instruments Co., Ltd., Liaoning, China). Each rumen fluid sample was filtered through 4 layers of gauze and divided into 6 parts for the analysis of volatile fatty acids (VFA), NH3–N, rumen urea nitrogen (RUN), LA, ruminal bacteria, and metabolites, respectively.
2.4.5. Rumen fermentation index measurement
The concentrations of VFA were detected by gas chromatography (Agilent 8890 GC, CA, USA) (Liu et al., 2019). A 10-mL rumen fluid sample from each cow was centrifuged at 10,000 × g for 10 min, and 1.5 mL supernatant was transferred to a centrifuge tube containing 0.15 mL of 25% metaphosphoric acid. The solution was held for 30 min and then centrifuged at 10,000 × g for 15 min. A 1.5-mL supernatant was pipetted into a sample bottle for gas chromatographic determination. The VFA standard solution was prepared according to Li et al. (2019). A 1.5 mL VFA standard solution was thoroughly mixed with 0.5-mL 25% metaphosphoric acid in a centrifuge tube. A 1.5-mL solution was used for gas chromatographic determination. Gas chromatographic conditions were as follows: chromatographic column: DB-FFAP (15 m × 0.32 mm, 0.25 μm); column temperature: 100 °C, 2 °C/min to 120 °C, maintained for 10 min; inlet temperature: 250 °C; detector temperature: 280 °C; constant voltage: 21.8 kPa; split ratio: 1:50 and injection volume: 2 μL. An enzyme-labeled instrument (Multiskan FC, Thermo Fisher, Beijing, China) was used to measure NH3–N concentrations (Hue et al., 2005). The concentrations of RUN and LA were detected by urea nitrogen kit (MS1812, Leigen Biotechnology Co., Ltd., Beijing, China) and LA detection kit (TC0733, Leigen Biotechnology Co., Ltd., Beijing, China), respectively. The ruminal microflora and metabolites were analyzed by 16S rRNA sequencing and untargeted metabolomics techniques.
2.4.6. Ruminal microorganism DNA extraction, PCR amplification and sequencing
The MagBeads Soil DNA Extraction Kit (SMDE-5010, Tiangen Biochemical Technology Co., Ltd., Beijing, China) was used to extract the total microbial DNA from 16 rumen fluid samples according to the manufacturer's instructions. DNA purity and concentration were detected with NanoDrop2000 (Thermo Fisher, New York, USA) and DNA integrity was assessed by 1% agarose gel electrophoresis. The 16S V3 and V4 regions were amplified with forward primer 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The PCR system (20 μL) included 4 μL 5 × FastPfu Buffer, 2 μL 2.5 mmol/L dNTPs, 0.8 μL forward primer (5 μmol/L), 0.8 μL reverse primer (5 μmol/L), 0.4 μL FastPfu Polymerase, 0.2 μL BSA, 10 ng template DNA, and ddH2O. Three PCR replicates for each sample were mixed, and 5 μL of the PCR products of each sample were detected by 2% agarose gel electrophoresis. The PCR products were purified using the AxyPrep DNA Gel Extraction Kit (AP-GX-250, Axygen Biosciences, Union City, USA) and quantified with a Quantus Fluorometer (E6150, Promega, WI, USA). The NEXTflex Rapid DNA-Seq Kit (5144-08, Bioo Scientific, Texas, USA) was used to construct the MiSeq libraries. Sequencing was conducted on the MiSeq PE300 platform (Illumina, San Diego, USA) (Liu et al., 2019).
2.4.7. Analysis of sequencing data
Trimmomatic software (version 0.36) was used for quality control (QC) of raw sequences, and FLASH software (version 1.2.11, https://ccb.jhu.edu/software/F LASH/index.shtml) was used for splicing: 1) The bases with tail mass value below 20 and the reads less than 50 bp length after QC were filtered, respectively, and the reads including N bases were removed; 2) According to the overlap relationship between PE reads, pairs of reads were merged into a sequence, with minimum overlap length of 10 bp; 3) The overlap area of the mosaic sequence was allowed the maximum error ratio of 0.2, and the unmatched sequences were filtered out; 4) Samples were distinguished and the sequence direction was adjusted according to the barcode and primer at both ends of the sequences. The allowed mismatches of barcode and primer mismatch were 0 and 2, respectively. UPARSE software (version 7.0.1090, http://drive5.com/uparse/) was used to perform operational taxonomic unit (OTU) sequence clustering, and select sequences with more than 97% similarity to the representative sequences of OTU. The chimeras were eliminated by UCHIME software (version 7.0, http://www.drive5.com/usearch/) and then the OTU table was generated. The ribosomal database project (RDP) classifier (http://rdp.cme.msu.edu/) was used to classify and annotate each sequence through comparison to Silva (Release132, http://www.arb-silva.de/) with a comparison threshold of 70% (Han et al., 2015).
The alpha diversity analysis at the OTU level was conducted by Mothur software (version 1.30.2, https://www.mothur.org/wiki/Download_mothur). The difference test of the alpha diversity index (Sobs, ACE, Chao, Shannon and Simpson) in the 2 groups was conducted by Wilcoxon rank-sum test. The abundance at various taxonomic levels and beta diversity distances were calculated using QIIME software (version 1.9.1, http://qiime.org/install/index.html). In the beta diversity analysis, the principal co-ordinates analysis (PCoA) and nonmetric multidimensional scaling analysis (NMDS) were analyzed at the OTU level with the distance algorithm of weighted normalized UniFrac. The differential bacteria were analyzed through linear discriminant analysis effect size (LEfSe) software (http://huttenhower.sph.harvard.edu/galaxy/root?Tool_id=lefse_Upload). Specifically, nonparametric factorial Kruskal–Wallis (KW) sum-rank test was applied to detect the taxa with significantly differential abundance. The P-value from the KW sum-rank test was adjusted by false discovery rate (FDR), with statistical significance declared at FDR-adjusted P < 0.05. Then, linear discriminant analysis (LDA) was used to estimate the influence of the abundance of each species on the difference effect (Ogunade et al., 2019).
2.4.8. Liquid chromatography–mass spectrometry (LC/MS) metabolomics analysis
One hundred microliters of thawed rumen fluid sample from each cow was transferred into a 1.5-mL centrifuge tube, and 400 μL extraction solution (acetonitrile:methanol = 1:1) was added and mixed thoroughly for 30 s. After treatment with low-temperature ultrasound for 30 min (5 °C, 40 kHz), the solution was held at 4 °C for 30 min and centrifuged at 13,000 × g for 15 min. The supernatant was removed and the precipitate was dried with nitrogen, and 120-μL reconstituted solution (acetonitrile:water = 1:1) was added to reconstitute the remaining material. The solution was treated with low-temperature ultrasonication for 5 min (5 °C, 40 kHz) and centrifuged again at 13,000 × g for 5 min. The supernatant was pipetted into a sample injection vial with an inner cannula for LC/MS analysis (Liu et al., 2019). The metabolites from all samples with equal volumes were mixed to prepare a QC sample. A QC sample was inserted into the queue for every 10 samples to check the repeatability of the entire analysis process.
LC/MS analysis was performed using a UPLC-Triple TOF system (Triple TOF5600, AB SCIEX, MA, USA). Ten microliters of sample from each cow was detected through mass spectrometry after separation by a BEH C18 chromatographic column (100 mm × 2.1 mm i.d., 1.8 μm) (Waters, Massachusetts, USA). LC-MS conditions were as follows: mobile phase A: water (containing 0.1% formic acid); mobile phase B: acetonitrile: isopropanol (1: 1) (containing 0.1% formic acid); separation gradient of mobile phase (A: B): 80%: 20% from 0 to 3 min, 5%: 95% from 3 to 9 min and maintained for 2 min, 95%: 5% from 13.0 to 13.1 min and then maintained for 3 min. The flow rate was 0.40 mL/min, and the column temperature was 40 °C. The signal acquisition of the mass spectrum employed positive and negative ion scanning modes with a scanning range of 50 to 1,000 (mass-to-charge ratio, m/z). The ion spray voltages in positive and negative ion modes were 5,000 V and 4,000 V, respectively. The ion source heating temperature was 500 °C with a voltage of 20 to 60 V (Artegoitia et al., 2017).
2.4.9. Metabolomics data processing and analysis
The raw data were processed by Progenesis QI (Waters, Milford, USA) software for baseline filtering, peak identification, integration, retention time (RT) correction, peak alignment, and finally, a data matrix including RT, m/z, and peak intensity was obtained. To reduce the error caused by sample preparation and instrument instability, the response intensity of the sample mass spectrum peak was normalized by the sum normalization method. Meanwhile, the variables in QC samples with a relative standard deviation (RSD) > 30% were deleted to obtain the final data matrix for subsequent analysis. The MS information was matched with the human metabolome database (HMDB) (http://www.hmdb.ca/) and Metlin database (https://metlin.scripps.edu/) to obtain metabolite information (Artegoitia et al., 2017).
The preprocessed data were uploaded to the Majorbio Cloud Platform (https://cloud.majorbio.com/) for further analysis. The ropls package of the R program (Version 1.6.2) was used to perform principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA), which were accompanied by 7 cycles of interactive verification to evaluate the stability of the model. In addition, Student's t-test (unpaired) was used to determine significant differences. The fold change (FC) was used to evaluate the change trend (upregulation or downregulation) of differential metabolites. The selection of significantly differential metabolites was based on the variable important in projection (VIP) obtained from the OPLS-DA model and the P-value from Student's t-test, which was adjusted by FDR. The metabolites with VIP >1 and FDR-adjusted P-value < 0.05 were identified as the significantly different metabolites between the 2 groups. The Venn diagram was printed by Venn Diagram in R packages (version 1.6.20). Hierarchical clustering analysis (HCA) for significantly different metabolites was performed by the Scipy package in Python software (version 1.0.0). The metabolite distance algorithm was Bray–Curtis. The hierarchical clustering method of metabolites was average. The metabolic pathways in which the differential metabolites clustered were annotated through the KEGG database (https://www.kegg.jp/kegg/pathway.html). The Scipy package in Python software (version 1.0.0) was used for pathway enrichment analysis (Artegoitia et al., 2017). Receiver operator characteristic (ROC) analysis was used to examine the metabolites that were critical to intergroup differentiation by ROC ANALYSIS of SPSS Statistics (version 22, IBM, New York, USA). The closer the area under curve (AUC) value was to 1, the higher was the accuracy of the prediction.
2.5. Correlation analysis
The correlation analysis was conducted using the OmicShare tools (http://www.omicshare.com/tools). The correlations between differential bacteria and metabolites, rumen fermentation parameters and milk compositions, as well as significantly differential metabolites and milk compositions, were calculated by Spearman's correlation coefficient, respectively. The range of the correlation coefficient (r) was from −1 to 1. r > 0 and < 0 represented a positive correlation and negative correlation, respectively. The |r| value denoted the degree of correlation between variables. In particular, r = −1, 0 and 1 meant a completely negative correlation, uncorrelated and a completely positive correlation, respectively. Correlation significance P-values below 0.05 and 0.01 were regarded as significant and extremely significant correlations, respectively.
2.6. Statistical analysis
Data statistics were performed by SPSS Statistics (version 22, IBM, New York, USA). The body condition information in dairy cows (parity, BW, DIM, and SCC), milk yield and composition, milk FA compositions, rumen fermentation parameters, and alpha diversity indexes were analyzed through one-way ANOVA and Student's t-test, with statistical significances declared at P < 0.05, and a tendency to be declared at 0.05 < P < 0.10.
3. Results
3.1. Effect of inulin supplementation on milk performance
The milk yield and compositions are shown in Table 1. With the supplementation of inulin, the dry matter intake (DMI) (P = 0.003), milk yield (P = 0.001), energy corrected milk (ECM), fat corrected milk (FCM) (P < 0.001), milk protein (P = 0.042) and lactose (P = 0.004) were increased, and milk fat showed a tendency of increase (P = 0.075). However, the MUN (P = 0.023), SCC (P = 0.036) and the fat-to-protein ratio (F/P) were decreased (P = 0.027), but the F/P was still in the normal range (1.12-1.36).
Table 1.
Effects of inulin supplementation in dietary on milk yield and compositions.
| Item | Con (n = 8) | Inulin (n = 8) | SEM | P-value |
|---|---|---|---|---|
| DMI | 25.1 | 25.4 | 0.07 | 0.103 |
| BW | 551 | 554 | 1.5 | 0.079 |
| Milk yield, kg/d | 34.4b | 37.2a | 0.55 | 0.001 |
| ECM1, kg/d | 34.0b | 37.8a | 0.79 | <0.001 |
| FCM2, kg/d | 34.8b | 38.0a | 0.70 | <0.001 |
| Milk fat, % | 4.08 | 4.14 | 0.034 | 0.075 |
| Milk protein, % | 3.12b | 3.39a | 0.068 | 0.032 |
| F/P | 1.31a | 1.22b | 0.033 | 0.027 |
| Milk lactose, % | 4.85b | 5.19a | 0.066 | 0.004 |
| MUN, mmol/L | 5.99a | 5.18b | 0.186 | 0.023 |
| SCC, × 103 cells/mL | 340a | 302b | 14.1 | 0.036 |
Con = control group; SEM = standard error of mean; DMI = dry matter intake; BW = body weight; ECM = energy corrected milk; FCM = fat corrected milk; F/P = fat-to-protein ratio; MUN = milk urea nitrogen; SCC = somatic cell counts. a, b Within a row, different letters mean differed significantly (P < 0.05).
ECM (kg/d) = milk yield (kg/d) × [383 × fat (%) + 242 × protein (%) + 783.2)/3,140 (Sjaunja et al., 1990).
FCM (kg/d) = 0.4 × milk yield (kg/d) + 15 × milk yield (kg/d) × fat (%) (Gaines, 1928).
The effect of adding inulin on the level of milk FA is listed in Table 2. Compared with the control group, the proportion of saturated FA (SFA) in the inulin group was increased (P < 0.001), mainly including an increase in C6:0 (P = 0.034), C8:0 (P = 0.006), C10:0 (P = 0.038), C12:0 (P = 0.001) and C16:0 (P = 0.001). Whereas, the proportions of C18:0 (P = 0.002) and C22:0 (P = 0.031) were reduced. On the other hand, the proportion of unsaturated FA (UFA) in the inulin group declined (P = 0.041), attributed mainly to the decrease in polyunsaturated FA (PUFA) (P = 0.013). Among these PUFA, the proportions of C18:2 cis-6 (P = 0.011) and C18:3n3 (P < 0.001) were reduced. Additionally, the levels of short- and medium-chain FA (SMCFA) in the inulin group were increased (P < 0.001).
Table 2.
Effect of inulin supplementation in dietary on milk fatty acids compositions.
| Item | Name | Con (n = 8) | Inulin (n = 8) | SEM | P-value |
|---|---|---|---|---|---|
| SFA | 65.9b | 70.1a | 1.37 | <0.001 | |
| C4:0 | Butyric acid | 3.11 | 3.38 | 0.064 | 0.99 |
| C6:0 | Methyl hexanoate | 2.08b | 2.72a | 0.060 | 0.034 |
| C8:0 | Methyl octanoate | 1.28b | 1.37a | 0.042 | 0.006 |
| C10:0 | Capric acid | 2.81b | 3.02a | 0.088 | 0.038 |
| C11:0 | Methyl undecanoate | 0.040 | 0.069 | 0.0103 | 0.175 |
| C12:0 | Dodecane lauric acid | 2.58b | 3.64a | 0.179 | 0.001 |
| C13:0 | Methyl tridecanote | 0.10 | 0.11 | 0.006 | 0.295 |
| C14:0 | Myristoic acid | 9.65 | 9.97 | 0.368 | 0.546 |
| C15:0 | Methyl pentadecanoate | 0.090 | 0.090 | 0.0305 | 0.427 |
| C16:0 | Palmitic acid | 33.2b | 37.3a | 0.65 | 0.001 |
| C17:0 | Methyl heptadecanoate | 0.47 | 0.43 | 0.007 | 0.54 |
| C18:0 | Octadecanoic acid | 9.77a | 7.45b | 0.421 | 0.002 |
| C20:0 | Eicosanoic acid | 0.095 | 0.068 | 0.0059 | 0.273 |
| C21:0 | Methyl heneicosanoate | 0.42 | 0.33 | 0.015 | 0.101 |
| C22:0 | Behenic acid | 0.17a | 0.13b | 0.013 | 0.031 |
| UFA | 27.8a | 26.4b | 0.65 | 0.041 | |
| MUFA | 24.1 | 23.3 | 0.64 | 0.066 | |
| C14:1 | Methyl myristoleate | 1.24 | 1.29 | 0.056 | 0.410 |
| C15:1 | Methyl cis-10-pentadecenoate | 0.27b | 0.32a | 0.009 | 0.039 |
| C16:1 | Palmitoleic acid | 1.43b | 1.57a | 0.065 | 0.031 |
| C17:1 | Methyl cis-10-heptadecenoate | 0.18 | 0.15 | 0.004 | 0.071 |
| C18:1 trans-9 | trans-9-octadecanoic acid (elaidic acid) | 0.53a | 0.48b | 0.012 | 0.043 |
| C18:1 cis-9 | cis-9-octadecanoic acid (oleicacid) | 19.9 | 19.0 | 0.66 | 0.533 |
| C20:1 | Cis-11-eicosenoic acid | 0.39 | 0.32 | 0.012 | 0.098 |
| C22:1 | Cis-13- decosahedaenoic acid | 0.19 | 0.15 | 0.010 | 0.064 |
| PUFA | 3.65a | 3.08b | 0.096 | 0.013 | |
| C18:2 trans-6 | transcis-9, 12-octadecadienoic acid (translinoleic acid) | 0.086 | 0.073 | 0.0078 | 0.407 |
| C18:2 cis-6 | Cis-9,12-octadecadienoic acid (linoleic acid) | 2.45a | 2.12b | 0.046 | 0.011 |
| C18:3n6 | Cis, cis, cis-6,9,12-octadecatrienoic acid (γ-linolenic acid) | 0.45 | 0.32 | 0.044 | 0.162 |
| C18:3n3 | Cis, cis, cis-9,12,15-octadecatrienoic acid (α-linolenic acid) | 0.34a | 0.29b | 0.017 | 0.031 |
| C20:2 | Cis, cis, 11, 14-eicosadienoic acid | 0.078 | 0.073 | 0.0068 | 0.076 |
| C20:3n3 | Cis-11, 14, 17-eicosapentaenoic acid | 0.12 | 0.09 | 0.005 | 0.054 |
| C20:4n6 | Arachidonic acid | 0.13 | 0.11 | 0.008 | 0.089 |
| SMCFA | 11.9b | 14.2a | 0.24 | <0.001 | |
| LCFA | 81.7 | 82.2 | 1.07 | 0.071 | |
Con = control group; SEM = standard error of mean; SFA = saturated fatty acid; UFA = unsaturated fatty acid; MUFA = monounsaturated fatty acid; PUFA = polyunsaturated fatty acids; SMCFA = short and medium-chain fatty acids; LCFA = long-chain fatty acids.
a, b Within a row, different letters mean differed significantly (P < 0.05).
3.2. Effect of inulin supplementation on serum indexes
As shown in Table 3, compared with the control group, the concentrations of TC (P = 0.008) and TG (P = 0.01) were declined in the inulin group, but the levels of TP, ALB, GLO and BUN were not significantly different (P > 0.05).
Table 3.
Effect of inulin supplementation in dietary on serum indexes.
| Item | Con (n = 8) | Inulin (n = 8) | SEM | P-value |
|---|---|---|---|---|
| TC, mmol/L | 8.01a | 6.49b | 0.434 | 0.008 |
| TG, mmol/L | 0.479a | 0.409b | 0.0152 | 0.01 |
| TP, g/L | 69.9 | 71.8 | 1.20 | 0.084 |
| ALB, g/L | 34.8 | 36.6 | 0.83 | 0.086 |
| GLO, g/L | 35.1 | 34.2 | 0.99 | 0.648 |
| BUN, mmol/L | 5.84 | 5.32 | 0.332 | 0.051 |
Con = control group; SEM = standard error of mean; TC = total cholesterol; TG = triglyceride; TP = total protein; ALB = albumin; GLO = globulin; BUN = blood urea nitrogen.
a, b Within a row, different letters mean differed significantly (P < 0.05).
3.3. Effects of inulin supplementation on rumen fermentation characteristics
The rumen fermentation parameters are listed in Table 4. The pH value in the rumen declined (P = 0.040) with the addition of inulin, which was accompanied by a significant increase in the concentrations of acetate (P < 0.001), propionate (P = 0.003), butyrate (P < 0.001), isobutyrate (P = 0.002), valetate (P = 0.001), isovaletate (P < 0.001) and LA (P = 0.043). Meanwhile, a significant decrease in the concentration of NH3–N (P = 0.024) was also observed in the inulin group.
Table 4.
Effect of inulin supplementation in dietary on rumen fermentation parameters.
| Item | Con (n = 8) | Inulin (n = 8) | SEM | P-value |
|---|---|---|---|---|
| pH | 6.56a | 6.30b | 0.015 | 0.040 |
| Acetate, mmol/L | 67.3b | 87.1a | 3.131 | <0.001 |
| Propionate, mmol/L | 25.8b | 31.1a | 0.98 | 0.003 |
| A/P | 2.61 | 2.80 | 0.08 | 0.179 |
| Butyrate, mmol/L | 11.3b | 17.5a | 0.86 | <0.001 |
| Isobutyrate, mmol/L | 1.05b | 1.25a | 0.037 | 0.002 |
| Valetate, mmol/L | 1.58b | 2.53a | 0.170 | 0.001 |
| Isovaletate, mmol/L | 2.08b | 2.69a | 0.101 | <0.001 |
| LA, mmol/L | 0.71b | 0.99a | 0.027 | 0.043 |
| RUN, mmol/L | 5.26 | 5.30 | 0.332 | 0.837 |
| NH3–N, mg/dL | 12.1a | 9.30b | 1.07 | 0.024 |
Con = control group; SEM = standard error of mean; A/P = acetate to propionate ratio; LA = lactic acid; RUN = rumen urea nitrogen.
a, b Within a row, different letters mean differed significantly (P < 0.05).
3.4. Effect of dietary supplementation with inulin on the richness, diversity and composition of ruminal bacteria
A total of 1,349,120 effective 16S rRNA sequences were detected in 16 rumen fluid samples and 2,337 OTU were obtained by performing OTU clustering on nonrepetitive sequences according to 97% similarity. Rarefaction curves (Appendix Fig. 1) showed that the current sequencing depth and sample size were sufficient to assess the microbial diversity, total species richness and core species number of rumen fluid samples. The α-diversity analysis revealed that the ACE (P = 0.031), Chao (P = 0.017) and Shannon (P = 0.026) indexes in the inulin group were increased, and the Simpson index also showed a tendency of rise (P = 0.071), which illustrated that the addition of inulin increased the ruminal microbial community richness and diversity (Table 5).
Table 5.
Effects of inulin supplementation in dietary on α-diversity of ruminal microbiota.
| Item | Con (n = 8) | Inulin (n = 8) | SEM | P-value |
|---|---|---|---|---|
| Sobs | 1446 | 1480 | 28.5 | 0.568 |
| ACE | 1,726b | 1,801a | 27.9 | 0.031 |
| Chao | 1,742b | 1,813a | 27.9 | 0.017 |
| Shannon | 5.66b | 6.19a | 0.044 | 0.026 |
| Simpson | 0.010 | 0.012 | 0.0006 | 0.071 |
| Coverage | 0.98 | 0.99 | 0.001 | 0.765 |
Con = control group; SEM = standard error of mean.
a, b Within a row, different letters mean differed significantly (P < 0.05).
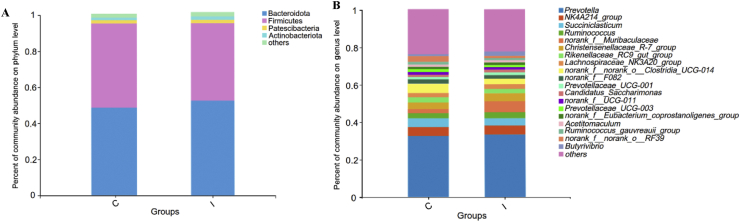
The RDP classifier Bayesian algorithm was used to perform taxonomic analysis on the OTU representative sequences, and 21 bacterial phyla and 304 genera were obtained from 16 samples (Appendix Tables 2 and 3). At the phylum level, Bacteroidota (48.4% and 51.7%), Firmicutes (46.3% and 42.3%), Patescibacteria (1.81% and 1.85%) and Actinobacteriota (1.43% and 1.82%) were the dominant bacteria in the control and inulin groups, respectively (Fig. 1A and Appendix Fig. 2A and B). At the genus level, Prevotella (36.8% and 39.5%), Oscillospirales_NK4A214_group (4.70% and 5.39%), Succiniclasticum (3.89% and 3.78%), Ruminococcus (4.15% and 3.90%) and Muribaculaceae (2.18% and 5.59%) were the predominant genera in the control and inulin groups, respectively (Fig. 1B and Appendix Fig. 2C and D).
Fig. 1.
The ruminal bacterial community compositions in control and inulin groups at (A) phylum and (B) genus level. C, control group. I, inulin group. The different colors of the bars represent different species, and the length of the bars represents the proportion of the species.
3.5. Significantly different ruminal bacteria between the control and inulin groups
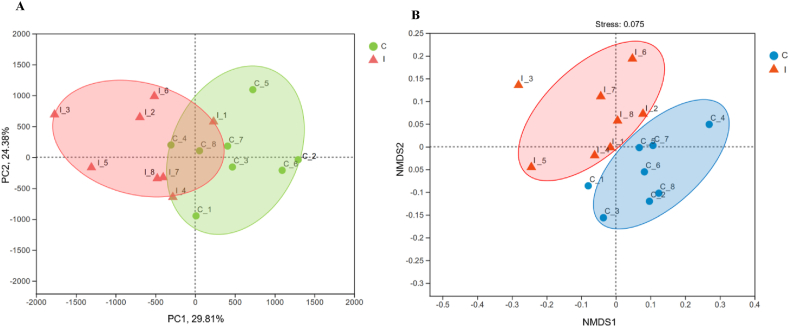
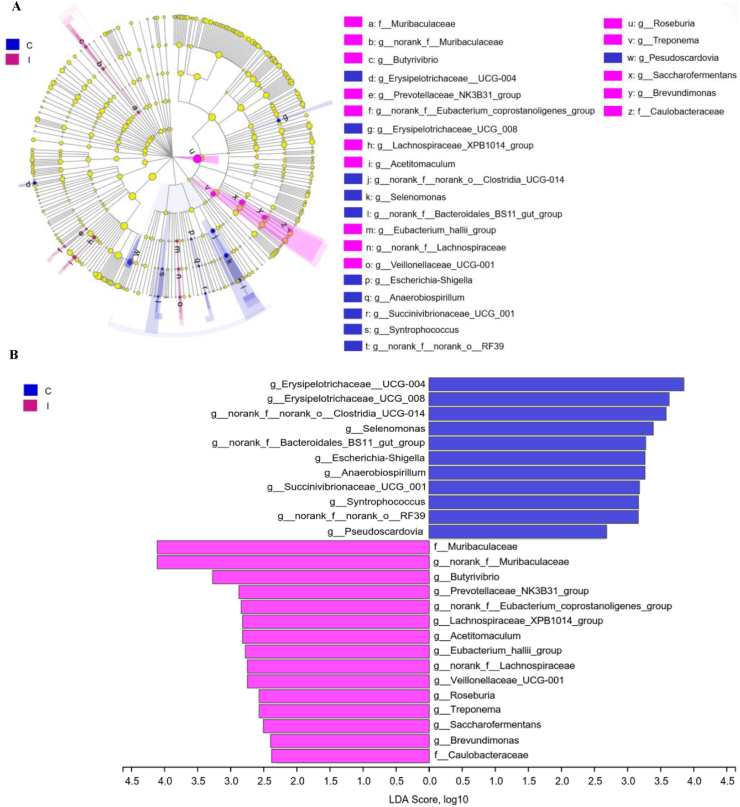
The β-diversity analysis was performed to explore differences in the rumen microbial community between the 2 groups (Fig. 2). The PCoA and NMDS plots based on the Bray–Curtis distance matrix showed that the points representing rumen microorganisms in the control and inulin groups respectively were significantly separated in different quadrants on the coordinate axis, which indicated that inulin intake had a distinct effect on rumen microbial species and abundance. A Venn diagram performed on the OTU samples with 97% identity identified 87 and 117 unique OTU in the control and inulin groups, respectively (Appendix Fig. 3). Furthermore, the significantly different abundant ruminal bacteria between the 2 groups were identified through LEfSe analysis and LDA (Fig. 3A and B). The relative abundances of Muribaculaceae (FDR-adjusted P < 0.01), Butyrivibrio (FDR-adjusted P = 0.036), Prevotellaceae_NK3B31_group (FDR-adjusted P = 0.032), Eubacterium_coprostanoligenes_group (P = 0.043), Lachnospiraceae_XPB1014_group (FDR-adjusted P = 0.035), Acetitomaculum (FDR-adjusted P = 0.043), Eubacterium_hallii_group (FDR-adjusted P = 0.031), Lachnospiraceae (FDR-adjusted P = 0.026) and Veillonellaceae_UCG-001 (FDR-adjusted P = 0.036) were increased in the inulin groups compared with the control group. In contrast, Erysipelotrichaceae__UCG-004 (FDR-adjusted P < 0.01), Erysipelotrichaceae__UCG-008 (FDR-adjusted P < 0.01), Clostridia_UCG-014 (FDR-adjusted P = 0.031), Selenomonas (FDR-adjusted P = 0.040), Bacteroidales_BS11_gut_group (FDR-adjusted P = 0.037), Escherichia-Shigella (FDR-adjusted P = 0.022), Anaerobiospirillum (FDR-adjusted P = 0.041), Succinivibrionaceae_UCG_001 (FDR-adjusted P = 0.042), Syntrophococcus (FDR-adjusted P = 0.037) and RF39 (FDR-adjusted P = 0.042) were decreased in rumen with the inulin addition (Appendix Table 3).
Fig. 2.
Beta diversity analysis of ruminal microbiota through (A) principal coordinate analysis (PCoA) and (B) non-metric multidimensional scaling analysis (NMDS). C, control group. I, inulin group. PC = principal components.
Fig. 3.
The linear discriminant analysis effect size (LEfSe) analysis of differential ruminal microorganisms in control and inulin group. (A) Cladogram showed the significantly different bacteria from phylum to genus level. The nodes with different color represent the microbes that are significantly enriched in the corresponding groups and have a significant influence on the difference between the 2 groups. The yellow nodes represent the microbes that have no significant difference between the 2 groups. (B) Linear discriminant analysis (LDA) bar showed the impact of the abundance of each species on the difference between the 2 groups. P-value > 0.05 and LDA score >2.5 were defined as significant difference. C, control group. I, inulin group.
3.6. Effect of dietary supplementation with inulin on ruminal metabolites
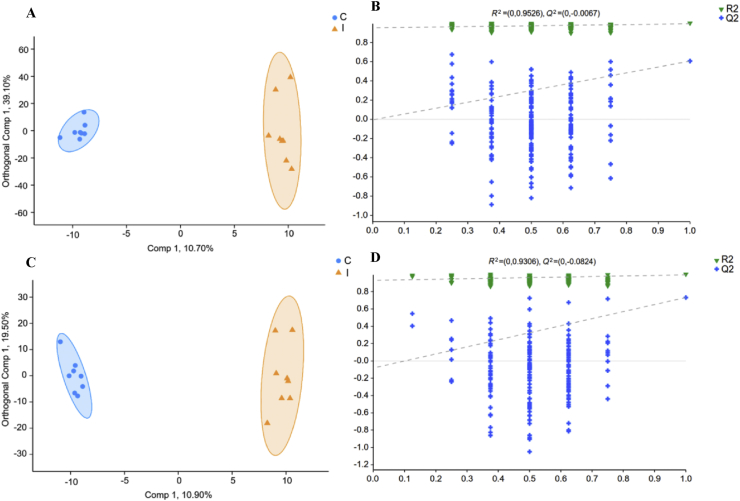
The rumen metabolites were analyzed through untargeted metabolomics techniques. The total ion chromatogram (TIC) plots of QC samples in positive and negative ion modes are shown in Appendix Fig. 4A and B. The overlap of QC samples revealed the good repeatability and high accuracy of the data. The unsupervised multivariate statistical analysis, PCA, generally reflected that a distinct difference existed in the ruminal metabolites between the 2 groups with a lower degree of variation among samples within a group in positive and negative ion modes (Appendix Fig. 5A and B). Further OPLS-DA provided a supervised discriminant analysis method, which could further distinguish the differences in ruminal metabolites between the control and inulin groups and improve the effectiveness and analytical capabilities of the model (Fig. 4). In the OPLS-DA plots, R2Y and Q2 were used to evaluate the modelling and prediction ability of the OPLS-DA model, respectively. The cumulative values of R2Y and Q2 in the positive (0.995 and 0.806) and negative (0.907 and 0.839) ion models were all above 0.80, which illustrated the stability and reliability of the model (Fig. 4A and C). Response permutation testing (RPT) is a randomized sequencing method used to evaluate the accuracy of OPLS-DA models. As shown in Fig. 4B and D, the value of R2 (0.953 and 0.931) and Q2 (−0.0067 and −0.082 < 0) in positive and negative ion models revealed the good accuracy of the OPLS-DA models.
Fig. 4.
Orthogonal partial least squares discriminant analysis (OPLS-DA) plot (A, C) and response permutation testing (RPT) (B, D) of rumen metabolites in comparisons of the control and inulin groups following (A, B) positive and (C, D) negative mode ionization. R2Y (cum) and Q2 indicates the cumulative interpretation power and predictive power of the model, respectively. C, control group; I, inulin group.
3.7. Significantly different ruminal metabolites between the control and inulin groups
A total of 99 differential metabolites in the rumen (64 in positive and 35 in negative ion models) between the control and inulin groups were detected with VIP >1 and FDR-adjusted P < 0.05 (Appendix Table 4). Among these metabolites, lipids and lipid-like molecules, organic acids and derivatives, organic oxygen compounds and organoheterocyclic compounds accounted for 37.0%, 22.2%, 16.7% and 14.8%, respectively (Appendix Fig. 6). The top 70 ruminal metabolites were selected for HCA analysis (Appendix Fig. 7). The differential metabolites between the 2 groups were divided into 2 clusters. The relative expression of phosphatidylcholine (PC, 16:0/0:0), lysophosphatidylcholine (LysoPC) (16:0), palmitoyl glucuronide, R-palmitoyl-(2-methyl) ethanolamide, 2-O-protocatechuoylalphitolic acid, sphinganine, LysoPC (18:2 (9Z, 12Z)) and LysoPC (18:1 (9Z)) etc. were higher in the control group than in the inulin group. However, the relative expression of deltonin, 3,4,5-trihydroxy-6-(2-oxoethoxy) oxane-2-carboxylic acid, L-proline, 2-hydroxycinnamic acid, L-tyrosine, L-phenylalanine, L-tyrosine, daidzein and L-lysine was elevated in the inulin group compared with the control group.
Furthermore, several significantly differential ruminal metabolites with VIP ≥2 and FDR-adjusted P < 0.05 between the 2 groups were screened out. Compared with the control group, LysoPC (18:1 (9Z)) (FDR-adjusted P = 1.03 × 10−3), LysoPC (16:0) (FDR-adjusted P = 0.0108), LysoPC (18:2 (9Z, 12Z)) (FDR-adjusted P = 1.65 × 10−3), phenylmethylglycidic ester (FDR-adjusted P = 1.19 × 10−3), N-acetylcadaverine (FDR-adjusted P = 0.0353) and 8-methylnonenoate (FDR-adjusted P = 1.60 × 10−3) were decreased in the inulin group. However, L-lysine (FDR-adjusted P = 4.24 × 10−3), L-proline (FDR-adjusted P = 0.0158), L-phenylalanine (FDR-adjusted P = 0.027), daidzein (FDR-adjusted P = 0.0130), uracil (FDR-adjusted P = 0.0396), deltonin (FDR-adjusted P = 0.0393) and L-tyrosine (FDR-adjusted P = 0.0353) were increased in the rumen with inulin supplementation (Appendix Fig. 8 and Appendix Table 4).
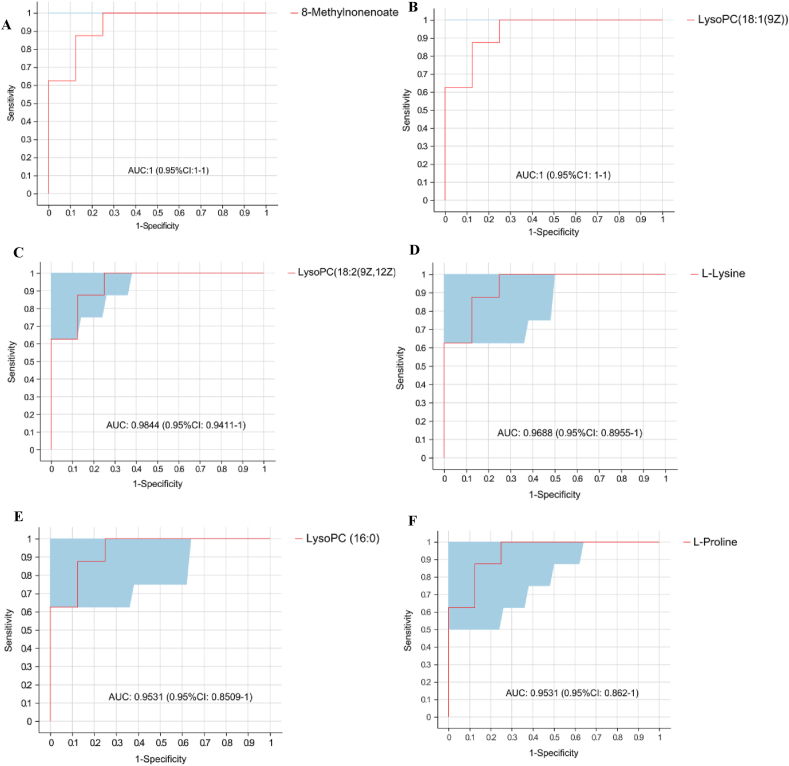
A receiver operator characteristic curve was generated to evaluate whether the differential metabolites screened were critical to intergroup differentiation. As shown in Fig. 5, the AUC of 8-methylnonenoate, LysoPC (18:1 (9Z)), LysoPC (18:2 (9Z, 12Z)), L-lysine, LysoPC (16:0) and L-proline were all above 0.95, which meant that these differential ruminal metabolites were representative to illustrate the effect of inulin on the shift of rumen metabolism.
Fig. 5.
Receiver operating characteristic (ROC) curves to evaluate the differential metabolites that have key impact on the differentiation between the 2 groups. ROC curve reflects the relationship between sensitivity and specificity. The x-axis is specificity (false positive rate). The closer the x-axis is to zero, the higher the accuracy will be. The y-axis is sensitivity (true positive rate). The larger the y-axis is, the better the accuracy is. The Area under curve (AUC) is used to indicate the accuracy of prediction. AUC value close to 1 suggested the higher the accuracy of prediction. (A) 8-Methylnonenoate; (B) lysophosphatidylcholine (LysoPC) (18:1 (9Z)); (C) LysoPC (18:2 (9Z, 12Z)); (D) L-Lysine; (E) LysoPC (16:0); (F) L-Proline.
3.8. Metabolic pathway enrichment analysis of differentially abundant metabolites
KEGG pathway enrichment analysis showed that inulin supplementation affected mainly the lipid and amino acid (AA) metabolism, vitamin metabolism, biosynthesis of plant secondary metabolites and protein metabolism in the rumen of dairy cows (Table 6).
Table 6.
Differential metabolic pathway enrichment analysis of significantly differential metabolites in rumen.
| Super pathway | Sub pathway description | Metabolites | P-value |
|---|---|---|---|
| Downregulated in inulin group | |||
| Lipid metabolism | Glycerophospholipid metabolism | LysoPC(18:1 (9Z)) | 0.006 |
| Choline metabolism | 0.011 | ||
| Glycerophospholipid metabolism | LysoPC(16:0) | 0.004 | |
| Choline metabolism | 0.014 | ||
| Glycerophosphocholines metabolism | LysoPC(18:2 (9Z,12Z)) | 0.018 | |
| Amino acid metabolism | /1 | Phenylmethylglycidic ester | 0.018 |
| Lysine degradation | N-Acetylcadaverine | 0.013 | |
| Leucine and valine pathway | 8-Methylnonenoate | 0.030 | |
| Upregulated in inulin group | |||
| Vitamin metabolism | Biotin metabolism | L-Lysine | 0.017 |
| Amino acid metabolism | Lysine biosynthesis | 0.023 | |
| Arginine and proline metabolism | L-Proline | 0.006 | |
| Aminoacyl-tRNA biosynthesis | 0.013 | ||
| Phenylalanine metabolism | L-Phenylalanine | 0.025 | |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 0.036 | ||
| Phenylpropanoid biosynthesis | 0.017 | ||
| Tyrosine metabolism | L-Tyrosine | 0.001 | |
| Biosynthesis of shikimate pathway | 0.013 | ||
| Nucleotide metabolism | Pyrimidine metabolism | Uracil | 0.008 |
| Vitamin metabolism | Pantothenate and CoA biosynthesis | 0.006 | |
| Biosynthesis of plant secondary metabolites | Steroid synthesis | Deltonin | 0.034 |
| Isoflavonoid biosynthesis | Daidzein | 0.029 | |
LysoPC = lysophosphatidylcholine.
The metabolic pathway was not annotated.
3.9. Correlation analysis among differential ruminal bacteria, metabolites, lactation and rumen fermentation performance
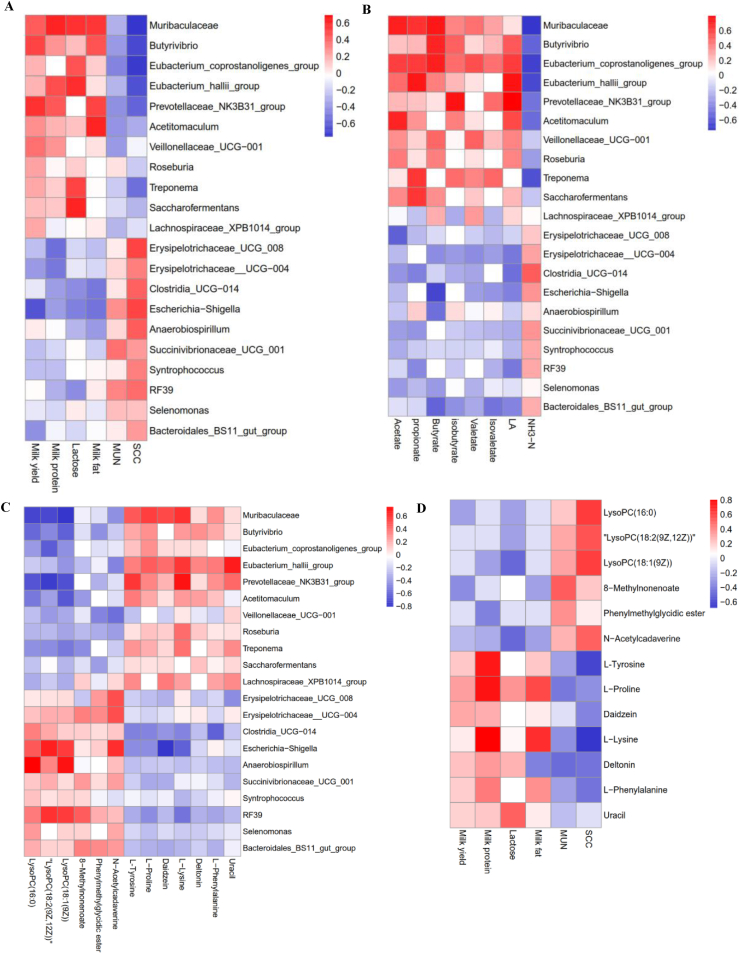
The Spearman correlation coefficient was used to calculate the correlation among several indicators (Fig. 6). Correlation analysis between significantly differential bacteria and milk components showed that the milk yield was positively associated with Muribaculaceae (r = 0.521, FDR-adjusted P = 0.042), Butyrivibrio (r = 0.602, FDR-adjusted P = 0.028) and Prevotellaceae_NK3B31_group (r = 0.607, FDR-adjusted P = 0.027) but negatively associated with Escherichia-Shigella (r = −0.613, FDR-adjusted P = 0.007). Milk protein was positively associated with Muribaculaceae (r = 0.595, FDR-adjusted P = 0.015), Eubacterium_hallii_group (r = 0.569, FDR-adjusted P = 0.037) and Prevotellaceae_NK3B31_group (r = 0.579, FDR-adjusted P = 0.039), but negatively associated with Erysipelotrichaceae_UCG_008 (r = 0.488, FDR-adjusted P = 0.044) and Erysipelotrichaceae_UCG_004 (r = 0.510, FDR-adjusted P = 0.044). Lactose was positively associated with Muribaculaceae (r = 0.585, FDR-adjusted P = 0.025), Eubacterium_coprostanoligenes_group (r = 0.565, FDR-adjusted P = 0.047), Eubacterium_hallii_group (r = 0.602, FDR-adjusted P = 0.023), Treponema (r = 0.573, FDR-adjusted P = 0.037) and Saccharofermentans (r = 0.582, FDR-adjusted P = 0.034), but negatively associated with Escherichia-Shigella (r = −0.481, FDR-adjusted P = 0.048). Milk fat was positively associated with Muribaculaceae (r = 0.543, FDR-adjusted P = 0.041), Butyrivibrio (r = 0.476, FDR-adjusted P = 0.042), Prevotellaceae_NK3B31_group (r = 0.468, FDR-adjusted P = 0.046) and Acetitomaculum (r = 0.584, FDR-adjusted P = 0.038). Milk SCC was negatively associated with Muribaculaceae (r = −0.759, FDR-adjusted P = 0.011), Butyrivibrio (r = −0.678, FDR-adjusted P = 0.014), Eubacterium_hallii_group and Eubacterium_coprostanoligenes_group (r = −0.743, FDR-adjusted P = 0.010) but positively associated with Erysipelotrichaceae_UCG_008 (r = 0.575, FDR-adjusted P = 0.031), Escherichia-Shigella (r = 0.594, FDR-adjusted P = 0.022) and Anaerobiospirillum (r = 0.594, FDR-adjusted P = 0.031) (Fig. 6A).
Fig. 6.
Correlation analysis between (A) significantly differential bacteria and milk components, (B) significantly differential bacteria and rumen fermentation parameters, (C) significantly differential bacteria and differential metabolites, as well as (D) significantly differential metabolites and milk compositions. LA, lactic acid; SCC, somatic cell counts; MUN, milk urea nitrogen. Red indicates a positive correlation; the blue indicates a negative correlation.
In addition, the correlation analysis between significantly differential bacteria and rumen fermentation parameters revealed that acetate was positively associated with Muribaculaceaec (r = 0.712, FDR-adjusted P = 0.002), Eubacterium_coprostanoligenes_group (r = 0.629, FDR-adjusted P = 0.014) and Acetitomaculum (r = 0.700, FDR-adjusted P = 0.003). Propionate was positively associated with Muribaculaceae (r = 0.649, FDR-adjusted P = 0.007), Eubacterium_coprostanoligenes_group (r = 0.622, FDR-adjusted P = 0.010), Eubacterium_hallii_group (r = 0.670, FDR-adjusted P = 0.021), Treponema (r = 0.608, FDR-adjusted P = 0.023) and Saccharofermentans (r = 0.626, FDR-adjusted P = 0.031). Butyrate was positively associated with Muribaculaceae (r = 0.739, FDR-adjusted P = 0.001), Eubacterium_coprostanoligenes_group (r = 0.719, FDR-adjusted P = 0.002) and Butyrivibrio (r = 0.700, FDR-adjusted P = 0.003) but negatively associated with Escherichia-Shigella (r = −0.681, FDR-adjusted P = 0.032). Prevotellaceae_NK3B31_group was positively associated with isovaletate (r = 0.644, FDR-adjusted P = 0.030) and isobutyrate (r = 0.602, FDR-adjusted P = 0.041). The LA was positively associated with Butyrivibrio (r = 0.639, FDR-adjusted P = 0.034), Prevotellaceae_NK3B31_group (r = 0.799, FDR-adjusted P < 0.001), Acetitomaculum (r = 0.678, FDR-adjusted P = 0.019) and Eubacterium_hallii_group (r = 0.653, FDR-adjusted P = 0.017). However, NH3–N was negatively associated with Muribaculaceae (r = −0.735, FDR-adjusted P = 0.001), Eubacterium_coprostanoligenes_group (r = −0.685, FDR-adjusted P = 0.030), Butyrivibrio (r = −0.648, FDR-adjusted P = 0.028), Eubacterium_hallii_group (r = −0.650, FDR-adjusted P = 0.036) and Treponema (r = −0.653, FDR-adjusted P = 0.021) (Fig. 6B).
The correlation between significantly differential metabolites and bacteria is shown in Fig. 6C. LysoPC (16:0), LysoPC (18:2 (9Z, 12Z)) and LysoPC (18:1 (9Z)) were negatively associated with Muribaculaceae (r = −0.806, FDR-adjusted P < 0.001; r = −0.740, FDR-adjusted P = 0.001; r = −0.717, FDR-adjusted P = 0.022) and Prevotellaceae_NK3B31_group (r = −0.730, FDR-adjusted P = 0.043; r = −0.676, FDR-adjusted P = 0.024; r = −0.649, FDR-adjusted P = 0.031) but positively associated with Escherichia-Shigella (r = −0.643, FDR-adjusted P = 0.030; r = −0.713, FDR-adjusted P = 0.024; r = −0.717, FDR-adjusted P = 0.046, FDR-adjusted P = 0.030; r = −0.713, FDR-adjusted P = 0.020; r = −0.675, FDR-adjusted P = -adjusted P = −0.044). N-acetylcadaverine were positively correlated with Escherichia-Shigella (r = −0.572, FDR-adjusted P = 0.042), Erysipelotrichaceae_UCG_008 (r = −0.566, FDR-adjusted P = 0.045) and Erysipelotrichaceae__UCG-004 (r = −0.562, FDR-adjusted P = 0.046). Uracil was positively correlated with Eubacterium_hallii_group (r = 0.673, FDR-adjusted P = 0.025). L-Lysine, L-proline and L-tyrosine were positively associated with Muribaculaceae (r = 0.602, FDR-adjusted P = 0.031; r = 0.585, FDR-adjusted P = 0.035; r = 0.607, FDR-adjusted P = 0.032), Eubacterium_hallii_group (r = 0.624, FDR-adjusted P = 0.042; r = 0.580, FDR-adjusted P = 0.024; r = 0.613, FDR-adjusted P = 0.031) and Prevotellaceae_NK3B31_group (r = 0.705, FDR-adjusted P = 0.032; r = 0.584, FDR-adjusted P = 0.047; r = 0.631, FDR-adjusted P = 0.040).
As shown in Fig. 6D, the relevance between significantly differential metabolites and milk compositions showed that the milk SCC was positively associated with LysoPC (16:0) (r = 0.772, FDR-adjusted P = 0.021), LysoPC (18:2 (9Z, 12Z)) (r = 0.752, FDR-adjusted P = 0.035), LysoPC (18:1 (9Z)) (r = 0.798, FDR-adjusted P = 0.023) and N-acetylcadaverine (r = 0.683, FDR-adjusted P = 0.041) but negatively associated with L-tyrosine (r = −0.677, FDR-adjusted P = 0.033) and L-lysine (r = −0.680, FDR-adjusted P = 0.037). Milk fat and protein were positively associated with L-proline (r = −0.741, FDR-adjusted P = 0.040; r = −0.769, FDR-adjusted P = 0.030) and L-lysine (r = −0.785, FDR-adjusted P = 0.036; r = −0.744, FDR-adjusted P = 0.040). Additionally, milk protein was also positively associated with L-tyrosine (r = −0.750, FDR-adjusted P = 0.035). Moreover, uracil was positively related to lactose (r = 0.679, FDR-adjusted P = 0.045).
4. Discussion
4.1. Milk composition
The effectiveness of inulin in improving human and monogastric animal health, especially the regulation of gut flora ecology, has motivated researchers to investigate its potential in ruminants (dairy cattle, beef, goats and sheep, etc.) (Biggs and Hancock, 1998; Zhao et al., 2016; Gndz, 2009; Umucalilar et al., 2010). However, limited information is currently available on the effect of inulin in ruminants, and the results are inconsistent (Samanta et al., 2013). In the present study, the increase in milk yield with dietary inulin supplementation might be attributed to the increase in the concentration of VFA in the rumen after inulin supplementation, which provided sufficient energy for lactation (Samanta et al., 2013). In the current study, the increase in the milk protein ratio might be due to the following 2 reasons: 1) inulin addition provided an energy substrate, which reduced the amount of AA used for the energy supply and thus increased microbial protein (MCP) synthesis in the rumen, along with the decreased MUN and NH3–N concentrations observed in this study, and nitrogen utilization by rumen microorganisms was enhanced. In addition, the increase in MCP entering the small intestine promoted the synthesis of milk proteins in the mammary gland (Emery, 1978), and 2) the increase in propionate concentration in the rumen after inulin supplementation could stimulate insulin secretion, which increased the absorption of AA by the mammary glands, thereby increasing the milk protein percentage (Samanta et al., 2013; Emery, 1978). An increase in AA levels was observed in the metabolomics results in our study. The uptake of glucose by the mammary glands directly affects the milk lactose content. After inulin supplementation, the increased precursor of gluconeogenesis in the rumen, such as propionate, LA and AA, was absorbed into the blood through the rumen wall, which may promote more glucose from the blood being obtained by the mammary gland (Sunehag et al., 2003).
Further analysis of milk FA composition showed that the proportion of SFA, especially SMCFA (C6:0, C8:0, C10:0 and C12:0), was significantly increased with the addition of inulin, but the ratio of C18:0 and C22:0 was decreased. The increase in SMCFA might be due to the increase in the concentration of acetate and butyrate in the rumen, which are substrates for de novo synthesis of milk FA by mammary epithelial cells (Cozma et al., 2013). However, approximately half of C16:0 and other long-chain FA in milk fat are directly absorbed from blood lipids (Cozma et al., 2013). Also, the metabolomics results in our study revealed that inulin supplementation significantly downregulated lipid metabolism in dairy cows. The increase in the ratio of SFA led to a decrease in the ratio of UFA (Cozma et al., 2013). The decrease in C18:1 trans-9 and C18:2 cis-6 might be related to the hydrogenation of rumen microorganisms, which could reduce the toxicity of UFA (Lock and Bauman, 2004). In addition, studies reported that milk fat percentage was negatively correlated with trans-C18:1 in milk (Matitashvili and Bauman, 2000). Trans-FA (TFA) could restrain the expression of milk fat synthase genes, such as acetyl-coenzyme A carboxylase and FA synthase (Matitashvili and Bauman, 2000).
4.2. Rumen microbiome
In accordance with the improvement effect of inulin on the intestinal flora in humans and monogastric animals, in the current study, inulin also showed a positive effect on the ruminal microbiome by increasing the relative abundance and diversity of rumen microflora. The possible reasons may be the increase in feed intake and the optimized rumen microbiota structure induced by inulin (Tian et al., 2019; Chambers et al., 2019). In the current study, inulin supplementation significantly increased the relative abundance of Bacteroides, which could regulate nutrient absorption and metabolism of exogenous substances, such as polysaccharides (Ivarsson et al., 2014). In addition, inulin intake increased the relative abundance mainly of several probiotics and SCFA-producing bacteria in the rumen. Muribaculaceae (also called S24-7), belonging to Bacteroides, has been found to act as a probiotic in multiple studies or is related to the innate immune system (Bunker et al., 2015). For example, homeostatic IgA responses could target Muribaculaceae residing in the small intestine (Bunker et al., 2015). In addition, the relative abundance of Muribaculaceae improved with increasing dietary fiber levels (Zhao et al., 2018). In addition, Muribaculaceae has the function of degrading complex carbohydrates and is positively correlated with the concentrations of intestinal acetate, propionate and butyrate (Zhao et al., 2018). Another increased bacterium induced by inulin addition was Butyrivibrio. Butyrivibrio was the main butyrate-producing bacterium, followed by LA (Kopecny, 2003). As a similar short-chain fatty acid producer, the main products of Prevotellaceae_NK3B31_group fermented FOSs are acetate and a small amount of isobutyrate, isovalerate and LA (Zhao et al., 2018). Furthermore, Acetitomaculum is another well-known acetate- and LA-producing bacterium (Rees et al., 1995). The increase in the abundance of these bacteria in the rumen was in accordance with the elevated SCFA ratio in blood, which might further promote milk fat synthesis (Lofgren and Warner, 1970). However, the increase in dietary fiber levels might promote the growth of a series of cellulolytic bacteria in the gastrointestinal tract. Eubacterium_hallii_group belongs to the Lacetospirillum family and can also degrade fructose and cellobiose, contributing to the formation of propionate and LA in the rumen (Engels et al., 2016). In addition, the Eubacterium hallii group could convert glycerol to 3-hydroxypropionaldehyde and form a multicompound system in aqueous solution called reuterin, which has antibacterial properties (Engels et al., 2016). Both Treponema and Saccharofermentans could degrade cellulose and hemicellulose to generate large amounts of propionate (Binek and Szynkiewicz, 1984; Perea et al., 2017). Therefore, the significant increase in the above bacteria may be the main reason for the increase in propionate concentration in the rumen and milk lactose rate.
In contrast, the significantly decreased ruminal microflora induced by inulin were mainly several conditional pathogens, highly abundant bacteria after high-fat feeding and microorganisms involved in FA hydrogenation. Escherichia-Shigella, Anaerobiospirillum and Clostridia have been reported to cause gastrointestinal infection or inflammation (Chen et al., 2014; Misawa et al., 2002; De La Serre et al., 2010). Similarly, as a conditioned pathogen, Erysipelotrichaceae that resides in the mouth and intestine of animals can result in endogenous infections and is significantly increased in the intestinal tract of colon cancer patients (Kaakoush, 2015). The current study showed that the above bacteria were positively correlated with milk SCC and negatively correlated with milk fat, protein and lactose, which implicated their adverse effects on milk composition. Moreover, Erysipelotrichaceae, Clostridia, Syntrophococcus and RF39 have been reported to increase significantly when animals are fed a high-fat diet. Studies have shown that the abundance of Erysipelotrichaceae in the intestines of rats fed long-term high-fat or Western diets was 2.5 times higher than the abundance of Erysipelotrichaceae in the intestines of the control group (Kaakoush, 2015). Martínez et al. (2009) proved that there was a positive correlation between the level of Erysipelotrichaceae and host cholesterol metabolites. The increase in the abundance of Clostridia in the intestines of rats fed a high-fat diet might be related to low fiber content (De La Serre et al., 2010). Additionally, a study on the growth requirements of Syntrophococcus in rumen fluid showed that TG and phospholipids could promote the proliferation of Syntrophococcus (Doré and Bryant, 1989). RF39 belongs to the Tenericutes phylum, and a high-fat diet could increase the abundance of Tenericutes, which made it easier for the host to absorb lipids (Niu et al., 2015). However, Selenomonas and Bacteroidales_BS11_gut_group were reported to be involved in the hydrogenation of C18:3n3 and C18:2n6c, respectively. The probable reasons were that they provided an energy substance, ATP, for the hydrogenation of these 2 FA (Benz et al., 1980; Solden et al., 2016).
In summary, after supplementation with inulin in the diet, the decrease in rumen pH and the increase in VFA concentration might be attributed to the proliferation of SCFA-producing bacteria. However, the acidic environment in the rumen effectively inhibited the proliferation of pathogens, which might be another reason for the decrease in milk SCC. In addition, more microorganisms increased the utilization of protein in the rumen and further increased the milk protein percentage, which might also explain the negative correlation between the dominant bacteria in the inulin group and MUN and NH3–N.
4.3. Rumen fluid metabolome
The metabolomics results further revealed that inulin supplementation significantly affected amino acid and lipid metabolism in dairy cows. In the current study, the decrease in the levels of LysoPC (18:1 (9Z)), LysoPC (16:0) and LysoPC (18:2 (9Z, 12Z)) in the rumen of cows fed inulin downregulated the glycerophospholipid and choline metabolism pathways. Lysophosphatidylcholine (LysoPC) is derived from the hydrolysis of lecithin or the oxidation of very low-density lipoprotein (VLDL). Inulin was reported to be able to reduce the number of VLDL particles in plasma and inhibit the expression of enzymes involved in lipid synthesis, such as acetyl-CoA carboxylase, fatty acid synthase and malic enzyme in the liver, which could reduce the de novo synthesis of FA and the levels of TG and cholesterol in serum (Beylot and Michel, 2005), consistent with our results. Another possible reason for the decrease in LysoPCs was that inulin could promote the proliferation of SCFA-producing bacteria and reduce the pH of the rumen, which led to a reduction in the solubility of bile acids and an increase in the excretion of bile acids and steroids in feces (Roberfroid and Delzenne, 1998), which might explain the negative correlation between LysoPC (18:1 (9Z)), LysoPC (16:0), LysoPC (18:2 (9Z, 12Z)), Muribaculaceae, Prevotellaceae_NK3B31_group and other SCFA-producing bacteria. However, in our study, the decline in the level of LysoPCs did not show a significant negative correlation with milk fat, possibly because the LysoPCs were not a key factor affecting milk fat synthesis. However, LysoPCs have been reported to have proinflammatory activity, which is related to cytosolic phospholipase A2 mediating phosphatidylcholine (PCs) to produce arachidonic acid (You et al., 2015), which might be the reason that the positive correlation between LysoPC(18:1 (9Z)), LysoPC(16:0), LysoPC(18:2 (9Z, 12Z)) and milk SCC as well as several pathogen abundances, including Escherichia-Shigella and Anaerobiospirillu, was observed in this study. Other downregulated lipids included phenylmethylglycidic ester and 8-methylnonenoate.
Phenylmethylglycidic ester is a sweet, fatty, and floral compound (Allen and Vanallan, 2003). However, its metabolic pathway needs to be further explored. 8-Methylnonenoic acid is a long-chain fatty acid derivative from the leucine/valine pathway and a component of fatty acyl groups (Markai et al., 2010). In the current study, the decrease in 8-methylnonenoic acid levels was consistent with the decrease in the proportion of long-chain fatty acids in milk. Therefore, the decreased levels of the above ruminal metabolites might imply the potential of inulin to downregulate lipid metabolism in dairy cows.
N-Acetylcadaverine is the acetylated form of the polyamine cadaverine, which is formed by the decarboxylation of lysine (Ma et al., 2017). In addition, N-acetylcadaverine was mainly a metabolite of Escherichia coli. Additionally, variants of N-acetylcadaverine have been found in Bacillus species (Ma et al., 2017). The accumulation of a large amount of N-acetylcadaverine in the body was related to the production of bacteriocins and toxin activity, which also had a specific induction effect on inflammatory factors (Ma et al., 2017) and might illustrate the positive correlation between N-acetylcadaverine and milk SCC, Escherichia-Shigella, and Erysipelotrichaceae (Bacillus class).
The AAs in the rumen were derived mainly from the degradation of feed proteins by rumen microorganisms. In the present study, the significant increase in the levels of L-lysine, L-proline, L-phenylalanine and L-tyrosine in cows fed inulin might be attributed to the proliferation of beneficial symbiotic bacteria in the rumen, which accelerated the decomposition of dietary protein and synthesis of MCP and further promoted the synthesis of milk protein (Liu et al., 2019). Consistent with Lykos and Varga (1997), the increase in nonstructural carbohydrates in the rumen could increase the level of nonessential AAs in the blood. However, inulin, regarded as a prebiotic, could provide substrates for intestinal flora to generate SCFA (Davila et al., 2013). Davila et al. proposed that AA could synthesize SCFA by prebiotic-activated intestinal flora (Davila et al., 2013). Lysine, phenylalanine and tyrosine could be used as precursors of acetate and butyrate. Butyrate is derived from glutamic acid and lysine, and propionate is synthesized from alanine and threonine (Davila et al., 2013). Meanwhile, isoacids, including isobutyrate, isovalerate and valerate, were generated by the degradation of proline (Andries et al., 1987), which might explain the positive correlation between L-lysine, L-proline, L-phenylalanine and L-tyrosine and SCFA-producing bacteria. Moreover, the positive correlation between L-lysine, L-proline, L-phenylalanine and L-tyrosine and milk fat might be because AA could enhance the uptake efficiency of milk fat precursors by the mammary gland and thus regulate the content of VFA in the udder, which promoted the de novo synthesis of FA in the mammary gland (Cant et al., 2002). Consistent with Maxin et al. (2011), the increase in AA content in milk could promote milk fat production. Among these AAs, the content of short- and medium-chain FA (C6:0-C14:0) tended to increase, but C16:0, C18:0, C18:2 and other long-chain FA were decreased.
In the present study, the increase in uracil upregulated nucleic acid metabolism could indicate that supplementation with inulin might promote the synthesis of rumen MCP (Liu et al., 2019). Additionally, the decomposed product of uracil was β-alanine, which could further synthesize propionate (West et al., 1985), which might explain the positive correlation between uracil and lactose. Deltonin and daidzein were reported to have antitumor activity and inhibit inflammatory factors in serum (Tong et al., 2011; Shen et al., 2019). The negative correlation between deltonin, daidzein and milk SCC might reveal the enhancement of immune and disease resistance in dairy cows. However, the effect of inulin on the secondary metabolites of plants in the rumen still needs to be further explored.
Throughout the study, the effects of dietary inulin supplementation on milk composition and serum lipid and rumen fermentation characteristics in dairy cows might be influenced mainly by the ruminal bacteria profile, which further affected the kinds and concentrations of rumen metabolites. The correlation among them indicated that the shifts in the composition and abundance of the ruminal microbiome as well as the concentration and activity of rumen metabolites caused by the increase in dietary fiber intake might be a key factor affecting performance and physiological indicators in dairy cows.
5. Conclusions
Dietary supplementation with 200 g/d inulin per dairy cow increased the abundance of commensal bacteria and SCFA-producing bacteria in the rumen, promoted AA synthesis and metabolism, inhibited lipid metabolism and decreased the levels of TC and TG in serum. The shift of the rumen inner environment further improved the lactation performance of dairy cows. However, the metabolic processes and mechanism of the action of inulin in the rumen of dairy cow merit further investigation. Overall, the current results might suggest the application potential of inulin in the production of ruminants. In practical production, inulin might be considered to be added to TMR in an appropriate manner.
Conflict of interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Author contributions
Yue Wang: Experimental designed, Experimental data analysis, Writing initial draft. Xuemei Nan and Yiguang Zhao: Manuscript modification. Hui Wang: Polishing the language of manuscript. Dengke Hua, Fan Zhang and Yapin Wang: Sampling, Analysis. Jun Liu: Providing inulin. Linshu Jiang and Junhu Yao: Providing the necessary experimental equipment and guidance. Benhai Xiong: Conceptualization, Funding acquisition.
Acknowledgments
This study was funded by the National Key R&D Program of China (Grant No. 2019YFE0125600) and Beijing Dairy Industry Innovation Team (bjcystx-ny-1). We thank the Beijing Key Laboratory for Dairy Cow Nutrition, Beijing University of Agriculture, Beijing, China for providing the experimental equipment.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2021.09.007
Contributor Information
Linshu Jiang, Email: jls@bua.edu.cn.
Benhai Xiong, Email: xiongbenhai@caas.cn.
Appendix ASupplementary data
The following is the Supplementary data to this article:
References
- Allen C.F.H., Vanallan J. Phenylmethylglycidic ester. Org Syn Coll. American Cancer Society. 2003;3(733):1944–1955. [Google Scholar]
- Andries J.I., Buysse F.X., Brabander D.L.D.e., Cottyn B.G. Isoacids in ruminant nutrition: their role in ruminal and intermediary metabolism and possible influences on performances-A review. Anim Feed Sci Technol. 1987;18(3):169–180. [Google Scholar]
- Artegoitia V.M., Foote A.P., Lewis R.M., Freetly H.C. Rumen fluid metabolomics analysis associated with feed efficiency on crossbred steers. Sci Rep. 2017;7(1):2864–2878. doi: 10.1038/s41598-017-02856-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz J., Wolf C., Wolfhart Rüdiger. Chlorophyll biosynthesis: hydrogenation of geranylgeraniol. Plant Sci Lett. 1980;19(3):225–230. [Google Scholar]
- Beylot, Michel Effects of inulin-type fructans on lipid metabolism in man and in animal models. Br J Nutr. 2005;93(S1):S163–S168. doi: 10.1079/BJN20041339. [DOI] [PubMed] [Google Scholar]
- Biggs D.R., Hancock K.R. In vitro digestion of bacterial and plant fructans and effects on ammonia accumulation in cow and sheep rumen fluids. J Gen Appl Microbiol. 1998;44(2):167–171. doi: 10.2323/jgam.44.167. [DOI] [PubMed] [Google Scholar]
- Binek M., Szynkiewicz Z.M. Physiological properties and classification of strains of Treponema sp. isolated from pigs in Poland. Comp Immunol Microb. 1984;7(3–4):141–148. doi: 10.1016/0147-9571(84)90019-5. [DOI] [PubMed] [Google Scholar]
- Bunker J.J., Flynn T.M., Koval J.C., Shaw D.G., Meisel M., McDonald B.D. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity. 2015;43(3):541–553. doi: 10.1016/j.immuni.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cant J.P., Trout D.R., Qiao F., Purdie N.G. Milk synthetic response of the bovine mammary gland to an increase in the local concentration of arterial glucose. J Dairy Sci. 2002;85(3):494–503. doi: 10.3168/jds.S0022-0302(02)74100-3. [DOI] [PubMed] [Google Scholar]
- Causey J.L., Feirtag J.M., Gallaher D.D., Tungland B.C., Slavin J.L. Effects of dietary inulin on serum lipids, blood glucose and the gastrointestinal environment in hypercholesterolemic men. Nutr Res. 2000;20(2):191–201. [Google Scholar]
- Chambers E.S., Byrne C.S., Morrison D.J., Murphy K.G., Preston T., Tedford C. Dietary supplementation with inulin-propionate ester or inulin improves insulin sensitivity in adults with overweight and obesity with distinct effects on the gut microbiota, plasma metabolome and systemic inflammatory responses: a randomised cross-over trial. Gut. 2019:1–9. doi: 10.1136/gutjnl-2019-318424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Chen H., Faas M.M., De Haan B.J., Li J., Xiao P. Specific inulin-type fructan fibers protect against autoimmune diabetes by modulating gut immunity, barrier function, and microbiota homeostasis. Mol Nutr Food Res. 2017;61(8):1601006. doi: 10.1002/mnfr.201601006. [DOI] [PubMed] [Google Scholar]
- Chen L., Wang W., Zhou R., Ng S.C., Li J., Huang M. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine. 2014;93(8):e51–e60. doi: 10.1097/MD.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozma A., Miere D., Filip L., Andrei S., Banc R., Loghin F. A review of the metabolic origins of milk fatty acids. Not Sci Biol. 2013;5(3):270–274. doi: 10.15835/nsb.5.3.9120. [DOI] [Google Scholar]
- Davila A.M., Blachier F., Gotteland M., Andriamihaja M., Benetti P.H., Sanz Y. Intestinal luminal nitrogen metabolism: role of the gut microbiota and consequences for the host. Pharmacol Res. 2013;68(1):95–107. doi: 10.1016/j.phrs.2012.11.005. [DOI] [PubMed] [Google Scholar]
- De La Serre C.B., Ellis C.L., Lee J., Hartman A.L., Rutledge J.C., Raybould H.E. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. AM J Physiol-Gastr L. 2010;299(2):G440–G448. doi: 10.1152/ajpgi.00098.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doré J., Bryant M.P. Lipid growth requirement and influence of lipid supplement on fatty acid and aldehyde composition of Syntrophococcus sucromutans. Appl Environ Microbiol. 1989;55(4):927–933. doi: 10.1002/bit.260330921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery R.S. Feeding for increased milk protein1. J Dairy Sci. 1978;61(6):825–828. doi: 10.3168/jds.S0022-0302(78)83656-X. [DOI] [Google Scholar]
- Engels C., Ruscheweyh H.J., Beerenwinkel N., Lacroix C., Schwab C. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front Microbiol. 2016;7(13):1–12. doi: 10.3389/fmicb.2016.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaines W.L. vol. 308. University of Illinois; Urbana: 1928. pp. 436–438. (The energy basis of measuring milk yield in dairy cows. Illinois agricultural experiment station bulletin). [Google Scholar]
- Gndz N. Effects of chicory inulin on ruminal fermentation in vitro. Ankara Univ Vet Fak Derg. 2009;56(3):171–175. [Google Scholar]
- Han X., Yang Y., Yan H., Wang X., Qu L., Chen Y. Rumen bacterial diversity of 80 to 110-day-old goats using 16s rRNA sequencing. PLoS One. 2015;10(2) doi: 10.1371/journal.pone.0117811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helrich K. 15th ed. AOAC; Arlington, VA: 1990. Official methods of analysis of the Association of Official Analytical Chemists: 2. Food composition; additives; natural contaminants. [Google Scholar]
- Hue W.L., Liu J.X., Ye J.A. Effects of tea saponin on rumen fermentation in vitro. Anim Feed Sci Technol. 2005;120(4):333–339. doi: 10.1016/j.anifeedsci.2005.02.029. [DOI] [Google Scholar]
- Ivarsson E., Roos S., Liu H.Y., Lindberg J.E. Fermentable non-starch polysaccharides increases the abundance of bacteroides-prevotella-porphyromonas in ileal microbial community of growing pigs. Animal. 2014;8(11):1777–1787. doi: 10.1017/S1751731114001827. [DOI] [PubMed] [Google Scholar]
- Kaakoush N.O. Insights into the role of Erysipelotrichaceae in the human host. Front Cell Infect Microbiol. 2015;5(84):1–4. doi: 10.3389/fcimb.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecny J. Butyrivibrio hungatei sp. nov. and Pseudobutyrivibrio xylanivorans sp. nov. butyrate-producing bacteria from the rumen. Int J Syst Evol Microbiol. 2003;53(1):201–209. doi: 10.1099/ijs.0.02345-0. [DOI] [PubMed] [Google Scholar]
- Liu C., Wu H., Liu S., Chai S., Meng Q., Zhou Z. Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front Microbiol. 2019;10:1–19. doi: 10.3389/fmicb.2019.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock A.L., Bauman D.E. Modifying milk fat composition of dairy cows to enhance fatty acids beneficial to human health. Lipids. 2004;39(12):1197–1206. doi: 10.1007/s11745-004-1348-6. [DOI] [PubMed] [Google Scholar]
- Lofgren P.A., Warner R.G. Influence of various fiber sources and fractions on milk fat percentage. J Dairy Sci. 1970;53(3):296–304. [Google Scholar]
- Lopez H.W., Coudray C., Levrat-Verny M.A., Feillet-Coudray C., Christian D., Christian R. Fructooligosaccharides enhance mineral apparent absorption and counteract the deleterious effects of phytic acid on mineral homeostasis in rats. J Nutr Biochem. 2000;11(10):500–508. doi: 10.1016/S0955-2863(00)00109-1. [DOI] [PubMed] [Google Scholar]
- Lykos T., Varga G.A. Varying degradation rates of total nonstructural carbohydrates: effects on nutrient uptake and utilization by the mammary gland in high producing Holstein cows. J Dairy Sci. 1997;80(12):3356–3367. doi: 10.3168/jds.S0022-0302(97)76311-2. [DOI] [PubMed] [Google Scholar]
- Ma W.C., Chen K.Q., Li Y., Wang X., Ouyang P.K. Advances in cadaverine bacterial production and its applications. Engineering. 2017;3:308–317. doi: 10.1016/J.ENG.2017.03.012. [DOI] [Google Scholar]
- Markai S., Marchand P.A., Mabon F., Baguet E., Billault I., Robins R.J. Natural deuterium distribution in branched-chain medium-length fatty acids is nonstatistical: a site-specific study by quantitative 2H NMR spectroscopy of the fatty acids of capsaicinoids. Chembiochem. 2010;3(2–3):212–218. doi: 10.1002/1439-7633(20020301)3:2/3<212::AID-CBIC212>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Martínez I., Wallace G., Zhang C., Legge R., Benson A.K., Carr T.P. Diet-induced metabolic improvements in a hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matitashvili E.A., Bauman D.E. Effeet of different isomers of C18:1 and C18:2 fatty acids on lipogenesis in bovine mammary eipthelial cells. J Dairy Sci. 2000;83(suppl.1):165. [Google Scholar]
- Maxin G., Rulquin H., Glasser F. Response of milk fat concentration and yield to nutrient supply in dairy cows. Animal. 2011;5(8):1299–1310. doi: 10.1017/S1751731111000206. [DOI] [PubMed] [Google Scholar]
- Misawa N., Kawashima K., Kondo F., Kushima E., Vandamme P. Isolation and characterization of Campylobacter, Helicobacter, and Anaerobiospirillum strains from a puppy with bloody diarrhea. Vet Microbiol. 2002;87(4):353–364. doi: 10.1016/s0378-1135(02)00086-x. [DOI] [PubMed] [Google Scholar]
- Niness K.R. Inulin and oligofructose: what are they? J Nutr. 1999;129(7):1402S–1406S. doi: 10.1038/sj.ijo.0800961. [DOI] [PubMed] [Google Scholar]
- Niu Q., Li P., Hao S., Hang Y., Kim S.W., Li H. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci Rep. 2015;5(9938):9938. doi: 10.1038/srep09938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (NRC) Seventh Revised Edition. The National Academies Press; Washington, DC: 2001. Nutrient requirements of dairy cattle. 2001. [Google Scholar]
- Ogunade I., Schweickart H., Mccoun M., Cannon K., Mcmanus C. Integrating 16s rRNA sequencing and LC-MS-based metabolomics to evaluate the effects of live yeast on rumen function in beef cattle. Animals. 2019;9(1):1–14. doi: 10.3390/ani9010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea K., Perz K., Olivo S.K., Williams A., Yeoman C.J. Feed efficiency phenotypes in lambs involve changes in ruminal, colonic, and small-intestine-located microbiota. J Anim Sci. 2017;95(6):2585–2592. doi: 10.2527/jas2016. [DOI] [PubMed] [Google Scholar]
- Rees E.M.R., David L., Williams A.G. The effects of co-cultivation with the acetogen Acetitomaculum ruminis on the fermentative metabolism of the rumen fungi neocallimastix patriciarum and neocallimastix sp. strain l2. FEMS Microbiol Lett. 1995;(1–2):175–180. doi: 10.1016/0378-1097(95)00367-E. [DOI] [PubMed] [Google Scholar]
- Roberfroid M.B., Delzenne N.M. Dietary fructans. Annu Rev Nutr. 1998;18(1):117–143. doi: 10.1146/annurev.nutr.18.1.117. [DOI] [PubMed] [Google Scholar]
- Roberfroid M.B. Inulin-type fructans: functional food ingredients. J Nutr. 2007;137(11):2493S–2502S. doi: 10.1201/9780203504932. [DOI] [PubMed] [Google Scholar]
- Ronkart S., Blecker C., Fougnies C., Herck J.C.V., Wouters J., Paquot M. Determination of physical changes of inulin related to sorption isotherms: an x-ray diffraction, modulated differential scanning calorimetry and environmental scanning electron microscopy study. Carbohydr Polym. 2006;63(2):210–217. doi: 10.1016/j.carbpol.2005.08.030. [DOI] [Google Scholar]
- Saengthongpinit W., Sajjaanantakul T. Influence of harvest time and storage temperature on characteristics of inulin from Jerusalem artichoke (Helianthus tuberosus L.) tubers. Postharvest Biol Technol. 2005;37(1):93–100. doi: 10.1016/j.postharvbio.2005.03.004. [DOI] [Google Scholar]
- Samanta A.K., Natasha J., Senani S., Kolte A.P., Manpal S. Prebiotic inulin: useful dietary adjuncts to manipulate the livestock gut microflora. Braz J Microbiol. 2013;44(1):1–14. doi: 10.1590/S1517-83822013005000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schley P.D., Field C.J. The immune-enhancing effects of dietary fibres and prebiotics. Br J Nutr. 2002;87(2):221–230. doi: 10.1079/BJNBJN/2002541. [DOI] [PubMed] [Google Scholar]
- Shen J.L., Li N., Zhang X. Daidzein ameliorates dextran sulfate sodium-induced experimental colitis in mice by regulating NF-κB signaling. J Environ Pathol Toxicol Oncol. 2019;38(1):29–39. doi: 10.1615/JEnvironPatholToxicolOncol.2018027531. https://doi. org/10.1615/JEnvironPatholToxicolOncol.2018027531. [DOI] [PubMed] [Google Scholar]
- Sjaunja L.O., Baevre L., Junkkarinen L., Pedersen J., Setälä J. Proceedings of the 27th biennial session of the international committee for animal recording. ICAR; 1990. A Nordic proposal for an energy corrected milk (ECM) formula; pp. 2–6. Paper presented at. [Google Scholar]
- Solden L., Hoyt D.W., Collins W., Plank J., Daly R., Erik C.H. New roles in hemicellulosic sugar fermentation for the uncultivated Bacteroidetes family BS11. ISME J. 2016;11(3):691–703. doi: 10.1038/ismej.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunehag A., Tigas S., Haymond M.W. Contribution of plasma galactose and glucose to milk lactose synthesis during galactose ingestion. J Clin Endocrinol Metab. 2003;88(1):225–229. doi: 10.1210/jc.2002-020768. [DOI] [PubMed] [Google Scholar]
- Tian K., Liu J., Sun Y., Wu Y., Chen J., Zhang R. Effects of dietary supplementation of inulin on rumen fermentation and bacterial microbiota, inflammatory response and growth performance in finishing beef steers fed high or low-concentrate diet. Anim Feed Sci Technol. 2019;258:114229. doi: 10.1016/j.anifeedsci.2019.114299. [DOI] [Google Scholar]
- Tong Q.Y., Qing Y., Shu D., He Y., Zhao Y.L., Li Y. Deltonin, a steroidal saponin, inhibits colon cancer cell growth in vitro and tumor growth in vivo via induction of apoptosis and antiangiogenesis. Cell Physiol Biochem. 2011;27(3–4):233–242. doi: 10.1159/000327949. [DOI] [PubMed] [Google Scholar]
- Umucalilar H.D., Glen N., Hayirli A., Alata M.S. Potential role of inulin in rumen fermentation. Rev Med Vet. 2010;161(1):3–9. doi: 10.1590/S1516-35982010000100030. [DOI] [Google Scholar]
- Wang X., Shen X., Han H., Zhao R., Jie C. Analysis of cis-9, trans-11-conjugated linoleic acid in milk fat by capillary gas chromatography. Chin J Chromatogr. 2006;24(6):645. doi: 10.1016/S1872-2059(06)60026-6. [DOI] [PubMed] [Google Scholar]
- Wang B., Tu Y., Zhao S.P., Hao Y.H., Liu J.X., Liu F.H. Effect of tea saponins on milk performance, milk fatty acids, and immune function in dairy cow. J Dairy Sci. 2017;100(10):8043–8052. doi: 10.3168/jds.2016-12425. [DOI] [PubMed] [Google Scholar]
- West T.P., Traut T.W., Shanley M.S., O'Donovan G.A. A salmonella typhimurium strain defective in uracil catabolism and β-alanine synthesis. Microbiology. 1985;131(5):1083–1090. doi: 10.1099/00221287-131-5-1083. [DOI] [PubMed] [Google Scholar]
- Williams C.M. Effects of inulin on lipid parameters in humans. J Nutr. 1999;129(7):1471–1473. doi: 10.1038/sj.ijo.0800961. [DOI] [PubMed] [Google Scholar]
- You J.C., Yang J., Fang R.P., Hu N., Ye L.H. Analysis of phosphatidylcholines (PCs) and lysophosphatidylcholines (LysoPCS) in metastasis of breast cancer cells. Prog Biochem Biophys. 2015;42(6):563–573. doi: 10.16476/j.pibb.2015.0031. [DOI] [Google Scholar]
- Young W., Hine B.C., Wallace O., Callaghan M., Bibiloni R. Transfer of intestinal bacterial components to mammary secretions in the cow. PeerJ. 2015;3:e888. doi: 10.7717/peerj.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zhang F., Ding X., Wu G., Lam Y.Y., Wang X. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359(6380):1151–1156. doi: 10.1126/science.aao5774. [DOI] [PubMed] [Google Scholar]
- Zhao X.H., Gong J.M., Zhou S., Liu C.J., Qu M.R. The effect of starch, inulin, and degradable protein on ruminal fermentation and microbial growth in rumen simulation technique. Ital J Anim Sci. 2016;13(1):189–195. doi: 10.4081/ijas.2014.3121. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.